Abstract
Current methods in comparative genomic analyses for metabolic potential prediction of proteins involved in, or associated with the Dsr (dissimilatory sulphite reductase)-dependent dissimilatory sulphur metabolism are both time-intensive and computationally challenging, especially when considering metagenomic data. We developed DiSCo, a Dsr-dependent dissimilatory sulphur metabolism classification tool, which automatically identifies and classifies the protein type from sequence data. It takes user-supplied protein sequences and lists the identified proteins and their classification in terms of protein family and predicted type. It can also extract the sequence data from user-input to serve as basis for additional downstream analyses. DiSCo provides the metabolic functional prediction of proteins involved in Dsr-dependent dissimilatory sulphur metabolism with high levels of accuracy in a fast manner. We ran DiSCo against a dataset composed of over 190 thousand (meta)genomic records and efficiently mapped Dsr-dependent dissimilatory sulphur proteins in 1798 lineages across both prokaryotic domains. This allowed the identification of new micro-organisms belonging to Thaumarchaeota and Spirochaetes lineages with the metabolic potential to use the Dsr-pathway for energy conservation. DiSCo is implemented in Perl 5 and freely available under the GNU GPLv3 at https://github.com/Genome-Evolution-and-Ecology-Group-GEEG/DiSCo.
Keywords: comparative genomics, dissimilatory sulphur oxidation, dissimilatory sulphate reduction, genotype-phenotype association, microbial physiology
Data Summary
The DiSCo tool is open source and available for Unix and Windows 10 systems at GitHub under the GNU General Public License version 3 or later as published by the Free Software Foundation: https://github.com/Genome-Evolution-and-Ecology-Group-GEEG/DiSCo.
Accession numbers of all genomic records screened in this paper, including taxonomic information, database of origin and download date, are provided in the supplementary tables.
Alignments, phylogenetic reconstructions and identity plots are available for download at figshare (DOI: 10.6084/m9.figshare.12206246).
This article contains five figures, one supplementary figure, supplementary information, and 13 supplementary tables.
Significance as a BioResource to the community.
Comparative genomics coupled with metagenomics is a powerful tool to explore and shed light onto microbial diversity. However, with the exponential increase in the amount of data available, current bioinformatics methods are often too time-consuming and computationally demanding. Here we present DiSCo, a classification tool able to screen protein data from genomes within seconds and to automatically identify proteins involved in, or associated with, Dsr-dependent dissimilatory sulphur metabolism. DiSCo also provides the user with the prediction of the metabolic potential and enzyme type, with high levels of accuracy, precision and recall. DiSCo can analyse thousands of genomes in a matter of hours on a personal computer with no need for high-performance servers. Therefore, DiSCo provides the scientific community with an easy-to-use tool that can be the basis of many independent studies regarding Dsr-dependent dissimilatory sulphur metabolism and metabolic diversity in general. This method is platform independent, freely available and open source.
Introduction
Advances in sequencing techniques, combined with their rapid decrease in cost, led to massive (meta)genomic datasets, which remain largely unexplored, both in terms of taxonomic wealth and metabolic diversity [1, 2]. This creates an urgent need for better and faster tools to perform, in an efficient way, (meta)genomic metabolic potential assignments and global analyses regarding the impact of microbial activities in the environment.
The biological sulphur cycle has been continuously changing the Earth’s history [3]. Micro-organisms from diverse taxonomic affiliations, with different sulphur metabolic solutions for energy conservation, participate in these biological processes [4]. In here, we address the Dsr (dissimilatory sulphite reductase)-dependent dissimilatory sulphur pathway, present in the two prokaryotic domains of life.
Long before the characterization of the first Dsr enzymes [5–7], several micro-organisms able to use them to reduce or oxidize sulphur compounds have been isolated and characterized ([8–10] and references therein). Decades of work, enabled the characterization of many of the Dsr enzymes involved in the cascade of redox reactions that allow the micro-organisms to perform this sulphur-based energy conservation pathway [see [11] and [12] for reviews of Dsr-dependent sulphate/thiosulphate/sulphite-reducing prokaryotes (dSRP) and Dsr-dependent sulphur-oxidizing bacteria (dSOB), respectively]. Being one of the best studied microbial energy conservation solutions [11, 13–15], new insights regarding the diversity of these pathways [16–18] are showing how much is still to be discovered regarding the biology of these micro-organisms.
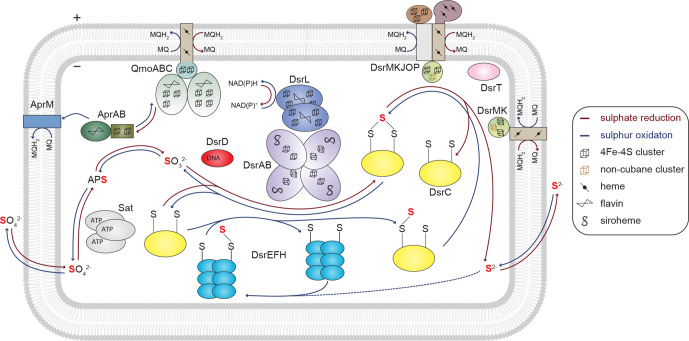
Based on the biology of known cultivated dissimilatory sulphate-reducing micro-organisms, currently, it is considered that a common set of proteins are involved in the reduction of sulphate to sulphide. After import to the cell, sulphate is first activated to APS (adenosine-5′-phosphosulfate) via a (dissimilatory) ATP sulfurylase/sulphate adenylyltransferase Sat (Fig. 1 red arrows) [19]. APS is then reduced to sulphite and AMP by AprAB (APS reductase), a dimeric complex containing one FAD (flavin adenine dinucleotide) and two [4Fe-4S] [20]. The reaction catalysed by AprAB is fueled by electrons received from the QmoABC membrane complex [21]. The subsequent formation of sulphide from sulphite occurs in two steps. First, a DsrC-trisulphide is formed by the action of the DsrAB complex, which contains bound siroheme and a [4Fe-4S] cofactor [7, 22]. The DsrC protein is then regenerated with the release of sulphide, possibly by the action of the DsrMK membrane complex [23, 24], which may contain the additional subunits DsrJOP in both dSRP and dSOB [25]. DsrC proteins belong to the DsrC/TusE/RspA protein family and are small proteins usually containing two conserved redox-active cysteines at their C-termini [24, 26]. In some micro-organisms, such as Desulfurella amilsii, it was shown that only one cysteine was necessary for the proper function of DsrC [27], albeit possibly with a slightly different mechanism.
Fig. 1.
Dsr-dependent dissimilatory sulphur species reduction and oxidation in prokaryotes. Schematic representation of the main complexes and reactions involved in Dsr-dependent dissimilatory sulphate reduction (red arrows) and/or sulphur oxidation (blue arrows) according to [24, 33]. Homologous proteins are represented with the same colour code. MQ – Menaquinone.
Many sulphate reducers are able to use as electron acceptors other sulphur-containing compounds such as thiosulphate, sulphite, or DMSO (Table S1, available in the online version of this article). Other micro-organisms such as Desulfitobacterium dehalogenans [28] or Pyrobaculum islandicum [29], mostly due to the absence of Sat, AprAB and AprAB’s interaction partner, are unable to use sulphate as an electron acceptor, starting the pathway with other sulphur compounds (Table S1). The DsrT protein, a paralogue of the RsbRD protein involved in the negative regulation of a stress transcription factor [30], was suggested as a possible marker for sulphite reduction [18]. This protein was mainly identified in (meta)genomes from sulphite reducers and some dSOB from the phylum Chlorobi and DsrT was found in synteny with the DsrMK(JOP) complex [18].
In the reverse or oxidative Dsr-pathway, which enables micro-organisms to oxidize several sulphur species, it is generally assumed that versions of the same enzymes used by sulphate reducers operate in the reverse direction [31–33]. However, several differences are worth noting, at the level of associated cytosolic and membrane processes. In some dSOB, the QmoABC complex is functionally replaced by the AprM membrane protein [34, 35]. Moreover, some dSOB [34, 35] or even Gram-positive dSRP [34, 36, 37], may replace the QmoC subunit with the heterodisulphide reductases HdrBC subunits [37], usually present in methanogens [38, 39], which, together with HdrA, form a complex involved in electron bifurcation [38, 40]. In dSOB, the oxidation of sulphite to sulphate is not strictly dependent on AprAB-Sat proteins. In Allochromatium vinosum both AprAB-Sat and the SoeABC complex (a membrane-bound iron-sulphur molybdoenzyme, omitted from Fig. 1 for simplicity) are present, providing a functional redundancy for the oxidation of sulphite to sulphate [41]. Similarly to AprAB, the Soe complex, is not specific of the oxidative Dsr-pathway, and SoeABC participate in additional sulphur oxidation pathways [42]. In contrast, in dSRP, the Sat, AprAB and Qmo proteins are strictly needed for the activation of sulphate to APS [43] and its reduction to sulphite [44, 45].
Another difference is the initial stage of sulphur oxidation. In A. vinosum, the involvement of three proteins in the delivery of sulphur to the DsrEFH complex was demonstrated: a membrane-bound DsrE-like protein, a rhodanese-like protein and a TusA protein [46] (omitted from Fig. 1 for simplicity). The DsrEFH proteins are a soluble complex present in dSOB that transfers sulphur to DsrC [47]. The persulphurated DsrC is oxidized by the DsrMK(JOP) complex and the DsrC-trisulphide will serve as a substrate for the DsrAB complex, similar to the mechanism of the Dsr-dependent sulphite reduction in dSRP. Recently, it was shown that, in A. vinosum, an additional protein (DsrL) mediates the electron transfer between NAD(P)H and the DsrAB complex [16]. The DsrL protein is not exclusively found in dSOB being also present in some (probable) sulphate/sulphur reducers [48].
