Key Points
Question
What is the effect of whole-genome sequencing (WGS) on clinical management in a diverse population of acutely ill infants?
Findings
In this randomized time-delayed clinical trial conducted at 5 children’s hospitals, a diverse population of 354 infants was randomized to receive WGS either 15 days or 60 days after enrollment. In both study groups, access to WGS doubled the proportion of patients with a precision diagnosis and a change of clinical management.
Meaning
These data support WGS implementation for acutely ill infants with a suspected genetic condition.
This randomized clinical trial investigates the effect of whole-genome sequencing on the clinical management of acutely ill infants with suspected genetic disease.
Abstract
Importance
Whole-genome sequencing (WGS) shows promise as a first-line genetic test for acutely ill infants, but widespread adoption and implementation requires evidence of an effect on clinical management.
Objective
To determine the effect of WGS on clinical management in a racially and ethnically diverse and geographically distributed population of acutely ill infants in the US.
Design, Setting, and Participants
This randomized, time-delayed clinical trial enrolled participants from September 11, 2017, to April 30, 2019, with an observation period extending to July 2, 2019. The study was conducted at 5 US academic medical centers and affiliated children’s hospitals. Participants included infants aged between 0 and 120 days who were admitted to an intensive care unit with a suspected genetic disease. Data were analyzed from January 14 to August 20, 2020.
Interventions
Patients were randomized to receive clinical WGS results 15 days (early) or 60 days (delayed) after enrollment, with the observation period extending to 90 days. Usual care was continued throughout the study.
Main Outcomes and Measures
The main outcome was the difference in the proportion of infants in the early and delayed groups who received a change of management (COM) 60 days after enrollment. Additional outcome measures included WGS diagnostic efficacy, within-group COM at 90 days, length of hospital stay, and mortality.
Results
A total of 354 infants were randomized to the early (n = 176) or delayed (n = 178) arms. The mean participant age was 15 days (IQR, 7-32 days); 201 participants (56.8%) were boys; 19 (5.4%) were Asian; 47 (13.3%) were Black; 250 (70.6%) were White; and 38 (10.7%) were of other race. At 60 days, twice as many infants in the early group vs the delayed group received a COM (34 of 161 [21.1%; 95% CI, 15.1%-28.2%] vs 17 of 165 [10.3%; 95% CI, 6.1%-16.0%]; P = .009; odds ratio, 2.3; 95% CI, 1.22-4.32) and a molecular diagnosis (55 of 176 [31.0%; 95% CI, 24.5%-38.7%] vs 27 of 178 [15.0%; 95% CI, 10.2%-21.3%]; P < .001). At 90 days, the delayed group showed a doubling of COM (to 45 of 161 [28.0%; 95% CI, 21.2%-35.6%]) and diagnostic efficacy (to 56 of 178 [31.0%; 95% CI, 24.7%-38.8%]). The most frequent COMs across the observation window were subspecialty referrals (39 of 354; 11%), surgery or other invasive procedures (17 of 354; 4%), condition-specific medications (9 of 354; 2%), or other supportive alterations in medication (12 of 354; 3%). No differences in length of stay or survival were observed.
Conclusions and Relevance
In this randomized clinical trial, for acutely ill infants in an intensive care unit, introduction of WGS was associated with a significant increase in focused clinical management compared with usual care. Access to first-line WGS may reduce health care disparities by enabling diagnostic equity. These data support WGS adoption and implementation in this population.
Trail Registration
ClinicalTrials.gov Identifier: NCT03290469
Introduction
Critically ill infants admitted to an intensive care unit (ICU) are at risk for high levels of morbidity and mortality.1,2 In the US, neonatal hospitalizations cost at least $17 billion annually,3,4 including approximately 400 000 newborns admitted to neonatal ICUs.5
Genetic disorders are a leading cause of ICU admission, and several recent investigations have used comprehensive genomic testing in populations of acutely ill infants, using either whole-exome sequencing, which surveys approximately 2% of the genome that codes for proteins, or whole-genome sequencing (WGS), which evaluates approximately 95% of nuclear and mitochondrial DNA. These studies have reported 20% to 50% diagnostic efficacy,6,7,8,9,10,11,12,13,14 with variability attributed to differing inclusion criteria and comprehensiveness of the genomic test. The highest yields were observed in cohorts of critically ill infants with clinical features strongly suggesting a genetic diagnosis.6,7,8,9,10,11,12,13 Observational and post hoc investigations of clinical utility have shown that up to two-thirds of molecularly diagnosed infants will receive an alteration in care6,7,8,9,10,11,12,13,14 and that genomic testing is perceived as having high utility in the acute care setting.15 The widespread implementation of WGS and other genomic tests in the ICU has been hampered, however, by a lack of controlled studies investigating their effect on change of management (COM) using matched comparator groups, real-world inclusion criteria, and diverse populations.16
Here, we report on the results of a multicenter, randomized, time-delayed investigation of the effect of clinical WGS on COM in infants admitted to an ICU at 5 US children’s hospitals. Patient selection emphasized clinician suspicion of a genetic disorder. Usual care, including molecular genetic testing, was continued throughout the study, which captured variation in infant management and enabled an assessment of WGS effect within a real-world clinical population. The primary objective of the study was to determine whether the introduction of WGS into an acutely ill infant population was associated with clinical utility as assessed by COM.
