Abstract
This study investigated the evolution and epidemiology of the community-associated and multidrug-resistant Staphylococcus aureus clone European CC1-MRSA-IV. Whole-genome sequences were obtained for 194 European CC1-MRSA-IV isolates (189 of human and 5 of animal origin) from 12 countries, and 10 meticillin-susceptible precursors (from North-Eastern Romania; all of human origin) of the clone. Phylogenetic analysis was performed using a maximum-likelihood approach, a time-measured phylogeny was reconstructed using Bayesian analysis, and in silico microarray genotyping was performed to identify resistance, virulence-associated and SCCmec (staphylococcal cassette chromosome mec) genes. Isolates were typically sequence type 1 (190/204) and spa type t127 (183/204). Bayesian analysis indicated that European CC1-MRSA-IV emerged in approximately 1995 before undergoing rapid expansion in the late 1990s and 2000s, while spreading throughout Europe and into the Middle East. Phylogenetic analysis revealed an unstructured meticillin-resistant S. aureus (MRSA) population, lacking significant geographical or temporal clusters. The MRSA were genotypically multidrug-resistant, consistently encoded seh, and intermittently (34/194) encoded an undisrupted hlb gene with concomitant absence of the lysogenic phage-encoded genes sak and scn. All MRSA also harboured a characteristic ~5350 nt insertion in SCCmec adjacent to orfX. Detailed demographic data from Denmark showed that there, the clone is typically (25/35) found in the community, and often (10/35) among individuals with links to South-Eastern Europe. This study elucidated the evolution and epidemiology of European CC1-MRSA-IV, which emerged from a meticillin-susceptible lineage prevalent in North-Eastern Romania before disseminating rapidly throughout Europe.
Keywords: CA-MRSA, European CC1-MRSA-IV clone, evolution, epidemiology, phylogenomics, transmission
Data Summary
This study generated sequencing data for 85 Staphylococcus aureus isolates and used published data for 122 isolates. All new sequence read sets are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the BioProject accession number PRJNA494507. References and BioSample accession numbers for all 207 isolates are detailed in supplementary Dataset S1.
Impact Statement.
Community-associated (CA) meticillin-resistant Staphylococcus aureus (MRSA) strains tend to spread in both healthcare and community settings, where they continue to pose a serious challenge to public health worldwide. Maintaining surveillance of these strains across borders enhances our understanding of CA-MRSA, enabling the development of effective strategies to limit its spread and, thus, reducing our dependence on antibiotics. European CC1-MRSA-IV is a multidrug-resistant and CA-MRSA strain. Previous work has shown that this strain may have originated in South-Eastern Europe, and is now endemic in certain regions of Germany, Ireland, Italy and Romania. Through the whole-genome analysis of 194 isolates, mostly from humans but also from animals, this study showed that European CC1-MRSA-IV emerged from a multidrug-resistant but meticillin-susceptible Staphylococcus aureus strain prevalent in North-Eastern Romania, in approximately 1995. In the late 1990s and 2000s, the strain then spread throughout Europe and into the Middle East, expanding particularly rapidly in 2004 and 2007/2008, and occasionally acquiring additional resistance or virulence determinants. Overall, this study has examined the evolutionary and epidemiological history of a multidrug-resistant and CA-MRSA strain that has received relatively little attention to date despite its importance in Europe, and significance to both humans and animals.
Introduction
Meticillin-resistant Staphylococcus aureus (MRSA) are a major cause of hospital-associated and community-associated (CA) infections worldwide. These infections are caused by clones that initially emerged in healthcare facilities, the community or livestock, but which tend to spread beyond these environments. The CA-MRSA population in Europe is remarkably diverse [1]. Continuous surveillance of this population is required to increase our overall understanding of CA-MRSA, and to develop effective strategies to limit its spread.
We recently described a Panton–Valentine leukocidin (PVL)-negative MRSA clone belonging to clonal complex (CC) 1, which is associated with the emergence of MRSA in the community, and harbouring a type IVa staphylococcal cassette chromosome mec (SCCmec) element [2, 3]. We showed that this clone is endemic in Ireland, Romania and the German state of Bavaria, and revealed that it likely emerged from a meticillin-susceptible CC1 lineage [2, 3] known to be highly prevalent in North-Eastern Romania [3]. Considering these findings, we refer to this clone as European CC1-MRSA-IV.
European CC1-MRSA-IV is a multidrug-resistant clone that typically encodes resistance to penicillin (blaZ), tetracycline [tet(K)], aminoglycoside-streptothricin antibiotics (aadE-sat-aphA3) and MLSB (macrolide, lincosamide and streptogramin B) compounds [erm(C)]. Additionally, the clone consistently harbours the enterotoxin gene seh (but usually lacks other enterotoxins), and often carries sak and scn, which are co-located on lysogenic hlb-converting bacteriophages [2–4]. This clone is further characterized by a SCCmec type IVa element that harbours an insertion of approximately 5350 nt in its downstream constant segment (dcs), adjacent to orfX [5]. To date, this clone has been associated predominantly with sequence type (ST) 1 and spa type t127 [2].
