Graphical abstract
Keywords: Chronotherapy, Irinotecan pharmacodynamics, Translational-transcriptional networks, Circadian rhythms, Molecular circadian profiles, Colorectal cancer
Abstract
Scheduling anticancer drug administration over 24 h may critically impact treatment success in a patient-specific manner. Here, we address personalization of treatment timing using a novel mathematical model of irinotecan cellular pharmacokinetics and –dynamics linked to a representation of the core clock and predict treatment toxicity in a colorectal cancer (CRC) cellular model. The mathematical model is fitted to three different scenarios: mouse liver, where the drug metabolism mainly occurs, and two human colorectal cancer cell lines representing an in vitro experimental system for human colorectal cancer progression. Our model successfully recapitulates quantitative circadian datasets of mRNA and protein expression together with timing-dependent irinotecan cytotoxicity data. The model also discriminates time-dependent toxicity between the different cells, suggesting that treatment can be optimized according to their cellular clock. Our results show that the time-dependent degradation of the protein mediating irinotecan activation, as well as an oscillation in the death rate may play an important role in the circadian variations of drug toxicity. In the future, this model can be used to support personalized treatment scheduling by predicting optimal drug timing based on the patient’s gene expression profile.
1. Introduction
Mammalian physiological and behavioural processes follow a daily rhythm of approximately 24 h, which is regulated by the circadian system. In mammals, the suprachiasmatic nuclei (SCN), a central pacemaker located in the brain, account for organismal entrainment to the geophysical time, primarily via light cues. The SCN pass on time information, in the form of physiological signals, to peripheral clocks, located in each nucleated cell of the organism. The cellular circadian clock is a molecular machinery of interconnected transcriptional-translational feedback loops that produces sustained 24 h-oscillations [1]. Via the regulation of clock-controlled genes (CCGs), the circadian clock controls the timing of multiple cellular and organismal processes, including the cell division cycle, DNA repair or energy metabolism and the immune system [2], [3], [4]. Large inter-individual differences have been observed in several endpoints aiming to measure circadian rhythms, from chronotype questionnaires to melatonin onset timing or circadian biomarkers measured by wearables [5]. Sex appears as a major determinant of circadian rhythms, as women, in general, have higher-amplitude behavioural rhythms than men [6]. The disruption of circadian rhythms leads to mis-regulation in the timing of cellular processes and organ functions, and accumulating evidence points to a negative impact on human health. Again, sex differences exist, as women tend to be more resilient to circadian disruption as compared to men [6]. Several pathologies have been associated to the mis-regulation of the circadian system including cardiovascular diseases, metabolism disorders and cancer [7], [8].
Also, most physiological processes involved in the transport and metabolism of xenobiotics are regulated in a time-dependent manner, which impacts the pharmacokinetics (PK) of numerous drugs that may vary largely depending on the administration timing [9], [10]. On the other hand, several drugs target circadian regulated genes. Recent findings showed that>50% of the top 100 best-selling drugs in the United States target products of circadian genes [11]. Thus, timing drug administration may also impact drug pharmacodynamics (PD) and eventually treatment outcome. In the field of cancer management, several clinical studies have addressed the effect of timing medications for treatment optimization – chronotherapy – with promising results [8], [12], [13], [14]. Giacchetti and colleagues reported data from three international Phase III clinical trials involving 842 patients (345 females and 497 males) treated with 5-fluorouracil, leucovorin and oxaliplatin administered as chronomodulated or conventional infusions [15]. The results showed that male patients lived significantly longer on chronomodulated chemotherapy compared to conventional chemotherapy. Yet, while this specific chronomodulated administration scheme showed a beneficial trend in males, leading to an increase in overall survival (OS), a decrease in OS was reported in females undergoing this chronomodulated regimen, in comparison to a control group receiving the conventional therapy [15]. Moreover, a recent international clinical trial concluded that irinotecan hematological and clinical toxicities were lower for early morning administration in male and for early afternoon infusion in female colorectal cancer patients receiving the drug in combination with 5-fluorouracil and oxaliplatin [16]. Such results highlight the need for more research in this field to understand inter-patient discrepancies and enable safe and efficient clinical application of chronotherapy. Given the reported alterations in circadian gene expression profiles of cancer cells [17], administering anticancer treatment at a time of least toxicity to healthy tissues is likely to provide a benefit to healthy cells while still targeting the cancer cells. In addition, by timing treatment, it would be possible to increase the tolerated dose, or prevent treatment discontinuation, to achieve a more effective toxicity to the tumor cells [8].
Chronotherapy might be more efficient when adapted to the internal time of the patient. Yet, the definition of a single internal time is challenging since the circadian timing system involves multiple inter-connected central and peripheral oscillating processes [18]. We suggest to base chronotherapy individualization on the patient's circadian profiles of selected genes including core-clock genes and genes involved in drug pharmacology. Several patient-friendly methods for measuring clock gene expression using saliva or blood sampling have been recently validated in the clinics [18]. Such patient datasets, combined with mathematical modeling and machine learning, may allow to predict the times of least toxicity to healthy tissues, and optimal antitumor efficacy for an individual patient [18]. In particular, computational models representing the chronopharmacology of a specific drug can help to predict therapy time windows of decreased toxicity and optimal efficacy [8], [18]. Such models can also be optimized for a given patient and used to generate personalized treatment timing indications. In the past years, several ODE (ordinary differential equation) models have been developed, which either aim to model the circadian clock network [19], [20], [21], [22], [23], [24], [25], [26] or the biochemical and biophysical interplay between the circadian timing system and a given drug [27], [28]. These chronoPK-PD models consider both the impact that the organism has on the drug, i.e. its PK, as well as the impact of the drug on the organism, i.e. its PD, and further include the control of the circadian time system on these processes.
Currently, there is a gap between existing mathematical models for the core-clock network and mathematical models of drug PK-PD. Here we aimed at merging the core-clock network with a model of the chronoPK-PD of irinotecan, an anticancer drug widely used against digestive malignancies. We generated a new mathematical model, which enables predictions of the cytotoxicity timing for irinotecan having as an input the circadian gene expression of a set of core clock and irinotecan metabolism-related mRNAs. For that, we refined and combined two previously published ODE mathematical models, a core clock from Relógio et al. [23] and a model of irinotecan chronoPK-PD from Ballesta et al. [27], [28], which have been successfully used for simulating the mammalian core clock and the time-dependent cytotoxicity of irinotecan, respectively. The core-clock model was refined using newly available quantitative circadian datasets of gene and protein expression in the liver of C57Bl6 male mice. Representing the clock of the liver is important in view of predicting the drug metabolism that mainly occurs in this organ in the whole-body scenario [29], [30]. To connect it with the PK-PD model, we extended the transcription-translation network of the core clock with a set of irinotecan-related genes. We fitted our new clock-irinotecan model with transcriptomic data from an in vitro colorectal cancer (CRC) experimental progression model and carried out time-dependent irinotecan treatment in both cell lines across 24 h. The CRC in vitro progression model includes two cell lines derived from the primary tumor (SW480) and from a metastasis site (SW620) of the same patient, which are known to display different circadian profiles [31].
Our mathematical model for timing of irinotecan cytotoxicity nicely reproduced mRNA circadian expression, as well as experimental data obtained via longitudinal monitoring of cytotoxicity for both cell lines. In addition, we found that particular parameters associated with BMAL1 and CLOCK (BMAL1 degradation rate, CLOCK activation rate, cytosolic BMAL1 degradation rate), showed high impact on drug toxicity emphasizing the relevance of the core clock for irinotecan PK-PD. Finally, we proposed possible candidates for molecular biomarkers of irinotecan chronotherapy, which were the prodrug activation enzyme and the enyzme responsible for deactivation of SN-38, the irinotecan main metabolite.
2. Material and methods
2.1. Cell culture
SW480 (ATCC® CCL-228™), SW620 (ATCC® CCL-227™) cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) low glucose (Lonza, Basel, CH) culture medium supplemented with 10% fetal bovine serum (FBS) (Life technologies, Carlsbad, CA, USA), 1% penicillin–streptomycin (Life technologies), 2 mM Ultraglutamine (Lonza) and 1% HEPES (Life technologies). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. The SW480 cell line originated from a surgical specimen of a primary tumour of a moderately differentiated colon adenocarcinoma (Dukes' type B) of a 51-year-old Caucasian male (blood group A, Rh + ). The SW620 cell line was derived from a lymph node metastasis (Dukes' type C) taken from the same patient one year later.
2.2. shRNA-mediated knockdown
For the knockdown of BMAL1, a TRC lentiviral shRNA glycerol set (Dharmacon, Lafayette, CO, USA) specific for BMAL1 was used consisting of five individual shRNAs. The construct that gave best knockdown efficiency was determined by gene expression analysis and used for further experiments.
2.3. RNA extraction
Total RNA isolation was performed using the RNeasy Mini kit (Qiagen, Venlo, NL) according to the supplier's manual. Medium was discarded and cells were washed twice with PBS and lysed in RLT buffer (Qiagen) prior to the purification procedure. RNA was eluted in 30 μL RNase-free water. Final RNA concentration measurement was performed using a Nanodrop 1000 (Thermo Fisher Scientific).
2.4. c-DNA and synthesis RT-qPCR
For Real Time quantitative PCR (RT-qPCR) analysis, the extracted RNA was reverse transcribed into cDNA (4 ng/µl) using random hexamers (Eurofins MWG Operon, Huntsville, AL, USA) and Reverse Transcriptase (Life technologies). RT-qPCR was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) in 96-well plates (see Table 1 for list of primers used). Human GAPDH (QuantiTect Primer, Qiagen) was used as reference housekeeping gene due to its high abundance and to the lack of circadian oscillations, as confirmed by a cosinor analysis carried out in microarray and RNA-seq data for SW480 cells (Supplementary Fig. 2d).
Table 1.
