Abstract
Acylation modifications, such as the succinylation of lysine, are post-translational modifications and a powerful means of regulating protein activity. Some acylations occur nonenzymatically, driven by an increase in the concentration of acyl group donors. Lysine succinylation has a profound effect on the corresponding site within the protein, as it dramatically changes the charge of the residue. In eukaryotes, it predominantly affects mitochondrial proteins because the donor of succinate, succinyl-CoA, is primarily generated in the tricarboxylic acid cycle. Although numerous succinylated mitochondrial proteins have been identified in Saccharomyces cerevisiae, a more detailed characterization of the yeast mitochondrial succinylome is still lacking. Here, we performed a proteomic MS analysis of purified yeast mitochondria and detected 314 succinylated mitochondrial proteins with 1763 novel succinylation sites. The mitochondrial nucleoid, a complex of mitochondrial DNA and mitochondrial proteins, is one of the structures whose protein components are affected by succinylation. We found that Abf2p, the principal component of mitochondrial nucleoids responsible for compacting mitochondrial DNA in S. cerevisiae, can be succinylated in vivo on at least thirteen lysine residues. Abf2p succinylation in vitro inhibits its DNA-binding activity and reduces its sensitivity to digestion by the ATP-dependent ScLon protease. We conclude that changes in the metabolic state of a cell resulting in an increase in the concentration of tricarboxylic acid intermediates may affect mitochondrial functions.
Keywords: DNA–protein interaction, lysine succinylation, mitochondria, mitochondrial DNA, mitochondrial nucleoid, post-translational modification (PTM), proteomics, succinylome, yeast
Abbreviations: ACN, acetonitrile; α-KG, α-ketoglutarate; 2HG, 2-hydroxyglutarate; GO, gene ontology; HMG box, high-mobility group box; IP, immunoprecipitation; Ksucc, 6-N-succinyl-l-lysine; mtDNA, mitochondrial DNA; mtHMG protein, mitochondrial high-mobility group box containing protein; mt-nucleoid, mitochondrial nucleoid; MTS, mitochondrial targeting sequence; non-Ksucc, nonsuccinylated lysines; PTM, post-translational modification; TCA, tricarboxylic acid
Mitochondria are semiautonomous organelles that must be flexible in their responses to changes in both extracellular and intracellular conditions. Mitochondrial functions, from basal metabolism to communication with other cellular compartments, are regulated on several levels (1, 2, 3). One of the most dynamic ways to affect the activity of proteins and nucleic acids is to add small chemical groups. For proteins, these modifications occur mostly post-translationally and are called post-translational modifications (PTMs).
The most studied PTM is phosphorylation catalyzed by protein kinases (4). Although the number of protein kinases localized to the yeast mitochondria does not seem to be high (5, 6, 7), an analysis of the mitochondrial phosphoproteomes of numerous eukaryotes, including the yeast Saccharomyces cerevisiae, revealed that mitochondrial proteins are relatively frequently phosphorylated (6, 7, 8, 9, 10, 11, 12). In addition to phosphate, many other chemical groups can be attached to the side chains of amino acids. In mitochondria, the most frequent PTM is the acylation (e.g., acetylation, propionylation, malonylation, glutarylation, succinylation) of lysine residues (13). Acyl groups are donated by the corresponding acyl-CoA, whose concentration is particularly high in mitochondria (14, 15). Moreover, the higher intramitochondrial pH favors the deprotonation of the lysine epsilon amino group, thus increasing its nucleophilicity and reactivity (13). Mitochondrial acylation is mostly nonenzymatic and is driven by increases in acyl-CoA concentration (16, 17).
The properties of the modified lysine are most dramatically changed by its malonylation, glutarylation, and succinylation, that is, acylations that change the charge of the affected residue. Succinylation of lysine results in the formation of 6-N-succinyl-l-lysine (Ksucc). Lysine succinylation was first shown to affect the active site of the Escherichia coli homoserine trans-succinylase (18); later, it became clear that succinylation is a frequently occurring PTM in both prokaryotes and eukaryotes (19, 20). Succinylation is particularly interesting in terms of mitochondrial regulation because the concentration of succinyl-CoA within the mitochondria varies significantly depending on the activity of the enzymes involved in the tricarboxylic acid (TCA) cycle (13, 19).
The succinylation of mitochondrial proteins is not only affected by natural physiological changes but can also be changed by mutations that cause changes in cellular metabolism. For example, the dominant mutations that occur in the genes encoding the NADP-dependent isocitrate dehydrogenases in some types of cancers (21) yield enzymes with altered substrate specificity, which catalyze the NADPH-dependent formation of 2-hydroxyglutarate (2HG) rather than the conversion of isocitrate to α-ketoglutarate (α-KG) (22). The increased concentration of 2HG in the affected cells leads to the inhibition of several key enzymes, including succinate dehydrogenase, which results in an increased concentration of succinyl-CoA (23). Because the addition of glycine, which channels conversion of 2HG into porphyrin, or the overexpression of the desuccinylase SIRT5 slows down the growth of tumor cells, it seems likely that succinylation plays a role in tumorigenesis (23).
There are numerous studies describing the cellular succinylome in a wide variety of prokaryotic and eukaryotic species (e.g., (19, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45)). In yeast, the pioneering analysis of Weinert et al. (19) provided the first glimpse into the succinylome of S. cerevisiae and showed that the succinylation of cellular proteins is affected by the growth conditions and genetic backgrounds that affect the concentration of succinyl-CoA. One of their conclusions was that “succinylation on abundant proteins occurs at a low level and that many succinylation sites remain unidentified”. As mitochondria are thought to be lysine succinylation hot spots, the proteins primarily affected by succinylation can naturally be expected to be the mitochondrial ones. However, with few exceptions (24, 43), most studies on eukaryotic succinylomes were performed on whole-cell extracts. This does provide a global picture of protein succinylation, but it may fail to detect some of the less-abundant protein targets specifically localized to distinct cellular compartments. To gain better insight into the yeast mitochondrial succinylome, we performed an MS analysis of proteins isolated from purified S. cerevisiae mitochondria, and we enriched the proportions of succinylated peptides by immunoprecipitating them with antibodies raised against Ksucc. We detected 314 succinylated mitochondrial proteins and identified 1763 novel succinylation sites. In particular, we found that the majority of the protein components of the mitochondrial nucleoid (mt-nucleoid), which consists of the mitochondrial DNA (mtDNA) compacted into a nucleoprotein structure, are succinylated in vivo. We also demonstrated that the succinylation of Abf2p, the principal component of the S. cerevisiae mt-nucleoid, inhibits its DNA-binding activity and reduces its sensitivity to the action of the mitochondrial ATP-dependent protease ScLon (Pim1p) in vitro. Finally, we discuss how changes in the concentration of succinyl-CoA can alter the compaction of mtDNA and thus might affect the biogenesis of mitochondrial respiratory complexes.
Results
Mitochondrial succinylome of S. cerevisiae
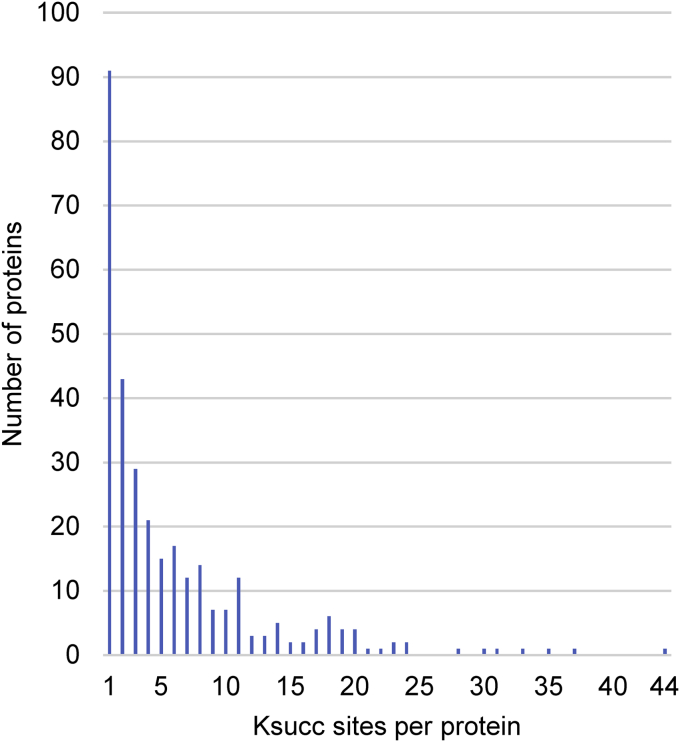
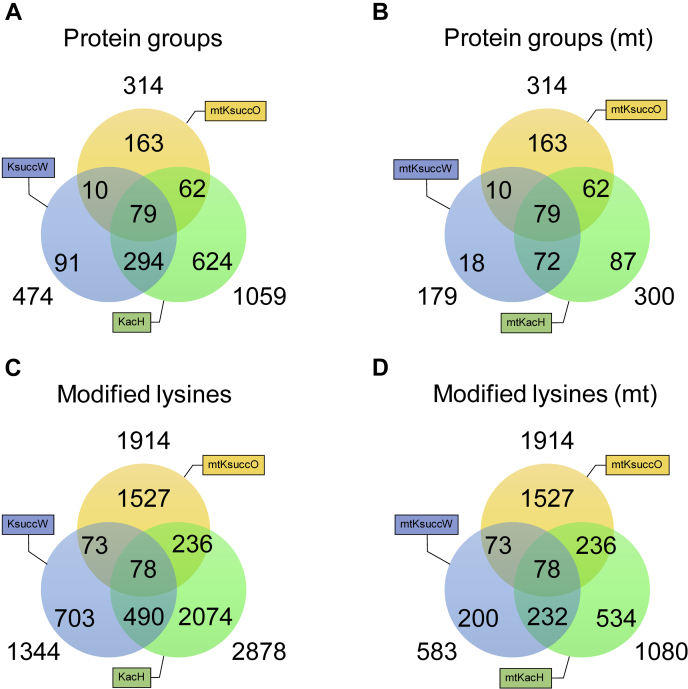
To carry out a comprehensive analysis of the S. cerevisiae mitochondrial succinylome in cells grown on a nonfermentable carbon source (glycerol + ethanol), we purified their mitochondria and prepared mitoplasts lacking the outer mitochondrial membrane and the proteins associated with the intermembrane space. To ensure that the sample to be analyzed by MS would contain a detectable proportion of the less-abundant succinylated proteins, we enriched the proportion of succinylated peptides by immunoprecipitating them with an anti-Ksucc antibody. To increase the number of identified succinylation sites in the WT mitoplasts, we also performed an anti-Ksucc immunoprecipitation (IP) experiment using mitoplasts isolated from a lsc1Δ strain. Disruption of Lsc1p, a succinyl-CoA ligase, leads to an approximate 3-fold increase in overall protein succinylation (19), allowing a larger set of modified peptides to be identified. The results of this experiment served as a template for identifying minor modified peptides in the WT strain using MaxQuant's ‘match-between-runs’ feature. Of 1436 identified protein groups (Table S1, protein groups), 865 were categorized as mitochondrial proteins (Table S2A), 314 of which were succinylated on at least one lysine residue (Table S2B). The number of different Ksucc sites within a single protein group ranged from 1 (91 protein groups) to 44 (the group containing aconitase (Aco1)) (Fig. 1, Table S2B). The identified Ksucc sites and the respective protein groups have been compared with the succinylome identified by Weinert et al. (19) and acetylome identified by Henriksen et al. (46), either globally or with respect to mitochondrial localization (Fig. 2, Table S1F). Our dataset contains 1763 novel Ksucc sites, 236 of which were shown to be acetylated, as well (46).
