Abstract
Pathogens of the Mycobacterium tuberculosis complex (MTBC) are considered to be monomorphic, with little gene content variation between strains. Nevertheless, several genotypic and phenotypic factors separate strains of the different MTBC lineages (L), especially L5 and L6 (traditionally termed Mycobacterium africanum) strains, from each other. However, this genome variability and gene content, especially of L5 strains, has not been fully explored and may be important for pathobiology and current approaches for genomic analysis of MTBC strains, including transmission studies. By comparing the genomes of 355 L5 clinical strains (including 3 complete genomes and 352 Illumina whole-genome sequenced isolates) to each other and to H37Rv, we identified multiple genes that were differentially present or absent between H37Rv and L5 strains. Additionally, considerable gene content variability was found across L5 strains, including a split in the L5.3 sub-lineage into L5.3.1 and L5.3.2. These gene content differences had a small knock-on effect on transmission cluster estimation, with clustering rates influenced by the selected reference genome, and with potential overestimation of recent transmission when using H37Rv as the reference genome. We conclude that full capture of the gene diversity, especially high-resolution outbreak analysis, requires a variation of the single H37Rv-centric reference genome mapping approach currently used in most whole-genome sequencing data analysis pipelines. Moreover, the high within-lineage gene content variability suggests that the pan-genome of M. tuberculosis is at least several kilobases larger than previously thought, implying that a concatenated or reference-free genome assembly (de novo) approach may be needed for particular questions.
Keywords: genomic diversity, gene presence/absence, H37Rv, lineage 5, L5.3.2, M. africanum, reference genome, within-lineage variability
Data Summary
Sequence data for the Illumina dataset are available at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ega/) under the study accession numbers PRJEB9003, PRJEB38656, PRJEB4884, PRJEB38317, PRJEB31139, PRJEB6273 and PRJEB31144. Individual run accession numbers are indicated in Table S8 (available in the online version of this article). PacBio raw reads for the L5 Benin genome are available on the ENA accession SAME3170744. The assembled L5 Benin genome is available at the National Center for Biotechnology Information (NCBI) with accession PRJNA641267. The two other complete genomes of L5 strains from Gambia (PcbL5Gam, WBB1453_11-00429-1 [1]) and Nigeria (PcbL5Nig, WBB1454_IB091-1 [1]) can be found at http://pathogenseq.lshtm.ac.uk/#tuberculosis (Tuberculosis section/Karonga Methylation study). To ensure that the naming conventions for the genes in the genomes of the three L5 strains can be followed, we have uploaded these annotated files to figshare [2].
The custom Python scripts used in this analysis can be found at https://github.com/conmeehan/pathophy.
Impact Statement.
The Mycobacterium tuberculosis complex (MTBC) consists of nine human-associated lineages. Although many of these have been described for decades, little is known about the gene content variation both between and within strains of these lineages. This is most pronounced for strains of lineage 5 (L5), once part of the Mycobacterium africanum species. We compared the genomes of over 350 clinical L5 strains, the largest dataset gathered to date, to each other and the H37Rv reference strain to look for gene content variation and the potential impact this would have on clinical use of genome sequence data. We found that multiple genes are differentially present or absent between H37Rv and L5 strains, and that there is high within-L5 gene content variability, resulting in the split of the sub-lineage L5.3 into L5.3.1 and L5.3.2. We quantified the potential impact of this gene content difference on transmission clustering estimation. We found that the current H37Rv-centric approach widely used in MTBC epidemiology would overestimate the clustering rate of L5 strains since it misses single-nucleotide polymorphisms present in L5-only genes. Thus, for high-resolution outbreak analysis, MTBC epidemiological studies may need to move away from the H37Rv-centric approach, especially when looking at transmission in countries where L5 is prominent.
Introduction
Tuberculosis (TB) is caused by pathogenic bacteria of the Mycobacterium tuberculosis complex (MTBC) that consists of strains of nine human-adapted lineages and several animal-adapted lineages [3–6]. This group is highly clonal with no detected horizontal gene transfer [7, 8]. Strains of particular lineages are primarily defined by large sequence polymorphisms (LSPs, the presence or deletion of genomic regions) such as the TbD1 region (MTBC-specific deletion 1) [9], other regions of difference (RDs) [10, 7, 11–13] and signature single-nucleotide polymorphisms (SNPs) [14]. RDs are particular long genomic regions deleted in some groups of MTBC strains but present in others; thus many MTBC lineages can be classified in groups using that LSP [10, 12, 13, 15]. Broadly, the lineages of the MTBC occur within three major clades: (1) L1–L4 and L7 form one group, (2) L5, L6, L9 and the animal lineages form another and (3) L8 sits within its own clade [4–6], based on the presence/absence of specific RDs, especially TbD1 [12, 16, 17]. L5 and L6, also called M. tuberculosis var. africanum (or historically M. africanum West-african 1 and 2, respectively) [18, 19] are primarily restricted to West Africa, where they cause up to 40 % of human TB [20, 21]. The reasons for the geographical restriction of L5 and L6 remain unclear, although adaptation to particular human subpopulations has been suggested [10, 22–24].
Several phenotypic and genotypic features separate L5 and L6 strains from strains of the other human-adapted lineages. Some TB diagnostics have a lower performance for L5 and L6 strains, compared to strains of other MTBC lineages [25, 26] and these lineages are less likely to grow in culture [27], with dysgonic appearance on solid medium [18, 26, 28, 29], despite the inclusion of pyruvate-supplemented medium [21, 27]. Mutations in genes essential for growth in culture were identified for L6 strains [30], yet for L5 strains the reasons for the difficulty in growth remain unclear. L6 strains and those of the closely related animal strains/lineages such as Mycobacterium bovis are reported to be less virulent in humans than those from other human-adapted MTBC lineages in population-based studies [20, 31, 32] and in genome studies of mutations in virulence regulation genes/systems [33, 34]. Infection with an L6 strain progresses slowly to TB disease, and is associated with impaired immunity in some settings but not all (e.g. HIV infection [20]). In contrast, although some authors have studied the genomics of L5 strains [35–37], protein secretion and in vivo immunogenicity [35], much remains to be learned on the genomics, virulence and disease progression of L5 strains.
Whole-genome sequencing (WGS) based on next-generation sequencing (NGS) of MTBC strains often involves data analysis by mapping of short sequence reads of the strains to a complete reference genome [38], usually H37Rv [39, 40] or a reconstructed ancestor with the same gene content as H37Rv [22, 41, 42]. The resulting SNPs are then used for drug resistance determination, subtyping and transmission analyses [38]. However, since H37Rv is a L4 strain, it might not be representative for the genome of strains of other MTBC lineages. Thus, if H37Rv is used as basis for genome analysis, several genes or larger genomic regions may be missed, resulting in an underestimation of genome diversity among the strains of other lineages that may, however, provide additional information, e.g. for transmission analysis.
The members of the MTBC evolved from an environmental organism to an obligate pathogen through genome reduction and acquisition of new genes [43]. In addition, it is known that some differences in gene content exist between strains of different lineages [10, 38, 44–48]. Furthermore, it was reported that genes had a higher genetic diversity among L6 strains compared to L5 ones [36], but the difference in gene content between strains of these two lineages was not investigated, but had been alluded to in an earlier abstract publication [37]. Further, little is known about the gene differences between genomes of L4 and L5 strains and the potential limitations this may impose for in-depth analysis of genome studies of L5 strains (e.g. sub-lineage detection and transmission tracking). Similarly, little is known about within-lineage diversity in terms of gene content.
To address these questions, we assessed the gene content diversity of L5 strains in the context of WGS and reference selection. To this end, we analysed 358 genomes of L5 strains, including 3 complete genomes, and compared them to H37Rv and strains of closely related lineages [L6 and M. tuberculosis var. bovis (hereafter called M. bovis) [19]. Our main focus was to determine particularities in the genomes of L5 strains compared to the MTBC strain type (H37Rv, a L4) and strains of MTBC lineages phylogenetically closely related to L5 (L6 and M. bovis); deduce hypotheses for L5 phenotype/biology; and define the level of within-lineage gene content differences among strains of this lineage of the MTBC.
Methods
Genomes
Complete genomes (PacBio-sequenced, complete long reads)
Three complete genomes from L5 clinical isolates, all sequenced with the PacBio SMRT technology, were analysed. One genome was from a Benin isolate [PcbL5Ben; sequenced in this study, National Center for Biotechnology Information (NCBI) accession PRJNA641267], one from an isolate from The Gambia (PcbL5Gam, WBB1453_11-00429-1) [1] and one from Nigeria (PcbL5Nig, WBB1454_IB091-1) [1]. These genomes also represent disparate parts of L5’s diversity, representing a broad range of L5 sub-lineages, as recently defined [6]. The previously published reference/complete genomes H37Rv (L4) (NC_000962.3) [39, 40, 49], L6 (GM041182, GenBank accession: FR878060.1, GCF_0001593225.1_ASM159322v1) [50] and M. bovis (AF2122/97, accession: LT708304.1) [51] were also included.
Whole genomic DNA extraction
Genomic DNA extraction was performed on growth from fresh Löwenstein–Jensen slants using the semi-automated Maxwell 16 Cell DNA purification kit in the Maxwell 16 machine, or from the late exponential phase of growth in 7H9 medium, using the CTAB method [6, 52].
Assembly and annotation of the complete (PacBio-sequenced) genomes
The Benin PacBio-sequenced genome was assembled using HGAP [53] and Quiver [53] and checked for sufficient quality and coverage (10 000 bp sliding window coverage was always above 65× and average coverage across entire genome was 107×). The other two PacBio-sequenced genomes were already assembled as described previously [1]. The genome sequences of all three complete (PacBio-sequenced) genomes were annotated using Prokka-1.12 [54] based on the reference genome H37Rv annotation.