Primers used for the RT-qPCR analysis of SW480 and SW620 cell lines. The primers for mouse can be found in the original publication [32], in the Supplementary File S2.
| Primer | Sequence (5′->3′) |
|---|---|
| PER2 forward | AGCCAAGTGAACGAACTGCC |
| PER2 reverse | GTTTGACCCGCTTGGACTTC |
| NR1D1 forward | CTCCATCGTCCGCATCAATC |
| NR1D1 reverse | AACGCACAGCGTCTCG |
| ARNTL forward | AACCTTCCCACAGCTCACAG |
| ARNTL reverse | CTCTTTGGGCCACCTTCTCC |
| TOP1 forward | CCAAGCATAGCAACAGTGAAC |
| TOP1 reverse | GAGGCTCGAACCTTTTCCTC |
The qPCR reaction was performed using a CFX Connect Real-Time PCR Detection System (Biorad). Relative gene expression was calculated using the 2-ΔΔCt method [33]. Biological and technical replicates were included into the analysis.
2.5. Time-dependent treatment with irinotecan
SW480 and SW620 cells were seeded in 96-well plates at 5000 cells and total volume 150 µL per well. The cells were synchronized by medium change at 4 different time points (6 h, 12 h, 18 h and 24 h) before treatment with 2 µM of irinotecan. Cells (at 60% confluence at the start of measurements) were incubated at 37 °C in a humidified atmosphere with 5% CO2. The corresponding untreated control condition was measured in parallel with the treated cells. Cytotoxicity was evaluated in real time with the IncuCyte® S3 Live-Cell system. Abundances of dead cells are measured experimentally as red florescent objects. Cytotox dyes are inert, non-fluorescent and do not enter viable cells, when added to the cell culture. In dying cells, the membrane integrity is lost, the cytotox dye enters the cells and fluorescently labels the nuclei. To prevent dependence on initial conditions, the cytotoxicity curves are shifted along the cytotoxicity axis such that the first value of all curves overlaps with the control curve. The cells are then identified and quantified by the appearance of red labelled nuclei.Because confluency saturated after 84.5 h for the control conditions, the analysis was restricted to 84.5 h, compare with Supplementary Fig. 7.
2.6. Omics data
The models were fitted to microarray time series data of 24 h sampled with an interval of 3 h for the SW480 and SW620 cells and of 48 h sampled with an interval of 2 h for the liver, which was scaled to concentrations based on RNA-seq data. The microarray data and RNA-seq data for liver tissue was published by Zhang et al. 2014, accession numbers GSE54650 and GSE54652 [11]. For the SW480 and SW620 cells, the microarray time series data was published by El-Athman et al. 2018, accession number E-MTAB-5876 [34], and the RNA-seq time series data was published by El-Athman et al. 2019, accession number E-MTAB-7779 [35]. To relate the microarray data with concentrations, the following steps were done for each gene separately. RNA-seq transcript data was used to calculate the temporal mean of the expression in TPM, which was then converted into mean concentrations in mol/L by a simple rescaling, see Supplementary Information for details. The microarray data was first unlogged (2values) as the data was given in fold change. Then the data was rescaled such that its mean expression matched that of the RNA-seq derived mean concentration, i.e. for a time series × of the original microarray data, we used C * 2x/less than2x>, where < . > denotes a temporal mean, and C is the concentration calculated for this gene based on the RNA-seq data. For gene families, genes with good oscillations were selected as representative gene for the gene family, as denoted in the figures.
2.7. Mathematical models
Model equations and parameters of the core clock are listed in the Supplementary Information, Supplementary Equations (1.1–18) and Supplementary Table 2, model equations and parameters of the clock-irinotecan model are listed in Supplementary Equations (3.1–30) and Supplementary Table 6. Parameter optimization was done using the evolutionary algorithm CMA-ES[36]. Computations for the core clock were carried out on a laptop with i5 2.9 gHz dual core processor using Python's pycma for the optimization and Python's scipy.integrate.odeint for the numerical integration (method: lsoda, relative tolerance = absolute tolerance = 10-12). Computations for the clock-irinotecan network were carried out on a compute cluster with the same Python packages. Model fits are restricted to oscillating mRNAs, with a minimum relative amplitude of 5%, i.e. (max–min)/max > 0.05 for each gene expression time series. The fit of the clock-irinotecan network uses the same algorithm and constraints as the core-clock model, see Supplementary Information. The cost function is extended to account for the additional genes in the network.
The model variables representing proteins relevant for irinotecan PK-PD, i.e. UGT, CES, ABCB and ABCC, do not regulate other genes' expression within our transcription-translation network and are thus not constrained by the available experimental data used for the model fit. Maximal protein concentration for UGT, CES, ABCB and ABCC are scaled to the maximal concentrations used in the original model by Dulong et al. 2015 [28]. Hence, the transcription-translation network predicts toxicity based on the estimated relative amplitude and phase of the protein oscillations, and formerly determined mean absolute levels. The model of Dulong et al. 2015 [28] explicitly involves ABCG2, which is in our case replaced by the dynamical variable ABCC with an appropriate rescaling. The existing model of irinotecan chronoPK-PD uses protein dynamics as inputs to ultimately predict irinotecan toxicity. We replaced the cosine fit with the dynamics that result from the clock-irinotecan network.
For the cell line Caco-2 (cell line derived from a human colorectal adenocarcinoma), the PK-PD model was fitted to cell death following irinotecan treatment [27], [28]. To fit the circadian variation in toxicity, we change the PK-PD model output by replacing the equation modelling apoptosis with two equations for the time series of alive and dead cells, see Supplementary Equations (3.29) and (3.30). As it turns out, this current model is not sufficient to reproduce the large timing-related differences in cytotoxicity observed experimentally, likely due to the small relative amplitudes of the protein oscillations, which in our model, as defined by Supplementary Equations (3.1) to (3.19), cannot be larger than the fitted mRNA oscillations. To relax this constraint, we replaced constant protein degradation for UGT, CES, ABCB and ABCC with oscillatory degradation in the final model [37].
For convenience, acrophases are rescaled to the range from 0 to 1 instead of 0 to 2π.
2.8. Statistical analysis
The experimental toxicity profile is fitted by a harmonic regression using Matlab, significance is set to p ≤ 0.05 [18]. For the cytotoxicity data, the Area Under the Curve (AUC) is calculated using the linear trapezoidal method, using as weights wk a vector with n elements (where n is the number of time points considered), with 1 h for the first and last element, and 2 h for the other elements, with the error associated calculated as , where SEMk is the standard error at the time point related to time point k, and var is the variance of the AUC calculated as .
3. Results
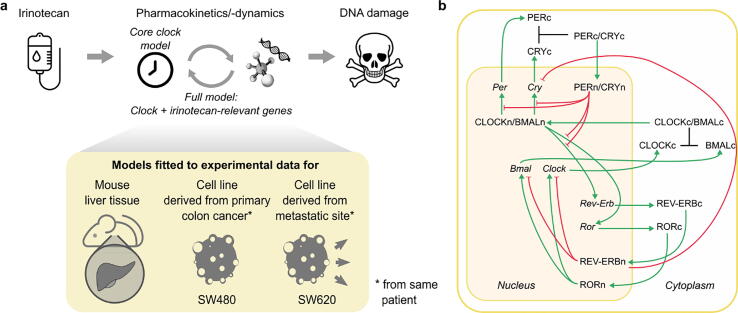
The effect of a drug results from an intricate interplay between its metabolites and the organism, which is under circadian control. Regarding the anticancer agent irinotecan, multiple genes and proteins involved in its PK-PD are directly or indirectly regulated by the cellular core clock. The aim of the study was to design a mathematical model combining the core clock and irinotecan PK-PD-related elements to investigate possible cellular biomarkers predicting irinotecan chronotoxicity rhythms (Fig. 1a, top). This model was developed and calibrated for three biological systems: the healthy mouse liver, and two cell lines derived from human colorectal cancer (CRC) (Fig. 1a bottom). The combined model was trained for the CRC cell lines using circadian datasets of mRNA levels and with experimental results on time-related irinotecan cytotoxicity. We first present a quantitative version of the core-clock model (Fig. 1b), followed by its extension to account for the clock-controlled regulation of genes involved in irinotecan PK-PD. Finally, this model was connected to a representation of irinotecan chronoPK-PD.
Fig. 1.
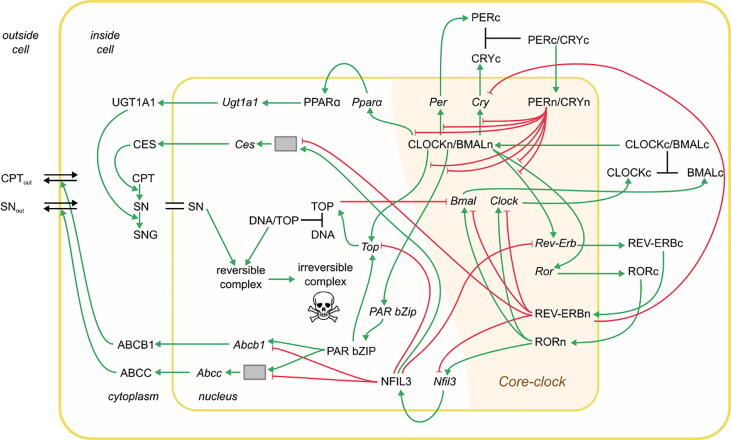
The action of the drug irinotecan involves the core clock and a set of clock-regulated genes, experimentally assessable in different cell types. a Workflow of the clock-irinotecan model construction. Irinotecan induces DNA damage and potentially cell death via its interaction with clock-controlled proteins. Mathematical models were fitted to different datasets in healthy mouse liver, and in human cancer cell lines. b Network representation of the core-clock model. Inhibitory interactions are presented in red with flat arrowheads, activating interactions in green with pointed arrowheads, and complex formation in black.
