SUMMARY
STING modulates immunity by responding to bacterial and endogenous cyclic dinucleotides (CDNs). Humans and mice with STING gain-of-function mutations develop a syndrome known as STING-associated vasculopathy with onset in infancy (SAVI), which is characterized by inflammatory or fibrosing lung disease. We hypothesized that hyperresponsiveness of gain-of-function STING to bacterial CDNs might explain autoinflammatory lung disease in SAVI mice. We report that depletion of gut microbes with oral antibiotics (vancomycin, neomycin, and ampicillin [VNA]) nearly eliminates lung disease in SAVI mice, implying that gut microbes might promote STING-associated autoinflammation. However, we show that germ-free SAVI mice still develop severe autoinflammatory disease and that transferring gut microbiota from antibiotics-treated mice to germ-free animals eliminates lung inflammation. Depletion of anaerobes with metronidazole abolishes the protective effect of the VNA antibiotics cocktail, and recolonization with the metronidazole-sensitive anaerobe Bacteroides thetaiotaomicron prevents disease, confirming a protective role of a metronidazole-sensitive microbe in a model of SAVI.
Graphical abstract
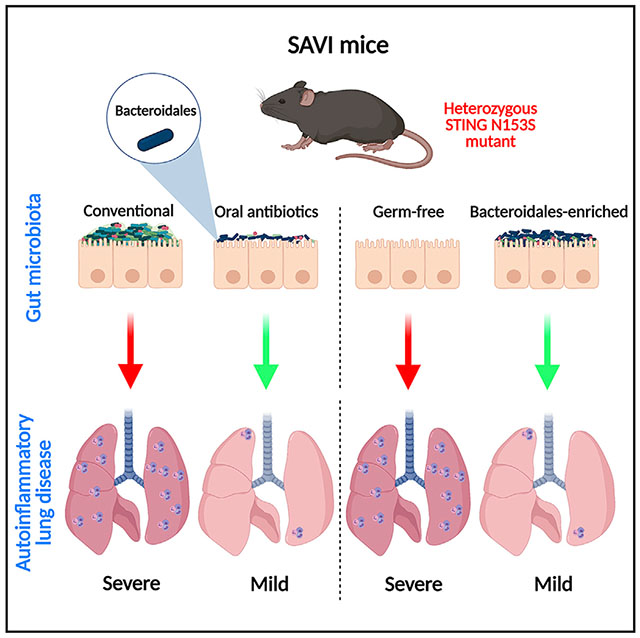
In brief
Platt et al. report that oral antibiotics but not germ-free conditions prevent autoinflammatory lung disease in a mouse model of STING-associated vasculopathy with onset in infancy (SAVI). Recolonization of SAVI mice with either Bacteroidales-enriched stool or Bacteroides thetaiotaomicron is protective in this model of STING-associated autoinflammatory lung disease.
INTRODUCTION
Stimulator of interferon genes (STING) regulates immune responses to bacteria and viruses by responding to cyclic dinucleotides (CDNs), including the endogenous STING ligand 2′3′-cyclic GMP-AMP (cGAMP) that is produced by cyclic GMP-AMP synthase (cGAS) (Sun et al., 2013). Bacteria also produce STING-activating CDNs, including c-di-GMP, c-di-AMP, and 3′3′-cGAMP (Davies et al., 2012). Humans and mice with gain-of-function mutations in STING develop spontaneous inflammation in the lungs, hypercytokinemia, and T cell cytopenia (Liu et al., 2014; Warner et al., 2017). Because STING gain-of-function mutants may exhibit enhanced ligand sensitivity (Liu et al., 2014), we reasoned that CDNs produced by commensal bacteria might regulate autoimmune lung disease pathogenesis in a mouse model of STING-associated vasculopathy with onset in infancy (SAVI).
STING responds to CDNs by activating the type I interferon (IFN) response. Bacterial CDNs were first described nearly 30 years ago (Ross et al., 1990) and can act as second messengers to regulate bacterial processes such as biofilm formation, virulence, antibiotic production, and other physiological processes (Ahmad et al., 2020; Nuzzo et al., 2020; Tang et al., 2016). In addition to activating the type I IFN response, CDNs are broadly immunostimulatory molecules that regulate innate and adaptive immunity (Elahi et al., 2014; Karaolis et al., 2007). Due to their immunostimulatory properties, CDNs have been studied extensively as adjuvants for vaccines and were effective in preclinical studies as cancer immunotherapy (Chandra et al., 2014; Fu et al., 2015; Nakamura et al., 2015). However, less is known about the role of bacterial and endogenous CDNs in autoimmune diseases, including those caused by gain-of-function mutations in STING.
The mammalian gastrointestinal tract is colonized by trillions of commensal bacteria, more in number and in species diversity than any other region of the body (Peterson et al., 2009; Savage, 1977). These commensal bacteria serve multiple functions that benefit the host, including facilitation of nutrient metabolism and other aspects of digestion (Hooper et al., 2002) and development of gut-associated lymphoid tissue (Lanning et al., 2000). Notably, gut bacteria and their metabolites can modulate allergic lung disease in mice (Trompette et al., 2014). Unlike models of asthma or allergic airway inflammation, SAVI mice develop inflammatory lung disease with perivascular immune cell infiltration (Warner et al., 2017). Whether gut microbes play a role in SAVI-associated lung disease was not previously studied.
Here, we report that gut anaerobes can modulate autoinflammatory lung disease in a mouse model of SAVI. Treatment of SAVI mice with an antibiotic cocktail, which spares some gut anaerobes, led to near-complete protection against lung disease in SAVI mice. Metagenomic sequencing revealed species of Bacteroidales, including Muribaculum and Bacteroides, in animals that were protected against systemic autoimmunity. Protection against autoimmunity was eliminated by adding the antibiotic metronidazole, which acts on anaerobic bacteria. The immunoprotective effects of gut anaerobes were transferrable to germ-free animals that otherwise develop severe autoinflammatory lung disease even in germ-free conditions. Furthermore, recolonization of microbe-depleted animals with Bacteroides thetaiotaomicron (B. theta) prevents autoinflammatory lung disease in SAVI mice, suggesting a role for specific commensal bacteria in regulating autoinflammatory lung disease. Our findings may lead to further studies of microbe-host interactions in SAVI patients.
RESULTS
Bone-marrow-derived macrophages (BMDMs) from SAVI mice are hyperresponsive to a bacterial CDN
We previously found that SAVI mice develop perivascular inflammation in the lung, but not airway disease (Warner et al., 2017). The perivascular lung pathology develops independently of cGAS, suggesting that the endogenous STING ligand cGAMP is not required to initiate disease (Luksch et al., 2019). Because STING also detects bacterial CDN ligands (Burdette et al., 2011), we hypothesized that STING N153S BMDMs might exhibit enhanced responses to a bacterial CDN. To test this hypothesis, we performed a dose-response experiment by transfecting wild-type (WT) and STING N153S BMDMs with c-di-GMP. We found that transfection with c-di-GMP results in a ~2- or 3-fold higher expression of interferon-stimulated genes (ISGs) in STING N153S BMDMs compared with WT control cells (Figures 1A and 1B), suggesting that the STING mutant is hyperresponsive to a bacterial CDN. STING signaling in antigen-presenting cells can cause upregulation of major histocompatibility complex (MHC) class II (Karaolis et al., 2007), but the impact of STING gain of function on MHC class II expression was not previously tested. In BMDMs, STING gain of function was associated with higher MHC class II expression at baseline and, in response to c-di-GMP, a ~1.5-fold increase in MHC class II upregulation compared with WT BMDMS (Figures 1C–1E). In contrast, MHC class II was not upregulated in response to c-di-GMP in STING Goldenticket (GT) BMDMs, which lack functional STING signaling (Figures 1C–1E). Similarly, the costimulatory molecule CD86 was upregulated in both WT and STING N153S cells, but not in STING GT macrophages (Figure 1F). In contrast, CD80 expression was not impacted by c-di-GMP under these conditions (Figure 1G). Collectively, these results demonstrate that the STING N153S mutation renders BMDMs more responsive to c-di-GMP stimulation.
Figure 1. STING N153S gain-of-function bone-marrow-derived macrophages (BMDMs) are hyper-responsive to cyclic di-GMP, a bacterial cyclic dinucleotide.