Although with exceptions, the specialization to use sulphite as a substrate or to have it as a product is imprinted at the level of the primary proteins sequence and can be observed in phylogenetic analyses of DsrA and DsrB proteins [18, 49]. The reductive-type DsrAB complex tends to be found in dSRP while the oxidative-type DsrAB is found in dSOB. However, several micro-organisms, such as Desulfurivibrio alkaliphilus [17] or Desulfobulbus propionicus [50], are able to perform sulphur-disproportionation and adapted the reductive version of the pathway to work in reverse (Table S1). Thus, the presence of a reductive-type DsrAB complex per se, is not a good proxy for the use of oxidized sulphur compounds as electron acceptors. Regardless of the type of Dsr-pathway, five proteins, DsrABCMK, are so far conserved in all dSRP, Dsr-dependent sulphur-disproportionating micro-organisms (dSDM), and dSOB.
The (automatic) prediction of the type of Dsr-dependent sulphur energy-yielding metabolism (i.e. is this micro-organism a dSOB or a dSRP) from sequence data is frequently based on two different approaches. One method is the classification of the DsrAB marker proteins according to types, based on their position in phylogenetic reconstructions [18, 49]. A second strategy relies on the presence of additional proteins, usually present in one type of metabolism, such as DsrD or the DsrEFH complex usually only found in dSRP [51] and dSOB [52], respectively. However, DsrD is not essential to perform sulphate reduction, albeit its conserved genomic arrangement across dSRP indicates a possible regulatory function in this type of metabolism [53, 54]. Some cultured micro-organisms are able to use sulphate as an electron acceptor, without the aid of the DsrD protein [45]. Thus, using this protein as a single marker for the distinction between dSRP from other dSOB might lead to incorrect assignments. Additional problems arise when dealing with metagenomic data, since not all of the genes present in the micro-organism’s genomes are sequenced and the corresponding translated protein records have different degrees of completeness and contamination. The careful distinction between the different paralogous families is hampered by the small size (~100 amino acids) of some proteins traditionally used as markers such as DsrEFH and their homology with other protein families [55]. On the other hand, phylogenetic reconstructions are time-consuming, especially when considering the exponential increase of (meta)genomic records being deposited daily to public databases. Additionally, phylogenies might lead to incorrect assignments due to the existence of micro-organisms with the reductive-type Dsr-system that in vivo are dSDM [17, 50].
Here we present DiSCo, a Dsr-dependent dIssimilatory Sulphur metabolism Classification tOol, able to automatically identify and predict the enzyme types involved in, and associated with biological Dsr-dependent dissimilatory sulphur metabolism. This mapping will guide future analyses such as cultivation of presently uncultivated prokaryotes, in order to determine the mechanisms used by these microorganisms to conserve energy.
Development and application of DiSCo
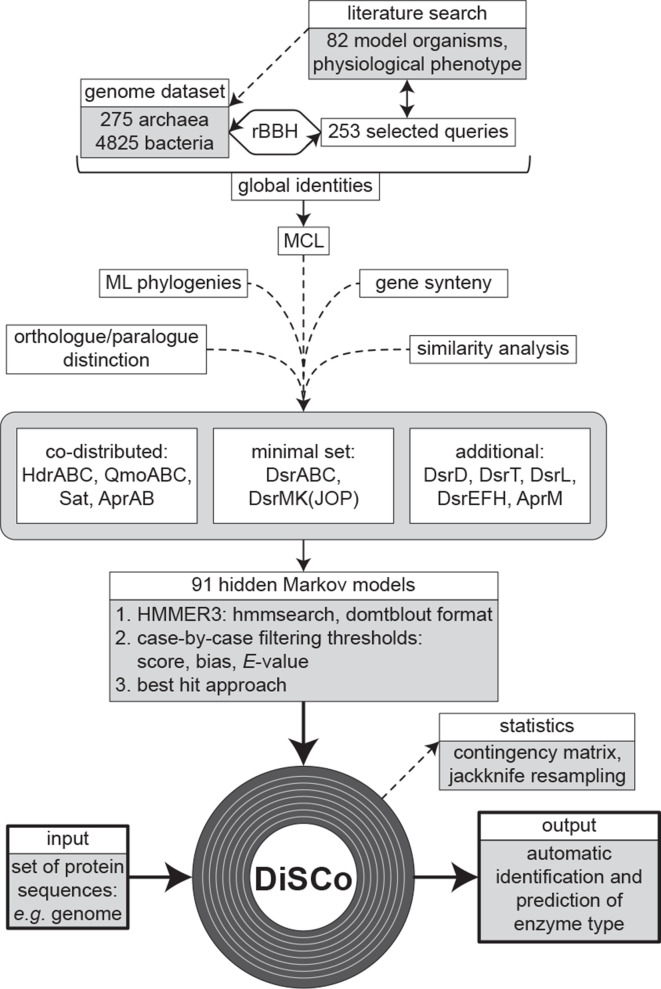
The development process of the DiSCo tool will be explained in the following sections. This includes the steps of model creation, development and the application of DiSCo against genomic datasets.
Genomic datasets
Publicly available complete genomic assemblies of 4825 bacteria and 253 archaea were downloaded from NCBI. Twenty-two assemblies belonging to new, uncultivated archaeal lineages were added to this dataset. The protein sequences of these 5100 assemblies constitute the complete genomes dataset (Table S2).
A second dataset (from now on referred to as the metagenomic dataset) composed of the protein sequences of 193 978 prokaryotic (meta)genomic assemblies and 1900 (meta)genomes belonging to potential new lineages with the ability to use the Dsr-dependent dissimilatory sulphur metabolism [14, 18, 45, 56–72] was created (Table S3). These additional 1900 (meta)genomes were collected from a survey of recent publications describing potential micro-organisms containing Dsr proteins in the following manner: all assemblies related to the publication BioProject (described or not in the text as a micro-organism having the Dsr pathway) were downloaded and added to the metagenomic dataset, if not already present.
Both datasets were mapped to, and sorted by, the NCBI taxonomy and are listed in Tables S2 and S3, which also include information regarding the database of origin and download date.
Genome quality estimation
Genome completeness and redundancy were estimated by domain-specific single copy maker proteins as in [2]. Pfam domains [73] were assigned with hmmsearch (hmmer v. 3.1b2, default parameters) [74] using the profile-specific cutoffs from [2]. The completeness was calculated based on the ratio between the number of marker proteins identified and the total number of domain-specific makers (162 archaeal profiles, 139 bacterial profiles [2]). Genomic redundancy (i.e. contamination) was defined as the proportion of distinct protein sequences per genome, in which marker proteins were detected multiple times, over the total number of domain-specific markers (Tables S2 and S3).
Literature search
A thorough literature search allowed gathering information regarding 82 micro-organisms known to be able to conserve energy using Dsr-dependent dissimilatory sulphur pathways. The Dsr-enzyme types (reductive or oxidative) of these micro-organisms were mapped to the corresponding genomic assembly of the complete genomes dataset (Table S1). Information regarding the micro-organism’s capability to perform S-disproportionation and the nature of the sulphur species used as electron donors/acceptors was also collected. The corresponding protein representatives and their genomic arrangements provided reference for the exploratory analysis and development of DiSCo.
Similarity searches
The complete genomes dataset was analysed and queried for selected proteins involved in or co-distributed with the Dsr-dependent dissimilatory sulphur metabolism and functional replacements thereof. Protein sequences obtained from the literature search were mapped to their respective genome using blastp (version 2.10.0, default parameters) [75], with an identity of ≥25 % and an E-value of ≤10−10 as thresholds. Selected paralogous sequences, based on blast results, genomic neighbourhood and KEGG pathways [76], as well as further accessory proteins, such as the pyrophosphatases HppA, and PpaC proposed to be essential at least in dSRP [36], were added. These sequences were used to query genomes from the micro-organisms described in Table S1, since not all of the accession numbers of the descripted proteins are given. In total, 253 query proteins belonging to 16 micro-organisms were collected (Table S4, Fig. 2). Protein representatives in the complete genomes dataset were acquired by using the reciprocal best blast hit (rBBH) approach [75] with the thresholds of E-value ≤10−10 and a global identity of ≥25 %, calculated based on the local identity considering sequence and alignment length. The proteins fulfilling the threshold were blasted against their respective genome and copies included if the threshold (≥70 % local identity, ≥70 % query coverage) was fulfilled. An all-versus-all blast of the protein pairs fulfilling the threshold was performed. All pairs with a local identity of ≥25 % and an E-value of ≤10−10 were globally aligned with needleall [77, 78] (Emboss package 6.6.0, default gap penalties). The pairwise global alignments were filtered using a ≥25 % global identity threshold and clustered into protein families using MCL [79] (version 14.137, default parameters, inflation rate 2.0). At this stage, 75 protein clusters were obtained, which were used for phylogenetic analysis. The clusters were functionally annotated by using the protein’s rBBH classification and the protein sequence annotation of the members of each cluster. Further annotations were obtained using HMM (Hidden Markov Model) assignments (hmmsearch v. 3.1b2) of the TIGRFAM [80] (release 15 using NCBI’s improvements from 2018), PFAM [73] (release 32) and KEGG Orthologs [81] (version 201904) databases as well as the corresponding profile-specific thresholds (Table S5).
Fig. 2.
DiSCo development and algorithm. Schematic representation of the strategy used to create the classification tool and example of DiSCo operation.
Phylogenetic, similarity and synteny analysis
For each protein cluster, a multiple sequence alignment was generated with Clustal Omega [82] (both 100 guide tree and HMM iterations, output format guide tree order, otherwise default parameters) and maximum-likelihood (ML) phylogenies reconstructed with IQ-TREE [83, 84] (version 1.6.12, best model selection and LG+I+G4 model, 1000 ultrafast bootstraps, SH-like approximate likelihood ratio test: 1000). Phylogenetic reconstructions were rooted with the minimal ancestor deviation method [85]. Intra-cluster protein identities were calculated and their heatmap representations analysed. This allowed the identification of sequences from paralogous proteins with a high sequence similarity to known enzymes used in this study. Synteny analysis was performed by mapping retrieved protein hits to the respective gene location files, and visualized in R-Studio (version 1.0.153) using the R-package genoPlotR [86] (version 0.1).
