Abstract
NUAK family kinase 2 (NUAK2) has been reported to be involved in various cancer cell processes, including proliferation, apoptosis and invasion, by targeting multiple genes. However, to the best of our knowledge, its biological function in cervical cancer (CC) has not yet been elucidated. Therefore, the present study aimed to measure the expression of NUAK2 and to evaluate its functions in CC. The expression levels of NUAK2 and cytoplasmic FMRP-interacting protein 2 (CYFIP2) were detected in CC tissues and cell lines. In addition, the effects of NUAK2 and CYFIP2 knockdown on CC cell proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) were evaluated in vitro using Cell Counting Kit-8, immunofluorescence, wound healing assay, Transwell assay and western blotting, respectively. Furthermore, co-immunoprecipitation was performed to determine the interaction between NUAK2 and CYFIP2. The results revealed that the expression levels of NUAK2 were upregulated in CC tissues and cells, whereas CYFIP2 expression was reduced. In addition, knockdown of NUAK2 reduced cell proliferation, migration, invasion and EMT. Notably, NUAK2 was found to bind directly to CYFIP2. Furthermore, CYFIP2 inhibition reversed the effects of NUAK2 on CC cells. In summary, NUAK2 may regulate CYFIP2 expression to promote CC cell proliferation, migration, invasion and EMT.
Keywords: NUAK family kinase 2, cytoplasmic FMRP-interacting protein 2, cervical cancer, proliferation, migration, epithelial-to-mesenchymal transition
Introduction
Cervical cancer (CC) is the fourth most common type of cancer in women worldwide and the leading cause of death, with ~530,000 new cases and 275,000 deaths occurring each year (1,2). Although CC treatments and screening methods continue to evolve, with early diagnosis and treatment reducing its mortality rate, the 5-year cancer-specific survival rate remains poor (3). Therefore, it is of great importance to recognize the lack of effective therapies and to identify novel methods for improving the accuracy of diagnosis and the prognosis of patients with CC. Discovering effective cancer treatments is highly challenging due to tumor invasion and metastasis (4). Therefore, investigation of biomarkers that can influence the pathogenesis and progression of cancer is of great interest for the diagnosis and treatment of CC.
NUAK family kinase 2 (NUAK2) is a member of the AMP-activated protein kinase family, which is located at 1q32 and can be suppressed by the tumor suppressor hepatic kinase B1, as well as by NF-κB, which inhibits death receptor signaling activation (5–7). NUAK2 has been reported to serve a key role in cancer development and tumor progression. It has been reported that NUAK2 is upregulated in gastric cancer tissues, and that it promotes the proliferation of gastric cancer cells and regulates the cell cycle, resulting in upregulation of proliferation and cancer stem cell marker expression (8). A previous study revealed that NUAK2 expression is increased in glioma tissues and is associated with advanced disease stage, whereas in vitro experiments demonstrated that overexpression of NUAK2 promoted the proliferation, migration and invasion of A172 glioblastoma cells (9).
The cytoplasmic FMR1-interacting protein (CYFIP) gene family has two highly conserved members, CYFIP1 and CYFIP2, which are ~145-kDa proteins with high homology (88% identity and 95% similarity) in their amino acid sequences (10). It has been reported that CYFIP2 mRNA expression is significantly reduced in gastric cancer tissues compared with that in non-cancerous tissues, and knockdown of CYFIP2 has been shown to promote cell proliferation and colony formation, and inhibit apoptosis (11). Additionally, overexpression of CYFIP2 has been demonstrated to promote apoptosis-like death of colorectal cancer cells (12).
The present study was undertaken to investigate the biological functions of NUAK2 in CC. The expression of NUAK2 in CC tissues and cells was examined, and further experiments were conducted to determine whether interference with NUAK2 expression could suppress the proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) of CC cells. The role of CYFIP2 in this process was also investigated.
Materials and methods
Bioinformatics analysis
The interaction between NUAK2 and CYFIP2 was predicted using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (version 11.0, http://string-db.org/) (13). The UALCAN cancer database (http://ualcan.path.uab.edu) was used to retrieve data regarding the expression levels of NUAK2 and CYFIP2 in the primary tumor tissues (n=305) of patients with cervical squamous cell carcinoma (CESC) and normal tissues (n=3) from healthy controls (14); these databases were from The Cancer Genome Atlas.
Tissues and cell lines
CC tissues and pair-matched non-cancerous cervical tissues (2 cm from the lesions) were obtained from 36 patients (age, 30–65 years) diagnosed with CC at Wuhan Children's Hospital (Wuhan, China) from April 2018 to September 2019. Patients were excluded from the study if they received chemotherapy or radiotherapy prior to surgery. Written informed consent was obtained from all patients.
The HeLa cervical adenocarcinoma cell line, C-33A, SiHa, CaSKi and HCC-94 cervical squamous cell carcinoma cell lines, and Ect1 human normal cervical cell line were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine calf serum, 100 µg/ml penicillin and 100 µg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated at 37°C in an incubator containing 5% CO2. The medium was changed every 2 days and cells were grown to logarithmic phase for use in subsequent experiments.