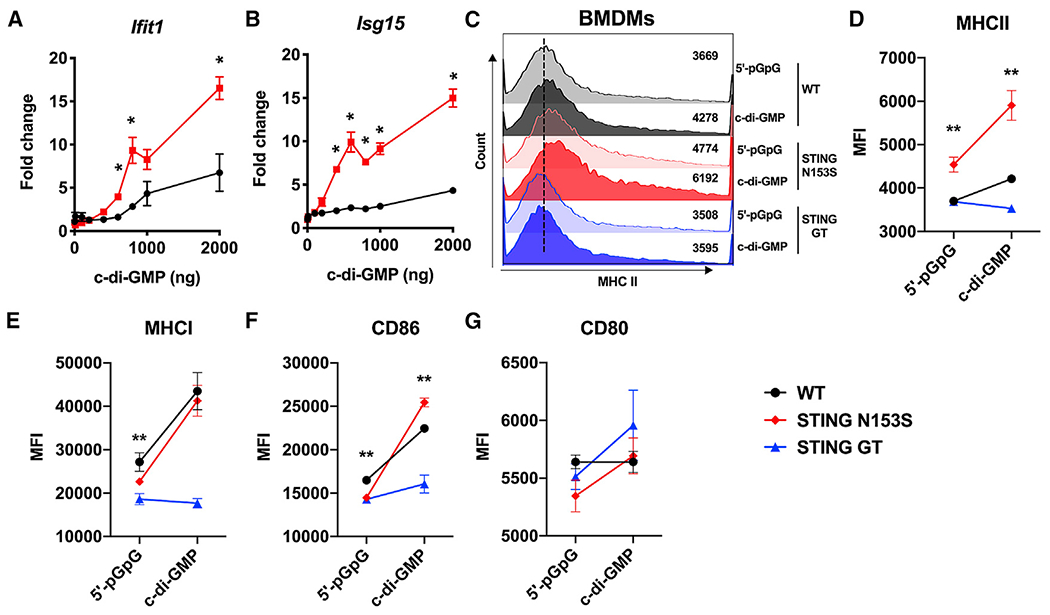
BMDMs were isolated from WT, STING N153S (SAVI), or STING Goldenticket mice following stimulation with cyclic di-GMP or control.
(A and B) Gene expression levels of IFIT1 and ISG15 were measured in WT and STING N153S BMDMs.
(C–G) Geometric mean of (C and D) MHC class II, (E) MHC class I, (F) CD86, and (G) CD80 on WT, STING N153S, or STING Goldenticket mice was measured by flow cytometry following stimulation with cyclic di-GMP or control.
Data represent the mean ± SEM of 4 biological replicates per group, which were pooled from 2 independent experiments. Results were analyzed by Mann-Whitney test. *p < 0.05; **p < 0.01.
Treatment with an antibiotic cocktail nearly eliminates lung disease in SAVI mice
Because STING N153S BMDMs are more responsive to bacterial c-di-GMP, we reasoned that commensal bacteria may contribute to spontaneous autoimmunity and lung disease in SAVI mice. To test our hypothesis that bacteria contribute to perivascular lung inflammation in our model of SAVI, we administered an antibiotic cocktail of vancomycin, neomycin, and ampicillin (VNA) to 4-week-old co-housed WT and SAVI mice. Treatment with VNA antibiotics almost completely eliminated histological evidence of lung disease in SAVI animals (Figures 2A and 2B). Treatment of mice for 4 weeks and subsequent removal of antibiotics led to recurrence of lung disease, suggesting a need for continuous depletion of microbes to prevent lung disease (Figure S1).
Figure 2. Treatment with the vancomycin, neomycin, and ampicillin (VNA) antibiotics cocktail reduces perivascular inflammation and ISGs in the lungs of SAVI mice.
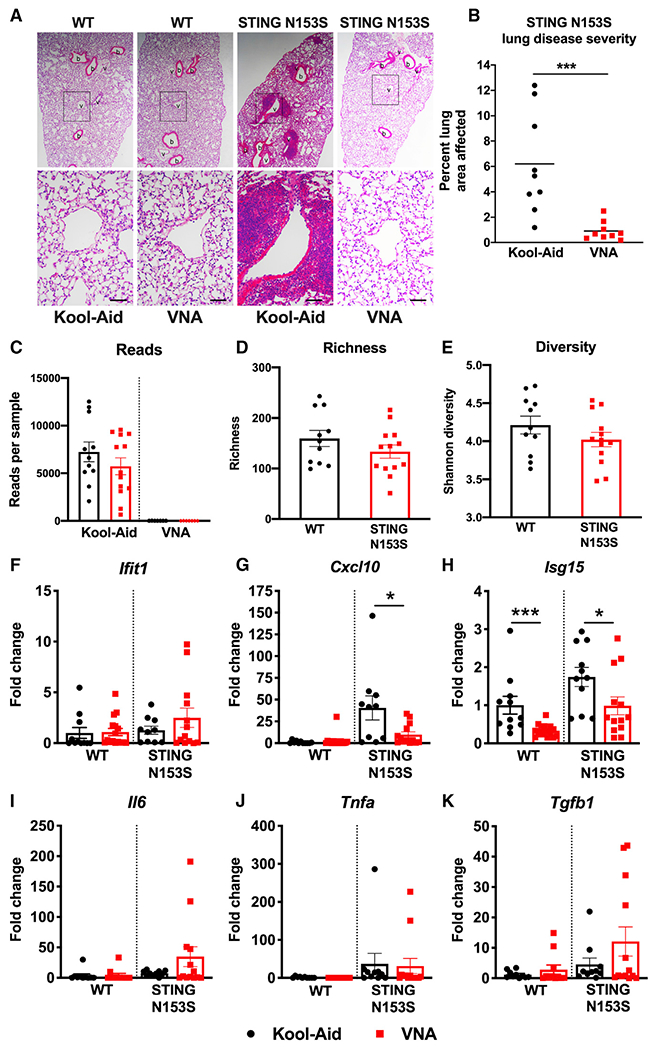
Four-week-old STING N153S (SAVI) mice were administered VNA cocktail or vehicle control ad libitum and euthanized at age 12 weeks.
(A) Representative hematoxylin and eosin-stained images of lung sections from age-matched 12-week-old STING N153S mice and littermate control mice. Top panel images were taken under low magnification, and bottom panel images were taken under high magnification. Scale bars: 100 μm.
(B) Quantitation of affected lung area of STING N153S mice following administration of VNA or control.
(C–E) 16S analysis on stool samples collected from 12-week-old, co-housed WT and STING N153S littermates.
(F–K) Gene expression of ISGs, cytokines, and chemokines following administration of VNA or control. Data represent the mean ± SEM of n = 5–13 mice per genotype pooled from 3 independent experiments. Results were analyzed by Mann-Whitney test. *p < 0.05; ***p < 0.001.
Because various bacteria differentially produce CDNs in response to phages (Cohen et al., 2019), we reasoned that differences in microbial populations may explain the effect of antibiotics on lung disease. To begin to characterize the gut microbiota of our animals, we performed 16S and phageome analysis of fecal samples from cohoused WT and SAVI mice. Treatment with the VNA antibiotics cocktail almost completely depleted the gut of bacteria based on 16S analysis (Figure 2C), although Bacteroides species remained detectable by PCR (data not shown). In untreated, co-housed WT and SAVI animals, mouse genotype had no effect on bacterial richness or diversity (Figures 2D, 2E, and S2A). Furthermore, virome analysis revealed similar bacteriophage compositions in co-housed WT and SAVI mice (Figures S2B–S2E).
Levels of pro-inflammatory cytokines, chemokines, and ISGs are elevated in the lungs of SAVI mice (Luksch et al., 2019). Treatment with VNA antibiotics caused a ~2-fold reduction in Isg15 and a ~4-fold reduction in Cxcl10 expression in the lungs of SAVI mice compared with WT control animals (p < 0.05) (Figures 2F–2K). In addition to almost completely eliminating histological evidence of lung disease (Figures 2A and 2B), multiplex cytokine analysis of lung homogenates revealed that pro-inflammatory cytokine and chemokine levels were returned to levels of healthy WT control mice (Figures 3A–3C). Thus, antibiotics can eliminate autoinflammatory lung disease in SAVI mice, but this protective effect requires continuous treatment with antibiotics.
Figure 3. VNA treatment reduces pro-inflammatory cytokines, chemokines, and Th1 cytokine levels in the lungs of SAVI mice.