3.1. A quantitative model of the core clock in mouse liver
To investigate the interactions between the circadian clock and irinotecan cellular PK-PD, we started by designing a quantitative model of the cellular core clock (Fig. 1b). We refined the previously published ODE model by Relógio et al. [23], which represents the molecular mechanisms of the core clock at the cellular level based on experimental data for the mammalian SCN. Clock gene paralogs and isoforms were merged into the following model variables for mRNA elements: Per (Per1, Per2, Per3), Cry (Cry1, Cry2), Ror (Rora, Rorb, Rorc), Rev-Erb (Rev-Erbα, Rev-Erbβ) and Bmal1. We applied the same principle for model variables representing proteins and protein complexes. The dynamical variable CLOCK/BMAL representing the CLOCK/BMAL1 dimer is assumed to activate the transcription of the core-clock genes Rev-Erb, Ror, Per, and Cry and the PER/CRY complex to inhibit this transcriptional activity. The model includes two main negative feedback loops. The first one involves the self-inhibition of the dynamical variables Per and Cry through the inhibition of CLOCK/BMAL by the PER/CRY complex. In addition, REV-ERB inhibits the transcription of Cry, thus inhibiting its own inhibition through the regulation of PER/CRY. The second feedback loop is induced by the self-repression of the dynamical variable Bmal through the activation of its repressor REV-ERB by CLOCK/BMAL. On the contrary, ROR, which is transcriptionally activated via CLOCK/BMAL, acts positively on Bmal regulation.
The Relógio et al. model differentiated between phosphorylated and unphosphorylated PER proteins [23]. However, in the absence of time-dependent quantitative data on PER phosphorylation, we opted to simplify the PER/CRY (PC) loop and to merge the phosphorylated/unphosphorylated variables (Fig. 1b). Similarly, the equations for the dynamical variables CLOCK/BMAL and PER/CRY cytoplasmic complexes originally included both a term for complex dissociation into free proteins and for complex degradation, which were not identifiable from the available data so that the degradation terms were removed (Supplementary Information, Section 1).
We further refined the core-clock model to represent the core clock in organs relevant for irinotecan pharmacology, in particular the liver, where the drug is processed. The Relógio et al. model did not explicitly consider Clock given its lack of rhythmicity in the SCN [38]. However, this is not the case in the liver [32]. Moreover, CLOCK/BMAL1 is a key transcriptional regulator of genes involved in the irinotecan network [39], [40]. Thus, we expanded the initial model by explicitly including Clock as follows. Similarly to the dynamical variable Bmal, Clock transcription is assumed to be positively regulated by ROR and negatively impacted by REV-ERB [41]. The cytoplasmic protein CLOCKC dimerizes with the dynamical variable BMALC and translocates to the nucleus to form the heterodimer complex CLOCK/BMALN. The dynamical variable CLOCKC representing the cytosolic CLOCK protein is assumed not to be able to enter the nucleus, as it was not detected in the nucleus of cells not expressing Bmal1 [42]. Of note, BMAL1 and CLOCK nuclear protein expressions shared the same circadian phase and amplitude experimentally, suggesting that both species exist mostly as dimers in the nucleus [43] (Fig. 1b). One last modification was made to the model structure to increase the accuracy of cytoplasm/nucleus transport terms. The equations now account for the ratio between the compartment volumes to ensure that the quantity of matter is conserved during transport (Supplementary Information, Section 1.1.5). The cytoplasm/nucleus volume ratio was set for mouse hepatocytes to 14 [44].
Our new core-clock model allows to quantitatively simulate gene and protein levels, expressed in mol/L, thus allowing for the fitting of quantitative datasets informing on absolute concentrations (Table 2). Parameter estimation was done using the time series data reported by Narumi et al. [32] (Supplementary Information, Section 1.3.4). Starting from the Relogio et al. model, we performed a linear change of variables, mapping the original model variables to their scaled versions with respect to the maximum of the observed data (Supplementary Information, Section 1.3.2). The obtained scaled parameter values were then used as an initial guess for the subsequent parameter estimation procedures.
Table 2.
Resources for the data used in the current study.
| Dataset | Organ / Cell line | Used for model | Acquisition technique | Accession number |
|---|---|---|---|---|
| Narumi et al. 2016 [32] | C57B16 male mouse liver (WT mouse and Bmal1-/-) | Liver core-clock | mRNA: RT-qPCR proteins: mass spectrometry |
NA |
| Zhang et al. 2014 [11] | C57B16 male mouse liver | Liver transcription-translation network | mRNA: microarray and RNA-seq | GSE54650 and GSE54652 |
| Wang et al. 2017 [43] | C57B16 male mouse liver | Liver core-clock | Nuclear proteins: mass spectrometry | E-MTAB-5876 |
| El-Athman et al. 2018 [34] | SW480 / SW620 human CRC cell lines | CRC core-clock CRC full model | mRNA: microarray | E-MTAB-7779 |
| El-Athman et al. 2019 [35] | SW480 / SW620 human CRC cell lines | CRC core-clock CRC full model | mRNA: RNA-seq | E-MTAB-7779 |
| In this publication | SW480 / SW620, control and siRNA Bmal1 | CRC core-clock CRC full model | mRNA: RT-qPCR | NA |
| Dulong et al. 2015; Ballesta et al. 2011 [27], [28] | Caco-2 human colorectal cancer cell line | CRC full model | CPT11 and SN38 cellular PK: HPLC TOP1 activity: DotBlot CPT11 and SN38 cytotoxicity: viability assays |
NA |
| In this publication | SW480 / SW620 | CRC full model | CPT11 cytotoxicity | NA |
| Zheng et al. 2019 [42] | C57B16 male mouse liver | Liver core-clock | Cytoplasmic and nuclear proteins CLOCK and BMAL abundance: immunoprecipitation | NA |
| Aryal et al. 2017 [46] | C57B16 male mouse liver | Liver core-clock | Cytoplasmic proteins PER and CRY: immunodepletion | NA |
| Schwanhäusser et al. 2011 [47] | Mouse fibroblasts NIH3T3 | Liver core-clock | mRNA transcription rates: RNA-seq | SRA030871 |
The liver dataset also included protein expression for Bmal1-/- mice [32]. Assuming that Bmal1 knockout (KO) led to a loss of oscillations in the clock [45], this data could be seen as a glance at the system at steady state. This enabled us to derive functional relationships to compute three transcription rate parameters as a function of the KO mice data and other parameters (Supplementary Information, Section 1.3.3), thus decreasing the number of parameters to estimate. This led to a simplification in the parameter estimation. We further reduced the number of parameters by assuming that Hill power coefficients were equal for all activators (parameter b) and all inhibitors (parameter c) of the transcription across genes. This led to a decrease of 8 parameters to be estimated while producing next to no change in the goodness of fit as expected from the argument of unidentifiability. Only Cry kept separated Hill coefficients due to its transcription being regulated by 3 species (the dynamical variables CLOCK/BMAL, PER/CRY and REV-ERB). The final core-clock model has 18 state variables and 58 parameters to be estimated.
The parameter estimation procedures consisted in a numerical minimization of a cost function, which was the sum of two terms (Supplementary Information, Section 1, Supplementary Equation (1.28)). The first term is the least square error between the data and the model’s simulation, while the second term accounts for biological constraints. These constraints were derived from co-immunoprecipitation experiments and provided bounds for complex concentrations with respect to free protein concentrations [42], [46]. Additional constraints were specified on the bounds of parameter search intervals including those of degradation or transcription rates based on mRNA and protein half-lives and levels [47]. Fig. 2 shows the model best-fit, which convincingly reproduced the data (R2 = 0.86). Bmal1 and Clock mRNA model-predicted profiles presented a similar phase but different mean levels (5.3 and 19.5 pmol/L, respectively) and relative amplitude (84% and 62% of the mean, respectively). Differences were also observed at the protein level as free BMAL1 and CLOCK protein mean levels were equal to 13.9 and 8.85 nmol/L respectively, with relative amplitudes of 35% and 25%. These differences came as a justification to the addition of the Clock gene into the core-clock model.
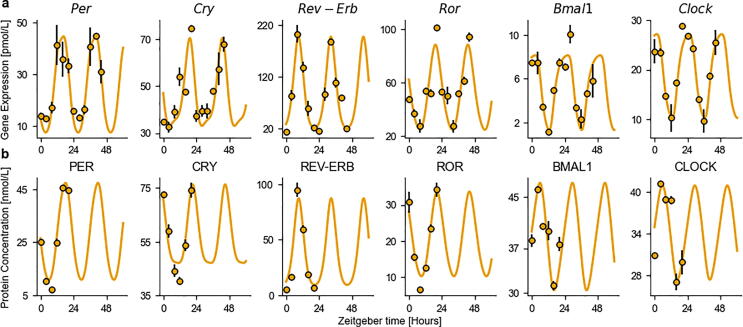
Fig. 2.
