Abstract
SAR11 bacteria dominate ocean surface bacterioplankton communities, and play an important role in marine carbon and nutrient cycling. The biology and ecology of SAR11 are impacted by SAR11 phages (pelagiphages) that are highly diverse and abundant in the ocean. Among the currently known pelagiphages, HTVC010P represents an extremely abundant but under-studied phage group in the ocean. In this study, we have isolated seven new HTVC010P-type pelagiphages, and recovered 77 nearly full-length HTVC010P-type metagenomic viral genomes from marine metagenomes. Comparative genomic and phylogenomic analyses showed that HTVC010P-type pelagiphages display genome synteny and can be clustered into two major subgroups, with subgroup I consisting of strictly lytic phages and subgroup II mostly consisting of phages with potential lysogenic life cycles. All but one member of the subgroup II contain an integrase gene. Site-specific integration of subgroup II HTVC010P-type pelagiphage was either verified experimentally or identified by in silico genomic sequence analyses, which revealed that various SAR11 tRNA genes can serve as the integration sites of HTVC010P-type pelagiphages. Moreover, HTVC010P-type pelagiphage integration was confirmed by the detection of several Global Ocean Survey (GOS) fragments that contain hybrid phage–host integration sites. Metagenomic recruitment analysis revealed that these HTVC010P-type phages were globally distributed and most lytic subgroup I members exhibited higher relative abundance. Altogether, this study significantly expands our knowledge about the genetic diversity, life strategies and ecology of HTVC010P-type pelagiphages.
Keywords: distribution patterns, genomic diversity, life strategies, metagenomic viral genomes, pelagiphages, SAR11
Data Summary
The genome sequences of HTVC010P-type pelagiphages have been deposited in GenBank under accession numbers MW273920–MW273926.
Impact Statement.
Pelagiphages that infect SAR11 bacteria are highly abundant and diverse in the ocean. The HTVC010P-type pelagiphage group represented by HTVC010P is known as one of the most abundant and prevalent phage groups in the ocean. However, the genetic diversity and life strategies of HTVC010P-type phages remain largely unexplored. In this study, we have expanded the study of HTVC010P-type phages by analysing the sequences of newly isolated HTVC010P-type pelagiphages and HTVC010P-type metagenomic viral genomes. We have illustrated that HTVC010P-type pelagiphages are diverse in the ocean and differentiate into two major subgroups with distinct life cycles. A proportion of HTVC010P-type pelagiphages may also have a lysogenic lifestyle. We also performed metagenomic recruitment analysis, which reveals that the lytic subgroup I HTVC010P-type pelagiphages are more abundant than the lysogenic subgroup II HTVC010P-type pelagiphages in the ocean. Our study suggests that both lytic and lysogenic phage predation strategies are prevalent in HTVC010P-type pelagiphages, providing more insight concerning the genomic diversity, evolution, host–phage interactions and distribution patterns of these important marine phages.
Introduction
Viruses are extremely abundant and genetically diverse in marine environments [1–3]. The ecological importance of marine viruses is highlighted by their impact on bacterial mortality and evolution, microbial community dynamics, and ecosystem biogeochemistry [1, 3–5]. Most marine viruses are bacteriophages that infect bacteria cells [1, 2, 4]. Bacteriophages infect hosts using two major life strategies: lytic and lysogenic infection cycles [6]. A viral infection model has recently been established that virus–host interactions often differ along a continuum of infection strategies ranging from lysis to persistent lysogeny [7]. As significant agents of microbial mortality, lytic bacteriophages kill bacteria and release virions; thus, can impact the composition and diversity of microbial communities [8]. Temperate bacteriophages can also replicate through the lysogenic life cycle, by which more complicated influences are exerted on the microbial community. For example, lysogenized host cells can acquire super-infection immunity and new phenotypic characteristics, resulting in niche expansion [6, 9–11]. Meanwhile, temperate phages have the opportunity to establish a long-term association with their bacterial hosts until an inducing event triggers the lytic cycle [6, 9, 10].
In marine environments, bacteriophages infecting major marine bacterial groups, such as SAR11 phages (pelagiphages), RCA phages and SAR116 phages, have been found to be ubiquitous and highly abundant [12–16]. Among the major marine bacterial groups, the SAR11 clade bacteria of Alphaproteobacteria (order Pelagibacterales) are known to be the most numerically abundant and ubiquitous, representing 30–40 % of total cell counts in the surface oceans [17]. SAR11 bacteria contribute significantly to marine biomass and the oxidation of marine dissolved organic matter, thereby strongly affecting the global carbon cycle [18]. Because of the extremely high abundance and ecological importance of SAR11 bacteria, pelagiphages have gained increasing attention in recent years. Thus far, 37 pelagiphage isolates belonging to 8 distinct phage groups have been characterized in the literature [12, 15, 16, 19]. In addition, the findings of Zhao et al. provided important insights into the life strategies of HTVC019P-type group pelagiphages of the family Podoviridae [19], revealing that most HTVC019P-type pelagiphage isolates contain an integrase gene and have both lytic and lysogenic life cycles. A very recent study reported on the first SAR11 prophages identified in two marine SAR11 strains, revealing that phage lysogeny occurs in some SAR11 genomes [20]. In recent years, surveys of metagenomic data have been shown to be powerful in recovering phage genomes [21–24]. This culture-independent approach has also been employed to study marine pelagimyophages, adding more insights into the understanding of pelagimyophages and their potential functions [25].
