Abstract
Whole-genome sequencing (WGS) is becoming the de facto standard for bacterial typing and outbreak surveillance of resistant bacterial pathogens. However, interoperability for WGS of bacterial outbreaks is poorly understood. We hypothesized that harmonization of WGS for outbreak surveillance is achievable through the use of identical protocols for both data generation and data analysis. A set of 30 bacterial isolates, comprising of various species belonging to the Enterobacteriaceae family and Enterococcus genera, were selected and sequenced using the same protocol on the Illumina MiSeq platform in each individual centre. All generated sequencing data were analysed by one centre using BioNumerics (6.7.3) for (i) genotyping origin of replications and antimicrobial resistance genes, (ii) core-genome multi-locus sequence typing (cgMLST) for Escherichia coli and Klebsiella pneumoniae and whole-genome multi-locus sequencing typing (wgMLST) for all species. Additionally, a split k-mer analysis was performed to determine the number of SNPs between samples. A precision of 99.0% and an accuracy of 99.2% was achieved for genotyping. Based on cgMLST, a discrepant allele was called only in 2/27 and 3/15 comparisons between two genomes, for E. coli and K. pneumoniae, respectively. Based on wgMLST, the number of discrepant alleles ranged from 0 to 7 (average 1.6). For SNPs, this ranged from 0 to 11 SNPs (average 3.4). Furthermore, we demonstrate that using different de novo assemblers to analyse the same dataset introduces up to 150 SNPs, which surpasses most thresholds for bacterial outbreaks. This shows the importance of harmonization of data-processing surveillance of bacterial outbreaks. In summary, multi-centre WGS for bacterial surveillance is achievable, but only if protocols are harmonized.
Keywords: bacterial typing, harmonisation, whole genome sequencing, ring-trial, nosocomial pathogens, antimicrobial resistance
Data Summary
The authors confirm that all supporting data, code and protocols have been provided within the article. All raw sequencing data were deposited at EBI-ENA under BioProject PRJEB40571.
Impact Statement.
Whole-genome sequencing (WGS) for typing bacterial outbreaks has surged in recent years. We performed an inter-laboratory ring-trial by sending out 30 bacterial isolates to assess the reproducibility of WGS. We demonstrated that the use of different de novo assemblers for a single outbreak analysis will lead to bacterial isolates being misclassified as not related to the outbreak cluster. Additionally, we show that implementing WGS for regional or (inter)national surveillance of bacterial pathogens is feasible if identical laboratory procedures and data analysis workflows are used.
Introduction
The dissemination of antimicrobial resistance (AMR) has grown to an issue of worldwide proportions. Routine surveillance by molecular typing can aid in the fight against AMR, as outlined by the global action plan of the World Health Organization [1]. ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) are of major interest as they are the leading cause of hospital-related infections and outbreaks. Furthermore, reports show that the number of infections by resistant micro-organisms have been on the rise in recent years. Infections by multi-drug-resistant (MDR) bacteria are associated with an increase in economic burden [2] and negative patient outcomes such as morbidity and mortality [3, 4].
To determine the spread of resistance and of resistant microbes, different molecular typing methods are being applied. Older, established typing methods for outbreak surveillance, such as pulsed field gel electrophoresis (PFGE), amplification fragment length polymorphism (AFLP), multi-locus sequencing typing (MLST) and multi-locus variable-number tandem repeat analysis (MLVA) are slowly being replaced by whole-genome sequencing (WGS). The introduction of WGS to the field of bacterial typing and spread of AMR has set a new standard for discriminatory power and accuracy, as it encompasses a comprehensive view of the bacterial core and accessory genome. This gives rise to the possibility to determine clonal relatedness in a more discriminatory fashion, and at the same time provide data on resistance genes, plasmids and virulence-potential, which would otherwise require a combination of other methods [5–8]. Current methods to determine phylogeny are based on core-/whole-genome multi-locus sequence typing (cgMLST, wgMLST) [9, 10] or SNPs [11–13].
Approaches like cgMLST and wgMLST determine the phylogeny among bacterial isolates based on differences in allelic profile in either the core genome or the entire genome, respectively. All coding sequences (CDS) or loci are identified using tools such as Prodigal [14]. Then, all variants of each locus are assigned a unique allele number and the complete set of allele numbers is called the allelic profile. The genetic distance is calculated by counting the number of discrepant alleles between two isolates. A relative genetic distance can also be calculated by dividing the number of discrepant alleles by the number of alleles that were compared. Next to commercial packages for cgMLST and wgMLST analyses, such as BioNumerics or SeqSphere, open source options are available as well, such as ChewBBACA [10] and Enterobase [15].