Cell transfection
Two small interfering RNA (siRNA) sequences against CYFIP2 (si-CYFIP2-1/si-CYFIP2-2) and a nontargeting siRNA used as a negative control (si-NC) were designed by Shanghai GenePharma Co., Ltd. Targeting and nontargeting sequences were as follows: si-CYFIP2-1, 5′-AGGCTAACTTTGACACAAACT-3′; si-CYFIP2-2, 5′-GGCTAACTTTGACACAAACTT-3′; si-NC, 5′-GCGTTCTTAACTTTGAACC-3′. siRNAs were transfected into cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, HeLa cells were seeded at a density of 2×105 cells/ml in 24-well plates and cultured for 24 h, after which they were transfected with siRNAs targeting CYFIP2 and si-NC; the knockdown efficiency was verified 48 h after transfection by western blotting and reverse transcription-quantitative PCR (RT-qPCR). All the siRNAs were used at a final concentration of 5 nM. In addition, two NUAK2-specific short hairpin RNAs (shRNAs) (shRNA-NUAK2-1/shRNA-NUAK2-2) and a scrambled shRNA used as a negative control (shRNA-NC) were obtained from Shanghai GenePharma Co., Ltd. The sequences were as follows: shRNA-NUAK2-1, sense 5′-CCATAAGATCCTAGTGAAA-3′, antisense 5′-TTTCACTAGGATCTTATGG-3′; shRNA-NUAK2-2, sense 5′-GCATGACCATAAGATCCTA-3′, antisense 5′-TAGGATCTTATGGTCATGC-3′; shRNA-NC, sense 5′-GATCCCCTTCTCCGAACG-3′, antisense 5′-AGCTAAAAATTCTCCGAAC-3′. The corresponding shRNA oligonucleotides (synthesized by Guangzhou RiboBio Co., Ltd.) were cloned into the pLentiLox 3.7 lentiviral plasmid [American Type Culture Collection (ATCC)]. Lipofectamine 3000 was used to transfect 293T cells (ATCC) at a ratio of 3 µg lentiviral construct and 6 µg package mix [pLP1 (3): pLP2 (2): pLP/VSVG (3); (Invitrogen; Thermo Fisher Scientific, Inc.)] cultured at 37°C, according to the manufacturer's recommendations. The supernatants were collected at 48 and 72 h then mixed for ultracentrifugation (4°C, 72,000 × g, 2 h) to obtain lentiviral particles. The lentivirus (multiplicity of infection, 10) and HeLa cells were incubated in DMEM supplemented with 10% fetal bovine serum containing 8 µg/ml polybrene (MineBio Life Sciences Ltd.) at 37°C for 12 h and then washed with PBS. The medium was then replaced with fresh medium and cells were cultured for a total of 2 days prior to subsequent experiments.
RT-qPCR analysis
The mRNA expression levels of NUAK2 and CYFIP2 were determined via RT-qPCR. Total RNA was extracted from tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the PrimeScript™ RT kit (Takara Bio, Inc.) under the following conditions: 15 min at 37°C and 5 sec at 85°C. The synthesized cDNA was used as a template for PCR amplification. The reactions were performed using SYBR Green Taq Mix (Takara Bio, Inc.) in a Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) under the following conditions: 45°C for 3 min, 95°C for 10 sec, 40 cycles at 95°C for 15 sec then 58°C for 1 min. Relative mRNA expression levels were quantified using the 2−ΔΔCq method (15) and normalized to GAPDH. All RT-qPCR experiments were performed in triplicate. The primer sequences used were as follows: NUAK2, forward, 5′-TGAGAAACGACGGAGACAAGCTGCT-3′ and reverse, 5′-GTCTGGAGGTTTTGCTGCAGGTCTG-3′; CYFIP2 forward, 5′-TGGCGTCATCATTCCGTATCC-3′ and reverse, 5′-GTCAGGTCCTCACTCAAGC-3′; and GAPDH forward, 5′-TGTGTCCGTCGTGGATCTGA-3′ and reverse, 5′-CCTGCTTCACCACCTTCTTGA-3′.
Western blotting
The protein expression levels of NUAK2, CYFIP2, E-cadherin, N-cadherin, Snail and ZEB1 were determined via western blotting. Total protein was extracted from harvested cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). The protein concentration was measured using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). A total of 30 µg protein per lane was separated via SDS-PAGE on 15% gels and separated proteins were subsequently transferred to polyvinylidene fluoride membranes (EMD Millipore). Membranes were then blocked with 10% non-fat milk for 2 h at room temperature, and incubated with the following primary antibodies overnight at 4°C: Anti-NUAK2 (1:500; cat. no. ab107287; Abcam), anti-CYFIP2 (1:500; cat. no. sc-134308; Santa Cruz Biotechnology, Inc.), anti-E-cadherin (1:500; cat. no. sc-8426; Santa Cruz Biotechnology, Inc.), anti-N-cadherin (1:500; cat. no. sc-8424; Santa Cruz Biotechnology, Inc.), anti-Snail (1:500; cat. no. sc-271977; Santa Cruz Biotechnology, Inc.), anti-ZEB1 (1:500; cat. no. sc-10572; Santa Cruz Biotechnology, Inc.), anti-GAPDH (1:2,000; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.) and anti-β-actin (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.). Subsequently, the membranes were washed with TBS-0.1% Tween-20 (TBST) for 5 min at room temperature. After three washes, the membranes were incubated at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.), HRP-conjugated goat anti-mouse IgG (1:2,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) or horseradish peroxidase (HRP)-conjugated mouse anti-goat IgG (1:2,000; cat. no. sc-2354; Santa Cruz Biotechnology, Inc.) for 2 h. After washing with TBST three times (10 min/wash), the protein bands were visualized using Western Blotting Luminescent Reagent (Santa Cruz Biotechnology, Inc.) and analyzed using ImageJ software (version 1.43; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8) assay (cat. no. C0037; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. Briefly, HeLa cells were seeded at a quantity of 1×104 cells/well in 96-well plates and cultured for 6 h, after which they were incubated with 15 µl/well CCK-8 solution at 37°C for 24, 48 or 72 h. Subsequently, the optical density (OD) was calculated at a wavelength of 450 nm to determine cell proliferation.
