Abstract
Head and neck squamous cell carcinoma (HNSCC) has been associated with poor prognosis, due to its strong invasive ability and resistance to chemotherapy. Thus, there is an urgent requirement to identify effective biomarkers for the early diagnosis and prognostic evaluation of HNSCC. COP9 signalosome (COPS) regulates numerous cancer-associated biological processes in various malignancies. The aim of the present study was to investigate the association between COPS and HNSCC. The mRNA expression profiles of COPS in HNSCC were analyzed using UALCAN, Oncomine and UCSC Xena databases. The association between overall survival time in patients with HNSCC and the COPS genes was investigated using the Kaplan-Meier plotter database. The CERES score was obtained and evaluated to determine the importance of the COPS genes for survival of the HNSCC cell lines. Functional analysis for Gene Ontology and Gene Set Enrichment Analysis (GSEA) was performed using The Database for Annotation, Visualization and Integrated Discovery and GSEA software, respectively. After knocking down COPS5 and COPS6, cell Counting Kit-8 and wound healing assays were used to detect cell growth and migration of the CAL27 and SCC25 cell lines, respectively. Among the 10 COPS genes examined, most COPS subunits were upregulated in HNSCC samples compared with that in normal tissues, except for COPS9. Increased mRNA expression level of COPS5, COPS6, COPS7B, COPS8 and COPS9 was associated with TNM stage in patients with HNSCC. High mRNA expression level of COPS2, COPS5, COPS6, COPS7A, COPS7B, COPS8 and COPS9 had prognostic significance of patients with HNSCC. Knockdown of COPS5 and COPS6 inhibited cell growth and migration of the CAL27 and SCC25 cell lines. The results from the present study suggested that COPS subunits could be potential biomarkers in patients with HNSCC. COPS5 and COPS6 were important for cell survival and migration of the HNSCC cells.
Keywords: COPS9, head and neck squamous cell carcinoma, prognostic factor, biomarker, cell survival
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the major pathological subtype of head and neck cancer (1). Every year, >600,000 new patients are diagnosed worldwide, and 300,000 individuals succumb to the disease (2). The methods for the diagnosis and treatment of HNSCC have developed rapidly; however, the prognosis of patients with HNSCC remains poor (3). Identifying biomarkers in body fluids is considered important for the earlier diagnosis of HNSCC (4). For example, salivary IL-8 and −6 and tumor necrosis factor (TNF) have been beneficial for the diagnosis of oral squamous cell carcinoma (5). However, the conclusions from different studies are still incomplete (4). Therefore, identifying novel biomarkers for HNSCC detection is important for definitive and effective diagnosis leading to timely prevention, precise treatment and alleviating the recurrence of HNSCC.
The COP9 signalosome (COPS) is an evolutionarily conserved complex of 8 protein subunits (COPS1-8) (6). The known biochemical activity of the COPS is to serve as the deneddylase to remove NEDD8 from a neddylated cullin, in the cullin-RING-E3 ligases (CRLs) via deneddylation, which is further enhanced by linking with the 9th subunit, COPS9 (7,8). COPS regulates numerous cellular and biological processes, such as embryonic development, cell cycle, DNA damage repair, checkpoint repair control, signal transduction, circadian rhythm, T-cell development and autophagy (9). COPS accomplishes these various functions by controlling CRLs, the most prominent class of E3-enzymes, which promote ubiquitination of a variety of regulatory proteins subsequently targeted for proteasomal degradation (10). In addition, the catalytic center of the COPS is harbored in COPS5 (11).
The COPS subunits, which form the lid complex (19S) of the 26S proteasome, are paralogous (12). The 19S complex can specifically recognize ubiquitinated substrates and induce their degradation (13). Given the characteristics of the COPS, numerous tumor-associated genes have been identified, including hypoxia inducible factor 1 (HIF1), p27, runt related transcription factor 3 and p53 (14–17). The oncoprotein Jab1/COPS5 is associated with the pathogenesis of nasopharyngeal carcinoma (11). COPS5 is overexpressed in thyroid carcinoma cell lines and associated with low expression of p27 (18). Epidermal growth factor receptor-extracellular regulated MAP kinase signaling pathway may upregulate COPS6, which inhibited programmed cell death ligand-1 (PD-L1) degradation and subsequently maintains PD-L1 stability in glioblastoma (19,20). In ultraviolet-irradiated cells, COPS1 is associated with both the DNA repair process and cell apoptosis in HeLa cells (21). COPS3 is associated with cell growth arrest mediated by myeloid leukemia factor 1 and impairs the activation of p53, which promotes cell proliferation and blocks constitutively photomorphogenic 1-mediated p53 degradation (22). COPS2 overexpression upregulates the production of vascular endothelial growth factor and promotes the stability of cyclin A (23). COPS4 deficiency facilitates the downregulation of S-phase kinase associated protein 2, which is associated with the degradation of p27 (24).
Compared with normal tissues, COPS5 and COPS6 have both been found to be upregulated >40% at the mRNA level in several types of human cancer tissues, including myeloma, lung cancer, colon adenocarcinoma, breast cancer, glioblastoma and leukemia (25). Using a proteome microarray method, Yang et al (26) found that COPS2 expression was significantly higher in the serum of patients with gastric cancer compared with that in healthy individuals, and the mRNA expression level of COPS2 was associated with poor prognosis. The amplification of COPS3 was found in 31% of patients with osteosarcoma and was significantly associated with the size of the tumor (27). In addition, COPS3 is upregulated in clear cell renal cell carcinoma tissues and cell lines and patients with high expression of COPS3 have lower overall survival (OS) rate (28). The mRNA level of COPS8 is upregulated and predicts a poor clinical outcome in patients with cutaneous melanoma (29). However, to the best of our best knowledge, the association between the COPS subunits and HNSCC remains unknown.
Therefore, the present study aimed to investigate the association between the expression profiles of the COPS subunits and HNSCC. It was found that the COPS subunits were highly expressed in the HNSCC samples compared with that in normal tissues, except for COPS9. High mRNA expression levels of COPS2, COPS5, COPS6, COPS7A, COPS7B, COPS8 and COPS9 had prognostic significance in patients with HNSCC. COPS5 and COPS6 could be important for the process of HNSCC.
Materials and methods
Cell culture
The human tongue squamous cell carcinoma cell lines, CAL27 and SCC25, were kindly provided by Professor Liang Jiang (Tongji Hospital, Hubei, China). The CAL27 and SCC25 cell lines were cultured in complete DMEM (Hyclone; Cytiva) containing 10% FBS (Lonza Group, Ltd.) with 100 U/ml streptomycin and penicillin (Thermo Fisher Scientific, Inc.). Both the cell lines were cultured at 37°C in a humidified incubator with 5% CO2. The CAL27 and SCC25 cell lines were harvested using 0.25% trypsin-EDTA (Thermo Fisher Scientific, Inc.) when it reached 80% confluence.
Cell transfection
The CAL27 and SCC25 cell lines were transfected with small interfering (si)RNA targeting COPS5 (5′-UUCUCAUACUGUCUUUCAGGUCUGAAAGACAGUAUGAGAAAA-3′) or COPS6 (5′-UUGAUUAUAUCAAUGACAGACCUGUCAUUGAUAUAAUCAAUG-3′) or a non-specific control (50 nM) using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. After cell transfection with siRNA for 4 h at 37°C, the medium was replaced. After 48 h transfection, cells were used for western blot analysis and wound healing assay. After 24 or 48 h transfected, cell viability was tested.