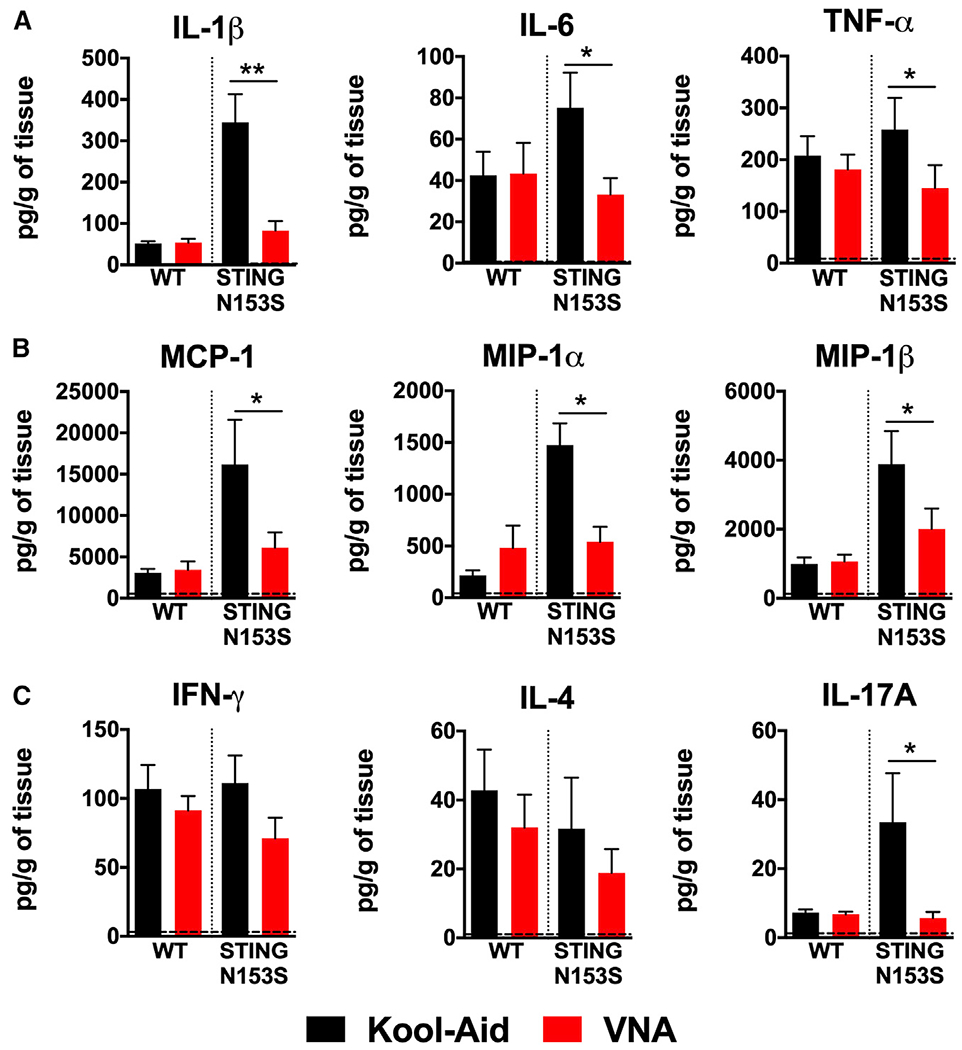
Four-week-old STING N153S (SAVI) mice were administered VNA cocktail or vehicle control ad libitum and euthanized at 12 weeks.
(A–C) Cytokine and chemokine levels were assessed by a BioPlex assay of lung homogenates. Data represent the mean ± SEM of n = 5–8 biological replicates group, which were pooled from 2 independent experiments. Results were analyzed by Mann-Whitney test. *p < 0.05; **p < 0.01.
Treatment with oral antibiotics alters immune cell populations in SAVI mice
We previously found that αβ T cells are required for severe lung disease in SAVI mice (Luksch et al., 2019), so we hypothesized that treatment with the VNA antibiotics cocktail may alter immune cell populations. Treatment of SAVI mice with oral antibiotics increased the percent of naive T cells while decreasing the percent and number of T effector cells (Figure S3). However, these relatively small alterations in T cell populations do not necessarily explain the near-complete elimination of lung disease in SAVI animals. Because bacterial CDNs can regulate numbers of myeloid cell populations in the peritoneum and the spleen (Karaolis et al., 2007), we reasoned that depletion of gut microbes may have the opposite effect on myeloid cell populations in mice. Indeed, we found that the VNA antibiotics cocktail led to a ~4-fold decrease in the number of dendritic cells (CD11c+ MHCII+), a ~2-fold decrease in inflammatory monocytes (CD11b+ Ly6Chi), and a ~3-fold reduction in neutrophils (CD11b+ Ly6G+) in the spleens of SAVI, but not WT littermate control animals (Figure S4). Although certain antibiotics have the capacity to alter immune cell populations via effects on hematopoiesis (Josefsdottir et al., 2017), we observed changes only in SAVI mice and not in WT littermate control animals, suggesting a specific effect of antibiotics in STING N153S gain of function. These results are consistent with the idea that STING N153S is hyperresponsive to bacterial CDNs.
Gut anaerobes but not germ-free conditions can prevent STING-associated autoinflammatory lung disease in mice
Because the VNA antibiotics cocktail nearly eliminates lung disease in SAVI mice, we hypothesized that deleterious bacteria are required to induce autoimmunity, and that autoinflammatory lung disease would not develop in the absence of commensal microbes. To test whether commensal microbes are required for lung disease, we generated germ-free SAVI mice. At 4 weeks of age, we recolonized half of the germ-free SAVI animals with stool from mice housed under enhanced specific-pathogen-free (ESPF) conditions. Because antibiotics prevent lung disease in SAVI mice, we anticipated that germ-free SAVI animals would not develop lung disease. Unexpectedly, we found that germ-free SAVI mice still develop severe lung disease. Indeed, the severity of disease was the same in re-colonized and germ-free SAVI mice, demonstrating that complete elimination of commensal bacteria does not prevent lung disease (Figures 4A and 4B).
Figure 4. Transplant of stool from VNA antibiotics-treated mice into germ-free SAVI mice nearly eliminates STING-associated lung disease.
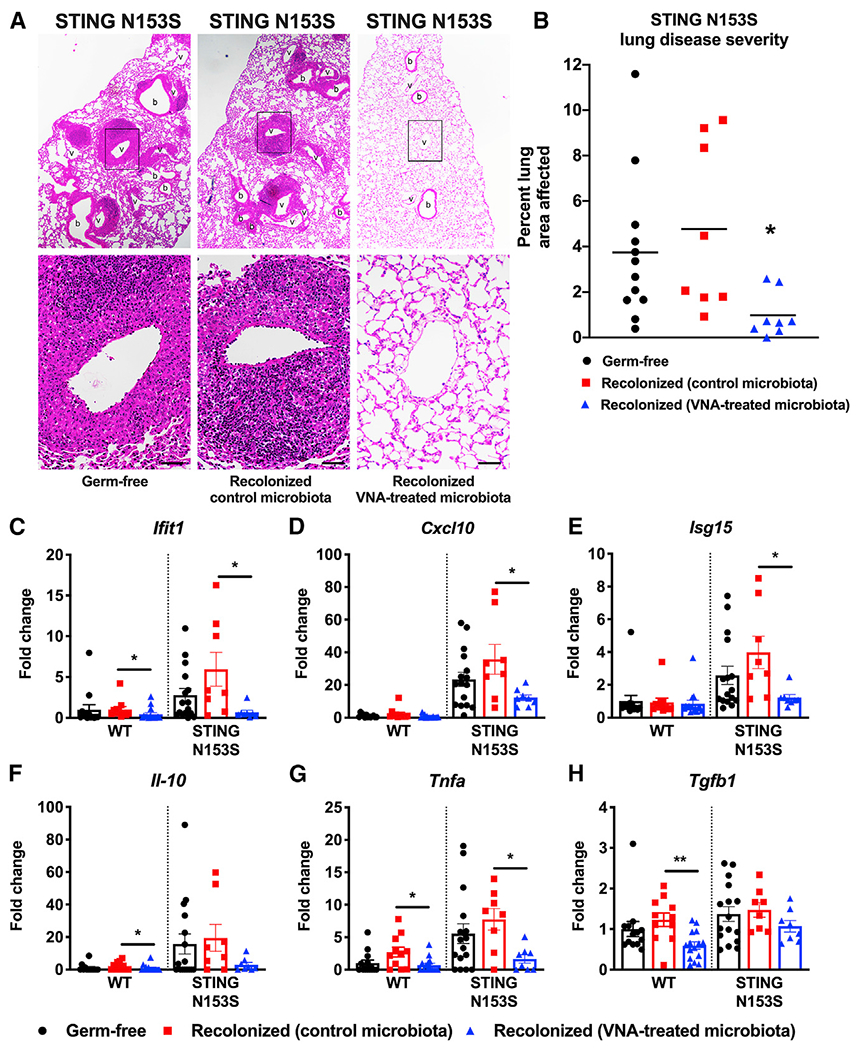
Germ-free, co-housed WT and STING N153S (SAVI) mice were recolonized with stool from either untreated STING N153S mice or VNA-treated STING N153S mice.