Figure 1.
Number of Ksucc sites per protein. The succinylated mitochondrial proteins identified in this study were sorted according to the number of Ksucc sites per protein group. Ksucc, 6-N-succinyl-l-lysine.
Figure 2.
Protein lysine acylation dataset comparison. Acyl-modified protein groups (A and B) or sites (C and D) identified in this study (mtKsuccO) were compared with succinylome data of Weinert et al. (19) ((mt)KsuccW) and acetylome data of Henriksen et al. (46) ((mt)KacH). Comparison was general (A and C) or of mitochondrial proteins groups/sites only (B and D). Positions of acylation sites were unaccounted for within the protein group overlap analyses. Ksucc, 6-N-succinyl-l-lysine.
Protein structural determinants of lysine succinylation
To date, no protein lysine succinyltransferase or desuccinylase has been found in yeast mitochondria, nor has any distinct sequence motif been identified. Indeed, as shown in Figure S1, it seems that no such motif exists. Because yeast mitochondrial succinylation is likely to proceed nonenzymatically, with succinyl-CoA serving as the primary donor of the succinyl group, the secondary structural context of the modified residue might have an impact on the likelihood of succinylation (17, 20, 47). To explore this hypothesis, NetSurfP-2.0 (48) was used to predict the structural characteristics of all succinylated (Ksucc) and nonsuccinylated lysines (non-Ksucc) found in this study and these were compared (Table 1). Ksucc sites were found to be significantly more abundant in α-helical regions, whereas their presence in coiled and disordered regions was clearly lower. Reflecting this, the average psi (ψ) angle for Ksucc sites also shows a slight decrease (in the direction of the right-handed α-helix region of the Ramachandran plot (49)) for succinylated residues. Acylation was previously predicted computationally to have the potential to disrupt α-helices, with a consequent effect on enzyme activity (17, 20). To account for possible biases in these characteristics introduced by including the mitochondrial targeting sequences (MTSs), which are normally cleaved upon protein import to mitochondria, the predictions, the analyses, and the statistical tests were also performed using only the Ksucc sites from the regions present in the cleaved mature proteins (the MTSs were predicted by TargetP-2.0 (50)). Apart from the surface accessibility category, the differences between the Ksucc and non-Ksucc characteristics remained statistically significant. Surface exposure is an intuitive predictor of nonenzymatic modification (47), but our data do not confirm its role in mitochondrial lysine succinylation. The most pronounced difference between the Ksucc and non-Ksucc residues was their presence in disordered regions: the likelihood of finding a succinylated lysine was more than 2-fold lower than finding a nonsuccinylated lysine (Table 1). Interestingly, lysine N6-acetylation, unlike some other PTMs, was not found to be correlated with sequence (dis)order (51). Intrinsically disordered proteins and protein regions are known to be involved in the regulation of many processes, including protein–protein and protein–nucleic acid interactions and in protein degradation (for a review, see (52)), but the case for lysine succinylation remains open.
Table 1.
Protein structural determinants of lysine succinylation
| Parameter | Value non-Ksucc (n = 7044) | Value Ksucc (n = 1908) |
Value Ksucc (mt) (n = 1331) |
p-value Ksucc | p-value Ksucc (mt) |
|---|---|---|---|---|---|
| Surface accessibility (relative) | 0.516076 | 0.524961 | 0.521774 | 2.27E-02 | 2.00E-01 |
| Helix (q3) | 0.445350 | 0.521543 | 0.507872 | 2.25E-11 | 2.95E-06 |
| Sheet (q3) | 0.114046 | 0.106577 | 0.117029 | 2.62E-01 | 7.07E-01 |
| Coil (q3) | 0.440603 | 0.371880 | 0.375099 | 1.63E-11 | 3.88E-08 |
| 310 helix (q8) | 0.038865 | 0.035246 | 0.035238 | 1.56E-01 | 2.37E-01 |
| α-helix (q8) | 0.404213 | 0.485332 | 0.472161 | 1.90E-12 | 4.73E-07 |
| π-helix (q8) | 0.003918 | 0.003664 | 0.003419 | 7.18E-01 | 5.73E-01 |
| β-bridge (q8) | 0.006241 | 0.006129 | 0.006593 | 8.32E-01 | 5.94E-01 |
| Extended strand (q8) | 0.107690 | 0.099686 | 0.109870 | 2.21E-01 | 7.80E-01 |
| Bend (q8) | 0.067766 | 0.070762 | 0.072459 | 4.00E-01 | 2.61E-01 |
| H-bonded turn (q8) | 0.099616 | 0.094841 | 0.097013 | 2.99E-01 | 6.38E-01 |
| Coil (q8) | 0.271690 | 0.204339 | 0.203246 | 9.54E-18 | 3.86E-14 |
| ϕ angle | −78.512599 | −77.455463 | −77.554781 | 8.26E-02 | 1.87E-01 |
| ψ angle | 26.115213 | 17.358207 | 20.789474 | 6.22E-06 | 1.97E-02 |
| Disorder | 0.125166 | 0.059944 | 0.055479 | 1.18E-36 | 4.30E-33 |
NetSurfP-2.0 was used to predict a probability value (or angular value) for each lysine within all proteins with at least one Ksucc site identified in this study. The average values for all lysines that were not found to be succinylated (value non-Ksucc) were compared with the average values for all lysines that were found to be succinylated (value Ksucc) and to all lysines that were found to be succinylated in the mature forms of the proteins (i.e., without the MTS, which were predicted using TargetP-2.0 (value Ksucc (mt))). p-values are listed. A significance threshold of p < 0.05 was used, and statistically significant categories are in bold. Surface accessibility was statistically significant in the “Ksucc” category only.
Gene ontology analysis of succinylated proteins
Gene ontology (GO) analysis of succinylated proteins using the PANTHER classification system revealed that components of some of the mitochondrial complexes exhibit >2-fold enrichment. These include respiratory chain complexes (2.13-fold), oxidoreductase complexes (2.26-fold), and mt-nucleoid (2.36-fold). The latter was chosen for a more detailed analysis as (i) it contains a distinct set of well-defined protein subunits (53), (ii) its in vivo properties can be assessed by a direct visualization by means of fluorescence microscopy, (iii) its principal component, the DNA-binding protein Abf2, contains multiple lysine residues that are potential targets of succinylation, (iv) the effect of succinylation on biochemical properties of Abf2p can be monitored by DNA-binding assays in vitro, and (v) changes in DNA-binding properties of Abf2p were shown to correlate with the state of mt-nucleoid.
The possibility that PTMs can play a role in regulation of bacterial and mt-nucleoids has already been discussed elsewhere (54, 55). To test the hypothesis that succinylation may play a role in regulation of mt-nucleoids in S. cerevisiae, we first checked whether a subset of the succinylated mitochondrial proteins is GO-enriched for a biological process, cellular component, or molecular function linked to mtDNA metabolism. A GO overrepresentation test for cellular component using PANTHER did give a statistically significant result for “mt-nucleoid” (raw p-value <0.001, FDR <0.01). However, our experimental procedure (enrichment of Ksucc followed by MS) is likely to be biased toward more abundant proteins, as was demonstrated previously (19). Thus, it is not clear whether this result arises because mt-nucleoid proteins are more frequently succinylated or because the mt-nucleoid proteins are among the most abundant ones in the mitochondria. The proteins associated with the dominant mitochondrial processes also suffer from the same problem (data not shown). This problem might be alleviated by leaving out the Ksucc enrichment step, but without this step, the succinylated peptides are too scarce for reliable measurements.
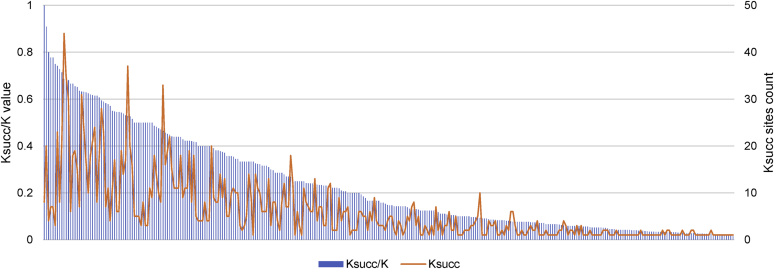
To address this issue, we calculated the ratio between the identified Ksucc sites divided by the total number of lysine residues in each protein (Ksucc/K) and the median abundance of each protein given in the meta-analysis by Ho et al. (56). Ksucc/K of the mitochondrial proteins ranged from 1 (inhibitor of F1FO-ATPase Inh1) to 0 (no Ksucc site detected). The results for each protein with at least one Ksucc site are plotted in Figure 3. Normalizing this number by the number of molecules per cell should allow us to distinguish between those proteins that were detected to be highly succinylated because they are frequently succinylation targets and those proteins that were detected to be highly succinylated simply because they are present in great abundance. Of the mt-nucleoid proteins, the subunits of α-KG dehydrogenase, Kgd2p and Kgd1p, have the highest Ksucc/K to molecules-per-cell ratio (see Fig. S2 for a scatter plot). With a moderate molecules-per-cell count (6516 and 11221, respectively; the median of the Ksucc proteins is 5911) (56) and a relatively high number of lysines (43 and 70, respectively; the median of the Ksucc proteins is 26), this observation is less likely to be biased. In general, the average Ksucc/K to molecules-per-cell ratio of the mt-nucleoid proteins is not higher than that of an average mitochondrial succinyl-protein (Figs. S2 and S3). Importantly, of the 24 annotated mt-nucleoid proteins, only 3 (Cha1p, Rpo41p, and Sls1p) had no detectable Ksucc sites.
Figure 3.
Plot of Ksucc site abundance in succinylated proteins. The value of Ksucc site-per-lysine (Ksucc/K) is plotted for all succinylated mitochondrial proteins identified in this study. The total number of Ksucc sites for each protein is displayed as well. Ksucc, 6-N-succinyl-l-lysine.