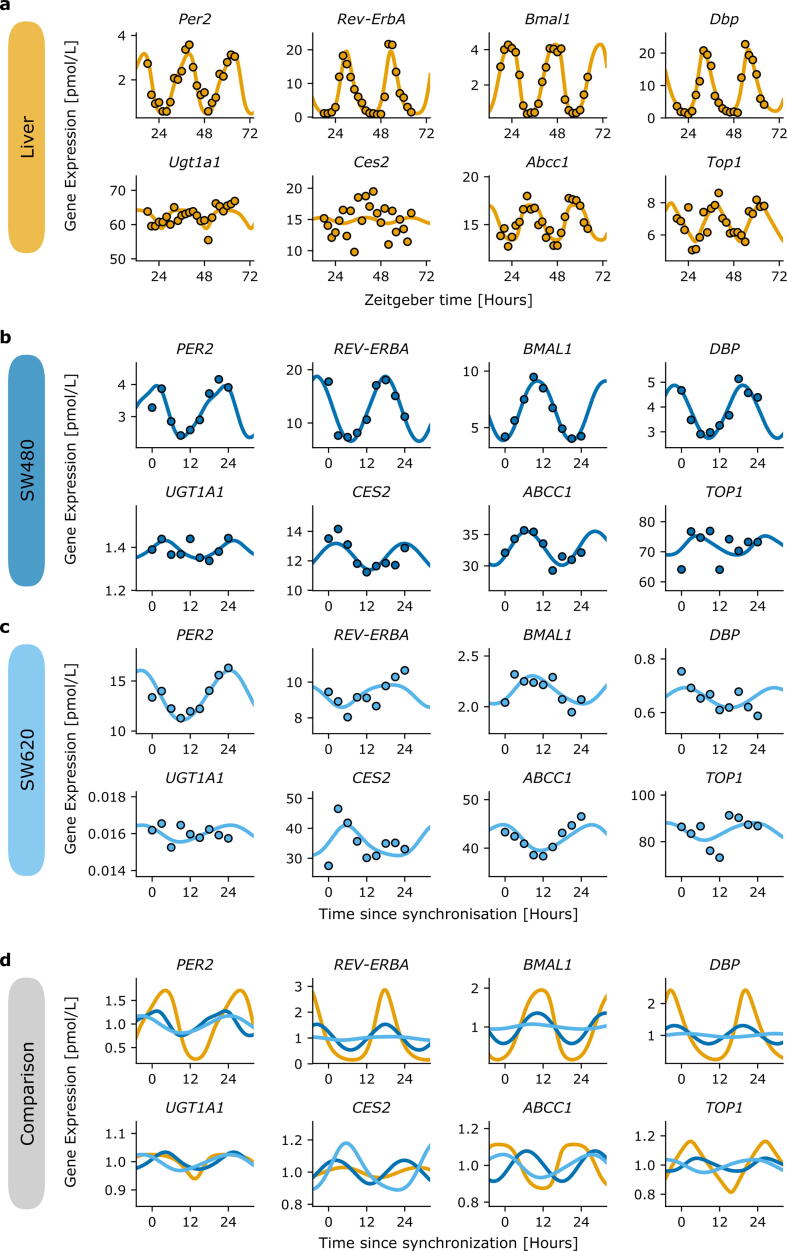
Best fit of the quantitative core-clock model to mRNA and protein circadian datasets in the mouse liver. a mRNA expression for core-clock elements in pmol/L. b Protein levels for core-clock elements in pmol/L. Model simulation (orange lines), experimental data used for calibration (black circles). Depicted are mean values (n = 2 biological replicates) ± SEM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Validation of the model was done using an external time course dataset from mouse hepatocytes, which was not used for the model design and calibration [43]. This study reports a phase between 8.5 h and 10.8 h for the circadian rhythm of REV-ERB nuclear expression and a relative amplitude of 98%, while the model simulation for phase and relative amplitude were 9.7 h and 90%, respectively. Similarly, for ROR nuclear expression, the reported phase was 20.8 h, as compared to 21.1 h for model predictions, and its relative amplitude was 80% as compared to 69% for the model. Both predictions are in close agreement with the study and serve as a validation of the model. For the other clock proteins, as our model only tracked them as complexes in the nucleus, the comparison to single-protein data was not possible. A subsequent robustness analysis was performed by analyzing whether the model could maintain sustained oscillations upon parameter perturbation. Gaussian noise was added to the best-fit parameter vector with a standard deviation of 10%, leading to oscillating simulations in 73% of the cases, thus demonstrating the model robustness (Supplementary Fig. 1).
3.2. The clock model reproduces the expression profiles of core-clock genes in CRC cell lines
To test our mathematical model in a colon cancer context, we chose a well-known in vitro cellular model of CRC progression, which includes two cell lines from the same patient (SW480, SW620), derived from the primary tumour and from the metastasis, respectively. We carried out a time course of 45 h (9 h − 54 h, after synchronization) with a 3 h sampling interval, for the gene expression analysis of PER2, REV-ERBα and BMAL1 via RT-qPCR, in either control or shBMAL1 conditions. This dataset was combined with microarray data for the expression of Cry, Ror and Clock in order to calibrate the core-clock model for each of the CRC cell lines (see Section 2.6). Quantitative mean concentration levels expressed in mol/L for the clock genes of CRC cell lines were derived from an already published RNA-seq transcriptomic dataset [11]. Thus, in total, three datasets were combined for the calibration of the core-clock model for the CRC cell lines. The cytoplasm/nucleus volume ratio of the CRC cell lines was set to 5 (manual curation, using snapshots of SW480 and S620 cells from Abdulrehman et al. [48], Fig. 2, the cytoplasm/nucleus ratio was computed for each cell of the figure and an average value close to 5 was found for both cell lines). The transcription-translation network was assumed to be similar in either control or shBMAL1 conditions, yet with a single different parameter to account for shBMAL1 activity. Accordingly, the RT-qPCR datasets obtained from the control and shBMAL1 conditions were fitted simultaneously to their respective models using the same set of parameter values with the exception of BMAL1 basal transcription, which was allowed to differ between both conditions (Supplementary Fig. 3). The shBMAL1 condition provided a view of a dampened circadian clock, due to the knockdown of Bmal1, which induced a lower activation power of the transcription factor CLOCK/BMAL1. Upon model calibration, a 375-fold reduction in the BMAL1 estimated basal transcription rate was necessary to allow for a good fit of both conditions. This demonstrates the ability of the model to reproduce two different physiological scenarios by tuning a single parameter.
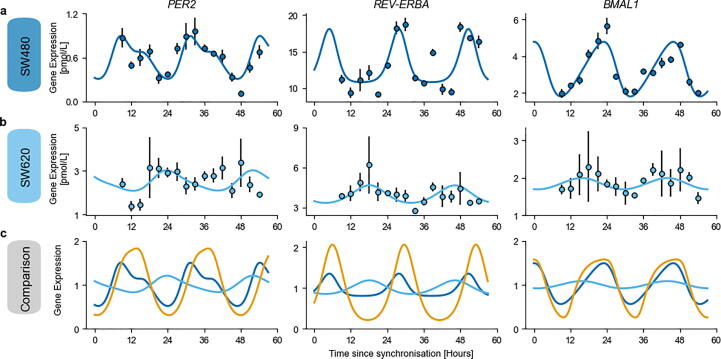
Concerning the SW480 cell line, the model achieved an excellent fit of the data (R2 = 0.75) (Fig. 3a, Supplementary Fig. 2a). The fit for the SW620 was reasonable as well (R2 = 0.67), but lacked a proper fit of CRY, ROR and CLOCK expression reported in the microarray dataset (Fig. 3b, Supplementary Fig. 2b). Oscillations of the core-clock genes, normalized to the mesor, showed larger relative amplitudes in the healthy mouse liver than in CRC-derived cell lines, with the circadian rhythms in SW620 cells being largely dampened as compared to both other systems (Fig. 3c). The peaks of BMAL1 mRNA levels of the best-fit model for mouse liver and SW480 cells were aligned to allow for an in vitro/in vivo systems comparison. This highlighted a moderate phase shift of 5 h (respectively 1 h) for PER2 (respectively REV-ERBα) between the SW480 cell line and liver tissue. On the opposite, larger phase delays were observed in the case of the SW620 cell line. Although the three models represent different organs in different conditions, their comparison exhibited a moderate agreement between the clocks of the healthy liver and of the SW480 colorectal cancer cell line, and large differences in terms of oscillations dampening and phase differences in comparison to the clock of the SW620 metastatic colon cancer cell line.
Fig. 3.
Comparison of the core-clock models fitted to healthy mouse liver or human cancer cell lines. Best fit of the quantitative core-clock model to (a) the SW480 cell line and (b) the SW620 cell line. Model simulation (line) against the RT-qPCR data used for calibration (dots), depicted as mean values (n = 3) ± SEM. c Comparison of the model fit for liver (orange), SW480 (dark blue) and SW620 (sky blue). Bmal1 circadian phases were aligned for mouse liver and SW480 cell line and all gene expression profiles were normalized to the mesor to allow for comparison. See Supplementary Fig. 2 for the other genes of the core clock model. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For most core-clock genes, the oscillations displayed a non-cosine shape, with different intervals of high versus low gene expression, see Fig. 3c and Supplementary Fig. 2c. Overall, the here presented core-clock model, based on cellular mRNA and protein concentrations, reproduced the circadian gene expression profiles for different sets of experimental data with good precision. Thus, this model provided a reasonable starting point for the following extension with irinotecan PK-PD-related genes.
3.3. Filling the gap: Connecting the core clock with irinotecan PK-PD related genes
We extended the core-clock network with eight clock-controlled genes relevant for irinotecan pharmacology as depicted in Fig. 4, named in the following clock-irinotecan network. The elements added to the core-clock model are involved in irinotecan metabolism, transport, and pharmacodynamics. Irinotecan is a prodrug, which needs to be converted into its active metabolite, SN-38, through the enzymatic activity of CES2 (Carboxylesterase 2) [49]. Subsequently, UGT1A1 (uridine diphosphate glucuronosyltransferase 1A1) regulates the conversion of SN-38 into its inactivated form, SN-38G [29]. The ATP-Binding Cassette (ABC) transporters ABCB1, ABCC1, ABCC2 and ABCG2 control the efflux of these molecules out of the cells [50]. Central to irinotecan action, SN-38 binds to the protein TOP1 (DNA topoisomerase 1), which under normal conditions releases the supercoiling and torsional tension of DNA by transiently cleaving and rejoining one strand of the DNA, thereby controlling DNA topology during replication and transcription. SN-38 binds to DNA-TOP1 complexes to stabilize them. This leads to double-stranded breaks, erroneous transcription and likely cell death [51], [52]. Besides these proteins directly relevant for irinotecan PK-PD, the clock-irinotecan network contains three elements that act as transcription factors for the above-mentioned genes, DBP (D site of albumin promoter (albumin D-box) binding protein), which is also considered as a core-clock element, NFIL3 (Nuclear factor, interleukin 3 regulated), and PPARα (Peroxisome proliferator-activated receptor alpha), a regulator of liver lipid metabolism that also acts as transcription factor for UGT1A1, which deactivates SN38 [53], [54], [55]. For the clock-irinotecan network, we only consider the ABC transporter ABCB1 for irinotecan efflux and ABCC1 for SN38 and SN38G efflux as ABCC2 showed less clear circadian oscillations and ABCG2 did not appear in our RNA-seq data for the studied cell lines. The mathematical description of the clock-irinotecan network contains 39 equations and 115 parameters (Supplementary Table 5 and 6, Supplementary Equations (3.1) - (3.19)). In the clock-irinotecan network, oscillations are inherited from the core clock to genes outside of the core-clock network via the CLOCK/BMAL1 complex, ROR and REV-ERB. Accordingly, most connections go from the core clock to the remaining elements. Only the inhibition of REV-ERB by NFIL3 and the inhibition of BMAL1 by TOP1 provides feedback from the irinotecan-related genes to the core clock [56], [57]. From the clock-irinotecan network fitted to experimental mRNA expression data, we use the mRNA dynamics of UGT1A1, CES2, ABCB1 and ABCC1 as an input to the protein dynamics and the PK-PD model to predict irinotecan toxicity, see below.