Immunofluorescence (IF) staining assay
To observe the expression of Ki67, IF staining was performed. HeLa cells were seeded onto glass coverslips in 6-well plates. Cells were incubated with lentiviruses containing shRNA-NUAK2 at 37°C for 2 days to silence NUAK2 gene expression. The cells were then incubated with si-CYFIP2 for an additional 36 h to obtain cells with low expression of NUAK2 and CYFIP2. After that, the cells were fixed with 4% paraformaldehyde (PFA; Millipore Sigma) at room temperature for 10 min, washed in PBS and blocked for 15 min in QuickBlock™ Blocking Buffer for Immunol Staining (Beyotime Biotechnology) at room temperature. After incubation with a primary antibody against anti-Ki67 (1:200; cat. no. sc-23900; Santa Cruz Biotechnology, Inc.) at 4°C overnight, cells were washed in PBS and incubated with a secondary antibody conjugated with Alexa Fluor 488 (1:1,000; cat. no. A32723; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Cells were then washed in PBS, incubated for 5 min with DAPI (1 µg/ml; cat. no. D1306; Invitrogen; Thermo Fisher Scientific, Inc.), and observations were performed using a fluorescence microscope (magnification, ×100; Olympus Corporation) and were analyzed with ImageJ version 1.43 software.
Wound healing assay
HeLa cells were seeded at 6×104 cells/well in 6-well plates, cultured until they reached 80–90% confluence and were subjected to serum starvation for 4 h. Subsequently, a sterile 10-µl pipette tip was used to create a linear scratch in the cell monolayer, the cells were washed with PBS to remove debris, and the medium was replaced with serum-free DMEM/F12 1:1. Images were captured under a light microscope (Olympus Corporation) prior to and at 24 h after incubation.
Transwell invasion assay
HeLa cells (2×104 cells/well) were cultured in the upper chamber of Transwell plates (8-µm pore size; Corning, Inc.). The surface of the upper chamber was pre-coated with Matrigel™ (BD Biosciences) at 37°C for 1 h. Complete medium (500 µl) supplemented with 10% bovine calf serum (Gibco; Thermo Fisher Scientific, Inc.) was added into the lower chamber. After 24-h incubation at 37°C, cells remaining on the upper membrane were wiped away, whereas cells that had invaded across the membrane were fixed with 4% PFA for 10 min at room temperature, stained with 0.1% crystal violet for 15 min at room temperature and counted under a light microscope.
Co-immunoprecipitation (Co-IP)
The Pierce Co-IP Kit (cat. no. 26149; Thermo Fisher Scientific, Inc.) was used to explore the interaction between proteins, according to the manufacturer's protocol. Briefly, the HeLa cells cultured in a 10-cm plate were washed twice with pre-cooled PBS, and the PBS was discarded. Subsequently, 1 ml lysis buffer was used to fully lyse the cells, the lysate was transferred into 1.5-ml microcentrifuge tubes, centrifuged for 15 min (4°C, 14,000 × g), and the supernatant protein was taken for quantification. The total volume was made up to 500 µl with lysis buffer after 500 µg protein was taken. Subsequently, 30 µl Agarose A + G was used to pre-clear lysate at 4°C for 2 h. The pre-cleared lysate was centrifuged at 4°C for 5 min at 1,000 × g and the supernatant was removed and incubated overnight with shaking at 4°C by adding the corresponding antibody. The following antibodies were used: NUAK2 (1:10; cat. no. sc-374348; Santa Cruz Biotechnology, Inc.), CYFIP2 (1:50; cat. no. sc-134308; Santa Cruz Biotechnology, Inc.) and IgG (1:50; cat. no. sc-69786; Santa Cruz Biotechnology, Inc.), which was used as a control. After 12 h of antibody conjugation, Agarose A + G (30 µl/tube) was added and shaken at 4°C for 4 h. The supernatant was discarded after centrifugation at 1,500 × g for 5 min to obtain the sediment. The precipitate was then washed three times with pre-chilled PBS to obtain the protein sample for western blotting.
