Abstract
Members of the Bifidobacterium longum species have been shown to possess adaptive abilities to allow colonization of different mammalian hosts, including humans, primates and domesticated mammalian species, such as dogs, horses, cattle and pigs. To date, three subspecies have formally been recognized to belong to this bifidobacterial taxon, i.e. B. longum subsp. longum, B. longum subsp. infantis and B. longum subsp. suis. Although B. longum subsp. longum is widely distributed in the human gut irrespective of host age, B. longum subsp. infantis appears to play a significant role as a prominent member of the gut microbiota of breast-fed infants. Nevertheless, despite the considerable scientific relevance of these taxa and the vast body of genomic data now available, an accurate dissection of the genetic features that comprehensively characterize the B. longum species and its subspecies is still missing. In the current study, we employed 261 publicly available B. longum genome sequences, combined with those of 11 new isolates, to investigate genomic diversity of this taxon through comparative genomic and phylogenomic approaches. These analyses allowed us to highlight a remarkable intra-species genetic and physiological diversity. Notably, characterization of the genome content of members of B. longum subsp. infantis subspecies suggested that this taxon may have acquired genetic features for increased competitiveness in the gut environment of suckling hosts. Furthermore, specific B. longum subsp. infantis genomic features appear to be responsible for enhanced horizontal gene transfer (HGT) occurrences, underpinning an intriguing dedication toward acquisition of foreign DNA by HGT events.
Keywords: bifidobacteria, bacterial evolution, comparative genomics, infant gut microbiome
Data Summary
Decoded genome sequences of 11 newly isolated B. longum strains were deposited at NCBI database under BioProject code PRJNA692178. A full listing of NCBI accession data for B. longum strains described in this paper is available in Table S1 (available in the online version of this article).
Supplementary material can be found at 10.6084/m9.figshare.14448249.
Impact Statement.
In this study, through comparative genomic analyses and phylogenomic reconstruction of 261 publicly available B. longum genomes, we gained insight into intra-species genetic and physiological diversity, identifying specific B. longum subsp. infantis genomic features, which appear to be linked with its enhanced ability to acquire foreign DNA. This remarkable genome plasticity may contribute to explain the specific adaptation of B. longum subsp. infantis toward colonization of the gut of suckling mammals.
Introduction
The human gut harbours at least 100 trillion (1014) microbial cells [1], collectively organized in a complex and dynamic microbial community that plays a fundamental role in defining the human health status [2]. It is well known that members of the gut microbiota engage in complex microbe–microbe and microbe–host interactions, with physiological consequences, including participation in metabolic activities such as (sometimes syntrophic) degradation of non-digestible carbohydrates, with consequent production of short-chain fatty acids (SCFAs) [3, 4]. The assembly of the human gut microbiota is believed to commence during delivery when the newborn passes through the mother’s birth canal [5]. During the developmental period following birth, the early gut microbiota is influenced by various factors, including mode of delivery, duration of gestation, antibiotic exposure, as well as feeding type [6, 7]. This latter factor is particularly noteworthy since breast-feeding can shape the gut-microbiota composition of the newborn by promoting a microbial community enriched by members of the Bifidobacterium genus [8]. In addition to the fermentation of non-digestible food compounds, especially glycans, the (bifido)bacterial consortia also engage with the host immune system, stimulating and modulating both innate and adaptive host immune responses, ultimately influencing overall intestinal functionality and homeostasis [9, 10]. Interestingly, it has been reported that particular bifidobacterial species, such as Bifidobacterium longum subsp. infantis, Bifidobacterium bifidum and Bifidobacterium breve, are able to efficiently utilize (certain) human milk oligosaccharides (HMOs) [11–15]. HMOs constitute complex milk glycans known to elicit prebiotic activity by allowing the above-mentioned bifidobacterial species to establish and persist in the infant gut, thereby representing a clear example of host-microbe co-evolution in humans [16–20].
Members of the Bifidobacterium longum species have been identified as very common inhabitants of the mammalian gut, reaching a prevalence of 95.5 %, representing the percentage of individuals harbouring this species within the population, as shown by a recent survey conducted in 67 assessed mammalian hosts [21]. In recent decades, members of the B. longum species have been grouped into three distinct subspecies, i.e. Bifidobacterium longum subsp. longum, Bifidobacterium longum subsp. infantis and Bifidobacterium longum subsp. suis [22], the latter isolated from the gut microbiota of swine [22, 23]. Despite the progressive reduction in the relative abundance of bifidobacteria in the human gut starting from 1/2 years of age [7], members of B. longum subsp. longum are known to commonly inhabit the infant, adult and elderly human gut [24], thereby perhaps exerting their positive health footprint throughout the human lifespan [24, 25]. In contrast, B. longum subsp. infantis is most frequently isolated from breast-fed infant faeces [26, 27]. Consistently, the decoding of B. longum subsp. infantis ATCC15697 genome sequence, which was published in 2008, revealed a genome that is dedicated to the degradation and utilization of a wide range of HMOs [15, 28].
Due to the substantial scientific and commercial interest in members of this species, which are able to colonize different hosts at different stages of life, during which they may contribute to host health, a large number of B. longum strains have been sequenced. Nevertheless, a comprehensive dissection of the genetic potential of B. longum and its subspecies is still lacking. For this reason, we decided to investigate the genomic diversity of and phylogenetic relationships between members of the B. longum species. This prompted a complete revision of subspecies classification and allowed a detailed dissection of their genetic features presumed to be responsible for efficient niche adaptation.
Methods
Ethical statement
Animal research was performed in compliance with the rules, regulations and recommendations of the Ethical Committee of the University of Parma. The corresponding protocols were approved by the ‘Comitato di Etica Università degli Studi di Parma’, Italy. All animal procedures were carried out in accordance with national guidelines (Decreto legislativo 26/2014).
Furthermore, the human study protocol (protocol number 2016/0028558) was approved by the Ethics Committee of the ‘Azienda Unità Sanitaria Locale di Reggio Emilia ‐ IRCCS’ in Reggio Emilia, Italy, as well as by the Ethics Committee of the University of Parma, Italy, and informed written consent was obtained from all participants or their legal guardians.
B. longum genome sequences
At the time of writing (November 2020), 363 publicly available B. longum genomes (complete and draft genome sequences) were retrieved from the National Center for Biotechnology Information (NCBI) public database and then subjected to genome quality-based selection. In detail, genome sequences showing a genome size less than 2.20 Mb or/and with a number of predicted CDSs less than 1600 as well as those exhibiting low sequencing quality (genome coverage lower than 30-fold or containing more than 100 contigs) were manually identified and discarded. Furthermore, duplicated bacterial genomes (ANI value >99.99 %) were removed, resulting in a final collection of 261 high-quality B. longum genomes encompassing 243, 7 and 11 chromosomes belonging to B. longum subsp. longum, B. longum subsp. suis and B. longum subsp. infantis subspecies, respectively. Furthermore, we decoded the chromosomes of 11 newly isolated B. longum strains that were also included in this study (Table S1). Notably, these latter isolates were obtained from human, bovine and canine faecal samples within the context of a bifidobacterial strain isolation project aimed at exploring the genetic variability of the Bifidobacterium genus.
Identification of novel B. longum strains and chromosomal DNA extraction
Based on a previous cultivation effort aimed at isolating Bifidobacterium pseudolongum strains from faecal samples of various mammalian species [29], bifidobacterial strains that did not belong to the above-mentioned bifidobacterial species were further subjected to species-specific PCR-based characterization in order to identify novel B. longum strains. Briefly, bifidobacterial strains were incubated in an anaerobic atmosphere (2.99 % H2, 17.01 % CO2 and 80 % N2) in a chamber (Concept 400, Ruskinn) in de Man-Rogosa-Sharpe (MRS) (Sharlau Chemie) supplemented with 0.05 % (wt/vol) l-cysteine hydrochloride and incubated at 37 °C for 16 h. Subsequently, cells were harvested by centrifugation at 3500 g for 8 min, and the obtained cell pellet was used for DNA extraction using the GenEluteTM Bacterial Genomic DNA kit (Merck, Germany), following the manufacturer’s instructions. The extracted DNA was then subjected to a B. longum species-specific identification protocol through a PCR-based methodology using primers Blong1 5′-TCCCAGTTGATCGCATGGTC-3′ and Blong2 5′-GGGAAGCCGTATCTCTACGA-3′, which are based on the 16S rRNA gene sequences of this taxon [30]. PCR amplification was carried out according to the following protocol: one cycle of 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s and 72 °C for 50 s, and a final cycle of 72 °C for 5 min. Furthermore, the DNA of strains identified as B. longum ssp. were further subjected to a genotyping PCR using primers ERIC1 5′-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC2 5′-AAGTAAGTGACTGGGGTGAGCG-3′ in order to sequence the genome of only one representative per genotype [31]. PCR amplification was performed according to a previous protocol: one cycle at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 48 °C for 30 s and 72 °C for 4 min, and a final cycle at 72 °C for 10 min [31].
B. longum genome sequencing and assemblies
Chromosomal DNA of the 11 newly identified B. longum strains was sequenced by GenProbio Srl (http://genprobio.com) using a MiSeq platform (Illumina, San Diego, CA, USA) according to the supplier’s protocol employing the Nextera XT DNA Library Prep Kit (Illumina), resulting in fragments of about 500–900 bp. The library samples obtained were then pooled into a Flow Cell V3 600 cycle (Illumina) in order to retrieve paired-end reads of 250 bp resulting from sequencing of fragment ends. Fastq files of paired-end reads generated from each genome sequencing effort were used as input for the genome assembly through the MEGAnnotator pipeline (https://github.com/GabrieleAndrea/MEGAnnotator) [32]. The SPAdes v3.14.0 program included in the MEGAnnotator platform was used for de novo assembly of each bifidobacterial genome sequence with the pipeline option ‘--careful’ and a list of k-mer sizes 21,33,55,77,99,127 as suggested in the SPAdes’ manual [33]. MEGAnnotator then employed contigs greater than 1000 bp to predict protein-encoding ORFs using Prodigal v2.0 (Linux command line ‘./prodigal -f gff -a [protein_translation_to_selected_file] -i [input_filename.fasta] -o [output_filename]”) [34]. Predicted ORFs were then functionally annotated using RAPSearch2 (reduced alphabet-based protein similarity search) (cutoff e-value of 1×10−5 and minimum alignment length 20) employing the NCBI reference sequences (RefSeq) database [35] together with hidden Markov model profile (HMM) searches (http://hmmer.org/) performed against the manually curated Pfam-A database (cutoff e-value of 1×10−10).