In recent years, European CC1-MRSA IV has been recognized as a somewhat problematic clone. A variant harbouring plasmid-encoded iles2/mupA and qacA (conferring mupirocin and chlorhexidine resistance, respectively) was responsible for a protracted outbreak spanning multiple hospitals and involving the community, in Ireland [6]. Furthermore, the clone is common in a children’s hospital in Italy [7], and its prevalence increased from 1 % in 2010 to 11 % in 2019 in Bavaria, Germany (our unpublished observations). In addition, we have speculated that this clone is common in the German state of North Rhine-Westphalia and among livestock in Italy [2, 8, 9]. Finally, the clone’s characteristic SCCmec insertion has recently been shown to give rise to false-negative results with a widely used, PCR-based commercial MRSA identification assay [5].
Considering the above points, we sought to further examine European CC1-MRSA-IV. Specifically, we aimed to investigate the evolution and epidemiology of the clone in order to better understand its expansion and spread. This study has built upon our previous work by including a larger number of isolates from more countries, several isolates from non-human hosts, and a more advanced and comprehensive phylogenetic analysis. Together, these factors have enabled us to estimate the year this clone emerged, and identify the dissemination pattern it has followed to date.
Methods
Isolates
Two hundred and four study isolates, and three outgroup isolates, were investigated. All isolates are described in detail in Dataset S1. Species identification and meticillin-resistance detection was undertaken as described previously [10]. Isolates were stored at −80 °C on cryogenic beads.
Study isolates
The study isolates comprised (i) 109 isolates definitively identified previously as European CC1-MRSA-IV, (ii) 10 meticillin-susceptible S. aureus (MSSA) isolates previously identified as precursors of European CC1-MRSA-IV, and (iii) 85 MRSA isolates tentatively identified as European CC1-MRSA-IV. The 109 isolates definitively identified previously as European CC1-MRSA-IV included 14 isolates investigated by Manara et al. [7], 38 isolates examined by Monecke et al. [5] and 57 isolates from our previous study [2]. While the isolates investigated by Manara et al. and Monecke et al. had been acquired directly from hospital collections, the 57 isolates from our previous study had been selected from an international S. aureus microarray database. This database comprises genotypic profiles for over 28 000 MRSA and MSSA isolates recovered from humans and animals worldwide [11]. By analysing these profiles, it is possible to determine the clone or lineage to which an isolate belongs [11]. The 57 isolates investigated had constituted a representative selection of all European CC1-MRSA-IV in the database; they had been recovered in Germany, Ireland and Romania [2].
The 10 MSSA isolates previously identified as precursors of European CC1-MRSA-IV had also been selected from the microarray database [2, 3]. Romania had been the only country from which MSSA precursor isolates could be selected; this lineage had not been detected among the isolates of any other country represented in the database.
The present study included these previously examined isolates, but aimed to expand the collection by acquiring isolates from additional countries. Accordingly, 85 additional MRSA isolates were obtained by requesting that clinical laboratories or research groups throughout Europe, and in Saudi Arabia and the UAE, provide isolates (or sequence reads of isolates) matching the description of the clone. Isolates were required to fit the description of PVL-negative CC1-MRSA-IV, harbour the resistance gene cluster aadE-sat-aphA3 and/or belong to spa type t127. A representative selection (based on year of recovery, recovery site and/or genotypic variations) of isolates fitting this description underwent whole-genome sequencing at the Dublin Dental University Hospital microbiology laboratory (Ireland). This selection comprised 80 human isolates and 5 animal isolates, from 10 different countries. Four of the animal isolates were previously described, but not sequenced [12]. Other previously described animal isolates that fulfilled the selection criteria were also identified [8]; however, these were unavailable for inclusion in this study. As South-Eastern Europe was underrepresented in our final isolate collection, the Staphopia database was searched for ST1 genomes from this region; however, no such genomes were identified.
Outgroup isolates
The outgroup isolates were 124889, MW2 and MSSA476; these were included in the analysis to represent the other well-known CC1 clones/lineages. Isolate 124 889 belongs to the PVL-negative CC1-MRSA-IV clone known as Western Australian MRSA-1 [2], MW2 belongs to the PVL-positive CC1-MRSA-IV clone known as USA400 [13], and MSSA476 is a PVL-negative CC1-MSSA that was recovered in the UK [14].
Whole-genome sequencing and sequence read processing
Genomic DNA was extracted from isolates using the S. aureus Genotyping Kit 2.0 (Abbott, Alere Technologies), and the Qiagen DNeasy blood and tissue kit (Qiagen). DNA quality was assessed as previously described [10]. The Nextera DNA Flex library preparation kit (Illumina) was used according to the manufacturer’s instructions, and libraries underwent paired-end sequencing using the 500-cycle MiSeq reagent kit v2 (Illumina). Libraries were scaled to exhibit at least 50× coverage and the quality of each sequencing run was assured following cluster density and Q30 assessment. Sequence read sets were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (BioProject accession number: PRJNA494507).