Gene presence–absence analysis
Gene content differences were assessed with an all-vs-all blastn approach, using blast+ version 2.8.1 [55, 56]. For a specific genome–genome comparison, the following procedure was used: the gene sequences of genome 1 (ffn file; coding regions only) were compared to those of genome 2 using an e-value cut-off of 1e−05 and a minimum similarity of 70 % to look for any homology for each gene. Those genes found in genome 1 and not in genome 2 were then compared to the complete (PacBio-sequenced) genome (fna file; coding and non-coding regions) of genome 2 to look for pseudogenes (herein qualified as ‘suspected pseudogenes’) using blastn with the same cut-offs. These suspected pseudogenes were then confirmed using tblastn of the genome 1 protein sequences (faa file) compared to the complete (PacBio-sequenced) genome (fna file) of genome 2. This procedure was used to compare the complete (PacBio-sequenced) genomes of all three L5 strains to each other as well as each to H37Rv. Those genes present or absent in H37Rv (or pseudogenes in either) were compared in a similar manner to the L6 and M. bovis reference strains to determine whether these genes/pseudogenes are L5-specific (i.e. present in L5, but not in H37Rv, L6 and M. bovis).
The ffn files of H37Rv, the complete (PacBio-sequenced) genomes of the three L5 strains and the complete genome of the L6 reference strain were also aligned using progressiveMauve (v20150226) [57] to identify rearrangements and examine synteny.
L5 Illumina-sequenced (short reads) genomes
In total, whole genomes from 355 L5 strains from various countries sequenced on the Illumina platform were included in the study. These genomes were derived from a larger study on the genetic diversity of L5 and L6 [6]. After reducing isolate redundancy (i.e. 1 representative retained for those that were extremely closely related), genomes from 205 L5 strains formed a non-redundant dataset. These 205 strains (genomes) originated from West, South, East and Central Africa (Table S8), but primarily (n=155) from two regions within West Africa: the western part of West Africa (including The Gambia, Sierra Leone, Ivory Coast, Liberia, Guinea and Mali) and the eastern part of West Africa (including Ghana, Benin and Nigeria) [6]. In general, L5 and L6 are geographically restricted to West and Central Africa, making this genome selection from L5 strains representative of most of the geographical regions affected.
Mapping of L5 Illumina reads to H37Rv and L5 complete (PacBio-sequenced) genomes
Raw reads (fastq files) of the 205 non-redundant Illumina-sequenced genomes were mapped respectively to H37Rv and each of the PacBio-sequenced genomes of the 3 L5 strains using the MTBseq pipeline [58]. The depth mapping coverage of the samples in the clinical Illumina-sequenced genomes against the reference genomes (percentage read mapping, unambiguous coverage mean) was compared between the PacBio-sequenced genomes of the three L5 strains and between the PacBio-sequenced genome of each L5 strain and H37Rv. Mapping statistics parameters such as percentage unambiguous total base, uncovered bases, SNPs, deletions, insertions, substitutions and percentage genes mapped to reference were also compared using the PacBio-sequenced genome of each L5 strain or H37Rv as a reference.
Checking unique versus missing genes in Illumina-sequenced genomes of L5 strains and SNPs in those confirmed as L5-specific
The genes found to be present or absent in L5 based on genome comparisons of the PacBio-sequenced genomes of the three L5 strains with H37Rv were checked for their expected presence or absence in the Illumina-sequenced genomes of L5 strains. Using the position tables produced by the MTBseq pipeline, a gene was considered absent if 95 % of its position in the genome had fewer than eight reads covering them. From these data, a gene presence/absence matrix was generated for Illumina-sequenced genomes of L5 strains mapped to H37Rv and each of the three complete (PacBio-sequenced) genomes of L5 strains. Genes found to be L5-specific (present in complete PacBio- and Illumina-sequenced genomes) were also checked for SNPs in these genes using each of the complete (PacBio-sequenced) genomes of the three L5 strains as a reference. The script used for undertaking this analysis can be found at https://github.com/conmeehan/pathophy.
Calculating the effect of reference genome selection on pairwise SNP distances
Transmission analysis of MTBC strains often involves clustering strains together based on specific SNP cut-offs [38, 59, 60]. We assessed whether the selection of the reference genome (H37Rv, PcbL5Ben, PcbL5Gam or PcbL5Nig) changed the clustering rate of L5 strains. For this we used the non-redundant set of L5 genomes from a previous study on L5 diversity [6], which included the 205 detailed above and a further 145 strains that were closely related to at least 1 of these 205 strains. Additionally, the TB-Profiler online SRA search tool (https://tbdr.lshtm.ac.uk/sra) was used to identify a further 5 isolates that were not in this dataset, resulting in 355 strains being included in this clustering study.
SNP alignments of the Illumina-sequenced genomes were created by first mapping to each of the four reference genomes (H37Rv and three PcbL5). The Amend function of MTBseq does this automatically for H37Rv, including masking of repetitive regions, accounting for 10 % of the genome [40], and exclusion of columns with 95 % ambiguous calls. To undertake this for the reference genomes from the three L5 strains, repetitive regions were first determined from the annotations of the genes. All genes whose description contained one of the following words was excluded: integrase, PE family, PE-PGRS, phage, transposase. SNP alignments were then created using MTBseq as done for H37Rv. Pairwise distance matrices, one for each alignment based on each reference genome, were created using a Python script that can be found at https://github.com/conmeehan/pathophy. Loose transmission clusters were created from these matrices at a cut-off of 1, 5 and 12 SNPs, as described previously [59]. Clustering rates and the presence of a strain in a transmission cluster was then compared across the four reference genome mapping approaches.
Determination of putative function of genes differentially present or absent in the complete (PacBio-sequenced) genomes of L5 strains
To find the (putative) function of genes present or absent only in L5 strains, and genes present in L5 strains but pseudogenes in H37Rv/L6/M. bovis or vice versa, the fasta sequences of the genes were searched against the NCBI’s NR database using blastx2.8.1+ [61] as well as the Tuberculist database (http://tuberculist.epfl.ch), Mycobrowser (https://mycobrowser.epfl.ch/) [62] and the literature. Furthermore, the gene function group/class was found using the COG database [63] and Mycobrowser.
Determination of the sub-lineage of the L5 strains
The sub-lineage of the strains was determined by looking for sub-lineage-defining SNPs in the genomes of L5 strains as previously described [6, 35]. The sub-lineage-defining SNPs were searched for in the SNP data generated using the MTBseq pipeline with H37Rv as the reference genome.
Results
Comparative analysis of the complete (PacBio-sequenced) genomes of L5 and other lineages strains reveals differences in gene content
The number of genes, including paralogues, in the complete L5 (PacBio-sequenced) genomes of the three L5 strains was: 4189 in the PcbL5Ben genome, 4162 in PcbL5Gam and 4134 in PcbL5Nig versus 4126 in H37Rv, 4126 in the reference L6 genome (GM041182) and 4059 in the reference M. bovis genome (AF2122/97). Hence, the maximum gene count difference was 55 genes among the 3 L5 strains from this study (4189–4134=55, including copies of genes with multiple copies), 63 genes among the human-adapted lineages (H37Rv, L5, L6; 4189–4126=63) and 130 genes among the human- and animal-adapted lineages (H37Rv, L5, L6 and M. bovis; 4189–4059=130).
The size of the complete genomes of L5 strains was respectively 4 438 262 bp for PcbL5Ben, 4 424 447 for PcbL5Gam and 4 417 534 for PcbL5Nig (data in Figshare [2]). The size was 4 411 532 bp for H37Rv and 4 493 502 for the M. bovis reference genome (data from annotation with Prokka).
The visualization of the structure of the genomes showed that all three complete genomes of all L5 strains had a region that was absent in H37Rv. Furthermore, the PacBio-sequenced genome of the Nigerian L5 strain (PcbL5Nig) was missing an additional region (herein called L5Nig-Del) that was present in the genomes of the Benin and the Gambian L5 strains and also present in H37Rv.
The genome of the Benin strain contained a syntenic block of three genes (1148, 1104, 2840 bp) that were present in neither the genome of the Nigerian strain nor the one of the Gambian strain (Tables 1 and 2). These increased the pangenome of L5 by at least 5092 bp. The Benin and Gambian strains each contained 33 genes that were not present in the Nigerian strain (Table 2 and S1). Interestingly, 32 of these genes were also present in H37Rv (Table 2 and S1). These 32 genes were thus missing in the Nigerian L5 strain exclusively, meaning that its ancestor has genes with a subsequent loss in the Nigerian L5 strain. Those 32 genes – absent in the Nigerian L5 strain only – were sequentially contiguous and formed 3 blocks of 19, 11 and 2 genes (Table S1). We found that 30 of those genes, the blocks of 19 and 11, were separated by 1 gene and represented the L5Nig-Del region (mentioned in the paragraph above, Table 3). Additionally, 11 genes were shared by the 3 PacBio-sequenced genomes of L5 strains, which were absent in H37Rv (Table S2), while 9 genes present in H37Rv were not present in any of the 3 complete (PacBio-sequenced) genomes of L5 strains (Table S3). Two (Rv2073c, Rv2074) of those nine genes were only present in H37Rv but absent in the complete genomes of L5, L6 and M. bovis strains. Note that for the gene presence/absence analysis, the genes are often referred to by the Rv designation but are actually putative orthologues of those H37Rv genes.
Table 1.
Presence in Illumina-sequenced genomes from 202 L5 strains of genes detected in only 1 of the complete genomes of the 3 L5 strains from Benin, The Gambia and Nigeria
|
Gene |
Co-ordinates in the specified genome |
Functional group |
Present in L5 Illumina-sequenced genomes % (n=202) |
|
|---|---|---|---|---|
|
PcbL5Ben |
PcbL5_01893 |
2004773–2005918 |
Cell wall and cell processes |
77.7 (157) |
|
|
PcbL5_01894 |
2006144–2007247 |
Intermediary metabolism and respiration |
77.7 (157) |
|
|
PcbL5_01895 |
2007448–2010285 |
Cell wall and cell processes |
77.7 (157) |
Table 2.