Effect of succinylation on mitochondrial HMG box protein Abf2
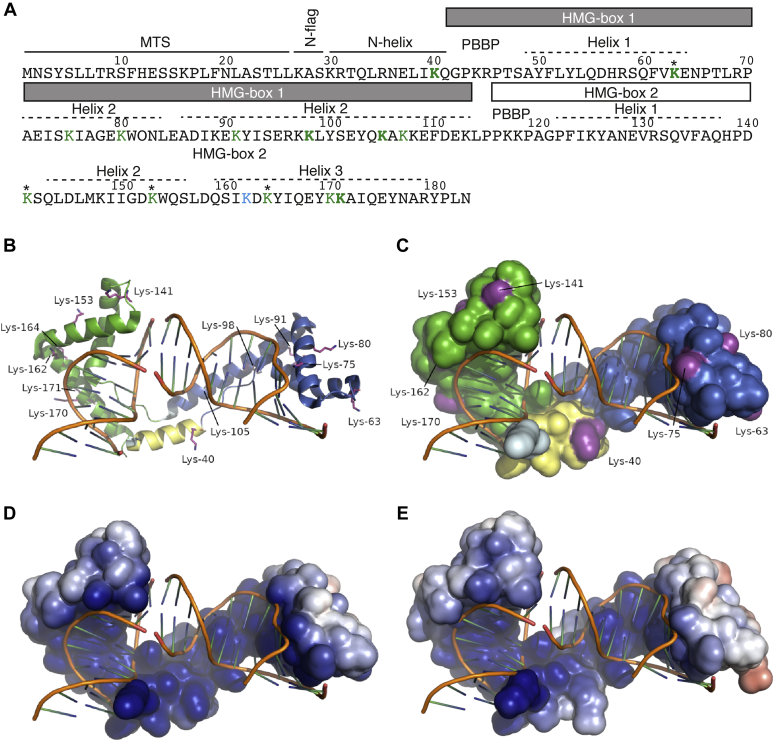
The fact that most mt-nucleoid proteins are succinylated prompted us to investigate the possible functional consequences of this PTM on mt-nucleoid characteristics. Most of the proteins known to associate with the mt-nucleoid are bifunctional (53, 57), so assessing the effect of their succinylation on mt-nucleoid-related functions is rather difficult. One protein that is mainly dedicated to compacting the mtDNA into the mt-nucleoid in S. cerevisiae is Abf2p, a mitochondrial high-mobility group box (HMG box) containing protein (mtHMG protein) that binds and bends mtDNA into higher-order structures (54). The previous proteomic study (19) identified a single succinylated lysine (K162) in Abf2p. We found twelve additional succinylated lysines distributed along the entire protein, but most of them occurred within its two HMG boxes (Fig. 4A). Specifically, as seen in Figure 4B, one of the Ksucc residues is found in the N-helix and six are found in each HMG box. It can also be seen that the second Ksucc in each box is separated from the first by 12 residues, although superimposing HMG box 2 on HMG box 1 does not show them to be structurally equivalent. This superposition does show that K80, K91, and K98 from HMG box 1 are structurally equivalent, respectively, to K153, K164, and K171 from HMG box 2. The succinylation sites are all found on the outer, convex surface of Abf2p; with the possible exception of K162, none are found on the concave, DNA-binding inner surface (Fig. 4C). Five of these residues (K40, K63, K98, K105, and K171) have in vitro–modified/–unmodified ratios of more than 0.25 (Table S3) and are also known to be succinylated in vivo; consequently, an Abf2p carrying Ksucc at these five locations might plausibly be expected to exist in vivo. Calculating the electrostatic surface area shows that the replacement of these five lysine residues with succinyllysine has a modest effect on the electrostatic potential of the protein. As seen in Figure 4, D and E, unsuccinylated Abf2p has a strongly positively charged surface, with an overall charge of +12 e, whereas the replacement of these five residues reduces the positive charge on the surface to only +5 e and leaves the DNA-binding cleft largely unchanged. If all 13 lysines were to be succinylated, the overall surface charge would actually become negative, −1 e. It should be noted here, however, that there is presently no data showing that all 13 succinylations occur at one time on a single molecule.
Figure 4.
An overview of a single monomer bound to DNA from the Abf2p complex structure and a model of its succinylated form. A, sequence of Abf2p with highlighted structural regions based on the 3D structure of the protein (58). Lysines identified in a succinylated form in vivo in this study are in green. K162 identified by Weinert et al. (19) is in blue. The lysine residues having in vitro modified/unmodified ratios of more than 0.25 are in bold (panel E). MS analysis of Abf2p succinylated in vitro identified peptides containing Ksucc corresponding to all lysine residues except K27, K30, and K45. Lysine residues marked with asterisks (∗) were also shown to be acetylated in vivo (46). B, an overview of half the Abf2p–DNA complex. The lysines found to be succinylated in this study are colored magenta, shown as sticks, and labeled. HMG box 1 is colored blue, HMG box 2 is green, the N-helix is yellow, and the N-flag is light cyan. This same coloring is used in the next panel. C, the same complex and the same view as before but showing the solvent-accessible surface area. Not all the succinylated lysines are visible. D, the solvent-accessible surface of Abf2p colored by electrostatic surface potential (the range is ±5 kT/e). E, the solvent-accessible surface of succinylated Abf2p colored by electrostatic surface potential (the same range as before). For the succinylated form, five residues were selected that had the greatest ratio of modified to unmodified forms in vitro and that were also found to be succinylated in vivo. B, basic amino acid; HMG box, high-mobility group box; MTS, mitochondrial targeting sequence.
K162, the previously identified Abf2p succinylation target (19), was also shown to interact with the DNA backbone (58), being the only one of the succinylation targets to do so. Although the remaining succinylated lysine residues do not appear to directly interact with the DNA backbone, they are close to some that do. In particular, K40 is close to K44 and R45, R69 is close to the succinylated K75, W81 is next to K80, K89 (not succinylated in the IP experiments) is close to K91 (which is), and I150 and W154 flank K153. Overall, it appears that succinylation of Abf2p is unlikely to prevent DNA binding sterically but is likely to disfavor it electrostatically.
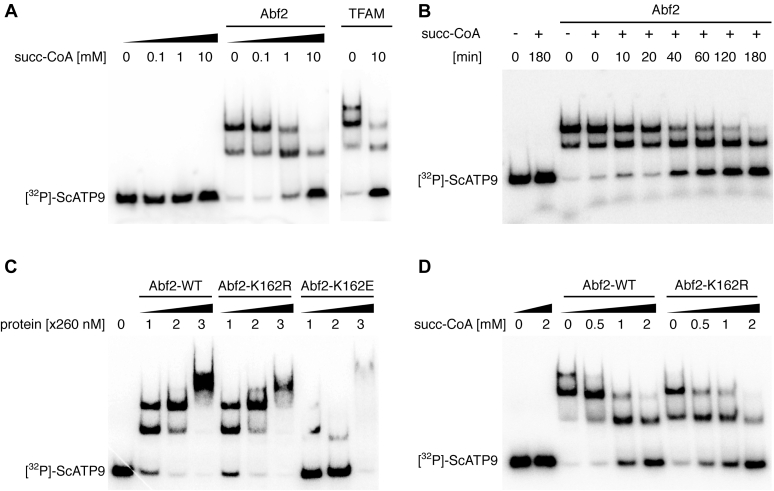
To test the effect of Abf2p succinylation on its DNA-binding properties, we succinylated recombinant Abf2p in vitro using succinyl-CoA as a succinyl group donor and found that it inhibited the DNA-binding activity of the protein in both a time- and concentration-dependent manner (Fig. 5, A and B). The incomplete inhibition may be due to unequal succinylation of the Abf2p molecules present in the reaction mixture and/or affecting dissociation constant of Abf2p to the dsDNA probe. The fact that the effect of succinylation on DNA binding starts to be apparent at concentrations of succinyl-CoA higher than 1 mM is in agreement with the physiological concentrations of this acyl-CoA (14, 15). We also found that the inhibition of Abf2p's DNA-binding is not caused simply by the mere presence of succinyl-CoA in the reaction mixture but is due to the actual modification of the protein (Fig. S4A). In addition, incubating Abf2p with DNA before the succinylation reaction did not affect the inhibitory effect of succinyl-CoA on its DNA-binding activity (Fig. S4B), indicating that, at least in vitro, the binding of Abf2p to DNA does not prevent its succinylation. The MS analysis of Abf2p succinylated in vitro revealed that the majority of the lysines are modified, including those identified in our immunoprecipitated samples (see the legend to Fig. 4A).
Figure 5.
Succinylation of Abf2p in vitro inhibits its DNA-binding activity. A, succinylation of Abf2p and TFAM in vitro inhibits their DNA-binding activity in a concentration-dependent manner. Succinylation was performed at the indicated concentrations of succinyl-CoA (succ-CoA) for 3 h at 30 °C, and then, the DNA-binding activity was assessed by EMSA using radioactively labeled ScATP9 as a probe. B, succinylation of Abf2p in vitro inhibits its DNA-binding activity in a time-dependent manner. Succinylation was performed in the presence of 10 mM succ-CoA at 30 °C for the indicated time periods. C, substitution of K162E affects binding of Abf2p to DNA in vitro. D, Abf2-K162R mutant protein is as sensitive to succinylation as the WT Abf2p.
As Abf2p and its orthologs from several fungal pathogens (including Gcf1p of Candida albicans) were shown to have several acetylated lysine residues (49, 59), we tested whether acetylation also inhibits the DNA-binding activity of Abf2p. To this end, we incubated purified Abf2p with 10 mM acetyl-CoA or 10 mM acetylphosphate, which were shown to be acetyl group donors in nonenzymatic reactions in vitro (60, 61). We observed that, in contrast to succinyl-CoA, preincubation of Abf2p with either acetyl-CoA or acetylphosphate did not result in the inhibition of its DNA-binding activity in vitro (Fig. S5).
Next, we tried to assess the effect of the succinylation of individual lysines on the DNA-binding activity of Abf2p. For the K162 identified by Weinert et al. (19), we generated two mutant versions. In the first version, K162 was substituted for arginine (Abf2-K162R), an amino acid that cannot be succinylated. The second version contained glutamate at this position (Abf2-K162E), which should mimic the succinylation of this site. We found that the Abf2-K162E mutant exhibited different DNA-binding properties than the WT: at lower concentrations, its binding to the DNA probe was negligible, and at higher concentrations, it produced a smear (Fig. 5C). This indicates that the replacement of the positively charged lysine with the negatively charged glutamate (mimicking a negatively charged succinylated lysine) strongly affects the DNA-binding properties of the protein, which is consistent with our hypothesis that Abf2p succinylation greatly disfavors DNA-binding electrostatically. In contrast to Abf2-K162E, the Abf2-K162R mutant bound DNA similarly to the WT protein (Fig. 5C). However, when the Abf2-K162R protein was succinylated in vitro, the extent of its DNA-binding inhibition was similar to that of the WT (Fig. 5D), indicating that the observed decrease in DNA-binding is most likely caused by the succinylation of lysines in different positions within the protein. With the aim of identifying the combination of lysines whose succinylation accounts for the inhibition of Abf2p in vitro, we prepared a series of Abf2p mutants carrying multiple K-to-R substitutions. However, the introduction of these mutations into Abf2p invariably decreased its DNA-binding activity, making it difficult to distinguish between the inhibitory effect of succinylation and that of the K-to-R substitution(s) of the corresponding mutants (Fig. S6) (62).
