The dominant focus of precision cancer medicine has been on predictive markers, linking specific therapeutic agents to biomarkers in order to select subsets of patients who are more likely to benefit from treatment, or to identify patients who would be resistant. Examples include Mismatch repair status, linked to use of immune checkpoint inhibitors; fusion proteins (BCR-ABL, NTRK, ROS-1, ALK and c-KIT), linked to a range of tyrosine kinase inhibitors; oestrogen, progesterone and androgen receptor assays, linked to selection of hormonal treatments; and HER2-neu and EGFR expression, used for a variety of antibody receptor inhibitors [1,2].
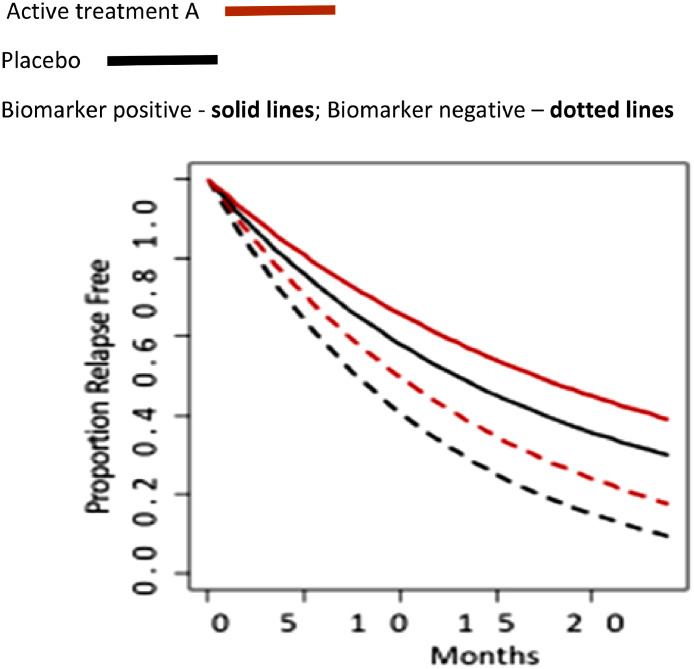
A prognostic biomarker is a clinical or biological characteristic that provides information on the likely patient health outcome (e.g. disease recurrence, progression free and overall survival), irrespective of the treatment. It is measured before treatment and identifies tumour-specific molecular or histopathological characteristics that are associated with long-term outcome or disease course, and is not treatment dependent (Figure 1a).
Figure 1.
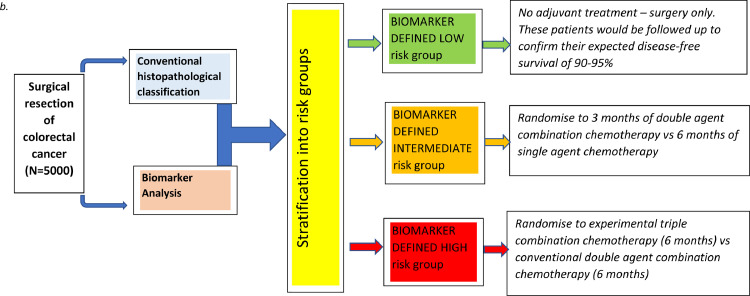
(a) Schematic representation of a cancer biomarker which is prognostic but not predictive. Tx, cohort received treatment; M+, biomarker positive; no Tx, cohort is treatment naïve; M-, biomarker negative. (b) Flow diagram summary of a potential trial structure.
Despite an enormous scientific literature, much less attention has been paid to the clinical application of prognostic markers. Despite hundreds of publications, relatively few prognostic markers find their way into routine clinical use. The majority of reported prognostic biomarker studies are underpowered, are not supported by analytical test systems with well-established performance characteristics and often fail to address the question of clinical applicability. Biomarker discovery has embraced many different technologies and can include estimates of protein expression or phosphorylation, somatic or germline mutations, changes in DNA methylation, multiplexed RNA signatures, micro-RNA levels, or circulating DNA or tumour cells in blood. The introduction of novel techniques has associated problems with reproducibility, validation, generalisability (central versus local laboratory delivery) and of course variation in tissue processing. Several studies have demonstrated the importance of data variability in relation to tissue processing techniques during and post-surgery with particular emphasis on tissue handling and time delays. Even well-established methods like immunohistochemistry are open to observer interpretation as evidenced by recent studies quantifying PD-L1 positive cells in lung cancer samples which showed low levels of interobserver concordance [3].
For a cancer biomarker to be clinically useful, we believe that it should fulfil the following characteristics – address a specific tumour stage; be clinically actionable and reliably estimate effect. The latter point can be described statistically in that well powered studies (n> 1,000) are more compelling, reducing the number of false positive associations, narrowing confidence intervals and providing greater clinical utility. At the risk of being somewhat arbitrary, one might argue that Hazard Ratios (HR) should be set at 2 (or greater) meaning that at any particular time, twice as many patients in the biomarker positive group are experiencing an event compared to the control group. The confidence Interval (CI) is the range of values that is likely to include the true population value and is used to measure the precision of the study's estimate (in this case, the precision of the HR). The narrower the confidence interval, the more precise the estimate. Precision is affected by the study's sample size.
