Abstract
Cell migration is an essential physiological process, and aberrant migration of epithelial cells underlies many pathological conditions. However, the molecular mechanisms governing cell migration are not fully understood. We report here that growth factor–induced epithelial cell migration is critically dependent on the crosstalk of two molecular switches, namely phosphorylation switch (P-switch) and transcriptional switch (T-switch). P-switch refers to dynamic interactions of deleted in liver cancer 1 (DLC1) and PI3K with tensin-3 (TNS3), phosphatase and tensin homolog (PTEN), C-terminal tension, and vav guanine nucleotide exchange factor 2 (VAV2) that are dictated by mitogen-activated protein kinase kinase 1/2–extracellular signal–regulated protein kinase 1/2–dependent phosphorylation of TNS3, PTEN, and VAV2. Phosphorylation of TNS3 and PTEN on specific Thr residues led to the switch of DLC1–TNS3 and PI3K–PTEN complexes to DLC1–PTEN and PI3K–TNS3 complexes, whereas Ser phosphorylation of VAV2 promotes the transition of the PI3K–TNS3/PTEN complexes to PI3K–VAV2 complex. T-switch denotes an increase in C-terminal tension transcription/expression regulated by both extracellular signal–regulated protein kinase 1/2 and signal transducer and activator of transcription 3 (STAT3) via interleukin-6–Janus kinase–STAT3 signaling pathway. We have found that, the P-switch is indispensable for both the initiation and continuation of cell migration induced by growth factors, whereas the T-switch is only required to sustain cell migration. The interplay of the two switches facilitated by the interleukin-6–Janus kinase–STAT3 pathway governs a sequence of dynamic protein–protein interactions for sustained cell migration. That a similar mechanism is employed by both normal and tumorigenic epithelial cells to drive their respective migration suggests that the P-switch and T-switch are general regulators of epithelial cell migration and potential therapeutic targets.
Keywords: cell migration, ERK, STAT3, IL-6, DLC1, PTEN, phosphorylation, PI3K, VAV2
Abbreviations: ABD, actin-binding domain; ChIP, chromatin immunoprecipitation; CTEN, C-terminal tension; DLC1, deleted in liver cancer 1; EGF, epithelial growth factor; EGFR, epithelial growth factor receptor; ERK1/2, extracellular signal–regulated protein kinase 1/2; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GF, growth factor; HGF, hepatocyte growth factor; IL-6, interleukin-6; IP, immunoprecipitation; JAK, Janus kinase; MEK1/2, mitogen-activated protein kinase kinase 1/2; P-switch, phosphorylation switch; PDGF, platelet-derived growth factor; pPTEN, phosphorylated PTEN; PTEN, phosphatase and tensin homolog; pTNS3, phosphorylated TNS3; qPCR, quantitative PCR; RTK, receptor tyrosine kinase; STAT3, signal transducer and activator of transcription 3; T-switch, transcriptional switch; TME, tumor microenvironment; TNS3, tensin-3; VAV2, vav guanine nucleotide exchange factor 2; WB, Western blot
Cell migration in response to motility cues provided by growth factors (GFs), cytokines, or chemokines plays a critical role in animal development, physiological processes such as wound healing and immune response, and pathological conditions such as cancer invasion and metastasis (1, 2, 3, 4). Cell migration involves the reorganization of the cytoskeleton, a process that is controlled by the Rho family of small GTPases, including RhoA, Rac1, and Cdc42 (5, 6, 7). The Rho GTPases, in turn, are activated by guanine nucleotide exchange factors (GEFs) that promote their binding to GTP (8, 9) and inactivated by GTPase-activating proteins (GAPs) that catalyze the hydrolysis of the bound GTP (10, 11). Understanding how GFs regulate the activity of the Rho GTPases is of great importance to decipher the mechanism of cell migration under physiological and pathological conditions.
The MCF-10A human mammary epithelial cell line is a widely used in vitro model to study the mechanism of cell migration (12, 13). Binding of the epithelial growth factor (EGF) to its receptor (epithelial growth factor receptor [EGFR]) leads to activation of the Ras–mitogen-activated protein kinase kinase 1/2 (MEK1/2)–extracellular signal–regulated protein kinase 1/2 (ERK1/2) and the PI3K–AKT signaling pathways essential for cell proliferation and survival (14, 15, 16, 17). We have found that components of the same signaling pathways are also involved in regulating cell migration through a phosphorylation switch (called P-switch herein) (13). The P-switch, in its original form, refers to the dynamic interactions of deleted in liver cancer 1 (DLC1) and PI3K with tensin-3 (TNS3) and phosphatase and tensin homolog (PTEN) that are dictated by the phosphorylation status of the latter two proteins (13). DLC1 is a Rho-specific GAP, whereas TNS3 is a member of the tensin family of focal adhesion molecules; both of which have been associated with cell migration. By catalyzing the conversion of phosphatidylinositol (4,5)-diphosphate to phosphatidylinositol (3,4,5)-trisphosphate, PI3K plays a critical role in Rac1 activation by facilitating the membrane recruitment and activation of GEFs, including Tiam-1 and vav guanine nucleotide exchange factor 2 (VAV2) (18) that contain a pleckstrin homology domain (19). By converting phosphatidylinositol (3,4,5)-trisphosphate back to phosphatidylinositol (4,5)-diphosphate, the tumor suppressor PTEN is believed to play a negative role in cell migration (20). We have shown previously that DLC1 binds to TNS3, whereas PI3K forms a complex with PTEN in serum-starved MCF-10A cells. However, EGF stimulation activates the MEK1/2–ERK1/2 kinase cascade, leading to TNS3 and PTEN phosphorylation on specific Thr residues (i.e., TNS3–Thr323 and PTEN–Thr319) (13). Remarkably, the phosphorylated PTEN (pPTEN) binds preferentially to DLC1 rather than PI3K, whereas the phosphorylated TNS3 (pTNS3) associates with PI3K instead of DLC1. Therefore, EGF promotes the DLC1–pPTEN and PI3K–pTNS3 interactions at the expense of the DLC1–TNS3 and PI3K–PTEN complexes.
The binding partner switch for DLC1 and PI3K, which is dependent on MEK1/2 activation, is necessary for the spatiotemporal activation of RhoA and Rac1 conducive to cell migration (13). In addition to having a Rho-specific GAP domain, DLC1 contains an N-terminal sterile alpha motif domain capable of binding to the GAP domain via an intramolecular interaction and thereby, keeping DLC1 in an inactive conformation (21). However, this autoinhibition can be relieved by TNS3 that binds to the DLC1 sterile alpha motif domain through the actin-binding domain (ABD) for the former (12, 13). Consequently, the DLC1–TNS3 interaction activates the DLC1 RhoGAP domain, resulting in RhoA inactivation. In contrast, the binding of PTEN to PI3K blocks Rac1 activation by the latter. Therefore, the DLC1–TNS3 and PI3K–PTEN complexes function, respectively, to keep the cellular RhoA-GTP and Rac1-GTP levels in check in the absence of a motility cue. However, following the activation of the MEK1/2–ERK1/2 axis by EGF stimulation and the subsequent phosphorylation of TNS3 and PTEN, the DLC1–TNS3 and PI3K–PTEN complexes are replaced by the DLC1–pPTEN and PI3K–pTNS3 complexes. Because PTEN does not contain an ABD domain and TNS3 has no phosphatidylinositol phosphatase activity, the phosphorylation-triggered binding partner switch for DLC1 and PI3K results in the spatiotemporal activation of RhoA and Rac1 required for cell migration.
While the P-switch, which is activated within 30 min of EGF stimulation, plays a pivotal role in initiating cell migration, its role in sustaining migration remains to be elucidated. We and others have shown that prolonged EGF treatment (>3 h), which is required for sustained cell migration, leads to a decrease in the transcript and protein levels for TNS3 and a concomitant increase for C-terminal tension (CTEN), the shorter isoform of TNS3 that lacks the ABD domain and is thus unable to activate DLC1 (4, 12). However, it is not known how the TNS3/CTEN transcriptional switch (i.e., the T-switch) is regulated and how it interacts with the P-switch to enable persistent cell migration in response to continuous GF stimulation.