For the 119 previously sequenced study isolates (109 MRSA, 10 MSSA), sequence read sets were acquired from our own collection, or contigs were downloaded from the NCBI GenBank (Dataset S1). For outgroup isolates MW2 and MSSA476, complete genomes were downloaded from the NCBI GenBank (accession numbers: NC_00392 and NC_002953, respectively). For outgroup isolate 124 889, the sequence read set was acquired from our own collection as, to our knowledge, there are currently no complete Western Australia MRSA-1 genomes publicly available. All sequence read sets were trimmed using fastp version 0.20.0 [15]. Trimmed reads were assembled using SPAdes version 3.13.1 [16], and contigs under 1000 bp were removed.
spa typing, MLST and genotypic analysis
Isolates underwent spa typing and multilocus sequence typing (MLST) using Ridom SeqSphere+ version 7.0 (Ridom). Genotypic analysis was performed in silico by mapping sequences to (i) virtual versions of the probes of the S. aureus Genotyping Kit 2.0 (Abbott, Alere Technologies) DNA microarray [17], and (ii) the virtual probes of a previously described SCCmec subtyping in silico array [18]. The in silico microarray method and the probe sequences used for both arrays have been previously described [17, 18]. The five animal isolate genomes were also searched for additional genes, namely, the animal-associated enterotoxin A gene variant, sea-320E (GenBank accession number: AY196686.1), the bovine-associated leukocidin genes, lukM/F-P83 (D83951.1), and the equid-associated leukocidin genes, lukP/Q (LT671578.1). Any unusual genotypic characteristics identified were quickly investigated using the in silico microarray approach. Alternatively, the sequence data were examined manually.
Phylogenetic analysis
Sequence alignment and SNP identification were performed using Snippy version 4.4.5 (https://github.com/tseemann/snippy). Default settings were used and MW2 was designated the reference genome. A maximum-likelihood tree based on core-genome SNPs was generated using iq-tree version 1.6.12 [19]. The general time reversible (GTR) model was used with an ascertainment bias correction and 1000 bootstraps. The phylogenetic tree was visualized using Interactive Tree of Life version 5.5.1 [20]. A SNP distance matrix was generated using SNP-dists (https://github.com/tseemann/snp-dists).
Time-scaled phylogenetic analysis
Molecular dating analysis of major divergences and the most recent common ancestor (MRCA) of the lineage was based on a recombination-pruned core-genome SNP alignment of a reduced set of 190 study isolates, including all MSSA, for which sequence read sets were available. Root-to-tip regression analysis and date randomization testing were used to confirm the data had a sufficient temporal signal (Fig. S1, available with the online version of this article) [21]. A time-scaled phylogenetic tree and effective population size over time (including the age of the MRCA) were estimated in beast-2 under a coalescent Bayesian skyline tree prior and a strict molecular clock model [22]. The GTR (general time reversible) substitution model with a gamma distribution for among site rate heterogeneity was used and a Markov chain Monte Carlo analysis of length 500 million steps was run; this was performed for various numbers of coalescent intervals of the Bayesian skyline (known as dimensions in the model) (see the Supplementary Information and Figs S2 and S3, available withe the online version of this article for details). Effective sample size values for all parameters were over 200 and sufficient chain mixing was confirmed using Tracer v1.7 [23].
Results
Isolates
The genomes of 204 study isolates and three outgroup isolates were investigated. The study isolates included 109 known European CC1-MRSA-IV and 10 MSSA previously identified as precursors of this clone, recovered in Germany, Ireland and Romania [2, 5, 7]. The present study included these 119 previously investigated isolates, but expanded the collection in order to help elucidate the evolutionary dynamics of European CC1-MRSA-IV. Hence, the study isolates also included 85 MRSA tentatively identified as European CC1-MRSA-IV, recovered in 10 different countries. The three outgroup isolates were included for comparative purposes, and represented the Western Australian MRSA-1 clone, USA400 clone and MSSA476 lineage. The majority (190/204; 93.1 %) of study isolates were assigned to ST1, with the remaining 14 assigned to 10 different single locus variants of ST1. A total of 14 different spa types were identified among the study isolates, although the majority (182/204; 89.2 %) exhibited spa type t127. None of the study isolates harboured PVL-encoding genes and all MRSA study isolates harboured SCCmec type IVa. All conventional molecular typing data are detailed in Dataset S1.