Western blot analysis
Proteins from CAL27 and SCC25 cells were extracted using RIPA lysate (Beyotime Biotechnology, China) containing 1× protease inhibitor (Thermo Fisher Scientific, USA) at 80–90% density. The protein concentration was determined by a BCA method (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. Total protein (20 µg) was separated using 10% SDS-PAGE and transferred to 0.2-µm PVDF membranes (Bio-Rad Laboratories) at 100 V for 80 min. After being blocked with TBS containing 0.1% Tween-20 (TBST) and 5% skimmed milk for 1 h at room temperature, the PVDF membrane was incubated with the primary antibody overnight at 4°C. The next day, after being washed 3 times with TBST, the membrane was incubated with the goat anti-mouse secondary antibody (cat. no. SA00001-1; 1:10,000; ProteinTech Group, Inc.) for 1 h at room temperature, then the bands were visualized using an ECL developing system (Beyotime Institute of Biotechnology). The following antibodies were used: Mouse monoclonal anti-COPS5 (cat. no. sc-13157; 1:200; Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-COPS6 (cat. no. sc-137153; 1:200; Santa Cruz biotechnology, Inc.) and mouse monoclonal anti-GAPDH (cat no. ab8245; 1:10,000; Abcam).
Cell viability
Following transfection with the siRNA, the cell lines (1×104) were collected and seeded in 96-well plates. The cells were then incubated with complete medium for 24 or 48 h and 10 µg Cell Counting Kit-8 reagent (MedChemExpress) was added into each well. The cells were incubated in 37°C for 30 min, then the samples were measured at 450 nm.
Wound healing assay
The CAL27 and SCC25 (1×105) cell lines were seeded in 12-well plate. Then, cells were transfected with siRNA or control. After the cells were cultured to 80–90% confluence with complete DMEM medium with 10% FBS, the cells were scratched slightly, then washed with PBS three times. Subsequently, the cells were incubated with DMEM without FBS for 48 h and images of the cells were captured with normal light microscope (BX51; Olympus Corporation). The wound was quantified by ImageJ with the Wound Healing Coherency Tool (version 1.8.0; national Institute of Health).
Datasets and samples
The RNA expression profile and clinical data sets were obtained from The Cancer Genome Atlas (TCGA) database (https://gdc.cancer.gov), which included 520 patients with HNSCC and 44 adjacent normal samples. After excluding the samples with survival times <30 days, missing clinical information and data integration, a total of 490 samples were used to perform the prognostic analysis.
mRNA profiles of the COPS subunits
The ONCOMINE database (www.oncomine.org) is the world's largest oncogene chip database and integrated data mining platform. A total of 11 datasets (Pyeon Multi-cancer, Sengupta Head-Neck, Toruner Head-Neck, Schlingemann Head-Neck, Cromer Head-Neck, Ginos Head-Neck, Giordano Thyroid, Peng Head-Neck, Ye Head-Neck, Vasko Thyroid and He Thyroid) were included in the analysis (30–35). The mRNA expression level of the COPS subunits in the clinical HNSCC specimens were compared with that in adjacent normal controls, using an unpaired Student's t-test. The fold change and P-value thresholds were defined as 1.5 and 0.01, respectively. TCGA gene expression data of the COPS subunits was determined using UALCAN (http://ualcan.path.uab.edu) using the ‘TCGA-HNSC’ module, which provides systematical and personalized analysis of TCGA database. University of California Santa Cruz (UCSC) Xena (http://xenabrowser.net/) is a comprehensive web analysis tool which provides customized analysis methods of TCGA data. To evaluate different mRNA expression levels of the COPS subunits and the TMN stages in primary HNSCC (n=765), UCSC Xena was used with a one-way ANOVA analysis followed by Bonferroni's post hoc test.
Prognosis evaluation of the COPS subunits in HNSCC
The Kaplan-Meier plotter (www.kmplot.com) includes the 21 types of TCGA datasets to evaluate the prognostic value of 54,000 genes in different types of cancer. The association between mRNA expression levels of all the COPS subunits and prognosis in patients with HNSCC was analyzed using the Kaplan-Meier plotter. All the patients were divided into two groups automatically using the best outcome setting. The hazard ratio (HR) with 95% confidence intervals (CI) and log-rank were obtained to evaluate the prognostic difference in patients with HNSCC.
Protein-protein interaction (PPI) network of the COPS subunits
To construct the PPI network of the COPS subunits, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org/), which includes both certified and predicated links, was used. In total, 10 COPS subunits (COPS1-6, 7A and B, 8 and 9) and Homo sapiens (organism) were selected for the analysis. Extra 50 associated nodes were included in the analysis.
Gene ontology (GO) enrichment analysis
GO enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/). The catalogues of GO analysis were selected as biological process (BP), cellular components (CC), molecular function (MF) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was also performed. All associated genes (36) were included in the analysis. P<0.05 was considered as significant. The top 12 affected pathways were included using R version 4.1.0. (R Development Core Team).
Cell survival analysis
To evaluate the importance of the COPS subunits in survival of the HNSCC cell lines, Project Achilles with CERES dependence scores was used. By accurately removing the expression of the corresponding gene with CRISPR-Cas9 system, Project Achilles systematically investigates and identifies the numbers of genes which are essential for survival of the cancer cell lines. The original data was downloaded from the Depmap portal (https://depmap.org/portal) (37). The inclusive criteria were as follow: i) The primary disease was head and neck cancer and ii) the pathological subtype was squamous cell carcinoma. A total of 21 cell lines were included in the analysis.
Gene set enrichment analysis (GSEA) of COPS subunits
GSEA (http://www.broad.mit.edu/gsea), with the annotated Hallmark effector gene sets, was performed according to the mRNA expression level of the COPS subunits in TCGA-HNSC dataset. The cut-off value of grouping was defined as 50%. The false discovery rate q-value <0.25 and P<0.05 was considered to indicate a significant difference (38). The visual results were performed using R version 4.1.0.
Protein expression of the COPS subunits
The protein expression of COPS subunits was evaluated using immunohistochemistry (IHC) data downloaded from the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/). The following primary antibodies against CCT5 and CCT6 were used CAB004242 and HPA044315.
Statistical analysis
The HR with log-rank test was used for survival analysis. Spearman's rank correlation test was used to evaluate the correlation of gene expression in tumor tissues compared with normal tissues, and the strength of the association was determined using the absolute values. One-way ANOVA followed by Bonferroni post hoc test was used to evaluate the difference in >2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
mRNA expression profiles of the COPS subunits in HNSCC
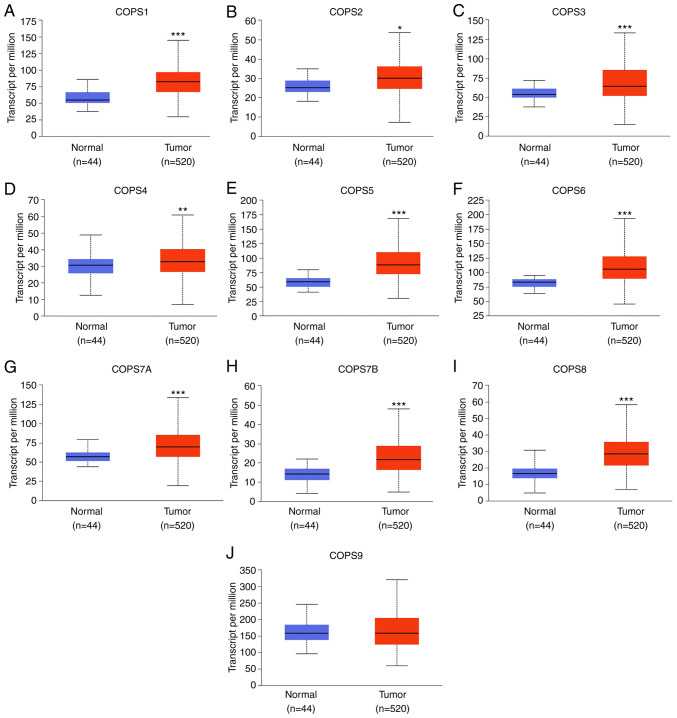
A total 10 COPS subunits (COPS1-9) have been identified in human cells (39,40). To evaluate the mRNA expression levels of the COPS subunits in HNSCC, TCGA dataset (TCGA-HNSC) was used within the UALCAN database. As shown in Fig. 1, the expression of COPS1-8, but not COPS9, was found to be upregulated in HNSCC tissues compared with that in normal tissues.