Methods
Trial Design
The randomized clinical trial protocol used a time-delayed study design17 that was reviewed and approved by a central institutional review board (Western Institutional Review Board, Olympia, Washington) and each study site ethics committee. The trial was sponsored by Illumina Inc as a collaboration between the sponsor and 5 academic medical centers (Children’s Hospital of Philadelphia, University of Nebraska Medical Center/Children’s Hospital, Children’s Hospital of Orange County/Rady Children’s Institute for Genomic Medicine, Washington University/St Louis Children’s Hospital, and Le Bonheur Children’s Hospital) and was supported by statistical analysis services from Precision for Medicine. Participants were recruited between September 11, 2017, and April 30, 2019, with an observation period extending to July 2, 2019. The trial was conducted in accordance with the principles of the Declaration of Helsinki. The study was proposed by the sponsor, with the design developed collaboratively, and was conducted in accordance with the Consolidated Standards of Reporting Trials (CONSORT) reporting guideline. The trial protocol including the statistical analysis plan is provided in Supplement 1, and the clinical services laboratory information can be found in the eAppendix in Supplement 2.
Participants
Patients were recruited from participating academic medical center ICUs. Eligible patients were aged between 0 and 120 days with a suspected genetic etiology of disease based on objective clinical findings for which genetic testing would be considered. At least 1 biological parent was required for participation. Exclusion criteria included an established genetic diagnosis; high clinical suspicion for trisomy 13, 18, 21, or monosomy X; or full explanation of the patient’s phenotype by complications of prematurity. Patients born prematurely with indications of a genetic disease, eg, those with multiple congenital anomalies, were considered for enrollment. Other affected family members, generally siblings, were considered if their phenotypic findings were consistent with the patient’s condition and lacked an etiologic diagnosis. Detailed inclusion and exclusion criteria are described in eMethods 1 in Supplement 2. Race and ethnicity were collected following US Department of Health and Human Services recommendations and classifications to assess differences in outcomes. Written informed consent was obtained for each patient and participating relatives, and pre- and posttest genetic counseling was offered to all participants.
Interventions
Owing to the potential benefit of WGS in the acute care population, a time-delayed study design was used to ensure that all participants received access to the diagnostic technology. Infants who met the inclusion criteria were randomized to receive WGS testing results either 15 or 60 days after enrollment—the early and delayed groups, respectively—with a total 90-day observation window. Staff at each site were blinded to the randomization group of each patient until 15 days after enrollment. To ensure that the intervention did not interfere with usual care in the ICU, the study directed health care professionals to order any imaging or molecular testing deemed appropriate. Whole-genome sequencing was performed on duo, trio, or quad family structures in the Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–approved Illumina Clinical Services Laboratory, which included assessment of single nucleotide variants, indels, copy number variants, mitochondrial variants, and spinal muscular atrophy gene status using a family-informed analysis pipeline (eMethods 2 in Supplement 2). Primary WGS findings were classified as positive, likely positive, inconclusive, or negative based on clinical concordance with the patient’s phenotype by the medical monitor and each site’s principal investigator. Secondary findings were reported in accordance with American College of Medical Genetics guidelines.18 Incidental findings were reported for pathogenic or likely pathogenic variants with medical actionability. Discrepancies were assessed in monthly team calls, and the final classification of the site principal investigator was considered final. Usual care was continued for all patients at all study sites for the duration of the study. Adverse events were classified and recorded for each patient, with unanticipated death independent of the study intervention the most common finding (eFigure 1 in Supplement 2).
Outcomes
The primary outcome was the difference in the proportion of patients who received a COM in the early and delayed groups at 60 days. Change of management was assessed using a state-change classification of patients as either having no change in care, a condition-specific intervention, condition-specific supportive care, palliative care, or a combination of the latter 3 (eTable 1 in Supplement 2). Change of management data were collected by each site’s study coordinators and attending clinicians, and COM assessments were reviewed with the participating clinicians at each site in study review meetings and by the medical monitor. In all cases, the site principal investigator made the final decisions about patient COM classification at each time point using the state-change rubric. Change of management was investigated within the delayed group by assessing the difference in the number of patients with COM in the interval between 60 days (return of WGS results) and the end of the observation period at 90 days after enrollment. Secondary outcome measures reported here include diagnostic efficacy (also frequently reported as diagnostic yield, ie, the proportion of positive diagnoses per group reported as a percentage) of both WGS and usual-care testing, and time to diagnosis relative to WGS and usual-care testing test outcomes and relative to enrollment, day of life, death, or discharge. An assessment of diagnostic accuracy was not performed as planned owing to elements of the study that involved active discussion of ambiguous WGS testing results. A planned analysis of change in care setting was not pursued owing to the high proportion of patients who were rapidly discharged. Physician and patient satisfaction survey findings and a health economic analysis of resource utilization will be presented in a subsequent manuscript.
Statistical Analysis
The null hypothesis of this study was that there would be no difference in COM between early and delayed groups 60 days after enrollment. The power analysis was based on a fixed sample size of 300 patients with 80% power to detect a significant increase in the proportion of cases with COM on the assumption of a 20% difference in diagnostic yield between the early and delayed groups—ie, that WGS would enable 20% more diagnoses when compared to the aggregate performance of all usual-care molecular diagnostic tests at 60 days. Change of management was treated as a categorical variable, and independence was assessed using a Cochran-Mantel-Haenszel test with the study site as the covariate. Diagnostic efficacy across groups was assessed by examining the difference in the count of patients with positive and likely positive test results with those who received a usual-care negative or inconclusive result using a Welch 2-sample t test. Within-patient assessment of diagnostic efficacy was analyzed using the McNemar test for paired data with continuity correction. A Cox proportional hazard model was used to assess time to diagnosis. A planned interim analysis took place at 150 patients enrolled. Statistical analysis of the primary outcome was performed by Precision for Medicine. P < .05 was considered significant. All statistical analyses were performed between January 14 and August 20, 2020. Additional details are available in the trial protocol in Supplement 1 and eMethods 3 in Supplement 2.
