Abstract
Alternative polyadenylation (APA) is an important RNA post-transcriptional process, which can generate diverse mRNA isoforms. Increasing evidence shows that APA is involved in cell self-renewal, development, immunity, and cancer. CPSF6 is one of the core proteins of CFIm complex and can modulate the APA process. Although it has been reported to play oncogenic roles in cancer, the underlying mechanisms remain unclear. The aim of the present study was to characterize CPSF6 in human gastric cancer (GC). We observed that CPSF6 was upregulated in GC. Knockdown of CPSF6 inhibited proliferation and enhanced apoptosis of GC cells both in vitro and in vivo. Global APA site profiling analysis revealed that knockdown of CPSF6 induced widespread 3′UTR shortening of genes in GC cells, including VHL. We also found CPSF6 negatively regulated the expression of VHL through APA and VHL short-3′UTR isoform enhanced apoptosis and inhibited cell growth in GC cells. Our data suggested that CPSF6-induced cell proliferation and inhibition of apoptosis were mediated by the preferential usage of poly(A) in VHL. Our data provide insights into the function of CPSF6 and may imply potential therapeutic targets against GC.
Keywords: alternative polyadenylation, gastric cancer, apoptosis, CPSF6, VHL
Introduction
Gastric cancer (GC), one of the top five prevalent cancers, is the third leading cause of cancer mortality across the world, owing to its poor prognosis and diagnoses only at advanced stage with median overall survival less than 1 year (Zhang and Zhang, 2017; Smyth et al., 2020). Many predisposing factors, such as Helicobacter pylori infection, age, environmental factors, dietary habits, and so on, assist in the development of GC (Chia and Tan, 2016; Petrovchich and Ford, 2016). Apart from that, genetic alterations are also reported to be involved in GC progression. Abnormal expression of HER2, CDH1, TP53, FGFR, MET, and other genes are frequently found in different GC types (Oue et al., 2019). The regulation of GC development and progression seems particularly complex; hence, a deeper understanding of the pathogenesis mechanism of GC at the molecular level may be beneficial to identify potential therapeutic targets.
Alternative polyadenylation (APA) is an RNA post-transcriptional process that produces distinct messenger RNA (mRNA) isoforms of a single gene through dictating the length of 3′ untranslated regions (UTRs). Modification of 3′UTR influences mRNA stability, translation, and cellular localization, as 3′UTRs provide major binding sites for protein factors or microRNAs (miRNAs) (Andreassi and Riccio, 2009; Fabian et al., 2010). Recent researches demonstrate that APA can affect various biological functions, including cell self-renewal, differentiation, activation of human immune cells, neuronal activation, and so on (Flavell et al., 2008; Sandberg et al., 2008; Ji and Tian, 2009; Lackford et al., 2014). Of note, APA is also involved in cancer. For example, Pik3ap1 displayed preferential loss of signal in the proximal 3′UTR in APC and APN cancer subtypes, uniform degradation in LPC cancer subtype, and no change in mature B cells (Singh et al., 2009). Another study also reported that lung (LUSC and LUAD), uterine (UCEC), breast (BRCA), and bladder (BLCA) cancers possessed the highest amount dynamic APA events than the other tumor types, such as kidney renal clear cell carcinoma (KIRC) and head and neck squamous cell carcinoma (Xia et al., 2014).
APA can be determined by over 80 proteins, and the core complex contains four subcomplexes: CPSF, CSTF, CFI, and CFII (Tian and Manley, 2017). Some of them are reported to play an important role in tumorigenesis. It was reported that CSTF2 enhanced oncogenic and metastatic abilities in urothelial carcinoma of the bladder cells (Chen et al., 2018). Intriguingly, NUDT21 (CFIm25) showed a dual role in cancers. For example, NUDT21 was downregulated and functioned as tumor suppressor in glioblastoma, hepatocellular carcinoma, and ovarian cancer (Masamha et al., 2014, 2016; Tan et al., 2018). However, other groups uncovered that knockdown of NUDT21 significantly inhibited cell proliferation and tumorigenicity (Lou et al., 2017; Zhang and Zhang, 2018). These observations implicate sophisticated regulation of APA factors. Meanwhile, how APA takes part in tumor formation is still unclear.
The polyadenylation complex cleavage factor I (CFIm) preferentially binds to UGUA subsequences in pre-mRNAs and controls 3′UTR length, which consists of NUDT21, CPSF6, and CPSF7 (Gruber et al., 2012). In addition, the recombinant NUDT21-CPSF6 subunit complex could also show cleavage activity in vitro, which was similar with full CFIm complex (Rüegsegger et al., 1998). As described earlier, NUDT21 regulated tumor growth negatively or positively (Tan et al., 2018). Also, to date, as its partner, the description of CPSF6’s function was mainly in HIV infection (Lee et al., 2010; Bejarano et al., 2019). Recent reports also discovered the oncological function of CPSF6, but not in an APA regulatory perspective (Binothman et al., 2017; Zhang et al., 2020). As one of the APA factors, CPSF6 plays an important role in regulation of 3′UTR length. It is reported that knockdown of CPSF6 significantly upregulated the usage of proximal PAS in C2C12 myoblast cells (Li et al., 2015). Besides that, another group also found that knockdown of CPSF6 led to a systematic and transcriptome-wide shift toward proximal poly(A) sites in HEK293 cells (Gruber et al., 2012; Martin et al., 2012). So, it is necessary to discover the roles of CPSF6 in GC progress.
Herein, to expand our knowledge on CPSF6 biological roles, we investigated the phenotypes of CPSF6 knockdown GC cells. We found CPSF6 promoted tumor growth and inhibited apoptosis in GC. Furthermore, we demonstrated CPSF6 negatively regulated von Hippel Lindau (VHL), the gene encoding a tumor suppressor, through selective poly(A) usage, thus contributing to tumor growth in GC. Our data provide insights into the function of CPSF6 and may imply potential therapeutic targets.
Materials and Methods
Cell Lines and Cell Cultures
Human normal gastric cell line GES1 and GC cancer cell lines (AGS, BGC-823, MKN-28) were purchased from the Chinese Academy of Sciences cell bank (Shanghai, China). All cells were cultured in RPMI-1640 medium (Gibco, United States) supplemented with 10% fetal bovine serum (FBS, Gibco, United States) and penicillin–streptomycin solution (Sangon, China) at 37°C in a 5% CO2 humidified atmosphere.
Quantitative Real Time RT-PCR (qRT-PCR)
Total RNA was isolated using Trizol (Life Technologies, United States) and reverse transcribed with PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Japan). Real-time qRT-PCR analysis was performed using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara), according to the manufacturer’s protocol. The primers used for RT-qPCR analysis are listed as follows: VHL 5′-CCCGTATGGCT CAACTTCG-3′ and 5′-GGTTAACCAGAAGCCCATCG-3′; GAPDH 5′-CACAGTCCATGCCATCACTG-3′ and 5′-CTTGGCAGCGCCAGTAAG-3′. The expression of genes was normalized to GAPDH.