There are very few prospective randomised trials of prognostic biomarkers, but this does not mean that they cannot guide clinical decision making. In our recently published study in EBiomedicine [4], we described how quantification of intratumoural CD8+ T-lymphocyte and stroma fractions can be combined with conventional prognostic markers to significantly improve patient stratification in early stage, resected colorectal cancer. These data, generated from a population of 1,500 patients could identify patients at very low risk of tumour recurrence at 5 years (TTR) of 86%. For conventionally staged high-risk patients, using TNM criteria, the biomarker defined by low CD8+ / high stroma fraction identified a very poor prognostic subgroup with 5-year TTR of 29%, whereas the high CD8+ / low stroma fraction subgroup had a TTR of 64% (HR =2.86, 95% CI 1.75-4.69; P < 0.001). These statistical parameters meet the utility criteria we described earlier, namely a large patient population, HR >2 and narrow confidence intervals.
Assuming, reasonably, that the proportional benefits of chemotherapy are the same across all these prognostic groups [5,6,7], the absolute benefits will be driven by prognostic stratification. That is to say, those with a worse clinical outlook will enjoy a greater absolute increase in survival. We have demonstrated that the proportional reduction in the odds of dying from recurrent colorectal cancer following the addition of chemotherapy to surgery is around 20%. If the 5-year survival rate was 90% with surgery alone, then the addition of chemotherapy would increase this to 92%. Considering the potential toxic death rate associated with chemotherapy of between 0.5-1%, the case for chemotherapy in addition to surgery is extremely weak. Conversely, for the biomarker-defined high-risk subgroup with a 5-year survival rate of 29%, the absolute benefits of chemotherapy would be an additional 14% of patients cured by adjuvant chemotherapy, tipping the therapeutic balance firmly in favour of recommending treatment.
The majority of clinicians, we believe, could use these data to inform clinical decisions about the type, intensity and duration of post-operative adjuvant chemotherapy. This would mean, no adjuvant therapy for those at very low risk of recurrence and dose intense, prolonged chemotherapy for patients in the especially high-risk group.
Of course, it is possible to design prospective trials to further verify the prognostic power of validated biomarkers. The biomarker would be applied to cancer tissue resected at the time of surgery, used to stratify patients into cohorts of low, intermediate and high risk of tumour recurrence and thereafter randomisation of most patients to a choice of chemotherapeutic regimens or observation would be performed to observe whether we can minimise toxicity and cost to those patients who are already at relatively low risk of relapse, whilst improving efficacy in those individuals at high risk of relapse. A potential trial structure is summarised in figure 1b.
Whilst attractive, the problem with this sort of trial is that we estimate that it would require recruitment of approximately 5,000 patients and take around 6-7 years to complete, a massive and expensive undertaking when considering that retrospective analysis of well curated biospecimens linked to high quality clinical outcome data can provide sufficiently robust data to support clinical decisions.
Contributors
All authors contributed to preparation and review of text.
Declaration of Competing Interest
Prof Li Yang – none
Prof David Kerr – Director, Oxford Cancer Biomarkers; Director, Celleron Therapeutics; Consultant, Indivumed; Consultant, Novartis; Consultant, Medscape
References
- 1.La Thangue NB, Kerr DJ. Predictive biomarkers: a shift towards personalised cancer medicine. Nature Reviews Clinical Oncology. 2011;8:587–596. doi: 10.1038/nrclinonc.2011.121. [DOI] [PubMed] [Google Scholar]
- 2.David KA, Unger FT, Uhlig P, Juhl H, Moore HM, Compton C. Surgical procedures and postsurgical tissue processing significantly affect expression of genes and EGFR-pathway proteins in colorectal cancer tissue. Oncotarget. 2014;5(22):11017–11028. doi: 10.18632/oncotarget.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheel AH, Dietel M, Heukamp LC, Johrens K, Kirchner T, Reu S. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol. 2016;29(10):1165–1172. doi: 10.1038/modpathol.2016.117. [DOI] [PubMed] [Google Scholar]
- 4.Jiang D, Hveem TS, Glaire M, Church DN, Danielsen HE, Li Y. Automated assessment of CD8+ T-lymphocytes and stroma fractions complement conventional staging of colorectal cancer. EBioMedicine. 2021 doi: 10.1016/j.ebiom.2021.103547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray R, Quirke P, Hardley K, Lopatin M, Magill L, Baehner FL. Validation Study of a Quantitative multi-gene RT-PCR assay as a predictor of recurrence in stage II colon cancer patient. J Clin Oncol. 2011;12:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 6.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr DJ. Genetic prognostic and predictive markers in colorectal cancer. Nature Reviews Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 7.Gray R, Barnwell J, McConkey C, Williams N, Kerr DJ. QUASAR: a randomised study of adjuvant chemotherapy versus observation including 3239 colorectal cancer patients QUASAR Collaborative Group. Lancet. 2007;370(9604):2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]