Among all known pelagiphages, HTVC010P represents a novel viral lineage that exhibits very distant relatedness to other known phages [12]. HTVC010P is a lytic phage affiliated with the family Podoviridae [12]. A recent study reported a HTVC010P-related SAR11 prophage PNP1, indicating that some HTVC010P-related pelagiphages also have a lysogenic lifecycle [20]. Metagenomics recruitment analyses revealed that HTVC010P-related sequences were abundant and widespread in the ocean [15, 26]. Quantitative PCR and digital droplet PCR techniques have been employed to investigate the absolute abundance of the isolate HTVC010P, which have revealed that the abundance of HTVC010P was within the range of 103–104 virus ml−1 in various oceanic regions [27, 28]. To date, only two HTVC010P-type isolates [12, 16], HTVC010P-type SAR11 prophage PNP1 [20], one HTVC010P-related contig from Western English Channel viral metagenome [29] and three reconstructed HTVC010P-related freshwater SAR11 phages (fonsiphages) [30] have been reported, limiting our understanding of this important phage group. Therefore, more HTVC010P-type pelagiphages need to be analysed to better understand their diversity and ecology.
In this study, the genomic diversity, life strategies and distribution of HTVC010P-type pelagiphages were investigated by analysing HTVC010P-type isolates and metagenomic viral genomes (MVGs). Genome comparison reveals the conservation and divergence of the HTVC010P-type pelagiphages. Phylogenetic analyses and genomic analyses reveal that HTVC010P-type pelagiphages contain two major subgroups with distinct life cycles. Finally, metagenomic recruitment analysis illustrates that the lytic subgroup I members are more abundant than the lysogenic subgroup II members.
Methods
Host strains and growth conditions
The SAR11 strains Pelagibacter HTCC7211 and Pelagibacter HTCC1062 were kindly provided by Professor Stephen Giovannoni, Oregon State University (USA). Both strains were grown in artificial seawater-based medium (ASM1) supplemented with 1 mM NH4Cl, 100 µM KH2PO4, 1 µM FeCl3, 100 µM pyruvate, 50 µM glycine, 50 µM methionine and excess vitamins [31]. HTCC7211 and HTCC1062 were grown at 20 and 17 °C, respectively. Pelagibacter FZCC0015 was isolated from the Pingtan coast (China) in 2017 and stored in our lab [19]. FZCC0015 was grown in natural seawater-based media [32] amended with 1 µM FeCl3, 100 µM pyruvate, 50 µM glycine, 50 µM methionine and excess vitamins at 23 °C.
Phage isolation and purification
The procedure for pelagiphage isolation and purification has been described previously [12, 15, 19]. Briefly, exponentially growing SAR11 bacteria cultures (~1×105 cells ml−1) were incubated with 0.1 µm filtered seawater samples. After 2 weeks of incubation, the cultures that underwent cell lysis, as detected by cell count, were observed by epifluorescence microscopy for phage particles. Phage clone purification was performed using the dilution-to-extinction method [15]. The purity of the pelagiphages was determined by whole-genome sequencing. However, in some pelagiphage cultures, a contamination appeared that had not been removed so far; therefore, further growth curve and phage infection experiments could not be performed. Approximately 25 % of the pelagiphage isolates we obtained display sequence similarity and genome synteny with pelagiphage HTVC010P, and seven pelagiphages were chosen for further analysis in this study.
Phage DNA preparation and sequencing
The preparation and concentration of pelagiphage lysates were carried out as described by Zhang et al. [14]. Briefly, 250 ml of each phage lysate was filtered through 0.1 µm Supor membrane to remove cells and cell debris. Phage lysates were concentrated by centrifugal filtration using Amicon Ultra-15 centrifugal filters (30 kDa; Merck Millipore) and Nanosep centrifugal devices (30 kDa; Pall Life Sciences). Phage genomic DNA was extracted using a formamide and phenol/chloroform extraction protocol [33], and sequenced on an Illumina HiSeq 2500 paired-end platform. The obtained raw reads were quality-filtered, trimmed and de novo assembled using CLC Genomic Workbench 11.0.1 with default settings. To complete the pelagiphage genome sequences, the remaining genomic gaps were closed by Sanger sequencing of the PCR products covering the gap regions.
Genome annotation
The programs GeneMark [34] and Prodigal v2.6.3 [35] were used to predict the ORFs. The translated ORFs were used as queries to search against the National Center for Biotechnology Information (NCBI) non-redundant (nr) and NCBI-RefSeq databases using blastp with default settings. Putative biological functions were assigned to ORFs based on their homology to proteins of known function. In this study, genes with ≥25 % amino acid identity, ≥50 % alignment coverage of the shorter protein and an E value cut-off ≤10−3 were considered to be putative homologues. The Pfam server [36], InterProScan program [37] (http://www.ebi.ac.uk/Tools/pfa/iprscan/), Conserved Domain search on the NCBI server [38] and HHpred server [39] (https://toolkit.tuebingen.mpg.de/tools/hhpred) were also used for the structure and function prediction. tRNA prediction was performed using tRNAscan-SE [40].