Figure 1.
Expression level of the COPS subunits was increased in the HNSCC samples. The mRNA expression profiles of (A) COPS1, (B) COPSS2, (C) COPS3, (D) COPS4, (E) COPS5, (F) COPS6, (G) COPS7A, (H) COPS7B, (I) COPS8 and (J) COPS9 in HNSCC and normal tissue samples were obtained from the UALCAN database. *P<0.05. **P<0.01. ***P<0.001. COPS, COP9 signalosome; HNSCC, head and neck squamous cell carcinoma.
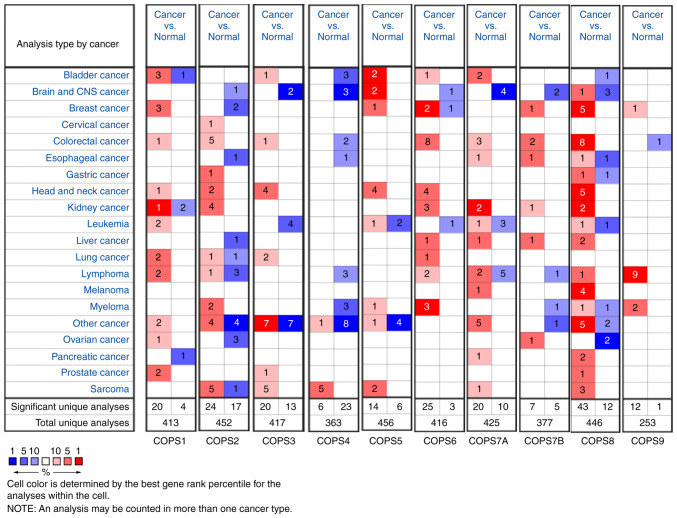
To further verify the results, the ONCOMINE database was used to compare the transcriptional levels of the COPS subunits in all types of cancer with the corresponding normal tissue (Fig. 2). The red color and number indicated the number of datasets with a statistically significant increase (P<0.01; fold change, 1.5) in mRNA expression levels in different types of cancers, while the blue color indicated a decrease in mRNA expression of the COPS subunits. There was a total of 11 HNSCC datasets in the ONCOMINE database. The overexpression status of COPS subunits in different parts of HNSCC are presented in Table I. In detail, one dataset indicated COPS1 was upregulated in floor of the mouth carcinoma, and COPS3 was increased in carcinoma from oropharyngeal, oral cavity, floor of the mouth and tongue (Pyeon Multi-cancer) (30). The expression of COPS2 was found increased in both NPC (Sengupta Head-Neck) (31) and floor of the mouth carcinoma (Pyeon Multi-cancer) (30). COPS5 (Pyeon Multi-cancer, Sengupta Head-Neck and Toruner Head-Neck Statistics) (30,31,34) and COPS6 (Pyeon Multi-cancer) (30) were upregulated in various sites of HNSCC. In addition, COPS8 was found highly expressed in HNSCC tissues in five datasets (Pyeon Multi-cancer, Sengupta Head-Neck, Schlingemann Head-Neck, Cromer Head-Neck and Ginos Head-Neck) (30–33,35). There was no significant difference in the expression of COPS7A, COPS7B and COPS9 between HNSCC and normal tissues in the ONCOMINE database.
Figure 2.
mRNA transcriptional levels of the COPS subunits in diverse types of cancer. The mRNA transcriptional levels of COPS in different types of cancer from the Oncomine database. The numbers in the box indicate statistically significant overexpression (red) or downregulation (blue) of the COPS subunits in cancer tissues compared with that in normal tissues. COPS, COP9 Signalosome.
Table I.
Expression of the COPS families in head and neck squamous cell carcinoma.
| Gene name | Cancer type (n) | P-value | Fold change | T-test | (Refs.) |
|---|---|---|---|---|---|
| COPS1 | Floor of the mouth carcinoma (5) | 4.00×10−3 | 1.543 | 3.837 | Pyeon Multi-cancer (50) |
| COPS2 | Nasopharyngeal carcinoma (31) | 2.80×10−6 | 2.095 | 5.746 | Sengupta Head-Neck (36) |
| Floor of the mouth carcinoma (5) | 2.07×10−4 | 2.261 | 5.383 | Pyeon Multi-cancer (50) | |
| COPS3 | Oropharyngeal carcinoma (6) | 3.32 ×10−4 | 2.295 | 4.225 | Pyeon Multi-cancer (50) |
| Oral cavity carcinoma (4) | 3.00×10−3 | 2.206 | 3.717 | Pyeon Multi-cancer (50) | |
| Floor of the mouth carcinoma (5) | 2.43×10−4 | 2.636 | 4.748 | Pyeon Multi-cancer (50) | |
| Tongue carcinoma (15) | 2.01×10−4 | 2.040 | 3.913 | Pyeon Multi-cancer (50) | |
| COPS5 | Tongue carcinoma (15) | 3.88×10−4 | 2.755 | 6.009 | Pyeon Multi-cancer (50) |
| Floor of the mouth carcinoma (5) | 1.71×10−6 | 4.533 | 8.121 | Pyeon Multi-cancer (50) | |
| Oral cavity carcinoma (4) | 2.26 ×10−4 | 2.401 | 4.971 | Pyeon Multi-cancer (50) | |
| Tonsillar carcinoma (6) | 9.52×10−4 | 1.719 | 3.509 | Pyeon Multi-cancer (50) | |
| Oropharyngeal carcinoma (6) | 1.99×10−4 | 2.762 | 4.910 | Pyeon Multi-cancer (50) | |
| Oral cavity squamous cell carcinoma (16) | 3.88×10−4 | 1.823 | 4.826 | Toruner Head-Neck Statistics (53) | |
| Nasopharyngeal carcinoma (31) | 1.17×10−4 | 1.595 | 4.233 | Sengupta Head-Neck Statistics (36) | |
| COPS6 | Tonsillar carcinoma (6) | 3.01×10−4 | 1.924 | 3.915 | Pyeon Multi-cancer (50) |
| Oral cavity carcinoma (4) | 2.00×10−3 | 2.634 | 3.902 | Pyeon Multi-cancer (50) | |
| Oropharyngeal carcinoma (6) | 4.81×10−4 | 2.118 | 3.820 | Pyeon Multi-cancer (50) | |
| Floor of the mouth carcinoma (5) | 1.49×10−4 | 2.665 | 4.656 | Pyeon Multi-cancer (50) | |
| COPS8 | Hypopharyngeal squamous cell carcinoma (4) | 5.00×10−3 | 2.533 | 3.794 | Schlingemann Head-Neck (52) |
| Head and neck squamous cell carcinoma (34) | 6.70×10−4 | 2.001 | 5.820 | Cromer Head-Neck (52) | |
| Head and neck squamous cell carcinoma (41) | 3.48×10−9 | 1.617 | 7.125 | Ginos Head-Neck (54) | |
| Oropharyngeal carcinoma (6) | 1.21×10−4 | 2.021 | 4.791 | Pyeon Multi-cancer (50) | |
| Nasopharyngeal carcinoma (31) | 2.47×10−4 | 1.523 | 3.957 | Sengupta Head-Neck (36) |
COPS, COP9 signalosome.