Inferring phylogeny based on SNPs can be performed by three different methods. (i) Alignment to a reference genome (Snippy [11]). (ii) (Core-) genome alignment (MAUVE [16] or Harvest Suite [17]). (iii) Alignment-free methods based on using the entire collection of subsequences of a sequence of length k: k-mer (kSNP [18] or SKA [13]).
Currently, only a few studies have described clonal-cluster thresholds definitions using cgMLST, wgMLST or SNP-based methods. Generally, these studies determine the thresholds based on either (i) previous or ongoing bacterial outbreaks in hospitals and in the food production chain, or (ii) by means of follow-up on human carriers of these pathogens over time. Furthermore, most of these studies only describe single-clone outbreaks, which can hamper the interpretation when these thresholds are applied to different lineages of a specific species. Some clinically relevant lineages might be more clonal than others, and so require different thresholds. One of the first reports on the use of WGS for bacterial outbreak analysis were on methicillin-resistant Staphylococcus aureus (MRSA) in 2013, in a neonatal intensive care unit. Next to standard assessment of epidemiological data and antibiograms, WGS was performed to resolve this putative outbreak [19]. In that study, a maximum of 20 SNPs was observed among the MRSA isolates found in the outbreak. For the foodborne pathogen E. coli O157:H7, the Public Health Agency Canada evaluated WGS for outbreak detection [20]. To this end, they retrospectively performed WGS for 250 isolates, from eight different outbreaks and analysed using wgMLST and SNP analyses. These 250 isolates were previously typed using MLVA or PFGE. WGS-based typing was in excellent concordance with MLVA and PFGE and also had higher discriminatory power to resolve outbreak clusters. Additionally, they reported that all isolates for each outbreak fell within a cutoff of 5 SNPs or 10 allele differences (on wgMLST basis). In their review, Schürch et al. suggested various clonal-cluster thresholds based on wgMLST or SNP analyses for a few common bacterial pathogens in outbreak situations [9].
Kluytmans-Van Den Bergh et al. recently determined clonal-cutoffs based on cgMLST and wgMLST for four extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-E): E. coli, K. pneumoniae, Citrobacter species and Enterobacter sp. [21]. In their study, isolates were classified as epidemiologically linked when these were cultured from a single patient in a 30 day time window and when they belonged to the same seven-gene sequence type. Subsequently, the genetic distance (here defined as number of discrepant alleles divided by the number of alleles compared) was compared among all isolates, and clonal thresholds were determined by the lowest genetic distance possible that included all epidemiologically linked isolates.
The goal of the i-4-1-Health study is to assess the prevalence and spread of resistant bacteria among humans and animals in the Dutch-Belgian border [22]. Across a 1 year period, we screened patients in hospitals and in long-term healthcare facilities, infants at day-care facilities, and broilers and weaned pigs for gut or rectal carriage of ESBL-producing, ciprofloxacin-resistant or carbapenemase-producing Enterobacteriaceae and vancomycin-resistant Enterococci. This One-Health approach could provide insights into the prevalence and spread of resistant bacteria between and within these separate domains. In the i-4–1-Health study, WGS data was generated in three independent locations, and thus inter-laboratory reproducibility needed to be assessed to allow the comparison of this data. To standardize the WGS results and interpretation, we made efforts to harmonize the WGS protocols, both for the wet-lab procedures and the bioinformatics analysis.
Here, we harmonized the inter-laboratory reproducibility of WGS for outbreak surveillance and genotyping of AMR and origin of replication (ORI) of plasmids for a selection of AMR bacteria frequently encountered in hospital-related infections and AMR surveillance within the i-4-1-Health project. As the implementation of WGS for routine outbreak surveillance is particularly dependent on standardized methodology, we evaluated the technical variation in phylogenetic comparison using a commercially available wgMLST tool in BioNumerics and an open-source reference-free SNP-based tool called SKA [13].