Pan-genome analyses of B. longum genomes
All 272 genome sequences of B. longum were employed for a core-genome analysis using the Pangenome Analysis Pipeline (PGAP) v1.1 (Linux command line ‘./PGAP.pl --strains [input_strain_list] --input input_path/ --output output_path/ --thread 20 --identity 0.5 --coverage 0.8 --cluster --method GF’) (http://pgap.sf.net) [36]. Predicted CDSs of each B. longum genome were classified into functional gene clusters through the gene family (GF) method, consisting of pairwise protein-similarity search employing blast software v2.2.28+ (cutoff e-value of 1×10−10 and exhibiting at least 50 % identity across at least 80 % of both protein sequences). Following this, using MCL (graph-theory-based Markov clustering algorithm) [37], the data obtained were used to assign proteins to so-called Clusters of Orthologous Groups (COGs). A pan-genome profile was then built using an optimized algorithm as part of the pgap software v1.1, based on a presence/absence matrix encompassing all COGs identified in the analysed genomes (Linux command line ‘./PGAP.pl --strains [input_strain_list] --input input_path/ --output output_path/ --thread 20 --identity 0.5 --coverage 0.8 --cluster --method GF --evolution --pangenome’). Subsequently, the core genome of B. longum species was obtained by selecting protein families, which are shared between all genomes, while truly unique genes (TUGs) encoded by a single genome were identified based on those protein families that are present in one B. longum genome yet absent in all other B. longum genomes. Separate pan- and core-genome analyses were performed on each B. longum subspecies as described above, involving genomes of 251 B. longum subsp. longum, 11 B. longum subsp. infantis and ten B. longum subsp. suis genomes.
Phylogenomic comparison between B. longum strains
In order to assess the genetic relatedness among the 272 members of B. longum species, the COGs constituting the core genome of each B. longum strain were concatenated, and they were then aligned using mafft v7.222 [38] through the Linux command line ‘mafft --thread 20 --retree 2 --clustalout --reorder [input_sequences.fasta] >output.aln’. The resulting phylogenomic tree was constructed using the neighbour-joining method in ClustalW v2.1 [39] through the Linux command line ‘clustalw -bootstrap=100 -seed=100 -bootlabels=NODE -outputtree=phylip -infile=file.aln’. Then, utilizing the graphical viewer of phylogenetic trees FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/), the core-genome-based visual tree was developed. Furthermore, a value for the average nucleotide identity (ANI) was calculated for each genome pair using the fastANI software v1.3 [40] through the Linux command line ‘./fastANI --ql [genome_list_path] --rl [genome_list_path] -t 20 --matrix -o output.txt’. Out of 272 obtained B. longum genomes, we selected 42 B. longum strains in order to perform downstream analyses (Fig. 1). For this purpose, we included all ten genomes that clustered with the B. longum subsp. suis type strain DSM20097 (seven publicly available and three newly isolated), 11 of the non-redundant identified B. longum subsp. infantis chromosomes with suitable quality (see above), along with 21 representative of B. longum subsp. longum. Notably, these latter comprised the type strain DSM 20219, eight newly isolated, and an additional 12 publicly available genome sequences, selected to maximize the description of the intra-subspecies diversity from the branch of the tree encompassing the whole selection of B. longum subsp. longum (Fig. S2).
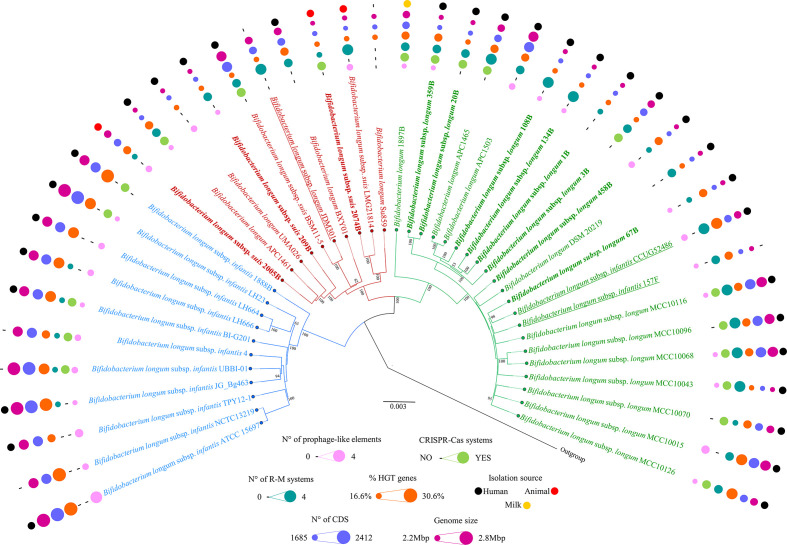
Fig. 1.
Phylogenomic tree based on the core genome of B. longum species. The phylogenomic tree, showing a selection of 42 representative genomes belonging to the B. longum species, was based on the concatenation of the 510 B. longum core genes and was built through the neighbour-joining method. Bootstrap percentages above 50 are shown at node points, based on 1000 replicates. Misclassified strains were underlined while the 11 new isolates were highlighted in bold. Phylogenetic clusters are highlighted with similarly coloured branches. Circles surrounding the tree represent the genome sizes (in dark pink), numbers of CDS (in purple), percentages of genes predicted to have undergone horizontal gene transfer (in orange), number of R-M systems (in dark green), occurrence of CRISPR-Cas systems (in light green), and isolation source (black=human, red=animal, yellow=milk).
Prediction of the mobilome of B. longum
The identification of the genes that may have been acquired by horizontal gene transfer (HGT) events was achieved using the suite colombo v3.8, with a sensitivity value of 0.7 (https://github.com/brinkmanlab/colombo/releases) [41]. Furthermore, the proteome of each B. longum strain was screened for the presence of restriction-modification (R-M) systems based on sequence similarity to genes classified in the rebase database [42] (http://rebase.neb.com/rebase/rebhelp.html; blast cutoff e-value of 1×10−5). The presence of transposable elements was performed through the IS Finder online tool with predefined parameters (https://isfinder.biotoul.fr/), while identification of clustered regularly interspaced short palindromic repeats (CRISPRs) was achieved through the web application CRISPRfinder (https://crispr.i2bc.paris-saclay.fr/Server/; default parameters were used) [43]. Prediction of prophage-like elements was conducted using a custom blast database (cutoff e-value of 1×10−5) encompassing previously bifidoprophage-validated sequences obtained from bifidobacterial type strains previously described [44]. Then, genomic regions encompassing predicted phage-related genes were manually examined to identify complete prophage-like sequences. Assessment of complete or partial plasmid sequences was carried out employing a combination of the PlasmidFinder 2.1 web service (https://cge.cbs.dtu.dk/services/PlasmidFinder/; minimum identity=50 % and minimum coverage=80 %) [45] and ABRicate software (https://github.com/tseemann/abricate).
B. longum type strains carbohydrate growth assays
In order to validate the in silico findings, we performed growth assays on selected carbon sources involving the type strains of each B. longum subspecies, i.e. B. longum subsp. longum DSM20219, B. longum subsp. suis DSM20097 and B. longum subsp. infantis ATCC15697. Notably, in silico analyses performed in this study generated predictions with regards to (carbohydrate) metabolic abilities of the above-mentioned strains and further discussed in Results. B. longum type strains were cultivated overnight on semisynthetic MRS medium supplemented with 0.05 % (w/vol) l-cysteine hydrochloride at 37 °C under anaerobic conditions. Subsequently, cells were diluted in MRS without glucose in order to obtain an OD600 nm=1 and 15 µl of the diluted cells were inoculated in 135 µl of MRS without glucose supplemented with 1 % (wt/vol) of a particular sugar in a 96-well microtitre plate and incubated in an anaerobic cabinet. Specifically, each carbohydrate was dissolved in MRS without glucose previously sterilized by autoclaving at 121 °C for 15 min. Subsequently, each obtained solution was filter sterilized using a 0.2 µm filter size prior to use. Cell growth was evaluated by monitoring the optical density at 600 nm with the use of a plate reader (Biotek, VT, USA). The plate was read in discontinuous mode, with absorbance readings performed at 3 min intervals for three times after 48 h of growth, and each reading was ahead of 30 s of shaking at medium speed. Cultures were grown in triplicates, and the resulting growth data were expressed as the average of these replicates. Carbohydrates tested in this study were purchased from Merck (Germany) and Carbosynth (Berkshire, UK), and include soluble starch from potato, amylopectin from maize, pullulan, maltotriose, maltodextrin, FOS, d-(+)-maltose, d-(+)-xylose, 2′-Fucosyllactose (2′-FL), 3′-Sialyllactose (3′-SL), and α-d-glucose.
Statistical analyses
All statistical analyses were performed with SPSS software v25 (www.ibm.com/software/it/analytics/spss/).
Results and Discussion
General genome features of B. longum genomes included in the comparative genomics analysis
In order to investigate the phylogenomic diversity of members belonging to the B. longum species, we undertook a comparative genomics analysis involving high-quality B. longum genome sequences selected amongst those publicly available (complete and draft genome sequences, see M and M section for the inclusion/exclusion criteria used). Remarkably, among the latter, B. longum subsp. infantis strains exhibited the highest number of suspected duplicated genomes (ANI ≥99.99 %). Accordingly, we removed such apparent copies of identical chromosomes, which had been deposited under different strain IDs, thereby allowing the generation of a curated B. longum subsp. infantis genome collection without duplicated chromosomal sequences (Table S1). The final collection of 272 B. longum genomes, including the 11 sequenced in this study, encompassed chromosomal sequences ranging in size from 2.2 Mb for B. longum APC1478 to 2.8 Mb for B. longum subsp. infantis ATCC 15697. As outlined in Table S1, the number of predicted coding DNA sequences (CDS) ranged from 1685 for B. longum subsp. longum 296B to 2412 for B. longum subsp. infantis ATCC 15697, with an average value of 1,927.17± 114.61 CDSs per genome (Table S1). Notably, the chromosomes belonging to the B. longum subsp. infantis subspecies emerged as the largest ones among the assessed B. longum genomes, ranging in size between 2.6 and 2.8 Mb (ANOVA P-value <0.05). These results showed that genome size might vary considerably even in closely related strains of the same species, thus indicating remarkable intra-species genetic and physiological diversity, unlike what was previously found for other bifidobacterial species such as Bifidobacterium bifidum and Bifidobacterium dentium [46, 47].