HTVC010P-type MVGs retrieval
To recover HTVC010P-type MVGs, amino acid sequences of HTVC010P-type pelagiphage ORFs were searched against the 515 588 MVG sequences from Global Ocean Viromes (GOV and GOV2.0) [22, 23], the Mediterranean DCM (MedDCM) fosmid library [21] and Station ALOHA assembly-free virus genomes (AFVGs) [24] using blastp (E value ≤10−3, ≥25 % amino acid identity). Orthologous groups were determined using OrthoMCL v2.0 [41, 42]. The criterion of ≥40 % of the shared genes was used to classify a phage group (approximately at genus level) [43]. MVGs that share ≥40 % genes with any HTVC010P-type phage and had a size ≥5 kb and G+C content between 29 and 36 mol% (the mol% G+C range of hosts and known pelagiphages) were designated as HTVC010P-type MVGs. Nearly full-length MVGs (90 % of the mean size of HTVC010P-type genome sizes, ≥32 kb) that contain a terL gene were used for further phylogenomic and comparative genomic analyses. Considering the position of the integrase gene (downstream of the DNA metabolism genes), the presence or absence of the integrase gene in HTVC010P-type MVGs can be determined.
Phylogenomic analysis
A total of seven core genes were selected for phylogenomic analysis (genes encoding nuclease, tail tube B, tail tube A, major capsid protein, capsid assemble protein, head-tail connector and terminase large subunit). Individual alignments for each gene were constructed using mafft v7.407 [44] and curated using trimAl v1.4 with default settings [45], and run with iq-tree v2.0.3 with 1000 bootstrap replicates. We also reconstructed a maximum-likelihood phylogenetic tree of integrase. Sequence alignments and editing were performed using mafft and trimAl, respectively. Maximum-likelihood phylogenetic tree was reconstructed using iq-tree.
Average amino acid identity (AAI) and whole-genome-based phylogeny analysis
AAI analysis was performed using CompareM v0.1.1 (https://github.com/dparks1134/CompareM). The heatmap was plotted by R package pheatmap with default clustering method ‘complete’. Whole-genome based phylogeny was built using victor (https://ggdc.dsmz.de/victor.php) [46].
Determination of HTVC028P integration sites
The site-specific integration between HTVC028P and HTCC1062 was confirmed using PCR assays, as previously described [19]. Briefly, two PCR primer sets targeting the phage integration site, attL and attR, were designed to be specific to lysogenic host bacteria. The location of each primer set is indicated in Fig. S1. The sequences of primers are listed in Table S1 (available with the online version of this article). PCR was performed in a 25 µl reaction volume containing 1×PCR Master Mix (TaKaRa), 0.2 µM each primer and 2 µl DNA template. DNA extracted from HTVC028P-infected host cells were used as the PCR template. The PCR program for both reactions included an initial denaturing step at 95 °C for 3 min, followed by 35 cycles of 95 °C for 1 min, annealing at 55 °C for 30 s and extension at 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min.
Integrase identification in HTVC010P-type MVG
Stockholm alignments of phage integrase and recombinase were downloaded to create the HMM (hidden Markov model) database (http://pfam.xfam.org) [47]. hmmsearch was then used to identify the putative integrase genes by searching HMM files against the HTVC010P-type MVGs [47]. HTVC010P-type MVGs containing the integrase gene were subjected to manual inspection and comparative genomic analyses. Putative ‘core sequences’ within the integration sites were identified by searching the MVGs against known SAR11 genome sequences using blastn.
Global Ocean Survey (GOS) metagenomic data search for SAR11-HTVC010P-type phage hybrid sequences
We identified the SAR11-pelagiphage hybrid sequences from the GOS metagenomic database following the strategy used in a previous study of HTVC019P-type pelagiphages [19]. The amino acid sequences of the genes located upstream and downstream of the core sequence (including int genes and other phage genes) from int-containing HTVC010P-type MVGs and HTVC028P were used as queries to search against the GOS database with tblastn (E value cut-off <10−3 and >40 % amino acid identity). This search resulted in GOS fragments containing homologues of integrase genes or other phage genes. The resulting fragments were then searched against the NCBI-RefSeq database and a dataset containing SAR11 and HTVC010P-type sequences using blastx. Only fragments containing the best hits to SAR11 genes and HTVC010P-type genes were retained for further analysis.
Viromic read recruitment analysis
Marine viromic datasets that were downloaded for read mapping include GOV [22] and GOV2.0 [23]. HTVC010P-type genomes that shared ≥95 % nucleotides were classified into a species, only the longest MVG within a species was retained for recruitment analysis. Viromic reads were recruited using blastn with ≥95 % identity and ≥90 % read coverage. The relative abundances of pelagiphages were normalized by total recruited nucleotides (kb) per kb of genome per gigabase of metagenome (KPKG). HTVC010P-type genomes for which <40 % of the genomes was covered by recruited reads in a given viromic dataset were given a KPKG value of 0 [16].
Results and Discussion
Genomic characterization of HTVC010P-type pelagiphages and MVGs
The seven pelagiphages isolated in this study ranged in size from 34.1 to 36.4 kb, encoding 52 to 65 ORFs (Table 1). The G+C content of the seven pelagiphages ranged from 31 to 33.1 mol %, similar to those of their hosts (29.0–29.7 mol%) and previously reported pelagiphages [12, 15, 16, 19]. No tRNA was found in any of the HTVC010P-type pelagiphage genomes. Genomic analysis indicated that these seven pelagiphages displayed obvious relationships with the previously reported pelagiphage HTVC010P (Fig. 1). A total of 31–62 % genes were shared between these pelagiphages and HTVC010P, and their overall genome architectures were similar to that of HTVC010P. In addition, no significant rearrangements were observed among these genomes (Fig. 1). We named this group the HTVC010P-type phage group after the first isolate HTVC010P. Genomic annotation reveals less than 40 % of all the predicted ORFs were assigned putative biological functions based on their similarity to proteins of known function or conserved domain analysis. Of all functionally annotated ORFs, most encode proteins were related to DNA metabolism, lysogeny control, phage morphogenesis, packaging and host lysis.