Western Blot Analysis
Cells were lysed in ice-cold RIPA Lysis and Extraction Buffer (Thermo, United States) supplemented with protease (Roche, Switzerland) and phosphatase inhibitor cocktail (Roche). The protein concentrations were determined using BCA Protein Assay Kit (Pierce, United States). Protein lysate was separated by 10% or 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis system (SDS-PAGE) and transferred onto PVDF membranes (Millipore, United States). The membrane was incubated in blocking buffer (1 × TBST with 5% BSA) for 2 h at room temperature and then incubated in diluted primary antibodies: anti-CPSF6 (1:1,000; Abcam, United Kingdom), anti-VHL (1:1,000, Sangon), anti-GAPDH (1:1,000; Proteintech, China), anti-β-actin (1:5,000; Sigma, United States), and anti-β-tubulin (1:1,000; Yeasen, China), and secondary antibody: peroxidase-conjugated goat anti-rabbit IgG (H+L) (1:5,000; Yeasen). The membrane was visualized using ECL start Western Blotting Substrate (GE Healthcare Life Sciences, United States). The intensities of the bands were qualified by ImageJ (National Institutes of Health, United States).
Generation of Stable Cell Lines
The lentiviral vector pLKO.1-puro-shRNA-CPSF6 was constructed for CPSF6 knockdown as described previously (Tan et al., 2021). The lentiviral vectors pCDH-PGK-VHL-CDS-short 3′UTR-SV40-NeoR and pCDH-PGK-VHL-CDS-long 3′UTR-SV40-NeoR were constructed for VHL overexpression. 293T cells were transfected for the lentiviral production. The supernatant was harvested after 48 and 72 h, respectively. Then AGS and BGC-823 cells were infected by supernatant for 72 h. Finally, cells were maintained in the presence of puromycin (2 μg/ml) and G418 (600 μg/ml or 1.2 mg/ml) for 2 weeks.
Cell Proliferation and Cell Viability Assays
For cell proliferation assay, cells (1 × 105 cells per well) were plated into 6-well plates. After a certain period of time, count the cell number of fields. Then, calculate the cell number of each well. For cell viability assay, cells (1 × 103 cells per well) were plated into 96-well plates. Five days later, 20 μl of MTT solution (5 mg/ml) was added into each well and cells were cultured for 2 h, and cells were rinsed by PBS and 100 μl DMSO was added into each well. Then the absorbance at 570 nm wavelength was examined.
Colony Formation Assays
GC cells were seeded into 6-well plates (500–1,000 cells/well) and cultured for 2 weeks. Cells were fixed with 4% methanol, stained with crystal violet (0.1%), and photographed.
Cell Apoptosis Assays
The apoptosis rate was evaluated using the Annexin V-FITC/PI Apoptosis Detection kit (Vazyme, China) according to the instructions from the manufacturer. The cells were seeded into 6-well tissue culture plates (1–4 × 105 cells/well). After cultured in serum-free medium for 36 h, the cells were collected, washed with ice-cold PBS, and resuspended in 500 μl binding buffer. Then, 5 μl Annexin V-FITC and 5 μl PI were added to the buffer and incubated at room temperature for 10 min in the dark. Cells were analyzed by BD LSRFortessa (BD Biosciences, United States) within 1 h. The TUNEL staining assay was performed using Fluorescein (FITC) Tunel Cell Apoptosis Detection Kit (Servicebio, China) according to the manufacturer’s instructions.
Xenograft Model in vivo
The design and protocol of in vivo experiments were approved by the Institutional Animal Care and Use Committee, Shanghai Jiao Tong University. For tumor growth assay, GC cells were resuspended in PBS/Matrigel Matrix mix at 1:1 ratio. Then 3 × 106 cells in 125 μl solution were injected into the flank of 4-week-old athymic male nude mice. Tumor volumes were calculated as follows: volume = ½ (L × W2), where L is the length and W is the width. Tumor weight was measured when the mice were executed. The IHC staining of xenograft tumors was performed following the manufacturer’s instruction. IHC labeling analysis was reviewed and scored by two independent pathologists.
Dual Luciferase Reporter Assays
The short or the long 3′UTR of the VHL gene was cloned into reporter plasmid psiCHECK2 (Promega, United States). GC cells were transfected with the reporter vector in 24-well plates (200 ng/well) using Lipofectamine 3000 (Invitrogen, United States). After 48 h, cells were collected and assayed using dual luciferase assay according to the manufacturer’s protocol (Transgen, China).
3T-Seq Analysis
3T-seq analysis was performed as described previously (Lai et al., 2015; Tan et al., 2018). Briefly, 50 μg total RNA was incubated with M280 beads, which were bound by Bio-oligo dT (Lou et al., 2017). After cDNA synthesis, the 3′UTR fragments were released by Gsu I digestion and then subjected to deep sequencing. The filtered reads were mapped to UCSC human reference genome (hg19) with bowtie2 (Langmead and Salzberg, 2012). The 3′UTR alteration for each gene in the cell lines was detected by linear trend test (the FDR-adjusted p-value < 0.05) (Fu et al., 2011). These sequence data have been submitted to the EMBL-EBI databases under accession number E-MTAB-9980 and available in the ArrayExpress database.1
Statistical Analysis
All experiments were repeated at least three times. The results of this study were analyzed with GraphPad statistical software (GraphPad Software, United States) and shown as mean ± SD. We downloaded the expression data of CPSF6 in normal and GC tissues from The Cancer Genome Atlas (TCGA) database2 and selected frozen sample data for analysis. To investigate the differential expression of CPSF6 between normal and GC tissues of distinct cancer stages or nodal metastasis status in TCGA, we used the website tool3.