Model reconstruction
Based on the previous analysis, sequences from enzymes of the oxidative and reductive Dsr-pathway, associated proteins, as well as from representatives of highly similar paralogues were selected to construct the DiSCo profiles. A second criterion for sequence selection was to use an as-small-as-possible sequence set from distant dSRP and dSOB derived from the literature search, ensuring an as-wide-as-possible taxonomic diversity (Table S6). Moreover, using sequences from dSDM, additional reductive-type models were created. These models do not intend to distinguish Dsr-dependent sulphur disproportionation from Dsr-reductive pathways. The aim is solely to flag those sequences in the DiSCo output, in case a user is interested in a deeper investigation.
The selected sequence sets were aligned in clustalw2 [87] (version 2.1, iteration set to tree and aligned output order, otherwise default parameters) and HMM were created using hmmbuild [74] (v. 3.1b2) with default parameters.
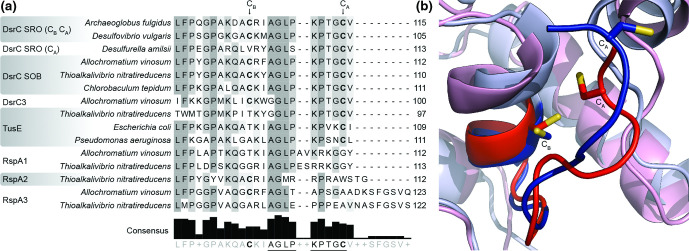
Model thresholds were optimized to retrieve proteins belonging to known lineages that use the Dsr-dependent dissimilatory sulphur pathway and to predict the corresponding enzyme type. The thresholds of the profile-specific scores were initially defined based on the score jumps of the retrieved HMM assignment and were furthermore manually fine-tuned to exclude potential false positives identified by similarity, phylogenetic and synteny analyses (Table S6). To enable the detection of DsrC proteins containing one or two cysteines [26, 27], in addition of fulfilling the model threshold, the presence of the two conserved C-terminal cysteines or the presence of the identified motif AGLPKPTG(not N)CA, allowing for one amino acid difference, was mapped onto each sequence (Fig. 3). This motif was identified by manual inspection of a multiple sequence alignment of DsrC, containing one or two cysteines at the C-termini, RspA and TusE proteins.
Fig. 3.
C-termini of DsrC proteins. (a) Alignment of the C-termini of the DsrC/TusE/RspA family. The strictly conserved cysteine and the identified conserved motif only present in DsrC(-like) proteins and absent in TusE/RspA proteins [26] are shown. (b) Structural alignment of oxidative-type DsrC protein from Allochromatium vinosum (light blue, PDB code 1YX3) and reductive-type DsrC protein from Archaeoglobus fulgidus (pink, PDB code 1SAU). The C-termini are indicated with red and dark blue; cysteines are represented as sticks and the sulphur atoms in yellow.
It was observed that some micro-organisms, besides the DsrMKJOP complex, contain an alternative DsrMK type [36]. Also, in Chlorobi, the oxidative DsrMKJOP complex is more similar to DsrMKJOP proteins from dSRP than to the ones found in dSOB [88]. Therefore, additional models for these proteins were created (Table S6). Similar criteria were used to create models for the DsrD, DsrL and DsrT proteins.
The combined results of phylogenetic reconstructions, similarity networks and genomic arrangements led to the identification of paralogous proteins, highly similar to enzymes from the Dsr-pathways. A QmoABC complex distinct from known QmoABCs and whose QmoAB subunits are both closely related with the HdrA subunit of the HdrABC complex was recently identified [45]. For a better orthologous to paralogous distinction, additional HMM profiles were created using representatives of heterodisulphide reductases from methanogens and Proteobacteria [89], assimilatory Sat as well as the AprAB-like paralogues previously identified in AprAB sequence similarity searches and/or phylogenies [45].
In total, 91 HMM models, 71 of them type-specific are included in DiSCo.
DiSCo hit assignments
The DiSCo tool can be executed with a single command in which all steps are performed automatically by using the wrapper Perl script DiSCo.pl. The first step of DiSCo.pl consists of an hmmsearch [74] of a given protein sequences set in FASTA format against the DiSCo HMM library. This hmmer process can be accelerated with the –n parameter using multiple threads. The hmmsearch raw output (hmmer ‘domtblout’ format [74]) is then automatically parsed with the filter_DiSCo.pl script. This filter step assigns DiSCo hits based on the best hit strategy: (i) a hit needs to fulfil the model specific cutoff; (ii) all hits to the same input sequence are compared and the model with the best score and E-value is assigned to a sequence (Fig. 2). DiSCo hits need to fulfil the cutoff of the hmmer-specific conditional and independent E-value of ≤10−10, an accuracy of ≥0.5 with the bias being one order of magnitude lower than the score. All hits fulfilling the thresholds are saved in a table whose delimiter can be changed by using the –s options. Furthermore, the location and file name prefix can be altered with the –d and –p parameters, respectively and an automatic extraction of the sequences corresponding to DiSCo assignments can be performed by using the –o flag. An option to apply user-defined thresholds is also available, but in this case, information regarding the type of enzyme cannot be provided.
This procedure was used to map Dsr-dependent dissimilatory sulphur enzymes within the protein sequence space of 195 878 (meta)genomic records.
Methods validation and comparison
The DsrA and DsrB proteins identified by DiSCo within the complete genomes dataset and their predicted type were compared with the results from the phylogenetic analysis and with literature knowledge (all sequences present within the dataset were identified by DiSCo; their predicted type agreed with their position within the reductive or oxidative clades of the phylogenetic reconstruction and the taxonomic affiliation of the microorganism agreed with the predicted enzyme type).
The reliability of DiSCo assignments for Dsr and co-distributed proteins was evaluated by calculating the contingency matrix in the following manner. A hit was only considered as a true positive for cases in which DsrAB proteins with the same type were found in the respective genome. A protein identified in a genome without the DsrAB complex, or with a predicted type different to the DsrAB complex, was considered a false positive. The absence of the proteins within genomes with DsrAB was considered a false negative. In the cases in which a protein family had models of only one type (e.g. DsrD and DsrEFH), a false negative was only considered by the absence of the protein in genomes containing a DsrAB complex of the same type. A true negative was defined as the absence of a DiSCo hit in a genome without DsrAB proteins.
A contingency matrix was also obtained for the results obtained with blast (best hit, cutoffs: ≥25 % local identity, E-value ≤10−10) and with the results from the rBBH approach (Table S7). These methods do not provide a type-assignment unless manual inspection and/or additional bioinformatic analyses (e.g. taxonomic comparison, similarity comparison with reductive and oxidative proteins) are performed. However, we assigned an enzyme type to all query sequences based on the physiology of the micro-organism, from which protein representatives were collected (Table S4).
We created an independent test set derived from the metagenomic dataset. The selection was based in one of two criteria. Firstly, a genome was reported in the literature to possess the DsrAB proteins [14, 18, 45, 56–72]. The presence of DsrAB proteins was validated using the rBBHs of DsrAB proteins following same rBBH strategy as in the complete genome dataset. Secondly, all genomes with a complete genome assembly level present in the metagenomic dataset, thus, not present in the complete genomes dataset, were grouped by the taxonomic family level and up to five genomes per family were randomly chosen. In total, the independent test set comprises 1187 genomes (Table S3). The DiSCo assignments, rBBH and simple best blast hits were used to create additional contingency matrices (Table S7). The determined contingency matrices of the three methods of both datasets were used to calculate, among others, the precision (PR), recall (RC), accuracy (AC), balanced accuracy (BA) [90] and false discovery rate (FDR) (Table S7).
To further validate the robust predictability of DiSCo models, a jackknife resampling was performed by excluding each one of the sequences used to create the DsrABCMK models. New models were created with the remaining sequences, and the previously defined DiSCo model’s thresholds were kept (Table S6). The new models were run against the complete genomes dataset and the results compared to the standard DiSCo assignments with regards to both the identification and predicted enzymatic type (Table S8).
DiSCo availability
DiSCo version 1.0 was developed as a standalone genome-mining tool using Perl 5 as the coding language preinstalled in Linux and Mac operative systems. The DiSCo dependency HMMER3 [74] is freely available for download at http://hmmer.org. DiSCo 1.0 code, operational details and examples (input/output data files) can be downloaded at https://github.com/Genome-Evolution-and-Ecology-Group-GEEG/DiSCo. DiSCo was tested on Windows 10 and Unix operating systems and runs in all platforms although Unix-based systems are recommended for efficient performance.
Benchmarking of DiSCo
Our aim was to create an automatic identifier and type-predictor of dissimilatory sulphur proteins from microbial protein data, to aid users to access in a fast way, the diversity of proteins involved in or associated with Dsr-dependent dissimilatory sulphur metabolism within metagenomic records. The DiSCo tool was compared to existing methods, applied to (meta)genomic records and its performance assessed.
Distribution of Dsr-dependent dissimilatory sulphur metabolism within the complete dataset
The use of completely sequenced genomes allowed an initial classification of DsrAB-containing micro-organisms and to determine the presence and absence of proteins involved in this type of metabolism per genome. This identification was based on the rBBH approach followed by MCL clustering, pathway completeness, synteny and phylogeny analyses as well as manual inspection of the results to distinguish paralogues from orthologues. Considering the presence of at least the DsrAB proteins, and besides the identification of the 82 micro-organisms whose phenotype was already known (Table S1), the Dsr-dependent dissimilatory sulphur pathway was identified in 21 additional micro-organisms (Tables S5 and S9). Similar to what previous studies reported, with respect to taxonomic ranks at the phylum/class level and enzyme types [11–13, 36, 49], and as expected, dSRP were found within the bacterial phyla Thermodesulfobacteria and Nitrospira, the classes Clostridia and Deltaproteobacteria and the actinobacterial family Coriobacteriaceaea. Within Archaea, reductive DsrAB proteins were identified in the order Archaeoglobales and the crenarchaeal family Thermoproteaceae. dSOB proteins were identified within the classes Chlorobi and the Alpha-, Beta-, and Gammaproteobacteria (Table S5). In total, the DsrAB complex was found to be present in eight out of the 48 phyla (eleven out of 93 classes) represented in the complete genome dataset.