Results
Patients
A total of 354 infants with features indicative of a genetic disease were enrolled from ICUs at 5 children’s hospitals from September 11, 2017, to April 30, 2019, with the date of observation extending to July 2, 2019. Approximately 35% of approached families declined to participate, consistent with previous studies and the challenges of recruitment in the acute care setting.19,20 Enrolled individuals matched the race and ethnicity distribution of the US population (19 Asian individuals [5.4%], 47 Black individuals [13.3%], 250 White individuals [70.6%], and 38 individuals of other race [10.7%]; and 81 Latino or Hispanic individuals [22.9%]) and were drawn from 4 US geographic regions (West Coast, Midwest, South, and Northeast) (Table 1; eTable 2 in Supplement 2).
Table 1. Demographic and Clinical Characteristics at Enrollment.
| Characteristic | No. (%) | ||
|---|---|---|---|
| Early (n = 176)a | Delayed (n = 178)b | All patients (n = 354) | |
| Age, mean (IQR), d | 14 (6-28) | 17 (7-37) | 15 (7-32) |
| Girls | 74 (42.0) | 79 (44.4) | 153 (43.2) |
| Boys | 102 (60.0) | 99 (55.6) | 201 (56.8)c |
| Race | |||
| Asian | 7 (4) | 12 (6.8) | 19 (5.4) |
| Black | 26 (14.8) | 21 (11.8) | 47 (13.3) |
| White | 125 (71.0) | 125 (70.2) | 250 (70.6) |
| Otherd | 18 (10.2) | 20 (11.2) | 38 (10.7) |
| Ethnicity | |||
| Latino or Hispanic | 47 (26.7) | 34 (19.1) | 81 (22.9) |
| Not Latino or Hispanic | 128 (72.7) | 141 (79.2) | 269 (76.0) |
| Unknown | 1 (0.6) | 3 (1.7) | 4 (1.1) |
| Family composition for testing | |||
| Trio | 130 (73.9) | 139 (78.1) | 269 (76.0) |
| Duo | 41 (23.3) | 35 (19.7) | 76 (21.5) |
| Quad | 4 (2.3) | 3 (1.7) | 7 (2.0) |
| Indication for testing | |||
| Congenital anomalies | |||
| Multiple | 109 (61.9) | 92 (51.7) | 201 (56.8) |
| Isolated major | 26 (14.8) | 36 (20.2) | 62 (17.5) |
| Neurologic disorder | 22 (12.5) | 30 (16.9) | 52 (14.7) |
| Single major feature | 18 (10.2) | 19 (10.7) | 37 (10.5) |
Results returned at visit 2, 15 days after enrollment.
Results returned at visit 3, 60 days after enrollment.
Includes a single patient with indeterminate sex.
Parents who selected “other” have indicated that they do not identify as Asian, Black, or White race.
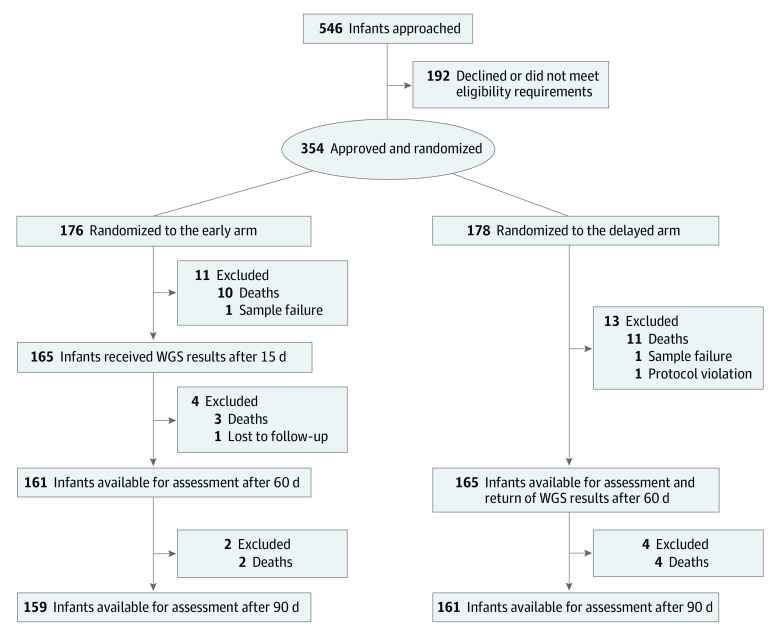
Patients were randomized to either early (n = 176) or delayed (n = 178) return of WGS results (Figure 1 and Table 1). The baseline characteristics of both groups were similar, including age (mean, 15 days; IQR, 7-32 days) and time from admission to enrollment (Table 1; eFigures 2 and 3 and eTable 2 in Supplement 2). A total of 296 of 354 infants (83%) were recruited from the neonatal ICU, 23 (7%) from the pediatric ICU, and 35 (10%) from the cardiovascular ICU (eTable 3 in Supplement 2). For the majority of enrolled infants, DNA samples from both parents were available, which enabled family-trio WGS analysis (Table 1). Consistent with previous reports,21 the most frequent clinical indication for testing was multiple congenital anomalies (Table 1; eTables 2 and 4, and eFigure 4 in Supplement 2; eTables 5 and 6 in Supplement 3). Deaths contributed a total of 32 losses (9%), highlighting the disease severity of the enrolled patient population (Figure 1). Taking into account other participants lost to follow-up, there were a total of 161 and 165 infants available for investigation in the early and delayed groups, respectively, before analysis of the primary outcome (COM) at day 60 (Figure 1). Assessments of diagnostic efficacy and other secondary outcome measures were calculated against the total enrolled in each group unless otherwise stated.
Figure 1. Consolidated Standards of Reporting Trials (CONSORT) Flow Diagram of Enrollment and Randomization of Patients in the NICUSeq Clinical Trial.
The NICUSeq study used a randomized time-delayed design to investigate the effect of whole-genome sequencing (WGS) on changes of management. The patient attrition noted here does not include 2 deaths in the early group that occurred after day 90 but within the study window.