Results
CPSF6 Is Upregulated in GC
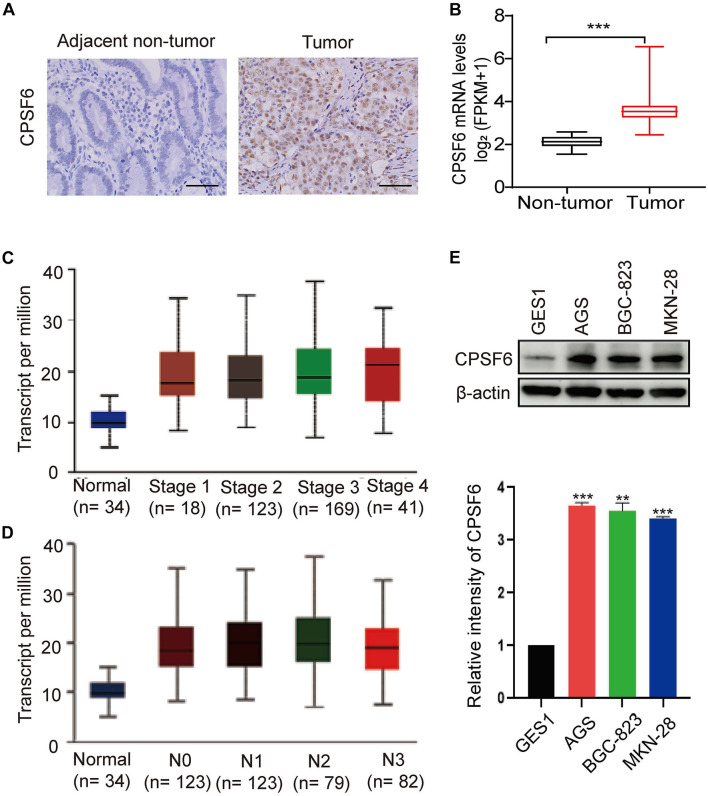
In this study, we first analyzed the CPSF6 levels using IHC assay in GC tissues and the paired non-tumor tissues. IHC staining assay showed that CPSF6 was upregulated in tumor tissues when compared with the surrounding non-tumor counterparts (Figure 1A). To explore the clinical relevance of CPSF6 in GC patients, we next compared the expression of CPSF6 mRNA, with the RNA-seq data derived from GC (n = 352) and non-tumor tissues (n = 32) in TCGA. We discovered that the expression of CPSF6 was significantly upregulated in GC tissues (Figure 1B). According to 7th edition of the AJCC Cancer Staging Manual, cancer stages and nodal metastasis were recommended to the TNM gastric cancer staging system (Washington, 2010). To further examine the levels of CPSF6 in distinct GC stages and nodal metastasis, we evaluated the expression of CPSF6 in normal and GC tissues based on individual cancer stages or nodal metastasis. The results showed that the levels of CPSF6 were higher in GC. However, the levels of CPSF6 in different stages and nodal metastasis were similar (Figures 1C,D and Supplementary Tables 1, 2). We also analyzed the overall survival curves of CPSF6, and the result showed no significant difference between low and high level of CPSF6 in GC patients (data not shown). The malignancy of GC exhibits the following characteristics: metastasis and uncontrolled proliferation (Hu et al., 2020). The results implied that involvement of CPSF6 in GC formation may be due to the cell growth, rather than the metastasis in GC. We then examined the protein expression of CPSF6 in non-tumorous gastric cell line GES1 and human GC cell lines (AGS, BGC-823, MKN-28). We found that the protein levels of CPSF6 increased in the GC cell lines compared with non-tumorous gastric cell line (Figure 1E). Together, our data suggested that CPSF6 was upregulated in GC tissue and cells.
FIGURE 1.
Expression of CPSF6 was upregulated in GC. (A) The IHC staining assay of CPSF6 in GC and adjacent tissues of the tumor (scale bar: 100 μm). (B) Analysis of CPSF6 mRNA levels of GC (n = 352) and non-tumor tissues (n = 32) in TCGA. (C) Expression of CPSF6 in GC based on individual cancer stages in TCGA. (D) Expression of CPSF6 in GC based on nodal metastasis status in TCGA. (E) Western blot assay for detecting CPSF6 expression in different cell lines. **0.001 < p < 0.01; ***p < 0.001.
CPSF6 Promotes Cell Growth and Inhibits Apoptosis in GC Cells in vitro
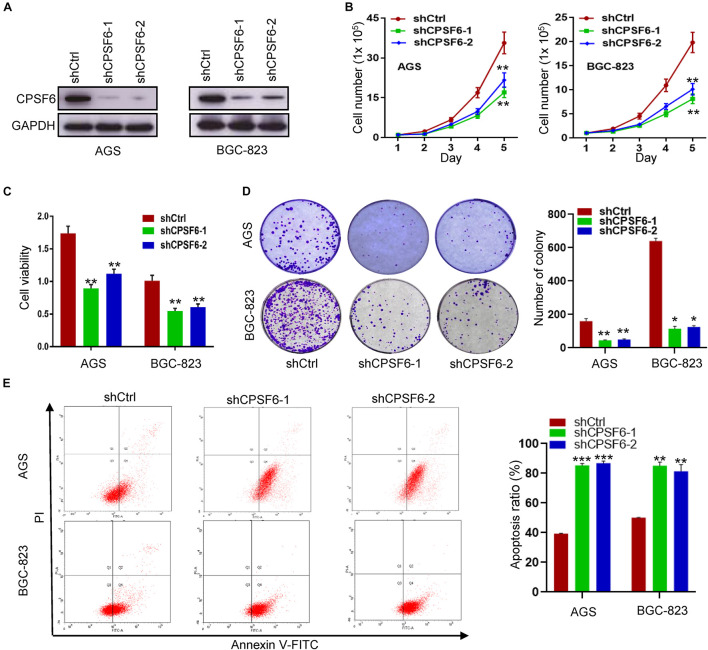
To investigate the function of CPSF6 in GC, we knocked down CPSF6 with short hairpin RNAs (shRNAs) in AGS and BGC-823 cells, which have higher levels of CPSF6 (Figure 1E). Expression of CPSF6 in these cell lines was obviously repressed (Figure 2A). Compared with the control, CPSF6 knockdown in AGS and BGC-823 significantly decreased cell proliferation (Figure 2B), viability (Figure 2C), and generated less and smaller colonies in colony formation assay (Figure 2D). It is reported that the cell cycle and apoptosis affect cell proliferation (Alenzi, 2004). We next investigated if CPSF6 regulated the cell growth through cell cycle and apoptosis in GC cells. The results showed that AGS-shCPSF6 cells increased fraction of S phase cells (p < 0.001), but BGC-823-shCPSF6 cells showed no significant changes (p > 0.05) (Supplementary Figure 1). Interestingly, both CPSF6 knockdown cell lines exhibited noteworthy increases in cell apoptosis (Figure 2E).
FIGURE 2.
CPSF6 promotes cell growth and inhibits apoptosis in GC cells in vitro. (A) Western blotting analysis of CPSF6 expression in AGS and BGC-823 cells stably transfected with shRNA plasmids. (B,C) Detection of cell proliferation and viability after knockdown of CPSF6 in AGS and BGC-823 cells. (D) Detection of colony formation after knockdown of CPSF6 in AGS and BGC-823 cells. (E) Detection of apoptosis in AGS and BGC-823 cells. *0.01 < p < 0.05; **0.001 < p < 0.01; ***p < 0.001.