Table 1.
General features of HTVC010P-type pelagiphages sequenced in this study
|
Phage |
Host |
Source water |
Latitude |
Longitude |
Sampling date |
Genome size (bp) |
G+C (mol%) |
No. of ORFs |
Reference |
|---|---|---|---|---|---|---|---|---|---|
|
HTVC028P |
HTCC1062 |
Guam coast, West Pacific |
13°28’N |
144°40’E |
Dec 2016 |
36 388 |
33.1 |
52 |
This study |
|
HTVC203P |
FZCC0015 |
Ningde coast, East China Sea |
26°31’N |
119°55’E |
Mar 2017 |
34 938 |
32.1 |
62 |
This study |
|
HTVC034P |
HTCC1062 |
Pattaya coast, Indian Ocean |
12°46’N |
100°53’E |
Feb 2018 |
35 450 |
32.6 |
63 |
This study |
|
HTVC035P |
HTCC1062 |
Pattaya coast, Indian Ocean |
12°46’N |
100°53’E |
Feb 2018 |
36 066 |
31.9 |
65 |
This study |
|
HTVC024P |
HTCC1062 |
Yantai coast, Bohai Sea |
37°28’N |
121°28’E |
Mar 2017 |
35 448 |
31.5 |
62 |
This study |
|
HTVC204P |
FZCC0015 |
Yantai coast, Bohai Sea |
37°28’N |
121°28’E |
Mar 2017 |
34 069 |
31.0 |
57 |
This study |
|
HTVC100P |
HTCC7211 |
Pingtan coast, East China Sea |
25°26’N |
119°47’E |
May 2017 |
34 605 |
31.8 |
62 |
This study |
Fig. 1.
Genomic map of nine HTVC010P-type pelagiphage isolates. Open reading frames (ORFs) are represented by arrows and the left or right depending on the transcription orientation. The number in the arrow indicates the number of ORFs. ORFs annotated with known functions are marked with different colors according to their functions. The asterisk represents the coregene. The red asterisk represents the core genes used for phylogenomic analysis. Shadows indicate similarities between homologous genes.
A search was performed to retrieve HTVC010P-type MVGs from environmental metagenomes. We obtained a total of 1447 HTVC010P-type MVGs (≥5 kb), 77 of which have a genome size larger than 32 kb (nearly full-length or completed genomes) (see Tables S2 and S3). It is noteworthy that, due to the high quality of ALOHA AFVGs and MedDCM metagenomic fosmid contigs, we were able to identify many nearly full-length HTVC010P-type genomes from these two datasets.
To investigate the evolutionary relationships among these HTVC010P-type pelagiphages, several analyses were performed. A total of seven core genes were used for phylogenomic tree reconstruction. This analysis included 8 HTVC010P-type pelagiphage isolates, SAR11 prophage PNP1 and 77 ≥32 kb HTVC010P-type MVGs. Based on this analysis, HTVC010P-type pelagiphages were clustered into two major subgroups (I and II) (Fig. 2), with subgroup I containing 8 HTVC010P-type isolates, Western English Channel viral metagenome contig Tig404 and 30 HTVC010P-type MVGs, and subgroup II containing HTVC028P, PNP1 and the remaining 47 HTVC010P-type MVGs. Subgroup II can be further divided into subgroup II-a and subgroup II-b. In addition, AAI-based phylogeny (Fig. S2) and whole-genome based phylogeny (Fig. S3) also show a similar topology, verifying the reliability of the phylogenomic clustering result.
Fig. 2.
Maximum-likelihood phylogenomic trees of HTVC010P-type pelagiphages and metagenomic viral genomes (MVGs) showing the two subgroups (I and II) defined in this study. The HTVC010P-type pelagiphage isolates and prophage PNP1 are shown in red. Genomes that contain an integrase gene are indicated by red triangles. The predicted bacterial attachment (attB) sites are shown. Bar represents 0.10 substitutions per site.
Analysis of genome structure and gene content of HTVC010P-type pelagiphages
Whole-genome alignment shows that all HTVC010P-type genomes can be roughly separated into early genomic and late genomic regions, with ORFs encoded on the opposite strands (Fig. 1). The early genomic regions contain genes that are mostly associated with DNA metabolism and replication, including genes encoding nuclease and chromosomal replication initiation protein (DnaA) (Fig. 1). Neither RNA polymerase nor DNA polymerase was identified in any of these HTVC010P-type genomes, suggesting that they are highly dependent on their host’s machinery for transcription and DNA replication. The only DNA metabolism gene found common to all genomes is a putative exodeoxyribonuclease gene that is likely involved in degrading bacterial DNA and phage genetic recombination. The gene encoding the DNA binding domain of the chromosomal replication initiator protein DnaA (PF08299) was identified in the majority of HTVC010P-type genomes. DnaA is a protein that activates the initiation of DNA replication in bacteria [48]. The Escherichia coli dnaA gene plays a role in the replication of bacteriophage λ [49]. In HTVC010P-type pelagiphages, this protein may be involved in the initiation and regulation of phage DNA replication.