We show here that the P-switch and T-switch play indispensable yet distinct roles in the migration of MCF-10A and breast and lung cancer cells induced by a variety of different GFs. We have identified signal transducer and activator of transcription 3 (STAT3) as the regulator of the T-switch by controlling CTEN transcription (22). Intriguingly, we have found that both STAT3 and MEK1/2–ERK1/2 may regulate the P-switch or the T-switch, depending on the duration of GF stimulation. Although the two molecular switches control discrete phases of cell migration that coincide with their kinetic activation patterns, they crosstalk with each other in a highly interactive manner to ensure timely activation of RhoA and Rac1 via DLC1 and PI3K and their binding partners. We show that the preferred binding partner for PI3K is changed, in a phosphorylation-dependent manner, from PTEN to TNS3 and then to VAV2 with time in cells under continuous GF stimulation. In a parallel fashion, DLC1 is found to change binding partners from TNS3, to PTEN and then CTEN, the latter of which coinciding with increased CTEN expression when the T-switch is activated by STAT3. Furthermore, we have found that the interplay of the two switches is facilitated by the interleukin-6 (IL-6)–STAT3–IL-6 feedback loop in which both ERK1/2 and STAT3 play an important part as transcription factor or cofactor for IL-6. Pharmacological inhibition of STAT3 or MEK1/2 significantly reduced or abolished cell migration by disabling the T- and P-switches which, in turn, disrupts the temporal activation pattern of RhoA and Rac1. Taken together, our work has revealed the regulatory mechanism of the P- and T-switches in GF-induced epithelial cell migration and show how different signaling pathways may be integrated to control the spatiotemporal activation of the RhoA and Rac1 GTPases to initiate and sustain cell migration.
Results
EGF-induced epithelial cell migration is dependent on MEK1/2–ERK1/2 and STAT3
Continuous treatment of epithelial cells with GFs or persistent activation of Ras has been shown to activate both MEK1/2 and STAT3, but with distinct kinetic patterns. While the former is activated by transient treatment, the latter requires hours of sustained stimulation (23). To characterize the role of these dynamic signal outputs in cell migration, we cultured MCF-10A cells with U1026, an inhibitor of MEK1/2, the upstream kinase for ERK1/2, or with the STAT3 inhibitor S3I-201 (13, 24). Both inhibitors abrogated EGF-induced cell migration (Fig. 1A). Because the inhibitors had no significant effect on cell viability under the same conditions as used in the wound-healing assay (Fig. S1), this suggests that MEK1/2 and STAT3 play an essential role in MCF-10A cell migration. To find out if MEK1/2 or STAT3 plays a role in the T-switch, we treated the cells with EGF for 8 h with or without the corresponding inhibitor and determined, by Western blot (WB), the dynamic changes in TNS3 and CTEN expression. EGF treatment led to a marked decrease in the TNS3 and a concomitant increase in the CTEN protein level. While neither inhibitor had a significant impact on the reduction in the TNS3 level caused by sustained EGF stimulation, both inhibitors abrogated the increase in CTEN under the same condition (Fig. 1B). This suggests that both STAT3 and MEK1/2 may regulate the T-switch through CTEN.
Figure 1.
MEK1/2–RK1/2 and STAT3 play indispensable, yet distinct roles in mammary epithelial cell migration.A, both the STAT3 inhibitor S3I-201 and the MEK1/2 inhibitor U0126 blocked migration of the MCF-10A cells induced by EGF. Shown are percentage of wound healing (in 16 h) in the presence or the absence of EGF and/or the inhibitor. n = 3, ∗∗p < 0.001; Student's t test. B, both S3I-201 and U0126 abrogated EGF-induced upregulation of CTEN expression. Data shown are obtained from samples with 8 h of treatment. C, Western blot showing the dynamic changes in the TNS3 and CTEN protein during 8 h of EGF treatment. Total and phosphorylated ERK1/2 and STAT3 were detected using specific antibodies. n = 3, ∗p < 0.05, ∗∗p < 0.001; Student's t test. D, distinct binding profiles for STAT3 and ERK1/2 to the cten gene promoter at different time points of EGF treatment, graphed from the corresponding ChIP–PCR data. E, U0126 abrogated ERK1/2 phosphorylation at all time points but inhibited STAT3–Tyr705 phosphorylation only at late time points of EGF stimulation (i.e., 3 and 8 h). F, S3I-201 blocked STAT3–Tyr705 phosphorylation but had no effect on ERK1/2 phosphorylation. ChIP, chromatin immunoprecipitation; CTEN, C-terminal tension; EGF, epithelial growth factor; ERK1/2, extracellular signal–regulated protein kinase 1/2; MEK1/2, mitogen-activated protein kinase kinase 1/2; STAT3, signal transducer and activator of transcription 3; TNS3, tensin-3.
To define the underlying mechanism, we determined the changes in TNS3, CTEN, STAT3, and ERK1/2 expression as well as STAT3 and ERK1/2 phosphorylation in MCF-10A cells treated with EGF for different durations. CTEN expression showed a noticeable increase in 3 h of EGF stimulation, whereas TNS3 expression started to decrease in 60 min (Fig. 1C) (4, 12), suggesting that the expression of the two proteins is not coupled. Activation of ERK1/2 is controlled by MEK1/2-mediated phosphorylation of Thr202 and Tyr204 within its kinase domain, whereas the activity of STAT3 is regulated by the phosphorylation of Tyr705 at its C-terminal tail (25, 26). Using phosphor-specific antibodies, we found that ERK1/2 and STAT3–Y705 exhibited markedly different phosphorylation dynamics. While the phosphorylation of both proteins peaked in 10 min of EGF stimulation, the pERK1/2 level decreased slowly with continuous EGF treatment. In contrast, the STAT3–pY705 level declined sharply in 30 min and dropped below the basal level (i.e., when EGF was absent) in 60 min. Curiously, STAT3–pY705 started to increase again afterward and reached a new plateau in 3 h of continuous EGF stimulation. Therefore, unlike ERK1/2, STAT3 went through two waves of phosphorylation—the first within 30 min and the second in 3 h of EGF stimulation. Importantly, the second wave of STAT3–pTyr705 coincided with a marked increase in the CTEN protein (Fig. 1C), suggesting STAT3 may regulate cten expression. Indeed, chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) showed that, compared with ERK1/2, significantly more STAT3 was found associated with the cten promoter, with more robust binding detected at the third hour compared with 10 min of EGF stimulation (Fig. 1D). In contrast, neither STAT3 nor ERK1/2 bound the promoter region of tns3 (Fig. S2), suggesting that tns3 transcription is not regulated by either protein.
To determine if STAT3 and MEK1/2 crosstalk with each other, we examined the effect of S3I-201 and U0126 on STAT3–Tyr705 and ERK1/2 phosphorylation in a time course of EGF treatment. As expected, S3I-201 effectively blocked STAT3 phosphorylation, and U0126 abrogated ERK1/2 phosphorylation at all time points examined. Intriguingly, S3I-201 had no effect on pERK1/2, whereas U0126 abolished STAT3 phosphorylation at the later time points (e.g., 3 or 8 h; Fig. 1, E and F) of EGF stimulation. These results suggest that ERK1/2 regulates STAT3 activity at late time points of EGF stimulation.
Dynamic evolvement of the P-switch in sustained cell migration
We have previously shown that the P-switch is activated within 30 min of EGF stimulation in MCF-10A. What happens to the P-switch when the cells are continuously treated with EGF as is required for sustained cell migration (Fig. S3)? To address this question, we examined the interactions of DLC1 and PI3K with TNS3 and PTEN at discrete time points of EGF treatment. Although DLC1 and PI3K exchanged binding partners between time 0 and 30 min of EGF treatment, DLC1 reverted to TNS3-binding within 1 h and bound both TNS3 and PTEN in 2 h. In contrast, PI3K appeared to partition between TNS3 and PTEN following 1 h of EGF treatment. The dynamic interactions for DLC1 and PI3K correlated with the activation/inactivation pattern for RhoA and Rac1, respectively. Specifically, the cellular RhoA-GTP level was dictated by DLC1 and its binding partner, with PTEN promoting and TNS3 impeding RhoA activation. In contrast, the Rac1 activity was governed by PI3K and fine tuned by TNS3 and PTEN binding in an opposite manner to RhoA (13) (Fig. 2A). Correlated approximately with the ratio of the DLC1–PTEN/DLC1–TNS3 complexes and the PI3K–TNS3/PI3K–PTEN complexes, the RhoA-GTP and Rac1-GTP levels peaked in 30 min of EGF stimulation, dropped to their respective basal levels (i.e., time 0) in 1 h but rose again to a level between the basal and peak levels with continuous EGF treatment (Fig. 2A).
Figure 2.