At this stage, the proteins involved in or associated with Dsr-dependent dissimilatory sulphur metabolism were categorized into minimal (necessary for both dissimilatory sulphate/sulphite/thiosulphate reduction, disproportionation, or sulphur oxidation processes [36, 88]), additional (typically exclusively present in either dSRP or dSOB) or co-distributed (e.g. enabling the use of sulphate as electron acceptor or having sulphate as a final product [13]). Within co-distributed proteins, besides dissimilatory Sat, AprAB and QmoABC, also non-specific enzymes (e.g. HdrABC) are included to allow a better orthologous to paralogous distinction (Fig. 2).
DiSCo assignments within the complete dataset
DiSCo was run against the complete genome dataset and the results from DsrA and DsrB models were compared with the previous analyses and the relative position of these sequences within the phylogenetic DsrA and DsrB reconstructions. Without exception, the DsrA and DsrB sequences belonging to the 103 genomes were found by DiSCo (Table S9, Fig. 4), and their classification into types congruent with the position in the respective phylogeny. No additional DsrA/B hits were found (Fig. S1, Table S9), showing that DsrA and DsrB models were consistent with the previous approach. Overall, and with few exceptions, the predicted protein types were congruent within a genome, indicating that each model is able to correctly and, most importantly, independently predict the protein’s type (Fig. 4). As in previous studies [17, 18, 91], the direct identification from sequence data of dSDM was not possible. Although some genomes showed an enrichment of DiSCo assignments with sulphur-disproportionating models (Table S9) not all of them have information regarding their ability to disproportionate S-species. So far, only cultivation and experimental characterization can enable the identification of dSDM.
Fig. 4.
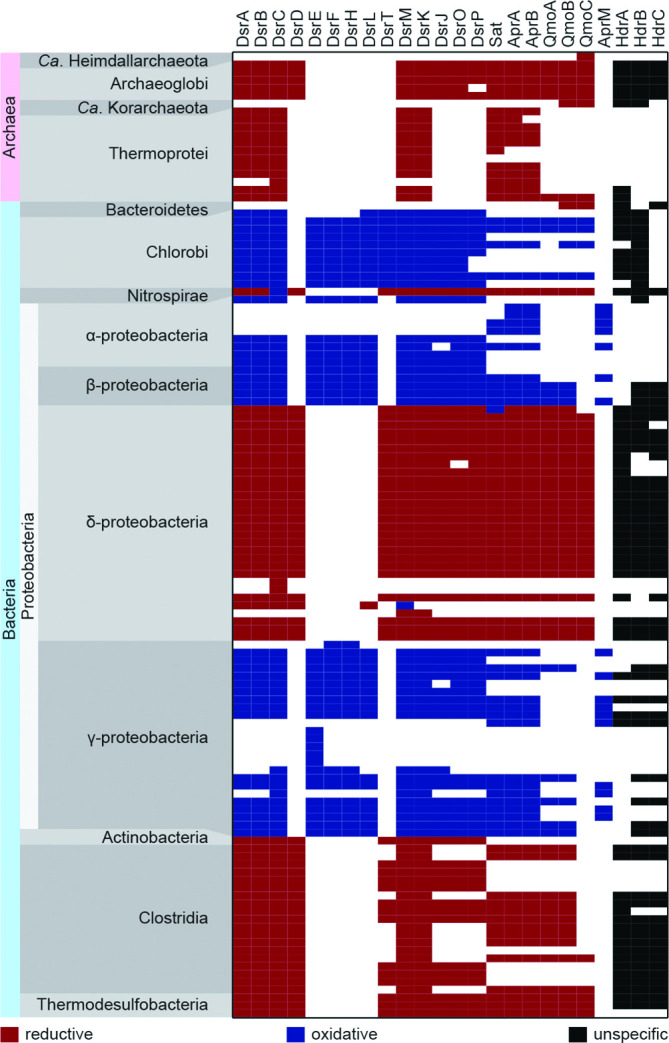
Distribution of DiSCo hits across the complete genomes dataset. Each column represents a protein and each row corresponds to a genome. Colour code represents the predicted type; red: reductive type, blue: oxidative type, black: unspecified hit, and white: no hit found. Only genomes with at least one type-specific hit, excluding genomes with only a dissimilatory Sat hit, are represented.
Interestingly, our AprA-like and AprB-like models led to the identification of this alternate complex in 63 micro-organisms belonging mostly to the class Clostridia, but also to Negativicutes, Deltaproteobacteria, Nitrospira and Archaeoglobi (Table S9), including nine micro-organisms known to contain the Dsr-pathway. A comparison of the AprAB-like proteins with the fold regions of bona-fide AprAB proteins [92] showed that while the AprB-like protein shares the ferredoxin domain with bona-fide AprB, it lacks its terminal tail responsible for interactions with the AprA subunit [92]. The alignment of selected AprA-like protein with bona-fide AprA proteins displayed gaps in the capping domain and in the C-terminal, helical domain (for definition of the domains see [92]). A closer inspection of the genomic neighbourhood of the aprAB-like genes showed an enrichment in sulphur assimilation and processing genes such as assimilatory sulphate adenylyltransferases (sat, cysD, cysC), anaerobic sulphite reductase (either asrABC or only the asrC subunit) and genes of several proteins involved in intracellular sulphur trafficking such as TusA, ThiI/F, IscU and IscI. TusA has been proposed to play a role in the synthesis of sulphur-containing cofactors [93] and in dissimilatory sulphur oxidation processes [46]. In addition, genes of several sulphate/thiosulphate transporters (sbp, cysW, cysU, cysA, sodium:sulphate symporters) or ABC transporters (nitT/tauT) are found in the proximity of the aprAB-like genes. In the iron-respiring Ferroglobus placidus both the aprA-like and sat genes showed increased mRNA transcripts when grown on insoluble Fe(III) oxide [94]. These findings, combined with our analysis lead to the proposal of a role for the AprAB-like complex in sulphur assimilation processes. Further experimental validation is necessary to fully elucidate the functional role of this complex.
DiSCo validation and comparison with other methods
The accuracy, balanced accuracy [90], recall, precision and false discovery rate of DiSCo assignments, rBBH and simple blast hits were calculated and compared for both the complete genome dataset and independent test set (Table S7). Of note, due to possible assembly contaminations (i.e. false positives), true positives were only considered in (meta)genomes containing both DsrAB proteins and for proteins whose predicted enzyme type was in agreement with the DsrAB complex. In the case of the independent test set, the non-identification of proteins due to genome incompleteness (i.e. false negatives) may inflate the statistical measurements. Since the complete genomes dataset consists of only closed genomes, in which all protein-coding genes, and no contaminations are present, the influence of the quality of metagenomes is excluded within this dataset. In addition, the strategy in here employed to assign true positives (correctly identified sequences that indeed perform the expected function) favour methods based in similarity such as simple blast, due to the identification of paralogous proteins (sequence evolutionary related to the protein of interested that evolved to perform a different function). Consequently, the differences of determined false positives and false negatives caused by the use of blast-based methods result in an overall higher accuracy, precision and recall favouring DiSCo over similarity searches (Table S7). The full analysis of the results of the complete genomes dataset is given in Supplementary Material.
Within the independent test set, while both DiSCo and the rBBH method identified the DsrAB proteins in 143 micro-organisms, blast led to the additional identification of 12 assemblies with hits for both DsrAB proteins and 57 cases where only one protein was identified. This result is not unexpected; many paralogous sequences fulfil the ≥25 % local identity cutoff commonly used in blast similarity searches. A direct comparison between the recall, accuracy and balanced accuracy of the three methods shows a small difference for Dsr proteins (DiSCo=0.99–0.59, rBBH=0.98–0.56), with simple blast having worse results (0.98–0.3). However, regarding precision (how many times the correct sequence was identified), both rBBH and blast have identified a much higher number of paralogues (lowest PC: DiSCo=0.56, rBBH=0.25, blast=0.04). For instance, regarding DsrO, an iron-sulphur protein, blast has a precision of 0.2, implying that only 20 out of 100 identified sequences are indeed potential DsrO proteins. This is a clear indication of the retrieval of paralogous sequences, which can also be observed with the worse (higher) false discovery rate of simple blast for DsrO (FDR=0.8) versus the other methods (rBBH=0.6, DiSCo=0.06). The DsrC protein was not identified by DiSCo in 11 of the DsrAB genomes and the predicted type of DsrC differed from the one of the DsrAB proteins in 25 cases. In addition, in four cases the protein was identified in genomes devoid of the DsrAB complex. This is reflected in a balanced accuracy of 0.94 (PR=0.79, RC=0.1, AC=0.97) (Table S7). DsrC proteins were identified by both blast-based methods in four DsrAB metagenomes, for which no DsrC protein was assigned by DiSCo. These sequences are often fragmented, mostly with incomplete C-termini. Thus, the DsrC-specific motif (Fig. 3) was not found by DiSCo, which resulted in differences in recall values (DiSCo=0.91, rBBH=0.95, blast=0.95). On the other hand, DsrC proteins were found in 50 genomes (rBBH) and 90 genomes (blast) devoid of DsrAB proteins. These determined false positives affect the reliability of both blast methods in terms of precision of 0.69 for rBBH, and 0.6 for blast (DiSCo=0.79), while a comparison of, e.g. only balanced accuracy, accuracy or recall of both blast-based methods would not show the incorrectly assigned rBBH/blast hits of DsrC paralogues (all values >0.90).
Similar values for the balanced accuracy (0.95–0.96) were determined for the DsrEFH proteins identified by DiSCo. DsrEFH proteins were found in 15 genomes with reductive-type DsrAB proteins and in ten genomes without the DsrAB proteins. Both cases were considered as false positives resulting in a precision of 0.56–0.63 (RC=0.94–0.91). The co-distribution of DsrEFH with deeply branching reductive-type DsrAB lineages was already reported [18], which could reflect a wider metabolic potential of micro-organisms possessing a chimeric Dsr system. On the other hand, DsrEFH are small proteins with homology to TusBCD [55], thus, some were potentially identified. All of those instances were in here classified as false positives. In any case, the precision of DiSCo for DsrEFH is higher than the one from blast-based methods (PC=0.27–0.38), in which many paralogous sequences are identified.