To test whether the effect of succinylation on the DNA-binding activity in vitro is specific for S. cerevisiae Abf2p, or universal for mtHMG proteins, we purified recombinant versions of similar proteins from three additional yeast species, Candida parapsilosis (CpGcf1p) (63), C. albicans (CaGcf1p) (64), and Yarrowia lipolytica (YlMhb1p) (65). We observed that the effect of succinylation on their DNA-binding activities varied (Fig. S7). Whereas Abf2p and CpGcf1p were significantly inhibited by succinylation in the presence of 10 mM succinyl-CoA, the DNA binding of CaGcf1p and YlMhb1p was not substantially affected (Fig. S7). Furthermore, we found that in vitro succinylation of human mtHMG protein TFAM inhibits its DNA-binding activity to a similar degree as in case of Abf2p (Fig. 5A). Three lysine residues (K-95, K-190, and K-205) were found to be succinylated on TFAM in HeLa cells (19), indicating that lysine succinylation mediated by an increased level of succinyl-CoA may provide another link between metabolism and mt-nucleoid maintenance not only in S. cerevisiae but also in human cells. In contrast, regulation of TFAM via fumarate-mediated succination of a cysteine residue (66, 67) is not reflected by this means of regulation in case of Abf2p that lacks cysteines (Fig. 4A). All these data illustrate that regulation of mtHMG proteins by PTMs exhibits species-specific characteristics.
Based on the Abf2p–DNA complex shown in Figure 4, succinylation decreases the charge of the structure, with most of the effect occurring on the outer surface of the complex, where it could disrupt the formation of higher oligomers. It was previously demonstrated that Abf2p acts as a dimer (68). Because succinylation mostly affects lysines located on the surface of the molecule (Fig. 4), we tested whether it decreased the ability of Abf2p to form dimers. Using size-exclusion chromatography, we found that succinylated Abf2p did not lose the ability to dimerize (Fig. S8).
Succinylated Abf2p is resistant to degradation by ScLon protease in vitro
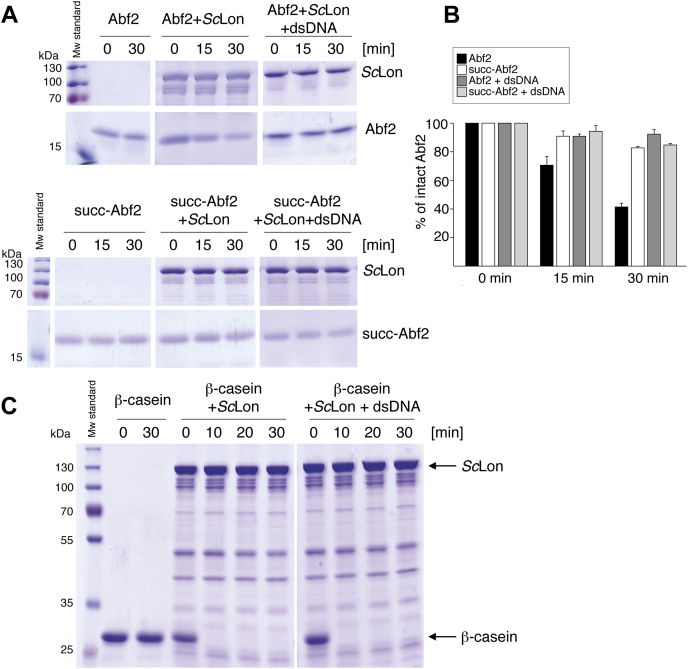
Previously, it was demonstrated that, when not bound to mtDNA, Abf2p is degraded by the ATP-dependent protease ScLon, which is another component of yeast mt-nucleoids (68). We tested the effect of succinylation on the sensitivity of Abf2p to degradation by ScLon. We observed that Abf2p succinylated in vitro was more resistant to ScLon-mediated proteolysis than unmodified Abf2p (Fig. 6, A and B). Namely, the treatment of Abf2p with ScLon protease resulted in about 60% reduction of the amount of the protein (black bars in Fig. 6B), whereas succinylated Abf2p was mostly protected from degradation by the protease (white bars). Moreover, the addition of dsDNA, which protects unsuccinylated Abf2p from ScLon-mediated proteolysis, did not affect the susceptibility of the succinylated protein to degradation (Fig. 6, A and B). Because succinylated Abf2p is predominantly in a DNA-free form regardless of the presence of DNA, its resistance to proteolysis must be due to the succinylation. Moreover, the presence of dsDNA did not affect the protease activity of ScLon toward β-casein (Fig. 6C).
Figure 6.
Succinylated Abf2p is less sensitive to digestion by the ATP-dependent ScLon protease. Nonsuccinylated (Abf2p) or in vitro–succinylated (succ-Abf2p) protein was incubated with purified ScLon in the presence or absence of a dsDNA probe. A, the course of an Abf2p ScLon-mediated digestion in reaction mixtures containing 2 mM ATP and 10 mM MgCl2 on Coomassie Brilliant Blue–stained gels. The reactions were terminated at the indicated times and the samples loaded onto a 12% SDS-polyacrylamide gel. B, quantification of the digestion experiments shown in panel A. Band intensity evaluation of undigested Abf2p (Abf2 (%)) was performed with ImageJ (v.1.53) (104) separately for each studied protein. The height of a given bar represents the mean value, and the error bars show ± one SD from at least three separate measurements. C, the course of β-casein digestion by ScLon. β-casein was incubated with ScLon in the presence or absence of a dsDNA in a reaction mixture containing 2 mM ATP and 10 mM MgCl2 for indicated times, and the proteins were separated on a 12% SDS-polyacrylamide gel.
Discussion
Since its discovery in 2004 (18), lysine succinylation was found to affect a substantial part of the prokaryotic and eukaryotic proteomes. Because it is driven by the intracellular concentration of succinyl-CoA, it links metabolism to global changes in the PTMs of cellular proteomes, including those of cancer cells. The proteome of S. cerevisiae is not an exception, and many of its members were shown to be succinylated (19). The effects of succinylation were studied primarily on yeast histones, as this PTM provides another set of epigenetic marks regulating the chromatin state (69, 70, 71).
Mitochondria are the predominant sites of succinyl-CoA production, which arises from the decarboxylation of α-KG during the TCA cycle; it is therefore expected that mitochondrial proteins are highly prone to succinylation. However, a study of the yeast mitochondrial succinylome based on a proteomic analysis of purified mitochondria has been lacking. To observe the effects of protein succinylation on respiring mitochondria, we analyzed the succinylation state of proteins extracted from yeast mitoplasts that were prepared from cells grown on nonfermentable (glycerol + ethanol) carbon sources rather than on the fermentable (glucose or galactose) ones used by previous studies. As a result, we produced the first comprehensive list of S. cerevisiae mitochondrial proteins modified by succinylation. We identified 314 mitochondrial proteins containing 1914 succinylated sites, 92.1% of which were not previously reported. From our data, it is not possible to assess the stoichiometry of succinylation even in the case of proteins containing a single Ksucc. Proteins with ≥2 Ksucc sites may constitute a heterogeneous population consisting of polypeptides carrying different combinations of Ksucc residues. As different patterns of succinylation (and PTMs including phosphorylation and other acylations) can result in a given protein having different biochemical properties, this may contribute to a significant increase in the complexity of the mitochondrial proteome.
Although we expanded the list of yeast succinylated mitochondrial proteins, it is likely that a substantial number of succinylation sites remain unidentified. This might be due in part to the low abundance of some proteins, whose succinylation may be below detection limit of our procedure. The succinylation pattern may also be influenced by the physiological conditions of the cell population, leading to different local concentrations of succinyl-CoA. Furthermore, the availability of potential acceptor lysine residues may differ because of their interactions with other intramitochondrial components (proteins, DNA, membrane), which may hinder their accessibility to succinyl-CoA. One possibility for increasing the overall level of succinylation is to boost the intracellular concentration of succinyl-CoA. To this end, we determined the mitochondrial succinylome of a S. cerevisiae strain lacking the α-subunit of the succinyl-CoA ligase Lsc1p. It was shown previously that a lsc1Δ mutant exhibits about a 3-fold higher level of succinylated proteins (19). Although succinylated proteins were indeed enriched in the mitoplast extract obtained from the lsc1Δ mutant, the list of succinylated proteins overlapped the WT mitochondrial succinylome to a large extent (Table S1). Combined with the fact that the increase in succinyl-CoA concentration in lsc1Δ is achieved by nonphysiological means, we decided to focus on the dataset obtained from the WT cells.
Based on our analysis, succinylation occurs predominantly on lysine residues located in ordered and α-helical regions, whereas lysines within coiled regions are less likely to be affected. As expected for a nonenzymatic modification, lysines exposed to the surface are slightly more prone to succinylation (47). Indeed, when we mapped the succinylated lysines on the 3D structure of Abf2p, all of them (with the possible exception of K162) were found on the outer surface of the protein (Fig. 4, B–E, see also below). This illustrates that it is physical proximity rather than a specific amino acid sequence motif that makes particular lysines susceptible to succinylation, which supports ongoing efforts to develop succinylation prediction algorithms based on secondary structures and evolutionary information rather than on simply trying to identify sequence motifs (72).
Because the majority of the components of the mt-nucleoid were succinylated, it is likely that this PTM plays a role in their metabolism. To investigate the possible functional importance of succinylation in the regulation of the mt-nucleoid, we analyzed the effect of succinylation on the DNA-binding activity of Abf2p, known to mediate the bending and compacting of mtDNA in S. cerevisiae (reviewed in (54)). Of the twenty-five lysine residues present in the mature form of Abf2p (lacking the MTS) (Fig. 4A), thirteen were succinylated in vivo. This prompted us to investigate the effect of succinylation on the DNA-binding activity of Abf2p in vitro. We found that incubation of Abf2p with succinyl-CoA, but not with acetyl-CoA or acetylphosphate, inhibited its binding to the DNA probe in EMSA experiments (Figs. 5, A and B and S5) and that this inhibition is due to the modification of the protein and not to the presence of succinyl-CoA in the reaction mixture (Fig. S4). MS analysis revealed that all the succinylated lysines identified by analyzing immunoprecipitated Abf2p are also modified in vitro (Fig. 4A). To assess whether there are any specific lysine residues whose succinylation affects DNA-binding properties of Abf2p, we tested several mutant versions carrying lysine to arginine substitutions, which cannot be succinylated. Almost all of these mutants exhibited decreased DNA-binding activity in vitro, even without succinyl-CoA treatment (62). This excluded the possibility of investigating the effect of succinylation on individual lysine residues on the ability of Abf2p to compact mt-nucleoids in vivo, as it would be impossible to distinguish between the phenotypic effects of the K-to-R substitutions themselves from the effects arising from their lack of succinylation. On the other hand, the substitution of a single lysine (K162) with glutamate (mimicking the negative charge of Ksucc) in the Abf2-K162E mutant greatly affected its DNA-binding activity (Fig. 5C). This indicates that the succinylation of only one or a very few lysines can have a profound effect on binding of Abf2p to mtDNA.