mRNA expression levels of the COPS subunits are associated with TNM stage in patients with HNSCC
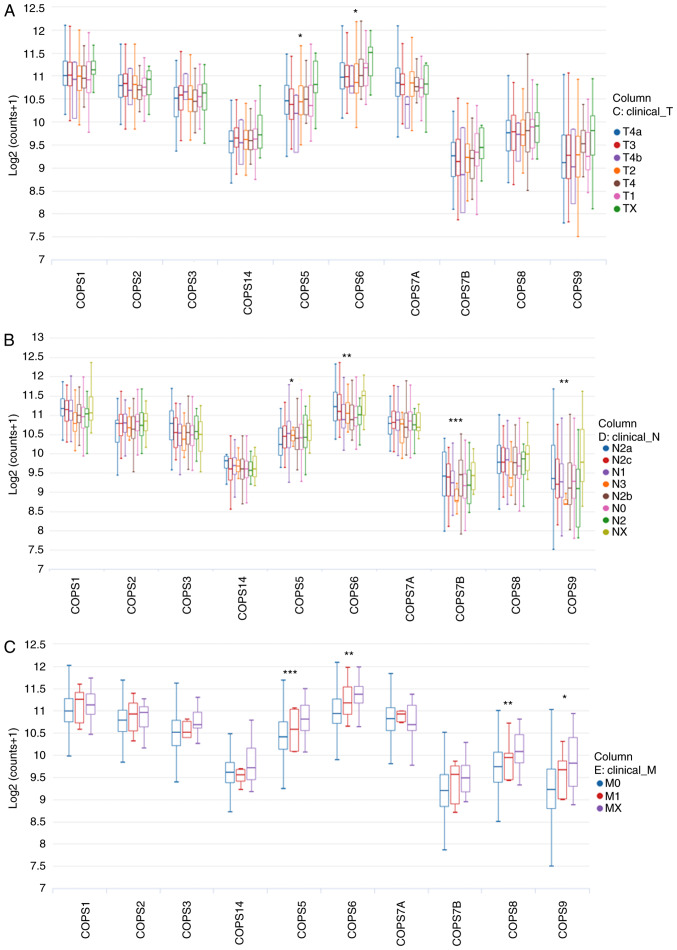
As most of the COPS subunits were found to be upregulated in HNSCC tissues, the association between the mRNA expression level of the COPS subunits and TNM stage was investigated. The UCSC Xena web tool was used for the analysis. As shown in Fig. 3A, the mRNA expression levels of COPS5 and COPS6 were significantly increased with increasing tumor size. With respect to lymph node metastasis, an increase in the expression level of COPS5, COPS6, COPS7B and COPS9 were found in patients with HNSCC and N stage (Fig. 3B). In addition, increased expression of COPS5, COPS6, COPS8 and COPS9 was associated with distant metastasis of HNSCC (Fig. 3C).
Figure 3.
Analysis of COPS subunits and clinical characteristics. Subgroup analysis of the COPS subunits based on (A) tumor size, (B) nodal metastasis and (C) distant metastasis in head and neck squamous cell carcinoma samples was evaluated using the UCSC Xena database. *P<0.05. **P<0.01. ***P<0.001. COPS, COP9 signalosome.
Prognostic values of the COPS subunits in patients with HNSCC
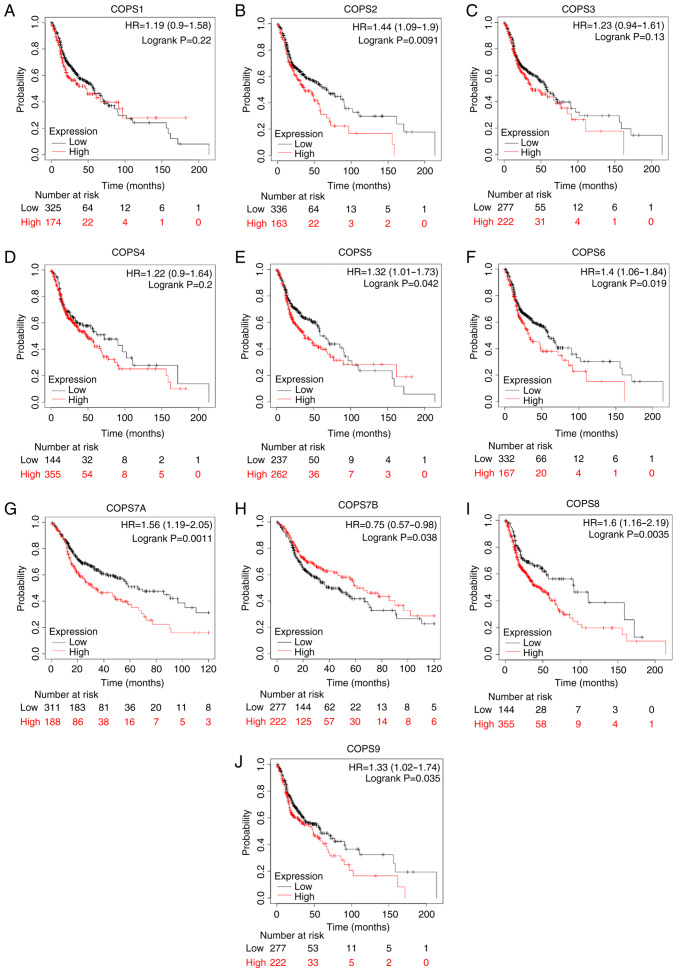
The Kaplan-Meier plotter database and TCGA-HNSC dataset was used to investigate whether the COPS subunits were associated with the prognosis of HNSCC. High mRNA expression level of COPS2 (HR, 1.44; CI, 1.09–1.9; P=0.0091; Fig. 4B), COPS5 (HR, 1.32; CI, 1.01–1.73; P=0.042; Fig. 4E), COPS6 (HR, 1.4; CI, 1.06–1.84; P=0.019; Fig. 4F), COPS7A (HR, 1.56 (1.19–2.05); P=0.0011; Fig. 4G), COPS8 (HR, 1.6; CI, 1.16–2.19; P=0.0035; Fig. 4I) and COPS9 (HR, 1.33; CI, 1.02–1.74; P=0.035; Fig. 4J) was associated with poor prognosis in patients with HNSCC. However, high expression of COPS7B (HR, 0.75 (0.57–0.98); P=0.038; Fig. 4H) was associated with improved outcome in patients with HNSCC.
Figure 4.
COPS subunit expression and prognostic value in patients with HNSCC. The prognostic value of (A) COPS1, (B) COPS2, (C) COPS3, (D) COPS4, (E) COPS5, (F) COPS6, (G) COPS7A, (H) COPS7B, (I) COPS8 and (J) COPS9 in the head and neck squamous cell carcinoma tissues was determined using the Kaplan-Meier plotter database and the mRNA expression profiles. COPS, COP9 signalosome; HR, hazard ratio.