Pan-genome and core genome of B. longum species
In recent years, computation of the pan genome has been employed as an approach to investigate overall genomic differences and infers the precise phylogenomic relationships between (bifido)bacterial taxa [29, 48–51]. Accordingly, the genomes of B. longum strains were subjected to pan-genome analysis, allowing the identification of a total of 22 591 COGs. Analysis of the rate of size increase of the pan genome observed as genomes are sequentially included showed an average of 49.7 newly added COGs at the last three iterations (see Supplementary Material for details). This trend is indicative of a pan genome that has not yet fully reached its completion, though approaching a saturation plateau (Fig. S1). Moreover, a total of 510 COGs were classified as a collection of genes shared by all assessed strains, thereby representing the core genome of the B. longum species. Furthermore, the truly unique genes (TUGs) for each B. longum strain were also identified, revealing an average of 48.5 TUGs per genome (see Supplementary Material for details). The relatively small number of core genes observed suggests the presence of rather high intra-species variability, particularly when compared to other previously investigated bifidobacterial species, such as B. bifidum, Bifidobacterium breve (1295 and 1307 conserved COGs, respectively) [46, 52]. On the other hand, the relatively small number of TUGs is comparable with that previously observed for the genomes of Bifidobacterium pseudolongum and B. dentium (41 and 60 average TUGs, respectively) [29, 47], implying that a large part of the genetic diversity resides in the dispensable gene pool, i.e. those genes that are shared by a subgroup of strains, possibly due to adaptation to specific ecological niches/hosts. Interestingly, B. pseudolongum species, for which the subspecies pseudolongum and globosum are recognized, showed a much larger number of core genes, i.e. 1069 COGs, when compared to those identified in B. longum genomes. Therefore, these findings suggest that the latter taxon is characterized by a relatively high intra-specific variability, which may be imputed to distinct genetic traits possessed by each B. longum subspecies.
Phylogenetic analyses the B. longum taxon
The pairwise percentage ANI is currently considered to represent the gold standard for inference of close phylogenetic relationships and (sub)species classification of bacterial genomes [40]. Evaluation of the overall genomic differences between the 271 B. longum genomes through ANI analysis resulted in values ranging from 94.2 to 98.9 % (Table S2). Notably, previous Bifidobacterium phylogenomic studies showed that an ANI threshold value of 94 % properly discriminates between bifidobacterial species [51, 53], being consistent with what has been observed for other phylogenetically related taxonomic groups in the Bifidobacteriaceae family, such as Gardnerella [54]. Accordingly, the finding that this phylogenomic analysis generated ANI values above 94.2 % indicates that the included genome sequences correctly fall within the boundaries of a single species, i.e. B. longum. Nonetheless, based on the ANI matrix (Table S2), it was possible to identify three subgroups corresponding to the three so far recognized subspecies of B. longum, within which the observed ANI values ranged from 96.3 to 98.9 % (Table S2). Furthermore, in order to precisely track the phylogenetic relationships between the strains of this species, we computed a phylogenetic tree based on the amino acid sequence alignment of the 510 COGs that constitute the core genome of this species (Fig. S2). Due to the high number of analysed genomes belonging to the B. longum subsp. longum subspecies, we decided to generate an additional tree encompassing a pool of 42 representative genomes of this taxon, chosen to maximize the genetic diversity coverage, in order to obtain a clearer graphical visualization of the complete B. longum phylogeny (Fig. 1) (see Supplementary Material for details). As expected, the resulting B. longum-based phylogenetic tree revealed the presence of three main clades (Figs 1 and S2), consisting of the B. longum subsp. longum taxonomic group (Bll), the B. longum subsp. infantis taxonomic group (Bli) and the B. longum subsp. suis (Bls) taxonomic group (Fig. 1). In-depth analysis of the tree revealed that strains B. longum subsp. infantis 157F [55], B. longum subsp. infantis CCUG 52486 [56] and B. longum subsp. longum JDM301 [57] had been misclassified. Specifically, consistent with what had previously been observed through ANI analysis (Table S2), strains 157F and CCUG 52486 had been assigned to the B. longum subsp. longum subspecies, while JDM301 had been classified as a member of the B. longum subsp. suis subspecies. Interpretation of the phylogenomic tree suggests a clear phylogenetic separation between members of B. longum subsp. infantis cluster and the other B. longum strains, indicative of earlier speciation with respect to B. longum subsp. longum and B. longum subsp. suis, which showed a closer phylogenetic relationship (Figs 1 and S2, Table S2).
Moreover, the phylogenomic-based approach, combined with ANI value assignment, was applied to taxonomically classify the 11 newly isolated B. longum strains in order to include them in subspecies-specific analyses (see below). Specifically, three genomes were shown to belong to B. longum subsp. suis subspecies, i.e. 209B, 2015B and 2074B, while the remaining eight were classified as members B. longum subsp. longum subspecies (Fig. 1). Interestingly, B. longum subsp. longum 1897B, which had been isolated from human milk, was shown to belong to a separate branch with respect to all other B. longum subsp. longum strains (Fig. 1), denoting a different evolutionary history compared to the other assessed B. longum members isolated from the mammalian gut.
The pan and core- genome of the B. longum subspecies
Evolutionary processes have shaped bacterial genomes by driving changes in their genetic repertoire in order to facilitate adaptation to a specific environmental niche [58, 59], thus leading to (sub)speciation events. Pan-genome reconstruction may provide insights into these evolutionary events by unveiling genomic peculiarities and shared genetic traits that characterize a given bacterial taxon [60]. In the context of a B. longum subspecies-focused comparative analysis, we separately analysed subspecies-specific pan genomes (Fig. S3) (see Supplementary Material for details). The 251 B. longum subsp. longum genomes and the ten members of B. longum subsp. suis used in these analyses showed similar average genome sizes, i.e. of 2.39 and 2.43 Mb (Table S1). The latter are significantly smaller compared to that observed for B. longum subsp. infantis (average of 2.65 Mb) (Table S1), which also showed an average of 253 additional CDSs when compared to those found in B. longum subsp. longum and B. longum subsp. suis genomes (ANOVA P-value <0.001) (details in Supplementary Material). This finding suggests that members of the B. longum subsp. infantis taxon may have evolved as a result of progressive acquisition of new genetic features [58]. The subspecies-specific pan-genome analyses also allowed the definition of the Bll-, Bls- and Bli-core genome (CG), intended as the subspecies-specific core-genes' repertoire. In detail, these subspecies-specific core genomes were defined by taking into account those COGs shared by at least 85 % of the strains belonging to a given B. longum subspecies while being absent in the other two subspecies. The decision to consider an 85 % gene-sharing level, rather than the typically employed 100 %, was motivated by the presence of a high number of draft genomes within the analysed genome collection, which therefore could influence the accuracy of the calculation of subspecies-specific core genomes. In this manner, a total of 24 and five core genes represented the Bll-CG and Bls-CG, respectively, whereas 53 genes were identified as constituting Bli-CG (Fig. S3) (details are reported in Supplementary Material). The relatively small size of the Bll-CG and Bls-CG may, at least in part, be due to their close phylogenetic relationship and to the high number of analysed Bll genomes. However, it suggests that the evolutionary path taken by these subspecies may not have led to the acquisition of a substantial number of subspecies-specific competencies compared to their common B. longum ancestor. In contrast, the higher number of genes constituting Bli-CG suggests that this subspecies was subject to a higher evolutionary pressure that instigated the acquisition of novel genetic traits. Interestingly, 31 (58 %) of Bli-CG, 17 (71 %) of Bll-CG and four of the five (80 %) of Bls-CG were found in other bifidobacterial species with identity >50 % and coverage >80 % by blastp search in currently available bifidobacterial genomes (Table S3). These data suggest, at first glance, that a subgroup of subspecies-specific core genes may have been acquired by a common bifidobacterial ancestor (as indicated by presence in other bifidobacteria) and subsequently lost at subspecies level. Nevertheless, each subspecies seems to have independently acquired new genetic features, with B. longum subsp. infantis showing the highest number of genes acquired by presumed HGT events (18.8 % of the Bli-CG) (Table 1).
Table 1.