The late genomic region in all HTVC010P-type genomes contain a set of conserved structural and packaging genes (Fig. 1). ORFs encoding the capsid, head-tail connected protein, internal virion protein, tail proteins and tail fibre were detected in this region. The majority of these genes show very weak sequence identity to other known phage structural genes. The terminase large subunit-encoding gene (terL) was identified in all HTVC010P-type pelagiphages, with the closest known homologues found in Rhodospirillaceae bacterium SYSU D60014 (49–51 % amino acid identity).
HTVC028P, PNP1 and all but one HTVC010P-type MVGs in subgroup II contain an additional lysogeny control module located in the early region (Fig. 1), which includes an integrase gene (int) and a putative transcriptional regulator gene. Phage-encoded integrase is the key enzyme that catalyses the site-specific recombination between the phage genome and bacterial chromosome [50]. Therefore, subgroup II HTVC010P-type pelagiphages are likely to be able to integrate their genome into the host genome. In contrast, the int gene is not present in any subgroup I HTVC010P-type genomes, suggesting that they may have an obligate lytic lifecycle. Most subgroup II genomes contain a helix-turn-helix motif, showing weak sequence identity to the transcriptional regulator Xre in Bacillus subtilis prophage PBSX (36.07–36.51 % amino acid identity). Xre is necessary for the maintenance of the lysogenic state of prophages [51]. It is likely that this transcriptional regulator is also important for the maintenance of integrated phage in the SAR11 chromosome. With respect to cell lysis, a gene encoding peptidase M5 (Peptidase_M15_3, PF08291) was identified in almost all HTVC010P-type pelagiphage genomes, located upstream of the phage structural genes (Fig. 1).
Other identifiable functional genes
Several additional functional genes were identified in HTVC010P-type genomes, which may be involved in other cellular and metabolic functions. All HTVC010P-type isolates encode a putative GCN5-related N-acetyltransferase (GNAT) with distant homology to other bacteria GNAT proteins. GNAT proteins are responsible for the acetylation of various substrates; thus, playing a role in diverse cellular processes [52]. The GNAT-encoding gene has been identified in marine RCA phages and has been suggested to play an important role in regulating host metabolism [14].
HTVC100P and several HTVC010P-type MVGs contain a gene encoding DNA N-6-adenine-methyltransferase, the homologues of which were also found in many other bacteriophage genomes [14, 53]. This enzyme catalyses DNA methylation at the N6 position of adenine. It may be involved in phage genome protection against cleavage by host restriction-modification systems [54].
A gene encoding phosphoadenosine phosphosulfate reductase (CysH, PF01507) was identified in the HTVC034P genome and several HTVC010P-type MVGs. CysH protein is involved in step 3 of the subpathway that synthesizes sulfite from sulfate [55]. The presence of the cysH gene suggests that HTVC034P is probably involved in sulfur cycling. Interestingly, all known SAR11 genomes, including the host of HTVC034P, lack the cysH gene and other genes involved in assimilatory sulfate reduction [56]. Therefore, this cysH gene was possibly transferred from other bacteria or phages. The function of the cysH gene during phage infection is still unclear.
Identifying the HTVC028P integration sites
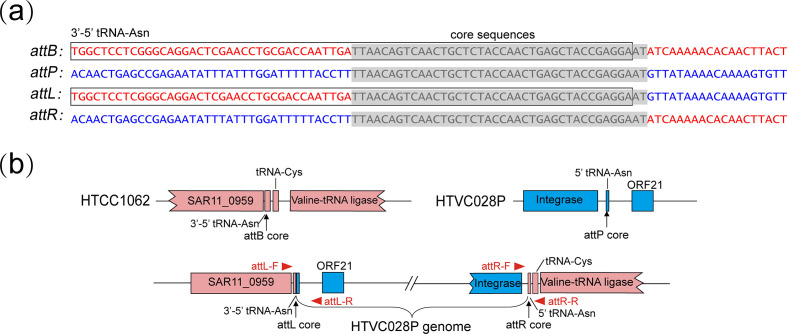
All but one subgroup II HTVC010P-type pelagiphages possess an int gene, suggesting that they may also have a lysogenic life cycle. The integration sites of isolate HTVC028P in subgroup II were predicted by sequence comparison and confirmed by experiment. Sequence analysis identified a hypothetical phage attachment site attP site located within the intergenic region downstream of the int gene in HTVC028P, which contains a 40 bp core sequence identical to the 5′ end of the tRNA-Asn site in the HTCC1062 genome (Fig. 3a). Direct evidence for phage integration can be obtained by PCR amplification and high-throughput sequencing of the phage-infected cells [19]. We verified the site-specific integration of HTVC028P into the HTCC1062 tRNA-Asn site by PCR amplification of the hybrid left and right integration sites (attL and attR) (Fig. S1). Analysis of the sequences of the integration sites revealed that upon HTVC028P integration, the HTCC1062 tRNA-Asn gene is disrupted and can be complemented by the identical core sequence in HTVC028P (Fig. 3a, b). Consequently, this reconstituted tRNA-Asn did not show any alteration. This result suggests that HTVC028P is a temperate phage with the potential to establish a lysogenic relationship with HTCC1062. The HTVC010P-type prophage PNP1 has been reported to use tRNA-Met as an integration site [18]. These results suggest that these HTVC010P-type phage-encoded int genes are functional in catalysing site-specific phage integration.