PPI network construction and GO analysis of the COPS subunits
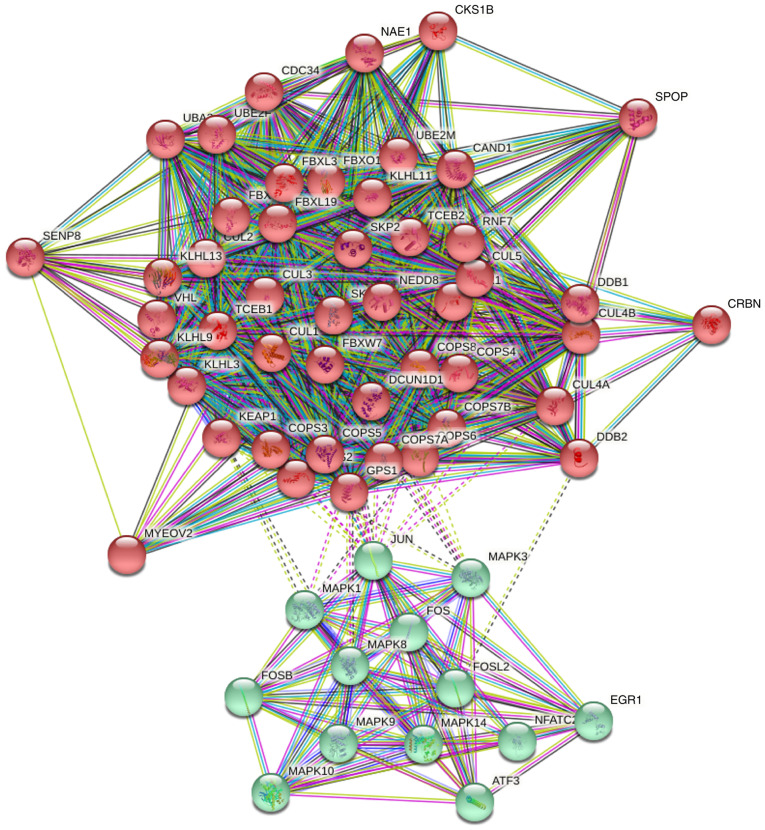
To predict the functions of the COPS subunits in humans, a PPI network was constructed with the 50 most relevant genes using STRING. The analysis included 60 nodes and 946 edges (Fig. 5). The PPI enrichment value was P<1.0×10−16.
Figure 5.
PPI network of the COPS subunits. The PPI network of the COPS subunits was constructed using STRING with the 50 most frequently altered neighbor genes and includes 60 nodes and 956 edges. COPS, COP9 signalosome; PPI, protein-protein interaction.
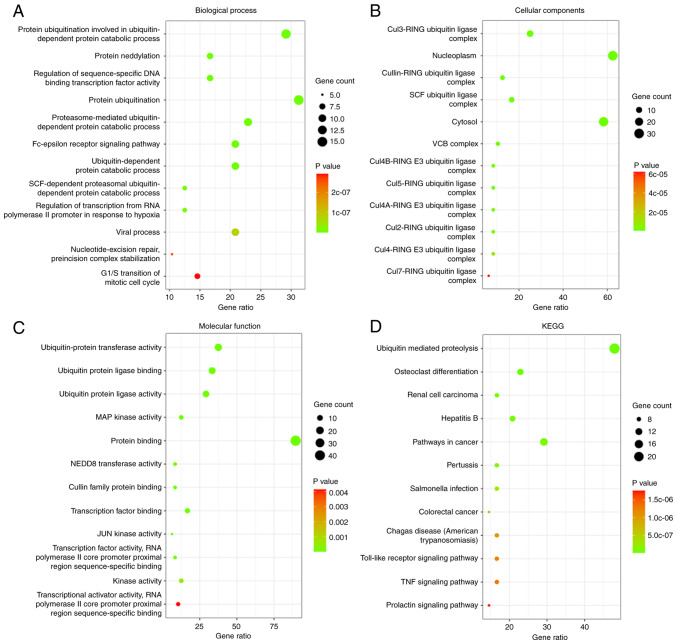
Then, the biological functions and pathways of the associated genes with the COPS subunits were analyzed using GO and KEGG analysis, respectively. The top 12 GO terms and KEGG pathways are shown in Fig. 6. With respect to BP, ‘protein ubiquitination involved in ubiquitin-dependent protein catabolic process’, ‘Fc-epsilon receptor signaling pathway’, ‘nucleotide-excision repair, preincision complex stabilization’ and ‘G1/S transition of mitotic cell cycle’ were significantly associated with the COPS subunits (Fig. 6A; Table SI). For CC, the genes were associated with ‘Cul3-RING ubiquitin ligase complex’, ‘nucleoplasm’ and ‘cytosol’ (Fig. 6B; Table SII). The genes associated with MF were ‘ubiquitin-protein transferase activity’, ‘MAP kinase activity’, ‘NEDD8 transferase activity’ and ‘JUN kinase activity’, which were cancer relative pathways (Fig. 6C; Table SIII). KEGG pathway analysis identifies the functions of the COPS subunits and the pathways they are involved in. The top 12 pathways are shown in Fig. 6D and Table SIV. Among these pathways, ‘ubiquitin mediated proteolysis’, ‘renal cell carcinoma’, ‘pathways in cancer’, ‘colorectal cancer’, ‘Toll-like receptor signaling pathway’ and ‘TNF signaling pathway’ were observed.
Figure 6.
Gene Ontology functional enrichment analysis of the COPS subunits. The bubble diagrams display the enrichment results of the COPS subunits and the top 50 genes altered in the COPS subunits in head and neck squamous cell carcinoma using Database for Annotation, Visualization and Integrated Discovery. (A) Biological processes. (B) Cellular components. (C) Molecular functions. (D) KEGG pathways. COPS, COP9 signalosome.
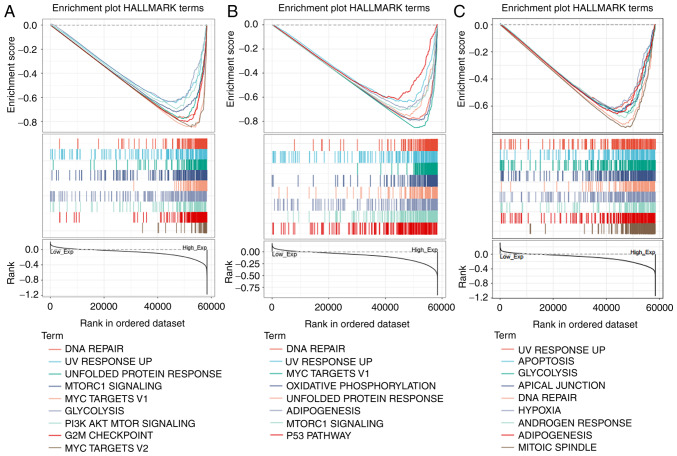
GSEA analysis of COPS5, COPS6 and COPS8
In HNSCC, COPS5, COPS6 and COPS8 are essential for cell growth and, as aforementioned, associated with tumor TNM stages and prognosis. Hence, it is important to evaluate the functions of these three genes and GSEA was performed to evaluate Hallmark annotation. As shown in Fig. 7A and Table SV, high expression of COPS5 was associated with tumorigenesis-associated pathway including ‘DNA repair’, ‘unfolded protein response’, ‘MTORC1 signaling’, ‘glycolysis’ and ‘PI3K AKT mTOR signaling’. In addition to these pathways, increased COPS6 was also associated with ‘oxidative phosphorylation’ and ‘p53 pathway’ (Fig. 7B and Table SVI). Similarly, COPS8 was additionally associated with ‘apoptosis’ (Fig. 7C and Table SVII).
Figure 7.
GSEA of the COPS subunits in head and neck squamous cell carcinoma. GSEA analysis was performed with Hallmark annotation based on expression level of (A) COPS6, (B) COPS6 and (C) COPS8. The most (top 9) affected signal pathways are shown. COPS, COP9 signalosome; GSEA, Gene Set Enrichment Analysis.