Statistical analysis
All experimental data are presented as the mean ± standard deviation of at least three independent experiments. Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software, Inc.). In order to assess whether data obtained from the tissue samples followed a normal distribution, a Shapiro-Wilk test was used. Based on this information, data were further analyzed using parametric tests. To compare differences among multiple groups, one-way ANOVA followed by Tukey's post hoc test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of NUAK2 in CC tissues and cells
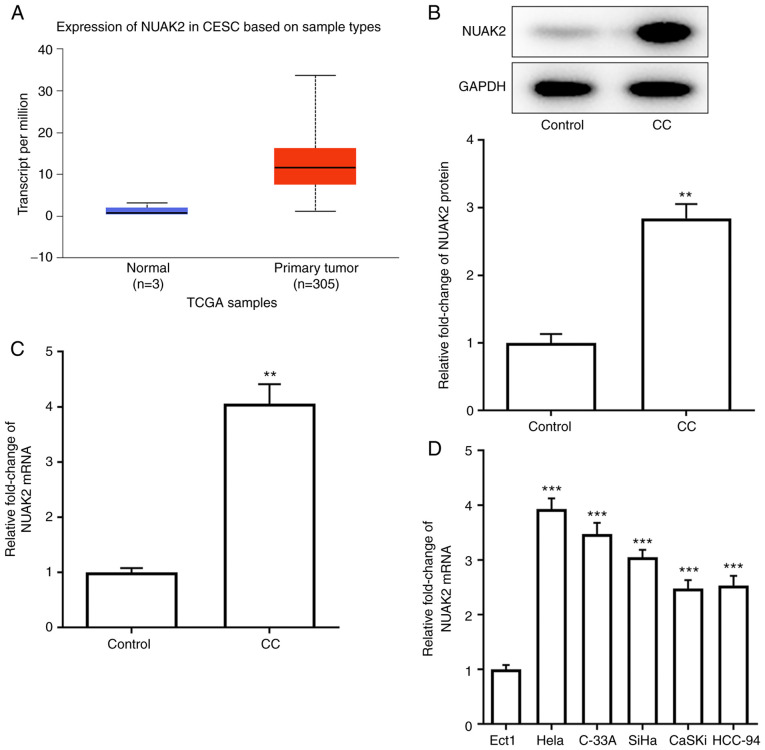
UALCAN website predicted the expression levels of NUAK2 in CESC (Fig. 1A). The expression of NUAK2 was detected in CC tissues by RT-qPCR and western blotting. As shown in Fig. 1B and C, the protein and mRNA expression levels of NUAK2 were significantly increased in CC tissues compared with those in the control group. Similarly, NUAK2 exhibited higher expression in CC cell lines compared with that in Ect1 cells, with the maximum expression observed in HeLa cells (Fig. 1D). Therefore, HeLa cells were selected for subsequent experiments. These results indicated that NUAK2 was highly expressed in CC tissues and cell lines.
Figure 1.
NUAK2 is upregulated in CC tissues and cell lines. (A) UALCAN website predicted the expression levels of NUAK2 in CESC. (B) NUAK2 protein expression in CC samples was assessed via western blotting. (C) NUAK2 mRNA expression in CC samples was assessed via RT-qPCR. **P<0.01 vs. Control. (D) mRNA expression levels of NUAK2 in CC cell lines were detected by RT-qPCR. ***P<0.001 vs. Ect1. CC, cervical cancer; RT-qPCR, reverse transcription-quantitative PCR; NUAK2, NUAK family kinase 2; TCGA, The Cancer Genome Atlas.
Effect of NUAK2 knockdown on CC cell proliferation
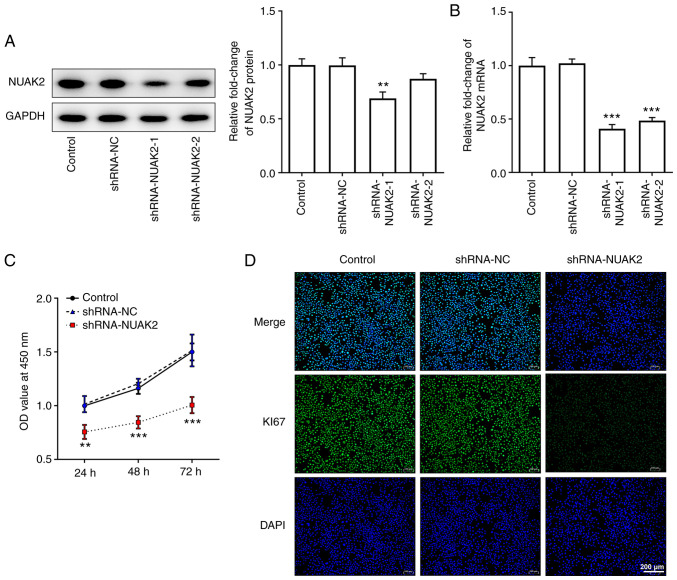
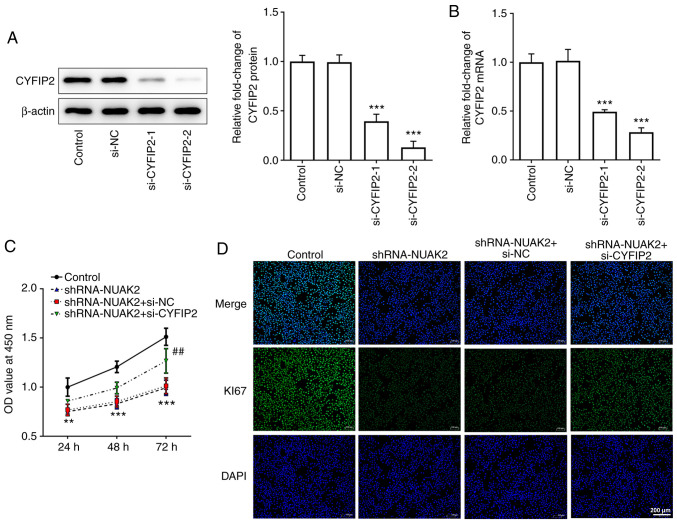
In order to determine the role of NUAK2 expression in CC, shRNA-NUAK2 was used. As shown in Fig. 2A and B, transfection of HeLa cells with shRNA-NUAK2-1 silenced NUAK2 expression more efficiently compared with shRNA-NUAK2-2; therefore, shRNA-NUAK2-1 was selected for subsequent experiments. As determined by CCK-8 assay, the OD value at 450 nm in the shRNA-NUAK2 group was significantly attenuated compared with that in the shRNA-NC group, whereas there were no changes between the untransfected control group and the shRNA-NC group (Fig. 2C). Furthermore, the protein expression levels of Ki67 were markedly decreased in the shRNA-NUAK2 group compared with those in the control group and the shRNA-NC group (Fig. 2D). These results indicated that knockdown of NUAK2 inhibited CC cell proliferation.