In vitro succinylated Abf2p not only exhibits decreased DNA-binding activity but also is more resistant to the ScLon protease (Fig. 6). It has been shown that the initial cleavage sites of human Lon protease are preferentially between hydrophobic amino acids located within highly charged environments on the surface of the folded protein (73). Assuming that ScLon cleaves its substrates in a similar manner, succinyl groups on lysines found predominantly on the surface of the Abf2p dimer might prevent ScLon from recognizing its characteristic surface sites and thus inhibit the initiation of proteolysis.
The mature form of Abf2p is a relatively basic protein (predicted pI = 9.68) containing in total 33 lysines plus arginines. Lysines are the prevalent (78.8%) basic residues in its HMG boxes. This bias toward lysines is not a common feature of all basic DNA-binding proteins, however. For example, S. cerevisiae histones Hho1p (H1), Htb1p (H2B), and Htb2p (H2B) exhibit this bias, but in Hht1p (H3), Hht2p (H3), Hta1p (H2A), and Hta2p (H2A), the lysine-to-arginine ratio is approximately 1. Similarly, there is no bias toward lysines in E. coli nucleoid–associated proteins HU, IHF, H-NS, or FIS, yet lysines constitute still at least 7% of total amino acid residues of these DNA-binding proteins. Perhaps, lysine-rich DNA-binding proteins are more susceptible to modifications such as succinylation, which may affect their DNA binding and interactions with other proteins. Because a substantial fraction of lysines is located on the surface of Abf2p, the addition of a succinyl group to a lysine residue would change the Abf2p surface, thereby potentially affecting its interactions with other mt-nucleoid proteins.
Because the in vitro succinylation of mtHMG proteins from four different yeast species has different effects on their DNA-binding activity (Fig. S7), this PTM is not universally adopted as a means of regulating their activity. Even mtHMG proteins from relatively closely related species (CaGcf1 and CpGcf1) differ in the effect that succinylation has on their DNA-binding ability. Furthermore, the lysines identified as succinylated on Abf2p in vivo are not conserved in species closely related to S. cerevisiae (see Fig. S1 in (54)). Although it is impossible to generalize based on this single example, it is conceivable that this kind of species-specific regulation of a protein by succinylation might be more common.
An increase in the activity of the TCA cycle results in changes in the concentration of its intermediates, including succinyl-CoA. For example, in E. coli, the concentrations of succinyl-CoA range from 0.233 mM, when cells are grown on glucose, to 1.44 mM, when cells are grown on glycerol (74). S. cerevisiae is a Crabtree-positive yeast species that when grown on glucose, first ferments the sugar to ethanol and then, after a short metabolic adaptation called diauxic shift, enters a respiratory phase of growth. This transition is accompanied by several changes in mitochondrial function and structure and reflected by reorganization of mitochondrial metabolism including the activation of the TCA cycle (75). When we inspected mt-nucleoids in cells taken from fermentative, diauxic shift and respiratory phases of the growth, we observed dramatic changes in both their number per cell and their appearance (Fig. S9). Of note, the subunits of α-KG dehydrogenase (Kgd1p and Kgd2p), the principal enzymes of the TCA cycle, were shown to be components of the S. cerevisiae mt-nucleoid (53, 57, 76), and thus, succinyl-CoA could be generated in its close proximity, thereby promoting succinylation of the mt-nucleoid-associated proteins. In fact, it was shown that α-KG dehydrogenase may act as a catalyst of the succinylation reaction (71, 77), so it may directly facilitate the modification of those proteins in its vicinity, thus affecting their biochemical properties. In the case of Abf2p, succinylation would decrease its ability to bind mtDNA and thus promote changes in mt-nucleoids observed during the diauxic shift. Of note, succinylation of several nucleoid-associated proteins was suggested to affect their DNA binding, structural integrity, or interaction with other proteins in E. coli (55), indicating that this model might be also relevant for the maintenance of the bacterial chromosome. Alternatively (or in parallel), succinylation of components of the respiratory complexes and/or the ATP synthase might result in their altered activity that could counterbalance the overactive TCA cycle during the respiratory growth phase. Future comparative studies aimed at careful quantification of succinylome of cells under defined metabolic states will shed more light on the functional significance of this PTM for linking metabolism and other mitochondrial (or cellular) functions.
By providing a comprehensive list of succinylated S. cerevisiae mitochondrial proteins and functional analysis of one of its putative targets, our study represents a primer for the systematic investigation of the functional importance of succinylation in mitochondria-related activities.
Experimental procedures
Microbial strains
S. cerevisiae strain SCY325 (MATα, ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1) (78) and its lsc1Δ derivative (MATα, ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1 lsc1Δ::LEU2) were used for the purification of mitoplasts. An lsc1Δ strain lacking the entire ORF for the α-subunit of succinyl-CoA ligase Lsc1p was derived from the SCY325 strain by one-step recombinational deletion using an LEU2 cassette. The deletion cassette was amplified from the genomic DNA of S. cerevisiae strain GG595 (MATα/MATa HO/HO ura3/ura3 (provided by H. Yde Steensma, Leiden University, The Netherlands)) by PCR using the LSC1_DC_LEU2_F and LSC1_DC_LEU2_R primers (Table S4) and Phusion Hot Start II DNA Polymerase (Thermo Fisher Scientific). Yeast cells were transformed using the LiAc/PEG/ssDNA method (79, 80), selected based on leucine prototrophy, and the cassette insertion was verified by colony PCR using the pLSC1_5_F and pLEU2_5_R (5′ region) and pLEU2_3_F and pLSC1_3_R (3′ region) primers (Table S4). The ScATP9 probe was amplified using a DNA template prepared from the S. cerevisiae strain W303-1A (MATa, leu2-3112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15), which was from the collection of the Departments of Genetics and Biochemistry, Comenius University in Bratislava. For ScLon purification, the strain pSEY c68-ScLON expressing the WT ScLon gene from the GAL1 promoter of the centromere-based plasmid pSEYc68 was used (81). E. coli strains BL21 Star (F−, ompT, hsdS, r−m−, gal, dcm, rne131(DE3)), Rosetta 2 (DE3) (F−, ompT, hsdSB, rB−m−, gal, dcm (DE3), pRARE2 (CamR)), and DH5α (F−, ϕ80dlacZΔM15, Δ(lacZYA-argF) U169 deoR, recA1, endA1, hsdR17 (rk−, mk+), λ, thi-1, gyrA96, relA1, glnV44, nupG) were purchased from Life Technologies.
Mitoplast preparation
S. cerevisiae strain SCY325 was cultivated in YPGE medium (yeast extract (1% (w/v)), Bacto Peptone (2% (w/v)), glycerol (3% (v/v)), ethanol (2% (v/v))). Mitochondria were isolated as described (82). Mitoplasts were prepared by the addition of 20 mM Hepes-KOH (pH 7.4) in a 10:1 volume ratio to the mitochondrial pellet. The suspension was incubated on ice for 30 min, centrifuged at 12,000g at 4 °C, and washed twice with ice-cold 0.6 M sorbitol buffer with 20 mM Hepes-KOH (pH 7.4). Freshly prepared mitoplasts were stored at −80 °C for further use.
For diauxic shift experiment, the cells of the S. cerevisiae strain SCY325 were inoculated at density of A600 = 0.2 into YPD medium (yeast extract (1% (w/v)), Bacto Peptone (2% (w/v)), glucose (2% (w/v))). The growth was monitored by measuring A600, and simultaneously, the glucose levels were determined as described (83).
Peptide preparation for Ksucc enrichment and MS analysis
Six milligrams of mitoplasts per WT and lsc1Δ biological triplicates were freeze-thawed; proteins were precipitated with 5 volumes of −20 °C acetone and sedimented (16,100g, 30 min, 4 °C). Pellets were washed 3 times and air-dried. Proteins were dissolved in 6.4 M urea, reduced with 5 mM DTT, and alkylated with 40 mM chloroacetamide. After dilution in 3 volumes of triethylammonium bicarbonate buffer, pH 8.0 (50 mM final concentration), the proteins were trypsin-digested (1:20 w/w) overnight. The acidified (with 0.5% (v/v) TFA) and oxidized (with 100 mM hydrogen peroxide) peptide solution was clarified by centrifugation and purified on a reversed-phase column (Supelclean LC-18 SPE; pre-washed with acetonitrile (ACN) followed by 0.1% (v/v) TFA). TFA-washed peptides (0.1%) were eluted from the matrix with TA70 (70% (v/v) ACN, 0.03% (v/v) TFA). After pH adjustment using Tris-HCl, pH 8, the eluate was divided, so that 5% of the sample could be used for the MS analysis of the protein background and 95% for Ksucc enrichment. The peptide aliquots were vacuum dried.
Enrichment of Ksucc-containing peptides for MS analysis
Peptides were dissolved in the IP buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, 0.5% (v/v) NP-40) with cOmplete Protease Inhibitor Cocktail (Roche), and the suspension was cleared by centrifugation. Peptides were incubated overnight at 6 °C with 30 μl of drained and PBS-washed Pan anti-succinyllysine antibody agarose conjugated resin (PTM Biolabs). The resin-bound peptides were washed using the IP buffer with and without 0.5% (v/v) NP-40 and using 50 mM ammonium bicarbonate. Succinylated peptides were eluted with 0.1% (v/v) TFA and vacuum-dried.
MS analyses
Peptide samples in biological triplicates, from both total WT and lsc1Δ mitoplast protein digests and their corresponding Ksucc enrichments, were dissolved in 0.1% (v/v) TFA and 2% (v/v) ACN. The samples were loaded onto a trap column (PepMap100 C18, 300 μm × 5 mm, Dionex) and separated with a C18 column (Acclaim PepMap C18, 75 μm × 500 mm, Dionex) on an UltiMate 3000 RSLCnano system (Dionex) in a 120-min gradient (3–43% B), curve 7, and a flow rate of 300 nl/min. The two mobile phases used were (A) 0.1% (v/v) formic acid and (B) 80% (v/v) ACN with 0.1% (v/v) formic acid. Eluted peptides were sprayed directly into an Orbitrap Elite mass spectrometer (Thermo Scientific), and spectral datasets were collected in a data-dependent mode using the Top15 strategy to select precursor ions for higher-energy collisional dissociation fragmentation (84). Each of the samples was analyzed in three technical replicates. The resulting datasets were processed by MaxQuant, version 1.5.3.30 (85), using the built-in Andromeda search engine with carbamidomethylation (C) and oxidation (M) set as permanent modifications. For the protein background, unmodified lysine and protein N-terminal acetylation were set as variable modifications permitting 2 missed tryptic cleavages. The variable search parameters for the Ksucc-enriched samples were lysine succinylation and protein N-terminal acetylation permitting four missed tryptic cleavages. The search was performed against a S. cerevisiae protein database downloaded on May 25, 2019, from https://www.uniprot.org/ containing 6049 entries. Identified protein groups, succinylation sites, and a summary of the analysis are in Table S1.