Change of Management
To assess whether receiving clinical WGS altered clinical management, the difference in the proportion of infants in the early and delayed groups with a COM was investigated. The study used a COM rubric that categorized infants into those with generally supportive care, management directed to the primary genetic etiology, supportive care specific to the genetic condition, palliative care, or a combination thereof. Change of management was assessed by comparing patient categorization at sequential visits (eTable 1 in Supplement 2).
At the time of the primary outcome, day 60, the early group showed 2-fold more infants with a COM compared with the delayed arm (34 of 161 [21.1%; 95% CI, 15.1%-28.2%] vs 17 of 165 [10.3%; 95% CI, 6.1%-16.0%]; P = .009; odds ratio, 2.3; 95% CI, 1.22-4.32) (Table 2; eTable 7 in Supplement 3 and eTable 8 in Supplement 2). Additionally, within-group analysis of the delayed group showed that COM increased by more than 2-fold after the return of WGS findings at 90 days to 45 of 161 patients receiving a COM (28.0%; 95% CI, 21.2%-35.6%) (Table 2) despite more than 60 days elapsing before results were returned. Patients with a pathogenic or likely pathogenic WGS finding were more than 3-fold more likely to receive a COM compared with those with uncertain or negative findings (eTable 9 in Supplement 2). Overall, 83 of 326 patients (25.0%; 95% CI, 20.8%-30.6%), or two-thirds of those who received a genetic diagnosis by any means, had a COM (Table 2).
Table 2. Diagnostic Efficacy and Change of Managementa.
| Variable | Early | Delayed | All patients | |||
|---|---|---|---|---|---|---|
| No./total No. | % (95% CI) | No./total No. | % (95% CI) | No./total No. | % (95% CI) | |
| At 60 d | ||||||
| Diagnostic efficacyb | 55/176 | 31.0 (24.5-38.7) | 27/178 | 15.0 (10.2-21.3) | 82/354 | 23.0 (18.9-27.0) |
| Change of managementc | ||||||
| Change | 34/161 | 21.1 (15.1-28.2) | 17/165 | 10.3 (6.1-16.0) | 51/326 | 15.6 (11.9-20.1) |
| No change | 127/161 | 78.9 (71.8-84.9) | 148/165 | 89.7 (84.0-93.9) | 275/326 | 84.4 (79.9-88.1) |
| At 90 d | ||||||
| Diagnostic efficacy | 55/176 | 31 (24.5-38.7) | 56/178 | 31.0 (24.7-38.8) | 111/354 | 31.0 (26.6-36.5) |
| Change of management | ||||||
| Change | 38/159 | 23.9 (17.5-31.3) | 45/161 | 28.0 (21.2-35.6) | 83/326 | 25.0 (20.8-30.6) |
| No change | 123/159 | 76.1 (70.1-83.6) | 120/161 | 73.0 (67.1-81.1) | 243/326 | 75.0 (69.4-79.2) |
Denominators for change of management account for transfers, deaths, and other losses to follow-up; diagnostic efficacy denominators are the total enrolled for each arm.
Difference in diagnostic efficacy between the 2 arms: P < .001.
Difference in change of management: P = .009; odds ratio, 2.3 (95% CI, 1.22-4.32).
The most frequent COMs at 60 days were associated with condition-supportive care and included subspeciality referral (21 of 354; 6.0%) and changes to medication (5 of 354; 1.4%), followed by condition-specific management, which included therapeutics specific to the primary genetic etiology (7 of 354; 2.0%) and surgical interventions (12 of 354; 3.4%) (Figure 2A; eTable 7 Supplement 3). All COMs were more frequent in the early cohort at day 60 and increased in the delayed cohort after the return of WGS findings (Figure 2A). A minority of patients (4 of 354; 1.1%) had redirection to palliative or other supportive care. The greatest number of patients to receive a COM were those with multiple congenital anomalies (45 of 182; 24%), but the highest proportion were patients with neurologic disorders (17 of 45; 35%) (Figure 2B). Whole-genome sequencing findings were frequently returned after discharge from the ICU and associated with COM (Figure 2C).
Figure 2. Change of Management (COM) Types, Effect by Clinical Classification, and Occurrence Relative to Discharge.