(A) Representative hematoxylin and eosin-stained images of lung sections from age-matched 12- to 13-week-old STING N153S mice and littermate control mice. Top panel images were taken under low magnification, and bottom panel images were taken under high magnification. Scale bars: 100 μm.
(B) Quantitation of affected lung area of STING N153S mice with or without recolonization.
(C–H) Gene expression of ISGs, cytokines, and chemokines with or without recolonization. Data represent the mean ± SEM of n = 8–16 mice per group pooled from 2 independent experiments. Results were analyzed by Mann-Whitney test. *p < 0.05; **p < 0.01.
The VNA antibiotics cocktail would not be expected to eliminate all commensal microbes, especially because these antibiotics would not fully eradicate anaerobic bacteria. For example, we used a pan-bacteroides PCR assay to confirm that at least some Bacteroides species likely persist after treatment with VNA (data not shown). Because germ-free SAVI mice develop severe autoinflammatory lung disease, we reasoned that some of these gut anaerobes not sensitive to VNA might protect against STING-associated lung disease. To determine whether the protective factor is transferrable, we performed a fecal transplant of stool from VNA-treated SAVI animals into germ-free SAVI mice using stool from VNA-treated SAVI animals. Two weeks after recolonization, we histologically examined lungs and found that recolonization using microbes from antibiotics-treated mice resulted in prevention of autoinflammatory lung disease in SAVI animals (Figures 4A and 4B). Recolonization with stool from VNA antibiotics-treated mice also led to a reduction in expression of Ifit1, Cxcl10, Isg15, and Tnfa, but not Il10 or Tgfb1 compared with control microbiota recolonized mice (Figures 4C–4H). However, unlike antibiotics treatment, germ-free conditions had no effect on numbers or subsets of lymphoid and myeloid cell populations other than a small increase in CD11b+ F4/80+ myeloid cells in recolonized animals (Figure S5). Similarly, recolonization with VNA-treated microbes had no effect on immune cells in SAVI animals (Figure S6). Thus, although bacteria are not required for lung disease pathogenesis in SAVI mice, transplant of gut microbiota from VNA antibiotics-treated animals almost completely protects against STING-associated autoinflammatory lung disease.
Because the VNA antibiotics cocktail spares some gut anaerobes that are sensitive to metronidazole, we hypothesized that metronidazole-sensitive microbes mediate protection against STING-associated autoimmunity. Indeed, addition of metronidazole to the VNA cocktail eliminated the protective effect of those antibiotics (Figure 5). This suggests that metronidazole-sensitive microbes can protect against STING-associated autoinflammatory lung disease in mice.
Figure 5. Metronidazole eliminates the lung-protective effects of other antibiotics.
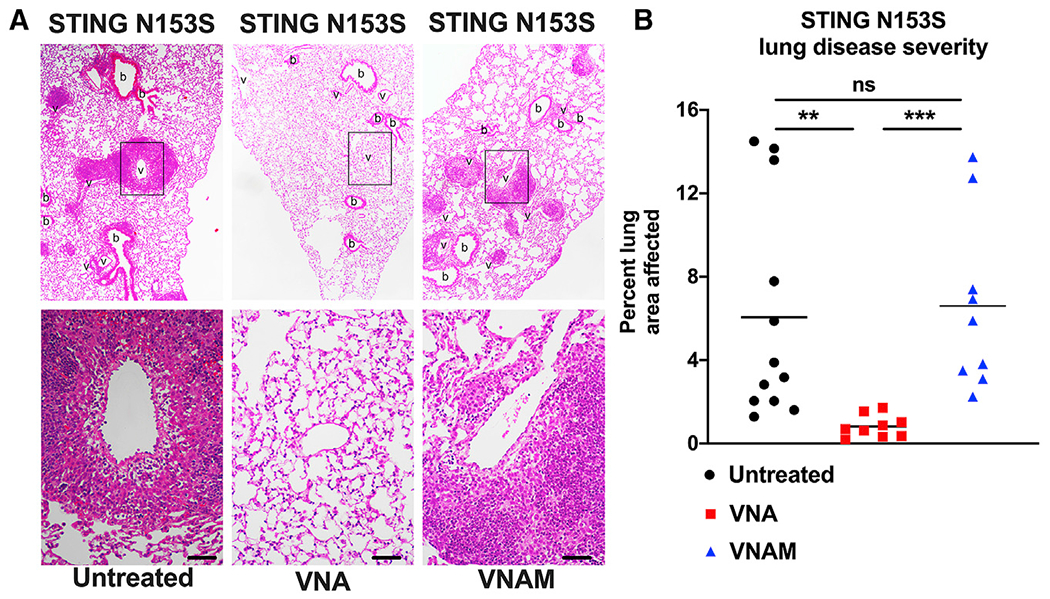
Ten-week-old STING N153S (SAVI) mice were administered a VNA cocktail or vehicle control ad libitum and euthanized at 12 weeks.
(A) Representative hematoxylin and eosin-stained images of lung sections from age-matched 12-week-old STING N153S mice. Top panel images were taken under low magnification, and bottom panel images were taken under high magnification. Scale bars: 100 μm.
(B) Quantitation of affected lung area of STING N153S mice following administration of VNA or control. Data represent the mean of n = 9–12 mice per group, pooled from 3 independent experiments. Results were analyzed by Mann-Whitney test. **p < 0.01; ***p < 0.001.
An isolated presence of Bacteroidales is associated with protection against STING-associated lung disease in mice
Because protozoa are susceptible to metronidazole, we considered that the protective, metronidazole-sensitive microbe might be eukaryotic. However, 18S analysis of stool from VNA-treated and untreated SAVI mice revealed no differences in the composition of the eukaryotic gut microbiota (Figure S7). Subsequent metagenomic analysis of stool from recolonized and germ-free animals revealed a dramatically reduced number of bacterial reads in germ-free animals, as well as in germ-free mice recolonized with microbiota from mice treated with the VNA antibiotics cocktail (Figure 6A), while recolonization with control gut microbiota resulted in a population of commensals similar to that of ESPF mice (Figures 6A and 6B). Shotgun metagenomic data of ESPF samples were concordant with 16S results (Figure S2A), reflecting classification of prior Porphyromonadaceae as Muribaculaceae in more recently updated databases (Lagkouvardos et al., 2016). In contrast, the relatively sparsely populated gut of recolonized mice that received microbiota from antibiotics-treated mice demonstrated a predominance of metronidazole-sensitive anaerobes of the order Bacteroidales, including gut anaerobes such as Muribaculum (Figures 6C and 6D). Collectively, these results suggest that a relative enrichment of Bacteroidales, combined with depletion of other gut bacteria, may protect against STING-associated autoinflammatory lung disease.
Figure 6. VNA-recolonized SAVI mice have a reduction in bacterial reads compared with ESPF and control recolonized mice and an outgrowth of Bacteroidales not found in germ-free samples.
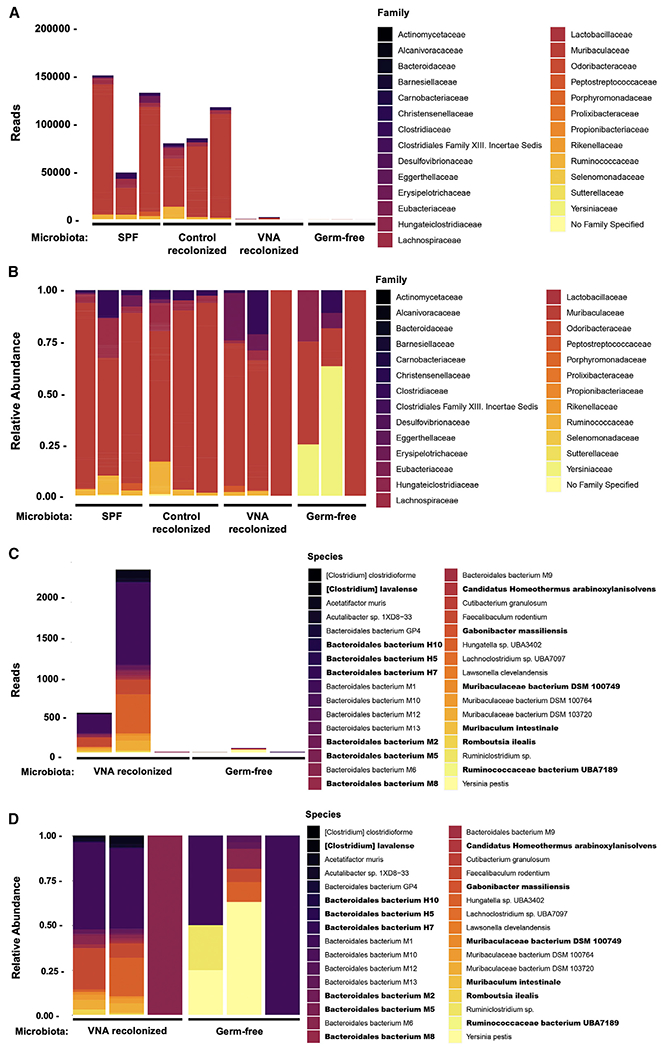
Ten-week-old STING N153S (SAVI) mice were recolonized with stool from VNA-treated STING N153S animals or left germ-free, and stool was collected at 12 weeks.