Methods
Selection of isolates
In total 30 resistant bacterial isolates were selected based on their extended-spectrum beta-lactamase (ESBL) or carbapenemase activity, or based on ciprofloxacin or vancomycin resistance phenotype. The complete collection of isolates consisted of nine E. coli; five K. pneumonia; four Citrobacter sp.; four Enterobacter sp.; two Klebsiella oxytoca; two Klebsiella aerogenes; two Enterococcus faecalis and two E. faecium. Six isolates (two E. coli, two K. pneumoniae and two Enterobacter sp.) were collected previously [21] and kindly provided by the SoM study-group, and 20 isolates were collected during the i-4-1-Health study [22]. The E. faecium and E. faecalis isolates were from a previous collection, stored at Antwerp University. The isolates were collected from perianal swabs of hospitalized patients (21) and clients in nursing homes (6), and from faeces from broilers (2) and weaned pigs (1) by selective culturing. The culturing methods are described elsewhere [21, 22]. An overview of isolates and their origin is available in Table 1. Isolates were inoculated from −80 °C on Mueller–Hinton II agar (BD, Franklin Lakes, NJ, USA) and sent to the participating institutes. The 30 isolates were divided in three sets of ten isolates. Each set was sequenced once by each centre, with a 6 month interval between each set.
Table 1.
Metadata of all isolates used in this study
|
Name |
Species |
Origin |
Study |
County |
Accession no. centre 1 |
Accession no. centre 2 |
Accession no. centre 3 |
|---|---|---|---|---|---|---|---|
|
Citrobacter sp. 1 |
Citrobacter sp. |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219870 |
ERS5219871 |
ERS5219872 |
|
Citrobacter sp. 2 |
Citrobacter sp. |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219873 |
ERS5219874 |
ERS5219875 |
|
Citrobacter sp. 3 |
Citrobacter sp. |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219876 |
ERS5219877 |
ERS5219878 |
|
Citrobacter sp. 4 |
Citrobacter sp. |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219879 |
ERS5219880 |
ERS5219881 |
|
Enterobacter sp. 1 |
Enterobacter sp. |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219882 |
ERS5219883 |
ERS5219884 |
|
Enterobacter sp. 2 |
Enterobacter sp. |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219885 |
ERS5219886 |
ERS5219887 |
|
Enterobacter sp. 3 |
Enterobacter sp. |
Hospital |
SoM |
Netherlands |
ERS5219888 |
ERS5219889 |
ERS5219890 |
|
Enterobacter sp. 4 |
Enterobacter sp. |
Hospital |
SoM |
Netherlands |
ERS5219891 |
ERS5219892 |
ERS5219893 |
|
E. coli 1 |
Hospital |
SoM |
Netherlands |
ERS5219828 |
ERS5219829 |
ERS5219830 |
|
|
E. coli 2 |
Hospital |
SoM |
Netherlands |
ERS5219831 |
ERS5219832 |
ERS5219833 |
|
|
E. coli 3 |
Hospital |
i-4-1-health |
Netherlands |
ERS5219834 |
ERS5219835 |
ERS5219836 |
|
|
E. coli 4 |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219837 |
ERS5219838 |
ERS5219839 |
|
|
E. coli 5 |
Broiler |
i-4-1-Health |
Netherlands |
ERS5219840 |
ERS5219841 |
ERS5219842 |
|
|
E. coli 6 |
Weaned pig |
i-4-1-Health |
Netherlands |
ERS5219843 |
ERS5219844 |
ERS5219845 |
|
|
E. coli 7 |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219846 |
ERS5219847 |
ERS5219848 |
|
|
E. coli 8 |
Broiler |
i-4-1-Health |
Netherlands |
ERS5219849 |
ERS5219850 |
ERS5219851 |
|
|
E. coli 9 |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219852 |
ERS5219853 |
ERS5219854 |
|
|
E. faecalis 1 |
Hospital |
Belgium |
ERS5219894 |
ERS5219895 |
ERS5219896 |
||
|
E. faecalis 2 |
Hospital |
Belgium |
ERS5219897 |
ERS5219898 |
ERS5219899 |
||
|
E. faecium 1 |
Hospital |
Belgium |
ERS5219900 |
ERS5219901 |
ERS5219902 |
||
|
E. faecium 2 |
Hospital |
Belgium |
ERS5219903 |
ERS5219904 |
ERS5219905 |
||
|
K. aerogenes 1 |
Hospital |
i-4-1-Health |
Belgium |
ERS5219912 |
ERS5219913 |
ERS5219914 |
|
|
K. aerogenes 2 |
Hospital |
i-4-1-Health |
Belgium |
ERS5219915 |
ERS5219916 |
ERS5219917 |
|
|
K. oxytoca 1 |
Hospital |
i-4-1-Health |
Belgium |
ERS5219906 |
ERS5219907 |
ERS5219908 |
|
|
K. oxytoca 2 |
Hospital |
i-4-1-Health |
Netherlands |
ERS5219909 |
ERS5219910 |
ERS5219911 |
|
|
K. pneumoniae 1 |