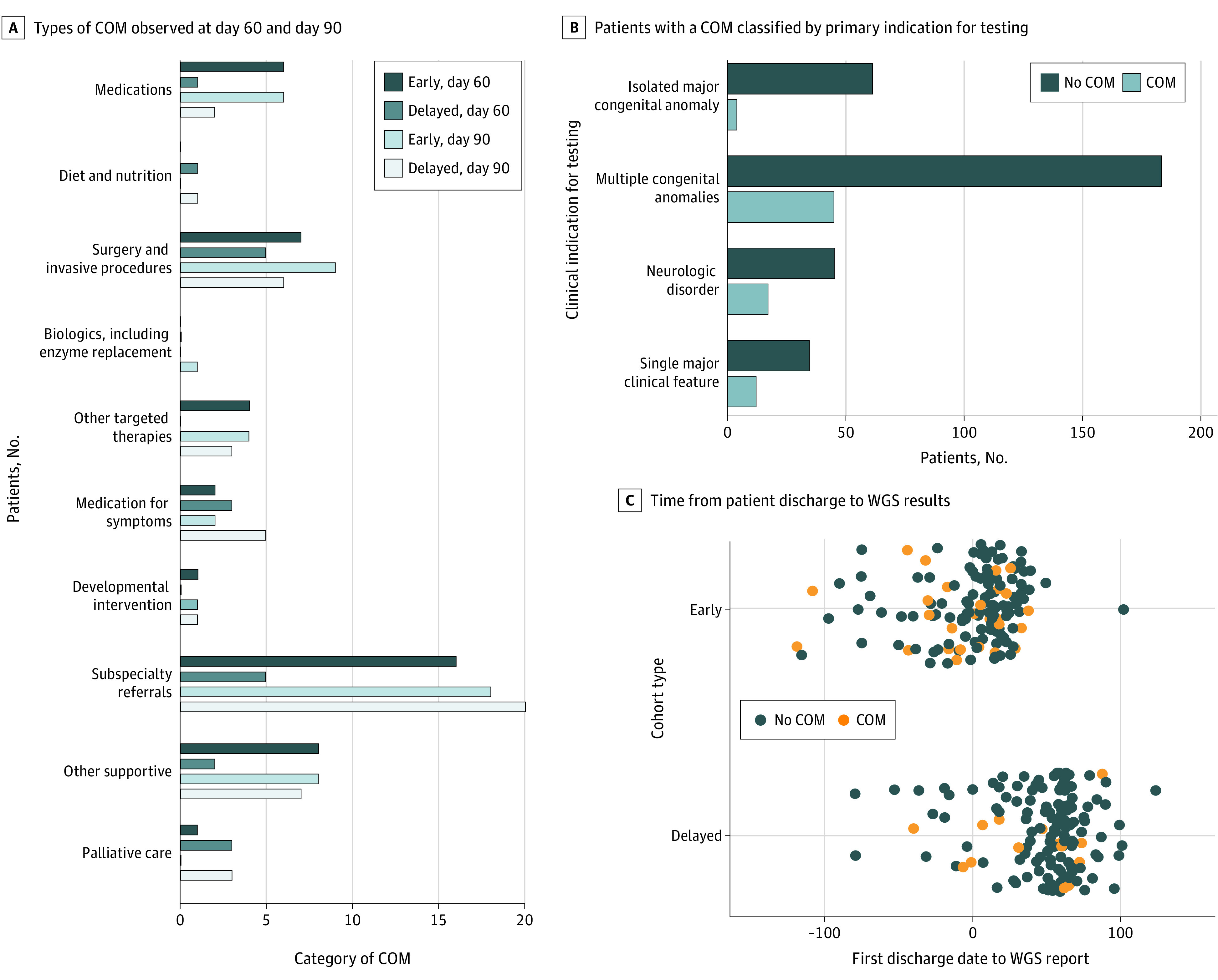
A, Types of COM observed at day 60 (black and dark blue bars) and at day 90 (light blue and white bars). COM categories are provided as horizontal x-axis labels. B, The number of patients with a COM across sites classified by the primary indication for testing. The greatest number of patients with a COM were observed in patients with multiple congenital anomalies, and the greatest proportion in those with a neurologic disorder. C, The time from a patient’s first discharge to the return of whole-genome sequencing (WGS) results, with the majority showing short length of stays followed by WGS return of results outside the NICU setting. Orange and blue dots denote patients with a COM (orange) or no COM (blue).
Several diagnoses led to COMs with notable clinical effect. For example, a male infant aged 29 days randomized to the early group received a diagnosis of Wiskott-Aldrich syndrome (OMIM 301000) by WGS and received a corrective bone marrow transplant (patient 909). In some cases, inappropriate interventions were halted as a result of diagnosis, including in a female infant aged 11 days with epilepsy who was stratified to the delayed group and diagnosed by WGS with an unsuspected KCNQ2 (OMIM 602235) variation; as a result, an ongoing metabolic work-up was stopped, including the ineffective administration of pyridoxine (patient 922).
A total of 4 of 354 infants (1%) received a COM as a result of nondiagnostic WGS results in the absence of any other molecular findings. Two infants (0.5%) with secondary findings in MYBPC3 and TP53, and 2 (0.5%) with incidental findings in PRSS1 and ABCA4 were referred to specialty clinics (eTables 10-12 in Supplement 3).
Diagnostic Efficacy and Other Secondary Outcome Measures
Consistent with the COM findings, the diagnostic rate in the early group was 31.0% (95% CI, 24.5%-38.7%; 55 of 176 patients diagnosed) compared with 15.0% (95% CI, 10.2%-21.3%; 27 of 178 patients diagnosed) in the delayed group at day 60 (P < .001), indicating that systematic WGS deployment led to a 2-fold increased diagnostic efficacy compared with aggregate usual-care testing. Similarly, the diagnostic rate doubled within the delayed group to 31.0% (95% CI, 24.7%-38.8%; 56 of 178 patients diagnosed; within-subject P < .001) (Table 2) at day 90 after the return of WGS results. The largest number of diagnoses were in infants with multiple congenital anomalies (63 of 191; 33%), and the highest proportion were those with a single major clinical feature (19 of 35; 54%) (Figure 3A). Whole-genome sequencing revealed a wide range of causal variant types, including terminal and interstitial chromosomal copy number variants, complex compound heterozygous variants, mitochondrial variants, and SMN1 copy loss leading to spinal muscular atrophy (Figure 3B; eTable 5 in Supplement 3). In the majority of resolved cases, de novo variants associated with autosomal dominant disorders were observed (Figure 3B; eTable 5 in Supplement 3). Among those diagnosed by WGS, 9 recurrent diagnoses were observed (eTable 5 in the Supplement 3), including 5 infants with type 1 Kabuki syndrome (OMIM 147920). Secondary and incidental findings were reported in 20 infants (5%) and 23 infants (6%) infants, respectively (eTables 10 and 11 in Supplement 3).
Figure 3. Secondary Outcomes Including Whole-Genome Sequencing (WGS) Findings, Length of Stay, and Survival.