Fig. 4.
Model of the interplay between irinotecan PK-PD and the core clock. Irinotecan treatment is simulated by a transcriptional-translational network that comprises the core clock, irinotecan-relevant genes, and the PK-PD of irinotecan. Different types of interactions are represented among the elements of the network: inhibition (red arrows with flat arrowhead) and activation interactions (green arrows); complex formation (black lines). Grey boxes represent post-transcriptional sub-networks necessary for a model fit to the data. The double black line indicates equal concentrations between nucleus and cytoplasm, the double black line with arrowheads indicates CPT-11 and SN-38 cellular transport inside and outside of the cell. All indicated molecular interactions are based on experimental evidence from a number of different sources and corresponding references are provided in Supplementary Table 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The clock-irinotecan network was fitted to experimental time-series datasets of mRNA concentrations [35], available for mouse liver [11] and extracted from microarray and RNA-seq data for CRC cell lines, see Section 2 for details. We fitted the clock-irinotecan model to the temporal dynamics of mouse liver, as well as to untreated SW480 and SW620 cells, which resulted in R2 scores of 0.72, 0.61 and 0.40, respectively (Fig. 5a-c), see Supplementary Fig. 4 for an example with all genes. For acrophases and relative amplitudes of the model fits see Supplementary Fig. 5. Periods predicted by the model are 23.5 h for the liver (in accordance with literature values for the circadian period of mice [58]), 21.6 h for the SW480 cells and 28.8 h for SW620 cells (within the range of previously reported values [17], [31]).
Fig. 5.
Comparison of the fitted clock-irinotecan network for healthy mouse liver and human cancer-derived cells. Selected gene expression and fit of the clock-irinotecan network for (a) mouse liver data, (b) SW480 and (c) SW620, all data without treatment. d Comparison of the model output when fitted to liver (orange), SW480 (dark blue) and SW620 (sky blue). Profiles were normalized to the mesor, and BMAL1 phases were aligned between mouse liver and the SW480 cell line to allow for comparison. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A first version of the model assumed direct (i.e. one step) regulation of irinotecan-related gene transcription by elements of the core clock (Supplementary Equations (3.1) to (3.12), i.e. Fig. 4 without the grey boxes). While this restriction did not hamper the fitting of most genes, the resulting best-fit curves for CES2 and ABCC1 mRNA levels were phase shifted compared to corresponding microarray data in SW cell lines. This originated from large phase delays between the clock-controlled regulators and the expression of the regulated genes. For example, CES2, which showed clear circadian oscillations in the SW480 cell line (harmonic regression for a 24 h period results in p-value = 0.014, acrophase = 0.09 rad/2π, relative amplitude = 30%), peaked seemingly before its two regulators, REV-ERBA and NFIL3, see Supplementary Fig. 5. Thus, more intermediate elements might play a role in the network. As a simple solution, we extended the model for CES2 clock-controlled transcription by a simple chain of post-transcriptional modifications (compare Supplementary Equations (3.13) to (3.19) and see Fig. 4, grey boxes). To cover the phase delay between Ces2 and NFIL3 of 0.60 rad/2π (12.9 h, phase delay between Ces2 and REV-ERBα is 0.85 rad/2π, i.e. 18.2 h), three unidirectional activation steps, with one parameter for both activation and degradation rates, were required. As ABCC1 showed a similar problem, we added an analogue set of intermediate reactions for ABCC1 transcription, for which two steps were sufficient, as the phase delay with its regulators was smaller (phase delay between Ces2 and NFIL3 of 0.39 rad/2π, i.e. 8.4 h, phase delay between Ces2 and REV-ERBα of 0.64 rad/2π, i.e. 13.8 h), see Supplementary Fig. 5. The fit of the additional genes does not reduce the quality of the fit of the core-clock model, with R2 scores for the core clock of the full fit of 0.93, 0.78 and 0.57 compared to 0.84, 0.67 and 0.52 for a fit of only the core clock using the rescaled microarray data (see Section 2), for liver tissue, SW480 and SW620 cell lines, respectively. Lower R2 scores for the full fit likely result from the longer optimization required for a good fit of all genes, as compared to the optimization required for fitting only the core clock genes. From liver to SW480 to SW620, the relative amplitude of the oscillation was reduced for genes of the core clock and for genes directly regulated by the core clock, whereas this amplitude reduction was relaxed for genes only indirectly controlled by the core clock (Fig. 5d and Supplementary Fig. 5b).
From the fit of the gene expression data, we obtained a calibrated model computing clock and irinotecan-related mRNA circadian rhythms. However, mRNAs need to be translated into proteins that eventually interact with the drug. Hence, the link between the clock-irinotecan network and the PK-PD model for treatment toxicity was assumed via the protein dynamics and allows us to investigate the interplay between the circadian clock and irinotecan action. We designed a new model of protein dynamics of UGT1A1, CES2, ABCB1 and ABCC1, which are the inputs for the irinotecan chronoPK-PD model (Supplementary Equation (3.3) with (3.29)), replacing the forced cosine function utilized in the original model by Dulong et al. 2015 [28]. The protein dynamics contained a term for ptotein translation including mRNA levels computed by the clock-irinotean model, together with a circadian process of degradation as suggested for many proteins [37]. Magnitude, amplitude and phase of the circadian degradation are fitted to cytotoxicity data; the translation rate is set to 1 as protein abundances are re-scaled in the PK-PD model, see Section 2 [37].
3.4. The full clock-irinotecan model recapitulates different chronotoxicity rhythms for CRC cells
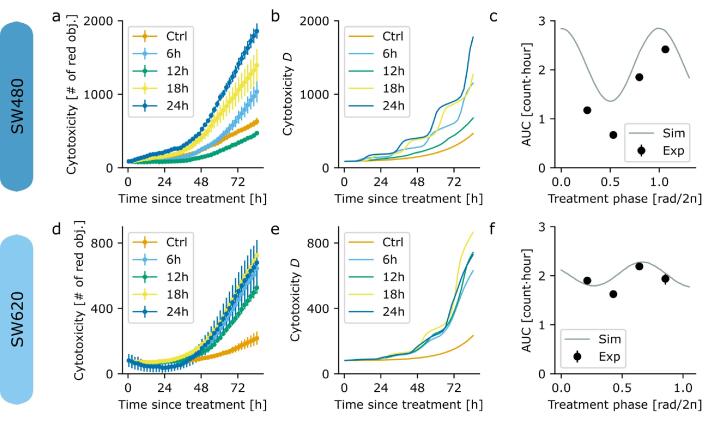
To investigate the putative effects of time-dependent treatment in CRC, SW480 and SW620 cells were synchronized by media change and treated with 2 µM of irinotecan at four different circadian times (CT after synchronization: 6 h, 12 h, 18 h and 24 h). The SW480 cell line exhibited a circadian cytotoxicity response to treatment (harmonic regression with the period of the model fit, p = 0.043 for SW480, see Supplementary Fig. 7; not significant for SW620). SW480 cells showed the highest toxicity when irinotecan was administered 24 h after synchronization, while the lowest toxicity was observed when irinotecan was administered 12 h post synchronization (acrophase of 0.006 ± 0.03 rad/2π, Fig. 6 c). The differences in cytotoxicity values between different treatment time points where higher in SW480 as compared to SW620 cells, which resulted in larger circadian amplitudes for the SW480 toxicity rhythm (Fig. 6b). Here it is relevant to notice that, in the absence of treatment, the number of dead cells in SW480 cultures is higher than for SW620 cell cultures (ratio of the area under the curve of SW480 and SW620 is 2.46 ± 0.07, Fig. 6 a and d) pointing to cell death and cell cycle differences between the tumor and the metastasis-derived cells.
Fig. 6.
Fitting of the time-dependent treatment from human cancer cell lines. a Experimentally measured cytotoxicity curves, estimated by measuring red fluorescent objects (see Methods), for SW480 cells that are untreated (Ctrl), or treated at indicated time points with irinotecan (6 h, 12 h, 18 h or 24 h after synchronization). Time is aligned to treatment onset. b Best-fit of the extended PK-PD model (shown is the number of dead cells, the dynamical variable D of Supplementary Equation (3.31)) to the experimental cytotoxicity data of the SW480 cell line. c Area Under the Curve (AUC) of treated SW480 cells normalized by the untreated control (dots), compared with the area under the curve of the best-fit model (grey line). d Experimentally measured cytotoxicity curves for SW620 cells untreated (Ctrl), or treated at indicated time points with irinotecan (6 h, 12 h, 18 h or 24 h after synchronization). e Best-fit of the extended PK-PD model to the experimental cytotoxicity data of the SW620 cell line. f Area under the curve of treated SW620 cells normalized by the untreated control (dots), compared with the area under the curve of the best-fit model (grey line). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To allow for the comparison of the model with this experimental data, we supplemented the model by Dulong et al. 2015 [28] with two cell population equations that explicitly track the number of alive and dead cells. An exponential growth and a first-order natural cell death were assumed in both control and treated conditions. Irinotecan was assumed to act negatively on cell proliferation and survival through DNA damage formation, and a circadian oscillation in the cell death rate was added, see Supplementary Equations (3.30) and (3.31). Parameters of the original model were kept unchanged apart from the formation rate of the irreversible complex which had to be adapted for a successful fit, and which ended up being reduced as compared to its former estimation.