Gene content difference between the M. tuberculosis H37Rv (L4) genome and the complete (PacBio-sequenced) genomes of three L5 strains from Benin, The Gambia and Nigeria
|
Present in |
|||||
|---|---|---|---|---|---|
|
|
PcbL5Ben |
PcbL5Gam |
PcbL5Nig |
H37Rv |
|
|
Absent in |
PcbL5Ben |
9† |
|||
|
PcbL5Gam |
2+3* |
2+9† |
|||
|
PcbL5Nig |
34+3* |
34 |
32+9† |
||
|
H37Rv |
10‡+3* |
10‡ |
10‡ |
||
|
|
|
4189 |
4162 |
4134 |
4126 |
*, includes three genes only present in PcbL5Ben.
†, includes nine genes only present in H37Rv.
‡, includes 10 genes shared by the PcbL5 (Benin, The Gambia, Nigeria) and absent in H37Rv.
Table 3.
Genes in the L5.3.2-Del region (PcbL5Nig-Del) and their function category. None of the 30 genes is an essential gene (Mycobrowser). The 30 genes formed 2 regions: Rv1493 through Rv1509 (L5.3.2-Del region 1) and Rv1511 through Rv1521 (L5.3.2-Del region 2), separated by the gene Rv1510, which is present in the L5.3.2 isolate. All genes in the table are absent from all L5.3.2 genomes (both PacBio- and Illumina-sequenced), except those marked with an asterisk (*), which are present in the PacBio-sequenced genome (PcbL5Nig) but absent in all six Illumina-sequenced genomes, and the one marked with a hash (#) (Rv1492), which is a gene present in all L5.3.2 strains and flanking the L5.3.2-specific deletion
|
Gene name |
Size |
Co-ordinates in H37Rv |
Functional category |
Present in L5 Illumina-sequenced genomes % (n=202) |
|---|---|---|---|---|
|
Rv1492# (mutA) |
1848 bp |
1 682 157–1 684 004 |
Lipid metabolism |
100 (202) |
|
Rv1493 (mutB) |
2253 bp |
1 684 005–1 686 257 |
Lipid metabolism |
97 (196) |
|
Rv1494 (mazE4) |
303 bp |
1 686 271–1 686 573 |
Virulence, detoxification, adaptation |
97 (196) |
|
Rv1495 (mazF4) |
318 bp |
1 686 570–1 686 887 |
Virulence, detoxification, adaptation |
97 (196) |
|
Rv1496 |
1005 bp |
1 686 884–1 687 888 |
Cell wall and cell processes |
97 (196) |
|
Rv1497 (lipL) |
1290 bp |
1 687 941–1 689 230 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1498c |
618 bp |
1 6893 03–1 689 920 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1498A |
213 bp |
1 690 134–1 690 346 |
Conserved hypothetical protein |
97 (196) |
|
Rv1499 |
399 bp |
16 900 407–1 690 805 |
Conserved hypothetical protein |
97 (196) |
|
Rv1500 |
1029 bp |
1 690 850–1 691 878 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1501 |
822 bp |
1 691 890–1 692 711 |
Conserved hypothetical protein |
97 (196) |
|
Rv1502 |
900 bp |
1 692 924–1 693 823 |
Unknown |
97 (196) |
|
Rv1503c |
549 bp |
1 693 996–1 694 544 |
Conserved hypothetical protein |
97 (196) |
|
Rv1504c |
600 bp |
1 694 545–1 695 144 |
Conserved hypothetical protein |
97 (196) |
|
Rv1505c |
666 bp |
1 695 281–1 695 946 |
Conserved hypotheticals |
97 (196) |
|
Rv1506c |
501 bp |
1 695 943–1 696 443 |
Unknown |
97 (196) |
|
Rv1507A |
504 bp |
1 697 356–1 697 859 |
Unknown |
97 (196) |
|
Rv1507c |
696 bp |
1 696 727–1 697 422 |
Conserved hypotheticals |
97 (196) |
|
Rv1508c |
1800 bp |
1 698 095–1 699 894 |
Cell wall and cell processes |
97 (196) |
|
Rv1508A |
636 bp |
1 699 866–1 700 228 |
Conserved hypotheticals |
97 (196) |
|
Rv1509 |
882 bp |
1 700 212–1 701 093 |
Unknown |
97 (196) |
|
Rv1510 * |
1299 bp |
1 701 295–1 702 593 |
Cell wall and cell processes |
97 (196) |
|
Rv1511 (gmdA) |
1023 bp |
1 703 074–1 704 096 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1512 (epiA) |
969 bp |
1 704 093–1 705 061 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1513 |
732 bp |
1 705 058–1 705 789 |
Conserved hypothetical protein |
97 (196) |
|
Rv1514c |
789 bp |
1 705 807–1 706 595 |
Conserved hypothetical protein |
97 (196) |
|
Rv1515c |
897 bp |
1 706 630–1 707 526 |
Conserved hypothetical protein |
97 (196) |
|
Rv1516c |
1011 bp |
1 707 529–1 708 539 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1517 |
765 bp |
1 708 871–1 709 635 |
Cell wall and cell processes |
97 (196) |
|
Rv1518 |
960 bp |
1 709 644–1 710 603 |
Conserved hypothetical protein |
97 (196) |
|
Rv1519 |
270 bp |
1 710 733–1 711 002 |
Conserved hypothetical protein |
97 (196) |
|
Rv1520 |
1041 bp |
1 711 028–1 712 068 |
Intermediary metabolism and respiration |
97 (196) |
|
Rv1521 (fadD25) |
1752 bp |
1 712 302–1 714 053 |
Lipid metabolism |
97 (196) |
|
Rv1522c* (mmpL12) |
3441 bp |
1 714 172–1 717 612 |
Cell wall and cell processes |
97 (196) |
Six of the genes shared by the three complete (PacBio-sequenced) genomes of L5 strains were confirmed pseudogenes in H37Rv (Table S4). Three genes present in H37Rv were confirmed to be pseudogenes in the three complete (PacBio-sequenced) genomes of the L5 strains (Table S5).
Gene presence/absence and related SNPs in lineage-specific genes in a wider set of clinical strains
The mapping estimates of the Illumina-sequenced genomes are presented in Table 4. Three (ERR502505, ERR751302, ERR1215478) of the 205 Illumina-sequenced genomes were identified as mixed infection strains based on their MTBseq output, and thus excluded from the analysis. Mapping quality and coverage against a complete L5 reference was superior to the H37Rv reference approach (Table 3, Fig. S1), as expected. Using the complete (PacBio-sequenced) genomes from Benin and The Gambia strains, yielded similar mapping estimates that were better than those of the genome of the Nigerian L5 strain, likely due to the large deletion in this genome. The genes specific to PcbL5Ben [3, region PcbL5Ben_1893 through PcbL5Ben_1895; none specific to PcbL5Gam (0), PcbL5Nig (0)] were each found in 77.7 % (157/202, Table 1) of the 202 Illumina-sequenced genomes of L5 strains.
Table 4.
Mapping of Illumina-sequenced genomes of the 202 L5 strains to the M. tuberculosis H37Rv (L4) genome and complete genomes of 3 L5 strains from Benin, The Gambia and Nigeria (mapping statistics/estimates). The best mapping results (numbers) are written in bold. When the best mapping result has been obtained for PcbL5Nig as the reference, the next best result is also written in bold (as PcbL5Nig compared to the other 2 PcbL5 genomes missed a 30-gene region)
|
H37Rv |
PcbL5Ben |
PcbL5Gam |
PcbL5Nig |
|
|---|---|---|---|---|
|
Reads |
||||
|
Mean of percentage L5 reads mapped to |
96.9 |
97.5 |
97.3 |
96.5 |
|
Mean of unambiguous coverage mean |
122.3 |
123.2 |
123.3 |
123.0 |
|
Bases |
||||
|
Mean of percentage unambiguous total bases |
98.0 |
98.8 |
98.9 |
98.2 |
|
Mean uncovered |
30 599.0 |
18 916.7 |
14 766.4 |
35 340.4 |
|
Mean SNP |
2209.7 |
529.5 |
513.0 |
503.3 |
|
Mean deletions |
374.1 |
96.5 |
96.4 |
97.9 |
|
Mean insertions |
239.5 |
77.0 |
107.6 |
60.8 |
|
Mean substitutions (including stop codons) |
1193 |
0 |
0 |
0 |
|
Genes |
||||
|
Mean of percentage gene mapped (presence) |
99.59 |
99.71 |
99.79 |
99.76 |
|
L5 illumina having all the ref. genome genes, % ( n =202) |
0 |
2 (4) |
7.4 (15) |
0 |
|
Mean gene count difference between L5 Illumina-sequenced genomes (gene count per Illumina-sequenced genome minus minimum gene count) |
30.3 |
38.2 |
34.4 |
32.8 |
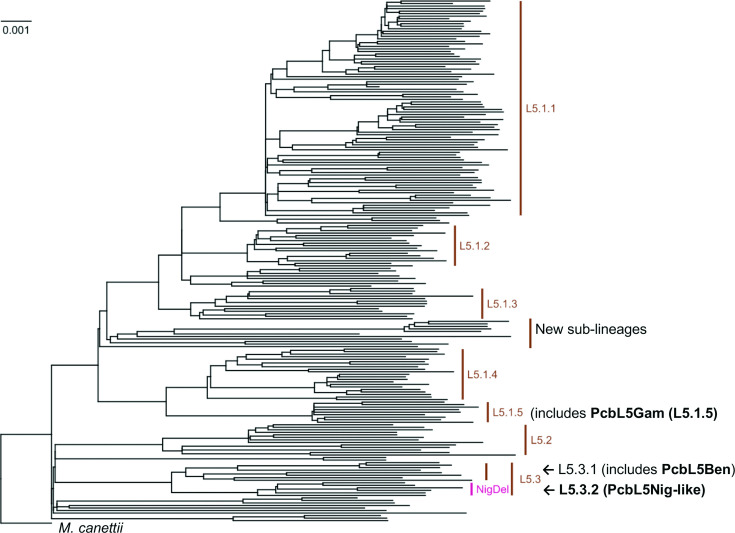
Interestingly, 3 % of the Illumina-sequenced genomes of L5 strains (6/202) had similar patterns of large gene loss like the Nigerian complete (PacBio-sequenced) genome, as they missed 30 of the 32 genes present in the genomes of the Benin and Gambian strains and H37Rv. These six PcbL5Nig-like Illumina-sequenced genomes of L5 strains formed a monophyletic group within the L5 clade (Fig. 1), suggesting a single loss of these gene clusters, although those six L5 strains genomes originated from several different countries, including Benin, Ghana and Nigeria. The 2 blocks of 19 (15 984 bp, Rv1493 through Rv1509) and 11 genes (10 209 bp, Rv1511 through Rv1521), separated by 1 gene (3441 bp, Rv1510), amounted in total to 26 193 bp. The two blocks contain genes whose annotations include mutB, mazE4, mazF4 and others with various putative functions (Table 3, Table S6); none of them were essential genes (Mycobrowser).