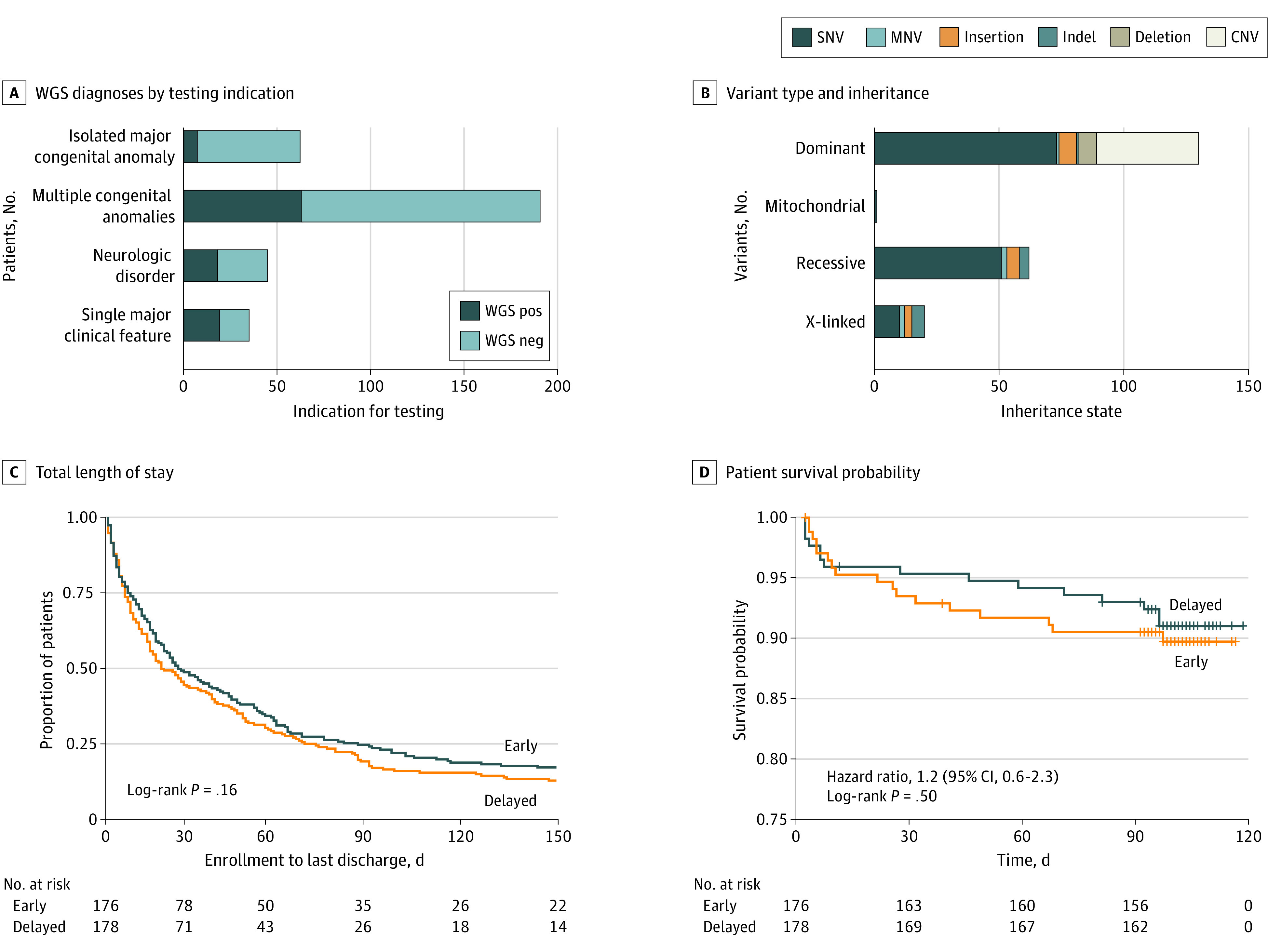
A, The distribution of positive WGS diagnoses by indication for testing. Dark blue shading indicates patients with a positive WGS finding, and light blue shading indicates those with no WGS finding. B, The distribution of variant types and their associated inheritance state detected by WGS (eTable 5 in Supplement 3). C, Total length of stay from the time of enrollment in the NICUSeq study to discharge (eFigure 7 in Supplement 2). D, Patient survival probability stratified by arm. Data collection extended to 115 days to complete final study visit assessments (eFigure 8 in Supplement 2). CNV indicates copy number variation; Indel, insertion/deletion polymorphism; MNV, multinucleotide variant; SNV, single-nucleotide variant.
The overall time to diagnosis was broadly associated with time to return of WGS testing results (eFigure 5 in Supplement 2). Unexpectedly, WGS returned positive findings in 9 of 32 extremely and very preterm infants (28.1%) (eTable 13 in Supplement 2), suggesting that WGS testing may have broad applicability in premature neonates.
Usual-care testing during the observation period varied substantially by site, as anticipated, and ranged from karyotyping to WGS with the negative microarray testing making up the majority of tests results (eFigure 6 in Supplement 2). For the 63 of 354 patients (17%) who received both positive WGS and usual-care molecular testing, there was concordance in 53 of 63 (85%). Among the 10 differences, 8 were attributable to differences in variant classification or reporting practices, and 2 were associated with genomic alterations that are currently undetectable with WGS (ie, methylation abnormalities and low-level somatic mosaicism) (eTable 5 in Supplement 3 and eTable 14 in Supplement 2). Usual-care testing in both the early and delayed groups was concentrated in the first approximately 40 days of life, after which few additional diagnoses were achieved (eFigure 5 in Supplement 2).
Investigation of survival and length of stay stratified by arm, test outcome, or observational period revealed no significant differences between groups (Figure 3C and D; eFigures 7 and 8 in Supplement 2). A negative correlation, however, was observed between gestational age and both time to enrollment (adjusted R2, 0.1199; P = 1.52 × 10−11) and length of stay (early group adjusted R2, 0.03382; P = .009; delayed group adjusted R2, 0.04512; P = .003) supporting early WGS deployment in this population (eFigure 9 in Supplement 2).