(A) Bacterial family reads obtained from metagenomic sequencing of stool DNA from ESPF, control recolonized, VNA-recolonized, or germ-free STING N153S mice.
(B) Bacterial family relative abundance obtained from metagenomic sequencing of stool DNA from ESPF, control recolonized, VNA-recolonized, or germ-free STING N153S mice.
(C) Bacterial species reads obtained from metagenomic sequencing of stool DNA from VNA-recolonized or germ-free STING N153S mice.
(D) Bacterial species relative abundance obtained from metagenomic sequencing of stool DNA from VNA-recolonized or germ-free STING N153S mice. Species unique to VNA-recolonized samples are shown in boldface. Data represent samples collected from n = 3 mice per condition pooled from 2 independent experiments.
Recolonization with B. theta protects against autoinflammatory lung disease in SAVI animals
To confirm that bacteria from the order Bacteroidales can protect against lung disease in SAVI mice, we recolonized VNAM-treated SAVI mice with B. theta as a representative species for Bacteroidales and histologically assessed perivascular lung inflammation 2 weeks after recolonization. We discovered that recolonization with B. theta was sufficient to reduce lung disease in SAVI mice (Figures 7A and 7B). Gene expression analysis of select ISGs and cytokines revealed a ~2-fold reduction in Il10, as well as trends toward diminished expression of Cxcl10 and Ifit1 (Figures 7C–7G). These data confirm the capacity of a Bacteroides species to protect against autoinflammatory lung disease in SAVI animals.
Figure 7. Recolonization with Bacteroides thetaiotaomicron and treatment with propionate and butyrate protect against STING-associated lung disease in mice.
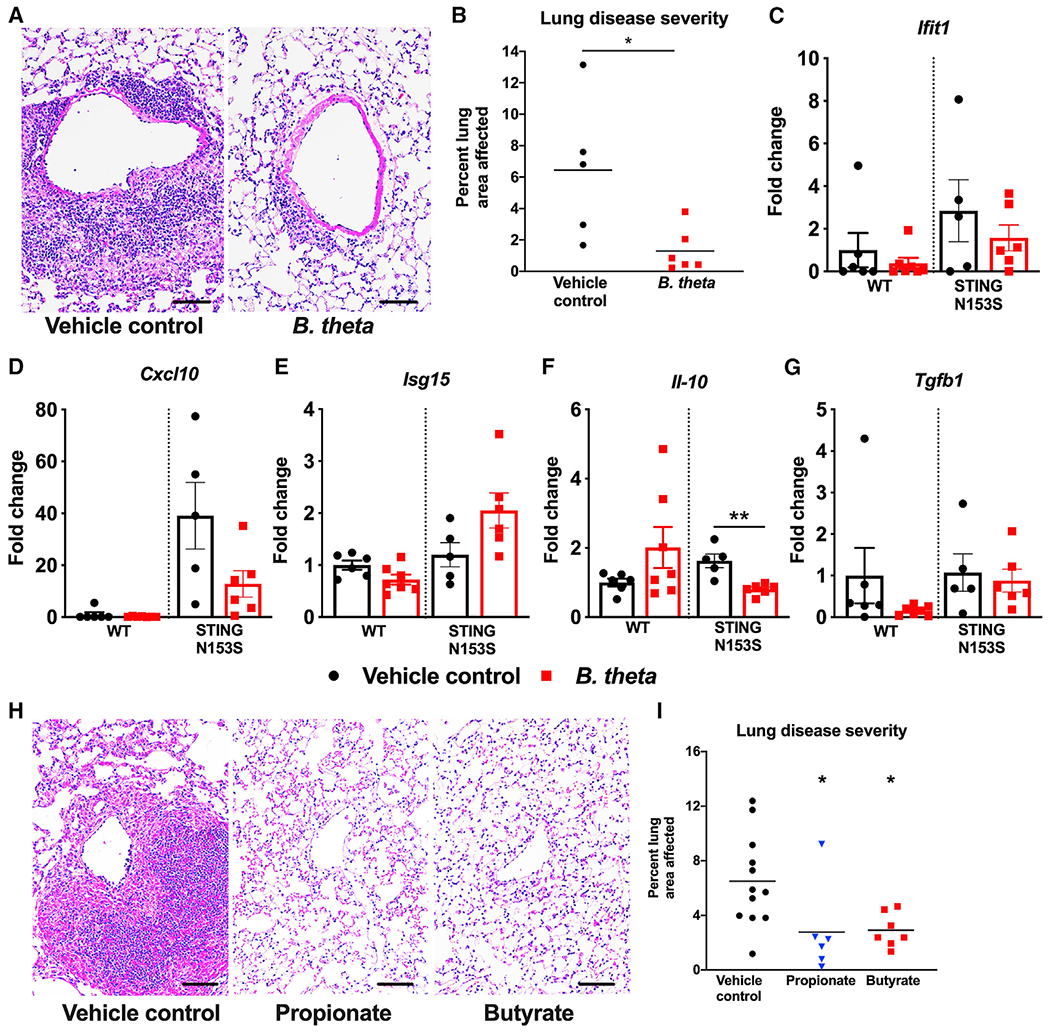
Ten-week-old co-housed WT and STING N153S (SAVI) mice were recolonized with either B. theta or vehicle control.
(A) Representative hematoxylin and eosin-stained images of lung sections from age-matched 12- to 13-week-old STING N153S mice and littermate control mice. Top panel images were taken under low magnification, and bottom panel images were taken under high magnification. Scale bar: 100 μm.
(B) Quantitation of affected lung area of STING N153S mice with or without recolonization.
(C–G) Gene expression of ISGs, cytokines, and chemokines with or without recolonization. Data represent the mean ± SEM of n = 5–6 mice per group pooled from 2 independent experiments and were analyzed using Mann-Whitney test. Ten-week-old co-housed WT and STING N153S mice were given vehicle control, 1 mg/kg propionate, or 1 mg/kg butyrate daily for 2 weeks via intraperitoneal injection and were euthanized at 12 weeks.
(H) Representative hematoxylin and eosin-stained images of lung sections from age-matched 12-week-old STING N153S mice and littermate control mice. Scale bars: 100 μm.
(I) Quantitation of affected lung area of STING N153S mice following administration of propionate or vehicle control.
Data in (H) and (I) represent the mean of n = 6–12 mice per group pooled from 2 independent experiments. Data were analyzed using Mann-Whitney test. *p < 0.05; **p < 0.01.
The short-chain fatty acids propionate and butyrate can protect against STING-associated lung disease
Short-chain fatty acids serve as an energy source for bacteria (Xie et al., 2009) but also have the capacity to regulate host immune responses (Arpaia et al., 2013; Meyer et al., 2020). Many bacterial species from the genus Bacteroides produce short-chain fatty acids, including propionate (Reichardt et al., 2014), and species of genus Muribaculum also are equipped with fermentation pathways to produce propionate (Smith et al., 2019). Since recolonization with B. theta protects against severe autoinflammatory lung disease, we hypothesized that treatment with short-chain fatty acids would protect against lung disease in SAVI animals. We found that treatment with propionate and butyrate led to a decrease in STING-associated autoinflammatory lung disease, highlighting a potential role for short-chain fatty acids produced by Bacteroidales (Figures 7H and 7I). Short-chain fatty acids derived from anaerobes might be one microbial factor that mediates protection against STING-associated autoinflammatory lung disease.
DISCUSSION
STING gain of function causes severe lung disease in patients with SAVI. Because STING responds to bacteria-derived CDN ligands, we tested whether bacteria contributes to autoinflammatory disease pathogenesis in a mouse model of SAVI. Unexpectedly, we discovered that STING gain of function causes autoinflammatory lung disease in germ-free animals that lack commensal microbes. In addition, we discovered that modulating the composition and abundance of commensal microbes almost completely eliminates systemic autoimmunity in SAVI mice. Furthermore, transfer of metronidazole-sensitive Bacteroidales bacteria to germ-free SAVI animals was associated with protection against STING-associated lung disease.