The dynamic and continuous evolvement of the P-switch depends on both MEK1/2 and STAT3.A, the P-switch within the first 3 h of EGF stimulation in MCF-10A cells. Western blots showing that dynamic changes in binding partners for DLC1 and PI3K and RhoA and Rac1 activation in response to EGF stimulation. B, U0126 blocked the Thr phosphorylation of PTEN and TNS3 induced by EGF at all the time points examined. C, U0126 abrogated the dynamic interactions of PI3K with TNS3, PTEN, and VAV2 during continuous EGF treatment. The inhibitor also abrogated Ser phosphorylation of VAV2. D, Western blot showing that the STAT3 inhibitor S3I-201 affected the P-switch by disabling the effect of EGF on CTEN expression. CTEN, C-terminal tension; DLC1, deleted in liver cancer 1; EGF, epithelial growth factor; MEK1/2, mitogen-activated protein kinase kinase ½; P-switch, phosphorylation switch; PTEN, phosphatase and tensin homolog; STAT3, signal transducer and activator of transcription 3; TNS3, tensin-3; VAV2, vav guanine nucleotide exchange factor 2.
Because the P-switch is regulated by phosphorylation, we used a pThr-specific antibody to monitor PTEN and TNS3 Thr phosphorylation at different time points of EGF stimulation. We found that the phosphorylation of both PTEN and TNS3 was markedly reduced, but not completely eliminated, in the MCF-10A cells treated with EGF for more than 30 min (Fig. 2B). It is noted that the level of TNS3 assessed by immunoprecipitation (IP) did not change significantly likely because of saturation of the anti-TNS3 antibody under the IP condition. Nevertheless, phosphorylation was no longer detectable for either protein in the presence of U0126 regardless of the length of EGF stimulation, suggesting that MEK1/2 plays a pivotal role in the phosphorylation of PTEN and TNS3. To substantiate this notion, we examined binding of PI3K to TNS3 or PTEN in cells stimulated with EGF for 16 h. Prolonged treatment (e.g., ≥3 h) of the cells led to loss of binding of PI3K to both PTEN and TNS3, suggesting that other protein(s) may be involved in binding PI3K under the condition. To identify the alternate binding partner(s) for PI3K in the later phase of EGF treatment, we immunoprecipitated PI3K (via the p85 subunit) from MCF-10A without EGF or with EGF stimulation for 30 min or 8 h and identified the bound proteins by mass spectrometry. While TNS3 was detected in the PI3K IP at 30 min of EGF treatment as expected (Fig. 2, A and C), VAV2 was identified as one of the most abundant proteins in the 8 h IP (Fig. S4), suggesting that VAV2 is a binding partner for PI3K. Subsequent WB experiments not only confirmed the PI3K–VAV2 interaction but also revealed its dynamic nature. Specifically, markedly more VAV2 was immunoprecipitated with PI3K in cells treated with EGF for 3 h or longer when the TNS3/PTEN–PI3K interaction was lost or drastically reduced (Fig. 2C). Furthermore, VAV2 was Ser-phosphorylated by MEK1/2 as U0126 abrogated VAV2 phosphorylation and markedly reduced its binding to the PI3K (Fig. 2C). Therefore, the PI3K arm of the P-switch, which controls Rac1 activity, is comprised of a series of dynamic protein–protein interactions such that PI3K changes binding partner from PTEN to pTNS3 and then to pVAV2 with continuous EGF stimulation. Importantly, the Thr/Ser phosphorylation of PTEN, TNS3, and VAV2 and their dynamic interactions with PI3K were abolished by U0126, indicating a pivotal role for MEK1/2 in regulating the P-switch (Fig. 2C).
Contrary to U0126, the STAT3 inhibitor S3I-201 did not affect the dynamic interactions of PI3K and DLC1 with PTEN and TNS3. However, at the third hour of EGF stimulation when CTEN expression started to increase, DLC1 was found to bind CTEN in addition to TNS3 and PTEN. At the eighth and 16th hour, when the cellular CTEN level was further increased, the DLC1–CTEN interaction became dominant, apparently at the expense of the DLC1–TNS3 complex. Intriguingly, the DLC1–PTEN interaction remained essentially unchanged at these time points. Therefore, the DLC1 arm of the P-switch, which controls RhoA activity, undergoes at least four distinct phases during continuous EGF stimulation. The first phase, occurring from 0 to 30 min of EGF stimulation, is characterized by the switch of binding partner for DLC1 from TNS3 to PTEN. The second phase, occurring between 0.5 and 1 h, DLC1 is found predominantly in a complex with TNS3. In the third phase, from 1 h to approximately 3 h of EGF treatment, DLC1 partitions between TNS3 and PTEN. And in the final phase, starting approximately at the third hour, DLC1 is engaged in binding TNS3, PTEN, and CTEN simultaneously. Nevertheless, the DLC1–CTEN complex becomes more predominant with time accompanied by increased CTEN expression. S3I-201 affected specifically phase 4 of the DLC1 arm and had no effect on the PI3K arm of the P-switch (Fig. 2D).
Crosstalk between the P- and T-switches is mediated by the IL-6–STAT3 signaling axis
Because EGF stimulation can trigger IL-6 release in an autocrine fashion or a paracrine fashion (23, 27, 28, 29), we wondered if IL-6 would play a role in promoting cell migration through the IL-6– Janus kinase 1/2 (JAK1/2)–STAT3 signaling pathway. In support of this possibility, we found that the IL-6 mRNA increased significantly between 3 and 8 h of EGF stimulation in MCF-10A (Fig. S5A). The same dynamic pattern of IL-6 expression was observed in the MDA-MB-231 breast cancer cells under continuous platelet-derived growth factor (PDGF) treatment (Fig. S5B). To find out if IL-6 could promote cell migration, we added recombinant IL-6 to the MCF-10A cell culture with or without EGF. While IL-6 alone had no effect on the cell migration (Fig. S6A), it synergized with EGF to promote cell migration when the two were applied together. Conversely, the addition of an IL-6 antibody reduced the promoting effect of EGF on cell migration (Fig. 3A). Because serum-starved cells were used to measure migration, the contribution of cell proliferation to migration in the wound-healing assay was negligible (Fig. S6B). The same synergistic effect between IL-6 and PDGF was observed on the MDA-MB-231 cells (Fig. S7). Moreover, the speed of wound healing increased with time during continuous EGF treatment (Fig. S8) likely because of IL-6 accumulation in the medium via the IL-6–JAK1/2–STAT3–IL-6 feedback loop (22). These results indicate that IL-6 can promote cell migration together with EGF. To determine if IL-6 indeed exerts its effect through STAT3, we immunoblotted for STAT3–pY705 in cells treated with recombinant IL-6 or the anti-IL-6 antibody and found that the former promoted, whereas the latter inhibited STAT3 phosphorylation. In contrast, neither IL-6 nor the anti-IL-6 antibody affected ERK1/2 phosphorylation (Fig. 3, B and C).
Figure 3.
The IL-6–STAT3–IL-6 feedback loop facilitates cell migration in response to continuous EGF treatment.A, IL-6 promoted EGF stimulated cell migration, whereas an anti-IL-6 antibody (αIL-6) significantly reduced cell migration. n = 3, ∗p < 0.05, Student's t test. B, IL-6 promoted STAT3–Y705 phosphorylation but had no effect on ERK1/2 phosphorylation. C, αIL-6 markedly reduced STAT3 phosphorylation but had no effect on ERK1/2 activation induced by EGF stimulation for 3 h. D and E, pharmacological inhibition of the EGFR, JAK, and Src kinases reduced or blocked STAT3 phosphorylation. F, relevant kinase inhibitors and anti-IL-6 antibody all decreased CTEN expression induced by EGF. G, nuclear translocation of STAT3 (STAT3-pY705) and ERK1/2 (pERK1/2) during continuous EGF stimulation. H, IL-6 transcription was regulated by ERK1/2 in the early phase (e.g., 10 min), but by STAT3 in the later phase (e.g., 3 h) of EGF stimulation. n = 3, ∗p < 0.05; ∗∗p < 0.001, Student's t test. I, both S3I-201 and U0126 effectively blocked EGF-induced IL-6 expression in MCF-10A cells. EGF, epithelial growth factor; EGFR, epithelial growth factor receptor; ERK1/2, extracellular signal–regulated protein kinase 1/2; IL-6, interleukin-6; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3.
STAT3 phosphorylation within the first 30 min of EGF stimulation is likely mediated by the EGFR and the associated JAK1/2 or Src kinases (30). To confirm this prediction, we incubated MCF-10A with the corresponding kinase inhibitors. Pharmacological inhibition of the EGFR, JAK1/2, or Src led to a significant reduction or complete blockade of EGF-induced cell migration (Fig. S9). Mechanistically, we found that blocking the EGFR or JAK with the corresponding inhibitor EGFRi or JAKi decreased the STAT3–Y705 phosphorylation while the combination of the two inhibitors abolished the phosphorylation (Fig. 3D). Although the JAKi markedly reduced STAT3 phosphorylation, it had no effect on ERK1/2 (Fig. 3D and Fig. S10), suggesting that the inhibitory effect of the JAKi on cell migration was exerted through STAT3. Similarly, the Src kinase inhibitor Src-I completely blocked STAT3 phosphorylation but had no effect on pERK1/2 (Fig. 3E). Furthermore, all the kinase inhibitors and the anti-IL-6 antibody decreased CTEN expression induced by EGF (Fig. 3F). Together, these data suggest that the EGFR–JAK–STAT3, EGFR–Src–STAT3, and IL-6–JAK–STAT3 signaling pathways converge on STAT3 to promote CTEN expression. Consequently, blocking these signaling pathways stalled EGF-induced cell migration by disabling the upregulation of CTEN.