IHC analysis of the protein expression level of the COPS subunits in HNSCC
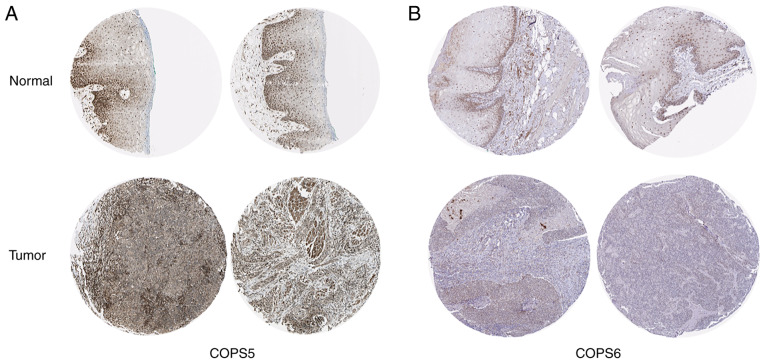
The protein expression level of the COPS subunits in HNSCC was investigated using the HPA database. Consistent with the mRNA expression level, COPS2 (Fig. S1A), COPS3 (Fig. S1B), COPS4 (Fig. S1C), COPS5 (Fig. 8A), COPS6 (Fig. 8B), COPS7A (Fig. S1D) and COPS7B (Fig. 1E) were notably upregulated in both the cytoplasm and the nucleus in the HNSCC tissues compared with that in normal oral epithelial tissues. COPS9 was increased in the normal oral epithelial tissue compared with that in the tumor tissues of HNSCC (Fig. S1F). COPS1 and COPS8 were not detected.
Figure 8.
Protein expression of the COPS subunits in HNSCC. Immunohistochemical staining images from the Human Protein Atlas portal was used to analyze protein expression of (A) COPS5 and (B) COPS6 in normal oral tissues and HNSCC. COPS, COP9 signalosome.
Role of COPS subunits in the survival of the HNSCC cell lines
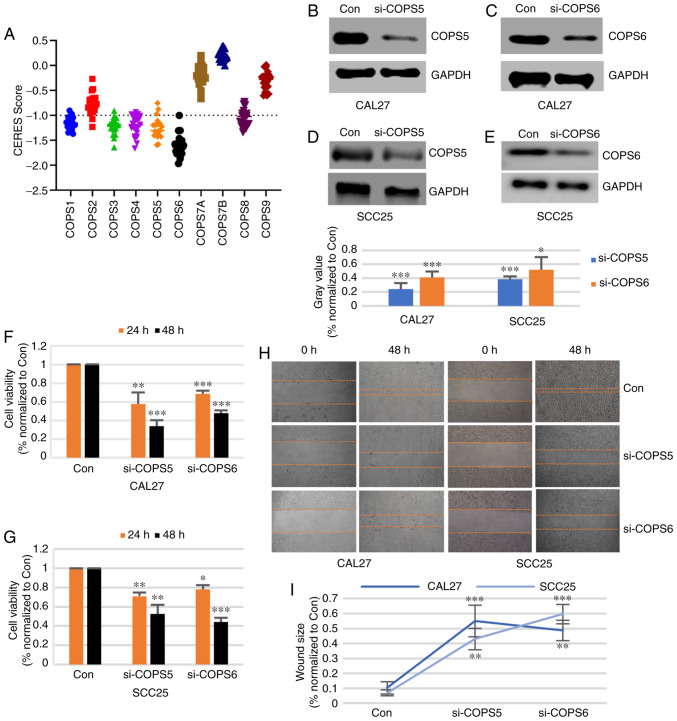
To evaluate the importance of the COPS subunits, Project Achilles with CERES dependence scores was used. When the score was >-1, it indicated that the gene was not essential for cell survival. Otherwise, the gene was considered important for cell survival. As shown in Fig. 9A and Table SVIII among all the HNSCC cell lines, COPS6 was important for cell survival. COPS1, COPS3, COPS4, COPS5 and COPS8 were critical for cell survival in most HNSCC cell lines. COPS2 was significantly essential for only 2 HNSCC cell lines. COPS7A, COPS7B and COPS9 were not important for HNSCC cell survival. These results suggested that COPS1, COPS3, COPS4, COPS5, COPS6 and COPS8 are essential for HNSCC cell survival.
Figure 9.
COPS5 and COPS6 are important for cell growth and migration in HNSCC cells. (A) CERES score was downloaded from Depmap and used to evaluate the significance of the COPS subunits for survival of the HNSCC cell lines. The CAL27 (B and C) and SCC25 (D and E) cell lines were transfected with control siRNA, si-COPS5 or si-COPS6 for 48 h, then western blot analysis was used to detect the protein expression of COPS5 or COPS6. A Cell Counting Kit-8 assay was performed to detect cell viability of the (F) CAL27 and (G) SCC25 cell lines following transfection with siRNA. (H) A wound healing assay was performed to determine the cell migration of the CAL27 and SCC25 cells following transfection with siRNA and the results were (I) quantified. *P<0.05, **P<0.01 and ***P<0.001. COPS, COP9 signalosome; HNSCC, head and neck squamous cell carcinoma.
COPS5 and COPS6 are essential for cell growth and invasion of HNSCC cells
Taken together, COPS5 and COPS6 were associated with the survival of the HNSCC cells and could be detected in tumor samples using IHC. To confirm the biological function of COPS5 and COPS6 in HNSCC, the CAL27 and SCC25 cell lines were transfected with a control siRNA, si-COPS5 and si-COPS6. Western blot analysis showed that the protein expression level of COPS5 (Fig. 9B and D) and COPS6 (Fig. 9C and E) was notably decreased by the siRNA. As shown in Fig. 9F and G, the cell viabilities of the CAL27 and SCC25 cell lines were significantly decreased by both si-COPS5 and si-COPS6 at 24 and 48 h time points (P<0.05). To detect the cell migration effect, the CAL27 and SCC25 cell lines were transfected with control siRNA, si-COPS5 or si-COPS6. The results from the wound healing assay suggested that si-COPS5 and si-COPS6 inhibited the migration of the CAL27 and SCC25 cells (Fig. 9H and I). These results suggested that COPS5 and COPS6 could be essential for growth and migration of the HNSCC cell lines.
Discussion
In the present study, ONCOMINE and TCGA databases were analyzed and it was found that the COPS subunits were upregulated in HNSCC samples compared with that in normal tissues, except for COPS9. Using subgroup analysis, based on TNM stage, high mRNA expression level of COPS5, COPS6, COPS7B, COPS8 and COPS9 was associated with progression of HNSCC. Prognosis analysis, using Kaplan-Meier plotter and TCGA-HNSC dataset suggested that mRNA expression level of COPS2, COPS5, COPS6, COPS7A, COPS7B, COPS8 and COPS9 could have prognostic significance. Survival analysis in the HNSCC cell lines suggested that COPS1, COPS3, COPS4, COPS5, COPS6 and COPS8 were necessary for tumor cell survival. COPS5 and COPS6 were also found to be essential for growth and migration of the HNSCC cell lines. Taken together, the COPS subunits may be potential diagnostic biomarkers in the future, and COPS5 and COPS6 could be important genes in the progression of HNSCC.
COP9 is a component of a large, multi-subunit nuclear protein complex (39). As COPS interacts and forms supercomplexes with cullin-RING ubiquitin-ligases, COPS could regulate the process of protein ubiquitination and maintain protein homeostasis (39). Increasing evidence have hypothesized that tumor cells have a large burden of unfolded proteins due to numerous mutations in protein-coding sequences, which leads to aberrant process of ubiquitination (41). Hence, abnormal expression of COPS subunits could contribute to the carcinogenesis and progression of cancer (42). The deregulated expression of several COPS subunits has been found in different types of tumor (14–17). However, not all the COPS subunits were investigated in most of these studies, and it cannot be ruled out that the expression of the COPS holo complex was changed instead of a single subunit. To the best of our best knowledge, the present study is the first study to investigate the expression and prognostic values of the different COPS9 subunits in HNSCC.