The DsrD protein was identified by DiSCo only in reductive-type DsrAB genomes, and no additional hits (neither in oxidative-type DsrAB nor in non-DsrAB genomes) were found (BA=0.93, AC=0.99, PC=1, FDR=0.0). The DsrD protein was absent in 16 genomes with reductive-type DsrAB proteins. These metagenomes belong either to Archaea, and potentially reduce sulphur compounds without DsrD as in [45], or belong to new reductive-type DsrAB-lineages, such as Ca. Rokubacteria, which lack the DsrD protein [18]. These absences were categorized as false negatives impacting DiSCo’s recall of 0.85. DsrD is a small protein (~80 amino acids) and similarity searches often miss such small proteins due to the stringent E-value cutoff of ≤10−10 leading to false negatives. DsrD proteins, detected by DiSCo, were not identified in 16 (rBBH) and 13 (blast) genomes (RC=0.71 rBBH, RC=0.74 blast). No false positives were found by both blast-based methods with balanced accuracy, accuracy, and precision ranging between 0.85–1 for rBBH, and 0.87–1 for blast.
The DsrMK(JOP) complex was absent or incomplete in multiple DsrAB genomes resulting in lower values for recall (DsrMK=0.89–0.9, DsrJOP=0.76–0.78). Most of these false negatives were found in metagenomes, in which missing DiSCo assignments are affected by the incompleteness of the genomes. Nevertheless, DiSCo showed a stable prediction for all proteins, with a balanced accuracy for DsrABCMK, the minimal set of Dsr proteins, of 0.94 (Table S7). The rBBH and blast recall is higher (rBBH=0.78–0.92, blast=0.78–0.95) in the case of DsrMKOP (but not DsrJ) since in most of the affected genomes, only the DsrMK complex is present and several incorrectly identified DsrO and DsrP proteins were found (11 genomes rBBH, 16 genomes blast). The DsrO protein is an iron-sulphur protein while DsrP is cofactor-less and member of the NrfD/PsrC protein family. Together they are part of a widespread redox-loop module, involved in redox transfers for the quinone pool [95]. This is also shown by the high number of false positives identified in genomes devoid of DsrAB. DsrO/DsrP proteins were identified in 535 genomes (blast, FDR=0.8/0.41) and 162 (rBBH, FDR=0.6/0.32) genomes without DsrAB proteins, when compared to DiSCo (three genomes, FDR=0.06/0.07).
The DsrL protein was identified by DiSCo in 28 oxidative-type DsrAB genomes and in 25 reductive-type DsrAB genomes, similar to what was reported before (see [48] and below). Both blast-based methods detected DsrL proteins in more DsrAB genomes than DiSCo (rBBH: 73 genomes, blast: 137) and even in genomes without the Dsr system, 46 genomes for rBBH and 724 genomes for blast. These additional blast hits are caused by the similarity of DsrL proteins to pyridine nucleotide:disulphide oxidoreductases [96], while DiSCo distinguished these homologous sequences and identified DsrL proteins only in DsrAB genomes. Both blast-based method have a higher number of false positives (rBBH=89, blast=828), resulting in lower values for precision (rBBH=0.25, blast=0.04). This also affected the accuracy, and led to a decreased reliability, in particular for blast (AC=0.3, BA=0.63, RC=0.97), while rBBH resulted in better values for the detection of DsrL (AC=0.92, BA=0.9, RC=0.88). DiSCo’s predictability of DsrL proteins shows a high accuracy of 0.98 (BA=0.91), while precision (0.6) and recall (0.84) are affected by predicted oxidative-type DsrL proteins present in reductive-type DsrAB genomes. Further, not all dSRP posses the DsrL protein, and the number of false negatives is overestimated (FDR=0.4).
Within both datasets, DiSCo outperformed the rBBH as well as simple blast approaches in the distinction of true positives over false positives (Table S7).
The comparison of all DiSCo hits and the jackknife resampling assignments in the complete genomes dataset showed the robust predictability of DiSCo models. Briefly, DsrC hits were not identified in two Thermoproteus species (two runs in which an archaeal sequence was removed) and in one Desulfurella species (single event). This can be explained due to lower similarities of the excluded sequence with canonical bacterial sulphite reductases such as the one from Desulfovibrio vulgaris [49]. Similar reasoning can explain the fact that a DsrK hit was not found in Pelobacter propionicus (one run) and a DsrM hit in Caldivirga maquilingensis (one run). The removal of sequences from one DsrM model led to the identification of eight additional sequences, present in a total of 13 genomic assemblies. A close inspection of the sequences revealed that they were respiratory nitrate reductases from different Staphylococcus species (five unique protein sequences, in total present in ten genomes), one Sulfobobus islandicus strain, a Geobacillus species and Ferroglobus placidus. This shows that the models and their cutoffs are efficient in the removal of paralogues. No additional hits were found in the case of DsrA, DsrB, DsrC and DsrK resampled models and none of the identified hits changed its type-prediction.
Overall, the resampled profiles provided the same results and only 0.9 % (five out of 545 sequences) of the assignments were lost showing a stable predictability not only in terms of Dsr-dependent dissimilatory sulphur metabolism identification but also regarding the predicted enzyme type (Table S8).
DiSCo performance
A time and memory consumption test was performed on a standard laptop [MacBook Pro (13-inch, Mid 2012), OS X ‘El Capitan’ 10.11.16, Intel Core i7-3520M @ 2.9 GHz, 8 GB RAM]. DiSCo was run against the complete genome dataset using one thread. The DiSCo process required, on average, 27 MB RAM and took approximately 3.4 s per genome.
Expanded distribution of the Dsr-dependent dissimilatory sulphur metabolism in prokaryotes
Running DiSCo against a dataset composed of 195 878 (meta)genomes allowed the expansion of this type of dissimilatory sulphur metabolism from the initial 103 to 1738 micro-organisms, in which at least one hit of DsrA and/or DsrB protein was found (Tables S10 and S11). Besides the distribution of dSOB and dSRP found in the complete genomes dataset, (Fig. 4, Table S9), the screening of metagenomic records allowed the identification of DsrA and/or DsrB proteins in 28 additional phyla (55 additional distinct classes). The full taxonomic diversity is summarized in Table S12. Below, this diversity will be discussed in detail, excluding assemblies with a contamination ≥5 % or a completeness ≤85 %.
In bacteria, reductive-type proteins were identified in Ignavibacteriae, Bacteroidetes/Chlorobi, candidate division LCP-89m, candidate division Zixibacteria, Ca. Hydrogenedentes, Nitrospirae, Ca. Omnitrophica, Actinobacteria, Armatimonadetes, Chloroflexi, Firmicutes, Thermodesulfobacteria and Proteobacteria (Deltaproteobacteria, Ca. Lambdaproteobacteria), lineages for which the metabolic potential had been reported [18]. Within Archaea, and excluding the known cultivated lineages, reductive-type proteins were identified in Ca. Korarchaeota, Ca. Hydrothermarchaeota, Aigarchaeota, and Diaforarchaea. Previous reports, also based on genomic content, reported the potential for sulphate/sulfite reduction within these taxa [60, 65, 71, 72].
The oxidative-type DsrAB proteins were identified in micro-organism belonging to Chlorobi, in proteobacterial metagenomes from several classes, including Alpha-, Beta-, Gamma-, and Deltaproteobacteria, Hydrogenophilalia, Ca. Muproteobacteria, Acidithiobacillia, as well as in some unclassified bacteria. With very few exceptions, the remaining proteins of the oxidative pathway were also identified (same type), as for instance DsrMK(JOP), DsrEFH, QmoABC/AprM, AprAB and Sat. Interestingly, both dSRP and dSOB metabolic proteins were found across Nitrospira (meta)genomes.
In some Actinobacteria, Nitrospinae, a few deltaproteobacterial assemblies, Ca. Desantisbacteria, Ca. Rokubacteria and Ca. Lambdaproteobacteria, DiSCo identified mixed profiles, with proteins being predicted to belong to different types (Fig. 5). For example, in Ca. Lambdaproteobacteria, DiSCo identified the reductive DsrAB, DsrD, DsrT, DsrM and DsrJP proteins as well as oxidative DsrC, DsrEFH, DsrK, DsrO and DsrL proteins. This could be for four reasons: (1) incorrect assignment of the type by DiSCo, (2) an incomplete metagenomic assembly in which only some of the genes of both enzyme types are sequenced and made available as protein sequences, (3) potential contamination within the assembly or (4) a broader diversity of the modular enzymatic scheme within the environment, due to extensive horizontal gene transfer events [97, 98]. This patchwork organization, albeit without type for the majority of the proteins, was already reported to occur in several of these genomes [18], which favours a combination of the second, third and fourth hypothesis over DiSCo misassignments.
Fig. 5.