B. longum subspecies-specific core genes
|
B. longumsubsp.longum |
||||||
|
Core gene |
Prevalence across the subspecies |
Function |
Transporter classification database |
HGT events |
||
|
Interpro Database |
Refseq Database |
Function |
Family |
|||
|
B1_0665 |
99 % |
Selenoprotein, putative |
YbdD/YjiX family protein |
Native |
||
|
B1_0666 |
98 % |
5TM C-terminal transporter carbon starvation CstA |
Carbon starvation protein A |
Peptide Transporter Carbon Starvation CstA (CstA) Family |
2.A.114.- |
Native |
|
B1_1343 |
98 % |
Protein of unknown function (DUF3073) |
DUF3073 domain-containing protein |
Native |
||
|
B1_0094 |
98 % |
NADH Oxidase |
Nitroreductase |
Native |
||
|
B1_0106 |
98 % |
Periplasmic binding protein-like II |
Extracellular solute-binding protein |
Native |
||
|
B1_0884 |
98 % |
– |
Aldo/keto reductase family protein |
Native |
||
|
B1_1277 |
97 % |
Glycosidases |
Pullulanase type I |
Native |
||
|
B1_0628 |
97 % |
l,d-transpeptidase YCIB-related |
l,d-transpeptidase |
Native |
||
|
B1_0345 |
97 % |
Metal-dependent hydrolase |
Amidohydrolase family protein |
Native |
||
|
B1_1278 |
97 % |
Alpha-amylase |
Native |
|||
|
B1_0156 |
95 % |
– |
DUF2400 domain-containing protein |
Native |
||
|
B1_1275 |
95 % |
ABC transporter permease protein MG189-related |
ABC transporter permease subunit |
It binds α-(1,6)-linked glucosides and galactosides |
3.A.1.1.53 |
Native |
|
B1_0738 |
94 % |
– |
DUF1846 domain-containing protein |
Native |
||
|
B1_1795 |
93 % |
Acyl-CoA N-acyltransferases (Nat) |
GNAT family N-acetyltransferase |
Native |
||
|
B1_0431 |
91 % |
Uncharacterized protein conserved in bacteria C-term(DUF2220) |
DUF3322 and DUF2220 domain-containing protein |
Native |
||
|
B1_1294 |
90 % |
– |
Substrate-binding domain-containing protein |
Native |
||
|
B1_0735 |
90 % |
– |
DUF87 domain-containing protein |
Foreign |
||
|
B1_0883 |
90 % |
Transcriptional dual regulator hcar-related |
LysR family transcriptional regulator |
Native |
||
|
B1_0737 |
89 % |
Type VII secretion system protein EsaG-like |
– |
Foreign |
||
|
134B_0607 |
88 % |
– |
DNA/RNA non-specific endonuclease |
Native |
||
|
134B_0472 |
88 % |
MATE_MepA_like |
MATE family efflux transporter |
Native |
||
|
B1_1296 |
88 % |
K+potassium transporter |
KUP/HAK/KT family potassium transporter |
Native |
||
|
B1_0107 |
86 % |
Glycosidase family 31 |
Alpha-xylosidase |
Native |
||
|
B1_0786 |
86% |
zinc-ribbon domain |
Zinc ribbon domain-containing protein |
Native |
||
|
Core gene |
Prevalence across the subspecies |
Function |
Transporter classification database |
HGT events |
||
|
Interpro Database |
Refseq Database |
Function |
Family |
|||
|
100 % |
MFS general substrate transporter domains |
MFS transporter |
Glucose Transporter (GT) Family |
2.A.1.68.1 |
Native |
|
|
100 % |
Tetratricopeptide-like helical domain |
DUF4037 domain-containing protein |
Native |
|||
|
100 % |
Ttransporter solute:sodium symporter family |
Sodium/solute symporter |
Glucose or galactose:Na +symporter |
2.A.1.68.1 |
Native |
|
|
100% |
Response regulator receiver domain |
Response regulator transcription factor |
Foreign |
|||
|
100 % |
– |
– |
Native |
|||
|
100 % |
Lantibiotic immunity protein Spa1 |
NisI/SpaI family lantibiotic immunity |
Native |
|||
|
100 % |
Pyridoxal-phosphate dependent enzyme |
Pyridoxal-phosphate dependent enzyme |
Native |
|||
|
100 % |
High-affinity nickel-transport protein |
Nickel/cobalt transporter |
Native |
|||
|
100 % |
Bacteriocin (Lactococcin_972) |
Lactococcin 972 family bacteriocin |
Foreign |
|||
|
100 % |
Nitrate/nitrite sensor protein narx-related |
Histidine kinase |
Native |
|||
|
100 % |
metallo-dependent hydrolases |
Guanine deaminase |
Native |
|||
|
100 % |
GDSL-like Lipase/Acylhydrolase family |
Lipase |
Native |
|||
|
100 % |
MFS multidrug transporter |
MFS transporter |
Tet38 tetracycline-resistance protein |
2.A.1.3.22 |
Native |
|
|
100 % |
– |
– |
Foreign |
|||
|
100 % |
RecG, C-terminal domain superfamily |
Transcriptional regulator, partial |
Native |
|||
|
100 % |
– |
– |
Native |
|||
|
100 % |
RelB antitoxin/Antitoxin DinJ |
Type II toxin-antitoxin system family |
Foreign |
|||
|
100 % |
– |
– |
Native |
|||
|
100 % |
– |
5'-nucleotidase C-terminal domain |
Native |
|||
|
100 % |
Response regulatory domain profile. |
Response regulator transcription factor |
Native |
|||
|
100 % |
Nucleoside triphosphate hydrolases |
ATP-binding domain-containing protein |
Foreign |
|||
|
100 % |
Lantibiotic protection ABC transporter permease |
Lantibiotic immunity ABC transporter permease |
3-component subtilin immunity exporter |
3.A.1.124.2 |
Foreign |
|
|
100 % |
ABC-2 family transporter protein |
Lantibiotic immunity permease |
CprABC antimicrobial peptide resistance ABC exporter |
3.A.1.124.6 |
Foreign |
|
|
100 % |
– |
– |
Native |
|||
|
90 % |
Antitoxin |
– |
Foreign |
|||
|
90 % |
ABC superfamily metabolite uptake |
ABC transporter permease |
Putative macrolide-specific efflux system, MacAB |
3.A.1.122.16 |
Native |
|
|
90 % |
Metallophosphoesterase, calcineurin family |
Metallophosphoesterase |
Native |
|||
|
90 % |
Sialidase |
Exo-alpha-sialidase |
Native |
|||
|
90 % |
MFS_MefA_like |
MFS transporter |
The tetracycline resistance determinant, TetV |
2.A.1.21.3 |
Native |
|
|
90 % |
HAMP domain-containing histidine kinase |
– |
– |
Native |
||
|
90 % |
lantibiotic, protection ABC transporter ATP binding protein |
– |
– |
Foreign |
||
|
90 % |
Protein/nucleic acid deglycase dj-1-related |
DJ-1/PfpI family protein |
Native |
|||
|
90 % |
Beta-lactamase superfamily domain |
MBL fold metallo-hydrolase |
Native |
|||
|
90 % |
PBP2_UgpB |
ABC transporter substrate-binding protein |
Involved in maltose and maltodextrin uptake |
Native |
||
|
90 % |
ABC transporter integral membrane type-1 |
Phosphonate ABC transporter, permease |
Putative phosphonate/phosphite/phosphate porter |
3.A.1.9.2 |
Native |
|
|
90 % |
type II toxin-antitoxin system |
BrnT family toxin |
Native |
|||
|
90 % |
ABC transporter-type domain profile |
Phosphonate ABC transporter ATP-binding |
Putative phosphonate/phosphite/phosphate porter |
3.A.1.9.2 |
Native |
|
|
90% |
Transport system inner membrane component |
Carbohydrate ABC transporter permease |
ABC transporters for maltose/maltotriose and trehalose |
3.A.1.1.23 |
Native |
|
|
90 % |
phosphonate ABC transporter, permease |
ABC transporter, permease protein |
Putative phosphonate/phosphite/phosphate porter |
3.A.1.9.2 |
Native |
|
|
90 % |
SIS_RpiR |
MurR/RpiR family transcriptional regulator |
Native |
|||
|
90 % |
Periplasmatic phosphonate-binding protein |
ABC transporter substrate-binding protein |
Putative phosphonate/phosphite/phosphate porter, PhnDCE |
3.A.1.9.2 |
Native |
|
|
90 % |
MFS_MdtG_SLC18_like |
MFS transporter |
Copper Uptake Porter |
2.A.1.81.- |
Native |
|
|
90 % |
HAD-like superfamily |
HAD family hydrolase |
Native |
|||
|
90 % |
5′-Nucleotidase/apyrase |
Metallophosphoesterase |
Native |
|||
|
90 % |
Haloacid dehalogenase-like hydrolase |
HAD family hydrolase |
Native |
|||
|
90 % |
ABC transporter-type domain profile. |
ABC transporter ATP-binding protein |
Involved in the uptake of pectin oligosaccharides |
3.A.1.1.34 |
Native |
|
|
90 % |
ABC transporter integral membrane type-1 |
Sugar ABC transporter permease |
The fructooligosaccharide porter |
3.A.1.1.20 |
Native |
|
|
90 % |
Glycosidases |
Family 20 glycosylhydrolase |
Native |
|||
|
90 % |
MetI-like |
Sugar ABC transporter permease |
The xylobiose porter; BxlEFG(K) |
3.A.1.1.21 |
Native |
|
|
90% |
– |
Tyrosine-type recombinase/integrase |
Foreign |
|||
|
90 % |
Maltose transport system permease |
ABC transporter permease subunit |
N-Acetylglucosamine/N,N'-diacetyl chitobiose porter |
3.A.1.1.18 |
Native |
|
|
90 % |
Duplicated hybrid motif |
PTS glucose transporter subunit IIA |
Native |
|||
|
90 % |
Carbohydrate substrate-binding protein |
Carbohydrate ABC transporter |
xylobiose porter |
3.A.1.1.21 |
Native |
|
|
Core gene |
Prevalence across the subspecies |
Function |
Transporter classification database |
HGT events |
||
|
Interpro Database |
Refseq Database |
Function |
Family |
|||
|
100 % |
ABC transporter, atp-binding protein |
ABC transporter ATP-binding protein |
The Macrolide Exporter (MacB) Family |
3.A.1.122.- |
Native |
|
|
100 % |
ABC transporter permease |
MacB-like periplasmic core domain |
Exports macrolide antibiotics |
3.A.1.122.18 |
Native |
|
|
70 % |
Heavy metal transporter |
ABC transporter ATP-binding protein |
Native |
|||
|
70 % |
ABC-2 type transporter |
FHA domain-containing protein |
ABC exporter involved in bacterial competitiveness |
3.A.1.105.19 |
Native |
|
|
70 % |
vWA-like |
VWA domain-containing protein |
Native |
|||
Functional assessment of B. longum subspecies-specific core genomes
In order to gain further insight into the physiological characteristics of each B. longum subspecies, we investigated the Bll-CG, Bls-CG and Bli-CG from a functional perspective by similarity searches in the NCBI RefSeq nr database [61] and protein domain prediction by InterProScan [62].