Fig. 1.
Phylogenetic tree showing the Illumina-sequenced genomes of the six L5 strains (L5.3.2) similar to the complete (PacBio-sequenced) genome of the Nigerian L5 strain (PcbL5Nig) and the position of the other two PacBio-sequenced L5 genomes (PcbL5Ben and PcbL5Gam). NigDel=L5Nig-Del=L5.3.2-Del=region of 30 genes (2 blocks of 19 and 11 genes: Rv1493 through Rv1509 and Rv1511 through Rv1521) missing in L5.3.2 strains but present in all other L5 strains (L5.1, L5.2, L5.3.1 and new sub-lineages).
Four of the 11 genes present in the 3 complete (PacBio-sequenced) genomes of the L5 strains but absent in H37Rv were found in all 202 Illumina-sequenced genomes of the L5 strains (Table 5), while the others were found in variable amounts [80.2–98 % (162–198)] (Table S2). These four genes include: Mb2048c (of unknown function, belonging to the RvD1 deletion in H37Rv), a PE/PPE gene, a hypothetical protein (possibly an IS256 transposase) and a hypothetical protein possibly related to the CAAX conserved domain (Table 5). Two of these four genes were only present in L5 genomes (a PE/PPE gene and a hypothetical protein, possibly CAAX conserved domain), while the remaining two [Mb2048c (RvD1) and hypothetical protein, possibly IS256 transposase] were found in L6 and M. bovis as well (Tables 5 and S2). Importantly for phylogenetic purposes, SNPs were detected in all 4 genes in 1.5–3.5 % of the 202 Illumina-sequenced genomes of L5 strains (Table S2). Predicted functions for all L5-specific genes are listed in Table S2.
Table 5.
Genes present or absent in all L5 genomes compared to H37Rv. The genes are ordered in the table along with their flanking genes, as in the genome. The RD regions are those reported by Gordon et al. [13]
|
Gene name |
Present in |
Size |
Co-ordinates in H37Rv |
Co-ordinates in PcbL5Ben |
Functional category |
Present in L5 Illumina-sequenced genomes % (n=202) |
Belongs to RD# (no. total of genes forming the RD) |
|---|---|---|---|---|---|---|---|
|
PcbL5Ben_2128 (Rv1976c) |
|
2218844–2219251 |
2244287–2244565 |
Conserved hypothetical protein |
100 (202) |
RD7 |
|
|
PcbL5Ben_2129 |
L5 |
648 bp |
– |
2244695–2245342 |
PE/PPE |
100 (202) |
|
|
PcbL5Ben-2130 |
L5 |
600 bp |
– |
2246122–2246721 |
Hypothetical protein |
100 (202) |
|
|
Rv1977 |
H37Rv, L6, M. bovis |
1047 bp |
2219754–2220800 |
– |
Conserved hypothetical protein |
0 (0) |
RD7 (14 genes) |
|
Rv1978 |
H37Rv, L6, M. bovis |
849 bp |
2220908–2221756 |
– |
Conserved hypothetical protein |
1 (2) |
RD2 |
|
Rv1979c |
H37Rv, L6, M. bovis |
1446 bp |
2221719–2223164 |
– |
Cell wall and cell processes |
0 (0) |
RD2 (10 genes) |
|
PcbL5Ben_2131 (Rv1980 (mpt64)) |
|
2223343–2224029 |
2246867–2247553 |
Cell wall and cell processes |
100 (202) |
RD2 |
|
|
………//……… |
|
|
|
||||
|
PcbL5Ben_2149 (Rv1992 (ctpG)) |
|
2234991–2237306 |
2258516–2259319 |
Cell wall and cell processes |
100 (202) |
|
|
|
Rv1993c |
H37Rv, L6, M. bovis |
273 bp |
2237303–2237575 |
– |
Conserved hypothetical protein |
0 (0) |
|
|
Rv1994c (ctmR) |
H37Rv, L6, M. bovis |
357 bp |
2237628–2237984 |
– |
Regulatory proteins |
0.5 (1) |
|
|
Rv1995 |
H37Rv, L6, M. bovis |
729 bp |
2238141–2238908 |
– |
Conserved hypothetical protein |
0 (0) |
|
|
PcbL5Ben_2150 (Rv1996) |
|
2239004–2239957 |
2259333–2260169 |
Virulence, detoxification, adaptation |
100 (202) |
|
|
|
………//……… |
|
|
|
||||
|
PcbL5Ben_2180 (Rv2023A) |
|
2268268–2268726 |
2289843–2290475 |
Conserved hypothetical protein |
100 (202) |
|
|
|
PcbL5Ben-2181 |
L5, L6, M. bovis |
237 bp |
– |
2290472–2290708 |
Hypothetical protein |
100 (202) |
|
|
PcbL5Ben-2182 (Mb2048c, RvD1) |
L5, L6, M. bovis |
897 bp |
– |
2290915–2291811 |
Unknown function |
100 (202) |
|
|
PcbL5Ben_2183 (Rv2024c) |
|
2268693–2270240 |
2291995–2296815 |
Conserved hypothetical protein |
100 (202) |
|
|
|
………//……… |
|
|
|
||||
|
PcbL5Ben_2231, PcbL5Ben_2232 (Rv2072 (cobL)) |
|
2328974–2330146 |
2355547–2356422, 2356373–2356561 |
Intermediary metabolism and respiration |
100 (202) |
RD9 (4 genes, including Rv2072 truncated) |
|
|
Rv2073c |
H37Rv |
750 bp |
2330214–2330963 |
– |
Intermediary metabolism and respiration |
0 (0) |
RD9 |
|
Rv2074 |
H37Rv |
408 bp |
2330993–2331406 |
Intermediary metabolism and respiration |
0 (0) |
RD9 |
|
|
PcbL5Ben_2233 (Rv2075c) |
|
2331416–2332879 |
2356636–2357388 |
Cell wall and cell processes |
100 (202) |
RD9 (4 genes, including Rv2075c truncated) |
Six (Rv1977, Rv1979c, Rv1993c, Rv1995, Rv2073c, Rv2074) of the 9 genes that were present in H37Rv and absent in the 3 complete (PacBio-sequenced) genomes of L5 strains were absent in all 202 Illumina-sequenced genomes of L5 strains (Table 5 and S3). Rv1977 is a conserved hypothetical, probably a peptidase; Rv1979c is involved in ‘cell wall and cell processes’, probably a permease; Rv1993c and Rv1995 are conserved hypotheticals; and Rv2073c and Rv2074 are involved in ‘intermediary metabolism and respiration’, with Rv2073c probably a dehydrogenase and Rv2074 a pyridoxamine-5-phosphate oxidase (Mycobrowser, Table 5). Four of these genes (Rv1977, Rv1979c, Rv1993c, Rv1995) were absent (did not have orthologues) in L5 strains only (i.e. present in L6 and M. bovis reference genomes), while the other two (Rv2073c and Rv2074) were also absent in L6 and M. bovis (Table 5). The three other genes absent from the complete (PacBio-sequenced) genomes were present in a minority of the L5 Illumina-sequenced genomes (Table S3, Fig. S1). Predicted functions for all nine genes are listed in Table S3.
Sub-lineage of the strains (PacBio and Illumina-sequenced genomes)
The determination of the strains sub-lineage revealed that the PcbL5Ben strain is an L5.3 strain that contains the 30 genes (2 blocks of 19 and 11 genes: Rv1493 through Rv1509 and Rv1511 through Rv1521), whereas the PcbL5Nig strain is also a L5.3 strain, but placed in a different clade (Fig. 1). Based on this new RD (L5Nig-Del), we classified PcbL5Ben as a L5.3.1 strain, the PcbL5Nig as a L5.3.2 strain. The PcbL5Gam strain is a L5.1.5 strain. The distribution of the Illumina-sequenced genomes is outlined in Table S7, amounting to a total of 146 L5.1 strains (72.3 %); 13 (6.4 %) L5.2 strains; 14 L5.3 strains (6.9 %); and 29 (14.4 %) strains of unknown (potentially new) sub-lineages.