Previous studies have reported that some COPS subunits are overexpressed in various types of cancer (14–17). For example, COPS5 and COPS6 were highly expressed in myeloma, lung cancer, colon adenocarcinoma, breast cancer, glioblastoma and leukemia (25). In the present study, the conservative COPS (COPS1-COPS8) were significantly upregulated in HNSCC tissues compared with that in normal tissues from TCGA database. However, the COPS9 was not differentially expressed in both TCGA and Oncomine databases. Notably, high expression of COPS5 and COPS6 were associated with tumor size, nodal metastasis and distant metastasis. These results suggested that COPS5 and COPS6 may play an important role in the progression of carcinogenesis. In fact, COPS5 and COPS6 regulate diverse signaling pathways to promote the development of other types of cancer. COPS5 collaborated with the oncogene, Myc, to contribute to cell invasion in breast cancer cell lines (43). By promoting the degradation of p27, COPS5 regulated the cytoplasm transport of p27, which assisted with HER2-activated cell growth, CDK2 and transformation efficiently in ovarian cancer (44,45). In addition, overexpression of COPS5 lead to reduced MDM2 self-ubiquitination and nuclear export of p53, which caused a dysregulation of Mdm2-P53 axis (46–49). Similarly, overexpression of COPS6 binds to MDM2, and prevents the process of its auto-ubiquitination and degradation, which leads to the accumulation of MDM2 and decrease of p53 in breast cancer (49). The results from GSEA analysis suggested that overexpression of COPS5 or COPS6 was associated with p53 signaling pathway in HNSCC, which is consistent with previous reports (15). Notably, following knockdown of COPS5 and COPS6 using siRNA, these genes were found to be essential for growth and migration of the HNSCC cell lines. These results suggested the critical roles played by COPS5 and COPS6 in HNSCC.
A recent study indicated that overexpression of COPS8 induced the progression of epithelial-mesenchymal transition in colorectal cancer cells, which lead to migration and invasion of the cancer cells (50). By promoting the expression of hypoxia response genes (solute carrier family 2 and HIF1) and key dormancy markers, COPS8 arrested cell proliferation and notably enhanced cell survival under serum deprivation, with hypoxia or the chemotherapy drug, 5-fluorouracil (50). Consistent with these results, in the present study, high mRNA expression level of COPS8 was found in HNSCC tissues, and was associated with distant metastasis of cancer and indicated poor prognosis of HNSCC. In addition, COPS8 was important for cell survival in most HNSCC cell lines. Mechanically, high expression of COPS8 was associated with apoptosis, DNA repair and hypoxia response. Notably, COPS8 was not detected in HNSCC tissue using an IHC assay. We hypothesized that the insensitive primary antibody caused the inaccurate detection.
With respect to the other COPS subunits, the high mRNA expression level of COPS1, COPS3 and COPS4 was found in HNSCC tissues, but was not associated with TMN stages or prognosis. However, high expression of COPS1 was associated with poor prognosis in patients with advanced esophageal squamous cell carcinoma (36). Notably, COPS1, COPS3 and COPS4 were all necessary for cancer cell survival. A previous study also found that knockdown of COPS3 expression with a specific shRNA in hepatocellular carcinoma cell lines significantly suppressed the cancer growth both in vitro and in vivo (51). COPS4 expression was essential for growth of breast cancer and prostate cancer cell lines (52,53). The results from the present study suggested that COPS2 was highly expressed in tumor tissues and upregulated COPS2 was associated with poor prognosis in patients with HNSCC. Zhao et al (54) found that DDA1 promoted the progression of colorectal cancer by activating the NFκB-COPS2-GSK3β pathway. COPS7A and COPS7B were highly expressed in the HNSCC samples; however, the prognostic significance was different. High mRNA expression level of COPS7A was associated with poor prognosis in patients with HNSCC, but the exact opposite was found with COPS7B.
There are some limitations to the present study. First and most important, the majority of the study is bioinformatic analysis, and a lack of clinical and functional experiments to support the results, which may restrict the conclusion. Second, a few results are marginally significant in this study (HR values are close to 1), which make the results not very convincing. Third, in Fig. 3, TX, NX, and MX are included in the sub-analysis. These ambiguous clinical data may lead some results unstable. Therefore, additional clinical and experimental studies should be performed in the future to focus on the association between the COPS subunits and HNSCC.
In conclusion, COPS1-8 mRNA expression level was upregulated in HNSCC tissues compared with that in normal tissues. The expression level of COPS2, COPS5, COPS6, COPS7A, COPS7B, COPS8 and COPS9 might be associated with the prognosis of patients with HNSCC. Notably, COPS5 and COPS6 were critical for the progression of HNSCC.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Liang Jiang (the Department of Oral and Maxillofacial Surgery, Tongji Hospital, Hubei, China) for providing the cell lines.
Funding Statement
This research was supported by a grant from the Youth Science Support Project of the Central Hospital of Wuhan (grant no. 201807632).
Funding
This research was supported by a grant from the Youth Science Support Project of the Central Hospital of Wuhan (grant no. 201807632).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Authors' contributions
HZ designed the study and performed the bioinformatic analysis. JZ performed the western blot, CCK-8 and wound healing assays. WS analyzed the data. HZ and JZ wrote the manuscript. All authors approve the final version of the manuscript. HZ and JZ confirm the authenticity of all the raw data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Forastiere AA, Trotti A, Pfister DG, Grandis JR. Head and neck cancer: Recent advances and new standards of care. J Clin Oncol. 2006;24:2603–2605. doi: 10.1200/JCO.2006.07.1464. [DOI] [PubMed] [Google Scholar]
- 2.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–1154. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 3.Lo Nigro C, Denaro N, Merlotti A, Merlano M. Head and neck cancer: Improving outcomes with a multidisciplinary approach. Cancer Manag Res. 2017;9:363–371. doi: 10.2147/CMAR.S115761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU. Role of salivary biomarkers in oral cancer detection. Adv Clin Chem. 2018;86:23–70. doi: 10.1016/bs.acc.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Sahibzada HA, Khurshid Z, Khan RS, Naseem M, Siddique KM, Mali M, Zafar MS. Salivary IL-8, IL-6 and TNF-α as potential diagnostic biomarkers for oral cancer. Diagnostics (Basel) 2017;7:21. doi: 10.3390/diagnostics7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubiel W, Chaithongyot S, Dubiel D, Naumann M. The COP9 signalosome: A multi-DUB complex. Biomolecules. 2020;10:1082. doi: 10.3390/biom10071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavadini S, Fischer ES, Bunker RD, Potenza A, Lingaraju GM, Goldie KN, Mohamed WI, Faty M, Petzold G, Beckwith RE, et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 8.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thomä NH. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 9.Qin N, Xu D, Li J, Deng XW. COP9 signalosome: Discovery, conservation, activity, and function. J Integr Plant Biol. 2020;62:90–103. doi: 10.1111/jipb.12903. [DOI] [PubMed] [Google Scholar]
- 10.Rao F, Lin H, Su Y. Cullin-RING ligase regulation by the COP9 signalosome: Structural mechanisms and new physiologic players. Adv Exp Med Biol. 2020;1217:47–60. doi: 10.1007/978-981-15-1025-0_4. [DOI] [PubMed] [Google Scholar]
- 11.Ebina M, Tsuruta F, Katoh M, Kigoshi Y, Someya A, Chiba T. Myeloma overexpressed 2 (Myeov2) regulates L11 subnuclear localization through Nedd8 modification. PLoS One. 2013;8:e65285. doi: 10.1371/journal.pone.0065285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Deng XW. The COP9 signalosome: An alternative lid for the 26S proteasome? Trends Cell Biol. 2003;13:507–509. doi: 10.1016/j.tcb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Karniol B, Chamovitz DA. The COP9 signalosome: From light signaling to general developmental regulation and back. Curr Opin Plant Biol. 2000;3:387–393. doi: 10.1016/S1369-5266(00)00101-1. [DOI] [PubMed] [Google Scholar]