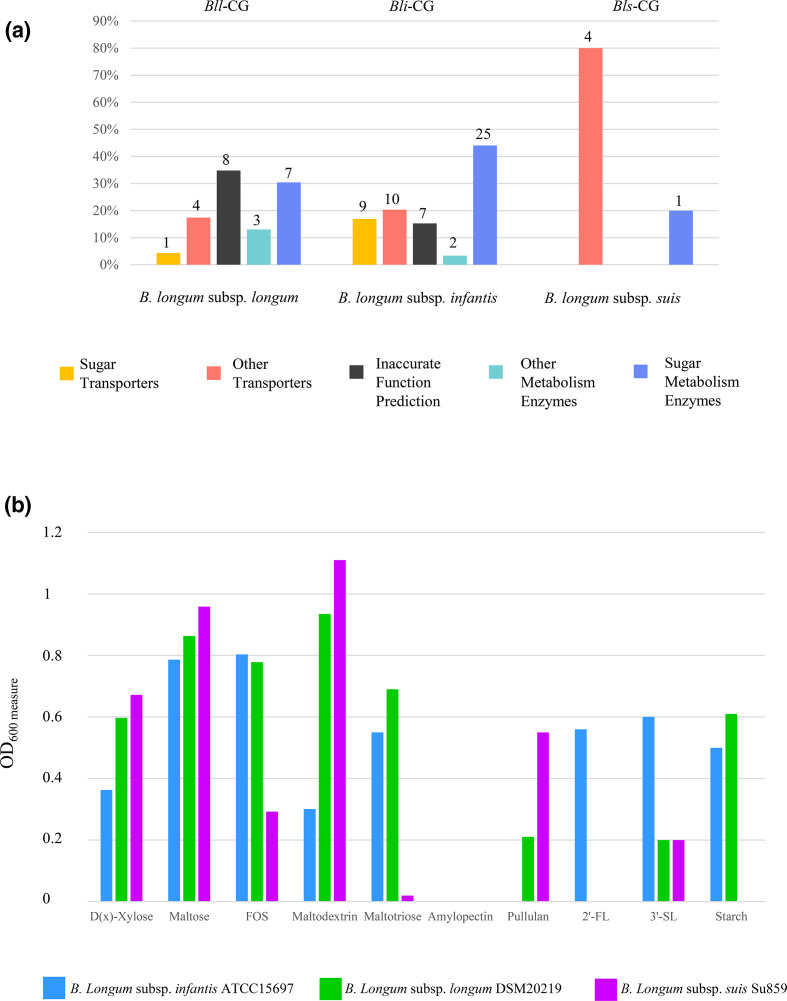
Of the 24 core genes unique to B. longum subsp. longum, eight could not be functionally annotated due to the absence of homologues with known function in the RefSeq nr database and known protein domains. In contrast, four were predicted to encode carbohydrate-utilization enzymes (Fig. 2a). Specifically, genes encoding pullulanase type I belonging to glycoside hydrolase family 13 (GH13), alpha-amylase (GH57), and a member of the amidohydrolase family proteins were found to be present in 97 % of the analysed genomes. Moreover, a gene whose protein product resembles members of glycosyl hydrolase family 31, representing enzymes such as alpha-glucosidase, glucoamylase, alpha-xylosidase and sucrase-isomaltase, was detected among 86 % of the analysed B. longum subsp. longum genomes (Table 1). Interestingly, the above-mentioned enzymes are typically involved in the utilization of plant-related carbohydrates, which, being undigested by the host, are thus available as a carbon source by the microbiota resident in the colon. This finding corresponds with B. longum subsp. longum being commonly present in faecal samples of human adults, whose diet includes such plant carbohydrates [14].
Fig. 2.
Functional annotation of core genes and growth performance of B. longum subspecies. Panel (a) shows the distribution in functional categories of the core genes identified in each B. longum subspecies. The number of genes assigned to each category is reported at the top of the columns. Panel (b) displays the growth performance of each B. longum type strain on different carbohydrates expressed through measurement of OD600 nm.
The functional dissection of genes attributed to Bls-CG revealed the ubiquitous presence of two genes encoding ATP-binding cassette (ABC) transporters that are implicated in macrolide resistance. At the same time, among 90 % of the strains, an additional ABC transporter was found to be involved in the detoxification of heavy metals (Table 1, Fig. 2a), allowing us to infer that these transporters play a critical role in niche adaptation of this subspecies. Notably, macrolides have been reported to be among the most frequently used classes of antimicrobials in pig breeding [63]. Therefore, they may have facilitated the development of antimicrobial resistance in bacteria present in the porcine gut microbiota [64].
Focusing on the B. longum subsp. infantis-specific core genetic repertoire, the most noticeable difference with respect to B. longum subsp. longum is the presence of genes involved in transporting a broad range of carbohydrates (Fig. 2a). Specifically, all assessed B. longum subsp. infantis genomes encompass genes that are predicted to encode a glucose transporter and a glucose/galactose-Na+ symporter. Interestingly, since glucose and galactose are the building blocks constituting lactose through glycosidic linkage, this finding suggests the presence of an extracellular (bifido)bacterial β-galactosidase (GH42), as also reported in previous genomic surveys [65], and improved specialization toward the uptake of released simple sugars. Moreover, we also identified transporters related to Tetracycline resistance, including the tetracycline resistance determinant Tet(V) and Tet38 gene. Tetracyclines are one of the most widely used groups of antibiotics worldwide, and resistance to this class of antibiotics is widespread even among bacteria that colonize the infant gut [66, 67]. Therefore, it may represent a trait that increases competitiveness of B. longum subsp. infantis in the gastrointestinal tract.
Progressively extending the analysis of Bli-CG by decreasing the level of gene sharing amongst members of this subspecies, i.e. prevalence, we identified genes that were involved in the uptake of pectic oligosaccharides (POSs), fructooligosaccharides (FOSs), maltose/maltotriose, and xylobiose with a prevalence of 90 % (Table 1). Intriguingly, these observations corroborated the well-known bifidogenic properties exhibited by POSs and FOSs, routinely used in commercial prebiotics due to their beneficial impact on the gut microbiota [68, 69]. Furthermore, within 90 % of B. longum subsp. infantis genomes, we identified an exo-alpha sialidase (GH33) and a member of glycosyl hydrolase family 20 (GH20) (Table 1), which represent enzymes known to be implicated in the metabolism of HMOs. These latter glycans are not processed by human digestive enzymes, thus reaching the colon intact where they are metabolized by certain members of the resident microbial community, such as B. bifidum, B. breve as well as B. longum, which encode gene clusters specifically dedicated to HMO metabolism [13, 70]. In particular, sialidases (GH33) catalyse the removal of terminal sialic acid residues, thus playing a critical role in the degradation of sialylated HMOs, such as 3′- and 6′-sialyllactose [71]. In line with previous publications, these findings show that degradation of sialylated HMOs is an ability that seems to be distinctive for B. longum subsp. infantis, thus being a characterizing genotypic and phenotypic feature of this subspecies [15]. In contrast, the above-described GH20 family comprises enzymes with β-hexosaminidase and lacto-N-biosidase activities, which act on substrates that form part of the HMO backbone, thereby releasing N-acetylglucosamine and lactose molecules, respectively [15, 72]. Previous investigations of the bifidobacterial glycobiome have highlighted that most B. longum subsp. longum strains (75–100 % of the strains) are predicted to encode the β-hexosaminidase (GH20), the lacto-N-biose phosphorylase (GH112), as well as an extracellular lacto-N-biosidase (GH136) [14, 73]. Intriguingly the finding of additional genes belonging to GH20 family encoded only by members of the B. longum subsp. infantis suggests that this subspecies has been subject to specific evolutionary selection. Remarkably, the latter seems to have driven B. longum subsp. infantis towards the acquisition of HMO-metabolizing genes, in addition to those shared with other members of the B. longum species.
Overall, the observed uneven distribution of the carbohydrate-active enzyme arsenal may reflect the distinct colonization strategy adopted by each B. longum subspecies, indicating that B. longum subsp. longum is more adapted to a (human) adult diet, as also supported by previous findings [15]. In contrast, members of the B. longum subsp. infantis subspecies may have evolved from a plant-derived glycan utilization gene-makeup towards a genomic repertoire that aims to achieve efficient colonization of the suckling mammalian gut.
To validate these in silico results, which indicate a more dedicated commitment of B. longum subsp. longum toward the breakdown of plant-related carbohydrate when compared to B. longum subsp. infantis, growth of the type strains of each B. longum subspecies, namely B. longum subsp. longum DSM20219, B. longum subsp. suis DSM20097 and B. longum subsp. infantis ATCC15697, was evaluated on ten different carbohydrates. In detail, for growth-profiling experiments, we used a carbohydrate-free basic MRS medium, which was supplemented with either amylopectin, pullulan, starch, maltotriose, maltodextrin, xylose, 2′-FL, 3′-SL, FOS or maltose as the sole carbon source (Table S4, Fig. 2b). Based on our analyses, B. longum subsp. suis was the only subspecies able to grow on pullulan-based medium (final OD above 0.5). Appreciable growth was also observed on xylose, maltose and maltodextrin (final OD ranging from 0.67 to 1.11). Conversely, both B. longum subsp. longum and B. longum subsp. infantis was shown to be able to grow on starch and starch-like glycans (final OD above 0.5), with the exception of amylopectin and pullulan for which no appreciable growth was noticed (Table S4, Fig. 2b). Nevertheless, as is displayed in Fig. 2(b). longum subsp. infantis was shown to exhibit a reduced level of metabolic abilities on various assessed plant-related glycans when compared to those elicited by B. longum subsp. longum. Furthermore, B. longum subsp. longum was shown to possess the most elaborate plant-related carbohydrate degrading activities among the B. longum species, being consistent with the above-described in silico reports (Table S4, Fig. 2b) [74]. Furthermore, B. longum subsp. infantis appears to be the only subspecies type strain capable of metabolizing 2′-FL and 3′-SL (Table S4, Fig. 2b). Consistently, the pronounced ability of B. longum subsp. infantis to metabolize a wide range of HMO compounds has been extensively reported [75–77]. However, carbohydrate metabolism data available in the literature have also highlighted specific HMO-utilizing abilities for certain members of the B. longum subsp. longum. While all strains can efficiently metabolize lacto-N-tetraose (LNT) and lacto-N-biose (LNB), only certain strains have shown growth capabilities on fucosylated HMOs and Lacto-N-neotetraose (LNnt) [76, 78]. In fact, growth profiles of the latter subspecies resemble that of Bifidobacterium adolescentis [74], which represents a gut-resident bifidobacterial taxon typical of the post-weaning period [79].