Data analyses
Only MS/MS-identified protein groups belonging to the high-confidence mitochondrial proteomes from Di Bartolomeo et al. (75), Morgenstern et al. (86), and Vögtle et al. (87) were considered. The same datasets were also used to annotate the localization of succinylated proteins by Weinert et al. (19) and acetylated proteins by Henriksen et al. (46) for comparative analysis. GOs were analyzed using either the PANTHER Overrepresentation Test tool (binomial test) (88) or YeastMine GO enrichment (Holm-Bonferroni correction) (89). The mitochondrial proteins detected in this study served as the reference. The medians for protein-per-cell abundances are from (56). Systematic gene name annotations and amino acid sequences were taken from the Saccharomyces Genome Database (90). The Saccharomyces Genome Database list of “mt-nucleoid” cellular component genes (GO:0042645) was also used. The sequence logos of the regions adjacent to succinylated lysine sites (the sequence windows were imported from MaxQuant) were analyzed using WebLogo 3.7.4 (91) and Seq2Logo 2.0 (92). Protein structural features were predicted using the MMseqs method of NetSurfP-2.0 (48). The differences between the predicted values of the properties of succinylated and nonsuccinylated lysines within succinylated proteins were evaluated using two-tailed t-tests with unequal variances (Microsoft Excel). MTSs were predicted by TargetP-2.0 (50).
DNA manipulations
Oligonucleotides (Table S4) were synthesized by Microsynth AG. The plasmids pGEX-6P-2-ScABF2noMP, pGEX-6P-2-YlMHB1noMP, and pGEX-6P-2-CpGCF1noMP are described in (65, 93), and the plasmid pGEX-6P-2-CaGCF1noMP in (64). Plasmid expression vectors carrying substitutions within the ABF2 sequence were constructed by inverse PCR using the corresponding pairs of 5′-end phosphorylated primers and pGEX-6P-2-ScABF2noMP (for Abf2_K162R, Abf2_K162E, and Abf2_K117,118R), pGEX-6P-2-ScABF2noMP-K117,118R (for Abf2_K44,117,118R), and pGEX-6P-2-ScABF2noMP-K162R (for Abf2_4K-4R) as templates (Table S4). After amplification by Phusion HF DNA polymerase (Thermo Scientific), the PCR fragments were ligated by T4 DNA ligase (Thermo Scientific) and transformed into E. coli DH5α, and the plasmids were isolated using the plasmid QIAprep Spin Miniprep kit from Qiagen. The sequence of each construct was verified by Sanger DNA sequencing in Microsynth AG.
Effect of succinylation on the DNA-binding activity of yeast mtHMG proteins assessed by EMSA
Recombinant Abf2p, its mutant variants, other yeast mtHMG proteins, and human TFAM used for DNA-binding assays were purified from extracts from E. coli BL21 Star (DE3) cells transformed with the corresponding plasmid construct as described (68, 93). Protein concentration was determined using the Bradford assay (94), and the purity was assessed by 12% SDS-PAGE (95). The 50-bp-long DNA substrate derived from the S. cerevisiae atp9 gene was amplified by PCR from the genomic DNA of S. cerevisiae strain W303-1A using the DreamTaq DNA polymerase (Thermo Scientific) and primers ScATP9_15_D and ScATP9_50_R (Table S4); the resulting dsDNA was 5′ end-labeled using T4 polynucleotide kinase (Thermo Scientific) and [γ32P]ATP (3000 Ci/mmol). EMSA experiments using 260 nM of the tested DNA-binding proteins (unless stated otherwise) were performed as described (93). Succinylation in vitro was performed at 30 °C in the presence of the indicated concentrations of succinyl-CoA (Sigma-Aldrich).
Expression and purification of ScLon
For ScLon isolation and purification, the S. cerevisiae Δlon strain carrying the plasmid pSEY c68-ScLON (81) was grown to the early stationary phase in yeast synthetic medium (0.67% (w/v) yeast nitrogen base with ammonium sulfate) supplemented with 0.2% (w/v) glucose, 1.5% (w/v) galactose, 0.5% (w/v) casamino acids (Difco), 0.002% (w/v) adenine, and 0.002% (w/v) tryptophan. Mitochondria were isolated and purified as described (82) and ScLon carrying a C-terminal 6× His tag was purified as described previously (81). In brief, mitochondria corresponding to 10 mg/ml of proteins were resuspended in buffer A (20 mM Hepes-KOH, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 20% (v/v) glycerol) containing 1 mM ATP and 1.6 mg of Lubrol/mg of mitochondrial protein. The lysate was centrifuged at 200,000g at 4 °C for 10 min, and the supernatant was applied to a Ni-NTA Agarose resin (Qiagen). The column was washed with five column volumes of buffer A containing 40 mM imidazole, and bound protein was then stepwise eluted with buffer A containing 0.1, 0.2, and 0.3 M imidazole. The enzymatic activity of the purified ScLon was then determined by β-casein degradation as described (68).
ScLon proteolytic assay
For proteolytic assays, recombinant S. cerevisiae Abf2p lacking the MTS (first 26 amino acid residues) and carrying an N-terminal His tag (6× His-GST) (96) was purified from E. coli Rosetta 2 (DE3) as described previously (68). In brief, the cells were resuspended in buffer B (50 mM ammonium acetate, pH 7.3, 1 M KCl, 10% (v/v) glycerol) with the addition of 0.1% (v/v) Tween 20 and 0.2 mg/ml lysozyme followed by a short incubation at room temperature (RT) and sonication on ice. The cell lysate was centrifuged (30,000g, 30 min, 6 °C), and the supernatant was loaded onto a Ni-Sepharose 6 Fast Flow column (GE Healthcare). The eluted protein was incubated overnight with PreScission protease (GE Healthcare) according to the manufacturer's instructions and then applied to a Glutathione Sepharose 4 Fast Flow column (GE Healthcare). Abf2p was eluted in the unbound fraction, diluted (2.5×) with buffer B (without KCl), and loaded onto a Heparin Sepharose 6 Fast Flow column (GE Healthcare). The bound protein was eluted with a stepwise KCl gradient, concentrated, and purified on a Superdex 75 10/300GL column (GE Healthcare). Succinylated Abf2p for proteolytic assays was prepared by incubating purified Abf2p with 10 mM succinyl-CoA in 20 mM Tris HCl (pH 7.4), 50 mM NaCl, and 1 mM EDTA for 2 h at RT. Free succinyl-CoA was removed by gel filtration on a Superdex 75 10/300GL column (GE Healthcare) equilibrated with buffer C (50 mM ammonium acetate (pH 7.3), 500 mM KCl, 5% (v/v) glycerol). The given amounts of substrate proteins (Abf2p, succinylated Abf2p, casein) were incubated in an ScLon reaction buffer (50 mM Tris HCl, pH 8.5, 10 mM MgCl2, 2 mM ATP). A 750-bp DNA fragment (KpnI/HindIII) from the pOPINM-XRCC6BP1 plasmid construct (4 μM) was used as the dsDNA probe. Each reaction was carried out at 30 °C for 30 min with aliquots withdrawn at the indicated times. Reaction products were analyzed by SDS-PAGE in 12% polyacrylamide gels and Coomassie Brilliant Blue R250 staining.
Succinylated Abf2p modeling
The model of succinylated Abf2p was built by manually combining the coordinates of the succinyl moiety of Ksucc from a succinylated histone 3 peptide in complex with sirtuin-5 (PDB ID: 3RIY) (97) with the side chains of those lysines from the previously reported 2.18 Å Abf2p structure (PDB ID: 5JH0) (58) that were found to be succinylated in our IP assays. After torsion angle adjustments to remove steric clashes introduced by these modifications, this structure's geometry was regularized by Refmac5 (98) using a geometry database derived from the Ksucc of 3RIY. The resulting structure was then minimized against the AMBER 14 forcefield (99) in Chimera (100). The geometry of the resulting structure was checked using MolProbity (101) and was found to be sufficient for the purposes of this study. The electrostatic surface of both succinylated and nonsuccinylated Abf2p was calculated using PDB2PQR and APBS (102). Coordinate manipulation and structure visualization were done using the PyMOL Molecular Graphics System, version 2.3.0, Schrödinger, LLC.
Data availability
The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (103) partner repository with the dataset identifier PXD023604 (username: reviewer_pxd023604@ebi.ac.uk; password: zMPT8wo0).
Supporting information
This article contains supporting information (19, 46, 56, 75, 83, 86, 87, 91, 92, 105).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Filip Tomáška for quantitative analysis of mitochondrial nucleoids and Dr Martin Valachovič (Institute of Animal Biochemistry and Genetics, Centre of Biosciences, Slovak Academy of Sciences, Bratislava, Slovakia) and Dr H. Yde Steensma (Leiden University, The Netherlands) for kindly providing the S. cerevisiae strains SCY325 and GG595, respectively.
This study was supported by the Operation Program of Integrated Infrastructure for the project Advancing University Capacity and Competence in Research, Development and Innovation, ITMS2014+: 313021X329, cofinanced by the European Regional Development Fund and with the support of the Operational Program Integrated Infrastructure for the project Center for Biomedical Research–BIOMEDIRES II stage, ITMS: 313011W428, and with the support of the Interreg V-A Slovakia-Austria program (www.sk-at.eu) for the project StruBioMol, ITMS: 305011X666 and cofinanced by the European Regional Development Fund. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2018127, is gratefully acknowledged for the financial support of the measurements at the CEITEC Proteomics Core Facility.
Author contributions
J. F. and L. T. conceptualization; J. F., B. K., J. B., N. K., N. C., K. H., J. A. B., G. O., V. L., K. P., P. B., and L. T. investigation; J. F., N. K., N. C., J. A. B., K. P., and L. T. visualization; J. F., B. K., J. B., N. K., K. H., J. A. B., V. P., P. B., and L. T. methodology; J. F., B. K., N. K., and L. T. writing–original draft; J. F., B. K., J. B., N. K., N. C., K. H., J. A. B., G. O., V. L., B. S., V. P., K. P., J. N., P. B., E. K., and L. T. writing–review and editing; J. B., V. L., B. S., and Z. Z. formal analysis; Z. Z., V. P., J. N., P. B., E. K., and L. T. resources; Z. Z., V. P., J. N., P. B., E. K., and L. T. funding acquisition; V. P., P. B., E. K., and L. T. supervision; V. P., P. B., E. K., and L. T. project administration.
Funding and additional information
Funding was provided by the Slovak Research and Development Agency (APVV) (APVV-15-0022 and APVV-19-0068 [to L. T.], APVV-18-0239 [to J. N.], APVV-15-0375 and APVV-19-0298 [to E. K.]) and the Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic (VEGA) (1/0061/20 [to L. T.], 1/0027/19 [to J. N.], 2/0075/18 [to E. K.]).
Edited by John Denu
Footnotes
Present address for Nikola Čanigová: Institute of Science and Technology Austria, Klosterneuburg, Austria.
Contributor Information
Peter Baráth, Email: chempeto@savba.sk.
Eva Kutejova, Email: eva.kutejova@savba.sk.
Lubomir Tomaska, Email: lubomir.tomaska@uniba.sk.