The dynamic expression of IL-6 may be directly regulated by STAT3 and ERK1/2, both of which may function as transcription factor or cofactor (31). To explore this notion, we determined, via cell fractionation followed by WB, nuclear STAT3 and ERK1/2 in EGF-treated MCF-10A cells. Similar to the dynamic profile of STAT3–pY705, STAT3 was detected in the nucleus at the highest level in 10 min of EGF stimulation but at lower levels in samples taken in 30 min, 3 h, and 8 h, and was undetectable at 1 h. In contrast, ERK1/2 phosphorylation and nuclear localization peaked in 10 min and decreased to a lower level afterward (Fig. 3G). Moreover, STAT3 was found to bind robustly to the IL-6 promoter following 3 h of EGF treatment when the IL-6 mRNA level started to rise. Intriguingly, significantly more ERK1/2 was found associated with the IL-6 promoter than STAT3 in 10 min (Fig. 3H). These data suggest that IL-6 transcription is controlled primarily by ERK1/2 in the early phase (<3 h) but by STAT3 in the late phase (>3 h) of EGF-induced cell migration. In agreement with this assertion, both S3I-201 and U0126 blocked IL-6 transcription induced by EGF. Intriguingly, U0126 inhibited IL-6 transcription more strongly than S3I-201 in cells incubated with EGF for 30 min or longer (Fig. 3I).
STAT3 and MEK1/2 play distinct roles in RhoA and Rac1 activation and reorganization of the actin cytoskeleton
Because cell migration is dependent on the dynamic activation of the Rho family of small GTPases, we determined the RhoA-GTP and Rac1-GTP levels in cells treated with EGF for different durations. Although the total cellular level of RhoA or Rac1 did not change significantly with time, the RhoA-GTP or Rac1-GTP levels underwent large dynamic shifts. Within the first hour of EGF stimulation, the activation or inactivation of RhoA and Rac1 correlated with the “on” or “off” states of the P-switch—that is, the highest RhoA-GTP and Rac1-GTP levels were detected in MCF-10A cells stimulated with EGF for 30 min when the DLC1–PTEN and PI3K–TNS3 complexes were formed, and the lowest (basal) levels were observed in 1 h when the DLC1–TNS3 and PI3K–PTEN complexes were detected (Figs. 2 and 4). Intriguingly, the activities of RhoA and Rac1 were markedly higher than their corresponding basal levels at later time points (e.g., at the third, eighth, and 16th hour). The role of MEK1/2 and STAT3 on the dynamic activation/inactivation of RhoA and Rac1 was interrogated using U0126 and S3I-201. Treatment of the cells with U0126 completely abolished RhoA and Rac1 activation at all time points (Fig. 4A), consistent with an essential role for MEK1/2–ERK1/2 in EGF-driven cell migration. In contrast, S3I-201 had no effect on Rac1 activation, yet it reduced RhoA activation at later time points (e.g., at the third, eighth, and 16th hour) of EGF stimulation. This latter observation agrees with the increased CTEN expression in response to prolonged (>3 h) EGF stimulation (Fig. 2D). Apparently, the resulting DLC1–CTEN interaction (Fig. 2D) promotes RhoA activation by maintaining DLC1 RhoGAP in an autoinhibited state (12). On the other hand, we predicted that the enhanced interaction of VAV2 with PI3K at the same time points would contribute to Rac1 activation. Indeed, depletion of VAV2 by siRNA significantly reduced cell migration (Fig. S11), decreased the Rac1-GTP level in cells stimulated with EGF for 3, 8, or 16 h, but had no impact on RhoA activation (Fig. 4C). These data are consistent with our earlier observation that the PI3K–VAV2 interaction was augmented by continuous EGF treatment (Fig. 2C).
Figure 4.
Dynamic RhoA and Rac1 activation is required to sustain EGF-induced epithelial cell migration.A, U0126 eliminated both RhoA and Rac1 activation in EGF-stimulated MCF-10A cells. B, S3I-201 reduced RhoA activity at later time points (>3 h) of EGF treatment but had no significant effect on Rac1 activity. C, VAV2 is required for Rac1 activation at later time points (>3 h) of EGF treatment. Numbers are signal intensities relative to the first lane in each Western blot. EGF, epithelial growth factor; PD, pull down; VAV2, vav guanine nucleotide exchange factor 2.
The distinct roles of the MEK1/2 and STAT3 inhibitors on RhoA and Rac1 activity were manifested in their different effects on F-actin and focal adhesion dynamics. EGF stimulation of the MCF-10A cells induced large dynamic changes in the actin cytoskeleton that correlated with the RhoA and Rac1 activation status. Specifically, an extensive network of actin stress fibers and focal adhesions were formed in cells stimulated with EGF for 30 min when the RhoA-GTP and Rac-GTP levels were at the highest. In contrast, actin filament and focal adhesion were greatly attenuated in cells treated with EGF for 1 h when the P-switch was off and RhoA-GTP and Rac-GTP were at their lowest levels (Fig. 5 and Fig. S12). However, the F-actin and focal adhesion network returned to a moderate level at the third hour and became more prominent at the eighth hour, which again, are consistent with the corresponding RhoA and Rac1 activity (Fig. 4). Compared with the vehicle (dimethyl sulfoxide) control, S3I-201 had no apparent effect on formation of actin filament and focal adhesion in cells in the early phase of EGF stimulation (e.g., 30 min). However, for the later time points (i.e., from the third hour and onward), the S3I-201–treated cells displayed a drastic defect in the formation of actin stress fibers. In contrast, U0126 treatment led to a marked reduction in actin stress fiber and focal adhesions throughout the course of EGF treatment (Fig. 5 and Figs. S12–S14).
Figure 5.
Distinct effects of STAT3 or MEK1/2 inhibition on the dynamic formation of actin stress fiber and focal adhesions during EGF-induced epithelial cell migration.A, representative confocal immunofluorescence images taken of MCF-10A cells at the indicated time points of EGF stimulation in the absence or the presence of S3I-201 or U0126. Red, rhodamine phalloidin for actin; green, paxillin for focal adhesion; and blue, Hoechst for nuclei. The scale bar represents 10 μm. B, a bar graph showing the percentage of cells displaying migratory characteristics (i.e., actin assembly at the leading edge) at the indicated time points of EGF stimulation in the absence or the presence of the inhibitors or vehicle (DMSO). Error bars correspond to standard deviation, n = 3. DMSO, dimethyl sulfoxide; EGF, epithelial growth factor; MEK1/2, mitogen-activated protein kinase kinase 1/2; STAT3, signal transducer and activator of transcription 3.
The T-switch and P-switch control epithelial cancer cell migration
The tumor microenvironment (TME) is enriched with cytokines (such as IL-6) and GFs that may promote their migration and eventual metastasis to a distal site. To investigate if the same mechanism that we have identified for MCF-10A also regulates the migration of epithelial cancer cells, we extended our study to MDA-MB-231, a highly metastatic breast cancer line and HCC-78, a non–small cell lung cancer line (32, 33). We found that the PDGF and the hepatocyte growth factor (HGF) significantly promoted the migration of MDA-MB-231 and HCC-78, respectively. Similar to what was shown for EGF-treated MCF-10A cells, the stimulatory effect of PDGF/HGF on the cancer cells was markedly diminished or eliminated by S3I-201 or U0126 (Fig. 6, A and B). Prolonged treatment of the cancer cells with PDGF/HGF caused a drastic increase in IL-6 expression, which, however, was abolished by the STAT3 or MEK1/2 inhibitor (Fig. S15), suggesting that the IL-6–STAT3–IL-6 amplification loop plays an important role in the migration of MDA-MB-231 or HCC-78. Accordingly, we found that CTEN expression was increased, whereas TNS3 decreased by PDGF/HGF in the cancer cells. Similar to earlier observations, the GF-induced increase in CTEN expression was largely abolished by S3I-201 and U0126 (Fig. 6C).
Figure 6.