Impact of the reference genome selection on genetic distances used for transmission clustering rates
Transmission analysis was undertaken on an expanded set of 352 L5 strains using each of the four reference genomes (H37Rv, PcbL5Ben, PcbL5Gam and PcbL5Nig) for the SNP mapping. The current gold standard is to use the H37Rv genome (NC_000962.3) for calling SNPs and then creating transmission clusters at a specific SNP cut-off differentiating two and more strains. For this dataset, using a 12 SNP cut-off, it was found that 40.6 % (n=144) of strains were within a transmission cluster with at least 1 other isolate (Table 6), for a total of 55 clusters. Using the PcbL5Ben genome instead of the H37Rv genome as the reference reduced the number of clusters to 54, resulting in a clustering rate of 39.7 % at a 12 SNP cut-off (Table 6). A similar reduction in transmission clustering was observed when using the PcbL5Nig genome as a reference but not when using the PcbL5Gam (Table 6). When a more conservative SNP cut-off was used, such as one SNP or five SNPs, the reduction in transmission clustering rates was more pronounced and was observed for all the PcbL5 genomes (Table 6). This demonstrates that even with this small dataset, transmission cluster estimations are affected by the selection of the reference genome, with potential overestimation of recent transmission when using H37Rv as the reference genome.
Table 6.
Comparison of transmission clustering rates based on choice of reference genome. Short-read data from 355 L5 strains were mapped against each of the 4 reference genomes for SNP calling. Distance matrices between all strains were constructed per the reference approach and transmission clusters were defined based on specific SNP cut-offs
|
1 SNP | |||
|---|---|---|---|
|
Reference used |
In transmission cluster |
Percentage of total dataset |
No. of clusters |
|
H37Rv |
100 |
28.17 |
44 |
|
PcbL5Ben |
94 |
26.48 |
43 |
|
PcbL5Gam |
95 |
26.76 |
43 |
|
PcbL5Nig |
95 |
26.76 |
43 |
|
5 SNP | |||
|
Reference used |
In transmission cluster |
Percentage of total dataset |
No. of clusters |
|
H37Rv |
129 |
36.34 |
53 |
|
PcbL5Ben |
124 |
34.93 |
51 |
|
PcbL5Gam |
124 |
34.93 |
51 |
|
PcbL5Nig |
124 |
34.93 |
51 |
|
12 SNP | |||
|
Reference used |
In transmission cluster |
Percentage of total dataset |
No. of clusters |
|
H37Rv |
144 |
40.56 |
55 |
|
PcbL5Ben |
141 |
39.72 |
54 |
|
PcbL5Gam |
144 |
40.56 |
55 |
|
PcbL5Nig |
141 |
39.72 |
54 |
Discussion
Comparison of complete (PacBio-sequenced) genomes and Illumina-sequenced genomes from clinical strains revealed gene content diversity both within L5 strains and between L5 strains and strains of other lineages. This diversity also had an impact on clinical epidemiology analysis of L5 strains, with the choice of reference genome affecting the estimation of recent transmission.
Several genes were found to be absent from the genome of either L5 strains only or also from strains of the closely related lineages L6 and M. bovis. Most of these genes are located within RDs previously described as being absent in one or all of these lineages.
RD9, consisting of Rv2073c and Rv2074 and partial Rv2072 and Rv2075c [13], has previously been reported to be absent in L5, L6 and M. bovis [10, 12, 15]. In this study, we confirmed that Rv2073c and Rv2074 are absent in strains of these lineages. Rv2072 and Rv2075c were present in their truncated form in the genomes of all L5 strains as previously described [13]. Rv2073c is an NAD(P)-dependent oxidoreductase (NCBI) that catalyzes a wide range of reactions and is involved in redox sensor mechanisms [64, 65]. Rv2074 was previously thought to be a pyridoxine (vitamin B6) oxidase, but is now known to be a F420-dependent biliverdin reductase, a cofactor of vitamin B6 synthesis [66, 67]. Vitamin B6 is essential for the survival and virulence of M. tuberculosis [68] and its cofactor (F420-dependent biliverdin reductase) is implicated in immune-evasive mechanisms to allow bacterial persistence [66, 67] (Table S3). Recently, it has been reported that the synthesis of F420 might be stimulated by phosphoenolpyruvate [69], which is the precursor to pyruvate, a supplement often added to culture medium to improve the in vitro growth of L5, L6 and M. bovis strains. Furthermore, there is a suggestion that F420 is needed for the activation of the antituberculosis drugs pretomanid and delamanid [69].
Another gene absent in L5 strains is Rv1977. This gene is one of the 14 genes contained in RD7 [13]), which is known to be lacking completely from strains of L6 and animal-adapted lineages such as M. bovis [12]. Similarly, Rv1979c, 1 of 10 genes that make up RD2, was absent from genomes of L5 strains; this RD is also absent from late-generation M. bovis BCG such as M. bovis BCG Pasteur strains [12]. Rv1978, another of the 10 genes in RD2, and located in the region Rv1977-Rv1978-Rv1979c was absent in the 3 complete (PacBio-sequenced) genomes of L5 strains, but present in 2 L5 strains Illumina-sequenced genomes [2/202, 1 %; further confirmed using blast search and manual check of the gene in the reads of the 2 L5 strains genomes (ERR439931 and ERR4192386)], making up the region containing Rv1977, Rv1978 and Rv1979c, a region absent in most of L5 strains (99 %, 200/202). Rv1979c has been associated with clofazimine and bedaquiline resistance [70, 71], two of the drugs used for the treatment of rifampicin- and multidrug-resistant TB [72, 73], but minimal inhibitory concentration testing of clofazimine in five L5 strains did not find that the deletion conferred resistance [74]. Further studies and protein function discovery is needed to investigate the consequences of the absence of those genes in genomes of L5 strains. Rv1978 is required for bacterial survival in macrophages [75].
Rv1993c and Rv1995 were absent in all L5 genomes. Rv1994c a gene outside of known RDs, located in the region Rv1993c-Rv1994-Rv1994 was absent in the three complete (PacBio-sequenced) genomes of L5 strains, yet present in one L5 strain Illumina-sequenced genome [1/202, 0.5 %; further confirmed, using blast search and manual checking of Rv1994 in the reads of the concerned L5 strain genome (ERR439931)]. This resulted in the absence, in most of the L5 strains (99.5 %, 201/202) of the region containing Rv1993c, Rv1994c and Rv1995. Rv1994c is involved in the regulation and transport (efflux) of toxic metals, especially copper, which is toxic in excess and may hamper in vitro growth [76–80] (Table S3). Rv1993c forms with Rv1994 (cmtR) the operon cmtR-Rv1993c-ctpG, and Rv1995 is involved in oxygen transport (Table S3).
In combination, the absence of these genes in L5 strains suggests that they would be less likely to survive in macrophages (Rv1978), have reduced growth in vitro (Rv1994c, as previously found by [27]), and be less immune-evasive, less persistent and less virulent (Rv2074) than L4 strains (at least H37Rv), as previously suggested for L5 and L6 strains [5, 32]). In addition, the absence of Rv2074 in most L5 strains (and in the complete genomes of L6 and M. bovis as well) suggests that L6 and M. bovis strains would also be less likely to be immune-evasive, to be persistent and to survive than L4 strains. Despite the presence of vitamin B6 in boiled egg (thus Lowenstein–Jensen medium), the quantity of vitamin B6 in the Lowenstein–Jensen medium is probably insufficient to allow a high yield of growth of L5, L6 and M. bovis strains in culture (survival). Further studies, including vitamin B6 supplementation, could investigate the consequences of the absence of those genes in L5 strains (and L6 and M. bovis for Rv2074).
In contrast to the absence of some genes in RD2 and RD7, or all genes in RD9, RD5 was found to be present in its entirety in nearly all sequenced genomes of L5 strains. Some authors reported that up to 45 % of L5 strains missed RD5, while others reported that RD5 is present in most L5 strains [35, 81]. The RD5 region includes Rv2346 (truncated), Rv2347, Rv2348, Rv2349c (plcC), Rv2350c (plcB), Rv2351c (plcA) and a PPE gene [13]. Our findings showed that of the 202 Illumina-sequenced L5 strains, the majority (196, 97 %) had the RD5 region in its entirety, whereas 6 L5 strains (3 %) missed a part (one or 2 genes among the non-PPE genes) of the RD5 region. Likewise, in contrast to a previous report [10], our data did not confirm that all L5 strains lack RD711 (coordinates: 1 501 713–1 503 655 in H37Rv [82]) composed of Rv1333 (truncated), Rv1334, Rv1335 and Rv1336 (truncated). The lacking of RD711 was rather observed in all L5.1 strains in our dataset, which is in agreement with the report of Ates et al. [35] and also observed among 58.6 %(17/29) of L5 strains of unknown sub-lineages (Table S7).
We found that four genes, present in all of the L5 strains in our multi-country collection, were absent (no orthologues) in the H37Rv genome. Those genes represent 2382 bp, which is around 0.05 % of the total length (base pair) of a typical genome of an MTBC strain. Our findings are in line with those of other reports indicating that some genes present in genomes of clinical MTBC strains were absent (no orthologues) in the H37Rv genome [46, 47]. Although these gene differences are small, they did affect the distance matrices used for transmission clustering analyses (Table 6). Overall, H37Rv-based mapping was found to place a slightly higher percentage of strains in transmission clusters than any L5-based mapping approach, especially at lower SNP cut-offs. This has implications for molecular epidemiology in West Africa, where most L5 strains are found. Additionally, if similar scenarios exist for other lineages, this may result in changes to all non-L4 transmission analyses.
Our findings support past recommendations to use additional reference genomes that are different from H37Rv [46, 83, 84]. Although another study [85] concluded that there is no need to use a lineage-specific reference genome, their observation was only based on the analysis of L4 clinical strains and focused on SNPs and short indels, not larger gene deletions or SNPs within these regions. In contrast, our findings indicate that mapping of NGS data from L5 strains to L4 reference will have an impact in terms of both reference genome coverage and coverage of lineage-specific genes.