Overall, the findings related to the in vitro growth experiments corroborate our in silico data and may be a reflection of the ecological niche in which each B. longum subspecies dominate. Our data therefore suggest that B. longum subsp. longum plays an ecological role in the metabolism of dietary, plant-derived carbohydrates during weaning and post-weaning phases when infants are gradually introduced to a solid diet containing such complex carbohydrates [80–82]). Accordingly, the identified fermentation capabilities may provide an explanation as to how B. longum subsp. longum is able colonize both the infant and adult gut. In contrast, B. longum subsp. infantis is more adapted to colonization of the pre-weaning gut environment due to its particular HMO degradation abilities [15, 83].
Mobilome prediction in B. longum genomes
HGT is the process by which genetic material is exchanged between and within microbial taxa/taxon [84, 85]. This phenomenon of acquisition of new genomic properties is crucial for adaptation to new ecological niches [86], while it generates genetic diversity across bacterial taxa [87]. To a large degree, among (bifido)bacteria, HGT is assumed to occur through mobile genetic elements, such as plasmids, transposons or bacteriophages, with the latter considered one of the main vectors for gene transfer [88, 89]. To explore the possibility that HGT events are responsible for the substantial intra-specific genomic diversity observed between B. longum subspecies, the genomes of the representative 42 strains previously selected for phylogenetic analyses (Fig. 1) were screened using the software Colombo [41].
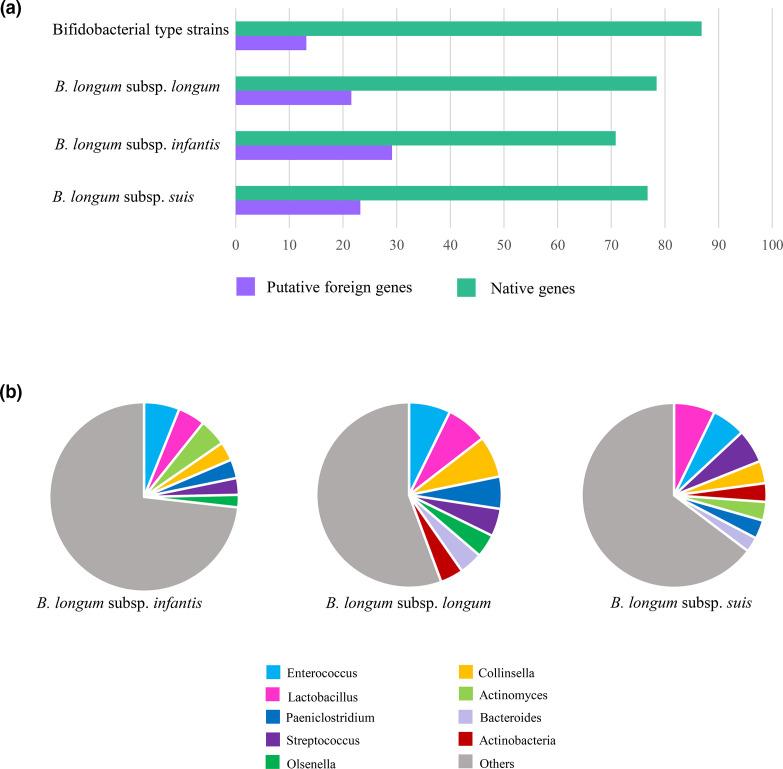
Following bioinformatic inspection of the B. longum subsp. longum and B. longum subsp. suis genomes, an average of 431 and 407 putative HGT genes, corresponding to an average of 22.5 and 20.9 % of the total number of CDS, respectively, were identified (Fig. 3a, Table S5). In contrast, an average of 640 CDS, corresponding to 29.5 % of the total number of predicted CDS, were identified as being potentially acquired by HGT in B. longum subsp. infantis (Fig. 3a, Table S5). To get an idea of the extent to which HGT events have contributed to shaping the genome architecture of B. longum subspecies, these values were compared to those obtained from 85 type strains belonging to different bifidobacterial species. Overall, the latter genomes showed an average of 12.8 % putative HGT-acquired genes, which was significantly lower than those identified in B. longum subspecies (ANOVA P-value <0.01) (Fig. 3a, Table S6), as was previously reported [90]. Furthermore, it is particularly noteworthy that B. longum subsp. infantis elicits the highest HGT gene numbers among the assessed bifidobacterial (sub)species, highlighting that this subspecies appears to be more suitable or to have been subject to higher selective pressure to acquire alien DNA when compared to not only other bifidobacterial species, but also compared to other B. longum subspecies (ANOVA P-value <0.01). Accordingly, these results provide an explanation for the higher average genome size of the B. longum subsp. infantis chromosomes.
Fig. 3.
Prediction of B. longum subspecies HGT events. Panel (a) shows the average percentages of the predicted foreign genes in each B. longum subspecies, compared with those obtained from 85 type strains belonging to different bifidobacterial species. Panel (b) displays the predominant non-bifidobacterial donor genera of the putative alien genes found in each B. longum subspecies.
Subsequently, the genes predicted to have been horizontally acquired by each of the B. longum subspecies were subjected to similarity searches in the NCBI refseq nr database in order to obtain an overview of the potential donor taxa. In particular, 124 (35 %) of the identified foreign genes of B. longum subsp. longum, 153 (30 %) of those of B. longum subsp. suis, and 280 (44 %) of the alien genes detected in B. longum subsp. infantis returned significant database hits in terms of similarity. Interestingly, of these identified HGT genes, 118 (95 %) of B. longum subsp. longum, 124 (81 %) of B. longum subsp. suis, and 191 (68 %) of B. longum subsp. infantis, corresponding respectively to 27, 30, and 30 % of the total HGT-acquired genes, appear to be derived from other bifidobacterial species, most frequently by B. bifidum, B. breve and Bifidobacterium adolescentis (Tables S7–S9). These latter species are also commonly found in the gastrointestinal tract of infants, thus representing a common niche that would facilitate horizontal transfer events. Furthermore, following the exclusion of hits corresponding to genera belonging to the Bifidobacteriaceae family, the analysis revealed a preferential origin of alien DNA from Enterococcus, Lactobacillus, Streptococcus, Collinsella, Bacteroides, Actinomyces as well as Paeniclostridium (Tables S7–S9). In particular, Enterococcus (7.2 %), Lactobacillus (7.2 %), and Collinsella (7.2 %) were identified as major donors of the B. longum subsp. longum horizontal genes (Fig. 3b, Table S8), while Lactobacillus (7.1 %), Enterococcus (5.8 %), and Streptococcus (5.8 %) were recognized as the prominent donors of the B. longum subsp. suis foreign genes (Fig. 3b, Table S9). In a similar fashion, the B. longum subsp. infantis genes putatively acquired by HGT were predicted to be originated mainly from Enterococcus (6.1 %), Lactobacillus (4.6 %) and Actinomyces (4.6 %) (Fig. 3b, Table S7). Interestingly, these donor genera, including the bifidobacterial ones, are known to share the human (infant) gut environment with members of B. longum species [7, 91], thus providing the opportunity for genetic transfer events, which can act as the driver of niche adaptation in members of the B. longum species [92].
To further investigate how HGT can contribute to differentially shape the B. longum subspecies, we assessed to what extent potential HGT events affect the specific core genome of each B. longum subspecies (Table 1). Notably, we found two alien core genes in the Bll-CG, and ten putative alien genes in the Bli-CG, corresponding respectively to 8.3 and 18.8 % of their own total number of core genes. Instead, no horizontal core genes were found among the five constituting the Bls-CG (Table 1). As expected, HGT seems to contribute only marginally to the core genome of the three B. longum subspecies. This observation is consistent with the notion that core genes are the most ancient genes, whose acquisition shaped the ancestors of each B. longum subspecies [93, 94]. Nevertheless, B. longum subsp. infantis was predicted to possess a higher number of foreign core genes compared to the other B. longum subspecies. Furthermore, based on RefSeq database annotation, horizontally acquired core genes in Bli-CG encompassed five genes putatively involved in the production of antimicrobial peptides, such as bacteriocins, and genes related to a toxin/antitoxin system (Table 1). Notably, bacteriocins are commonly produced by lactic acid bacteria, including Lactobacillus, Streptococcus and members of the Enterococcus genus [95, 96] that were consistently found amongst the major donor genera of foreign B. longum genes.