Discussion
This randomized clinical trial investigated the effect of WGS on COM, as a measure for clinical utility, using a time-delayed trial design and a predefined rubric of care classification. It included several elements designed to capture the effect of WGS on COM against a background of real-world variability in infant care, including simplified inclusion criteria, enrollment from 5 children’s hospitals, a racially and ethnically diverse patient population, and the continuation of all usual-care practices, including molecular testing, which were anticipated to vary among enrollment sites.
The primary end point was the difference in the number of patients who received a COM 60 days after enrollment, which showed that 2-fold more patients in the early group had received a COM and a molecular diagnosis compared with the delayed group. Within-group analysis of the delayed group replicated these findings, showing that even when the return of WGS results was delayed by 60 days, there was a further doubling in the number of patients with a COM and a precision molecular diagnosis. Although the analysis of diagnostic efficacy was restricted to those patients with primary etiologic findings, COM was observed for a small proportion (1%) of patients who only received WGS secondary or incidental findings during the observation period.
The majority of observed management changes were supportive, with specialty referrals being the most common alteration of care. In 8% of diagnosed patients, however, changes of management directly addressed their etiologic molecular alteration. Additionally, the proportion of patients with a COM at day 90 trended upward even for infants who received their WGS diagnosis early, suggesting that genetic findings may provide a foundation for long-term clinical decision-making. There were no observed differences in length of stay or mortality, suggesting that either the principal effect of WGS was more focused and refined clinical care or that a larger study with shorter WGS testing turnaround times may be necessary to see these effects.
Improved diagnostic efficacy associated with WGS was likely tied to the wide range of variant types investigated and its uniform application to all patients across all 5 study sites, in contrast to the use of a wide range of usual-care testing. Indeed, we found that two-thirds of patients received a COM regardless of the testing modality, suggesting that the 2-fold higher diagnostic efficacy of systematically applied first-line WGS in acute infant care could reduce health care disparities.
Limitations
The primary limitation of this study was an observation window limited to 90 days. Given that the results presented here indicate ongoing use of genetic data for clinical decision-making, it is likely that subsequent genetic testing–related COMs were not captured. Additional limitations included the lack of validated instruments to assess patient- and family-reported outcomes and the study size and structure, which limited investigation of the diagnostic efficacy of WGS compared with specific usual-care tests. The cost of WGS may be a barrier to implementation in some environments, but this may be ameliorated by 2030 if recent projections of a $20 WGS are correct.22
Conclusions
The results of this randomized clinical trial add to a growing body of literature demonstrating that comprehensive genomic testing of acute care infants can be implemented in health systems,6 affect clinical management,6,7,8,9,10,11,12,13,14 and is positively viewed by both clinicians15 and the parents23 of acutely ill infants. The findings reported here demonstrate that WGS leads to focused, and therefore improved, patient care and should be considered as a primary tool in the assessment of critically ill infants with a suspected genetic disease.
Trial Protocol
eAppendix. NICUSeq Contributors, Clinical Site Operational Teams and Other Contributors, and Illumina Laboratory Services / Illumina Clinical Services Laboratory
eMethods 1. Inclusion & Exclusion Criteria
eMethods 2. Clinical Whole-Genome Sequencing
eMethods 3. Statistical Analysis
eReferences
eTable 1. Change of Management Classification Rubric
eTable 2. Demographic and Clinical Characteristics by Site
eTable 3. Enrollment Care Setting
eTable 4. Clinical Classification Definitions
eTable 8. Change of Management Outcome by Site at Day 60 (Visit 3)
eTable 9. Change of Management Outcome by WGS Variant Pathogenicity Classification
eTable 13. Diagnostic Efficacy* in Preterm Infants
eTable 14. WGS and UC Molecular Diagnosis Concordance
eFigure 1. Adverse Events by Site
eFigure 2. Patient Age at Enrollment
eFigure 3. Time From Admission to NICUSeq Enrollment Across Sites
eFigure 4. Clinical Indications for Testing by Site
eFigure 5. Time From Enrollment or Birth to Diagnosis
eFigure 6. Variation in Usual Care Testing by Assay, Outcome, and Site
eFigure 7. Length of Stay Stratified by Arm, Measurement Duration, and Test Outcome
eFigure 8. Survival Stratified by Testing Outcome
eFigure 9. Gestational Age and Time to Enrollment or Length of Stay
eTable 5. Patient Demographics, WGS/UC Findings and COM Status
eTable 6. Expanded HPO Terms for all Enrolled Patients
eTable 7. Change of Management (COM) Outcomes by Patient
eTable 10. All Secondary Findings
eTable 11. Incidental Findings
eTable 12. Patient Subset Who Received Only Secondary or Incidental Findings and Had COM
Data Sharing Statement
References
- 1.Chow S, Chow R, Popovic M, et al. A selected review of the mortality rates of neonatal intensive care units. Front Public Health. 2015;3:225. doi: 10.3389/fpubh.2015.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Namachivayam P, Shann F, Shekerdemian L, et al. Three decades of pediatric intensive care: who was admitted, what happened in intensive care, and what happened afterward. Pediatr Crit Care Med. 2010;11(5):549-555. doi: 10.1097/PCC.0b013e3181ce7427 [DOI] [PubMed] [Google Scholar]