Figure 2.
Knockdown of NUAK2 inhibits the proliferation of cervical cancer cells. Transfection efficiency of shRNA-NUAK2-1 and shRNA-NUAK2-2 was determined by (A) western blotting and (B) reverse transcription-quantitative PCR. **P<0.01, ***P<0.001 vs. Control. (C) Proliferation of HeLa cells transfected with shRNA-NUAK2 was determined using Cell Counting Kit-8 assay at 24, 48 and 72 h. **P<0.01, ***P<0.001 vs. shRNA-NC. (D) Immunofluorescence was performed to detect Ki67 expression. shRNA, short hairpin RNA; NC, negative control; NUAK2, NUAK family kinase 2; OD, optical density.
Effect of NUAK2 knockdown on CC cell migration, invasion and EMT
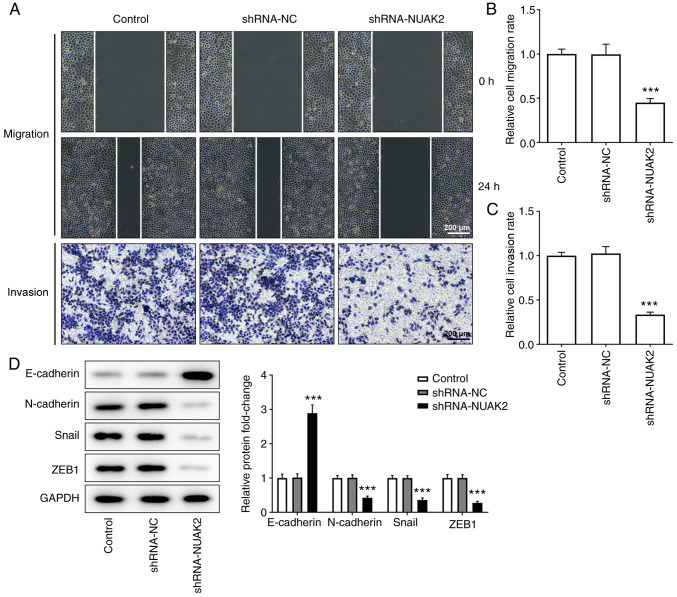
As shown in Fig. 3A-C, after 24 h of incubation, the scratches in the cell monolayer were wider and the relative cell migration rate was lower in the shRNA-NUAK2 group compared with that in the control group and the shRNA-NC group, suggesting that cell migration was inhibited when NUAK2 expression was knocked down. Similarly, the results of the Transwell invasion assay revealed that, compared with in the control and shRNA-NC groups, the invasive ability of HeLa cells was suppressed in the shRNA-NUAK2 group. Furthermore, western blotting revealed that NUAK2 knockdown elevated the expression levels of E-cadherin, and decreased the expression levels of N-cadherin, Snail and ZEB1 compared with those in the shRNA-NC group (Fig. 3D), suggesting that NUAK2 promoted EMT in CC cells. These data confirmed that NUAK2 knockdown suppressed the EMT of CC cells. Taken together, these data indicated that knockdown of NUAK2 expression decreased the mobility of HeLa cells after transfection.
Figure 3.
Knockdown of NUAK2 inhibits migration, invasion and epithelial-to-mesenchymal transition in cervical cancer cells. (A and B) Cell migration and (A and C) invasion of HeLa cells transfected with shRNA-NUAK2 were measured via wound healing and Transwell assays, respectively. (D) Protein expression levels of E-cadherin, N-cadherin, Snail and ZEB1 in HeLa cells transfected with shRNA-NUAK2 were determined by western blotting. ***P<0.001 vs. shRNA-NC; shRNA, short hairpin RNA; NC, negative control; NUAK2, NUAK family kinase 2.