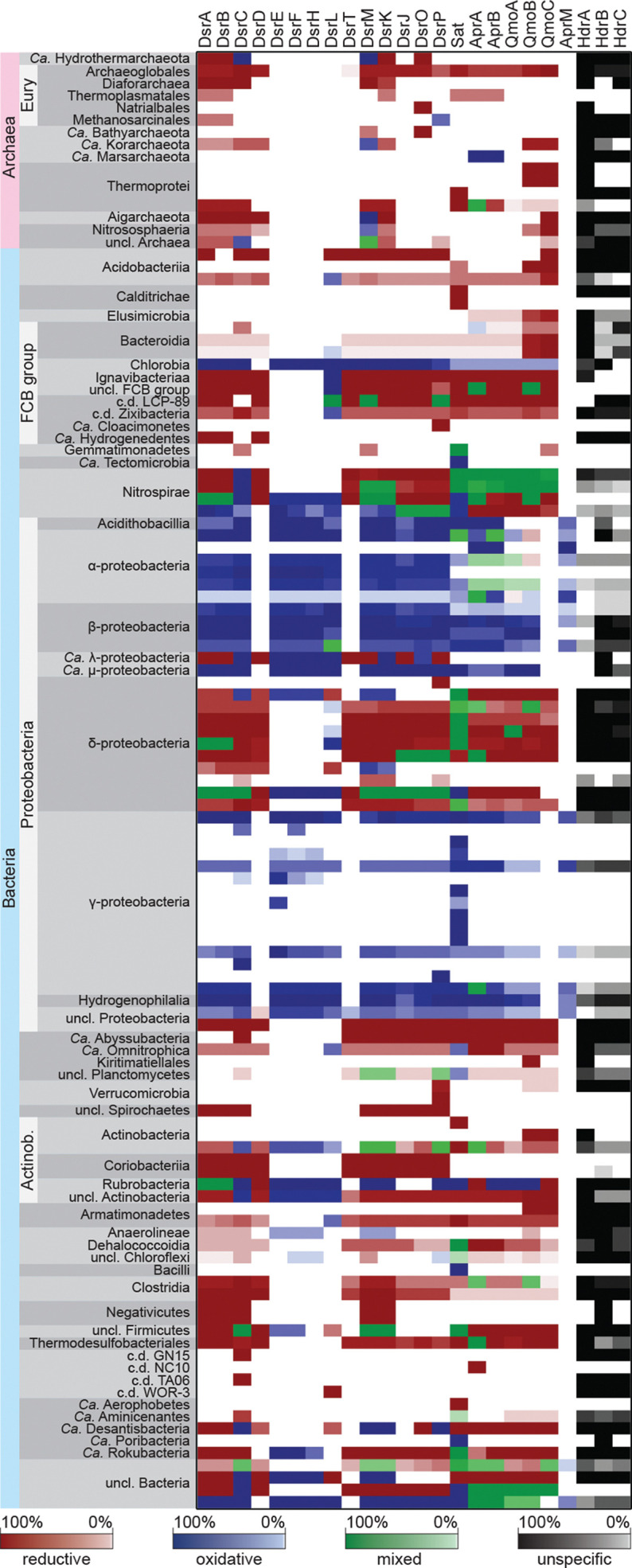
DiSCo screening of Dsr-dependent dissimilatory sulphur metabolism across metagenomic records. The distribution of DiSCo hits is shown as the percentage of genomes containing the respective protein per taxonomic group. Genomes were grouped by order or phylum. The colour code indicates DiSCo predicted enzyme type as in Fig. 4 with green representing cases in which multiple enzyme types were predicted. Only taxa containing high-quality genomes (≥85 % completeness and ≤5 % redundancy) with at least one type-specific hit are represented. Absolute values are listed in Table S13.
Recently, the existence of two types of DsrLs (DsrL1 and DsrL2) as well as the co-distribution of DsrL1 and oxidative DsrAB type as a confident indication for dissimilatory sulphur oxidizers was proposed [48]. Although DiSCo also has two models to cover the DsrL protein diversity, there is no direct correlation between the reported DsrL1 and DsrL2 types and DiSCo DsrL models. Our strategy was to create profiles able to automatically identify DsrL proteins from metagenomic data, independent of the (meta)genomic context, diminishing the need for extensive paralogous identification and manual curation.
Out of 195 878 metagenomic records, DiSCo automatically identified DsrL proteins in 813 assemblies. In 664 of those, DsrL proteins were co-distributed with oxidative DsrAB and in 73 cases with the reductive DsrAB complex. DsrL was also present in 15 assemblies, containing versions of both reductive and oxidative Dsr-pathways. Only in 61 cases was the DsrL found in micro-organisms devoid of DsrAB proteins. In 41 out of these, additional Dsr proteins were identified, suggesting that the non-identification of DsrAB proteins by DiSCo might be due to metagenomic incompleteness. The DsrL proteins associated with reductive-type DsrAB proteins were found in Acidobacteria, Bacteroidetes, Ignavibacteriae, Proteobacteria, Ca. Omnitrophica, Actinobacteria, Armatimonadetes, Firmicutes and Ca. Desantisbacteria. The DsrL protein co-distributed with oxidative-type DsrAB was found in Chlorobi, Proteobacteria and Nitrospirae. Comparing DiSCo results with the results from [48], with some minor differences due to differences in the genomic datasets used, a very similar DsrL diversity is recovered by both methods. We could identify DsrL proteins in 145 of the 155 DsrL sequences mentioned in [48]. Nine sequences were derived from genomic assemblies not present in our datasets, and only one sequence was not identified by DiSCo. This sequence from Acidobacteria bacterium 21-70-11 is annotated as a partial protein and represents a fusion of DsrN (C-terminus) and DsrL (N-terminus) with missing fragments of both proteins.
We also compared DiSCo assignments with the results from [18], where, based on metagenomic content, synteny, and phylogenetic analyses, 19 high rank lineages (phyla or classes) of micro-organisms with potential for sulphate/sulphite reduction or sulphur oxidization were discovered. DiSCo was able to identify Dsr proteins in these lineages. The DiSCo protein type classification is also in agreement with the metabolic potential classification given in [18]. In addition, as in [18], within Ca. Falkowbacteria metagenomes, only DsrD proteins were identified. However, using DsrT as a possible marker for dissimilatory sulphite-reducing micro-organisms as proposed [18] is problematic, since DsrT proteins are found in known dSDM, such as Desulfurivibrio alkaliphilus [17] and Desulfobulbus propionicus [50].
This distribution, with respect to the taxonomic affiliation and protein’s type is in line with previous studies [18, 65, 69, 71, 72]. As observed in the screening of the complete genomes dataset, most of the independent type predictions of Dsr proteins are congruent within a genome, even for metagenomic records (Fig. 5, Tables S10 and S11).
Previously unknown lineages identified by DiSCo
Several Dsr proteins were identified within Spirochaetes metagenomes with varying levels of contamination and completeness (24 –97 % completeness, up to ~23 % contamination). Particularly, in the high-quality assembly of Spirochaetes bacterium FW300 bin.19 (97 % completeness, 1.44 % contamination) the reductive-type proteins DsrAB, DsrC and a DsrMKJOP complex were found. Several Spirochaetes were isolated from sulphur ‘Thiodendron’ mats and, although not common within this phylum, at least in one micro-organism, Spirochaeta perfilievii, sulphur and thiosulphate (but not sulphate) were shown to support growth as electron acceptors in anaerobic conditions [99]. To our knowledge, so far, no Dsr proteins were identified in this phylum, and only the identification of Sat, AprAB and a potential Qmo complex were reported in Alkalispirochaeta odontotermitis JC202 (previously Spirochaeta odontotermitis JC202) [100]. Within this metagenome, DiSCo identified the DsrC, DsrT, DsrMKJOP, Sat, AprAB and QmoABC proteins but the high contamination level (23 %) refrains from any other conclusion. In particular, due to the co-existence of Spirochaetes with both sulphide oxidizers and sulphate reducers [101], the hypothesis of assembly artefacts can not be ruled out. Further experimental characterizations are needed to illuminate the predicted metabolic potential of this phylum. This argument is valid not only for the diversity herein depicted but also for the newly proposed diversity of Dsr-dependent dissimilatory sulphur metabolism recently described [18, 48, 60, 64, 65, 71, 72].
Surprisingly, reductive-type Dsr proteins were also identified in a group of metagenomes affiliated with the phylum Thaumarchaeota, class Nitrososphaeria. To our knowledge, the metabolic potential for Dsr-dependent dissimilatory sulphur reduction has not been described in Thaumarchaeota. In the most complete of those assemblies, (~87 % completeness, 0–1.9 % redundancy), reductive-type DsrABC proteins as well as a DsrMK complex were identified. In some, DsrD and two subunits of the Qmo complex (QmoB, QmoC) were also found. A closer inspection of DsrAB sequences of one of these assemblies (Nitrososphaeria archaeon SpSt-95) showed that outside this phylum, the highest identities (~71 %) are with korarchaeal DsrAB sequences, including the one from Ca. Methanodesulfokores washburnensis, recently proposed to have the potential to perform sulphite-dependent, anaerobic oxidation of methane to methanol [71]. In comparison with the DsrA from Archaeoglogus fulgidus and Desulfovibrio vulgaris, the identities were around 58 and 48 %, respectively. This is an indication that DiSCo’s models and cutoffs are able to retrieve previously unknown diversity.
Conclusion
To enable a faster and computationally more efficient method for the study of Dsr-dependent dissimilatory sulphur metabolism from (meta)genomic records, we developed DiSCo, a tool to identify and predict the enzyme type from protein data. When benchmarked against genomic records the tool proved to be more efficient as blast-based methods with a minimal accuracy of 0.98 for complete genomes and 0.93 for metagenomic records.
Overall, DiSCo was able to identify type-specific proteins in 66 different phyla. Thus, DiSCo has proven to be a valuable tool for future studies aimed at analysing and exploring Dsr-dependent dissimilatory sulphur metabolism diversity from protein data and can be used with complete genomic records, metagenomic data or a user-provided dataset. DiSCo also avoids the need of manual inspection of intermediate steps, filtering of similarity search results and genome mining to extract sequences. Moreover, DiSCo circumvents the need for performing computational demanding phylogenetic reconstructions with the sole aim of identifying an enzyme’s type. This automatic classification can aid in further down-stream analyses and provide guidance for the cultivation of micro-organisms.
DiSCo is fast (screening thousands of genomes per hour) and allows the use of personal computers for large-scale analyses in a computationally straightforward and more efficient manner (in terms of runtime and memory use) than traditional methods.
To summarize, DiSCo provides the scientific community with a Dsr-dependent dissimilatory sulphur metabolism specialized tool in which independent protein identification and assignment of the enzyme type is performed.
Supplementary Data
Funding information
This project has received funding from the Wiener Wissenschafts-, Forschungs- und Technologiefonds (grant agreement VRG15-007) to FLS.
Acknowledgement
We thank Joost van Ham for testing DiSCo on Windows 10 and the members of the Genome Evolution and Ecology Group, in particular Angus Hilts and Anastasiia Padalko, who contributed to valuable discussions.
Author contributions
F.L.S. and S.N. conceived the study. S.N. developed and implemented the method and performed the analyses with input from F.L.S., S.N. and F.L.S. analysed and interpreted the data. Both authors wrote and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AC, accuracy; Apr, adenosine-5'-phosphosulphate reductase; BA, balanced accuracy; DiSCo, Dsr-dependent dissimilatory sulphur metabolism classification tool; dSDM, Dsr-dependent sulphur-disproportionating micro-organisms; dSOB, Dsr-dependent sulphur-oxidizing bacteria; Dsr, dissimilatory sulphite reductase; dSRP, Dsr-dependent sulphate/thiosulphate/sulphite-reducing prokaryotes; FDR, false discovery rate; Hdr, heterodisulphide reductase; PR, precision; Qmo, quinone-interacting membrane-bound oxidoreductase; rBBH, reciprocal best blast hit; RC, recall.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure, supplementary files and thirteen supplementary tables are available with the online version of this article.