While the use of a single L5 genome as reference would have many benefits over H37Rv for particular study questions, several gene content differences were still observed within the L5 lineage. One (PcbL5Ben) of the three genomes of L5 strains sequenced with PacBio had genes that were not shared with the two others that were all found in Illumina-sequenced genomes of L5 strains (thus excluding exogenous contamination) (Table 1). The L5NigDel was also found in the genomes of six L5 strains from different countries. These six strains formed a monophyletic group of the (SNP-defined) L5.3 sub-lineage [6] (Fig. 1), suggesting that the loss of these 30 genes is a marker for strains of L5.3.2 sub-lineage, compared to the sub-lineage L5.3.1 strains that have these genes intact. Indeed, deletions, as described for MTBC lineages [10, 45, 48], may also be limited to sub-lineages.
The between- and within-lineage differences in both gene content and potential functionality indicate a need for more closed genomes of MTBC sub-lineages to be constructed. Due to the impact of reference choice on transmission studies, there is now a need for varying approaches to NGS data analysis to be considered. For instance, sub-lineage-specific reference genomes could improve resolution, although such ad hoc (e.g. outbreak-specific) reference genomes are only specific to that particular situation/outbreak/population/lineage and cannot be used in another context, making comparisons between lineages and settings difficult. Alternatively, a pangenome-based reference genome(s) capturing all the known diversity may be required instead. This can take two forms: a composite genome containing all the genes found in strains of all lineages and sub-lineages of the MTBC (i.e. both the core and accessory genome) [86], represented as a graph instead of a single sequence [87–92], or a selection of reference genomes, with mapping to all or a subset undertaken, as has been done with strains of Mycobacterium chimaera and other pathogens [93, 94]. Other authors have also reported that, because of the genetic variability between strains, using a single strain genome as reference genome lacks accuracy [95–97]. This MTBC-wide pangenome approach has been suggested before, including for other organisms [38, 47, 95–100]. However, such an approach also has its own drawbacks, including difficulty in mapping reads that bridge the boundaries between accessory genes and the rest of the genome, comparing strains of different lineages including phylogenetic analyses and retention of gene names and codes in clinical use, where H37Rv is deeply embedded [89]. The specific sequence of each orthologue gene would also need to be chosen for such a reference, with the inferred ancestral genome representative of MTBC lineages approach being the most likely method [22, 42, 101].
Another alternative approach is a de novo assembly reference-free approach [38, 96, 102, 103], where strains are either assembled into contigs without the use of a reference genome or compared to each other without first calling SNPs. This would allow for clustering of samples regardless of lineage, e.g. for genotyping or transmission analyses using Mash (software for fast/meta -genome distance estimation technique) [104] but would require new cut-offs for defining clusters and many additional steps for further gene annotation and between-sample comparisons, making its clinical use potentially confusing.
A limitation of this study is that the complete (PacBio) and short-read (Illumina) genomes were derived from positive cultures, excluding possible minority L5 strain diversity, as L5 strains are overrepresented in negative cultures [27]. WGS applied directly to sputum is increasingly needed, especially for ancestral lineages (including L5 and L6), where negative culture or dysgonic isolates are more common and are a challenge for DNA extraction [26, 27]. There is also a limitation regarding the comparison of gene presence/absence in strains of L5 versus L4 (including H37Rv), L6, M. bovis complete genomes; which is that our study only included a single complete genome of one strain of L4, L6 and M. bovis, respectively, while strains of these lineages may display similar variability to the intra-L5 variability we observed in our study. Of note, the genes that are present in L5 strains but not specific to L5 strains could be either active or inactive (mutated), requiring additional in vitro or in vivo validations to fully elucidate the metabolic profile of these strains.
In conclusion, the use of a (sub-)lineage reference genome can increase resolution for strain comparison in comparison to a H37Rv-based mapping approach for L5 genome analyses for epidemiology (transmission), phylogeny and sub-lineage determination. Still, the use of a (sub-)lineage reference genome may miss some within-lineage gene differences. For drug resistance detection in clinical L5 strains or strains of other lineages, H37Rv could still be used as a reference genome as resistance-related mutations are usually among the core genes (shared across all lineages). The high within-lineage gene content variability suggests that the pangenome of MTBC strains may be larger (at least by 5092 bp) than previously thought, implying that a reference-free genome assembly (de novo assembly) approach may be needed.
Supplementary Data
Funding information
This work was supported by funds from the Directorate General for Development (DGD), Belgium (FA4 to C. N. S., B. C. d. J., D. A. and L. R.); the European Research Council-INTERRUPTB starting grant (number 311725 to B. C. D. J., C. J. M. and L. R.) M. C. is supported by ESCMID, Ministerio de Ciencia (RYC-2015–18213 and RTI2018-094399-A-I00) and Generalitat Valenciana (SEJI/2019/011). S. G. is supported by the Swiss National Science Foundation (grants 310030_188888, IZRJZ3_164171, IZLSZ3_170834 and CRSII5_177163) and the European Research Council (883582-ECOEVODRTB). The funders had no role in study design, data collection and interpretation or the decision to submit the work for publication.
Acknowledgements
We would like to acknowledge Pim de Rijk Willem and Patrick Beckert for their technical input. We also thank Jody Phelan for providing us with information on retrieving the PacBio-sequenced L5 genomes from The Gambia and Nigeria online.
Author contributions
C. N. S. was involved in conceptualization, methodology, formal analysis, investigation, data curation, writing of the original draft, reviewing/editing the manuscript and visualization. M. C. was involved in conceptualization, methodology, investigation, data curation, reviewing/editing the manuscript and visualization. B. O. was involved in methodology, investigation and reviewing/editing the manuscript. I. D. O. was involved in resources and reviewing/editing the manuscript. M. A. was involved in resources. S. N. was involved in conceptualization, reviewing/editing the manuscript and supervision. J. P., was involved in investigation, data curation and reviewing/editing the manuscript. S. H. was involved in conceptualization, investigation, data curation and reviewing/editing the manuscript. D. Y. was involved in conceptualization, resources and reviewing/editing the manuscript. S. G. was involved in conceptualization and reviewing/editing the manuscript. L. R. was involved in conceptualization, resources, reviewing/editing the manuscript and supervision. D. A. was involved in resources, reviewing/editing the manuscript and supervision. B. C. d. J. was involved in conceptualization, methodology, writing of the original draft, reviewing/editing the manuscript and supervision. C. J. M. was involved in conceptualization, methodology, formal analysis, investigation, writing of the original draft, reviewing/editing the manuscript, supervision and visualization.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The PacBio-sequenced L5 strain from Benin was isolated during the multicentric OFLOTUB study approved by the National Ethics Committee in Cotonou, Benin and each of the participating countries (Merle et al. 2012 [105]).
Footnotes
Abbreviations: L4, lineage 4; L5, lineage 5 (Mycobacterium africanum lineage 5); L6, lineage 6 (Mycobacterium africanum lineage 6); L, lineage; L5Nig-Del, region deletion found in the PcbL5Nig; LSP, large sequence polymorphism; MTBC, Mycobacterium tuberculosis complex; NGS, next generation sequencing; PcbL5Ben, PacBio sequenced genome (completed genome) of a L5 strain from Benin; PcbL5Gam, PacBio sequenced genome (completed genome) of a L5 strain from The Gambia; PcbL5Nig, PacBio sequenced genome (completed genome) of a L5 strain from Nigeria; RD, region of difference; SNP, single nucleotide polymorphism; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and eight supplementary tables are available with the online version of this article.