CPSF6 Promotes Proliferation and Inhibits Apoptosis of GC Cells in vivo
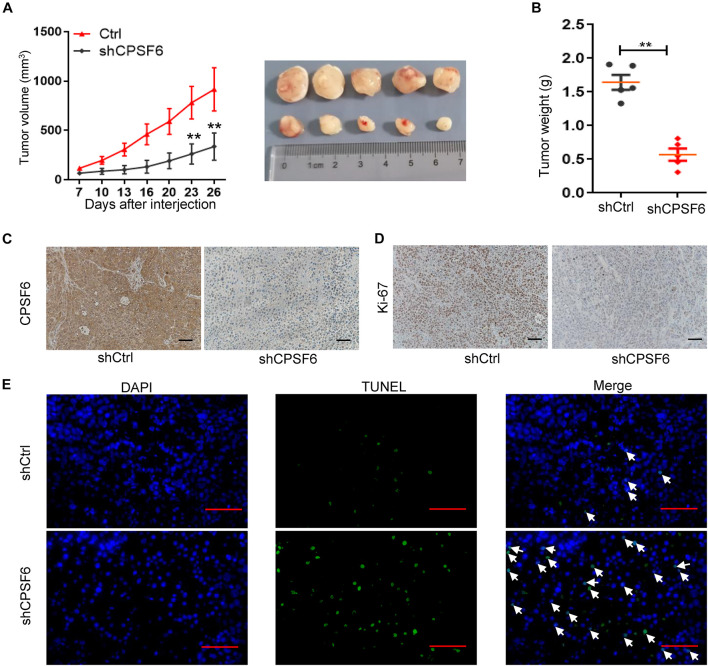
Considering that AGS cells have the highest levels of CPSF6 in the three GC cell lines, the AGS stably transfected cells (AGS-shCtrl, AGS-shCPSF6) were implanted subcutaneously into the flanks of nude mice to further assess the role of CPSF6 in vivo. CPSF6 silencing mouse model result also showed similar biological effects in GC cells. We observed a significant decrease in both tumor growth and weight (Figures 3A,B) in AGS-shCPSF6 mouse model compared with shCtrl tumors. Besides that, Ki-67 staining result displayed that tumors derived from AGS-shCPSF6 cells exhibited a significantly lower percentage of proliferative cells, as opposed to tumors derived from AGS-shCtrl cells (Figures 3C,D). In contrast, TUNEL assay result showed that CPSF6 knockdown increased the apoptosis percentage (Figure 3E). Collectively, our results demonstrated that knockdown of CPSF6 inhibited cell growth and increased apoptosis in GC cells.
FIGURE 3.
CPSF6 promotes cell growth and inhibits apoptosis in GC cells in vivo. (A,B) Xenograft nude mouse model of tumorigenesis was used to confirm cell growth and weight after knockdown of CPSF6. (C,D) CPSF6 and Ki-67 staining of the respective GC cell–derived tumor sections (scale bar: 100 μm). (E) TUNEL assay of the respective GC cell–derived tumor sections (scale bar: 100 μm). The white arrows represent apoptotic cells. (**0.001 < p < 0.01).
Knockdown of CPSF6 Modulates the Widespread 3′UTR Shortening in GC Cells
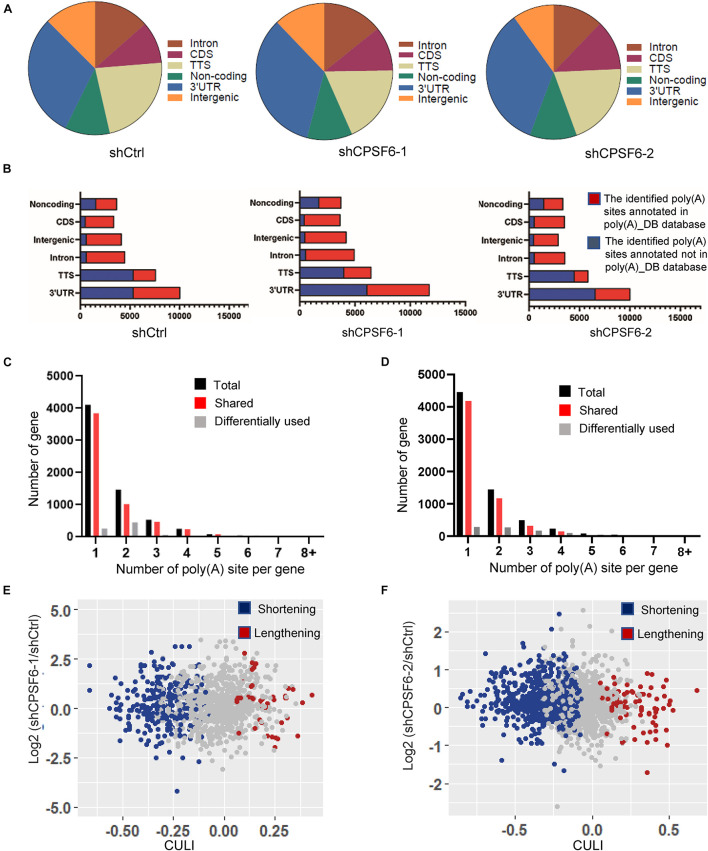
Given that CPSF6 involves in APA formation and its role in promoting cell growth and restraining apoptosis in GC cells as described earlier, we hypothesized that CPSF6 acts as a tumor promoter in GC, at least in part, by influencing APA and 3′UTR. To test this hypothesis, we performed transcriptome-wide APA profiling analysis in CPSF6 knockdown AGS cells and the control cells, using 3T-seq we reported previously (Lai et al., 2015). In total, we gained about 35.7 million reads in AGS-shCtrl cells, as well as 28.9 and 30.9 million reads in AGS-shCPSF6-1 and shCPSF6-2 cells, respectively. About 10.4, 6.6, and 9.1 million reads were uniquely mapped to the reference genome (Supplementary Table 3). The internal priming was determined through examining the presence of more than 12 “A”s or continuous 8 “A”s at the region of 1–20 nt downstream the mapping position. After filtering internal priming events, we found majority of qualified reads were mapped to the annotated transcription terminal sites (TTSs) or 3′UTRs (Figure 4A), yielding over 10,000 poly(A) sites (Supplementary Table 3).
FIGURE 4.
Characteristics of 3T-seq data. (A) Genomic locations of 3T-seq reads mapped to the reference genome after filtering internal priming events. (B) Genomic distribution of the poly(A) sites. (C,D) The statistics of genes with various number of detected poly(A) sites in different libraries. (E,F) Scatterplot of CULI in CPSF6 knockdown and control cells [false discovery rate (FDR) < 0.05] in different libraries.
Applying the snowball clustering method (Fu et al., 2011), we found that most of the identified poly(A) sites were located at the 3′ terminus of genes. Particularly, about 60–76% of these poly(A) sites were mapped to TTSs and 52–70% to the 3′UTR regions in each 3T-seq library (Figure 4B), according to PolyA_DB database (Wang et al., 2018). More than one third of these poly(A) sites have been annotated in the poly(A) database, and thus the remaining sites may be potential new sites (Figure 4B). Also, about 30% of genes harbor three or more poly(A) sites (Figures 4C,D). We measured the 3′UTR alteration for CPSF6 responsive genes by introducing cancer 3′UTR length index (CULI) (Fu et al., 2011). A positive CULI suggests that a gene harbors lengthened 3′UTR in CPSF6 knockdown cells compared with the control cells, and a negative CULI indicates the shortened one. Compared with AGS-shCtrl cells, we identified 494 and 633 genes with altered 3′UTR in two independent CPSF6 shRNA knockdown AGS cells, respectively, and 83% and 89% of which displayed a shift from distal to proximal poly(A) site usage and thus possessed shortened 3′UTRs (Figures 4E,F and Supplementary Tables 4, 5). This result revealed that knockdown of CPSF6 modulated the widespread 3′UTR shortening in GC cells, which was consistent with previous studies in different cell types (Gruber et al., 2012; Martin et al., 2012).