Survey of genetic features supporting HGT events
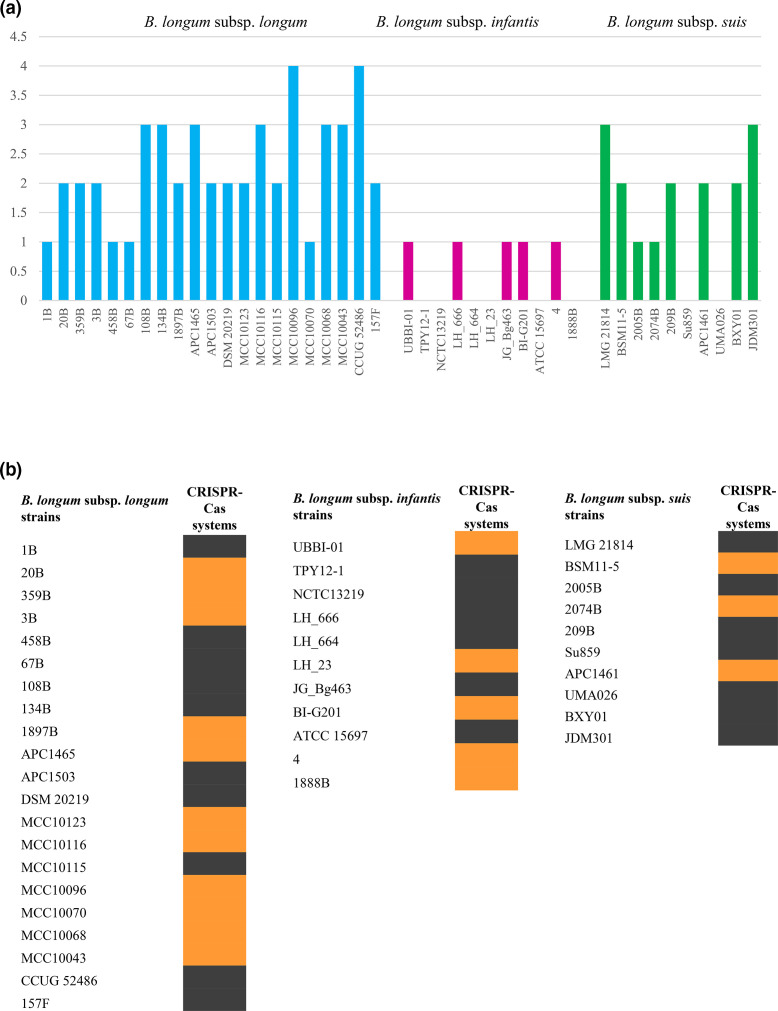
Mobile genetic elements, such as transposable elements and prophage-like elements, can promote DNA acquisition and facilitate the genetic material transmission between different bacterial taxa [87]. Conversely, CRISPR and R-M systems, which both represent microbial defence mechanisms against invasion of alien genetic material, are responsible for the degradation of nonself-DNA thereby preventing HGT events [97]. In order to investigate the genetic features of B. longum subspecies involved in the acquisition of foreign DNA, the representative 42 genomes were screened for R-M and CRISPR-Cas systems (Fig. 4a, b, Table S10). Overall, these analyses revealed that B. longum genomes mainly harbour type II and type I R-M systems, with a higher average number of R-M enzymes found in the subspecies B. longum subsp. longum. In detail, this latter subspecies exhibited an average of 2.2± 0.8 R-M genes (Fig. 4a, Table S10), while assessments of B. longum subsp. suis and B. longum subsp. infantis revealed the presence of 1.7± 1.1 and 0.45± 0.5 R-M genes, respectively (Fig. 4a, Table S9). Interestingly, these results negatively correlate with the number of alien genes as mentioned above for each B. longum subspecies (t-test P-value <0.05), corroborating the hypothesis that R-M systems counteract HGT events. As mentioned above, CRISPR-Cas systems represent another bacterial defence mechanism against invading alien DNA [98]. Based on our screening, out of the 21 representative B. longum subsp. longum strains considered, 11 were shown to contain at least one complete CRISPR-Cas locus in their genome (prevalence of 52.4 %) (Fig. 4b, Table S10). Besides, complete CRISPR-Cas systems were detected in 3 out of the 10 B. longum subsp. suis genomes, corresponding to a prevalence of 30 %, as well as within 3 out of the 11 B. longum subsp. infantis chromosomes, corresponding to a prevalence of 27 % (Fig. 4b, Table S10). Furthermore, the screening highlighted the occurrence of type I (subtypes I-C, I-E and I-U) and type II systems (subtypes II-C), characterized by the presence of cas3 and cas9 genes, respectively [99]. Specifically, a type II CRISPR-Cas system was detected only among B. longum subsp. longum strains, while such a system seems to be absent in B. longum subsp. suis and B. longum subsp. infantis. Overall, profiling of defence mechanisms highlighted that B. longum subsp. longum genomes seem to be equipped with a more efficient defence against foreign DNA invasion compared to those of both B. longum subsp. suis and B. longum subsp. infantis [100].
Fig. 4.
R-M and CRISPR-Cas systems in B. longum subspecies. Panel (a) shows the number of genomic R-M systems found in each of the 58 representative B. longum strains. Panel (b) depicts the presence (orange) or absence (black) of CRISPR-Cas systems in each of the 58 representative B. longum strains.
To obtain an overview of the B. longum genetic elements that may be implicated in HGT events, we screened for the presence of prophage-like and IS elements as well as plasmid sequences (Table S10). This allowed the identification of 21 (average of 1 per genome) and 8 (average of 0.8 per genome) prophage-like sequences in the inspected B. longum subsp. longum and B. longum subsp. suis genomes, respectively. In contrast, 22 (bifido)prophage, corresponding to an average of 2 integrated phages per genome, were observed in the chromosomes of B. longum subsp. infantis (Table S10). The genomic structural features of these retrieved bifidoprophages suggest that they represent members of the Siphoviridae family, consisting of lysogeny, DNA replication, DNA packaging, head and tail synthesis, and host lysis modules (Table S11). Furthermore, on average, 26.3±11.8 and 23.4±8.8 transposase genes per genome were found by inspecting B. longum subsp. longum and B. longum subsp. suis genomes, respectively, while B. longum subsp. infantis harbours 24.2±19.9 transposase genes per genome. In contrast, in silico prediction did not reveal any plasmid sequences among the inspected B. longum genomes.
Altogether, these results, coupled with data obtained from the analysis of HGT occurrences, suggests that B. longum subsp. infantis seems to be more prone to acquire alien genes than the other two B. longum subspecies, highlighting how HGT events may represent one of the key factors that shaped the genome of this taxon, thus contributing, to some extent, to provide it with specific ecological niche adaptations.
Conclusions
We investigated the genome diversity of B. longum species and its subspecies B. longum subsp. longum, B. longum subsp. infantis and B. longum subsp. suis through comparative genomic analyses and phylogenomic reconstruction of 261 publicly available and high-quality genomes, along with 11 novel strains sequenced as part of this study. These analyses revealed that members of B. longum subsp. infantis appear to contain a more extensive genetic repertoire than the other B. longum strains, highlighting how the former was shaped over the course of evolution through the acquisition of new genetic features. Notably, the functional analyses of the core genome unveiled that members of B. longum subsp. infantis possess unique carbohydrate utilization capabilities toward host glycans, particularly those for HMO degradation. When we investigated to what extent HGT events had been responsible for shaping B. longum subsp. infantis genomes, we revealed the increased frequency by which B. longum subsp. infantis had acquired alien DNA when compared to the other B. longum subspecies and to the type strains of other known bifidobacterial species. Notably, such higher genome plasticity, supported by specific genetic features such as lower number of restriction/modification and CRISPR-Cas systems coupled with a higher occurrence of prophage-like elements, appears to be a possible factor that allowed B. longum subsp. infantis to adapt to early life mammalian gut colonization. Furthermore, prediction of putative donor taxa of alien DNA revealed a preferential origin from other bifidobacterial and non-bifidobacterial species inhabiting the gut environment, suggesting that the extensive milk-related carbohydrate utilization capabilities that characterize the B. longum subsp. infantis subspecies may have been obtained through extensive gene harvesting from co-colonizing bacterial taxa. Though our findings provide insights into how the three B. longum subspecies probably developed at least in part through differential gene acquisition and subsequent niche occupation, it should be kept in mind that our conclusions are predominantly based on bioinformatic analyses. Our future efforts will therefore aim to further support these in silico data with experimental evidence.
Nevertheless, certain limitations of this study should be kept in mind. In particular, the fact that the number of publicly available sequenced chromosomes belonging to B. longum subsp. infantis and B. longum subsp. suis is significantly lower compared with that of B. longum subsp. longum subspecies. This may imply that the genetic variation within the first two subspecies may not have been completely disentangled, as also demonstrated by the identification of an open pan genome characterizing B. longum subsp. infantis as well as B. longum subsp. suis (see Supplementary Material). In addition, our study very much focused on the in silico assessment of the genetic traits distinguishing each B. longum subspecies, highlighting the need for experimental validation of our presented bioinformatics data.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
G.A. is supported by Fondazione Cariparma, Parma, Italy. We furthermore thank GenProbio Srl for the financial support of the Laboratory of Probiogenomics. This research benefited from the HPC (High-Performance Computing) facility of the University of Parma, Italy. D.v.S. is a member of APC Microbiome Ireland, which is supported by Science Foundation Ireland, through the Irish Government’s National Development Plan (SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ABC, ATP-binding cassette; ANI, average nucleotide identity; CDS, coding DNA sequences; COG, cluster of orthologous groups; CRISPR, clustered regularly interspaced short palindromic repeats; FOSs, fructo oligosaccharides; GH, glycoside hydrolases; HGT, horizontal gene transfer; HMOs, human milk oligosaccharides; LNB, lacto-N-biose; LNnt, lacto-N-neotetraose; LNT, lacto-N-tetraose; NCBI, National Center for Biotechnology Information; OD, optical density; POSs, pectic oligosaccharises; R-M systems, restriction modification systems; SCFA, short-chain fatty acids.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures and eleven supplementary tables are available with the online version of this article.