Using the mRNA dynamics computed by the clock-irinotecan model, the irinotecan PK-PD model allows to fit the circadian dynamics of cell death (Fig. 6). The best-fit full model generated a circadian cell death profile that agreed with the toxicity phase of the experimental data for the SW480 cell line. The model also recapitulated a different toxicity profile for the SW620 cell line, supporting the hypothesis that the same drug at the same concentration could lead to different responses based on the time of treatment administration and on the cancer clocks. Interestingly, while the highest and lowest cytotoxicity trends were the same in both cell lines, the overall response to the cytotoxic effect of the drug was higher in SW480 (derived from the primary tumour) in comparison to SW620 (derived from a metastasis, but from the same patient). This also alludes to a role of the cellular clock profile in treatment outcome, as the two cell lines have different oscillatory patterns. We further tested a simplified version of the equation for the protein dynamics assuming constant, i.e. non-circadian, protein degradation (Supplementary information, Equation (3.3) with constant degradation rate). As anticipated by the mathematical analysis, toxicity oscillation amplitudes were drastically reduced to approximately 1% of the mesor and were then much smaller than those observed experimentally. Yet, our simplified clockPK-PD model with minor adaptations to the experimental settings gave reasonable toxicity phases using non-circadian protein dynamics with an appropriately chosen degradation rate (Supplementary Fig. 6), without fitting the model to the circadian toxicity values obtained experimentally, see Supplementary Fig. 7.
To test for the sensitivity of our final model to parameter variations, we evaluated parameter sensibility of a set of 123 parameters with respect to the phase and amplitude of irinotecan circadian toxicity profiles (i.e. the curves depicted in Fig. 6c and f) by calculating Sobol sensitivity total order indices, see Supplementary Fig. 9. A close agreement was found between the parameter sensitivity on the phase and on the amplitude of the drug chronotoxicity rhythms. Our analysis highlighted the impact of the protein dynamics on the toxicity profile, most importantly the relevance of the phases of the circadian degradation of CES2 (parameter phiCes from Supplementary Fig. 9) and UGT1A1 (parameter phiUgt) and the amplitude of CES2 (parameter ampCes). Besides those parameters, several core-clock elements - in particular parameters associated with the loop formed by ROR, BMAL1 and CLOCK (maximal transcription rates, degradation rates, production rates) - showed high sensitivity, probably because existence of most oscillations depends on the core clock. The feedback from irinotecan-relevant genes to the core clock, through the inhibition of REV-ERB by NFIL3 (parameter i_RevNfil) and the inhibition of BMAL1 by TOP1 (parameter i_BmalTop), only showed a weak impact on the toxicity curve. All parameters associated with ABC transporters showed a low impact on the toxicity profile.
4. Discussion
The circadian clock regulates the timing of various crucial molecular pathways including drug metabolism, apoptosis, DNA damage repair and cell cycle [59], [60], [61]. The malfunctioning of these pathways is involved in cancer onset and progression. On the other hand, several drugs used in cancer treatment target genes, which are expressed in a circadian manner and also the metabolism of these drugs is carried out by circadian-regulated genes and proteins. Hence, timing treatment in accordance with the patient’s circadian timing system is likely to contribute to improved treatment outcome, and several studies have shown promising results using chronotherapy in cancer treatment [8].
We developed a novel mathematical model of irinotecan cellular PK-PD, which links the core clock with predicted treatment toxicity for CRC cells. The model simulations highlighted the existence of time-dependent toxicity for the different cells, which was different for the tumour-derived cell line as compared to the metastasis-derived cell line. Our results suggest that, in addition to gene expression, the dynamics of proteins, with circadian variation in their degradation, plays an important role in the timing of drug toxicity. In particular the phase (the time of maximum expression) and amplitude (difference between minimum and maximum) of the circadian oscillation in protein degradation of CES2, which controls the activation of irinotecan, seems to be relevant in shaping the toxicity profile. Moreover, elements associated with core-clock genes, such as BMAL1 or CLOCK showed high sensitivity which proved the importance of the core-clock parameters on irinotecan toxicity.
4.1. A comprehensive mathematical model for circadian regulation of irinotecan PK-PD
Our clock-irinotecan model can be fitted to different scenarios providing different circadian toxicity profiles for CRC cells. The core-clock model was initially developed from multiple datasets of mammalian SCN cells [23], and was successfully refined here using quantitative measurements of the clock of the mouse liver, and of SW480 and SW620 cell lines. Regarding the model of irinotecan PK-PD, it was designed based on extensive datasets in Caco-2 cells [28], and was further validated in both SW480 and SW620 cell lines. Using SW480 and SW620 cell lines here provided a proof of principle that a personalization of the model to other cell lines was possible. Hence, models of both transcription-translation clock network and irinotecan PK-PD were validated in several in vitro and in vivo experimental settings, which argues in favour of their reliability.
The reduction in fit quality from liver to SW480 to SW620 cells likely results from the decreasing amplitudes of circadian oscillations, see Supplementary Fig. 5b [17], [31]. Indeed, assuming that the experimental data of the same gene, yet from different biological sources (liver or CRC cell lines), shows the same level of noise, arising from biological stochasticity or from experimental constraints, larger oscillation amplitudes lead to higher signal to noise ratios, which facilitates fitting. In addition, the fact that the core-clock model did not fully fit the SW620 data was an indication that clock gene and protein interactions may be impaired in this cell line or at least different from the ones implemented in the model. It is also important to notice that each patient (or healthy individuals) and each cancer are unique. Accordingly, different cell lines, patients or healthy individuals [62] have specific clock phenotypes, and this requires personalization of treatment. Thus, any clinical application requires that our model is fitted to the individual patient, or to groups of patients with similar clock molecular profiles.
Another ODE-based model of the mouse liver clock was designed by Woller et al. to investigate the effect of feeding cycles on liver circadian rhythms [25], [26]. That model was developed to address a different question as compared to this study and could not be readily used here, as for instance the energy metabolism part was out of the scope here. In addition, that model did not include different compartments for the nucleus and the cytoplasm, which we were able to do thanks to the recent publication of data on clock-gene subcellular trafficking [43]. Further, we included the gene Clock to the model, to incorporate available data on this gene and investigate its importance in the clock machinery. Finally, our model integrates both mRNA and protein circadian datasets in a quantitative manner, meaning that it does not only predict the phase and relative amplitude of the gene expression data as existing models do [23], [26], but also computes the protein expression levels, such information being critical for the connection to PK-PD models. A very interesting perspective for future studies would be to consider coupling the model by Woller et al. [26] with ours to investigate the impact of feeding/fasting cycles on irinotecan chronopharmacology, as liver enzymes involved in the drug PK showed variations according to food intake [63].
Our core-clock model represents intracellular regulatory feedback loops that implicitly include extrinsic circadian regulators such as temperature or light/dark cycles. Such external synchronizers were not present in our cell culture setting, so that the SW480 and SW620 models are likely to represent the actual events at stake. On the opposite, external or systemic regulators have a great influence on the mouse liver clock. This precise question was the topic of another of our recent studies, in which we have explicitly modelled the influence of temperature cycles and food intake on the core clock in four classes of mice (2 strains, 2 sexes) [64].
Regarding CES2 modelling, we chose not to connect its protein degradation rate directly to the core clock, since there is no published data regarding the existence or absence of such molecular links. Thus, instead of including unreliable reactions to the model, we preferred to estimate the circadian rhythm of CES2 protein degradation directly from the data. The parameter sensitivity analysis evidenced the importance of this part of the model and strongly advocates the generation of additional biological results about the circadian control of CES2 protein degradation.
Our experimental results using a CRC in vitro cellular system highlight the existence of cytotoxicity differences resulting from time-dependent treatment, which were further emphasised by our simulations. Compared to the predicted toxicity acrophase of the SW480 cell line, the toxicity acrophase of the SW620 cell line is delayed by about four hours. In particular, the metastasis-derived cell line showed the lowest variations in cytotoxicity among different treatment times. Our data also shows a difference in terms of drug resistance between the two CRC cell lines, which, we hypothesize, could be overcome if using treatment times of high cytotoxicity with the same amount of drug, or potentially by increasing dosages in times of least toxicity. Yet, these are still speculative ideas, which need further studies and validation in a clinical setting. The small number of cell lines included in our experimental setup present some limitations to the generalization of our findings and thus further investigation of time-dependent treatment with a higher variety of cell lines and anticancer treatment agents needs to be carried out in future research.
4.2. Personalized models to optimize timing in cancer treatment
Chronotherapeutic studies aim at increasing treatment efficacy and minimizing toxicity for healthy cells leading to a reduction of the side effects for patients [65]. Previous clinical results have shown that personalization is a key element of successful chronotherapy outcome, for example males and females have shown different toxicities depending on treatment timing [6], [16], [66]. Sex should be considered as a relevant determinant of circadian rhythms and optimal drug timing in the light of recent preclinical and clinical findings [6], [16], [66]. Here, the mouse liver data was obtained from male mice, and the cell lines were derived from a human male, so that the sex specificity seems out of the scope of this study. However, we have started to investigate the impact of sex on the circadian timing system as mentioned above and did find significant sex-related differences in the shape and intensity of systemic controls on the core clock in a mouse study. Several sex-specific datasets related to irinotecan chronotoxicity are available in mice and in patients, and further modelling work would allow to investigate molecular determinants of male/female differences in irinotecan response with respect to timing [8].