Fig. 3.
(a) Alignment of DNA sequences around HTVC028P and HTCC1062 integration sites. The HTCC1062 genomic sequences and HTVC028P sequences are shown in red and blue, respectively. The identical core sequences are indicated with light gray boxes. The tRNA genes found in the integration sites are boxed. (b) Gene map of HTVC028P and HTCC1062 integration sites. Host and phage genes are shown in pink and blue, respectively. The positions of PCR primers are indicated by red arrows.
Prevalence of lysogenic life strategy in HTVC010P-type MVGs
Among all the HTVC010P-type MVGs (≥5 kb), 94 contain an int gene. All of the HTVC010P-type MVGs encoded int genes belonging to the tyrosine integrase family. The phylogeny of these int genes reveals at least four divergent subgroups (Fig. S4). Nineteen HTVC010P-type MVGs that contain an int gene were recovered from the MedDCM fosmid metagenome [21]. Many of these sequences were previously classified as pelagiphages, because they contain an int gene and a sequence identical to SAR11 tRNA [21]. The remaining 75 int-containing MVGs were recovered from GOV, GOV 2.0 and the ALOHA AFVGs. In order to identify putative integration sites of int-containing HTVC010P-type MVGs, sequence comparisons between HTVC010P-type pelagiphages and SAR11 genomes were performed, which revealed a common 14–43 bp core sequence in 86 of 94 MVGs. In HTVC010P-type MVGs, these core sequences are all located within the intergenic region in the vicinity of the int gene (Fig. 4). Eight MVGs lack the identified core sequences because their int genes are all located by the end of sequences; thus, the region containing the core sequence is possibly not covered. In the SAR11 genomes, the core sequences are located within various tRNA genes. A total of eight different SAR11 tRNA genes were identified as putative bacterial attachment sites (attB) for HTVC010P-type pelagiphages (see Table S4, Fig. 2). tRNA genes are highly preferred phage integration targets in a wide variety of bacteria [57]. All current known temperate pelagiphages use the SAR11 tRNA genes as integration sites [19, 20], and approximately half of the available SAR11 tRNA genes have been identified as phage integration sites. A GOS search was performed to obtain phage–host hybrid sequences, resulting in a total of 10 SAR11-pelagiphage hybrid fragments. These GOS fragments all contain homologues of a SAR11 gene, a tRNA gene and a HTVC010P-type phage gene, providing evidence for the in situ HTVC010P-type pelagiphage integration (Fig. 5). As Fig. 2 shows, all int-containing HTVC010P-type phages belong to subgroup II. It is notable that most of the phages located in the same branch have similar integrase genes and use the same tRNA genes as putative integration sites (Figs 2 and S4), suggesting that closely related phages share a highly conserved site-specific recombination system.
Fig. 4.
Gene map and the putative attP site of representative HTVC010-type MVGs. The genes are coloured according to the degree of amino acid sequence identity to the genes in HTVC028P. The red asterisks indicate the integrase genes. Core sequences that are identical to SAR11 tRNA genes are shaded.
Fig. 5.
SAR11-pelagiphage hybrid sequences identified from the GOS database. Host and phage genes are shown in pink and blue, respectively.
Lysogeny in a HTVC010P-type pelagiphages has been reported in a published study [18] and was further investigated in this study. The results of this study suggest that both the lytic and lysogenic phage predation strategies are prevalent in HTVC010P-type pelagiphages. Different types of bacteria–phage interactions have different impacts on host physiology and evolution. Lytic phage infection causes cell death and can serve as selective pressure on bacterial evolution, resulting in increased genetic divergence. During lysogenic infection, integrated phages can provide hosts with super-infection immunity and novel biological functions, and can also mediate horizontal gene transfer through transduction and lysogenic conversion [58]. The establishment of lysogeny can also protect phages from unfavourable environments [11]. Bacteriophage integration frequently occurs in marine bacteria genomes. It has been estimated that approximately half of sequenced marine bacterial genomes harbour prophage-like elements [11]. A previous study suggested that prophages are more frequently found in bacteria with small doubling times, fast growth rates and large genome sizes [59]. The genomes of SAR11 bacteria are streamlined, and only two prophages have been identified from sequenced SAR11 genomes so far [20]. In contrast, our present study and previous studies suggest that many pelagiphages can proceed to lysogenize the host; likewise, integrase genes have been identified in some marine cyanophage genomes [60–63]. In addition, prophage relics were found in some marine cyanobacteria genomes [64–66]. Together, these results suggest that lysogenic infections also widely occur in streamlined-genome marine bacteria and play a non-negligible role in shaping bacterial genomes. However, we were not able to compare the effect of different HTVC010P-type subgroups on the growth of host cells, as contamination persistently existed in some pelagiphage cultures. The potential advantages and ecological implications of HTVC010P-type phage integration warrant further evaluation.