CPSF6 Downregulates the Expression of VHL Through APA
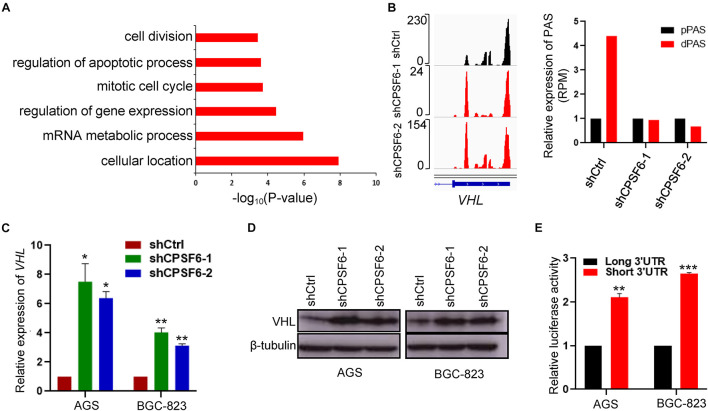
To ask how CPSF6-modulated APA contributes GC tumorigenesis, we assessed the functional consequence of CPSF6 responsive genes. Thus, to further understand the biological function of APA alternation in GC cells, we collected the 243 genes with shortened 3′UTR, which was simultaneous happened in the two independent AGS CPSF6 knockdown cells and performed the gene ontology (GO) enrichment analysis, using PANTHER classification system (p-value < 0.001, FDR < 0.005)4 (Supplementary Table 6). Notably, GO terms indicated that 3′UTR shortened genes are involved in regulation of apoptosis process and in other biological processes such as cellular and mRNA metabolic processes (Figure 5A).
FIGURE 5.
CPSF6 downregulates the expression of VHL. (A) GO analysis of genes with shortened 3′UTR after knockdown of CPSF6 in AGS cells. (B) CPSF6 induced APA shift of VHL. Left, Integrative Genomics Viewer (IGV) genome browser exhibited the poly(A) site usage of VHL 3′UTR. Right, histogram showed the relative expression of the isoform with distal polyadenylation site (dPAS) relative to the one with proximal PAS (pPAS). (C) The mRNA levels of VHL in CPSF6 knockdown AGS and BGC-823 cells. (D) The protein levels of VHL in CPSF6 knockdown AGS and BGC-823 cells. (E) The luciferase reporter assay for the expression activity of the isoform of VHL 3′UTR in AGS and BGC-823 cells. *0.01 < p < 0.05; **0.001 < p < 0.01; ***p < 0.001).
Next, we manually inspected the apoptosis-promoting or proliferation-inhibiting genes under the criterion that the proximal poly(A) site usage increased and the distal poly(A) site usage decreased in both AGS knockdown cells, which were visualized in Integrative Genomics Viewer (IGV) genome browser, yielding three candidates (IER3IP1, IGF1R, and VHL) with top 3′UTR usage fold changes (Figure 5B and Supplementary Figure 2A). Then, we detected the mRNA expression of the candidate genes by RT-qPCR, and identified VHL as CPSF6 responsive gene, which had highest increased mRNA levels in CPSF6 knockdown AGS and BGC-823 cells (Figure 5C and Supplementary Figure 2B). Moreover, western blotting results indicated that CPSF6 negatively regulated VHL, that is, VHL expression increased in case of CPSF6 knockdown (Figure 5D).
A recent study has shown that the gene isoform with short 3′UTR had higher protein expression than the longer one, via escaping the repression of miRNA (Sandberg et al., 2008). As predicted, most of the predicted miRNA binding sites lay outside the proximal poly(A) sites on VHL 3′UTR (Supplementary Figure 2C). To investigate if the distinct 3′UTR isoforms affect the expression of proteins, we examined expression the short or long VHL 3′UTR fragments through the dual luciferase reporter system. As expected, the vector with short 3′UTR showed significantly higher luciferase activities than that with the long 3′UTR, which means short 3′UTR had higher expression ability (Figure 5E). Taken together, these data suggested that the expression of VHL was downregulated by CPSF6 through APA in GC.
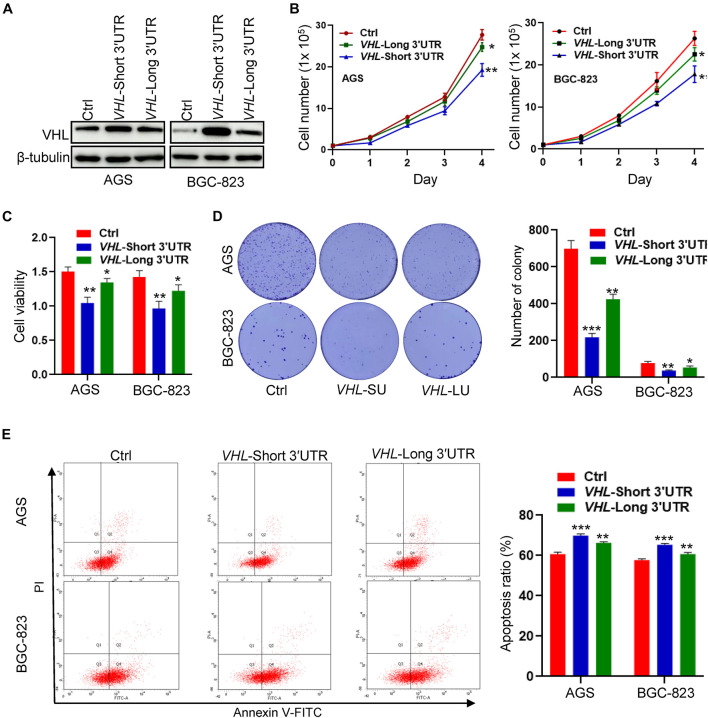
VHL Short−3′UTR Isoform Enhances Apoptosis and Inhibits Cell Growth in GC Cells
To study the biological function of distinct VHL isoforms in GC cells, we stably transfected short- or long−3′UTR isoform of VHL into two GC cell lines: AGS and BGC-823. The protein expression of VHL is shown in Figure 6A. We found that the short−3′UTR VHL isoform dramatically upregulated the protein expression of VHL, as compared with transfection with the long−3′UTR VHL isoform. We further observed that of the short−3′UTR isoform of VHL in GC cells significantly decreased cell growth, cell viability (Figures 6B,C), colony formation (Figure 6D), and enhanced cell apoptosis (Figure 6E) when compared with that of control cells with the expression of full-length VHL and control vector. These data, collectively, suggested high levels of protein produced by VHL with shorter 3′UTR may, at least partially, contribute to enhance apoptosis and inhibit cell growth in GC cells.