One aspect of personalized chronotherapy is an adaptation of medication timing to the patient’s internal time, which can be best assessed by a combination of mathematical modelling and machine learning. Existing models of irinotecan PK-PD offer to predict best drug timing based on circadian rhythms of proteins involved in irinotecan pharmacology, in the organ of interest (e.g. liver, or intestine) or in the tumour. However, such datasets are unlikely to be obtained in the clinics on an individual patient basis as it would involve multiple around-the-clock biopsies, which obviously raises questions of feasibility and ethics regarding benefit/risk ratios. Furthermore, circadian datasets on irinotecan-related proteins would not be informative for personalizing the timing of other drugs, in particular the ones usually combined with irinotecan (e.g. 5-fluorouracil, oxaliplatin). Instead, our new combined model provides the option of computing irinotecan best timing from circadian rhythms of core-clock mRNA levels. The major advantage of measuring core-clock genes - and not directly drug-related genes - is that it can be done in any organ, since the peripheral core clock is synchronized across healthy tissues as suggested by mouse and baboon studies [11], [35], [67]. Several patient-friendly methods for measuring clock-gene expression in saliva or blood samples have been recently validated in the clinics (see [12] for a review). Furthermore, strong oscillations, clearly above noise level, are expected in core-clock gene expression, which facilitates the characterisation of their circadian profiles and reduces the number of needed time points to do so [62]. In addition, a newly available statistical algorithm offers to derive clock gene mRNA circadian rhythms from a single-time-point measurement of 10 clock genes [68]. Such methodology could potentially be used to predict clock gene variations when only one time point is available, which is often the case for the tumour. Our combined model could then be used to infer irinotecan personalized best timing from clock-gene expression. As such chronoPK-PD models could be developed for any other drug, optimal timing could be derived for multiple compounds from a single dataset of clock gene mRNA circadian variations. In addition, instead of simplifying a patient’s complex circadian profile by an estimate of a value associated with their circadian time, our model has the potential to fit the circadian rhythms of the patient based on their personal gene expression data from peripheral tissues (e.g. saliva [62]). Thus, in a clinical application, the model can be fitted both to the tumour clock and to the healthy peripheral clock of the patient. Several therapeutic strategies may then be considered from maximizing efficacy, or minimizing side effects, of a given drug dose, to more advanced approaches aiming to optimize antitumor efficacy under strict tolerability constraints [27]. To exemplify the power of our model for personalization, we fit the model to two different cell lines derived from human CRC with cell line-specific toxicity profiles, which are different in the metastasis-derived cells as compared to the primary tumour cells likely due to a disruption of the circadian profile and an alteration of metabolism in the former cells [31]. Overall, our approach provides a promising direction for mechanism-based chronotherapy personalization in the clinical setting.
The sensitivity analysis of the circadian toxicity profile is in accordance with the previously published sensitivity analysis by Dulong et al. 2015 [28], highlighting especially the importance of CES2, which is responsible for activation of irinotecan. Using our fitted mathematical model, changes in toxicity in response to relevant alterations in core-clock or protein dynamics can in principle be predicted based on circadian data for core-clock and drug-pharmacology genes. Further, the model can also be adapted for patients with alterations in irinotecan PK-PD proteins, such as patients with increased sensitivity against irinotecan due to a reduced UGT1A1 activity (reduced deactivation of SN-38) [69], or patients with a decreased sensitivity to irinotecan due to an overexpression of ABC transporters, which leads to a faster drug removal from the cell [70].
The influence of the core clock on the toxicity profile supports a dependence of optimal treatment times on the personal circadian rhythm of patients, in accordance with previous reports [8]. In particular, several core-clock parameters associated with BMAL1 and CLOCK (BMAL1 degradation rate, CLOCK activation rate, cytosolic BMAL1 degradation rate), show high sensitivity in our model, highlighting the relevance of the core clock for irinotecan PK-PD. This is particularly relevant for cancer patients, who often show alteration in their circadian rhythms that might be further changed during hospitalization, as bedridden patients seem to have disrupted circadian rhythms [71], [72], [73], [74], [75]. This suggests that even during a treatment frame of a few weeks, optimal treatment times might be shifted by a flattening of the circadian rhythms. Light therapy might help to stabilize toxicity profiles, as it has been shown to improve circadian oscillations in breast cancer patients [72]. Also melatonin administration or pharmacological modulation of core-clock genes may have a positive impact on cancer therapy [76], [77]. We here report differently timed toxicity peaks for CRC cell lines. Naively, one would assume that cancer cells show less robust oscillations compared to healthy cells, but this remains to be shown in future research. While the toxicity of Caco-2 cells in the model of Dulong et al. was predicted following a repeated 2-hour treatment [28], we here predict toxicity phase following a 84.5-h long treatment. The situation in the patient is most likely somewhere between these values as irinotecan terminal half-life after a 30-min infusion to colorectal cancer patients in the morning was approximately equal to 12 h [65]. As the treatment administration scheme may present complex and chronomodulated shapes, irinotecan whole-body pharmacokinetics must be precisely modelled in order to faithfully predict plasma and tissue exposure concentrations. A mathematical model relating infusion pump and administration schedules to predict actual drug concentrations in the body has been developed [78], and a corresponding extension could be used to further improve predictions of the here presented cellular model in a whole-body context. Thus, for personalized medical treatments the personalization of mathematical models is key, using easily accessible patient data to predict unassessable information relevant for medication.
5. Conclusion
Our clock-irinotecan model can be further optimized in a personalized manner and may be used to predict the toxicity profile of a particular patient upon fitting his or her molecular circadian profile. The model can be additionally used to investigate whether the differential regulation of PK-PD elements, for example via additional medication with melatonin, can result in circadian toxicity profiles that would support chronotherapy in irinotecan-treated cancers [76]. Altogether, our findings highlight the relevance of investigating the effect of chronomodulated therapy in a clinical setting as it may contribute to providing better personalized medical treatment with higher efficacy and lower cytotoxicity, leading to a decrease of side effects and an increase of life quality for the patient.
6. Code availability
The code is available at biomodels (identifier: MODEL2109140001).
7. Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT Author Statement
Janina Hesse: Formal analysis, Methodology, Investigation, Visualization, Writing - original draft, Writing - review & editing. Julien Martinelli: Formal analysis, Methodology, Investigation, Visualization, Writing - original draft, Writing - review & editing. Ouda Aboumanify: Methodology, Investigation, Visualization, Writing - review & editing. Annabelle Ballesta: Conceptualization of computational part, Funding acquisition, Investigation, Supervision, Writing - original draft, Writing - review & editing. Angela Relógio: Conceptualization of computational and experimental parts, Funding acquisition, Investigation, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The work in A.R.'s group was supported by the German Federal Ministry of Education and Research (BMBF)—eBio-CIRSPLICE – [FKZ031A316] and by the Dr. Rolf M. Schwiete Stiftung. O.A. was additionally funded by the I4H program of the Charité - Universitätsmedizin Berlin. We acknowledge support from the German Research Foundation (DFG) and the Open Access Publication Fund of the Charité - Universitätsmedizin Berlin. The work in the group of A.B. was supported by INSERM, Inria, the ATIP-Avenir program and the French Plan Cancer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.051.
Contributor Information
Annabelle Ballesta, Email: annabelle.ballesta@inserm.fr.