References
- 1.Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 2.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 3.Colman DR, Lindsay MR, Amenabar MJ, Boyd ES. The intersection of geology, geochemistry, and microbiology in continental hydrothermal systems. Astrobiology. 2019;19:1505–1522. doi: 10.1089/ast.2018.2016. [DOI] [PubMed] [Google Scholar]
- 4.Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Annu Rev Microbiol. 2013;67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 5.Postgate JR. Cytochrome c3 and desulphoviridin; pigments of the anaerobe Desulphovibrio desulphuricans . J Gen Microbiol Microbiol. 1956;14:545–572. doi: 10.1099/00221287-14-3-545. [DOI] [PubMed] [Google Scholar]
- 6.Lee JP, LeGall J, Peck HD. Isolation of assimilatory- and dissimilatory-type sulfite reductases from Desulfovibrio vulgaris . J Bacteriol. 1973;115:529–542. doi: 10.1128/jb.115.2.529-542.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schedel M, Trüper HG. Purification of Thiobacillus denitrificans siroheme sulfite reductase and investigation of some molecular and catalytic properties. Biochim Biophys Acta -Enzymology. 1979;568:454–466. doi: 10.1016/0005-2744(79)90314-0. [DOI] [PubMed] [Google Scholar]
- 8.Butlin KR, Adams ME, Thomas M. The isolation and cultivation of sulphate-reducing bacteria. J Gen Microbiol. 1949;3:46–59. doi: 10.1099/00221287-3-1-46. [DOI] [PubMed] [Google Scholar]
- 9.van Niel CB. On the morphology and physiology of the purple and green sulphur bacteria. Arch Mikrobiol. 1931;3:1–112. [Google Scholar]
- 10.Perty M. Zur Kenntniss kleinster Lebensformen nach Bau, Funktionen, Systematik, mit Specialverzeichniss der in der Schweiz beobachteten. Bern: Verlag von Jent & Reinert; 1852. [Google Scholar]
- 11.Rabus R, Venceslau SS, Wöhlbrand L, Voordouw G, Wall JD, et al. In: Advances in Microbial Physiology. Poole R, editor. Academic Press; 2015. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes; pp. 55–321. [DOI] [PubMed] [Google Scholar]
- 12.Dahl C. In: Modern Topics in the Phototrophic Prokaryotes. Hallenbeck P, editor. Cham: Springer; 2017. Sulfur metabolism in phototrophic bacteria; pp. 27–66. [DOI] [Google Scholar]
- 13.Grein F, Ramos AR, Venceslau SS, Pereira IAC. Unifying concepts in anaerobic respiration: Insights from dissimilatory sulfur metabolism. Biochim Biophys Acta - Bioenerg Bioenerg. 2013;1827:145–160. doi: 10.1016/j.bbabio.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Hausmann B, Pelikan C, Herbold CW, Köstlbacher S, Albertsen M, et al. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 2018;12:1729–1742. doi: 10.1038/s41396-018-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loy A, Duller S, Wagner M. In: Microbial Sulfur Metabolism. Dahl C, Friedrich C, editors. Heidelberg: Springer; 2008. Evolution and ecology of microbes dissimilating sulfur compounds: Insights from siroheme sulfite reductases; pp. 46–59. [DOI] [Google Scholar]
- 16.Löffler M, Feldhues J, Venceslau SS, Kammler L, Grein F, et al. DsrL mediates electron transfer between NADH and rDsrAB in Allochromatium vinosum . Environ Microbiol. 2020;22:783–795. doi: 10.1111/1462-2920.14899. [DOI] [PubMed] [Google Scholar]
- 17.Thorup C, Schramm A, Findlay AJ, Finster KW, Schreiber L. Disguised as a sulfate reducer: Growth of the deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. MBio. 2017;8:1–9. doi: 10.1128/mBio.00671-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–1728. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sperling D, Kappler U, Wynen A, Dahl C, Trüper HG. Dissimilatory ATP sulfurylase from the hyperthermophilic sulfate reducer Archaeoglobus fulgidus belongs to the group of homo-oligomeric ATP sulfurylases. FEMS Microbiol Lett. 1998;162:257–264. doi: 10.1111/j.1574-6968.1998.tb13007.x. [DOI] [PubMed] [Google Scholar]
- 20.Fritz G, Roth A, Schiffer A, Büchert T, Bourenkov G, et al. Structure of adenylylsulfate reductase from the hyperthermophilic Archaeoglobus fulgidus at 1.6-Å resolution. Proc Natl Acad Sci U S A. 2002;99:1836–1841. doi: 10.1073/pnas.042664399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duarte AG, Santos AA, Pereira IAC. Electron transfer between the QmoABC membrane complex and adenosine 5′-phosphosulfate reductase. Biochim Biophys Acta - Bioenerg. 2016;1857:380–386. doi: 10.1016/j.bbabio.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira TF, Vonrhein C, Matias PM, Venceslau SS, Pereira IAC, et al. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J Biol Chem. 2008;283:34141–34149. doi: 10.1074/jbc.M805643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pires RH, Venceslau SS, Morais F, Teixeira M, Xavier A, et al. Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complex - A membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry. 2006;45:249–262. doi: 10.1021/bi0515265. [DOI] [PubMed] [Google Scholar]
- 24.Santos AA, Venceslau SS, Grein F, Leavitt WD, Dahl C, et al. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science. 2015;350:1541–1545. doi: 10.1126/science.aad3558. [DOI] [PubMed] [Google Scholar]
- 25.Grein F, Venceslau SS, Schneider L, Hildebrandt P, Todorovic S, et al. DsrJ, an essential part of the DsrMKJOP transmembrane complex in the purple sulfur bacterium Allochromatium vinosum, is an unusual triheme cytochrome c . Biochemistry. 2010;49:8290–8299. doi: 10.1021/bi1007673. [DOI] [PubMed] [Google Scholar]
- 26.Venceslau SS, Stockdreher Y, Dahl C, Pereira IAC. The ‘bacterial heterodisulfide’ DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta. 2014;1837:1148–1164. doi: 10.1016/j.bbabio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Florentino AP, Pereira IAC, Boeren S, van den Born M, Stams AJM, et al. Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ Microbiol. 2019;21:209–225. doi: 10.1111/1462-2920.14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 29.Huber R, Kristjansson JK, Stetter KO. Pyrobaculum gen. nov., a new genus of neutrophilic, rod-shaped archaebacteria from continental solfataras growing optimally at 100°C. Arch Microbiol. 1987;149:95–101. doi: 10.1007/BF00425072. [DOI] [Google Scholar]
- 30.Akbar S, Gaidenko TA, Min Kang C, O’Reilly M, Devine KM, et al. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis . J Bacteriol. 2001;183:1329–1338. doi: 10.1128/JB.183.4.1329-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pott AS, Dahl C. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology. 1998;144:1881–1894. doi: 10.1099/00221287-144-7-1881. [DOI] [PubMed] [Google Scholar]
- 32.Frigaard NU, Dahl C. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol. 2008;54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- 33.Weissgerber T, Watanabe M, Hoefgen R, Dahl C. Metabolomic profiling of the purple sulfur bacterium Allochromatium vinosum during growth on different reduced sulfur compounds and malate. Metabolomics. 2014;10:1094–1112. doi: 10.1007/s11306-014-0649-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer B, Kuever J. Phylogeny of the alpha and beta subunits of the dissimilatory adenosine-5′ -phosphosulfate (APS) reductase from sulfate-reducing prokaryotes - Origin and evolution of the dissimilatory sulfate-reduction pathway. Microbiology. 2007;153:2026–2044. doi: 10.1099/mic.0.2006/003152-0. [DOI] [PubMed] [Google Scholar]
- 35.Meyer B, Kuever J. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5’-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing. Microbiology. 2007;153:3478–3498. doi: 10.1099/mic.0.2007/008250-0. [DOI] [PubMed] [Google Scholar]
- 36.Pereira IAC, Ramos AR, Grein F, Marques MC, da Silva SM, et al. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol. 2011;2:1–22. doi: 10.3389/fmicb.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Junier P, Junier T, Podell S, Sims DR, Detter JC, et al. The genome of the Gram-positive metal- and sulfate-reducing bacterium Desulfotomaculum reducens strain MI-1. Environ Microbiol. 2010;12:2738–2754. doi: 10.1111/j.1462-2920.2010.02242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaster AK, Moll J, Parey K, Thauer RK. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci U S A. 2011;108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner T, Koch J, Ermler U, Shima S. Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science. 2017;357:699–703. doi: 10.1126/science.aan0425. [DOI] [PubMed] [Google Scholar]
- 40.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta - Bioenerg. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: Identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology. 2013;159:2626–2638. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe T, Kojima H, Umezawa K, Hori C, Takasuka TE, et al. Genomes of neutrophilic sulfur-oxidizing chemolithoautotrophs representing 9 proteobacterial species from 8 genera. Front Microbiol. 2019;10:316. doi: 10.3389/fmicb.2019.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/BR.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zane GM, Bill Yen HC, Wall JD. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 2010;76:5500–5509. doi: 10.1128/AEM.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chernyh NA, Neukirchen S, Frolov EN, Sousa FL, Miroshnichenko ML, et al. Dissimilatory sulfate reduction in the archaeon ‘Candidatus Vulcanisaeta moutnovskia’ sheds light on the evolution of sulfur metabolism. Nat Microbiol. 2020;5:1428–1438. doi: 10.1038/s41564-020-0776-z. [DOI] [PubMed] [Google Scholar]