MEK1/2 and STAT3 regulate the T- and P-switches to control growth factor–induced migration of MDA-231 and HCC-78 cells.A, S3I-201 and U0126 blocked PDGF-induced MDA-MB-231 cell migration. n = 3, ∗∗p < 0.001; Student's t test. B, both inhibitors blocked the HGF-induced HCC-78 cell migration. n = 3, ∗∗p < 0.001; Student's t test. C, both S3I-201 and U0126 abrogated the TNS3 to CTEN transcriptional switch induced by PDGF in MDA-MB-231 and HGF in HCC-78. Data shown were obtained from cells collected after 8 h of growth factor treatment. D, coimmunoprecipitation and Western blot showed that STAT3 activation was required for DLC1 binding to CTEN in response to prolonged (e.g., >3 h) PDGF treatment of MDA-MB-231 cells. CTEN, C-terminal tension; DLC1, deleted in liver cancer 1; HGF, hepatocyte growth factor; MEK1/2, mitogen-activated protein kinase kinase 1/2; P-switch, phosphorylation switch; PDGF, platelet-derived growth factor; STAT3, signal transducer and activator of transcription 3; T-switch, transcriptional switch; TNS3, tensin-3.
Similar to what was observed in MCF-10A, GF treatment led to dynamic protein–protein interactions associated with the P-switch in the cancer cells. Specifically, the changing of binding partners for DLC1 from TNS3 to PTEN and back to TNS3 occurred in the first hour of PDGF stimulation in MDA-MB-231. This early phase of the P-switch was insensitive to STAT3 inhibition by S3I-201. Nevertheless, the DLC1–CTEN interaction, which occurred in 3 h of PDGF treatment, was reduced by the inhibitor as was the RhoA-GTP level. Similar to earlier findings, S3I-201 only affected Rac1-GTP in cells under PDGF treatment for 3 h or longer (Fig. 6D). Collectively, these data suggest that the P-switch and T-switch are common mechanisms employed by nontumorigenic and tumorigenic epithelial cells to control their migration in response to a variety of different GFs, including EGF, HGF, and PDGF.
Discussion
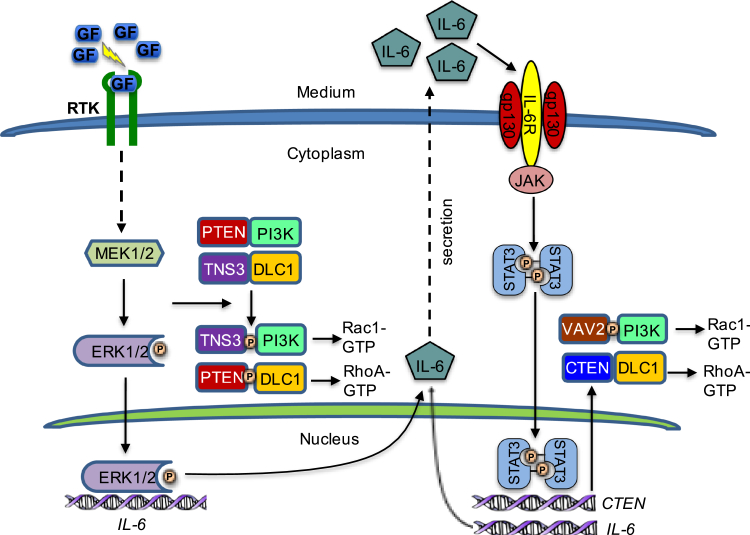
Our main finding from this study is that GF-induced migration of epithelial cells, including both mammary gland–derived epithelial cells and certain breast and lung cancer cells, involves a continuum of dynamic protein–protein interactions that are regulated at both the transcriptional (e.g., via the T-switch) and the post-translational (e.g., via the P-switch) levels. As depicted in Figure 7, epithelial cell migration may be arbitrarily divided into two phases—an early phase and a late phase—depending on the length of GF stimulation. For the cells examined herein, the transition from the early to late phase occurred in approximately 3 h of GF stimulation. We have shown that cell migration within the first 3 h is controlled primarily by the P-switch. GF binding to the corresponding receptor tyrosine kinase (RTK) activates the Ras–Raf–MEK1/2–ERK1/2 signaling cascade. This then leads to MEK1/2-dependent phosphorylation of PTEN and TNS3, resulting in binding-partner exchange for DLC1 and PI3K. Formation of the DLC1–pPTEN and PI3K–pTNS3 complexes within 30 min of GF stimulation is critical for RhoA and Rac1 activation to initiate migration. In addition to activating the P-switch, MEK1/2 also phosphorylates and activates ERK1/2. The phosphorylated ERK1/2, once entering the nucleus, can regulate IL-6 expression. Thus, the MEK1/2–ERK1/2 kinase axis plays a pivotal role not only in the P-switch but also in the transition from the P-switch to the T-switch. With regard to the latter, the ERK1/2-dependent expression of IL-6 plays a critical role in the T-switch as IL-6 can activate STAT3 via the IL6R/gp130–JAK–STAT3 signaling pathway in an autocrine fashion or a paracrine fashion. STAT3, upon pTyr705-mediated dimerization, translocates to the nucleus to further enhance IL-6 transcription, setting off the IL-6–STAT3–IL-6 amplification loop. Meanwhile, nuclear STAT3 may function as a transcription factor for cten and promote CTEN expression. Consequently, the RhoA arm of the P-switch is dominated by the DLC1–CTEN interaction during the second phase of GF-induced epithelial cell migration. Unlike TNS3, CTEN is incapable of activating the RhoGAP domain in DLC1. Therefore, the DLC1–CTEN complex functions to maintain a high level of RhoA-GTP in the cell. In contrast, the Rac1 aim of the P-switch in the second phase is dominated by PI3K binding to VAV2, a pleckstrin homology domain–containing GEF for Rac1. Intriguingly, the PI3K–VAV2 interaction was greatly enhanced by Ser phosphorylation of the latter, which is likely mediated by MEK1/2 as U0126 completely blocked VAV2 phosphorylation. Therefore, the MEK1/2–ERK1/2 and IL-6–STAT3 axes interact with each other throughout the course of GF stimulation to control the dynamic binding partner switch for DLC1 and PI3K, and thereby fine tuning the spatiotemporal activation of RhoA and Rac1 required for the initiation and continuation of cell migration.
Figure 7.
A model depicting the transition from the P-switch to the T-switch during growth factor (GF)–induced cell migration. GF-triggered activation of MEK1/2 leads to TNS3/PTEN phosphorylation and the exchange of binding partners for DLC1 and PI3K for enhanced RhoA and Rac1 activity to initiate cell migration. At the same time, MEK1/2 phosphorylates ERK1/2, and the latter translocates to the nucleus to promote IL-6 transcription, leading to activation of the IL-6-JAK–STAT3–IL-6 amplification loop and increased CTEN expression. In the later phase of GF-induced cell migration, DLC1 and PI3K bind preferentially to CTEN and VAV2 to maintain RhoA and Rac1 activity for sustained migration. CTEN, C-terminal tension; DLC1, deleted in liver cancer 1; ERK1/2, extracellular signal–regulated protein kinase 1/2; IL-6, interleukin-6; JAK, Janus kinase; MEK1/2, mitogen-activated protein kinase kinase 1/2; P-switch, phosphorylation switch; PTEN, phosphatase and tensin homolog; STAT3, signal transducer and activator of transcription 3; T-switch, transcriptional switch; TNS3, tensin-3; VAV2, vav guanine nucleotide exchange factor 2.
Because the migration of tumorigenic epithelial cells may also be regulated by the same signaling pathways and the associated molecular switches, our work may be relevant to understand the molecular basis of cancer invasion and metastasis. GFs, such as EGF, PDGF, HGF, and vascular endothelial growth factor, which may be produced by the tumor cells or by other cells in the TME, have been implicated in promoting cancer metastasis (34, 35, 36, 37). We have shown that both PDGF-induced MDA-MB-231 and HGF-induced HCC-78 cell migration are dependent on MEK1/2 and STAT3. What distinguishes physiological from pathological cell migration may not be the underlying signaling pathways but the specific GFs that trigger cell migration. For example, we found that EGF, but not PDGF, promoted the MCF-10A cell migration. In contrast, PDGF, but not EGF, effectively induced MDA-MB-231 migration. Compared with the nontumorigenic counterpart, the cancer cells may have altered profiles of RTK expression to better allow them to interact with the TME, including tumor-associated fibroblasts and tumor-infiltrating leukocytes, and respond favorably to GFs or cytokines produced in the TME.