Liu et al. (2014) found that SAVI patient fibroblasts with STING gain-of-function mutations are hyperresponsive to CDNs. We previously found that STING gain-of function mice develop autoimmunity, T cell cytopenia, and lung disease independently of the endogenous STING ligand cGAMP. Here, we found that STING N153S BMDMs are hyperresponsive to the bacterial STING ligand c-di-GMP, and so we hypothesized that bacterial CDNs may be responsible for lung disease in SAVI animals. However, our discovery that germ-free SAVI mice still develop autoinflammatory lung disease revealed that bacterial CDNs are not required for STING-associated autoimmunity. Nevertheless, we cannot exclude the possibility that bacterial CDNs may influence disease severity in SAVI, perhaps in ways that were beyond our ability to detect.
That gut microbes influence autoinflammatory lung disease confirms the presence of a gut-lung axis in a model of SAVI. How gut microbes influence autoimmunity in the lung is less clear. Organs previously thought to be sterile or relatively sterile have their own distinct microbial populations (Harris et al., 2007). These distinct microbial communities do not exist in isolation and can influence the microbiota in other organs (Bassis et al., 2015). Some have speculated that a gut-lung axis exists, such that intestinal and pulmonary environments can influence each other (Cait et al., 2018; Trompette et al., 2014). For example, Cooke et al. (2000) suggested a role for gut-derived lipopolysaccharide in modulating immune cell populations and inflammatory mediators in the lung during idiopathic pneumonia syndrome after bone marrow transplantation. Trompette et al. (2014) found that altering the ratio of Firmicutes to Bacteroidetes led to a reduction in Th2 responses and protection against allergic airway inflammation. Others have found that treatment with antibiotics that deplete gut microbes can alter vulnerability to bacterial infections of the lung (Robak et al., 2018; Schuijt et al., 2016). Segmented filamentous bacteria (SFB) can induce lung inflammation in transgenic mice expressing dual SFB-specific T cell receptors (TCRs) (Bradley et al., 2017), indicating that lung inflammation can be initiated by antigen-specific T cells directed against a gut microbe. Unlike other studies, which were primarily focused on the role of gut microbes in regulating infection or allergic inflammation, our work reveals a role for gut microbes in regulating autoinflammation in a lung disease caused by a human disease-associated mutation. Furthermore, in contrast with the work of Trompette et al. (2014), our data do not suggest any alteration in Th2 cytokines in association with the reduction in lung disease (Figure 3). This suggests that the mechanism of protection in our model of STING-associated autoimmunity is distinct from that which was shown in a different model of lung disease.
We found that antibiotics caused alterations in myeloid and lymphoid cell populations, and that those effects could not be transferred by recolonization of germ-free mice. However, we cannot exclude the possibility that antibiotics also have microbe-independent effects on immune cells (Gopinath et al., 2018), which may protect against lung disease in SAVI animals. Nevertheless, transfer of microbes into germ-free animals was protective even without appreciably altering immune cell populations. Without our germ-free mouse studies, we might have been misled to speculate that commensal microbes were required for STING-associated autoinflammatory lung disease. Our results underscore the value of studies in germ-free and recolonized animals.
Anaerobes regulate immunity in other models of autoimmunity, including experimental autoimmune encephalomyelitis (EAE). For example, Bacteroides fragilis produces molecules including polysaccharide A, which can promote expression of anti-inflammatory cytokine IL-10 and protect against demyelination in EAE (Ochoa-Repáraz et al., 2010). Lactobacillus reuteri also can suppress EAE (He et al., 2019). In other instances, bacterial metabolites such as short-chain fatty acids can modulate immunity and reduce allergic inflammation in the lung (Trompette et al., 2014). Because STING-associated lung disease is primarily mediated by αβ T cells, one reasonable hypothesis is that a gut anaerobe metabolite mediates the protective effect. Indeed, we confirmed that the short-chain fatty acids propionate and butyrate, which are common metabolites produced by anaerobes, can inhibit lung disease in SAVI animals. However, our results also suggest that metronidazole-sensitive anaerobes protect against STING-associated autoimmunity only upon depletion of bacteria sensitive to VNA. This implies that protective effects of anaerobes can be inhibited by VNA-sensitive bacteria. The precise roles and molecular mechanisms of these microbial community interactions remain to be determined.
Metagenomic analysis on animals recolonized with stool from VNA antibiotics-treated SAVI mice revealed several anaerobic species from the order Bacteroidales, most species of which have not been characterized. In recent years, Muribaculaceae species, such as Muribaculum intestinale, were identified as a core component of the commensal microbiome of several homeothermic species (Ormerod et al., 2016). However, little is known about M. intestinale outside of its existence as a commensal microorganism, and even less is known about many of the other Bacteroidales species that are enriched in mice recolonized with stool from VNA antibiotics-treated animals. Whether these bacterial populations have the capacity to modulate other autoimmune diseases has not been investigated. Recolonization with B. theta, which we chose as a representative species of Bacteroidales, was sufficient to diminish the severity of autoinflammatory lung disease in SAVI animals. Although treatment with VNA antibiotics led to the relative enrichment of several Bacteroidales species bacteria, our finding provides evidence that a single species can modulate perivascular inflammation in the lung, although the immunological mechanism has not yet been elucidated.
Commensal bacteria of the order Bacteroidales can produce short-chain fatty acids (Reichardt et al., 2014). We found that treatment of SAVI mice with the short-chain fatty acids propionate or butyrate can protect against STING-associated autoinflammatory lung disease, and that elimination of lung disease was associated with isolated colonization with Bacteroidales. Production of immune-modulating short-chain fatty acids might be one of several mechanisms by which relative enrichment of certain bacterial species can protect against autoinflammatory lung disease in SAVI animals. Short-chain fatty acid receptors GPR41 and GPR43 are expressed on immune cells (Maslowski et al., 2009; Sina et al., 2009) and modulate inflammatory responses through inhibition of histone deacetylase activity. Nevertheless, our studies did not definitively elucidate an immunological mechanism by which antibiotics modulate autoinflammatory lung disease in SAVI mice. An alternative hypothesis is that specific bacterial populations may produce small-molecule antagonists of STING that specifically modulate inflammatory responses in our model of STING-associated autoimmunity. Further characterization of protective gut anaerobes, including characterization of potentially protective metabolites, may lead to novel therapies for the treatment of SAVI and other autoimmune diseases associated with hyperactivation of the STING pathway.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jonathan J. Miner (miner@wustl.edu).