Supporting information
References
- 1.Cagin U., Enriquez J.A. The complex crosstalk between mitochondria and the nucleus: What goes in between? Int. J. Biochem. Cell Biol. 2015;63:10–15. doi: 10.1016/j.biocel.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 2.Guaragnella N., Coyne L.P., Chen X.J., Giannattasio S. Mitochondria-cytosol-nucleus crosstalk: Learning from Saccharomyces cerevisiae. FEMS Yeast Res. 2018;18:foy088. doi: 10.1093/femsyr/foy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfanner N., Warscheid B., Wiedemann N. Mitochondrial proteins: From biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 2019;20:267–284. doi: 10.1038/s41580-018-0092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves J.D., Krebs E.G. Protein phosphorylation and signal transduction. Pharmacol. Ther. 1999;82:111–121. doi: 10.1016/s0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 5.Tomáška Ľ. Mitochondrial protein phosphorylation: Lessons from yeasts. Gene. 2000;255:59–64. doi: 10.1016/s0378-1119(00)00315-2. [DOI] [PubMed] [Google Scholar]
- 6.Rao S., Gerbeth C., Harbauer A., Mikropoulou D., Meisinger C., Schmidt O. Signaling at the gate: Phosphorylation of the mitochondrial protein import machinery. Cell Cycle. 2011;10:2083–2090. doi: 10.4161/cc.10.13.16054. [DOI] [PubMed] [Google Scholar]
- 7.Frankovsky J., Vozáriková V., Nosek J., Tomáška Ľ. Mitochondrial protein phosphorylation in yeast revisited. Mitochondrion. 2021;57:148–162. doi: 10.1016/j.mito.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Guo X., Niemi N.M., Coon J.J., Pagliarini D.J. Integrative proteomics and biochemical analyses define Ptc6p as the Saccharomyces cerevisiae pyruvate dehydrogenase phosphatase. J. Biol. Chem. 2017;292:11751–11759. doi: 10.1074/jbc.M117.787341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo X., Niemi N.M., Hutchins P.D., Condon S.G.F., Jochem A., Ulbrich A., Higbee A.J., Russell J.D., Senes A., Coon J.J., Pagliarini D.J. Ptc7p dephosphorylates select mitochondrial proteins to enhance metabolic function. Cell Rep. 2017;18:307–313. doi: 10.1016/j.celrep.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinders J., Wagner K., Zahedit R.P., Stojanovski D., Eyrich B., van der Laan M., Rehling P., Sickman A., Pfanner N., Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol. Cell. Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Renvoisé M., Bonhomme L., Davanture M., Valot B., Zivy M., Lemaire C. Quantitative variations of the mitochondrial proteome and phosphoproteome during fermentative and respiratory growth in Saccharomyces cerevisiae. J. Proteomics. 2014;106:140–150. doi: 10.1016/j.jprot.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt O., Harbauer A.B., Rao S., Eyrich B., Zahedi R.P., Stojanovski D., Schönfisch B., Guiard B., Sickmann A., Pfanner N., Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 13.Ringel A.E., Tucker S.A., Haigis M.C. Chemical and physiological features of mitochondrial acylation. Mol. Cell. 2018;72:610–624. doi: 10.1016/j.molcel.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garland P.B., Shepherd D., Yates D.W. Steady-state concentrations of coenzyme A, acetyl-coenzyme A and long-chain fatty acyl-coenzyme A in rat-liver mitochondria oxidizing palmitate. Biochem. J. 1965;97:587–594. doi: 10.1042/bj0970587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansford R.G., Johnson R.N. The steady state concentrations of coenzyme A-SH and coenzyme A thioester, citrate, and isocitrate during tricarboxylate cycle oxidations in rabbit heart mitochondria. J. Biol. Chem. 1975;250:8361–8375. [PubMed] [Google Scholar]
- 16.Ghanta S., Grossmann R.E., Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: Chemical and metabolic logic of acetyl-lysine modifications. Crit. Rev. Biochem. Mol. Biol. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner G.R., Bhatt D.P., O'Connell T.M., Thompson J.W., Dubois L.G., Backos D.S., Yang H., Mitchell G.A., Ilkayeva O.R., Stevens R.D., Grimsrud P.A., Hirschey M.D. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab. 2017;25:823–837. doi: 10.1016/j.cmet.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen R., Becher D., Büttner K., Biran D., Hecker M., Ron E.Z. Probing the active site of homoserine trans-succinylase. FEBS Lett. 2004;577:386–392. doi: 10.1016/j.febslet.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 19.Weinert B.T., Schölz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waitkus M.S., Diplas B.H., Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 2018;34:186–195. doi: 10.1016/j.ccell.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang L., Su S.-S.M. Isocitrate dehydrogenase mutation and (R)-2-hydroxyglutarate: From basic discovery to therapeutics development. Annu. Rev. Biochem. 2017;86:305–331. doi: 10.1146/annurev-biochem-061516-044732. [DOI] [PubMed] [Google Scholar]
- 23.Li F., He X., Ye D., Lin Y., Yu H., Yao C., Huang L., Zhang J., Wang F., Xu S., Wu X., Liu L., Yang C., Shi J., He X. NADP+-IDH mutations promote hypersuccinylation that impairs mitochondria respiration and induces apoptosis resistance. Mol. Cell. 2015;60:661–675. doi: 10.1016/j.molcel.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Rardin M.J., He W., Nishida Y., Newman J.C., Carrico C., Danielson S.R., Guo A., Gut P., Sahu A.K., Li B., Uppala R., Fitch M., Riiff T., Zhu L., Zhou J. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaviard C., Broutin I., Cosette P., Dé E., Jouenne T., Hardouin J. Lysine succinylation and acetylation in Pseudomonas aeruginosa. J. Proteome Res. 2018;17:2449–2459. doi: 10.1021/acs.jproteome.8b00210. [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Li L., Chai R., Zhang Z., Qiu H., Mao X., Hao Z., Wang Y., Sun G. Succinyl-proteome profiling of Pyricularia oryzae, a devastating phytopathogenic fungus that causes rice blast disease. Sci. Rep. 2019;9:3490. doi: 10.1038/s41598-018-36852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B.M.M., Skinner M.E., Lombard D.B., Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G., Xu L., Yu H., Gao J., Guo L. Systematic analysis of the lysine succinylome in the model medicinal mushroom Ganoderma lucidum. BMC Genomics. 2019;20:585. doi: 10.1186/s12864-019-5962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C., Dai J., Lu H., Chen Z., Guo M., He Y., Gao K., Ge T., Jin J., Wang L., Tian B., Hua Y., Zhao Y. Succinylome analysis reveals the involvement of lysine succinylation in the extreme resistance of Deinococcus radiodurans. Proteomics. 2019;19 doi: 10.1002/pmic.201900158. [DOI] [PubMed] [Google Scholar]
- 30.Yuan H., Chen J., Yang Y., Shen C., Xu D., Wang J., Yan D., He Y., Zheng B. Quantitative succinyl-proteome profiling of Chinese hickory (Carya cathayensis) during the grafting process. BMC Plant Biol. 2019;19:467. doi: 10.1186/s12870-019-2072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X., Hu X., Wan Y., Xie G., Li X., Chen D., Cheng Z., Yi X., Liang S., Tan F. Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii. J. Proteome Res. 2014;13:6087–6095. doi: 10.1021/pr500992r. [DOI] [PubMed] [Google Scholar]
- 32.Yang M., Wang Y., Chen Y., Cheng Z., Gu J., Deng J., Bi L., Chen C., Mo R., Wang X., Ge F. Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis. Mol. Cell. Proteomics. 2015;14:796–811. doi: 10.1074/mcp.M114.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosono S., Tamura M., Suzuki S., Kawamura Y., Yoshida A., Nishiyama M., Yoshida M. Changes in the acetylome and succinylome of Bacillus subtilis in response to carbon source. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizuno Y., Nagano-Shoji M., Kubo S., Kawamura Y., Yoshida A., Kawasaki H., Nishiyama M., Yoshida M., Kosono S. Altered acetylation and succinylation profiles in Corynebacterium glutamicum in response to conditions inducing glutamate overproduction. Microbiologyopen. 2016;5:152–173. doi: 10.1002/mbo3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H., He Y., Zhou X., Qian G., Lv G., Shen Y., Liu J., Li D., Li X., Liu W. Systematic analysis of the lysine succinylome in Candida albicans. J. Proteome Res. 2016;15:3793–3801. doi: 10.1021/acs.jproteome.6b00578. [DOI] [PubMed] [Google Scholar]
- 36.Jin W., Wu F. Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum) PLoS One. 2016;11 doi: 10.1371/journal.pone.0147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen C., Xue J., Sun T., Guo H., Zhang L., Meng Y., Wang H. Succinyl-proteome profiling of a high taxol containing hybrid Taxus species (Taxus x media) revealed involvement of succinylation in multiple metabolic pathways. Sci. Rep. 2016;6:21764. doi: 10.1038/srep21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian L., Nie L., Chen M., Liu P., Zhu J., Zhai L., Tao S.C., Cheng Z., Zhao Y., Tan M. Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism. J. Proteome Res. 2016;15:2060–2071. doi: 10.1021/acs.jproteome.6b00264. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang G., Song L., Mu P., Wang S., Liang W., Lin Q. Global analysis of protein lysine succinylation profiles in common wheat. BMC Genomics. 2017;18:309. doi: 10.1186/s12864-017-3698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y.X., Shen C.J., Ma J.Q., Chen W., Mao J., Zhou Y.Y., Chen L. Quantitative succinyl-proteome profiling of Camellia sinensis cv. “Anji Baicha” during periodic albinism. Sci. Rep. 2017;7:1873. doi: 10.1038/s41598-017-02128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X., Liu T., Yang J., Chen L., Liu B., Wei C., Wang L., Jin Q. The first succinylome profile of Trichophyton rubrum reveals lysine succinylation on proteins involved in various key cellular processes. BMC Genomics. 2017;18:577. doi: 10.1186/s12864-017-3977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng S., Jiao K., Guo H., Jiang M., Hao J., Wang H., Shen C. Succinyl-proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway. BMC Genomics. 2017;18:598. doi: 10.1186/s12864-017-3978-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J., Li F., Liu Y., Shen W., Du X., He L., Meng Z., Ma X., Wang Y. Systematic identification of mitochondrial lysine succinylome in silkworm (Bombyx mori) midgut during the larval gluttonous stage. J. Proteomics. 2018;174:61–70. doi: 10.1016/j.jprot.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Ren S., Yang M., Yue Y., Ge F., Li Y., Guo X., Zhang J., Zhang F., Nie X., Wang S. Lysine succinylation contributes to aflatoxin production and pathogenicity in Aspergillus flavus. Mol. Cell. Proteomics. 2018;17:457–471. doi: 10.1074/mcp.RA117.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng X., Lv Y., Mujahid H., Edelmann M.J., Zhao H., Peng X., Peng Z. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta Proteins Proteom. 2018;1866:451–463. doi: 10.1016/j.bbapap.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Henriksen P., Wagner S.A., Weinert B.T., Sharma S., Bačinskaja G., Rehman M., Juffer A.H., Walther T.C., Lisby M., Choudhary C. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in saccharomyces cerevisiae. Mol. Cell. Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harmel R., Fiedler D. Features and regulation of non-enzymatic post-translational modifications. Nat. Chem. Biol. 2018;14:244–252. doi: 10.1038/nchembio.2575. [DOI] [PubMed] [Google Scholar]