Phylogenetic insights into the European CC1-MRSA-IV clone
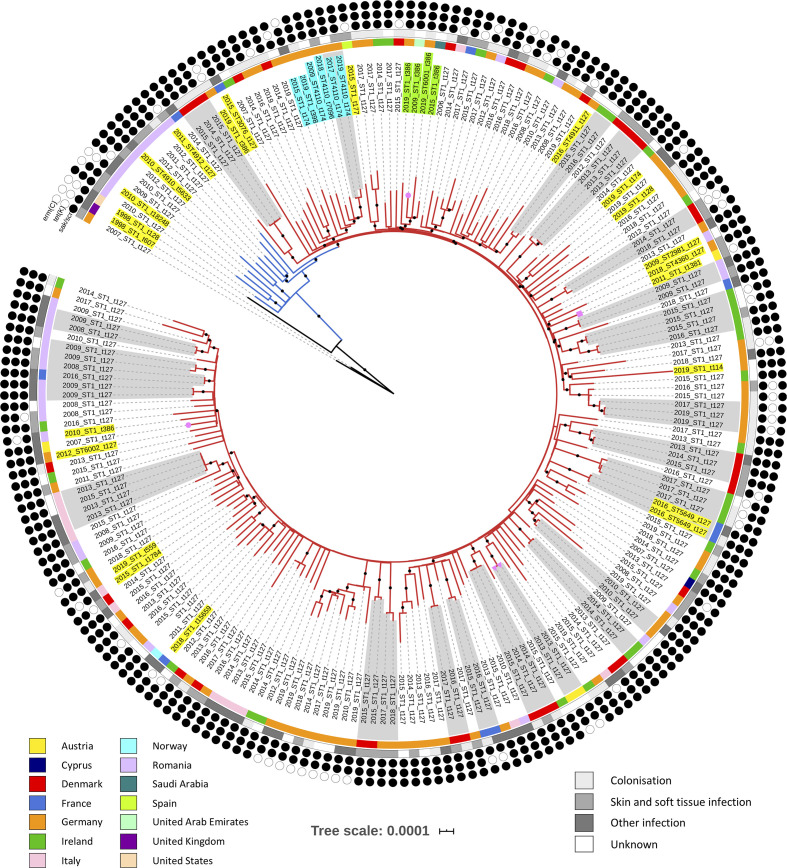
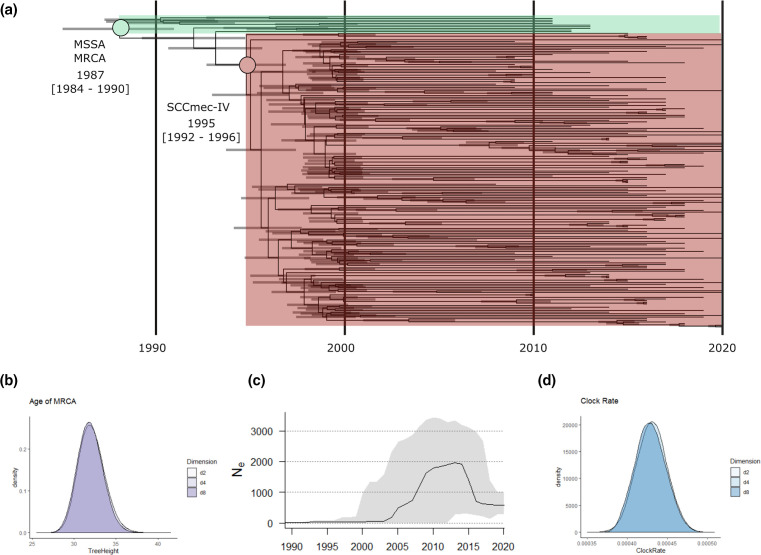
An in-depth phylogenetic analysis revealed significant details regarding the origin and dissemination of this clone. Using a maximum-likelihood approach, phylogenetic analysis based on 7066 SNPs grouped all 204 study isolates into a single clade (blue and red branches in Fig. 1). The 10 MSSA precursors of the European CC1-MRSA-IV clone formed the base of this large clade (blue branches in Fig. 1), while the remaining study isolates derived from the base (red branches in Fig. 1). This tree topology confirmed the identity of all 194 MRSA study isolates as European CC1-MRSA-IV. Typically, isolates did not group according to their country of origin or year of recovery; however, several small country-specific clusters were evident (Fig. 1). Similarly, the only two isolates from the Middle East investigated grouped into the same three-isolate sub-clade. Molecular clock analysis based on 7330 SNPs indicated that the study isolates shared their MRCA around 1987 [95 % highest posterior density (HPD): 1984–1990) (Fig. 2a, b). The European CC1-MRSA-IV clone then diverged from MSSA in approximately 1995 (95 % HPD: 1992–1996) before spreading throughout Europe, largely in the late 1990s and early 2000s (Fig. 2a). Correspondingly, reconstruction of the effective population size over time estimated a sharp rise in genetic diversity during this time (particularly in 2004 and 2007/2008) (Fig. 2c). The genomic clock rate of the clone was estimated at approximately 1.498×10−6 (95 % HPD: 1.368×10−6–1.626×10−6) substitutions per site per year, when assuming a core genome of 2.1 MB (Fig. 2d). This rate estimate is in line with previous estimates for other S. aureus lineages [24–27].