Materials availability
This study did not generate new unique reagents. Commercially available reagents are indicated in the Key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-CD3 | Biolegend | RRID:AB_2562555 |
| Mouse anti-CD4 | Biolegend | RRID:AB_2562557 |
| Mouse anti-CD8a | Biolegend | RRID:AB_2075239 |
| Mouse anti-CD11b | Biolegend | RRID:AB_2629529 |
| Mouse anti- CD11c | Biolegend | RRID:AB_389306 |
| Mouse anti-CD19 | Biolegend | RRID:AB_312825 |
| Mouse anti-CD45 | Biolegend | RRID:AB_2650656 |
| Mouse anti-F4/80 | Biolegend | RRID:AB_893481 |
| Mouse anti-I-A/I-E | Biolegend | RRID:AB_313323 |
| Mouse anti-Ly6C | Biolegend | RRID:AB_2562177 |
| Mouse anti-Ly6G | Biolegend | RRID:AB_2616999 |
| Mouse anti-NK-1.1 | Biolegend | RRID:AB_2783136 |
| Bacterial and virus strains | ||
| Bacteroides thetaiotaomicron | ATCC | Cat# 29148 |
| Chemicals, peptides, and recombinant proteins | ||
| Vancomycin hydrochloride | Sigma-Aldrich | Cat# V2002 |
| Neomycin trisulfate salt hydrate | Sigma-Aldrich | Cat# N1876 |
| Ampicillin sodium salt | Sigma-Aldrich | Cat# A9518 |
| Metronidazole | Sigma-Aldrich | Cat# M1547 |
| Dulbecco’s phosphate buffered saline | ThermoFisher | Cat# 14190136 |
| Paraformaldehyde 20% solution, EM grade | Electron Microscopy Sciences | Cat# 15713-S |
| Butyric acid sodium salt | Sigma-Aldrich | Cat# 303410 |
| Sodium propionate | Sigma-Aldrich | Cat# P1880 |
| Critical commercial assays | ||
| Bio-Plex Pro Mouse Cytokine 23-Plex | Bio-Rad | Cat# M60009RDPD |
| Assay | ||
| DNeasy 96 Blood and Tissue Kit | QIAGEN | Cat# 69581 |
| QIAamp Fast DNA Stool Mini Kit | QIAGEN | Cat# 51604 |
| Nextera DNA Library Prep Kit | Illumina | Cat# 20018795 |
| RNeasy Mini Kit | QIAGEN | Cat# 74106 |
| TaqMan RNA-to-Ct 1-step kit | Applied Biosystems | Cat# 4392653 |
| Deposited data | ||
| 16S and Metagenomics data | European Nucleotide Archive | PRJEB42379 |
| Experimental models: Organisms/strains | ||
| Mouse: STING N153S mice | Jackson Laboratory | Stock# 033543 |
| Mouse: Germ-free STING N153S mice | This paper | N/A |
| Oligonucleotides | ||
| Cxcl10 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.43575827 |
| Gapdh PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.39a.1 |
| Ifit1 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.32674307 |
| Isg15 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.41476392 |
| Il6 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.10005566 |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Il10 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.13531087 |
| Tnfa PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.12575861 |
| Tgfb1 PrimeTime qPCR Assay | Integrated DNA Technologies | Mm.PT.58.11254750 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com |
| FlowJo | FlowJo | https://www.flowjo.com |
| Microbial Ecology software (QIIME, version 1.8.0) |
QIIME | http://qiime.org/ |
| BBTools v38.26 | BBTools | https://jgi.doe.gov/data-and-tools/bbtools/ |
| One Codex Database | One Codex | https://www.onecodex.com/ |
Data and code availability
The accession numbers for the rDNA and metagenomic sequencing data reported in this paper are European Nucleotide Archive: PRJEB42379
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All protocols for animal studies were approved by the Institutional Animal Care and Use Committees at the Washington University School of Medicine (assurance no. A-3381-01). Mice were housed in specific-pathogen-free mouse facilities at the Washington University School of Medicine. STING N153S mice (SAVI mice) were generated on a C57BL/6 background using CRISPR/Cas9 as previously described (Warner et al., 2017). In all experiments using SAVI mice, co-housed WT littermates were used as controls. Germ-free and recolonized mice were housed in the gnotobiotic mouse facility at the Washington University School of Medicine. Confirmation of a germ-free environment is done by bi-weekly wet mount observation, anaerobic and aerobic culture, fungal culture, and monthly 16S and Transnetyx sequencing. For recolonization studies, co-housed WT and SAVI mice in ESPF conditions were given the VNA cocktail or Kool-Aid in drinking water for two weeks. At the end of two weeks, fecal pellets from VNA- or Kool-Aid-treated SAVI animals were collected, then pooled and homogenized anaerobically into a liquid mixture. Germ-free WT and SAVI mice were recolonized by oral gavage with the anaerobic mixtures or left germ-free for two weeks. At the two week time point, mice were sacrificed, and stool, lungs, and spleen were collected for subsequent processing. For Bacteroides thetaiotaomicron recolonization studies, WT and SAVI mice were given the VNAM cocktail in drinking water for two weeks. After stopping treatment for one day, mice were given 1 × 108 CFU B. theta or vehicle control via oral gavage. Two days later, the mice were given a second oral gavage of 1 × 108 CFU B. theta or vehicle control. Two weeks after recolonization, mice were sacrificed and organs were collected for subsequent processing. Power analysis was conducted for Institutional Animal Care and Use Committee-approved in vivo studies in order to determine the number of animals needed per experimental group. At least two independent experiments were conducted to replicate findings. No outliers were excluded from analyses. Approximately equal numbers of both sexes were used. The age and number of animals used for each experiment is listed in the figure legends.
Bone marrow-derived macrophages
Bone marrow-derived macrophages (BMDMs) were generated from bone marrow cells flushed from the femurs and tibias of age-matched SAVI mice, WT littermates, and STING Goldenticket mice. Then 2.5 × 106 cells were incubated at 37°C in 10-cm cell culture plates in complete medium containing Dulbecco’s modified Eagle medium (DMEM) (catalog no. D6429; Sigma), 10% fetal bovine serum (FBS) (catalog no. SH30070-03; HyClone), 1% penicillin and streptomycin (catalog no. 15140; GIBCO), 1% sodium pyruvate (catalog no. 11360; GIBCO), and 1% L-glutamate (catalog no. 35050; GIBCO), supplemented with 40 ng/mL of macrophage colony-stimulating factor (M-CSF) (catalog no. 300-25; PeproTech), for 6 days. On day 7, BMDMs were replated and thereafter were cultured in complete medium with 20 ng/mL of M-CSF. Cells from approximately equal numbers of both sexes were used.
METHOD DETAILS
Histologic staining analysis
Mice were perfused with 20 mL of PBS, then lung samples were isolated from mice, fixed in 4% paraformaldehyde for 48 hours at room temperature, and suspended in 70% ethanol before being embedded in paraffin. Tissue sections (thickness of 5 μm) were stained with hematoxylin and eosin, followed by image acquisition using a Nikon Eclipse E400 microscope and NIS Elements software. Using ImageJ software (Schneider et al., 2012), lung lesions were encircled by a blinded researcher, who quantitated the number of pixels within lung lesions divided by the total number of pixels in the lung tissue, excluding large airway spaces. For colon histology, the entire length of the colon was removed from mice and gently flushed with 5 mL of PBS, followed by 5 mL of 4% paraformaldehyde with blunt tipped needles. Next, scissors were used to cut a straight line along the length of the colon. With epithelial layer facing up, colon samples were pinned out and fixed in 4% paraformaldehyde for 16h at room temperature. The fixed tissue was washed with 70% ethanol three times and then embedded in 2% agar, followed by paraffin embedding, sectioning, and hematoxylin and eosin staining.
Flow cytometry
Spleens were crushed and filtered through a 70-um cell strainer to generate single-cell suspensions. Cells were incubated with antibodies in PBS/4% FBS for 30 minutes, including fluorescently-conjugated antibodies specific for CD3, CD4, CD8a, CD11b, CD11c, CD19, CD45, F4/80, Ly6C, Ly6G, MHCII, and NK1.1 (All antibodies from BioLegend). Next, splenocytes were washed twice with PBS/4% FBS and analyzed on a BD FACS Canto II or Thermo Fisher Scientific Attune NxT Flow Cytometer. Flow cytometry data were analyzed with FlowJo software.
Multiplex cytokine assay
Lungs were harvested and homogenized in 200 μL of PBS. Cytokine and chemokine levels were measured with the Bio-Plex Pro Mouse Cytokine Group I Panel 23-Plex Assay Kit (Bio-Rad Laboratories, Hercules, California) on a Luminex platform. Twenty-three cytokines and chemokines were examined.
16S rDNA sequencing and analysis
Fecal pellets were collected from co-housed WT and SAVI mice and stored at −80°C. DNA was extracted from fecal pellets using phenol:chloroform and purified using the DNeasy Blood and Tissue Kit. (QIAGEN) per manufacturer’s instructions. Primer selection and PCRs were performed as described previously (Caporaso et al., 2011). Samples were amplified in triplicate using Golay-bar-coded primers specific to the V4 region within the 16S rRNA gene. Amplified samples were then consolidated, and sample quality was determined using gel electrophoresis. Platinum High-Fidelity Taq (Invitrogen) was used to amplify genomic DNA by PCR and amplicons were pooled and purified using 0.6x Agencourt Ampure XP beads (Beckman-Coulter) according to the manufacturer’s instructions, and quantitated using Qubit (ThermoFisher). The DNA pool was then sequenced using the Illumina MiSeq platform. Sequencing data was analyzed using Quantitative Insights into Microbial Ecology software (QIIME, version 1.8.0) (Caporaso et al., 2010).
18S sequencing and analysis
Fecal pellets were collected from co-housed WT and SAVI mice following two-week administration of VNA or Kool-Aid in drinking water. Samples were stored at −80°C. DNA was extracted using the QIAamp Fast DNA Stool Mini Kit (QIAGEN) and submitted to CD Genomics for 18S sequencing and data analysis.