Expression of CYFIP2 in CC tissues and cells
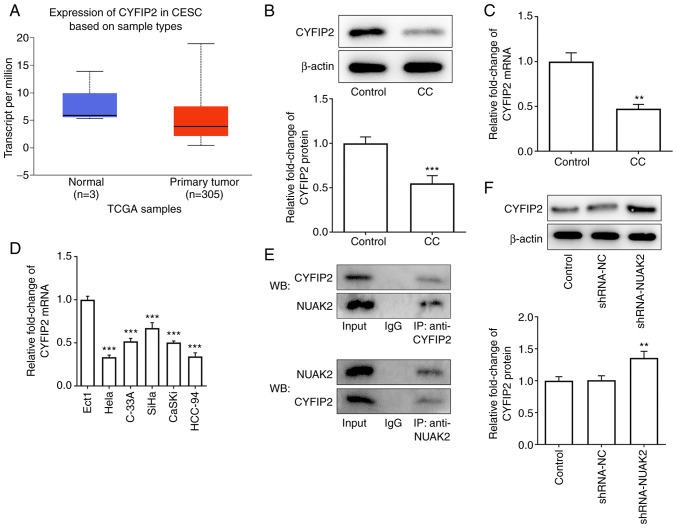
The UALCAN website predicted the expression levels of CYEIP2 in CESC (Fig. 4A). To reveal the molecular mechanisms underlying the role of NUAK2 in CC, the STRING database was used to identify whether NUAK2 directly binds to CYFIP2. The results of western blotting (Fig. 4B) and RT-qPCR (Fig. 4C) revealed that CYFIP2 expression was decreased in tissues from patients with CC compared with that in the control group. Subsequently, the mRNA expression levels of CYFIP2 were detected in several CC cell lines (HeLa, C-33A, SiHa, CaSKi and HCC-94), which demonstrated that CYFIP2 was downregulated at different levels in these CC cell lines compared with that in Ect1 cells (Fig. 4D). To verify the targeted binding of NUAK2 and CYFIP2, Co-IP was performed. The results of Co-IP demonstrated that NUAK2 could interact with CYFIP2 (Fig. 4E). The western blotting results confirmed that NUAK2 knockdown increased the expression levels of CYFIP2 (Fig. 4F). These results indicated that CYFIP2 was expressed at low levels in CC and was negatively associated with NUAK2.
Figure 4.
CYFIP2 is downregulated in CC tissues and cell lines, and is negatively associated with NUAK2. (A) UALCAN website predicted the expression levels of CYFIP2 in CESC. (B) CYFIP2 protein expression in CC samples was assessed via WB. (C) CYFIP2 mRNA expression in CC samples was assessed via RT-qPCR. **P<0.01, ***P<0.001 vs. Control. (D) mRNA expression levels of CYFIP2 in CC cell lines were detected by RT-qPCR. ***P<0.001 vs. Ect1. (E) Co-IP was performed to detect the levels of CYFIP2 in response to NUAK2. (F) CYFIP2 protein expression in HeLa cells transfected with shRNA-NUAK2. **P<0.01 vs. shRNA-NC. CC, cervical cancer; RT-qPCR, reverse transcription-quantitative PCR; shRNA, short hairpin RNA; NC, negative control; IP, immunoprecipitation; WB, western blotting; NUAK2, NUAK family kinase 2; CYFIP2, cytoplasmic FMRP-interacting protein 2; TCGA, The Cancer Genome Atlas.
Effects of CYFIP2 knockdown on CC cell proliferation
In order to further explore the role of CYFIP2 in CC, si-CYFIP2 was used in the following experiments. The results presented in Fig. 5A and B show that both si-CYFIP2-1 and si-CYFIP2-2 could reduce the protein and mRNA expression levels of CYFIP2 in HeLa cells, but si-CYFIP2-2 was more efficient. Thus, si-CYFIP2-2 was selected to knock down CYFIP2 expression. The CCK-8 assay and IF revealed that si-CYFIP2 could enhance cell proliferation compared with that in the shRNA-NUAK2+ si-NC group (Fig. 5C and D). These results suggested that the effect of NUAK2 knockdown on CC cell proliferation may be reversed by inhibition of CYFIP2 expression.
Figure 5.
Knockdown of CYFIP2 reverses the effect of NUAK2 knockdown on the proliferation of cervical cancer cells. (A) Transfection efficiency of si-CYFIP2-1 and si-CYFIP2-2 was determined via western blotting. (B) Transfection efficiency of si-CYFIP2-1 and si-CYFIP2-2 was determined via reverse transcription-quantitative PCR. ***P<0.001 vs. si-NC. (C) Proliferation of HeLa cells transfected with shRNA-NUAK2 and si-CYFIP2 was determined using a Cell Counting Kit-8 assay at 24, 48 and 72 h. **P<0.01, ***P<0.001 shRNA-NUAK2 vs. Control; ##P<0.01 shRNA-NUAK2 + si-CYFIP2 vs. shRNA-NUAK2 + si-NC. (D) Immunofluorescence was performed to detect the expression of Ki67. si, small interfering; shRNA, short hairpin RNA; NC, negative control; NUAK2, NUAK family kinase 2; OD, optical density.
Effect of CYFIP2 knockdown on CC cell migration, invasion and EMT
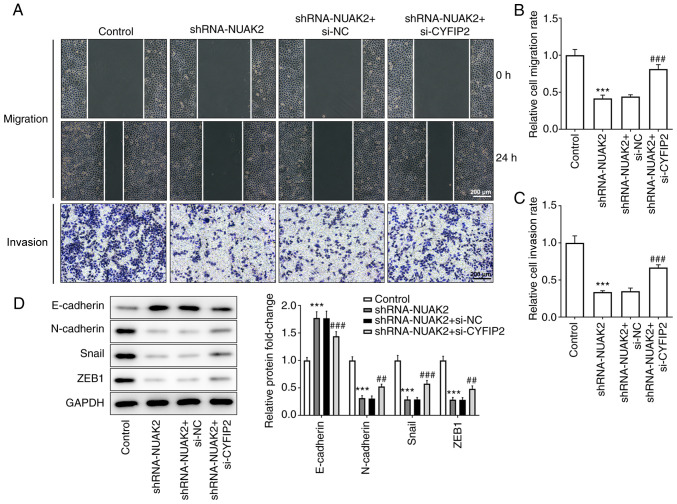
As presented in Fig. 6A-C, in HeLa cells transfected with shRNA-NUAK2, transfection with siRNA to interfere with CYFIP2 expression could accelerate gap closure in the cell monolayer and enhanced the invasive ability of HeLa cells compared with in the shRNA-NUAK2 group and the shRNA-NUAK2+ siRNA-NC group. In addition, inhibition of CYFIP2 expression reversed the effect of NUAK2 knockdown on the EMT phenotype of HeLa cells (Fig. 6D). These results demonstrated that interference with NUAK2 inhibited CC cell migration and the EMT process through upregulation of CYFIP2.