Fig. 1.
A maximum-likelihood phylogeny based on 7066 SNPs. The red branches represent the European CC1-MRSA-IV clone, the blue branches represent CC1-MSSA precursors of European CC1-MRSA-IV and the black branches represent CC1 outgroup isolates. The countries and sites of isolate recovery are indicated in the key. The presence or absence of erm(C), tet(K) and sak/scn is indicated by the black or white circles, respectively. Isolate pairs or clusters containing isolates that differed by 10 or less SNPs are shaded in grey. The pink shapes on branch tips indicate that the isolate was recovered from an animal: circle, horse; rectangle, cat; triangle, wild rook. The majority of isolates were ST1 (MLST profile: 1-1-1-1-1-1-1) and spa type t127 (repeats succession: 07-23-21-16-34-33-13); non-ST1 and/or t127 variants are highlighted. One variant cluster (highlighted in turquoise) comprised one t7096 (14-13), one t398 (26-33-13) and four t174 (14-21-16-34-33-13) isolates. Four of these six isolates were also ST4110 (1-1-1-1-1-1-558). Another variant cluster (highlighted in green) comprised two German isolates, a Saudi Arabian isolate and a UAE isolate, all of which were spa type t386 (07-23-13). One of these isolates was also ST6001 (1-1-794-1-1-1-1). The remaining variants (highlighted in yellow) did not cluster into groups. Branches with 100 % bootstrap support are indicated with black dots. The tree scale denotes substitutions per site.
Fig. 2.
Bayesian analysis of 180 European CC1-MRSA-IV isolates and 10 MSSA precursors of the clone. (a) A Bayesian consensus tree based on 7330 non-recombinant core-genome SNPs and including 95 % HPD intervals across different dimensional partitioning of population sizes. (b) Posterior density distribution showing tree height (MRCA). (c) Estimated effective population size of the clone over time (dark line) including 95 % HPD intervals (grey shading) denoting uncertainty. (d) Posterior density distribution showing clock-rate estimates for the core-genome SNP alignment.
Epidemiological characteristics of the European CC1-MRSA-IV clone
The epidemiological data available for each MRSA study isolate were examined. While the clone had been previously identified in Ireland, Germany, Romania and Italy, an expanded search led to its identification in six other European countries (Austria, Cyprus, Denmark, France, Norway and Spain) and two Middle Eastern countries (Saudi Arabia and the UAE). Additional European CC1-MRSA-IV isolates from Germany and Romania were also identified. The oldest study isolate was recovered in 2006, in Denmark. Fifty-five study isolates (26.9 %) were from colonization sites, 105 (51.5 %) from infection sites and 44 (21.6 %) from unknown sites. Fifty-five (52.4 %) of the infection isolates were from skin and soft tissue infections, and 50/105 (47.6 %) from other infections. The majority (199/204; 97.5 %) of study isolates were from humans, while 5 were from wild or domesticated animals. Two of these animal isolates were from wild rooks (Corvus frugilegus; both in Austria), two from horses (one in Austria; one in the UAE) and one from a cat (in Austria). The extent of metadata available for the study isolates varied; Danish isolates were associated with particularly detailed demographic data. The majority (25/35; 71.4 %) of Danish isolates were from samples taken by community general medical practitioners, while the remaining samples were taken in eight different hospitals. Thirteen (37.1 %) of the Danish isolates were recovered from patients with international links, e.g. they were known to have migrated from a specific country, or had a name associated with a specific country. Nine (25.7 %) of these links were to Romania (including the oldest study isolate), while the remaining four were to Afghanistan, Macedonia, Nigeria and Ukraine.
Genotypic analysis of European CC1-MRSA-IV
The genotypic profiles of the study isolates were examined to help determine the clone’s resistance and virulence potential, identify any significant patterns among the isolates, and aid identification of the clone. All genotypic data are shown in Fig. S4, specific points are highlighted in Fig. 1 and selected frequencies are detailed in Table 1.
Table 1.