FIGURE 6.
The VHL isoform with short 3′UTR inhibits cell growth and promotes apoptosis in GC cells. (A) Western blotting analysis of VHL expression in AGS and BGC-823 cells stably transfected with distinct VHL isoform with short or long 3′UTR. (B,C) Detection of cell proliferation and viability after stably enforced expression of the short- or long-3′UTR isoform of VHL in AGS and BGC-823 cells. (D) Detection of colony formation after stably enforced expression of the VHL isoform with short or long 3′UTR in AGS and BGC-823 cells. (E) Apoptosis ratio was analyzed in AGS and BGC-823 cells. *0.01 < p < 0.05; **0.001 < p < 0.01; ***p < 0.001.
Discussion
GC is the third leading cause of cancer mortality across the world (Zhang and Zhang, 2017; Smyth et al., 2020). However, the mechanisms of GC progression are poorly understood. Increasing evidence has proved that APA is involved in many biological functions, including tumorigenesis (Singh et al., 2009; Xia et al., 2014). The APA-mediated regulatory mechanism is determined by numerous APA factors (Tian and Manley, 2017), and some of them are reported to participate in the tumor process. For example, NUDT21 showed a dual role in different cancer types (Masamha et al., 2014; Zhang and Zhang, 2018). This reflects that the regulatory mechanism of APA factors is complex and cancer specific, and it is necessary to characterize the role of APA in different types of cancers. Although CPSF6 played roles in aggressive breast cancer behavior and exhibited a novel CPSF6-RARG fusion variant in acute myeloid leukemia patients (Binothman et al., 2017; Zhang et al., 2020), the role of CPSF6 in GC is still unknown.
In this study, we examined expression of CPSF6 in GC and normal tissues. We found that CPSF6 was upregulated in GC, and what is more, the levels of CPSF6 in different stages and nodal metastasis were similar. We also analyzed the survival curves of CPSF6, and the result showed no significant difference between low and high level of CPSF6 in GC patients (data not shown). This may imply CPSF6 is not involved in metastasis and may contribute to other biological processes (such as proliferation) during GC initiation and progress. Here, we focus on investigating the relationship between CPSF6 and GC growth. Through in vitro and in vivo assays, we demonstrated CPSF6 enhances the proliferation and inhibits apoptosis in GC.
Given that CPSF6 involves in APA formation and its relevance in GC cells as described previously, we hypothesized that CPSF6 acts as a tumor promoter in GC, at least in part, by influencing APA and 3′UTR. We next performed the APA profiling analysis and identified the majority of APA genes with shortened 3′UTR under the condition of CPSF6 knockdown. The data suggested that knockdown of CPSF6 modulates the widespread 3′UTR shortening in GC cells. In another of our study, we analyzed the usage of 3′UTR in GC relative to normal cells (Lai et al., 2015), and the results showed a global 3′UTR shortening and tendency to use shorter isoforms in GC relative to normal cells. It is very interesting that cancer is associated with global 3′UTR shortening relative to normal cells, and our results seem that CPSF6 promotes cancer via 3′UTR lengthening. Consider that APA factors involve in cancer and regulate the APA progress. We have analyzed the expression of 20 core APA factors in GC and normal tissues using a website tool (see text footnote 3). Beyond our expectation, we found that 19 of 20 core APA factors were upregulated in GC tissues, and only RBP1 was not regulated significantly (p = 0.109) (data not shown). Next, we have searched the function of the 19 APA factors on regulation of 3′UTR. We found that eight factors (CPSF1, CPSF3, CSTF3, CSTF1, PCF11, SYMPK, RBBP6, FIP1L1) prefer to produce the short 3′UTR, and the rest of the factors (WDR33, CPSF4, CPSF2, CSTF2, NUDT21, CPSF6, CPSF7, CLP1, PAPOLA, PABPN1, PABPC1, RBP1) prefer to produce the long 3′UTR (Gruber et al., 2012; Martin et al., 2012; Li et al., 2015). Different factors make different contributions to APA. So, the tendency to use which isoforms in GC is determined by multiple factors. We assumed that although CPSF6 could promote cancer via 3′UTR lengthening, the other APA factors still regulated 3′UTR shortening in GC cells and led GC cells to still prefer to use short 3′UTR.
To understand how CPSF6 exerts its proliferation-promoting and apoptosis-inhibiting function by APA in GC cells, we further characterized CPSF6 regulated genes with shortened 3′UTRs. VHL was demonstrated to contribute to the function of CPSF6-mediated tumor growth in GC cells. We also analyzed the expression of VHL in GC relative to normal tissues. The results showed that the levels of VHL were higher in GC tissues and the expression of VHL in different stages and nodal metastasis was similar (data not shown). As CPSF6 was upregulated in GC cells and downregulated in the VHL, how was VHL increased in GC tissues? As we all know, many genes are expressed in cancer abnormally, and each of these genes makes partial contribution to cancer. To figure out this question, we analyzed the expression of all genes in GC relative to normal tissues (data not shown). We found that about 3,740 genes were upregulated in GC tissues, including 19 APA factors as described earlier. The 19 APA factors may play an important role in regulating the 3′UTR of VHL in different ways and lead to higher expression of VHL in GC. Other genes were also increased in GC tissues, for example, ZNF350 (ZBRK1) was upregulated in GC tissues, and it was reported that ZNF350 could activate VHL gene transcription through formation of a complex with VHL and p300 in renal cancer (Chen et al., 2015). We also found that about 900 genes were downregulated in GC tissues, including Daam2. According to the report, Daam2 suppressed VHL expression and promoted tumorigenesis (Zhu et al., 2017). We supposed that although CPSF6 negatively regulated the VHL expression partly, other proteins might increase the level of VHL in GC tissues, and which was stronger than the effect of CPSF6 on expression of VHL, leading to a higher level of VHL in GC tissues.