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan SM. Introduction to the special focus issue on the impact of diet on gut microbiota composition and function and future opportunities for nutritional modulation of the gut microbiome to improve human health. Gut Microbes. 2017;8:75–81. doi: 10.1080/19490976.2017.1299309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81 doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci. 2018;75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mancabelli L, Tarracchini C, Milani C, Lugli GA, Fontana F, et al. Multi-population cohort meta-analysis of human intestinal microbiota in early life reveals the existence of infant community state types (Icsts. Comput Struct Biotechnol J. 2020;18:2480–2493. doi: 10.1016/j.csbj.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986. doi: 10.1111/j.1348-0421.1984.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 9.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, et al. Bifidobacteria and their health-promoting effects. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.BAD-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. 2017;8:2345. doi: 10.3389/fmicb.2017.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James K, Motherway MO, Bottacini F, van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugli GA, Duranti S, Milani C, Mancabelli L, Turroni F, et al. Investigating bifidobacteria and human milk oligosaccharide composition of lactating mothers. FEMS Microbiol Ecol. 2020;96 doi: 10.1093/femsec/fiaa049. [DOI] [PubMed] [Google Scholar]
- 15.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duranti S, Lugli GA, Milani C, James K, Mancabelli L, et al. Bifidobacterium bifidum and the infant gut microbiota: an intriguing case of microbe-host co-evolution. Environ Microbiol. 2019;21:3683–3695. doi: 10.1111/1462-2920.14705. [DOI] [PubMed] [Google Scholar]
- 18.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. 2015;81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, et al. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl Environ Microbiol. 2009;75:965–969. doi: 10.1128/AEM.02063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 21.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017;11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattarelli P, Bonaparte C, Pot B, Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58:767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 23.Matteuzzi D, Crociani F, Zani G, Trovatelli LD. Bifidobacterium suis n. sp.: a new species of the genus Bifidobacterium isolated from pig feces. Z Allg Mikrobiol. 1971;11:387–395. doi: 10.1002/jobm.3630110504. [DOI] [PubMed] [Google Scholar]
- 24.Odamaki T, Bottacini F, Kato K, Mitsuyama E, Yoshida K, et al. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci Rep. 2018;8:85. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oki K, Akiyama T, Matsuda K, Gawad A, Makino H, et al. Long-term colonization exceeding six years from early infancy of Bifidobacterium longum subsp. longum in human gut. BMC Microbiol. 2018;18:209. doi: 10.1186/s12866-018-1358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr. 2012;3:422S–429S. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, et al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zabel B, Yde CC, Roos P, Marcussen J, Jensen HM, et al. Novel genes and metabolite trends in Bifidobacterium longum subsp. infantis Bi-26 metabolism of human milk Oligosaccharide 2’-fucosyllactose. Sci Rep. 2019;9:7983. doi: 10.1038/s41598-019-43780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lugli GA, Duranti S, Albert K, Mancabelli L, Napoli S, et al. Unveiling genomic diversity among members of the species Bifidobacterium pseudolongum, a widely distributed gut commensal of the animal kingdom. Appl Environ Microbiol. 2019;85 doi: 10.1128/AEM.03065-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65:4506–4512. doi: 10.1128/AEM.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventura M, Meylan V, Zink R. Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences. Appl Environ Microbiol. 2003;69:4296–4301. doi: 10.1128/aem.69.7.4296-4301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett. 2016;363 doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 33.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Wu J, Yang J, Sun S, Xiao J, et al. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waack S, Keller O, Asper R, Brodag T, Damm C, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015;43:299–D298. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grissa I, Vergnaud G, Pourcel C. Crisprfinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurston NE, Kerr JC. A nutritional knowledge questionnaire for the elderly. Can J Public Health. 1983;74:256–260. [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, et al. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum . Environ Microbiol. 2015;17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 47.Lugli GA, Tarracchini C, Alessandri G, Milani C, Mancabelli L, et al. Decoding the genomic variability among members of the Bifidobacterium dentium species. Microorganisms. 2020;8:11. doi: 10.3390/microorganisms8111720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albert K, Rani A, Sela DA. Comparative pangenomics of the mammalian gut commensal Bifidobacterium longum . Microorganisms. 2019;8 doi: 10.3390/microorganisms8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freitas AC, Hill JE. Bifidobacteria isolated from vaginal and gut microbiomes are indistinguishable by comparative genomics. PLoS One. 2018;13:e0196290. doi: 10.1371/journal.pone.0196290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, et al. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis . Sci Rep. 2016;6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, et al. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics. 2017;18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell KJ, Serafini F, et al. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics. 2014;15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, et al. Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarracchini C, Lugli GA, Mancabelli L, Milani C, Turroni F, et al. Assessing the genomic variability of Gardnerella vaginalis through comparative genomic analyses: Evolutionary and ecological implications. Appl Environ Microbiol. 2020;87 doi: 10.1128/AEM.02188-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 56.Prasanna PH, Bell A, Grandison AS, Charalampopoulos D. Emulsifying, rheological and physicochemical properties of exopolysaccharide produced by Bifidobacterium longum subsp. infantis CCUG 52486 and Bifidobacterium infantis NCIMB 702205. Carbohydr Polym. 2012;90:533–540. doi: 10.1016/j.carbpol.2012.05.075. [DOI] [PubMed] [Google Scholar]
- 57.Wei YX, Zhang ZY, Liu C, Zhu YZ, Zhu YQ, et al. Complete genome sequence of Bifidobacterium longum JDM301. J Bacteriol. 2010;192:4076–4077. doi: 10.1128/JB.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koskiniemi S, Sun S, Berg OG, Andersson DI. Selection-driven gene loss in bacteria. Plos Genet. 2012;8:e1002787. doi: 10.1371/journal.pgen.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul S, Sokurenko EV, Chattopadhyay S. Corrected genome annotations reveal gene loss and antibiotic resistance as drivers in the fitness evolution of Salmonella enterica serovar typhimurium. J Bacteriol. 2016;198:3152–3161. doi: 10.1128/JB.00545-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 61.Maglott DR, Katz KS, Sicotte H, Pruitt KD. NCBI’s LocusLink and RefSeq. Nucleic Acids Res. 2000;28:126–128. doi: 10.1093/nar/28.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones P, Binns D, Chang HY, Fraser M, Li W, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarrazin S, Joosten P, Van Gompel L, Luiken REC, Mevius DJ, et al. Quantitative and qualitative analysis of antimicrobial usage patterns in 180 selected farrow-to-finish pig farms from nine European countries based on single batch and purchase data. J Antimicrob Chemother. 2019;74:807–816. doi: 10.1093/jac/dky503. [DOI] [PubMed] [Google Scholar]
- 64.Burow E, Rostalski A, Harlizius J, Gangl A, Simoneit C, et al. Antibiotic resistance in Escherichia coli from pigs from birth to slaughter and its association with antibiotic treatment. Prev Vet Med. 2019;165:52–62. doi: 10.1016/j.prevetmed.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Moller PL, Jorgensen F, Hansen OC, Madsen SM, Stougaard P. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl Environ Microbiol. 2001;67:2276–2283. doi: 10.1128/AEM.67.5.2276-2283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gueimonde M, Salminen S, Isolauri E. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol Med Microbiol. 2006;48:21–25. doi: 10.1111/j.1574-695X.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 67.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 68.Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, et al. Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8 doi: 10.3390/foods8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiciński M, Sawicka E, Gębalski J, Kubiak K, Malinowski B. Human milk oligosaccharides: Health benefits, potential applications in infant formulas, and pharmacology. Nutrients. 2020;12 doi: 10.3390/nu12010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duar RM, Casaburi G, Mitchell RD, Scofield LNC, Ortega Ramirez CA, et al. Comparative genome analysis of bifidobacterium longum Subsp. Infantis strains reveals variation in human milk oligosaccharide utilization genes among commercial probiotics. Nutrients. 2020;12:11. doi: 10.3390/nu12113247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, et al. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Intra J, Pavesi G, Horner DS. Phylogenetic analyses suggest multiple changes of substrate specificity within the glycosyl hydrolase 20 family. BMC Evol Biol. 2008;8:214. doi: 10.1186/1471-2148-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, et al. Varied pathways of infant gut-associated bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2019;12 doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, et al. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol. 2014;80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zabel BE, Gerdes S, Evans KC, Nedveck D, Singles SK, et al. Strain-specific strategies of 2’-fucosyllactose, 3-fucosyllactose, and difucosyllactose assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci Rep. 2020;10:15919. doi: 10.1038/s41598-020-72792-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, et al. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, et al. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep. 2016;6:35045. doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kato K, Odamaki T, Mitsuyama E, Sugahara H, Xiao JZ, et al. Age-related changes in the composition of gut bifidobacterium species. Curr Microbiol. 2017;74:987–995. doi: 10.1007/s00284-017-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sela DA, Mills DA. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelly SM, Munoz-Munoz J, van Sinderen D. Plant glycan metabolism by bifidobacteria. Front Microbiol. 2021;12:609418. doi: 10.3389/fmicb.2021.609418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kujawska M, La Rosa SL, Roger LC, Pope PB, Hoyles L, et al. Succession of bifidobacterium longum strains in response to a changing early life nutritional environment reveals dietary substrate adaptations. iScience. 2020;23:101368. doi: 10.1016/j.isci.2020.101368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012;3:415S–421S. doi: 10.3945/an.111.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bolotin E, Hershberg R. Horizontally acquired genes are often shared between closely related bacterial species. Front Microbiol. 2017;8:1536. doi: 10.3389/fmicb.2017.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonham KS, Wolfe BE, Dutton RJ. Extensive horizontal gene transfer in cheese-associated bacteria. elife. 2017;6 doi: 10.7554/eLife.22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gyles C, Boerlin P. Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet Pathol. 2014;51:328–340. doi: 10.1177/0300985813511131. [DOI] [PubMed] [Google Scholar]
- 87.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 88.Bottacini F, Motherway O, Casey M, McDonnell E, Mahony J B, et al. Discovery of a conjugative megaplasmid in Bifidobacterium breve . Appl Environ Microbiol. 2015;81:166–176. doi: 10.1128/AEM.02871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ventura M, Turroni F, Foroni E, Duranti S, Giubellini V, et al. Analyses of bifidobacterial prophage-like sequences. Antonie van Leeuwenhoek. 2010;98:39–50. doi: 10.1007/s10482-010-9426-4. [DOI] [PubMed] [Google Scholar]
- 90.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, et al. Genomic encyclopedia of type strains of the genus Bifidobacterium . Appl Environ Microbiol. 2014;80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mancabelli L, Milani C, Lugli GA, Turroni F, Cocconi D, et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol. 2017;93:12. doi: 10.1093/femsec/fix153. [DOI] [PubMed] [Google Scholar]
- 92.Woods LC, Gorrell RJ, Taylor F, Connallon T, Kwok T, et al. Horizontal gene transfer potentiates adaptation by reducing selective constraints on the spread of genetic variation. Proc Natl Acad Sci U S A. 2020;117:26868–26875. doi: 10.1073/pnas.2005331117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Collins RE, Higgs PG. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol Biol Evol. 2012;29:3413–3425. doi: 10.1093/molbev/mss163. [DOI] [PubMed] [Google Scholar]
- 95.Wackett LP. Lactic acid bacteria: An annotated selection of World Wide Web sites relevant to the topics in microbial biotechnology. Microb Biotechnol. 2016;9:525–526. doi: 10.1111/1751-7915.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McAuliffe O, Ross RP, Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. 2001;25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x. [DOI] [PubMed] [Google Scholar]
- 97.Oliveira PH, Touchon M, Rocha EP. Regulation of genetic flux between bacteria by restriction-modification systems. Proc Natl Acad Sci U S A. 2016;113:5658–5663. doi: 10.1073/pnas.1603257113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 99.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hidalgo-Cantabrana C, Crawley AB, Sanchez B, Barrangou R. Characterization and exploitation of CRISPR Loci in Bifidobacterium longum . Front Microbiol. 2017;8:1851. doi: 10.3389/fmicb.2017.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.