Prevalence of major antimicrobial-resistance and virulence-associated genes identified in European CC1-MRSA-IV and ten MSSA precursors
|
Gene(s) |
Frequency |
|||
|---|---|---|---|---|
|
MRSA (n=194) |
MSSA (n=10) |
|||
|
No. |
% |
No. |
% |
|
|
Resistance |
||||
|
aacA-aphD |
7 |
3.6 |
1 |
10 |
|
aad6 |
1 |
0.5 |
0 |
0 |
|
aadD |
1 |
0.5 |
0 |
0 |
|
aadE-sat-aphA3 |
193 |
99.5 |
8 |
80 |
|
blaZ |
194 |
100 |
10 |
100 |
|
cat |
2 |
1 |
1 |
10 |
|
dfrA |
3 |
1.5 |
1 |
10 |
|
erm(C) |
179 |
92.3 |
8 |
80 |
|
lnuA |
2 |
1 |
0 |
0 |
|
mph(C) |
1 |
0.5 |
0 |
0 |
|
msrA |
1 |
0.5 |
0 |
0 |
|
mupA |
3 |
1.5 |
0 |
0 |
|
qacA |
4 |
2 |
0 |
0 |
|
qacC |
1 |
0.5 |
1 |
10 |
|
tet(K) |
175 |
90.2 |
8 |
80 |
|
Virulence |
||||
|
sak-scn |
170 |
87.7 |
8 |
80 |
|
Undisrupted hlb |
24 |
12.3 |
2 |
20 |
|
seh |
194 |
100 |
10 |
100 |
|
sek-seq |
7 |
3.6 |
0 |
0 |
Antimicrobial-resistance genes
Genotypic data indicated that this clone is typically resistant to β-lactams, aminoglycosides, streptothricin, tetracycline and MLSB (macrolide, lincosamide and streptogramin B) compounds (Table 1). As indicated by the sporadic occurrence of variants in the phylogeny, the clone has a tendency to lose erm(C) and tet(K), and gain other resistance genes such as aacA-aphD, dfrA, mupA/ileS2 and qacA (Table 1, Figs 1 and S4). Notably, one European CC1-MRSA-IV isolate lacked the aadE-sat-aphA3 gene cluster (according to both microarray analysis and in silico microarray analysis). This unique isolate (2546-va69252) was recovered in Regensburg, Germany, in 2019. Excluding mecA, the MSSA precursor isolates harboured similar resistance genes to the European CC1-MRSA-IV isolates (Table 1).
Virulence-associated genes
This clone consistently harboured seh, and often harboured sak and scn (Table 1). The pattern of variants in the phylogeny suggests that the clone has lost sak and scn on many occasions, and has acquired sek and seq on three separate occasions (Figs 1 and S4). Isolates lacking sak and scn harboured an undisrupted β-haemolysin gene (hlb), indicating the absence of lysogenic phages that encode combinations of human-specific immune modulator genes (sea, sak, chp, scn). The animal-associated enterotoxin A gene variant, sea-320E, the bovine-associated leukocidin genes, lukM/F, and the equid-associated leukocidin genes, lukP/Q, were not detected in the animal isolate genomes investigated. The sek and seq genes were universally present in the three outgroup isolate genomes. The MSSA precursor isolates harboured similar virulence genes to the European CC1-MRSA-IV isolates (Table 1).
SCCmec element
An SCCmec type IVa element harbouring the mvaS-SCC gene and a characteristic dcs insertion was typically identified in isolates of the European CC1-MRSA-IV clone. This insertion encodes six hypothetical proteins and is approximately 5350 nt in size [5]. According to in silico microarray mapping analysis, however, three isolates lacked C5QAP8 (part of the dcs insertion) and three other isolates lacked mvaS-SCC. Upon further analysis, the C5QAP8 gene was identified split between contigs, for all three isolates. In the case of mvaS-SCC, no remnants of the gene were identified among the three contig sets and its absence was, therefore, considered representative of an authentic deletion (Fig. S4).
Discussion
Using a combination of maximum-likelihood, time-scaled phylogenetic, genotypic and epidemiological analyses, this study elucidated the evolutionary history of the epidemic CA-MRSA clone known as European CC1-MRSA-IV. Phylogenomic analyses indicated that European CC1-MRSA-IV emerged from an MSSA lineage prevalent in Romania, in approximately 1995. Upon acquiring SCCmec, the clonal population underwent rapid expansion, while spreading to at least 10 European and 2 Middle Eastern countries (Fig. 2a). During this time, it appears that no major temporal or geographical variants of the clone evolved within Europe. In fact, the CC1-MRSA-IV population investigated lacked any significant structure, resembling a star-like phylogeny (Fig. 1). Although the clone was typically identified as ST1-MRSA-IV-t127, variants such as ST4110-MRSA-IV-t174 were also identified (Fig. 1), and additional variants may have been overlooked if they did not fit the inclusion criteria during isolate collection.
The phylogenomic data were in line with the available epidemiological information, previous studies and known human migration patterns. Firstly, the oldest study isolate was recovered in 2006, shortly after a sharp clonal population increase was first observed according to our estimations (Fig. 2c). Secondly, the grouping of the Romanian MSSA isolates at the base of European CC1-MRSA-IV clade in both phylogenies (rather than among the outgroup isolates) was in agreement with previous work determining that European CC1-MRSA-IV may have emerged in South-Eastern Europe [2, 3]. Among other evidence, this previous work took into consideration that (i) the CC1-MSSA precursor lineage is common in North-Eastern Romania, and (ii) European CC1-MRSA-IV was the predominant clone in a hospital in Iaşi, Romania, by 2008, at which point it had been recognized in few other countries [3]. Finally, sharp population increases were observed in approximately 2004 and particularly in 2007/2008 (Fig. 2c), corresponding with periods of increased migration from Eastern Europe and South-Eastern Europe, respectively, into the rest of Europe [28].