Our work provides a potential mechanism by which cancer cells may interact with cells in the TME to facilitate metastasis through the IL-6/JAK/STAT3 or more generally, the cytokine–JAK–STAT3 signaling pathway. Indeed, IL-6 has been identified as an important mediator of metastasis in breast cancer (38, 39), gastric cancer (40), and colorectal cancer (29, 41). Because STAT3 may be activated by a number of cytokines via the JAK–STAT3 pathway, it may be a converging point of signaling between cancer cells and cells in the TME. In agreement with this assertion, a recent study has identified a critical role for STAT3 in tumorigenesis and metastasis of pancreatic ductal adenocarcinoma that is activated by the leukemia inhibitory factor secreted by the pancreatic stellate cells in a paracrine fashion (42). Similarly, the IL-6–JAK–STAT3 pathway is frequently hyperactivated in cancer, and several agents targeting components of this pathway, including IL-6, IL6R, and the JAKs, have been developed for the treatment of certain hematopoietic malignancies, and many more, including STAT3 inhibitors, are undergoing clinical trials or preclinical investigation for the treatment of solid tumors (22, 28, 43). The multiple targets identified from our study offer alternative means or combination strategies to curb cancer cell migration by inhibiting STAT3 or its upstream regulators, including IL-6, JAK1/2, MEK1/2, Src, and the RTK (44). Future studies using mouse models of cancer would help define the role of the P- and T-switches in cancer metastasis and establish the therapeutic relevance of targeting the MEK1/2–ERK1/2 and IL-6–STAT3 axes individually or in combination for the treatment of metastatic cancer.
It should be noted that cell migration may be regulated by a variety of different mechanisms depending on the cell type and physiological or pathological context. For example, many cancer cells are characterized with mutations or deletions in the DLC1 or PTEN gene. For these cells, the P-switch may be replaced by a different mechanism to regulate their migration. Moreover, DLC1 itself can be phosphorylated by a number of different kinases, including Src, ERK1/2, CDK5, and Akt, which may lead to changes in its Rho-GAP activity (45, 46, 47). By the same token, it is unlikely that VAV2 is the only GEF involved in regulating epithelial cell migration in the late phase. It is possible that the combination of different GEFs and GAPs underlies the migration of different cells in response to different motility cues. Future studies should focus on identifying the differences in the regulatory mechanism of cell migration between normal and cancer cells and understanding how diverse signaling inputs (e.g., GFs, cytokines, chemokines) may be integrated at the molecular level to control physiological and pathological cell migration.
Experimental procedures
Cell culture
MCF-10A, MDA-MB-231, and HCC-78 cells were obtained from American Type Culture Collection. The MCF-10A culture medium contained Dulbecco's modified Eagle's medium: F12 medium supplemented with antibiotics, EGF (20 ng/ml), insulin (10 μg/ml), cholera toxin (100 μg/ml), hydrocortisone (0.5 μg/ml), heat-inactivated horse serum (5%; Invitrogen), and 1% penicillin/streptomycin. MDA-MB-231 cells were maintained in Dulbecco's modified Eagle's medium: F12 containing antibiotics, 10% fetal bovine serum, and 1% penicillin/streptomycin. HCC-78 cells were maintained in RPMI1640 medium containing antibiotics, 10% fetal bovine serum (Sigma–Aldrich), and 1% penicillin/streptomycin.
Serum-free (starved) medium contained no serum or GF. GF treatment medium (serum free) contained 20 ng/ml EGF for MCF-10A, 30 ng/ml PDGF for MDA-MB-231, and 10 ng/ml HGF for HCC-78. Cells were incubated with the GF for the specified durations after 16-h serum starvation at 37 °C in 5% CO2.
Wound-healing assay
Cells at ∼100% confluency post 16 h serum starvation were scratched using a 200 μl pipette tip, and the cell debris was washed away with PBS. The cells were then incubated for 16 h with or without GFs or inhibitors. Images were captured at the beginning (0 h) and desired intervals (up to 16 h) using the Infinity Capture Imaging System (Lumenera Corporation) on a Motic AE31 Inverted Microscope (Matic Microscope). The migration data were quantified using ImageJ (National Institutes of Health; RRID: SCR_003070).
Cell proliferation assay
Cells were cultured in 100 μl medium in a 96-well plate. After 16 h of serum starvation, cells were incubated in serum-free medium with or without IL-6, EGF, or a mixture of IL-6 and EGF for 16 h. Vial cell number was determined using WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) cell proliferation assay (Sigma). Specifically, 10 μl of WST-8 solution was added to each 100 μl well. The plate was then incubated in the dark at 37 oC for 30 min before absorbance at 460 nm was measured.
Flow cytometry
Cells at ∼100% confluency post 16 h of serum starvation were incubated with EGF (20 ng/ml), IL-6 (20 ng/ml), or a mixture of EGF and IL-6 for 16 h. Subsequently, the cells were harvested and resuspended in annexin-binding buffer (10 mM Hepes [pH 7.4], 140 mM NaCl, and 2.5 mM CaCl2) and stained with annexin V-FITC (BioLegend) and SYTOX AADvanced (Thermo Fisher Scientific). The samples were analyzed using LSRII flow cytometer (BD Biosciences) and FlowJo, version 10 (FlowJo LLC). A minimum of 20,000 events was recorded.
IP and WB
Cells were lysed in cold lysis buffer (1% NP-40, 50 mM Tris, pH 7.4, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 10% glycerol, Halt Protease, and Phosphatase Inhibitor Cocktail) (Thermo Fisher Scientific; diluted at 1:100). To prepare cell lysate, cell pellets were sonicated in 0.2 ml lysis buffer on ice, and the lysate was spun down at 16,000g for 15 min at 4 °C. The supernatant was collected, and the protein concentration was determined using Pierce Protein Assay Kit (Thermo Fisher Scientific). After clearing the lysate with appropriate preimmune serum and protein G (Roche), IP was carried out using the indicated antibodies (3 μg antibody applied per reaction). The IP proteins were redissolved in SDS–PAGE loading buffer. After separation in SDS–PAGE gel, the proteins were transferred to polyvinylidene difluoride membrane, immunoblotted with appropriate secondary antibodies, and visualized by ECL. Specifically, rat antimouse nonreduced–specific IgG (Abcam; Ab131368) and mouse anti-rabbit light chain–specific IgG (Jackson ImmunoResearch; 211-032-171) was used as the secondary antibodies. Where appropriate, goat anti-rabbit/mouse or rabbit antigoat IgGs (Bio-Rad) was used as the secondary antibodies.
RNA preparation and real-time PCR analysis (RT-qPCR)
Total RNA was extracted using the QIAGEN RNeasy Mini Kit (74104), and the reverse transcription reaction was performed using QIAGEN QuantiTect Reverse Transcription Kit (205311) following the manufacturer's protocol. After mixing the extracted RNA with FastStart SYBR Green Master (04673492001; Roche) and specific primers, the RT-qPCR was performed on a Stratagene Mx3005P QPCR System (QIAGEN-SABiosciences). The sequences of the primers are listed in Table 1.
Table 1.