- 46.Stockdreher Y, Sturm M, Josten M, Sahl HG, Dobler N, et al. New proteins involved in sulfur trafficking in the cytoplasm of Allochromatium vinosum . J Biol Chem. 2014;289:12390–12403. doi: 10.1074/jbc.M113.536425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stockdreher Y, Venceslau SS, Josten M, Sahl HG, Pereira IAC, et al. Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: Evidence for sulfur transfer from DsrEFH to DsrC. PLoS One. 2012;7:e40785. doi: 10.1371/journal.pone.0040785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Löffler M, Wallerang KB, Venceslau SS, Pereira IAC, Dahl C. The Iron-Sulfur Flavoprotein DsrL as NAD(P)H:Acceptor Oxidoreductase in oxidative and reductive dissimilatory sulfur metabolism. Front Microbiol. 2020;11:1–15. doi: 10.3389/fmicb.2020.578209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller A, Kjeldsen KU, Rattei T, Pester M, Loy A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015;9:1152–1165. doi: 10.1038/ismej.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lovley DR, Phillips EJP. Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl Environ Microbiol. 1994;60:2394–2399. doi: 10.1128/AEM.60.7.2394-2399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hittel DS, Voordouw G. Overexpression, purification and immunodetection of DsrD from Desulfovibrio vulgaris Hildenborough. Antonie van Leeuwenhoek. 2000;77:271–280. doi: 10.1023/A:1002449227469. [DOI] [PubMed] [Google Scholar]
- 52.Dahl C, Schulte A, Shin DH. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of DsrEFH from Allochromatium vinosum. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:890–892. doi: 10.1107/S1744309107041188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuno N, Voordouw G, Miki K, Sarai A, Higuchi Y. Crystal structure of dissimilatory sulfite reductase D (DsrD) protein—possible interaction with B- and Z-DNA by its winged-helix motif. Structure. 2003;11:1133–1140. doi: 10.1016/s0969-2126(03)00156-4. [DOI] [PubMed] [Google Scholar]
- 54.Larsen LT, Birkeland NK. A novel organization of the dissimilatory sulfite reductase operon of Thermodesulforhabdus norvegica verified by RT-PCR. FEMS Microbiol Lett. 2001;203:81–85. doi: 10.1111/j.1574-6968.2001.tb10824.x. [DOI] [PubMed] [Google Scholar]
- 55.Numata T, Fukai S, Ikeuchi Y, Suzuki T, Nureki O. Structural basis for sulfur relay to RNA mediated by heterohexameric TusBCD complex. Structure. 2006;14:357–366. doi: 10.1016/j.str.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Pinnell LJ, Turner JW. Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front Microbiol. 2019;10:1252. doi: 10.3389/fmicb.2019.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmers PHA, Vavourakis CD, Kleerebezem R, Sinninghe Damsté JS, Muyzer G, et al. Metabolism and occurrence of methanogenic and sulfate-reducing syntrophic acetate oxidizing communities in haloalkaline environments. Front Microbiol. 2018;9:1–18. doi: 10.3389/fmicb.2018.03039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allioux M, Yvenou S, Slobodkina G, Slobodkin A, Shao Z, et al. Genomic characterization and environmental distribution of a thermophilic anaerobe Dissulfurirhabdus thermomarina SH388T. Microorganisms. 2020;8:1–14. doi: 10.3390/microorganisms8081132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiel V, Costas AMG, Fortney NW, Martinez JN, Tank M, et al. 'Candidatus Thermonerobacter thiotrophicus,’ a non-phototrophic member of the bacteroidetes/ chlorobi with dissimilatory sulfur metabolism in hot spring mat communities. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2018.03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colman DR, Lindsay MR, Amenabar MJ, Fernandes-Martins MC, Roden ER, et al. Phylogenomic analysis of novel Diaforarchaea is consistent with sulfite but not sulfate reduction in volcanic environments on early Earth. ISME J. 2020;14:1316–1331. doi: 10.1038/s41396-020-0611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umezawa K, Kojima H, Kato Y, Fukui M. Disproportionation of inorganic sulfur compounds by a novel autotrophic bacterium belonging to Nitrospirota . Syst Appl Microbiol. 2020;43:126110. doi: 10.1016/j.syapm.2020.126110. [DOI] [PubMed] [Google Scholar]
- 62.Wilkins LGE, Ettinger CL, Jospin G, Eisen JA. Metagenome-assembled genomes provide new insight into the microbial diversity of two thermal pools in Kamchatka, Russia. Sci Rep. 2019;9:3059. doi: 10.1038/s41598-019-39576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z, Tran PQ, Kieft K, Anantharaman K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020;14:2060–2077. doi: 10.1038/s41396-020-0669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kato S, Nakano S, Kouduka M, Hirai M, Suzuki K, et al. Metabolic potential of as-yet-uncultured archaeal lineages of Candidatus Hydrothermarchaeota thriving in deep-sea metal sulfide deposits. Microbes Environ. 2019;34:293–303. doi: 10.1264/jsme2.ME19021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Z, Liu Y, Xu W, Pan J, Luo ZH, et al. Genome- and community-level interaction insights into carbon utilization and element cycling functions of Hydrothermarchaeota in hydrothermal sediment. mSystems. 2020;5:e00795–e00819. doi: 10.1128/msystems.00795-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zecchin S, Mueller RC, Seifert J, Stingl U, Anantharaman K, et al. Rice paddy nitrospirae carry and express genes related to sulfate respiration: Proposal of the new genus “Candidatus Sulfobium”. Appl Environ Microbiol. 2018;84:e02224–17. doi: 10.1128/AEM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vavourakis CD, Andrei AS, Mehrshad M, Ghai R, Sorokin DY, et al. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments. Microbiome. 2018;6:1–18. doi: 10.1186/s40168-018-0548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato S, Shibuya T, Takaki Y, Hirai M, Nunoura T, et al. Genome-enabled metabolic reconstruction of dominant chemosynthetic colonizers in deep-sea massive sulfide deposits. Environ Microbiol. 2018;20:862–877. doi: 10.1111/1462-2920.14032. [DOI] [PubMed] [Google Scholar]
- 69.Tan S, Liu J, Fang Y, Hedlund BP, Lian ZH, et al. Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 2019;13:2044–2057. doi: 10.1038/s41396-019-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kato S, Itoh T, Yuki M, Nagamori M, Ohnishi M, et al. Isolation and characterization of a thermophilic sulfur- and iron-reducing thaumarchaeote from a terrestrial acidic hot spring. ISME J. 2019;13:2465–2474. doi: 10.1038/s41396-019-0447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKay L, Dlakic M, Fields M, Jay Z, Eren M, et al. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota . Nat Microbiol. 2019;4:614–622. doi: 10.1038/s41564-019-0362-4. [DOI] [PubMed] [Google Scholar]
- 72.Hua ZS, YN Q, Zhu Q, Zhou EM, YL Q, et al. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota . Nat Commun. 2018;9:1–11. doi: 10.1038/s41467-018-05284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42:199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 78.Rice P, Longden I, Bleasby A. EMBOSS: The european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 79.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41:387–395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Minh BQ, Nguyen MAT, Von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tria FDK, Landan G, Dagan T. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol. 2017;1:0193. doi: 10.1038/s41559-017-0193. [DOI] [PubMed] [Google Scholar]
- 86.Guy L, Kultima JR, Andersson SGE, Quackenbush J. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 88.Sander J, Engels-Schwarzlose S, Dahl C. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch Microbiol. 2006;186:357–366. doi: 10.1007/s00203-006-0156-y. [DOI] [PubMed] [Google Scholar]
- 89.Koch T, Dahl C. A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 2018;1–13 doi: 10.1038/s41396-018-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feldbauer R, Schulz F, Horn M, Rattei T. Prediction of microbial phenotypes based on comparative genomics. BMC Bioinformatics. 2015;16:S1. doi: 10.1186/1471-2105-16-S14-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finster K. Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem. 2008;29:281–292. doi: 10.1080/17415990802105770. [DOI] [Google Scholar]
- 92.Schiffer A, Fritz G, Büchert T, Herrmanns K, Steuber J, et al. In: Handbook of Metalloproteins. Messerschmidt A, editor. Chichester, UK: Wiley; 2011. Dissimilatory adenosine-5’-phosphosulfate reductase; pp. 183–194. [Google Scholar]
- 93.Dahl JU, Radon C, Bühning M, Nimtz M, Leichert LI, et al. The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J Biol Chem. 2013;288:5426–5442. doi: 10.1074/jbc.M112.431569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith JA, Aklujkar M, Risso C, Leang C, Giloteaux L, et al. Mechanisms involved in Fe(III) respiration by the hyperthermophilic archaeon Ferroglobus placidus . Appl Environ Microbiol. 2015;81:2735–2744. doi: 10.1128/AEM.04038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duarte AG, Barbosa ACC, Ferreira D, Manteigas G, Domingos RM, et al. Redox loops in anaerobic respiration - The role of the widespread NrfD protein family and associated dimeric redox module. BBA - Bioenerg. 2021;148416 doi: 10.1016/j.bbabio.2021.148416. [DOI] [PubMed] [Google Scholar]
- 96.Dahl C, Engels S, Pott-Sperling AS, Schulte A, Sander J, et al. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J Bacteriol. 2005;187:1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Omelchenko M, Makarova KS, Wolf YI, Rogozin IB, Koonin E. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 2003;4:R55. doi: 10.1186/gb-2003-4-9-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dubinina G, Grabovich M, Leshcheva N, Rainey FA, Gavrish E. Spirochaeta perfilievii sp. nov., an oxygen-tolerant, sulfide-oxidizing, sulfur- and thiosulfate-reducing spirochaete isolated from a saline spring. Int J Syst Evol Microbiol. 2011;61:110–117. doi: 10.1099/ijs.0.018333-0. [DOI] [PubMed] [Google Scholar]
- 100.Watanabe T, Kojima H, Fukui M. Identity of major sulfur-cycle prokaryotes in freshwater lake ecosystems revealed by a comprehensive phylogenetic study of the dissimilatory adenylylsulfate reductase. Sci Rep. 2016;6:1–9. doi: 10.1038/srep36262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blazejak A, Erséus C, Amann R, Dubilier N. Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (oligochaeta) from the Peru margin. Appl Environ Microbiol. 2005;71:1553–1561. doi: 10.1128/AEM.71.3.1553-1561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.