- 14.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 15.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–1639. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Choi JK, Cinghu S, Jang JW, Lee YS, Li YH, Goh YM, Chi XZ, Lee KS, Wee H, Bae SC. Jab1/CSN5 induces the cytoplasmic localization and degradation of RUNX3. J Cell Biochem. 2009;107:557–565. doi: 10.1002/jcb.22157. [DOI] [PubMed] [Google Scholar]
- 17.Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, Chen X, Stenmark K, Giaccia AJ, Zundel W. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 2004;18:739–744. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie P, Wang H, Fang J, Du D, Tian Z, Zhen J, Liu Y, Ding Y, Fu B, Liu F, et al. CSN5 promotes carcinogenesis of thyroid carcinoma cells through ANGPTL2. Endocrinology. 2021;162:bqaa206. doi: 10.1210/endocr/bqaa206. [DOI] [PubMed] [Google Scholar]
- 19.Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, Zhang W, Cui H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–1144. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 20.Su L, Guo W, Lou L, Nie S, Zhang Q, Liu Y, Chang Y, Zhang X, Li Y, Shen H. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol Carcinog. 2020;59:520–532. doi: 10.1002/mc.23176. [DOI] [PubMed] [Google Scholar]
- 21.Dubois EL, Gerber S, Kisselev A, Harel-Bellan A, Groisman R. UV-dependent phosphorylation of COP9/signalosome in UV-induced apoptosis. Oncol Rep. 2016;35:3101–3105. doi: 10.3892/or.2016.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneda-Kato N, Tomoda K, Umehara M, Arata Y, Kato JY. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 2005;24:1739–1749. doi: 10.1038/sj.emboj.7600656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollmann C, Huang X, Mall J, Bech-Otschir D, Naumann M, Dubiel W. The constitutive photomorphogenesis 9 signalosome directs vascular endothelial growth factor production in tumor cells. Cancer Res. 2001;61:8416–8421. [PubMed] [Google Scholar]
- 24.Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E. The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem. 2006;281:32188–32196. doi: 10.1074/jbc.M604746200. [DOI] [PubMed] [Google Scholar]
- 25.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Wang J, Li J, Zhang H, Guo S, Yan M, Zhu Z, Lan B, Ding Y, Xu M, et al. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol Cell Proteomics. 2016;15:614–623. doi: 10.1074/mcp.M115.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan T, Wunder JS, Gokgoz N, Gill M, Eskandarian S, Parkes RK, Bull SB, Bell RS, Andrulis IL. COPS3 amplification and clinical outcome in osteosarcoma. Cancer. 2007;109:1870–1876. doi: 10.1002/cncr.22595. [DOI] [PubMed] [Google Scholar]
- 28.Hong Y, Huang X, An L, Ye H, Ma K, Zhang F, Xu Q. Overexpression of COPS3 promotes clear cell renal cell carcinoma progression via regulation of Phospho-AKT(Thr308), cyclin D1 and caspase-3. Exp Cell Res. 2018;365:163–170. doi: 10.1016/j.yexcr.2018.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Sun L. COPS8 in cutaneous melanoma: An oncogene that accelerates the malignant development of tumor cells and predicts poor prognosis. Biosci Biotechnol Biochem. 2021;85:242–250. doi: 10.1093/bbb/zbaa017. [DOI] [PubMed] [Google Scholar]
- 30.Reis PP, Waldron L, Perez-Ordonez B, Pintilie M, Galloni NN, Xuan Y, Cervigne NK, Warner GC, Makitie AA, Simpson C, et al. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer. 2011;11:437. doi: 10.1186/1471-2407-11-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodd LE, Sengupta S, Chen IH, den Boon JA, Cheng YJ, Westra W, Newton MA, Mittl BF, McShane L, Chen CJ, et al. Genes involved in DNA repair and nitrosamine metabolism and those located on chromosome 14q32 are dysregulated in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:2216–2225. doi: 10.1158/1055-9965.EPI-06-0455. [DOI] [PubMed] [Google Scholar]
- 32.Schlingemann J, Habtemichael N, Ittrich C, Toedt G, Kramer H, Hambek M, Knecht R, Lichter P, Stauber R, Hahn M. Patient-based cross-platform comparison of oligonucleotide microarray expression profiles. Lab Invest. 2005;85:1024–1039. doi: 10.1038/labinvest.3700293. [DOI] [PubMed] [Google Scholar]
- 33.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 34.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, Dermody JJ. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 35.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.CAN-03-2144. [DOI] [PubMed] [Google Scholar]
- 36.Okuno T, Wakabayashi M, Kato K, Shinoda M, Katayama H, Igaki H, Tsubosa Y, Kojima T, Okabe H, Kimura Y, et al. Esophageal stenosis and the Glasgow Prognostic Score as independent factors of poor prognosis for patients with locally advanced unresectable esophageal cancer treated with chemoradiotherapy (exploratory analysis of JCOG0303) Int J Clin Oncol. 2017;22:1042–1049. doi: 10.1007/s10147-017-1154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18:565–575. doi: 10.1177/0962280209351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf DA, Zhou C, Wee S. The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 40.Füzesi-Levi MG, Fainer I, Ivanov Enchev R, Ben-Nissan G, Levin Y, Kupervaser M, Friedlander G, Salame TM, Nevo R, Peter M, Sharon M. CSNAP, the smallest CSN subunit, modulates proteostasis through cullin-RING ubiquitin ligases. Cell Death Differ. 2020;27:984–998. doi: 10.1038/s41418-019-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroha K, Ando K, Nakagawa R, Inada T. The Upf factor complex interacts with aberrant products derived from mRNAs containing a premature termination codon and facilitates their proteasomal degradation. J Biol Chem. 2013;288:28630–28640. doi: 10.1074/jbc.M113.460691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 43.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tai Y, Tokuda M. Jab1 expression is associated with inverse expression of p27(kip1) and poor prognosis in epithelial ovarian tumors. Clin Cancer Res. 2001;7:4130–4135. [PubMed] [Google Scholar]
- 45.Sutterlüty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Müller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 46.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 47.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 48.Oh W, Lee EW, Sung YH, Yang MR, Ghim J, Lee HW, Song J. Jab1 induces the cytoplasmic localization and degradation of p53 in coordination with Hdm2. J Biol Chem. 2006;281:17457–17465. doi: 10.1074/jbc.M601857200. [DOI] [PubMed] [Google Scholar]
- 49.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, Qu C, Zhang F, Chen YT, Lin YL, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–865. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju S, Wang F, Wang Y, Ju S. CSN8 is a key regulator in hypoxia-induced epithelial-mesenchymal transition and dormancy of colorectal cancer cells. Mol Cancer. 2020;19:168. doi: 10.1186/s12943-020-01285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu YS, Tang ZH, Pan QC, Chen XH, Liu XN, Zang GQ. Inhibition of Csn3 expression induces growth arrest and apoptosis of hepatocellular carcinoma cells. Cancer Chemother Pharmacol. 2012;69:1173–1180. doi: 10.1007/s00280-011-1810-x. [DOI] [PubMed] [Google Scholar]
- 52.Bhansali M, Shemshedini L. COP9 subunits 4 and 5 target soluble guanylyl cyclase α1 and p53 in prostate cancer cells. Mol Endocrinol. 2014;28:834–845. doi: 10.1210/me.2014-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu TL, Cai DL, Zhu GF, Ye XJ, Min TS, Chen HY, Lu DR, Chen HM. Effects of CSN4 knockdown on proliferation and apoptosis of breast cancer MDA-MB-231 cells. Yi Chuan. 2019;41:318–326. doi: 10.16288/j.yczz.18-278. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 54.Zhao S, Tang H, Yan D, Fan J, Sun H, Wen Y, Yu F, Cui F, Zhang D, Xue Y, et al. DDA1 promotes stage IIB-IIC colon cancer progression by activating NFκB/CSN2/GSK-3β signaling. Oncotarget. 2016;7:19794–19812. doi: 10.18632/oncotarget.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.