References
- 1.Phelan J, Sessions PFD, Tientcheu L, Perdigao J, Machado D. Methylation in mycobacterium tuberculosis is lineage specific with associated mutations present globally. Scientific Reports. 2018;8:1–7. doi: 10.1038/s41598-017-18188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meehan CJ. Mycobacterium Tuberculosis L5 Complete Genomes with Annonations. 2020. [DOI] [Google Scholar]
- 3.Brites D, Loiseau C, Menardo F, Borrell S, Boniotti MB, et al. A New Phylogenetic Framework for the Animal-Adapted Mycobacterium Tuberculosis Complex. Frontiers in Microbiology 9 (NOV. 2018;9 doi: 10.3389/fmicb.2018.02820. In. Vol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngabonziza CS, Jean CL, Marceau M, Jouet A, Menardo F, et al. A sister lineage of the Mycobacterium Tuberculosis complex discovered in the african great lakes region. Nature Communications. 2020;11:2917. doi: 10.1038/s41467-020-16626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coscolla M, Gagneux S. Consequences of genomic diversity in Mycobacterium Tuberculosis . Semin Immunol. 2014;26:431–444. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coscolla M, Gagneux S, Menardo F, Loiseau C, Ruiz-Rodriguez P, et al. Phylogenomics of Mycobacterium Africanum Reveals a new lineage and a complex evolutionary history. Microbial Genomics. 2021;7:477. doi: 10.1099/mgen.0.000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boritsch EC, Khanna V, Pawlik A, Honoré N, Navas VH, et al. Key experimental evidence of chromosomal DNA transfer among selected tuberculosiscausing mycobacteria. Proc Natl Acad Sci USA. 2016;113:9876–9881. doi: 10.1073/pnas.1604921113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiner-Oms L, Sánchez-Busó J, Corander S, Gagneux SR, Harris D, et al. Genomic determinants of speciation and spread of the Mycobacterium Tuberculosis complex. Science Advances. 2019;5 doi: 10.1126/sciadv.aaw3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottai D, Frigui W, Sayes F, Luca MD, Spadoni D, et al. TbD1 Deletion as a driver of the evolutionary success of modern epidemic Mycobacterium Tuberculosis lineages. Nature Communications. 2020;11:1. doi: 10.1038/s41467-020-14508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, et al. Variable Host-Pathogen comen Compatibility in Mycobacterium Tuberculosis. Proc Natl Acad Sci USA. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, et al. Comparative genomics of BCG Vaccines by Whole-Genome DNA Microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 12.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, et al. A new evolutionary scenario for the Mycobacterium Tuberculosis Complex. PNAS. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon S, Brosch R, Billault A, Garnier T, Eiglmeier K, et al. Identification of Variable Regions in the Genomes of Tubercle Bacilli Using Bacterial Artificial Chromosome Arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 14.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, et al. A Robust SNP barcode for typing Mycobacterium Tuberculosis Complex Strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the mycobacterium tuberculosis complex. JID. 2002 doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 16.Firdessa R, Berg S, Hailu E, Schelling E, Gumi B, et al. Mycobacterial lineages causing pulmonary and extrapulmonary Tuberculosis, Ethiopia. Emerg Infect Dis. 2013;19:460–463. doi: 10.3201/eid1903.120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nebenzahl-Guimaraes H, Yimer SA, Holm-Hansen C, Beer JD, Brosch R, et al. Genomic characterization of Mycobacterium Tuberculosis Lineage 7 and a Proposed Name: ‘Aethiops Vetus. Microbial Genomics. 2016;2 doi: 10.1099/mgen.0.000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niemann S, Kubica T, Bange FC, Adjei O, Browne EN, et al. The species mycobacterium Africanum in the light of new molecular markers. J Clin Microbiol. 2004;42:3958–3962. doi: 10.1128/JCM.42.9.3958-3962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riojas MA, Mcgough KJ, Rider-Riojas CJ, Rastogi N, Hazbón MH. Phylogenomic Analysis of the Species of the Mycobacterium Tuberculosis Complex Demonstrates That Mycobacterium Africanum, Mycobacterium Bovis, Mycobacterium Caprae, Mycobacterium Microti and Mycobacterium Pinnipedii Are Later Heterotypic Synonyms of Mycob. 2018. [DOI] [PubMed] [Google Scholar]
- 20.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, et al. Progression to active tuberculosis, but not transmission, varssion to Active Tuberculosis, but Not Transmission, Varies by M. Tuberculosis lineage in the gamLineage in The Gambia”. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum--review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis. 2010;4:e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, et al. Out-of-Africa migration and neolithic coexpansion of Mycobacterium Tuberculosis with Modern Humans. Nature Genetics. 2013;45:10. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Intemann CD, Thye T, Niemann S, Browne ENL, Chinbuah MA, et al. Autophagy Gene Variant IRGM −261T contributes to protection from tuberculosis caused by Mycobacterium Tuberculosis but Not by M. Africanum strains. Edited by William Bishai PLoS Pathogens. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thye T, Niemann S, Walter K, Homolka S, Intemann CD, et al. Variant G57E of mannose binding lectin associated with protection against tuberculosis caused by mycobacterium africanum but not by M. Tuberculosis” PLoS ONE. 2011;6:6. doi: 10.1371/journal.pone.0020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ofori-Anyinam B, Kanuteh F, Agbla SC, Adetifa I, Okoi C. Impact of the mycoBaterium africanum West Africa 2 lineage on tb diagnostics in West Africa: Decreased sensitivity of rapid identification tests in the Gambia. PLOS Neglected Tropical Diseases. 2016;10:1–12. doi: 10.1371/journal.pntd.0004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanoussi CN, de Jong BC, Odoun M, Arekpa K, Ligali MA, et al. Low Sensitivity of the MPT64 Identification Test to Detect Lineage 5 of the Mycobacterium Tuberculosis Complex. 2018. [DOI] [PubMed] [Google Scholar]
- 27.Sanoussi CN, Affolabi D, Rigouts L, Anagonou S, de Jong B. Genotypic characterization directly applied to sputum improves the detection of mycobacterium Africanum west African 1, under-represented in positive cultures. PLOS Neglected Tropical Diseases. 2017;11:e0005900. doi: 10.1371/journal.pntd.0005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leao S, Martin A, Mejia GI, Portaels F, Por- Taels F Leao SC. Practical handbook for the phenotypic and genotypic identification of mycobacteria. 2004. pp. 77–126. pp. [Google Scholar]
- 29.Pattyn SR, Portaels F, Spanoghe L, Magos J. Further studies on african strains of mycobacterium tuberculosis. Ann Soc belge Méd Trop. 1970 [PubMed] [Google Scholar]
- 30.Gehre F, Otu J, DeRiemer K, Sessions PF de, Hibberd ML, et al. Deciphering the growth behaviour of Mycobacterium Africanum . PLoS Neglected Tropical Diseases. 2013;7 doi: 10.1371/journal.pntd.0002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnus K. Epidemiological basis of tuberculosis eradication 3. Risk of pulmonary tuberculosis after human and bovine infection. Bull Org Mond Sante. 1966;Vol. 35 [PMC free article] [PubMed] [Google Scholar]
- 32.Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, et al. Clade-specific virulence patterns of mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. MBio. 2013;4 doi: 10.1128/mBio.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalo-Asensio J, Malaga W, Pawlik A, Astarie-Dequeker C, Passemar C, et al. Evolutionary History of Tuberculosis Shaped by Conserved Mutations in the PhoPR Virulence Regulator. Proc Natl Acad Sci USA. 2014;111:11491–11496. doi: 10.1073/pnas.1406693111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofori-Anyinam B, Riley AJ, Jobarteh T, Gitteh E, Sarr B, et al. Comparative genomics shows differences in the electron transport and carbon metabolic pathways of mycobacterium Africanum relative to mycobacterium tuberculosis and suggests an adaptation to low oxygen tension. Tuberculosis. 2020:101899.:101899. doi: 10.1016/j.tube.2020.101899. p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ates LS, Dippenaar A, Sayes F, Pawlik A, Bouchier C, et al. Unexpected genomic and phenotypic diversity of Mycobacterium Africanum lineage 5 affects drug resistance, protein secretion, and immunogenicity. Genome Biol Evol. 2018;10:1858–1874. doi: 10.1093/gbe/evy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otchere D, Isaac MC, Sánchez-Busó L, Asante-Poku A, Brites D, et al. Comparative genomics parative Genomics of Mycobacterium Africanum Lineage 5 and Lineage 6 from ghana suggests distinct ecological nm Ghana Suggests Distinct Ecological Niches. Scientific RepoRtS |. 2018;8:11269. doi: 10.1038/s41598-018-29620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otchere ID, Harris SR, Busso SL, Asante-Poku A, Osei-Wusu S. The first population structure and comparative genomics analysis of mycobacterium africanum strains from Ghana reveals higher diversity of lineage 5. Int J Mycobact. 2016;5:81.:S80. doi: 10.1016/j.ijmyco.2016.09.051. [DOI] [PubMed] [Google Scholar]
- 38.Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, et al. Whole genome sequencing of mycobacterium tuberculosis: Current standards and open issues. Nat Rev Microbiol. 2019;17:533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 39.Camus J-C, Pryor MJ, Me C, Cole ST. Re-Annotation of the Genome Sequence of Mycobacterium Tuberculosis H37Rv. 2019. [DOI] [PubMed] [Google Scholar]
- 40.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium Tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 41.Goig GA, Blanco S, Garcia-Basteiro AL, Comas I. Contaminant DNA in Bacterial Sequencing Experiments Is a Major Source of False Genetic Variability. BioRxiv. 2019 doi: 10.1186/s12915-020-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goig GA, Blanco S, Garcia-Basteiro AL, Comas I. Contaminant DNA in Bacterial Sequencing Experiments Is a Major Source of False Genetic Variability. BMC Biol. 2020;18:24. doi: 10.1186/s12915-020-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gagneux S. Ecology and evoology and Evolution of Mycobacterium Tuberculosis. Na Rev Microbiol. 2018 doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 44.Bifani P, Moghazeh S, Shopsin B, Driscoll J, Ravikovitch A, et al. Molecular characterization of Mycobacterium Tuberculosis H37Rv/Ra Variants: distinguishing the mycobacterial laboratory strain. J Clin Microbiol. 2000;38:3200–3204. doi: 10.1128/JCM.38.9.3200-3204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato-Maeda M, Rhee JT, Gingeras TR, Salamon H, Drenkow J, et al. Comparing Genomes within the Species Mycobacterium Tuberculosis. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Toole RF, Gautam SS. Limitations of the mycobacterium tuberculosis reference genome h37rv in the detection of virulence-related loci. Genomics. 2017;109:471–474. doi: 10.1016/j.ygeno.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Periwal V, Patowary A, Vellarikkal SK, Gupta A, Singh M, et al. Comparative Whole-Genome Analysis of Clinical Isolates Reveals Characteristic Architecture of Mycobacterium Tuberculosis Pangenome. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsolaki AG, Gagneux S, Pym AS, Yves Olivier L, Kreiswirth BN, et al. Genomic deletions classify the beijing/w strains as a distinct genetic lineage of mycobacterium tuberculosis. JClin Microbiol. 2005;43:3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList - 10 Years After. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Bentley SD, Comas I, Bryant JM, Walker D, Smith NH, et al. The genome of Mycobacterium Africanum West african 2 reveals a Lineage-Specific locus and genome erosion common to the M. Tuberculosis Complex.” Edited by Pamela L. C Small PLoS Neglected Tropical Diseases. 2012;6:e1552. doi: 10.1371/journal.pntd.0001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone KM, Farrell D, Stuber TP, Schubert OT, Aebersold R. Updated reference genome sequence and annotation of Mycobacterium bovis af2122/97. Genome Announcements. 2017;5:17–e00157. doi: 10.1128/genomeA.00157-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belisle JT, Sonnenberg MG. In Mycobacteria Protocols. New Jersey: Humana Press; 1998. Isolation of Genomic DNA from Mycobacteria; pp. 31–44. [DOI] [PubMed] [Google Scholar]
- 53.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, et al. Nonhybrid, finished microbial genome assemblies from Long-Read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 54.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: Architecture and Applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohl TA, Utpatel C, Schleusener V, Filippo MRD, Beckert P, et al. MTBseq: A Comprehensive Pipeline for Whole Genome Sequence Analysis of Mycobacterium Tuberculosis Complex Isolates. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meehan CJ, Moris P, Kohl TA, Pečerska J, Akter S, et al. The Relationship between Transmission Time and Clustering Methods in Mycobacterium Tuberculosis Epidemiology. EBioMedicine. 2018;37:410–416. doi: 10.1016/j.ebiom.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, et al. Whole-genome sequencing to delineate mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;Vol. 25 doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapopoulou A, Lew JM, Cole ST. The Mycobrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 63.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG Database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research. 2000;Vol. 28 doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kavanagh KL, Jçrnvall H, Persson B, Oppermann U. The SDR Superfamily: Functional and Structural Diversity within a Family of Metabolic and Regulatory Enzymes. 2008. p. 77. p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vidal S, Lara CLK, Mordaka PM, Heap JT. Review of NAD(P)H-Dependent Oxidoreductases: Properties, Engineering and Application. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1866. 2018;2:327–347. doi: 10.1016/j.bbapap.2017.11.005. In. Vol. pp. [DOI] [PubMed] [Google Scholar]
- 66.Ahmed FH, Elaaf Mohamed A, Carr PD, Lee BM, Condic-Jurkic K, et al. Rv2074 Is a Novel F 420 H 2-Dependent Biliverdin Reductase in Mycobacterium Tuberculosis. 2016 doi: 10.1002/pro.2975. [DOI] [PMC free article] [PubMed]
- 67.Selengut JD, Haft DH. Unexpected Abundance of Coenzyme F 420-Dependent enzymes in mycobacterium tuberculosis and other actinobacteria †. J Bacteriol. 2010;192:5788–5798. doi: 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dick T, Manjunatha U, Kappes B, Gengenbacher M. Vitamin B6 biosynthesis is essential for survival and virulence of Mycobacterium Tuberculosis . Mol Microbiol. 2010;78:980–988. doi: 10.1111/j.1365-2958.2010.07381.x. [DOI] [PubMed] [Google Scholar]
- 69.Grinter R, Ney B, Brammananth R, Barlow CK, Cordero PRF, et al. Cellular and structural basis of synthesis of the unique intermediate Dehydro-F420-0 in Mycobacteria . BioRxiv. 2020 doi: 10.1101/2020.02.27.968891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghodousi A, Rizvi AH, Baloch AQ, Ghafoor A, Masood F, et al. Acquisition of Cross-Resistance to Bedaquiline and Clofazimine Following Treatment for Tuberculosis in 2 Pakistan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S, Chen J, Cui P, Shi W, Zhang W, et al. Identification of Novel Mutations Associated with Clofazimine Resistance in Mycobacterium Tuberculosis. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.WHO WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment. 2019. [PubMed] [Google Scholar]
- 73.WHO The shorter mdr-tb regimen features of the shorter mdr-tb regimen regimen composition 4-6. 2016.