Certain characteristics of European CC1-MRSA-IV resemble those associated with the PVL-positive ST80-MRSA-IV clone known as European CA-MRSA. For example, the European CA-MRSA population also appears to lack any significant structure, exhibiting a star-like phylogeny reminiscent of that described here [24]. However, more structure may be revealed in the future as additional European CC1-MRSA-IV isolates are identified for analysis. Furthermore, evidence suggests that European CA-MRSA derived from a single multidrug-resistant progenitor [24]. Likewise, genotypic data from this study suggest that a single SCCmec element was acquired by an ancestral MSSA that had a resistance-gene and virulence-gene profile closely matching that of European CC1-MRSA-IV (Table 1). This is in contrast to the well-studied CA-MRSA clone, USA300, which does not inherently harbour antimicrobial-resistance genes, but has acquired them over time [29]. Worryingly, however, European CC1-MRSA-IV can also acquire additional resistance over time. Indeed, certain study isolates harboured extra antimicrobial-resistance genes, such as those encoding resistance to trimethoprim, mupirocin and quaternary ammonium compounds (Table 1, Figs 1 and S4). Furthermore, a multihospital outbreak of a mupirocin-resistant variant of European CC1-MRSA-IV occurred previously in Ireland [6].
Limitations of this study centre mainly around potential gaps in our isolate collection. Specifically, it is likely that the MSSA progenitor lineage exists also outside of Romania. Therefore, while the phylogenomic evidence indicates that European CC1-MRSA-IV emerged from this MSSA lineage, the country in which this happened cannot be definitively determined. Relevant MRSA isolates may also have been missed. Indeed, there is a general lack of MSSA data for Europe, and a lack of MRSA data for many parts of South-Eastern Europe. Furthermore, just five animal isolates were included, although we have previously speculated that European CC1-MRSA-IV is well-established among Italian livestock [2, 8].
This study demonstrated that European CC1-MRSA-IV spread rapidly throughout Europe in the late 1990s and early 2000s. Ongoing surveillance is essential to minimize its further spread.
Supplementary Data
Funding information
D. C. C. and M. R. E. were supported by the Microbiology Research Unit, Division of Oral Biosciences, Dublin Dental University Hospital, University of Dublin, Trinity College Dublin, Ireland. S. M. and R. E. were supported by the German Federal Ministry of Education and Research, within the framework of a project (ADA; 13GW0456C) on rapid tests for the detection of MRSA. The funders had no role in study design, data collection and interpretation, nor the decision to submit the work for publication.
Acknowledgements
We thank Peter Slickers at Abbott Rapid Diagnostics, Jena, Germany, for performing the in silico microarray analysis. D.C.C. and M.R.E. thank Grainne Brennan and the staff of the Irish National MRSA Reference Laboratory (NMRSARL) for technical assistance, and the hospitals in which Irish isolates were recovered for referring their isolates to the NMRSARL.
Author contributions
M. R. E., conceptualization, methodology, formal analysis, writing original draft preparation. E. J. S., methodology, formal analysis, writing original draft preparation. S. M., conceptualization, methodology, formal analysis, writing original draft preparation. J. A. S. C., formal analysis. A. S., W. S.-B., T. V., O. S. D., I. L., M. B., A. L., C. P. A., U. W., M. A.-P., A. B. data curation, reviewing and editing of manuscript. S. D., formal analysis, reviewing and editing of manuscript. M. D. B., data curation, formal analysis, writing original draft preparation. R. E., conceptualization, methodology, formal analysis, writing original draft preparation, supervision. D. C. C., conceptualization, methodology, formal analysis, writing original draft preparation, supervision, funding.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CA, community-associated; CC, clonal complex; HPD, highest posterior density; MRCA, most recent common ancestor; MRSA, meticillin-resistant Staphylococcus aureus; MSSA, meticillin-susceptible Staphylococcus aureus; NCBI, National Center for Biotechnology Information; PVL, Panton–Valentine leukocidin; SCCmec, staphylococcal cassette chromosome mec; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary figures and one dataset are available with the online version of this article.