Global distribution patterns of HTVC010P-type pelagiphages
Viromic read recruitment at the species level (≥95 % nucleotide identity) reveals that these HTVC010P-type pelagiphages were predominantly recruited from epipelagic viromes (0–200 m) (Fig. 6) and were not detected from deep ocean viromes (1000–4000 m) (data not shown). Overall, most subgroup I HTVC010P-type pelagiphages exhibited significantly higher relative abundance than subgroup II pelagiphages in most analysed datasets (P value <0.01, Mann–Whitney U test). It is noteworthy that the HTVC010P-type pelagiphages located at the same branch with HTVC010P and HTVC034P were the most abundant HTVC010P-type phages in most analysed viromes (Fig. 6). In addition, they were more widely distributed in the global ocean, from tropical and temperate regions to polar regions. Based on these observations, we hypothesize that these evolutionary closely related pelagiphages may have evolved specific attributes to have greater adaptability or wider host ranges; thus, dominating the HTVC010P-type phage group.
Fig. 6.
Heatmap displaying the relative abundance of each HTVC010P-type pelagiphage genome in different marine viromic datasets. Normalized relative abundance is depicted as total recruited nucleotides (kb) per kb of genome per gigabase of metagenome (KPKG). Pelagiphages located at the same branch with HTVC010P and HTVC034P are boxed in the phylogenomic tree. DCM, Deep chlorophyll maximum; MESO, mesopelagic; SRF, surface; MIX, bottom of mixed layer; NA, not available.
It is interesting that infection strategies of HTVC010P-type phages correlate with evolution and distribution patterns. The possession and inheritance of host integration function could be important to the survival of subgroup II HTVC010P-type phages in some respects. However, metagenomic recruitment reveals that lytic life strategy HTVC010P-type pelagiphages exhibited significantly higher relative abundance than pelagiphages with a lysogenic strategy. Lysogeny seems to make no significant contribution to the phage abundance. Although lysogeny can protect phages from unfavourable environments and, thus, enhance the survival of phages [2], the lower abundance subgroup II HTVC010P-type pelagiphages suggests that carrying an integrase gene may not increase the ecological fitness of HTVC010P-type pelagiphages.
Conclusions
In this study, cultivation-dependent and -independent approaches were combined to investigate the diversity, infection strategies, evolution and distribution patterns of HTVC010P-type pelagiphages. Our study proved that there are both strictly lytic and potential lysogenic HTVC010P-type pelagiphages. In addition, we found that infection strategies correlate with evolution and distribution patterns. Finally, our study raises questions for future studies on pelagiphage lysogeny features and the potential influence of prophages on SAR11 diversification and adaption. The SAR11-pelagiphages analysed in this study can serve as model systems for the study of phage ecological roles and applications, and phage–host interactions.
Supplementary Data
Funding information
This research was funded by National Natural Science Foundation of China, grant numbers 41 706 173 and 42 076 105.
Author contributions
Conceptualization, S. D. and Y. Z.; methodology, S. D. and F. Q.; software, Z. Z.; validation, F. Q., S. D. and Z. T.; formal analysis, S. D. and Z. Z; investigation, S. D., F. Q. and X. L; resources, Z. T., M. Y. and Q. X.; data curation, S. D. and G. Z.; writing – original draft preparation, S. D.; writing – review and editing, Y. Z.; visualization, S. D.; supervision, Y. Z.; project administration, Y. Z.; funding acquisition, Y. Z. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AAI, average amino acid identity; AFVG, assembly-free virus genome; GOS, Global Ocean Survey; GOV, Global Ocean Viromes; MVG, metagenomic viral genome; NCBI, National Center for Biotechnology Information.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary figures and four supplementary tables are available with the online version of this article.
References
- 1.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 2.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses – major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 5.Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the Sea. Bioscience. 1999;49:781–788. [Google Scholar]
- 6.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, et al. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 7.Correa AMS, Howard-Varona C, Coy SR, Buchan A, Sullivan MB, et al. Revisiting the rules of life for viruses of microorganisms. Nat Rev Microbiol. 2021 doi: 10.1038/s41579-021-00530-x. [DOI] [PubMed] [Google Scholar]
- 8.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Sime-Ngando T. Environmental bacteriophages: viruses of microbes in aquatic ecosystems. Front Microbiol. 2014;5:355. doi: 10.3389/fmicb.2014.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–1520. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas. ISME J. 2008;2:579–589. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, et al. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 13.Kang I, Oh HM, Kang D, Cho JC. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci U S A. 2013;110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Chen F, Chu X, Zhang H, Luo H, et al. Diverse, abundant, and novel viruses infecting the marine roseobacter RCA lineage. mSystems. 2019;4:e00494–19. doi: 10.1128/mSystems.00494-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Qin F, Chen F, Chu X, Luo H, et al. Culturing novel and abundant pelagiphages in the ocean. Environ Microbiol. 2021;23:1145–1161. doi: 10.1111/1462-2920.15272. [DOI] [PubMed] [Google Scholar]
- 16.Buchholz HH, Michelsen ML, Bolaños LM, Browne E, Allen MJ, et al. Efficient dilution-to-extinction isolation of novel virus-host model systems for fastidious heterotrophic bacteria. ISME J. 2021 doi: 10.1038/s41396-020-00872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 18.Giovannoni SJ. SAR11 bacteria: the most abundant plankton in the oceans. Ann Rev Mar Sci. 2017;9:231–255. doi: 10.1146/annurev-marine-010814-015934. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Qin F, Zhang R, Giovannoni SJ, Zhang Z, et al. Pelagiphages in the Podoviridae family integrate into host genomes. Environ Microbiol. 2019;21:1989–2001. doi: 10.1111/1462-2920.14487. [DOI] [PubMed] [Google Scholar]