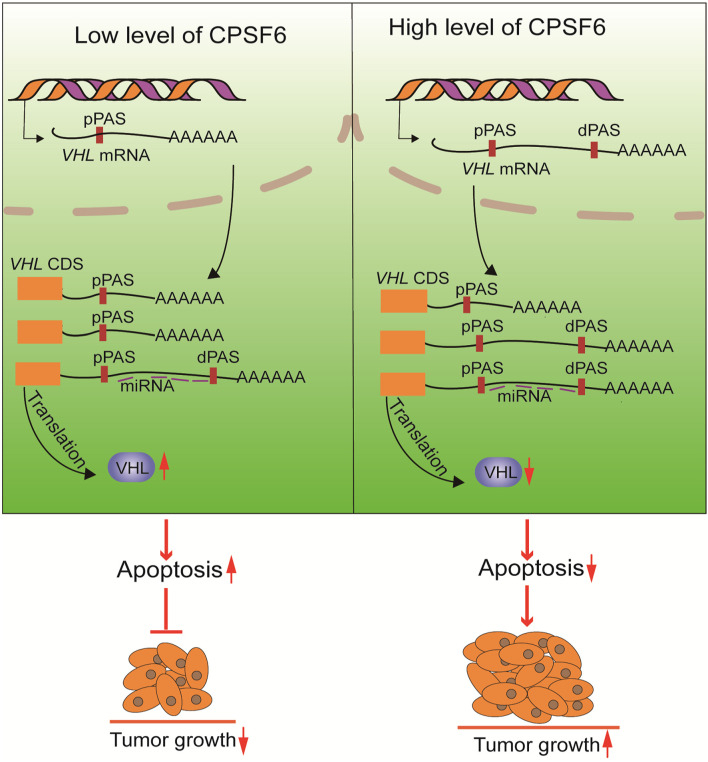
In this study, we validated that knockdown of CPSF6 leads to a shift from distal to proximal poly(A) site usage, the shortening of 3′UTR in VHL, which consequently causes the increased expression of VHL, and inhibition of proliferation in GC cells (Figure 7). VHL functions as a tumor suppressor gene in some tumors, such as renal cell carcinoma and pheochromocytoma (Richards, 2001), but few studies have reported its role in GC. Interestingly, it was reported that the genetic and epigenetic alterations of the VHL were not detected in GC (Cao et al., 2008). Thus, our study may offer a new perspective to understand the regulation of VHL. However, how CPSF6 regulates the 3′UTR length and whether other APA factors are involved this progress and the regulation of APA factors in GC and normal tissues still need to be explored. In any case, our data may provide new insights into the understanding of CPSF6’s role in GC cells and may imply potential therapeutic targets.
FIGURE 7.
The proposed mechanism of CPSF6 in tumorigenesis of GC progression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Biomedical Ethics Committee of Anhui Medical University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Institutional Animal Care and Use Committee, Shanghai Jiao Tong University.
Author Contributions
XS, KD, and ST performed most of the experiments and interpretation of the data. QZ, PL, and YK contributed to the analysis of the data and advised on the experimental design. ST and XS wrote the article. ST and JS critically revised the manuscript and contributed to the conception and design. All authors read and approved the final article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Xiaodong Zhao of Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University, China, for the helpful discussion on the conception and design of this study.
Footnotes
Funding
This research was supported by the Shanghai Jiao Tong University Scientific and Technological Innovation Fund (No. 2019TPA09), the Major Projects of Science and Technology Commission of Shanghai (No. 17JC1400800), the Interdisciplinary Program of Shanghai Jiao Tong University (No. YG2019ZDA25), the Special Fund Project for Youth Scientific Research of the Key Laboratory of Systems Biomedicine (Ministry of Education) (No. KLSB2020QN-07), and the Natural Science Foundation of Anhui Province (No. 2008085QH378).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.707644/full#supplementary-material
The cell cycle distribution after knockdown of CPSF6 in AGS and BGC-823 cells.
The candidate genes of CPSF6. (A) CPSF6 induced APA shift of candidate genes. UP, Integrative Genomics Viewer (IGV) genome browser exhibited the poly(A) site usage of candidate genes’ 3′UTR. Down, histogram showed the relative expression of the isoform with distal polyadenylation site (dPAS) relative to the one with proximal PAS (pPAS). (B) The mRNA levels of candidate genes in CPSF6 knockdown AGS cells (up) and BGC-823 cells (down). (C) Schematic illustration of the VHL isoform with long or short 3′UTR. Positions of the binding sites of miRNA were predicted by website tool (http://www.targetscan.org/) and indicated by purple horizontal lines. CR, CDS region. ∗0.01 < p < 0.05; ∗∗0.001 < p < 0.01; ∗∗∗p < 0.001.
Statistical significance of CPSF6 expression in GC of individual cancer stages.
Statistical significance of CPSF6 expression in GC based on nodal metastasis status.
The sequencing statistics of AGS 3T-seq libraries.
The list of genes with altered 3′UTR in AGS shCPSF6-1 cells.
The list of genes with altered 3′UTR in AGS shCPSF6-2 cells.
The list of genes with shortened 3′UTR shared by AGS shCPSF6-1 and shCPSF6-2 cells.
References
- Alenzi F. Q. (2004). Links between apoptosis, proliferation and the cell cycle. Br. J. Biomed. Sci. 61 99–102. 10.1080/09674845.2004.11732652 [DOI] [PubMed] [Google Scholar]
- Andreassi C., Riccio A. (2009). To localize or not to localize: mRNA fate is in 3 3tability. Trends Cell Biol. 19 465–474. 10.1016/j.tcb.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Bejarano D. A., Peng K., Laketa V., Börner K., Jost K. L., Lucic B., et al. (2019). HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. Elife 8:e41800. 10.7554/eLife.41800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binothman N., Hachim I. Y., Lebrun J. J., Ali S. (2017). CPSF6 is a Clinically Relevant Breast Cancer Vulnerability Target: role of CPSF6 in Breast Cancer. EBioMedicine 21 65–78. 10.1016/j.ebiom.2017.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Song J. H., Kim C. J., Cho Y. G., Kim S. Y., Nam S. W., et al. (2008). Genetic and epigenetic analysis of the VHL gene in gastric cancers. Acta Oncol. 47 1551–1556. 10.1080/02841860802001459 [DOI] [PubMed] [Google Scholar]
- Chen K., Yu G., Gumireddy K., Li A., Yao W., Gao L., et al. (2015). ZBRK1, a novel tumor suppressor, activates VHL gene transcription through formation of a complex with VHL and p300 in renal cancer. Oncotarget 6 6959–6976. 10.18632/oncotarget.3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang J. X., Luo J. H., Wu S., Yuan G. J., Ma N. F., et al. (2018). CSTF2-Induced Shortening of the RAC1 3′UTR Promotes the Pathogenesis of Urothelial Carcinoma of the Bladder. Cancer Res. 78 5848–5862. 10.1158/0008-5472.CAN-18-0822 [DOI] [PubMed] [Google Scholar]
- Chia N. Y., Tan P. (2016). Molecular classification of gastric cancer. Ann. Oncol. 27 763–769. 10.1093/annonc/mdw040 [DOI] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 79 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Flavell S. W., Kim T.-K., Gray J. M., Harmin D. A., Hemberg M., Hong E. J., et al. (2008). Genome-Wide Analysis of MEF2 Transcriptional Program Reveals Synaptic Target Genes and Neuronal Activity-Dependent Polyadenylation Site Selection. Neuron 60 1022–1038. 10.1016/j.neuron.2008.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sun Y., Li Y., Li J., Rao X., Chen C., et al. (2011). Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21 741–747. 10.1101/gr.115295.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Martin G., Keller W., Zavolan M. (2012). Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 9 1405–1412. 10.4161/rna.22570 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhang Y., Ding M., Xu R. (2020). LncRNA TMPO-AS1/miR-126-5p/BRCC3 axis accelerates gastric cancer progression and angiogenesis via activating PI3K/Akt/mTOR pathway. J. Gastroenterol. Hepatol. 36 1877–1888. 10.1111/jgh.15362 [DOI] [PubMed] [Google Scholar]