References
- 1.Rolo J, Miragaia M, Turlej-Rogacka A, Empel J, Bouchami O, et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earls MR, Shore AC, Brennan GI, Simbeck A, Schneider-Brachert W, et al. A novel multidrug-resistant PVL-negative CC1-MRSA-IV clone emerging in Ireland and Germany likely originated in South-Eastern Europe. Infect Genet Evol. 2019;69:117–126. doi: 10.1016/j.meegid.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 3.Monecke S, Müller E, Dorneanu OS, Vremerǎ T, Ehricht R. Molecular typing of MRSA and of clinical Staphylococcus aureus isolates from Iaşi, Romania. PLoS One. 2014;9:e97833. doi: 10.1371/journal.pone.0097833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll D, Kehoe MA, Cavanagh D, Coleman DC. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages φ13 and φ42. Mol Microbiol. 1995;16:877–893. doi: 10.1111/j.1365-2958.1995.tb02315.x. [DOI] [PubMed] [Google Scholar]
- 5.Monecke S, König E, Earls MR, Leitner E, Müller E, et al. An epidemic CC1-MRSA-IV clone yields false-negative test results in molecular MRSA identification assays: a note of caution, Austria, Germany, Ireland, 2020. Euro Surveill. 2020;25:pii=2000929. doi: 10.2807/1560-7917.ES.2020.25.25.2000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earls MR, Kinnevey PM, Brennan GI, Lazaris A, Skally M, et al. The recent emergence in hospitals of multidrug-resistant community-associated sequence type 1 and spa type t127 methicillin-resistant Staphylococcus aureus investigated by whole-genome sequencing: implications for screening. PLoS One. 2017;12:e0175542. doi: 10.1371/journal.pone.0175542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manara S, Pasolli E, Dolce D, Ravenni N, Campana S, et al. Whole-genome epidemiology, characterisation, and phylogenetic reconstruction of Staphylococcus aureus strains in a paediatric hospital. Genome Med. 2018;10:82. doi: 10.1186/s13073-018-0593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alba P, Feltrin F, Cordaro G, Porrero MC, Kraushaar B, et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: high genetic relatedness of isolates from Italian cattle herds and humans. PLoS One. 2015;10:e0137143. doi: 10.1371/journal.pone.0137143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheithauer S, Trepels-Kottek S, Häfner H, Keller D, Ittel T, et al. Healthcare worker-related MRSA cluster in a German neonatology level III ICU: a true European story. Int J Hyg Environ Health. 2014;217:307–311. doi: 10.1016/j.ijheh.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Earls MR, Coleman DC, Brennan GI, Fleming T, Monecke S, et al. Intra-hospital, inter-hospital and intercontinental spread of ST78 MRSA from two neonatal intensive care unit outbreaks established using whole-genome sequencing. Front Microbiol. 2018;9:1485. doi: 10.3389/fmicb.2018.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monecke S, Slickers P, Ehricht R. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol. 2008;53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 12.Loncaric I, Stalder GL, Mehinagic K, Rosengarten R, Hoelzl F, et al. Comparison of ESBL and AmpC producing enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA) isolated from migratory and resident population of rooks (Corvus frugilegus) in Austria. PLoS One. 2013;8:12–e84048. doi: 10.1371/journal.pone.0084048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 14.Holden MTG, Feil EJ, Lindsay JA, Peacock SJ, Day NPJ, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monecke S, Slickers P, Gawlik D, Müller E, Reissig A, et al. Molecular typing of ST239-MRSA-III from diverse geographic locations and the evolution of the SCCmec III element during its intercontinental spread. Front Microbiol. 2018;9:1436. doi: 10.3389/fmicb.2018.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monecke S, Jatzwauk L, Müller E, Nitschke H, Pfohl K, et al. Diversity of SCCmec elements in Staphylococcus aureus as observed in South-Eastern Germany. PLoS One. 2016;11:e0162654. doi: 10.1371/journal.pone.0162654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duchêne S, Duchêne D, Holmes EC, Ho SYW. The performance of the date-randomization test in phylogenetic analyses of time-structured virus data. Mol Biol Evol. 2015;32:1895–1906. doi: 10.1093/molbev/msv056. [DOI] [PubMed] [Google Scholar]
- 22.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, et al. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus . mBio. 2014;5:e01044–14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinig EJ, Duchene S, Robinson DA, Monecke S, Yokoyama M, et al. Evolution and global transmission of a multidrug-resistant, community-associated methicillin-resistant Staphylococcus aureus lineage from the Indian subcontinent. mBio. 2019;10:e01105–19. doi: 10.1128/mBio.01105-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duchêne S, Holt KE, Weill F-X, Le Hello S, Hawkey J, et al. Genome-scale rates of evolutionary change in bacteria. Microb Genom. 2016;2:e000094. doi: 10.1099/mgen.0.000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hal SJ, Steinig EJ, Andersson P, Holden MTG, Harris SR, et al. Global scale dissemination of ST93: a divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote northern Australia. Front Microbiol. 2018;9:1453. doi: 10.3389/fmicb.2018.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahanec M, Zimmerman KF. Migration in an enlarged EU: a challenging solution? Econ Pap. 2009;363:16–19. [Google Scholar]
- 29.Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus . Infect Genet Evol. 2014;21:563–574. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.