Figure 6.
Knockdown of CYFIP2 reverses the effect of NUAK2 knockdown on cell migration, invasion and epithelial-to-mesenchymal transition in cervical cancer cells. (A and B) Migration and (A and C) invasion of HeLa cells transfected with shRNA-NUAK2 and shRNA-NUAK2+ si-CYFIP2 were assessed via wound healing and Transwell assays, respectively. (D) Protein expression levels of E-cadherin, N-cadherin, Snail and ZEB1 in HeLa cells transfected with shRNA-NUAK2 and si-CYFIP2 were determined by western blotting. ***P<0.001 vs. Control; ##P<0.01, ###P<0.001 vs. shRNA-NUAK2 + si-NC. si, small interfering; shRNA, short hairpin RNA; NC, negative control; NUAK2, NUAK family kinase 2.
Discussion
CC has become a major health concern among women worldwide, and the survival rate of patients with advanced CC is poor (16). Therefore, the search for novel biomarkers is of great importance in the diagnosis and treatment of CC. The present study focused on NUAK2.
Previous studies have identified an important role for NUAK2 in human tumors. Amplification of NUAK2 has been reported to facilitate the development of melanoma (17). Furthermore, it was experimentally demonstrated that knockdown of NUAK2 inhibited the proliferation and migration of melanoma cells (18). NUAK2 upregulation was previously observed in the surface epithelium of ovaries in a large cohort of patients with ovarian plasmacytoma, and was predicted to have a role in driving mutations in ovarian cancer; furthermore, patients with lower NUAK2 expression levels were found to have longer overall survival rates (19). In addition, upregulation of NUAK2 in the maternal kidney has been demonstrated to accelerate cellular senescence in short-lived neonatal mice (20). These previous studies indicated that NUAK2 may play a key a role in cancer. However, to the best of our knowledge, there are no reports on the role of NUAK2 in CC; therefore, the present study was undertaken to investigate the role of NUAK2 expression in CC. Through the search of the UALCAN cancer database, NUAK2 expression was found to be elevated in the tissues of patients with CESC, which is consistent with the literature reporting that NUAK2 is highly expressed in gastric cancer and glioblastoma (8,9). The results were also validated in tissues collected from patients with CC and in CC cell lines.
It is commonly known that the developmental process of tumor cells, including cell proliferation, migration and invasion, is important in the study of cancer. EMT is the important biological process by which epithelial-derived malignant cells are transformed into cells with a mesenchymal phenotype with the ability to migrate and invade; EMT has an important role in cancer metastasis (21). Therefore, to investigate the role of NUAK2 in CC, the present study primarily observed its role in cell progression and EMT. The results demonstrated that NUAK2 knockdown inhibited CC cell proliferation, migration, invasion and EMT.
In order to further investigate the mechanism of action of NUAK2, the online STRING database was used to search for genes that bind to NUAK2, which determined that CYFIP2 could directly bind to NUAK2. CYFIP2 is often studied in neurological disorders and is associated with neuronal functions (10). CYFIP2 has also been proposed as a candidate gene for intellectual disability and autism (22). Furthermore, CYFIP2 has been suggested as a potential target in the treatment of Alzheimer's disease (23). In a previous study, CYFIP2 expression was reduced in gastric cancer, and inhibition of CYFIP2 promoted gastric cancer cell proliferation and chemotherapy resistance to 5-fluorouracil (11). A whole-exome sequencing study previously revealed that mutational dynamics and genetic variation, as well as aberrant DNA repair, tumor cell cycle control and apoptotic pathways were associated with CYFIP2 in endometrial cancer in the Taiwanese population (24). In the present study, it was predicted that the expression levels of CYFIP2 would be decreased in CC tissues, which was confirmed through analysis of CC tumor samples collected from patients and CC cell lines. A Co-IP assay was conducted to detect the direct binding between NUAK2 and CYFIP2.
Once the present study determined that CYFIP2 was negatively associated with NUAK2, si-CYFIP2 was selected to explore the mechanism of the inhibitory effects of NUAK2 knockdown on CC cell processes. The results revealed that inhibition of CYFIP2 expression could partially counteract the effects of NUAK2 knockdown on CC cell proliferation, migration and EMT.