Metagenomic sequencing and analysis
Fecal pellets were collected from germ-free mice and mice recolonized with stool from VNA-treated SAVI mice. Fecal samples were stored at −80°C. DNA was extracted from fecal pellets using phenol:chloroform and purified using the DNeasy Blood and Tissue Kit. (QIAGEN) per manufacturer’s instructions. Next, we performed tagmentation on the samples, followed by purification using 0.6x Agencourt Ampure XP beads (Beckman-Coulter) according to the manufacturer’s instructions. Samples were prepared for shotgun sequencing using a modified version of a previously described protocol (Baym et al., 2015) and makes use of the Nextera DNA Library Prep Kit (Catalog # FC-121-1030/1031). Raw sequencing reads were quality filtered and trimmed using BBTools v38.26. Samples then had any PhiX contamination removed and were screened against the mouse reference genome to remove any host reads. Paired-end reads were then interleaved into a single file for analysis. Samples were then uploaded to the One Codex analysis platform and screened against the One Codex Database. Differences between samples were first examined at the Family level selecting any taxa with > 0.1% Relative Abundance. Germ-free and VNA Re-colonized mice were compared at the Species level with the same cutoffs.
Gene expression analysis
Total RNA from lung homogenates or BMDMs was isolated using the RNeasy kit (QIAGEN) per the manufacturer’s protocol. TaqMan RNA-to-Ct 1-step kit (Applied Biosystems) was used to measure mRNA expression. Primer and probe assays were obtained from Integrated DNA Technologies. ΔΔCt values were calculated and then normalized to the untreated WT samples. All samples analyzed were normalized to the house keeping gene (GAPDH). Samples where the target gene did not amplify were assigned a CT value of 38 as the limit of detection.
Short-chain fatty acid treatment
Propionate and butyrate solutions were prepared in sterile PBS. Co-housed, WT and SAVI mice were given either PBS, propionate, or butyrate at a concentration of 1 mg/kg body weight and volume of 200 μL via intraperitoneal injection. Mice were injected daily for 14 days and euthanized on day 14. Lungs were fixed and submitted for H&E staining.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were analyzed using GraphPad Prism software by Mann-Whitney test as specified in the figure legends.
Supplementary Material
Highlights.
Oral antibiotics prevent lung disease in STING gain-of-function (SAVI) mice
Germ-free conditions do not protect against autoinflammatory disease
Transfer of Bacteroidales-rich microbiota to germ-free SAVI mice confers protection
ACKNOWLEDGMENTS
We thank the Washington University School of Medicine Morphology and Imaging Core for histology processing services. We thank Michael White and Summer Rucknagel of the Washington University in St. Louis Gnotobiotic Core for germ-free mouse generation and recolonization services. We also thank Brock G. Bennion for technical assistance with tissue harvesting. The Miner laboratory is supported by grants from the NIH (K08AR070918 and R01AI143982). D.J.P. is supported by the Washington University Chancellors Graduate Fellowship Program and the Initiative for Maximizing Student Development. M.T.B. is supported by grants from the NIH (R01AI141716 and R01AI139314) and from the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (MI-II-2019-790). D.L. is supported by T32HG000045.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109113.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
REFERENCES
- Ahmad I, Nygren E, Khalid F, Myint SL, and Uhlin BE (2020). A Cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep. 10, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, and Rudensky AY (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM,Young VB, Beck JM, Curtis JL, and Huffnagle GB (2015).Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, and Kishony R (2015). Inexpensive multiplexed library preparation for mega-base-sized genomes. PLoS ONE 10, e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE, Huang H, Knox KS, Littman DR, and Wu HJ (2017). Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 22, 697–704.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, Rey-nolds LA, Hacker L, Mohr J, Finlay BB, et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11,785–795. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R (2011). Global patterns of16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108 (Suppl 1), 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D, Quispe-Tintaya W, Jahangir A, Asafu-Adjei D, Ramos I, Sintim HO, Zhou J, Hayakawa Y, Karaolis DK, and Gravekamp C (2014). STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol. Res. 2, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, and Sorek R (2019). Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 574, 691–695. [DOI] [PubMed] [Google Scholar]
- Cooke KR, Hill GR, Gerbitz A, Kobzik L, Martin TR, Crawford JM, Brewer JP, and Ferrara JL (2000). Hyporesponsiveness of donor cells to lipopolysaccharide stimulation reduces the severity of experimental idiopathic pneumonia syndrome: potential role for a gut-lung axis of inflammation. J. Immunol. 165, 6612–6619. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, and Mekalanos JJ (2012). Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, Van Kessel J, Kiros TG, Strom S, Hayakawa Y, Hyodo M, Babiuk LA, and Gerdts V (2014). c-di-GMP enhances protective innate immunity in a murine model of pertussis. PLoS ONE 9, e109778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W, et al. (2015). STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 7, 283ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S, Kim MV, Rakib T, Wong PW, van Zandt M, Barry NA, Kaisho T, Goodman AL, and Iwasaki A (2018). Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat. Microbiol. 3, 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, and Pace NR (2007). Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc. Natl. Acad. Sci. USA 104, 20529–20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Hoang TK, Tian X, Taylor CM, Blanchard E, Luo M, Bhattachar-jee MB, Freeborn J, Park S, Couturier J, et al. (2019). Lactobacillus reuteri Reduces The Severity of Experimental Autoimmune Encephalomyelitis in Mice by Modulating Gut Microbiota. Front. Immunol. 10, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, and Gordon JI (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307. [DOI] [PubMed] [Google Scholar]
- Josefsdottir KS, Baldridge MT, Kadmon CS, and King KY (2017). Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, et al. (2007). Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 178, 2171–2181. [DOI] [PubMed] [Google Scholar]
- Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S, Ziegler C, et al. (2016). The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 1, 16131. [DOI] [PubMed] [Google Scholar]
- Lanning D, Zhu X, Zhai SK, and Knight KL (2000). Development of the antibody repertoire in rabbit: gut-associated lymphoid tissue, microbes, and selection. Immunol. Rev. 175, 214–228. [PubMed] [Google Scholar]
- Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GAM, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, et al. (2014). Activated STING in avascular and pulmonary syndrome. N. Engl. J. Med. 371,507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luksch H, Stinson WA, Platt DJ, Qian W, Kalugotla G, Miner CA, Bennion BG, Gerbaulet A, Rösen-Wolff A, and Miner JJ (2019). STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144, 254–266.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. (2009). Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Seibert FS, Nienen M, Welzel M, Beisser D, Bauer F, Rohn B, Westhoff TH, Stervbo U, and Babel N (2020). Propionate supplementation promotes the expansion of peripheral regulatory T-Cells in patients with end-stage renal disease. J. Nephrol. 33, 817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Miyabe H, Hyodo M, Sato Y, Hayakawa Y, and Harashima H (2015). Liposomes loaded with a STING pathway ligand, cyclic di-GMP, enhance cancer immunotherapy against metastatic melanoma. J. Control. Release 216, 149–157. [DOI] [PubMed] [Google Scholar]
- Nuzzo D, Makitrynskyy R, Tsypik O, and Bechthold A (2020). Cyclic di-GMP cyclase SSFG_02181 from Streptomyces ghanaensis ATCC14672 regulates antibiotic biosynthesis and morphological differentiation in streptomycetes. Sci. Rep. 10, 12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, and Kasper LH (2010). A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3, 487–495. [DOI] [PubMed] [Google Scholar]
- Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CG, Palfreyman RW, Nielsen LK, Cooper MA, et al. (2016). Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. ; NIH HMP Working Group (2009). The NIH Human Microbiome Project. Genome Res. 19, 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, and Louis P (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robak OH, Heimesaat MM, Kruglov AA, Prepens S, Ninnemann J, Gutbier B, Reppe K, Hochrein H, Suter M, Kirschning CJ, et al. (2018). Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 128, 3535–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, Benziman M, de Vroom E, Fidder A, de Paus P, Sliedregt LA, et al. (1990). The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265, 18933–18943. [PubMed] [Google Scholar]
- Savage DC (1977). Microbial ecology ofthe gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, et al. (2016). The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, et al. (2009). G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. 183, 7514–7522. [DOI] [PubMed] [Google Scholar]
- Smith BJ, Miller RA, Ericsson AC, Harrison DC, Strong R, and Schmidt TM (2019). Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 19, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Yin K, Qian H, Zhao Y, Wang W, Chou SH, Fu Y, and He J (2016). Cyclic di-GMP contributes to adaption and virulence of Bacillus thuringiensis through a riboswitch-regulated collagen adhesion protein. Sci. Rep. 6, 28807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, and Marsland BJ (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. [DOI] [PubMed] [Google Scholar]
- Warner JD, Irizarry-Caro RA, Bennion BG, Ai TL, Smith AM, Miner CA, Sakai T, Gonugunta VK, Wu J, Platt DJ, et al. (2017). STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 214, 3279–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Liu J, Li L, and Qiao C (2009). Biodegradation of malathion by Acinetobacter johnsonii MA19 and optimization of cometabolism substrates. J. Environ. Sci. (China) 21, 76–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the rDNA and metagenomic sequencing data reported in this paper are European Nucleotide Archive: PRJEB42379