Reagent or resource
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-TNS3 | Sigma | SAB4200296 |
| Mouse monoclonal anti-CTEN | Abnova | H00084951-M01 |
| Rabbit monoclonal anti-STAT3-pY705 | Cell Signaling Technology | D3A7 |
| Mouse monoclonal anti-STAT3-pS705 | Cell Signaling Technology | 9136 |
| Mouse monoclonal anti-STAT3 | Santa Cruz | Sc-8019 |
| Rabbit monoclonal anti-pERK1/2 | Thermo Fisher Scientific | MA515174 |
| Rabbit monoclonal anti-ERK1/2 | Thermo Fisher Scientific | MA515134 |
| Rabbit polyclonal anti-β-tubulin | Santa Cruz | Sc-9104 |
| Goat polyclonal anti-laminA/C | Santa Cruz | Sc-6215 |
| Mouse monoclonal anti-IL-6 | R&D Systems | MAB-206 |
| Rabbit monoclonal anti-PTEN | Cell Signaling Technology | 9559 |
| Mouse monoclonal anti-DLC1 | BD Biosciences | 612020 |
| Mouse monoclonal anti-PI3K | Millipore | 1981549 |
| Mouse monoclonal anti-RhoA | Cytoskeleton | ARH03 |
| Rabbit polyclonal anti-Rac1 | Santa Cruz | Sc-217 |
| Mouse monoclonal anti-VaV2 | Santa Cruz | Sc-271442 |
| Rabbit monoclonal anti-paxillin | Abcam | ab32084 |
| Mouse monoclonal anti-phospho-serine | Millipore | 05-1000 |
| Mouse monoclonal anti-phospho-threonine | Cell Signaling Technology | 9386 |
| Rat antimouse nonreduced–specific IgG | Abcam | Ab131368 |
| Mouse anti-rabbit light chain–specific IgG | Jackson Immuno Research | 211-032-171 |
| Goat antimouse IgG | Bio-Rad | 1706516 |
| Goat anti-rabbit IgG | Bio-Rad | 1706515 |
| Rabbit antigoat IgG | Bio-Rad | 1721034 |
| Chemicals and proteins | ||
| EGF | Sigma | E9644 |
| HGF | Millipore | GF116 |
| PDGF | Sigma | SRP3138 |
| EGFR inhibitor | Millipore | 324647 |
| JAK inhibitor | Millipore | 420097 |
| S3I-201 | Millipore | 573130 |
| Hydrocortisone | Sigma | H4001 |
| Cholera toxin | Sigma | C8052 |
| Insulin | Sigma | I0516 |
| Src inhibitor I | Sigma | 567805 |
| VAV2 siRNA | Thermo Fisher | AM16708 |
| Scramble siRNA | Thermo Fisher | AM4611 |
| U0126 | Promega | V1112A |
| IL-6 | Pepro Tech | 200-06 |
| Experimental models: cell lines | ||
| Human: MCF-10A cell line | ATCC | CRL-10317 |
| Human: MDA-MB-231 cell line | ATCC | HTB-26 |
| Human: HCC-78 cell line | DSMZ | ACC563 |
| Oligonucleotides | ||
| TNS3-forward (For ChIP–qPCR) GGGCCATCTGAATCTCAGGG | N/A | N/A |
| TNS3-reverse (For ChIP–qPCR) GGAGGAGGTCAGGGAAGTCT | N/A | N/A |
| CTEN-forward (For ChIP–qPCR) GCCCCTCTAGAACAGGGAGA | N/A | N/A |
| CTEN-reverse (For ChIP–qPCR) TTCCCTGAGGGGAGGACATT | N/A | N/A |
| IL-6-forward (For ChIP–qPCR) GGGCCGACTAGACTGACTTC | N/A | N/A |
| IL-6-reverse (For ChIP–qPCR) AACCCTCAGCTCATGCCAAA | N/A | N/A |
| IL-6-forward (For qRT–PCR) AGTGAGGAACAAGCCAGAGC | N/A | N/A |
| IL-6-reverse (For qRT–PCR) AGCTGCGCAGAATGAGATGA | N/A | N/A |
| Software | ||
| ImageJ | ImageJ | https://imagej.net/Welcome |
Abbreviations: ATCC, American Type Culture Collection; N/A, not available.
RhoA or Rac1 activation assay
RhoA or Rac1 activities were measured using the Rho or Rac Activation Assay Biochem Kit (Cytoskeleton). Cells at 40 to 60% confluency were treated with GFs. After treatment, the cells were washed in ice-cold PBS and lysed. Equal amounts of whole-cell lysate were incubated with 20 mg Rho-binding domain of rhotekin or p21 binding domain of p21 activated kinase 1 beads for 1 h at 4 °C. The beads were washed three times with PBS, and the bound RhoA or Rac1 proteins were analyzed by WBs using an anti-RhoA or anti-Rac1 antibody.
ChIP
Cells were washed twice with cold PBS and collected in 1-ml microcentrifuge tubes. After centrifugation for 5 min at 2000g, supernatants were removed and 1 ml PBS containing 0.1% NP-40 was added to the pellet. After triturating the pellet a few times with a pipette tip, the resuspended lysate was centrifuged for 5 min under 750g. The supernatant was removed, and the nuclear fraction in the pellet was collected. The ChIP experiment was performed essentially as described on Abcam's Web page (https://docs.abcam.com/pdf/chromatin/A-beginners-guide-to-ChIP.pdf). Specifically, 5 g antibody or IgG control was used for IP. The sequence of primers is listed in Table 1.
Confocal immunofluorescence microscopy
Serum-starved cells grown in 35-mm glass-bottom dishes (P35G-1.0-14-C; MatTek) were treated with GFs for the specified durations. Cells were then fixed in 10% neutral buffered formaldehyde at room temperature for 1 h. After being rinsed in PBS, cells were incubated with 0.1% Triton X-100 (in PBS) at room temperature for 30 min. After three PBS washes, the cells were incubated with 2% bovine serum albumin at room temperature for 60 min, followed by PBS wash, and incubation with anti-Paxillin antibody (ab32084; Abcam; 1:200) for 1 h at room temperature. Samples were then incubated with the corresponding Alexa Fluor-488 for 1 h followed by incubation with rhodamine phalloidin (Invitrogen; 1:200). Actin was stained with rhodamine phalloidin (Invitrogen; 1:50), and nuclei were stained with Hoechst in water for 5 min. The specimens were imagined on a ZEISS LSM 800 confocal microscope (Carl Zeiss MicroImaging) with pinhole set at 1 airy unit using 488, 564, and/or 633 nm excitation and an ×63/1.4 oil objective lens.
Mass spectrometry
After the PI3K IP, beads were resuspended in 20 μl 100 mM ammonium bicarbonate. The bound proteins were subjected to on-beads digestion overnight at 37 °C by adding 100 ng trypsin (Promega; catalog no. v5113). The next morning, 100 ng of extra trypsin was supplemented to each IP sample and the samples were incubated for four more hours at 37 °C, to complete the digestion. The digested peptides were separated from beads by a glass fiber filter, desalted by the ZipTip C18 column (Millipore), and were eluted in 5 μl 70% acetonitrile/0.1% formic acid. The peptide elution was diluted in 0.1% formic acid, and the diluted samples were injected into the mass spectrometry.
The peptides were separated on a 50 cm EASY_Spray C18 column (catalog no. ES803A; Thermo Fisher Scientific), over a 90-min gradient of 3% to 35% solvent B (0.1% formic acid/100% acetonitrile). The samples were analyzed by a Q Exactive instrument (Thermo Scientific) in the data-dependent acquisition mode. Full scans were acquired with resolution 70,000 at 200 m/z, and with 1E6 ions accumulated within a maximum injection time of 50 ms. The ten most intense ions with charge states 2, 3, or 4 were sequentially isolated to a target value of 5E4 with a maximum injection time of 50 ms, fragmented, and detected at 17,500 resolution.
The mass spectrometric data were analyzed in the MaxQuant environment, version 1.6.1.0. The MS/MS spectra were matched against the human Swiss-Prot database (20,238 entries; retrieved on May 22, 2018). The search included methionine oxidation and N-acetylation of protein as variable modifications. Up to two missed cleavages were allowed.
Statistical analysis
All statistical analyses were performed using Excel. All data based on statistical analysis were shown as means ± SD. Statistical significance was analyzed by paired Student's t test. All p values were two tailed, and the level of statistical significance was set as ∗p < 0.05 and ∗∗p < 0.001.
Data availability
The important data that support the findings of this study are contained within the article; other data are to be shared upon request to the corresponding author: Shawn S.-C. Li, Departments of Biochemistry and Oncology, Schulich School of Medicine and Dentistry, Western University. Canada. E-mail: sli@uwo.ca.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
X. C. and S. S.-C. L. conceptualization; L. Q. methodology; L. Q. and G. W. formal analysis; L. Q., T. K., C. V., and X. L. investigation; X. C. and G. W. resources; L. Q., T. K., and S. S.-C. L. data curation; L. Q. writing–original draft; S. S.-C. L. supervision L. Q. and S. S.-C. L. project administration; S. S.-C. L. funding acquisition.
Funding and additional information
This work was supported by a grant (to S. S.-C. L.) from the Canadian Cancer Society. S. S.-C. L. held a Canada Research Chair and Wolfe Medical Research Professorship in Molecular and Epigenetic Basis of Cancer.