- 20.Morris RM, Cain KR, Hvorecny KL, Kollman JM. Lysogenic host-virus interactions in SAR11 marine bacteria. Nat Microbiol. 2020;5:1011–1015. doi: 10.1038/s41564-020-0725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roux S, Brum JR, Dutilh BE, Sunagawa S, Duhaime MB, et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 23.Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019;177:1109–1123. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaulaurier J, Luo E, Eppley JM, Uyl PD, Dai X, et al. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 2020;30:437–446. doi: 10.1101/gr.251686.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaragoza-Solas A, Rodriguez-Valera F, López-Pérez M. Metagenome mining reveals hidden genomic diversity of pelagimyophages in aquatic environments. mSystems. 2020;5:00919–e00905. doi: 10.1128/mSystems.00905-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Hernandez F, Fornas O, Lluesma Gomez M, Bolduc B, de la Cruz Peña MJ, et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat Commun. 2017;8:15892. doi: 10.1038/ncomms15892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Hernandez F, Garcia-Heredia I, Lluesma Gomez M, Maestre-Carballa L, Martínez Martínez J, et al. Droplet digital PCR for estimating absolute abundances of widespread pelagibacter viruses. Front Microbiol. 2019;10:1226. doi: 10.3389/fmicb.2019.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eggleston EM, Hewson I. Abundance of two pelagibacter ubique bacteriophage genotypes along a latitudinal transect in the North and South Atlantic oceans. Front Microbiol. 2016;7:1534. doi: 10.3389/fmicb.2016.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warwick-Dugdale J, Solonenko N, Moore K, Chittick L, Gregory AC, et al. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ. 2019;7:e6800. doi: 10.7717/peerj.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L-X, Zhao Y, McMahon KD, Mori JF, Jessen GL, et al. Wide distribution of phage that infect freshwater SAR11 bacteria. mSystems. 2019;4:e00410–19. doi: 10.1128/mSystems.00410-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph “Candidatus Pelagibacter ubique” HTCC1062 on a defined medium. ISME J. 2013;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connon SA, Giovannoni SJ. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/aem.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Lukashin AV, Borodovsky M. Genemark.Hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zdobnov EM, Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 38.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Söding J, Biegert A, Lupas AN. The Hhpred Interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, et al. Using Orthomcl to assign proteins to orthomcl-db groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;35:6–12. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol. 2008;159:406–414. doi: 10.1016/j.resmic.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 45.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meier-Kolthoff JP, Goker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 48.Fuller RS, Funnell BE, Kornberg A. The dnAA protein complex with the E. Coli chromosomal replication origin (ORIC) and other DNA sites. Cell. 1984;3:889–990. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 49.Wegrzyn G, Szalewska-Pałasz A, Wegrzyn A, Obuchowski M, Taylor KJG. Transcriptional activation of the origin of coliphage λ DNA replication is regulated by the host DnaA initiator function. Gene. 1995;154:47–50. doi: 10.1016/0378-1119(94)00849-n. [DOI] [PubMed] [Google Scholar]
- 50.Fogg PC, Colloms S, Rosser S, Stark M, Smith MC. New applications for phage integrases. J Mol Biol. 2014;426:2703–2716. doi: 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonnell GE, McConnell DJ. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J Bacteriol. 1994;176:5831–5834. doi: 10.1128/jb.176.18.5831-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salah Ud-Din AI, Tikhomirova A, Roujeinikova A. Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT. Int J Mol Sci. 2016;17:1018. doi: 10.3390/ijms17071018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, et al. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc Natl Acad Sci USA. 2012;109:2037–2042. doi: 10.1073/pnas.1115467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murphy J, Mahony J, Ainsworth S, Nauta A, Van Sinderen D. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol. 2013;79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krone FA, Westphal G, Schwenn JD. Characterisation of the gene cysH and of its product phospho-adenylylsulphate reductase from Escherichia coli . Mol Gen Genet. 1991;225:314–319. doi: 10.1007/BF00269864. [DOI] [PubMed] [Google Scholar]
- 56.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, et al. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 57.Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, et al. Lytic to temperate switching of viral communities. Nature. 2016;531:466–470. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 59.Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–2754. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J Mol Biol. 2007;368:966–981. doi: 10.1016/j.jmb.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labrie SJ, Frois-Moniz K, Osburne MS, Kelly L, Roggensack SE, et al. Genomes of marine cyanopodoviruses reveal multiple origins of diversity. Environ Microbiol. 2013;15:1356–1376. doi: 10.1111/1462-2920.12053. [DOI] [PubMed] [Google Scholar]
- 63.Huang S, Zhang S, Jiao N, Chen F. Comparative genomic and phylogenomic analyses reveal a conserved core genome shared by estuarine and oceanic cyanopodoviruses. PLoS One. 2015;10:11. doi: 10.1371/journal.pone.0142962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, et al. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial “mobilome”. Environ Microbiol. 2009;11:2935–2951. doi: 10.1111/j.1462-2920.2009.02081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, et al. Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J. 2013;7:184–198. doi: 10.1038/ismej.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flores‐Uribe J, Philosof A, Sharon I, Fridman S, Larom S, et al. A novel uncultured marine cyanophage lineage with lysogenic potential linked to a putative marine Synechococcus ‘relic’ prophage. Environ Microbiol Rep. 2019;11:598–604. doi: 10.1111/1758-2229.12773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.