In conclusion, NUAK2 was revealed to serve a crucial role in CC cell proliferation, migration, invasion and EMT by regulating CYFIP2. These findings suggested that NUAK2 may be a potential therapeutic target for CC detection and clinical treatment. However, there are limitations to the present study. Firstly, the lack of validation of the results in additional cell lines and in vivo experiments. Secondly, the effects of NUAK2 overexpression on the progression/prognosis of CC were not determined. Thirdly, the mechanism underlying the negative regulation of CYFIP2 by NUAK2 was not deeply investigated. These issues require further in-depth investigations and will be addressed in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LY and YL designed the study and wrote the manuscript. YL and XS performed the experiments. LL collected, analyzed and interpreted the data. YL and LY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by Tongji Medical College (approval no. WHCH 2018047; Wuhan, China). Written informed consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Olusola P, Banerjee HN, Philley JV, Dasgupta S. Human papilloma virus-associated cervical cancer and health disparities. Cells. 2019;8:622. doi: 10.3390/cells8060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vu M, Yu J, Awolude OA, Chuang L. Cervical cancer worldwide. Curr Probl Cancer. 2018;42:457–465. doi: 10.1016/j.currproblcancer.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 4.Mei D, Zhu Y, Zhang L, Wei W. The Role of CTHRC1 in regulation of multiple signaling and tumor progression and metastasis. Mediators Inflamm. 2020;2020:9578701. doi: 10.1155/2020/9578701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zagorska A, Deak M, Campbell DG, Banerjee S, Hirano M, Aizawa S, Prescott AR, Alessi DR. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci Signal. 2010;3:ra25. doi: 10.1126/scisignal.2000616. [DOI] [PubMed] [Google Scholar]
- 6.Bright NJ, Thornton C, Carling D. The regulation and function of mammalian AMPK-related kinases. Acta Physiol (Oxf) 2009;196:15–26. doi: 10.1111/j.1748-1716.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Gao L, Chien HY, Li WC, Zhao J. The regulation and function of the NUAK family. J Mol Endocrinol. 2013;51:R15–R22. doi: 10.1530/JME-13-0063. [DOI] [PubMed] [Google Scholar]
- 8.Tang L, Tong SJ, Zhan Z, Wang Q, Tian Y, Chen F. Expression of NUAK2 in gastric cancer tissue and its effects on the proliferation of gastric cancer cells. Exp Ther Med. 2017;13:676–680. doi: 10.3892/etm.2016.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu TG, Wang L, Li W, Li JZ, Li J. miR-143 inhibits oncogenic traits by degrading NUAK2 in glioblastoma. Int J Mol Med. 2016;37:1627–1635. doi: 10.3892/ijmm.2016.2562. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Lee Y, Han K. Neuronal function and dysfunction of CYFIP2: From actin dynamics to early infantile epileptic encephalopathy. BMB Rep. 2019;52:304–311. doi: 10.5483/BMBRep.2019.52.5.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao S, Li N, Cai S, Guo H, Wen Y. Inhibition of CYFIP2 promotes gastric cancer cell proliferation and chemoresistance to 5-fluorouracil through activation of the Akt signaling pathway. Oncol Lett. 2017;13:2133–2140. doi: 10.3892/ol.2017.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson RS, II, Cho YJ, Stein S, Liang P. CYFIP2, a direct p53 target, is leptomycin-B sensitive. Cell Cycle. 2007;6:95–103. doi: 10.4161/cc.6.1.3665. [DOI] [PubMed] [Google Scholar]
- 13.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Sne B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Hung P, Zahnd WE, Brandt HM, Adams SA, Wang S, Eberth JM. Cervical cancer treatment initiation and survival: The role of residential proximity to cancer care. Gynecol Oncol. 2021;160:219–226. doi: 10.1016/j.ygyno.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Namiki T, Yaguchi T, Nakamura K, Valencia JC, Coelho SG, Yin L, Kawaguchi M, Vieira WD, Kaneko Y, Tanemura A, et al. NUAK2 amplification coupled with PTEN deficiency promotes melanoma development via CDK activation. Cancer Res. 2015;75:2708–2715. doi: 10.1158/0008-5472.CAN-13-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namiki T, Tanemura A, Valencia JC, Coelho SG, Passeron T, Kawaguchi M, Vieira WD, Ishikawa M, Nishijima W, Izumo T, et al. AMP kinase-related kinase NUAK2 affects tumor growth, migration, and clinical outcome of human melanoma. Proc Natl Acad Sci USA. 2011;108:6597–6602. doi: 10.1073/pnas.1007694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmanuel C, Gava N, Kennedy C, Balleine RL, Sharma R, Wain G, Brand A, Hogg R, Etemadmoghadam D, George J, et al. Comparison of expression profiles in ovarian epithelium in vivo and ovarian cancer identifies novel candidate genes involved in disease pathogenesis. PLoS One. 2011;6:e17617. doi: 10.1371/journal.pone.0017617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen JH, Tarry-Adkins JL, Matharu K, Yeo GS, Ozanne SE. Maternal protein restriction affects gene expression profiles in the kidney at weaning with implications for the regulation of renal function and lifespan. Clin Sci (Lond) 2010;119:373–384. doi: 10.1042/CS20100230. [DOI] [PubMed] [Google Scholar]
- 21.Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med. 2019;216:1016–1026. doi: 10.1084/jem.20181827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweier M, Begemann A, McWalter K, Cho MT, Abela L, Banka S, Behring B, Berger A, Brown CW, Carneiro M, et al. Spatially clustering de novo variants in CYFIP2, encoding the cytoplasmic FMRP interacting protein 2, cause intellectual disability and seizures. Eur J Hum Genet. 2019;27:747–759. doi: 10.1038/s41431-018-0331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiwari SS, Mizuno K, Ghosh A, Aziz W, Troakes C, Daoud J, Golash V, Noble W, Hortobágyi T, Giese KP. Alzheimer-related decrease in CYFIP2 links amyloid production to tau hyperphosphorylation and memory loss. Brain. 2016;139:2751–2765. doi: 10.1093/brain/aww205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang YS, Huang HD, Yeh KT, Chang JG. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. Int J Oncol. 2017;50:1778–1784. doi: 10.3892/ijo.2017.3919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.