- 3.Dukhovny D, Zupancic JAF. Economic evaluation with clinical trials in neonatology. Neoreviews. 2011;12(2):e69-e75. doi: 10.1542/neo.12-2-e69 [DOI] [Google Scholar]
- 4.Gonzaludo N, Belmont JW, Gainullin VG, Taft RJ. Estimating the burden and economic impact of pediatric genetic disease. Genet Med. 2019;21(8):1781-1789. doi: 10.1038/s41436-018-0398-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison W, Goodman D. Epidemiologic trends in neonatal intensive care, 2007-2012. JAMA Pediatr. 2015;169(9):855-862. doi: 10.1001/jamapediatrics.2015.1305 [DOI] [PubMed] [Google Scholar]
- 6.Lunke S, Eggers S, Wilson M, et al. ; Australian Genomics Health Alliance Acute Care Flagship . Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian public health care system. JAMA. 2020;323(24):2503-2511. doi: 10.1001/jama.2020.7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L, Liu X, Li Z, et al. Genetic aetiology of early infant deaths in a neonatal intensive care unit. J Med Genet. 2020;57(3):169-177. doi: 10.1136/jmedgenet-2019-106221 [DOI] [PubMed] [Google Scholar]
- 8.Kingsmore SF, Cakici JA, Clark MM, et al. ; RCIGM Investigators . A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am J Hum Genet. 2019;105(4):719-733. doi: 10.1016/j.ajhg.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrikin JE, Cakici JA, Clark MM, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med. 2018;3:6. doi: 10.1038/s41525-018-0045-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnaes L, Hildreth A, Sweeney NM, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 2018;3:10. doi: 10.1038/s41525-018-0049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171(12):e173438. doi: 10.1001/jamapediatrics.2017.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark Z, Schofield D, Martyn M, et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet Med. 2019;21(1):173-180. doi: 10.1038/s41436-018-0006-8 [DOI] [PubMed] [Google Scholar]
- 13.Stark Z, Tan TY, Chong B, et al. ; Melbourne Genomics Health Alliance . A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18(11):1090-1096. doi: 10.1038/gim.2016.1 [DOI] [PubMed] [Google Scholar]
- 14.French CE, Delon I, Dolling H, et al. ; NIHR BioResource—Rare Disease; Next Generation Children Project . Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019;45(5):627-636. doi: 10.1007/s00134-019-05552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimmock DP, Clark MM, Gaughran M, et al. ; RCIGM Investigators . An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management, and low perceived harm. Am J Hum Genet. 2020;107(5):942-952. doi: 10.1016/j.ajhg.2020.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips KA, Douglas MP, Marshall DA. Expanding use of clinical genome sequencing and the need for more data on implementation. JAMA. 2020;324(20):2029-2030. doi: 10.1001/jama.2020.19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spineli LM, Jenz E, Großhennig A, Koch A. Critical appraisal of arguments for the delayed-start design proposed as alternative to the parallel-group randomized clinical trial design in the field of rare disease. Orphanet J Rare Dis. 2017;12(1):140. doi: 10.1186/s13023-017-0692-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249-255. doi: 10.1038/gim.2016.190 [DOI] [PubMed] [Google Scholar]
- 19.Amendola LM, Robinson JO, Hart R, et al. Why patients decline genomic sequencing studies: experiences from the CSER Consortium. J Genet Couns. 2018;27(5):1220-1227. doi: 10.1007/s10897-018-0243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golec L, Gibbins S, Dunn MS, Hebert P. Informed consent in the NICU setting: an ethically optimal model for research solicitation. J Perinatol. 2004;24(12):783-791. doi: 10.1038/sj.jp.7211198 [DOI] [PubMed] [Google Scholar]
- 21.Prescott KR, Wilkie AOM. Genetic aspects of birth defects: new understandings of old problems. Arch Dis Child Fetal Neonatal Ed. 2007;92(4):F308-F314. doi: 10.1136/adc.2004.062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denny JC, Collins FS. Precision medicine in 2030-seven ways to transform healthcare. Cell. 2021;184(6):1415-1419. doi: 10.1016/j.cell.2021.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cakici JA, Dimmock DP, Caylor SA, et al. A prospective study of parental perceptions of rapid whole-genome and -exome sequencing among seriously ill infants. Am J Hum Genet. 2020;107(5):953-962. doi: 10.1016/j.ajhg.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial Protocol
eAppendix. NICUSeq Contributors, Clinical Site Operational Teams and Other Contributors, and Illumina Laboratory Services / Illumina Clinical Services Laboratory
eMethods 1. Inclusion & Exclusion Criteria
eMethods 2. Clinical Whole-Genome Sequencing
eMethods 3. Statistical Analysis
eReferences
eTable 1. Change of Management Classification Rubric
eTable 2. Demographic and Clinical Characteristics by Site
eTable 3. Enrollment Care Setting
eTable 4. Clinical Classification Definitions
eTable 8. Change of Management Outcome by Site at Day 60 (Visit 3)
eTable 9. Change of Management Outcome by WGS Variant Pathogenicity Classification
eTable 13. Diagnostic Efficacy* in Preterm Infants
eTable 14. WGS and UC Molecular Diagnosis Concordance
eFigure 1. Adverse Events by Site
eFigure 2. Patient Age at Enrollment
eFigure 3. Time From Admission to NICUSeq Enrollment Across Sites
eFigure 4. Clinical Indications for Testing by Site
eFigure 5. Time From Enrollment or Birth to Diagnosis
eFigure 6. Variation in Usual Care Testing by Assay, Outcome, and Site
eFigure 7. Length of Stay Stratified by Arm, Measurement Duration, and Test Outcome
eFigure 8. Survival Stratified by Testing Outcome
eFigure 9. Gestational Age and Time to Enrollment or Length of Stay
eTable 5. Patient Demographics, WGS/UC Findings and COM Status
eTable 6. Expanded HPO Terms for all Enrolled Patients
eTable 7. Change of Management (COM) Outcomes by Patient
eTable 10. All Secondary Findings
eTable 11. Incidental Findings
eTable 12. Patient Subset Who Received Only Secondary or Incidental Findings and Had COM
Data Sharing Statement