- 48.Klausen M.S., Jespersen M.C., Nielsen H., Jensen K.K., Jurtz V.I., Sønderby C.K., Sommer M.O.A., Winther O., Nielsen M., Petersen B., Marcatili P. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins. 2019;87:520–527. doi: 10.1002/prot.25674. [DOI] [PubMed] [Google Scholar]
- 49.Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 50.Armenteros J.J.A., Salvatore M., Emanuelsson O., Winther O., von Heijne G., Elofsson A., Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J., Xu D. Correlation between posttranslational modification and intrinsic disorder in protein. Biocomput. 2011;2012:94–103. [PMC free article] [PubMed] [Google Scholar]
- 52.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 53.Miyakawa I. Organization and dynamics of yeast mitochondrial nucleoids. Proc. Jpn. Acad. Ser. B. 2017;93:339–359. doi: 10.2183/pjab.93.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vozáriková V., Kunová N., Bauer J.A., Frankovský J., Kotrasová V., Procházková K., Džugasová V., Kutejová E., Pevala V., Nosek J., Tomáška Ľ. Mitochondrial HMG-box containing proteins: From biochemical properties to the roles in human diseases. Biomolecules. 2020;10:1193. doi: 10.3390/biom10081193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dilweg I.W., Dame R.T. Post-translational modification of nucleoid-associated proteins: An extra layer of functional modulation in bacteria? Biochem. Soc. Trans. 2018;46:1381–1392. doi: 10.1042/BST20180488. [DOI] [PubMed] [Google Scholar]
- 56.Ho B., Baryshnikova A., Brown G.W. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 2018;6:192–205.e3. doi: 10.1016/j.cels.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 57.Kaufman B.A., Newman S.M., Hallberg R.L., Slaughter C.A., Perlman P.S., Butow R.A. In organello formaldehyde crosslinking of proteins to mtDNA: Identification of bifunctional proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakraborty A., Lyonnais S., Battistini F., Hospital A., Medici G., Prohens R., Orozco M., Vilardell J., Soì M. DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p. Nucleic Acids Res. 2016;45:951–967. doi: 10.1093/nar/gkw1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Li H., Sui M., Li M., Wang J., Meng Y., Sun T., Liang Q., Suo C., Gao X., Li C., Li Z., Du W., Zhang B., Sai S. Fungal acetylome comparative analysis identifies an essential role of acetylation in human fungal pathogen virulence. Commun. Biol. 2019;2:154. doi: 10.1038/s42003-019-0419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paik W.K., Pearson D., Lee H.W., Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta. 1970;213:513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 61.Wolfe A.J. Bacterial protein acetylation: New discoveries unanswered questions. Curr. Genet. 2016;62:335–341. doi: 10.1007/s00294-015-0552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Čanigová N. Comenius University in Bratislava; 2017. Comparative Analysis of Mitochondrial HMG Box-Containing Proteins. M.Sc. Thesis. [Google Scholar]
- 63.Miyakawa I., Okamuro A., Kinsky S., Visacka K., Tomaska L., Nosek J. Mitochondrial nucleoids from the yeast Candida parapsilosis: Expansion of the repertoire of proteins associated with mitochondrial DNA. Microbiology. 2009;155:1558–1568. doi: 10.1099/mic.0.027474-0. [DOI] [PubMed] [Google Scholar]
- 64.Višacká K., Gerhold J.M., Petrovičová J., Kinský S., Jõers P., Nosek J., Sedman J., Tomáška Ľ. Novel subfamily of mitochondrial HMG box-containing proteins: Functional analysis of Gcf1p from Candida albicans. Microbiology. 2009;155:1226–1240. doi: 10.1099/mic.0.025759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bakkaiová J., Arata K., Matsunobu M., Ono B., Aoki T., Lajdova D., Nebohacova M., Nosek J., Miyakawa I., Tomaska L. The strictly aerobic yeast Yarrowia lipolytica tolerates loss of a mitochondrial DNA-packaging protein. Eukaryot. Cell. 2014;13:1143–1157. doi: 10.1128/EC.00092-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crooks D.R., Maio N., Lang M., Ricketts C.J., Vocke C.D., Gurram S., Turan S., Kim Y.Y., Cawthon G.M., Sohelian F., De Val N., Pfeiffer R.M., Jailwala P., Tandon M., Tran B. Mitochondrial DNA alterations underlie an irreversible shift to aerobic glycolysis in fumarate hydratase-deficient renal cancer. Sci. Signal. 2021;14 doi: 10.1126/scisignal.abc4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang M., Ternette N., Su H., Dabiri R., Kessler B., Adam J., Teh B., Pollard P. The succinated proteome of FH-mutant tumours. Metabolites. 2014;4:640–654. doi: 10.3390/metabo4030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kunová N., Ondrovičová G., Bauer J.A., Bellová J., Ambro L., Martináková L., Kotrasová V., Kutejová E., Pevala V. The role of Lon-mediated proteolysis in the dynamics of mitochondrial nucleic acid-protein complexes. Sci. Rep. 2017;7:631. doi: 10.1038/s41598-017-00632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao W., Wu M., Wang N., Zhang Y., Hua J., Tang G., Wang Y. Increased expression of mitochondrial transcription factor a and nuclear respiratory factor-1 predicts a poor clinical outcome of breast cancer. Oncol. Lett. 2018;15:1449–1458. doi: 10.3892/ol.2017.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J.D., Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Guo Y.R., Liu K., Yin Z., Liu R., Xia Y., Tan L., Yang P., Lee J.H., Li X.J., Hawke D., Zheng Y., Qian X., Lyu J., He J. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552:273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dehzangi A., López Y., Lal S.P., Taherzadeh G., Sattar A., Tsunoda T., Sharma A. Improving succinylation prediction accuracy by incorporating the secondary structure via helix, strand and coil, and evolutionary information from profile bigrams. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ondrovičová G., Liu T., Singh K., Tian B., Li H., Gakh O., Perečko D., Janata J., Granot Z., Orly J., Kutejová E., Suzuki C.K. Cleavage site selection within a folded substrate by the ATP-dependent Lon protease. J. Biol. Chem. 2005;280:25103–25110. doi: 10.1074/jbc.M502796200. [DOI] [PubMed] [Google Scholar]
- 74.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Bartolomeo F., Malina C., Campbell K., Mormino M., Fuchs J., Vorontsov E., Gustafsson C.M., Nielsen J. Absolute yeast mitochondrial proteome quantification reveals trade-off between biosynthesis and energy generation during diauxic shift. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7524–7535. doi: 10.1073/pnas.1918216117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen X.J., Butow R.A. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 77.Gibson G.E., Xu H., Chen H.-L., Chen W., Denton T.T., Zhang S. Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. J. Neurochem. 2015;134:86–96. doi: 10.1111/jnc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valachovič M., Bareither B.M., Bhuiyan M.S.A., Eckstein J., Barbuch R., Balderes D., Wilcox L., Sturley S.L., Dickson R.C., Bard M. Cumulative mutations affecting sterol biosynthesis in the yeast Saccharomyces cerevisiae result in synthetic lethality that is suppressed by alterations in sphingolipid profiles. Genetics. 2006;173:1893–1908. doi: 10.1534/genetics.105.053025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 80.Nosek J., Tomáška Ľ. Create Space Independent Publishing Platform, Create Space Independent Publishing Platform; Charleston, SC: 2013. Laboratory Protocols in Molecular and Cell Biology of Yeasts. [Google Scholar]
- 81.Van Dijl J.M., Kutejová E., Suda K., Perečko D., Schatz G., Suzuki C.K. The ATPase and protease domains of yeast mitochondrial Lon: Roles in proteolysis and respiration-dependent growth. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10584–10589. doi: 10.1073/pnas.95.18.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki C.K., Kutejová E., Suda K. Analysis and purification of ATP-dependent mitochondrial Lon protease of Saccharomyces cerevisiae. Methods Enzymol. 1995;260:486–494. doi: 10.1016/0076-6879(95)60160-0. [DOI] [PubMed] [Google Scholar]
- 83.Somogyi M. Notes on sugar determination. J. Biol. Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 84.Michalski A., Damoc E., Lange O., Denisov E., Nolting D., Müller M., Viner R., Schwartz J., Remes P., Belford M., Dunyach J.J., Cox J., Horning S., Mann M., Makarov A. Ultra high resolution linear ion trap orbitrap mass spectrometer (orbitrap elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 86.Morgenstern M., Stiller S.B., Lübbert P., Peikert C.D., Dannenmaier S., Drepper F., Weill U., Höß P., Feuerstein R., Gebert M., Bohnert M., van der Laan M., Schuldiner M., Schütze C., Oeljeklaus S. Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 2017;19:2836–2852. doi: 10.1016/j.celrep.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vögtle F.N., Burkhart J.M., Gonczarowska-Jorge H., Kücükköse C., Taskin A.A., Kopczynski D., Ahrends R., Mossmann D., Sickmann A., Zahedi R.P., Meisinger C. Landscape of submitochondrial protein distribution. Nat. Commun. 2017;8:1–10. doi: 10.1038/s41467-017-00359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balakrishnan R., Park J., Karra K., Hitz B.C., Binkley G., Hong E.L., Sullivan J., Micklem G., Cherry J.M. YeastMine-An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database. 2012;2012:bar062. doi: 10.1093/database/bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hirschman J.E., Hitz B.C., Karra K., Krieger C.J. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomsen M.C.F., Nielsen M. Seq2Logo: A method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bakkaiová J., Marini V., Willcox S., Nosek J., Griffith J.D., Krejci L., Tomáška Ľ. Yeast mitochondrial HMG proteins: DNA-binding properties of the most evolutionarily divergent component of mitochondrial nucleoids. Biosci. Rep. 2016;36 doi: 10.1042/BSR20150275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 95.Green M.R., Sambrook J. Cold Spring Harbor Laboratory Press; New York, NY: 2012. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 96.Berrow N.S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D.I., Owens R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H., Kim J., Woo J., Kim J.H., Choi B.H., He B., Chen W., Zhang S., Cerione R.A., Auwerx J. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kovalevskiy O., Nicholls R.A., Long F., Carlon A., Murshudov G.N. Overview of refinement procedures within REFMAC 5: Utilizing data from different sources. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:215–227. doi: 10.1107/S2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., Goetz A.W., Gusarov S., Homeyer N., Janowski P., Kaus J. University of California, San Francisco; San Francisco, CA: 2014. AMBER 14. [Google Scholar]
- 100.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 101.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., Jain S., Lewis S.M., Arendall W.B., Snoeyink J., Adams P.D. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D.W., Dolinsky T., Konecny R. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz Ş. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (103) partner repository with the dataset identifier PXD023604 (username: reviewer_pxd023604@ebi.ac.uk; password: zMPT8wo0).