- Ji Z., Tian B. (2009). Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4:e8419. 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackford B., Yao C., Charles G. M., Weng L., Zheng X., Choi E.-A., et al. (2014). Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 33 878–889. 10.1002/embj.201386537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D. P., Tan S., Kang Y. N., Wu J., Ooi H. S., Chen J., et al. (2015). Genome-wide profiling of polyadenylation sites reveals a link between selective polyadenylation and cancer metastasis. Hum. Mol. Genet. 24 3410–3417. 10.1093/hmg/ddv089 [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Ambrose Z., Martin T. D., Oztop I., Mulky A., Julias J. G., et al. (2010). Flexible Use of Nuclear Import Pathways by HIV-1. Cell Host Microbe 7 221–233. 10.1016/j.chom.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., et al. (2015). Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 11:e1005166. 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J. C., Lan Y. L., Gao J. X., Ma B. B., Yang T., Yuan Z. B., et al. (2017). Silencing NUDT21 Attenuates the Mesenchymal Identity of Glioblastoma Cells via the NF-κB Pathway. Front. Mol. Neurosci. 10:420. 10.3389/fnmol.2017.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gruber Andreas R., Keller W., Zavolan M. (2012). Genome-wide Analysis of Pre-mRNA 3′ End Processing Reveals a Decisive Role of Human Cleavage Factor I in the Regulation of 3′ UTR Length. Cell Rep. 1 753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Masamha C. P., Xia Z., Peart N., Collum S., Li W., Wagner E. J., et al. (2016). CFIm25 regulates glutaminase alternative terminal exon definition to modulate miR-23 function. RNA 22 830–838. 10.1261/rna.055939.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha C. P., Xia Z., Yang J., Albrecht T. R., Li M., Shyu A. B., et al. (2014). CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510 412–416. 10.1038/nature13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oue N., Sentani K., Sakamoto N., Uraoka N., Yasui W. (2019). Molecular carcinogenesis of gastric cancer: lauren classification, mucin phenotype expression, and cancer stem cells. Int. J. Clin. Oncol. 24 771–778. 10.1007/s10147-019-01443-9 [DOI] [PubMed] [Google Scholar]
- Petrovchich I., Ford J. M. (2016). Genetic predisposition to gastric cancer. Semin. Oncol. 43 554–559. 10.1053/j.seminoncol.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Richards F. M. (2001). Molecular pathology of von HippelLindau disease and the VHL tumour suppressor gene. Expert Rev. Mol. Med. 2001 1–27. 10.1017/s1462399401002654 [DOI] [PubMed] [Google Scholar]
- Rüegsegger U., Blank D., Keller W. (1998). Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1 243–253. 10.1016/s1097-2765(00)80025-8 [DOI] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320 1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Alley T. L., Wright S. M., Kamdar S., Schott W., Wilpan R. Y., et al. (2009). Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 69 9422–9430. 10.1158/0008-5472.CAN-09-2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E. C., Nilsson M., Grabsch H. I., van Grieken N. C. T., Lordick F. (2020). Gastric cancer. Lancet 396 635–648. 10.1016/S0140-6736(20)31288-5 [DOI] [PubMed] [Google Scholar]
- Tan S., Li H., Zhang W., Shao Y., Liu Y., Guan H., et al. (2018). NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene 37 4887–4900. 10.1038/s41388-018-0280-6 [DOI] [PubMed] [Google Scholar]
- Tan S., Zhang M., Shi X., Ding K., Zhao Q., Guo Q., et al. (2021). CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 40:85. 10.1186/s13046-021-01884-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Manley J. L. (2017). Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Nambiar R., Zheng D., Tian B. (2018). PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 46 D315–D319. 10.1093/nar/gkx1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington K. (2010). 7th Edition of the AJCC Cancer Staging Manual: stomach. Ann. Surg. Oncol. 17 3077–3079. 10.1245/s10434-010-1362-z [DOI] [PubMed] [Google Scholar]
- Xia Z., Donehower L. A., Cooper T. A., Neilson J. R., Wheeler D. A., Wagner E. J., et al. (2014). Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 5:5274. 10.1038/ncomms6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang W. (2018). Knockdown of NUDT21 inhibits proliferation and promotes apoptosis of human K562 leukemia cells through ERK pathway. Cancer Manag. Res. 10 4311–4323. 10.2147/cmar.S173496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Zhang P. Y. (2017). Gastric cancer: somatic genetics as a guide to therapy. J. Med. Genet. 54 305–312. 10.1136/jmedgenet-2016-104171 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Jiang M., Borthakur G., Luan S., Huang X., Tang G., et al. (2020). Acute myeloid leukemia with a novel CPSF6-RARG variant is sensitive to homoharringtonine and cytarabine chemotherapy. Am. J. Hematol. 95 E48–E51. 10.1002/ajh.25689 [DOI] [PubMed] [Google Scholar]
- Zhu W., Krishna S., Garcia C., Lin C. J., Mitchell B. D., Scott K. L., et al. (2017). Daam2 driven degradation of VHL promotes gliomagenesis. Elife 6:e31926. 10.7554/eLife.31926 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The cell cycle distribution after knockdown of CPSF6 in AGS and BGC-823 cells.
The candidate genes of CPSF6. (A) CPSF6 induced APA shift of candidate genes. UP, Integrative Genomics Viewer (IGV) genome browser exhibited the poly(A) site usage of candidate genes’ 3′UTR. Down, histogram showed the relative expression of the isoform with distal polyadenylation site (dPAS) relative to the one with proximal PAS (pPAS). (B) The mRNA levels of candidate genes in CPSF6 knockdown AGS cells (up) and BGC-823 cells (down). (C) Schematic illustration of the VHL isoform with long or short 3′UTR. Positions of the binding sites of miRNA were predicted by website tool (http://www.targetscan.org/) and indicated by purple horizontal lines. CR, CDS region. ∗0.01 < p < 0.05; ∗∗0.001 < p < 0.01; ∗∗∗p < 0.001.
Statistical significance of CPSF6 expression in GC of individual cancer stages.
Statistical significance of CPSF6 expression in GC based on nodal metastasis status.
The sequencing statistics of AGS 3T-seq libraries.
The list of genes with altered 3′UTR in AGS shCPSF6-1 cells.
The list of genes with altered 3′UTR in AGS shCPSF6-2 cells.
The list of genes with shortened 3′UTR shared by AGS shCPSF6-1 and shCPSF6-2 cells.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.