- 74.Merker M, Kohl TA, Barilar I, Andres S, Fowler PW, et al. Phylogenetically informative mutations in genes implicated in antibiotic resistance in mycobacterium tuberculosis complex. Genome Med. 2020;12:27. doi: 10.1186/s13073-020-00726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for mycobacterium tuberculosis adaptation and survival in macrophages. PNAS June. 2005;Vol. 7 doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cavet JS, Graham AI, Meng W, Robinson NJ. A cadmium-lead-sensing ARSR-SMTB repressor with novel sensory sites. Complementary metal discrimination by NMTR and CMTR in a common cytosol. J Bio Chem. 2003;278:44560–44566. doi: 10.1074/jbc.M307877200. [DOI] [PubMed] [Google Scholar]
- 77.Marcus SA, Sidiropoulos SW, Steinberg H, Talaat AM. CsoR Is Essential for Maintaining Copper Homeostasis in Mycobacterium Tuberculosis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rowland JL, Niederweis M. Resistance Mechanisms of Mycobacterium Tuberculosis against Phagosomal Copper Overload. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samanovic M, Ding C, Thiele DJ, Heran Darwin K. Copper in microbial pathogenesis: Meddling with the metal. Cell Host & Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parsons LM, Brosch R, Cole ST, Somoskövi A, Loder A, et al. Rapid and simple approach for identification of mycobacterium tuberculosis complex isolates by pcr-based genomic deletion analysis. J Clin Microbiol. 2002;40:2339–2345. doi: 10.1128/jcm.40.7.2339-2345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mostowy S, Onipede A, Gagneux S, Niemann S, Kremer K, et al. Genomic analysis distinguishes mycobacterium africanum. J Clin Microbiol. 2004;42:3594–3599. doi: 10.1128/JCM.42.8.3594-3599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee RS, Proulx JF, McIntosh F, Behr MA, Hanage WP. Previously undetected super-spreading of mycobacterium tuberculosis revealed by deep sequencing. ELife 9. 2020 doi: 10.7554/eLife.53245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Norman A, Folkvardsen DB, Overballe-Petersen S, Lillebaek T. Complete Genome Sequence of Mycobacterium Tuberculosis DKC2, the Predominant Danish Outbreak Strain. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee RS, Behr MA. Does choice matter? Reference-based alignment for molecular epidemiology of tuberculosis. J Clin Microbiol. 2016;54:1891–1895. doi: 10.1128/JCM.00364-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McInerney JO, Whelan FJ, Domingo-Sananes MR, McNally A, O’Connell MJ. In The Pangenome. Springer International Publishing; 2020. Pangenomes and Selection: The Public Goods Hypothesis; pp. 151–167. [DOI] [PubMed] [Google Scholar]
- 87.Church DM, Schneider VA, Steinberg KM, Schatz MC, Quinlan AR, et al. Extending reference assembly models. Genome Biology. 2015;16:1–5. doi: 10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colquhoun RM, Hall MB, Lima L, Roberts LW, Hunt M, et al. Nucleotide-Resolution bacterial pan-genomics with reference graphs. BioRxiv, 2020. 2020:11.12.380378. doi: 10.1186/s13059-021-02473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marschall T, Marz M, Abeel T, Dijkstra L, Dutilh BE. Computational Pan-genomics: Status, promises and challenges. Briefings in Bioinformatics. 2018;19:118–135. doi: 10.1093/bib/bbw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martiniano R, Garrison E, Jones ER, Manica A, Durbin R. Removing reference bias in ancient DNA data analysis by mapping to a sequence variation graph. BioRxiv. 2019;782755 doi: 10.1186/s13059-020-02160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paten B, Novak AM, Eizenga JM, Garrison E. Genome Research. Cold Spring Harbor Laboratory Press; 2017. Genome Graphs and the Evolution of Genome Inference. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rakocevic G, Semenyuk V, Lee WP, Spencer J, Browning J, et al. Fast and accurate genomic analyses using genome graphs. Nat Genet. 2019;51:354–362. doi: 10.1038/s41588-018-0316-4. [DOI] [PubMed] [Google Scholar]
- 93.Ingen J van, Kohl TA, Kranzer K, Hasse B, Keller PM, et al. Global outbreak of severe Mycobacterium Chimaera disease after cardiac surgery: a molecular epidemiological study. The Lancet Infect Dis. 2017;17:1033–1041. doi: 10.1016/S1473-3099(17)30324-9. [DOI] [PubMed] [Google Scholar]
- 94.Garimella K, Iqbal Z, Krause MA, Campino S, Kekre M, et al. Detection of simple and complex de novo mutations without, with, or with multiple reference sequences. BioRxiv. 2019;698910 doi: 10.1101/gr.255505.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ballouz S, Dobin A, Gillis JA. Is it time to change the reference genome?” Genome Biology. BioMed Central Ltd. 2019 doi: 10.1186/s13059-019-1774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li R, Li Y, Zheng H, Luo R, Zhu H, et al. Building the sequence map of the human pan-genome. Nat Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 97.Sherman RM, Forman J, Antonescu V, Puiu D, Daya M, et al. Assembly of a Pan-Genome from Deep Sequencing of 910 Humans of African Descent. Nature Genetics Nature Publishing Group. 2019 doi: 10.1038/s41588-018-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jandrasits C, Kröger S, Haas W, Renard BY. Computational pan-genome mapping and pairwise SNP-distance improve detection of Mycobacterium tuberculosis transmission clusters. PLoS Comput Biol. 2019;15:e1007527. :e1007527. doi: 10.1371/journal.pcbi.1007527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. Current Opinion in Genetics and Development. Elsevier Current Trends. 2005. The Microbial Pan-Genome. In. [DOI] [PubMed] [Google Scholar]
- 100.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. Genome analysis of multiple pathogenic isolates of streptococcus agalactiae: implications for the microbial ‘Pan-Genome. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goig GA, Blanco S, Garcia-Basteiro AL, Comas I. Pervasive Contaminations in Sequencing Experiments Are a Major Source of False Genetic Variability: A Mycobacterium Tuberculosis Meta-Analysis. 2018. [Google Scholar]
- 102.Iqbal Z, Caccamo M, Turner I, Flicek P, Mcvean G. De Novo Assembly and Genotyping of Variants Using Colored de Bruijn Graphs. Nature Genetics. 2012;44:226–232. doi: 10.1038/ng.1028. In. Vol. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maretty L, Jensen JM, Petersen B, Jonas andreas sibbesen, Liu siyang. Sequencing and de novo assembly of 150 genomes from Denmark as a population reference. Nature Publishing Group. 2017;548 doi: 10.1038/nature23264. [DOI] [PubMed] [Google Scholar]
- 104.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. MASH: Fast genome and metagenome distance estimation using minhash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merle CSC, Sismanidis C, Sow OB, Gninafon M, Horton J. A pivotal registration phase III, multicenter, randomized tuberculosis controlled trial: Design issues and lessons learnt from the gatifloxacin for TB (OFLOTUB) project. Trials 13 (May) 2012 doi: 10.1186/1745-6215-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.