Edited by Alex Toker
Supporting information
References
- 1.Jin K., Pandey N.B., Popel A.S. Simultaneous blockade of IL-6 and CCL5 signaling for synergistic inhibition of triple-negative breast cancer growth and metastasis. Breast Cancer Res. 2018;20:54. doi: 10.1186/s13058-018-0981-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrientos S., Stojadinovic O., Golinko M.S., Brem H., Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 3.Atretkhany K.N., Drutskaya M.S., Nedospasov S.A., Grivennikov S.I., Kuprash D.V. Chemokines, cytokines and exosomes help tumors to shape inflammatory microenvironment. Pharmacol. Ther. 2016;168:98–112. doi: 10.1016/j.pharmthera.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., Ben-Chetrit N., Tarcic G., Lindzen M., Avraham R., Liao Y.C. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe A.B., Hall A. Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 6.Hall A. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 7.Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J. Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 8.ten Klooster J.P., Jaffer Z.M., Chernoff J., Hordijk P.L. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 2006;172:759–769. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurin M., Huber J., Pelletier A., Houalla T., Park M., Fukui Y., Haibe-Kains B., Muller W.J., Cote J.F. Rac-specific guanine nucleotide exchange factor DOCK1 is a critical regulator of HER2-mediated breast cancer metastasis. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7434–7439. doi: 10.1073/pnas.1213050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haga R.B., Ridley A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases. 2016;7:207–221. doi: 10.1080/21541248.2016.1232583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahoz A., Hall A. DLC1: A significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao X., Voss C., Zhao B., Kaneko T., Li S.S. Differential regulation of the activity of deleted in liver cancer 1 (DLC1) by tensins controls cell migration and transformation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1455–1460. doi: 10.1073/pnas.1114368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X., Kaneko T., Li J.S., Liu A.D., Voss C., Li S.S. A phosphorylation switch controls the spatiotemporal activation of Rho GTPases in directional cell migration. Nat. Commun. 2015;6:7721. doi: 10.1038/ncomms8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y., Zhang C., Croucher D.R., Soliman M.A., St-Denis N., Pasculescu A., Taylor L., Tate S.A., Hardy W.R., Colwill K., Dai A.Y., Bagshaw R., Dennis J.W., Gingras A.C., Daly R.J. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H.W., Shin M.G., Lee S., Kim J.R., Park W.S., Cho K.H., Meyer T., Heo W.D. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol. Cell. 2012;47:281–290. doi: 10.1016/j.molcel.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cully M., You H., Levine A.J., Mak T.W. Beyond PTEN mutations: The PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 17.Young A., Lyons J., Miller A.L., Phan V.T., Alarcon I.R., McCormick F. Ras signaling and therapies. Adv. Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 18.Zhu G., Fan Z., Ding M., Zhang H., Mu L., Ding Y., Zhang Y., Jia B., Chen L., Chang Z., Wu W. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene. 2015;34:5971–5982. doi: 10.1038/onc.2015.45. [DOI] [PubMed] [Google Scholar]
- 19.Liu B.P., Burridge K. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol. Cell. Biol. 2000;20:7160–7169. doi: 10.1128/mcb.20.19.7160-7169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liliental J., Moon S.Y., Lesche R., Mamillapalli R., Li D., Zheng Y., Sun H., Wu H. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 21.Joshi R., Qin L., Cao X., Zhong S., Voss C., Min W., Li S.S.C. DLC1 SAM domain-binding peptides inhibit cancer cell growth and migration by inactivating RhoA. J. Biol. Chem. 2020;295:645–656. doi: 10.1074/jbc.RA119.011929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson D.E., O'Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toettcher J.E., Weiner O.D., Lim W.A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beebe J.D., Liu J.Y., Zhang J.T. Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we? Pharmacol. Ther. 2018;191:74–91. doi: 10.1016/j.pharmthera.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Huang G., Yan H., Ye S., Tong C., Ying Q.L. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32:1149–1160. doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair R.R., Tolentino J.H., Hazlehurst L.A. Role of STAT3 in transformation and drug resistance in CML. Front. Oncol. 2012;2:30. doi: 10.3389/fonc.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sriuranpong V., Park J.I., Amornphimoltham P., Patel V., Nelkin B.D., Gutkind J.S. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- 28.Rodriguez-Barrueco R., Yu J., Saucedo-Cuevas L.P., Olivan M., Llobet-Navas D., Putcha P., Castro V., Murga-Penas E.M., Collazo-Lorduy A., Castillo-Martin M., Alvarez M., Cordon-Cardo C., Kalinsky K., Maurer M., Califano A. Inhibition of the autocrine IL-6-JAK2-STAT3-calprotectin axis as targeted therapy for HR-/HER2+ breast cancers. Genes Dev. 2015;29:1631–1648. doi: 10.1101/gad.262642.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Hu F., Li G., Li G., Yang X., Liu L., Zhang R., Zhang B., Feng Y. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis. 2018;9:25. doi: 10.1038/s41419-017-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egloff A.M., Grandis J.R. Response to combined molecular targeting: Defining the role of P-STAT3. Clin. Cancer Res. 2011;17:393–395. doi: 10.1158/1078-0432.CCR-10-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter R.L., Lo H.W. STAT3 target genes relevant to human cancers. Cancers (Basel) 2014;6:897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergethon K., Shaw A.T., Ou S.H., Katayama R., Lovly C.M., McDonald N.T., Massion P.P., Siwak-Tapp C., Gonzalez A., Fang R., Mark E.J., Batten J.M., Chen H., Wilner K.D., Kwak E.L. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., Speed T., Spellman P.T., DeVries S., Lapuk A., Wang N.J. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G.F. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 35.Thoma C. Prostate cancer: Targeting the FGFR curbs bone metastasis. Nat. Rev. Urol. 2014;11:604. doi: 10.1038/nrurol.2014.270. [DOI] [PubMed] [Google Scholar]
- 36.Li W., Ng J.M., Wong C.C., Ng E.K.W., Yu J. Molecular alterations of cancer cell and tumour microenvironment in metastatic gastric cancer. Oncogene. 2018;37:4903–4920. doi: 10.1038/s41388-018-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Zhou W. The emerging regulation of VEGFR-2 in triple-negative breast cancer. Front. Endocrinol. (Lausanne) 2015;6:159. doi: 10.3389/fendo.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbieri I., Pensa S., Pannellini T., Quaglino E., Maritano D., Demaria M., Voster A., Turkson J., Cavallo F., Watson C.J., Provero P., Musiani P., Poli V. Constitutively active Stat3 enhances neu-mediated migration and metastasis in mammary tumors via upregulation of Cten. Cancer Res. 2010;70:2558–2567. doi: 10.1158/0008-5472.CAN-09-2840. [DOI] [PubMed] [Google Scholar]
- 39.Badache A., Hynes N.E. Interleukin 6 inhibits proliferation and, in cooperation with an epidermal growth factor receptor autocrine loop, increases migration of T47D breast cancer cells. Cancer Res. 2001;61:383–391. [PubMed] [Google Scholar]
- 40.Wu X., Tao P., Zhou Q., Li J., Yu Z., Wang X., Li J., Li C., Yan M., Zhu Z., Liu B., Su L. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget. 2017;8:20741–20750. doi: 10.18632/oncotarget.15119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun D., Shen W., Zhang F., Fan H., Xu C., Li L., Tan J., Miao Y., Zhang H., Yang Y., Cheng H. alpha-Hederin inhibits interleukin 6-induced epithelial-to-mesenchymal transition associated with disruption of JAK2/STAT3 signaling in colon cancer cells. Biomed. Pharmacother. 2018;101:107–114. doi: 10.1016/j.biopha.2018.02.062. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y., Gao W., Lytle N.K., Huang P., Yuan X., Dann A.M., Ridinger-Saison M., DelGiorno K.E., Antal C.E., Liang G., Atkins A.R., Erikson G., Sun H., Meisenhelder J., Terenziani E. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569:131–135. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Priego N., Zhu L., Monteiro C., Mulders M., Wasilewski D., Bindeman W., Doglio L., Martinez L., Martinez-Saez E., Ramon Y.C.S., Megias D., Hernandez-Encinas E., Blanco-Aparicio C., Martinez L., Zarzuela E. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018;24:1024–1035. doi: 10.1038/s41591-018-0044-4. [DOI] [PubMed] [Google Scholar]
- 44.Chua C.Y., Liu Y., Granberg K.J., Hu L., Haapasalo H., Annala M.J., Cogdell D.E., Verploegen M., Moore L.M., Fuller G.N., Nykter M., Cavenee W.K., Zhang W. IGFBP2 potentiates nuclear EGFR-STAT3 signaling. Oncogene. 2016;35:738–747. doi: 10.1038/onc.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tripathi B.K., Anderman M.F., Qian X., Zhou M., Wang D., Papageorge A.G., Lowy D.R. SRC and ERK cooperatively phosphorylate DLC1 and attenuate its Rho-GAP and tumor suppressor functions. J. Cell Biol. 2019;218:3060–3076. doi: 10.1083/jcb.201810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripathi B.K., Qian X., Mertins P., Wang D., Papageorge A.G., Carr S.A., Lowy D.R. CDK5 is a major regulator of the tumor suppressor DLC1. J. Cell Biol. 2014;207:627–642. doi: 10.1083/jcb.201405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi B.K., Grant T., Qian X., Zhou M., Mertins P., Wang D., Papageorge A.G., Tarasov S.G., Hunter K.W., Carr S.A., Lowy D.R. Receptor tyrosine kinase activation of RhoA is mediated by AKT phosphorylation of DLC1. J. Cell Biol. 2017;216:4255–4270. doi: 10.1083/jcb.201703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The important data that support the findings of this study are contained within the article; other data are to be shared upon request to the corresponding author: Shawn S.-C. Li, Departments of Biochemistry and Oncology, Schulich School of Medicine and Dentistry, Western University. Canada. E-mail: sli@uwo.ca.