Angela Relógio, Email: angela.relogio@charite.de, angela.relogio@medicalschool-hamburg.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ko, C.H., Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum Mol Genet; 2006. 15 Spec No 2(suppl_2): p. R271-7. [DOI] [PubMed]
- 2.Matsuo T. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 3.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheiermann C., Kunisaki Y., Frenette P.S. Circadian control of the immune system. Nat Rev Immunol. 2013;13(3):190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D.W., Zavala E., Kim J.K. Wearable technology and systems modeling for personalized chronotherapy. Current Opin Syst Biol. 2020;21:9–15. [Google Scholar]
- 6.Anderson S.T., FitzGerald G.A. Sexual dimorphism in body clocks. Science. 2020;369(6508):1164–1165. doi: 10.1126/science.abd4964. [DOI] [PubMed] [Google Scholar]
- 7.El-Athman R., Relogio A. Escaping Circadian Regulation: An Emerging Hallmark of Cancer? Cell Syst. 2018;6(3):266–267. doi: 10.1016/j.cels.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Ballesta A. Systems Chronotherapeutics. Pharmacol Rev. 2017;69(2):161–199. doi: 10.1124/pr.116.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bollinger T., Schibler U. Circadian rhythms - from genes to physiology and disease. Swiss Med Wkly. 2014;144(2930) doi: 10.4414/smw.2014.13984. [DOI] [PubMed] [Google Scholar]
- 10.Bicker J. Timing in drug absorption and disposition: The past, present, and future of chronopharmacokinetics. Br J Pharmacol. 2020;177(10):2215–2239. doi: 10.1111/bph.15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaspar L.S. The importance of determining circadian parameters in pharmacological studies. Br J Pharmacol. 2019;176(16):2827–2847. doi: 10.1111/bph.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Innominato P.F. Efficacy and safety of chronomodulated irinotecan, oxaliplatin, 5-fluorouracil and leucovorin combination as first- or second-line treatment against metastatic colorectal cancer: Results from the International EORTC 05011 Trial. Int J Cancer. 2020;n/a(n/a) doi: 10.1002/ijc.33422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levi F. Cetuximab and circadian chronomodulated chemotherapy as salvage treatment for metastatic colorectal cancer (mCRC): safety, efficacy and improved secondary surgical resectability. Cancer Chemother Pharmacol. 2011;67(2):339–348. doi: 10.1007/s00280-010-1327-8. [DOI] [PubMed] [Google Scholar]
- 15.Giacchetti S. Sex moderates circadian chemotherapy effects on survival of patients with metastatic colorectal cancer: a meta-analysis. Ann Oncol. 2012;23(12):3110–3116. doi: 10.1093/annonc/mds148. [DOI] [PubMed] [Google Scholar]
- 16.Innominato P.F. Sex-dependent least toxic timing of irinotecan combined with chronomodulated chemotherapy for metastatic colorectal cancer: Randomized multicenter EORTC 05011 trial. Cancer Med. 2020;9(12):4148–4159. doi: 10.1002/cam4.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relogio A. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hesse J. An Optimal Time for Treatment-Predicting Circadian Time by Machine Learning and Mathematical Modelling. Cancers (Basel) 2020;12(11):3103. doi: 10.3390/cancers12113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leloup J.C., Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc Natl Acad Sci U S A. 2003;100(12):7051–7056. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forger D.B., Peskin C.S. A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci U S A. 2003;100(25):14806–14811. doi: 10.1073/pnas.2036281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker-Weimann S. Modeling feedback loops of the Mammalian circadian oscillator. Biophys J. 2004;87(5):3023–3034. doi: 10.1529/biophysj.104.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirsky H.P. A model of the cell-autonomous mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106(27):11107–11112. doi: 10.1073/pnas.0904837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relogio A. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput Biol. 2011;7(12) doi: 10.1371/journal.pcbi.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J.K., Forger D.B. A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol. 2012;8:630. doi: 10.1038/msb.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furlan A. Mathematical models converge on PGC1α as the key metabolic integrator of SIRT1 and AMPK regulation of the circadian clock. Proc Natl Acad Sci U S A. 2019;116(27):13171–13172. doi: 10.1073/pnas.1907751116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woller A. A Mathematical Model of the Liver Circadian Clock Linking Feeding and Fasting Cycles to Clock Function. Cell Rep. 2016;17(4):1087–1097. doi: 10.1016/j.celrep.2016.09.060. [DOI] [PubMed] [Google Scholar]
- 27.Ballesta A. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput Biol. 2011;7(9) doi: 10.1371/journal.pcbi.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dulong S. Identification of Circadian Determinants of Cancer Chronotherapy through In Vitro Chronopharmacology and Mathematical Modeling. Mol Cancer Ther. 2015;14(9):2154–2164. doi: 10.1158/1535-7163.MCT-15-0129. [DOI] [PubMed] [Google Scholar]
- 29.Mathijssen R.H. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7(8):2182–2194. [PubMed] [Google Scholar]
- 30.de Man F.M. Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics. Clin Pharmacokinet. 2018;57(10):1229–1254. doi: 10.1007/s40262-018-0644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuhr L. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. EBioMedicine. 2018;33:105–121. doi: 10.1016/j.ebiom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narumi R. Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins. Proc Natl Acad Sci. 2016;113(24):E3461–E3467. doi: 10.1073/pnas.1603799113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.El-Athman R., Fuhr L., Relogio A. A Systems-Level Analysis Reveals Circadian Regulation of Splicing in Colorectal Cancer. EBioMedicine. 2018;33:68–81. doi: 10.1016/j.ebiom.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Athman R. A Computational Analysis of Alternative Splicing across Mammalian Tissues Reveals Circadian and Ultradian Rhythms in Splicing Events. Int J Mol Sci. 2019;20(16) doi: 10.3390/ijms20163977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen N., Ostermeier A. Completely derandomized self-adaptation in evolution strategies. Evol Comput. 2001;9(2):159–195. doi: 10.1162/106365601750190398. [DOI] [PubMed] [Google Scholar]
- 37.Luck S. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Reppert S.M., Weaver D.R. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 39.Yang F. The molecular mechanism regulating the autonomous circadian expression of Topoisomerase I in NIH3T3 cells. Biochem Biophys Res Commun. 2009;380(1):22–27. doi: 10.1016/j.bbrc.2008.12.186. [DOI] [PubMed] [Google Scholar]
- 40.Oishi K., Shirai H., Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J. 2005;386(Pt 3):575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preitner N. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X. RAE1 promotes BMAL1 shuttling and regulates degradation and activity of CLOCK: BMAL1 heterodimer. Cell Death Dis. 2019;10(2):62. doi: 10.1038/s41419-019-1346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J. Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab. 2017;25(1):102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters R. Nucleo-cytoplasmic flux and intracellular mobility in single hepatocytes measured by fluorescence microphotolysis. EMBO J. 1984;3(8):1831–1836. doi: 10.1002/j.1460-2075.1984.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimba S. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aryal R.P. Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell. 2017;67(5):770–782 e6. doi: 10.1016/j.molcel.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwanhausser B. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 48.Abdulrehman G., Xv K., Li Y., Kang L. Effects of meta-tetrahydroxyphenylchlorin photodynamic therapy on isogenic colorectal cancer SW480 and SW620 cells with different metastatic potentials. Lasers Med Sci. 2018;33(7):1581–1590. doi: 10.1007/s10103-018-2524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu G. Human carboxylesterase 2 is commonly expressed in tumor tissue and is correlated with activation of irinotecan. Clin Cancer Res. 2002;8(8):2605–2611. [PubMed] [Google Scholar]
- 50.Mathijssen R.H. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9(9):3246–3253. [PubMed] [Google Scholar]
- 51.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 52.Smith N.F., Figg W.D., Sparreboom A. Pharmacogenetics of irinotecan metabolism and transport: an update. Toxicol In Vitro. 2006;20(2):163–175. doi: 10.1016/j.tiv.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 53.Ciotti M. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38) by the human UDP-glucuronosyltransferases encoded at the UGT1 locus. Biochem Biophys Res Commun. 1999;260(1):199–202. doi: 10.1006/bbrc.1999.0453. [DOI] [PubMed] [Google Scholar]
- 54.Ueda H.R. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 55.Gascoyne D.M. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10(10):1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 56.Onishi Y., Kawano Y. Rhythmic binding of Topoisomerase I impacts on the transcription of Bmal1 and circadian period. Nucleic Acids Res. 2012;40(19):9482–9492. doi: 10.1093/nar/gks779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao M. E4bp4 regulates carboxylesterase 2 enzymes through repression of the nuclear receptor Rev-erbalpha in mice. Biochem Pharmacol. 2018;152:293–301. doi: 10.1016/j.bcp.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Ripperger J.A., Jud C., Albrecht U. The daily rhythm of mice. FEBS Lett. 2011;585(10):1384–1392. doi: 10.1016/j.febslet.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 59.Sancar G., Brunner M. Circadian clocks and energy metabolism. Cell Mol Life Sci. 2014;71(14):2667–2680. doi: 10.1007/s00018-014-1574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sancar A. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584(12):2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaucher J., Montellier E., Sassone-Corsi P. Molecular Cogs: Interplay between Circadian Clock and Cell Cycle. Trends Cell Biol. 2018;28(5):368–379. doi: 10.1016/j.tcb.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Basti A., Yalçin M., Herms D., Hesse J., Aboumanify O., Li Y. Diurnal variations in the expression of core-clock genes correlate with resting muscle properties and predict fluctuations in exercise performance across the day. BMJ open sport & exercise medicine. 2021;7(1):e000876. doi: 10.1136/bmjsem-2020-000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okyar A., Kumar S.A., Filipski E., Piccolo E., Ozturk N., Xandri-Monje H. Sex-, feeding-, and circadian time-dependency of P-glycoprotein expression and activity - implications for mechanistic pharmacokinetics modeling. Scientific reports. 2019;9(1):10505. doi: 10.1038/s41598-019-46977-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinelli J. Model learning to identify systemic regulators of the peripheral circadian clock. Bioinformatics. 2021;37(Suppl_1):i401–i409. doi: 10.1093/bioinformatics/btab297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levi F. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 66.Li X.M. A circadian clock transcription model for the personalization of cancer chronotherapy. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1528. [DOI] [PubMed] [Google Scholar]
- 67.Mure L.S., Le H.D., Benegiamo G., Chang M.W., Rios L., Jillani N. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science (New York, N.Y.) 2018;359(6381):eaao0318. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlachou D. TimeTeller: a New Tool for Precision Circadian Medicine and Cancer Prognosis. bioRxiv. 2019 [Google Scholar]
- 69.Fujii H. Dose adjustment of irinotecan based on UGT1A1 polymorphisms in patients with colorectal cancer. Cancer Chemother Pharmacol. 2019;83(1):123–129. doi: 10.1007/s00280-018-3711-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fletcher J.I. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist Updat. 2016;26:1–9. doi: 10.1016/j.drup.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 71.Rida P., Syed M.I., Aneja R. Time will tell: Circadian clock dysregulation in triple negative breast cancer. Front Biosci (Schol Ed) 2019;11:178–192. doi: 10.2741/S533. [DOI] [PubMed] [Google Scholar]
- 72.Neikrug A.B. Bright light therapy protects women from circadian rhythm desynchronization during chemotherapy for breast cancer. Behav Sleep Med. 2012;10(3):202–216. doi: 10.1080/15402002.2011.634940. [DOI] [PubMed] [Google Scholar]
- 73.Ikegami K. Interconnection between circadian clocks and thyroid function. Nat Rev Endocrinol. 2019;15(10):590–600. doi: 10.1038/s41574-019-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roenneberg T., Merrow M. The Circadian Clock and Human Health. Curr Biol. 2016;26(10):R432–R443. doi: 10.1016/j.cub.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 75.Innominato P.F. The circadian timing system in clinical oncology. Ann Med. 2014;46(4):191–207. doi: 10.3109/07853890.2014.916990. [DOI] [PubMed] [Google Scholar]
- 76.Gil-Martin E. The emergence of melatonin in oncology: Focus on colorectal cancer. Med Res Rev. 2019;39(6):2239–2285. doi: 10.1002/med.21582. [DOI] [PubMed] [Google Scholar]
- 77.Sulli G., Lam M.T.Y., Panda S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer. 2019;5(8):475–494. doi: 10.1016/j.trecan.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hill R.J.W. Optimizing circadian drug infusion schedules towards personalized cancer chronotherapy. PLoS Comput Biol. 2020;16(1):e1007218. doi: 10.1371/journal.pcbi.1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.