Hospital |
i-4-1-Health |
Belgium |
ERS5219855 |
ERS5219856 |
ERS5219857 |
|
|
K. pneumoniae 2 |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219858 |
ERS5219859 |
ERS5219860 |
|
|
K. pneumoniae 3 |
Long-term healthcare facility |
i-4-1-Health |
Netherlands |
ERS5219861 |
ERS5219862 |
ERS5219863 |
|
|
K. pneumoniae 4 |
Hospital |
SoM |
Netherlands |
ERS5219864 |
ERS5219865 |
ERS5219866 |
|
|
K. pneumoniae 5 |
Hospital |
SoM |
Netherlands |
ERS5219867 |
ERS5219868 |
ERS5219869 |
DNA isolation and WGS
The DNA isolation and WGS procedure was performed as follows: DNA was extracted using the MasterPure DNA isolation kit (Lucigen) or MasterPure Gram Positive DNA purification kit (Lucigen). Sequencing libraries were prepared using NexteraXT (Illumina). Libraries were sequenced on the Illumina MiSeq platform in paired end 2×250 base-pair (bp) reads using the MiSeq V2 cartridge. Where possible, each set of isolates was subjected to WGS in a single run. Acceptance criteria for WGS were a de novo assembly with an average coverage higher than 30 and less than 1000 contigs, as reported in BioNumerics (7.6.3). Samples not fulfilling acceptance criteria were re-sequenced. The accession numbers for the raw sequencing data are available in Table 1. Analysis of the generated datasets (n=90) was performed in one institute.
cgMLST and wgMLST allele calling and genotyping
Raw sequencing reads were assembled using a custom pipeline in BioNumerics (7.6.3) employing SPAdes [23] (v3.7.0) for itsde novo assembly. From the raw reads and the de novo assembly, alleles were called for core-genome and whole-genome MLST (cgMLST/wgMLST). In BioNumerics, cgMLST schemes were only available for E. coli and K. pneumoniae consisting of 2513 and 634 fixed loci, respectively. Pairwise allelic distance was determined by counting the number of discrepant allele variants between two datasets, ignoring loci that were not present in both datasets. Resistance genes and origins of replication (ORI) were determined using blast [ 24] and two custom databases based on Resfinder [25] and PlasmidFinder [26]. AMR genes were called with a using 90% identity and 60% length cutoff. ORIs were called using 95% identity and 60% length cutoff. In total, 90 WGS datasets were generated. As no gold standard with regard to true genotype of each isolate was available, the following rules were applied: (i) if either two or three out of three datasets of an isolate had a specific genotype, this was considered as a true positive observation; (ii) if only one out of three datasets of an isolate had a specific genotype, this was considered as a false positive observation; (iii) if a different allelic variant was observed (i.e.two blaTEM-1B and one blaTEM-116) this was noted as a discrepancy and counted as a false positive.
wgSNP analysis
To determine the best de novo assembler to use for wgSNP analysis, we chose the assembler generating the least amount of pairwise SNPs (using SKA), among assemblies of the same isolate. To avoid complexity, only the E. coli dataset of this study was used. The following assemblers were used: (I) SPAdes (v3.14.0) [23], (II) SKESA (parameters:‘—use-paired_end’, v2.3.0) [27], (III) Megahit [28] (v1.2.9). All tools were used in default settings, unless otherwise specified. Additionally, the assembly-free method to determine SNPs straight from the raw reads, using ‘SKA fastq’, was also used in this comparison. The complete workflow is available at ‘https://github.com/MUMC-MEDMIC/assemblercompare’ (v1.0). SKA [13] was used to determine SNPs on a whole-genome level, using a splitk-mer length of 31. In short, pairwise SNPs were determined by generating a profile of split k-mers, in which the middle base may vary (‘SKA fasta’ for assembly- or ‘SKA fastq’ for read based SNP profiling). The number of SNPs, between two datasets, was determined by comparing the split k-mer files (‘SKA distance’). All data preprocessing for the SNP-based data analysis was performed using Snakemake [29] as the workflow manager.
Statistical analysis
Statistical analyses were done using scipy.stats module (V1.3.1) [30] and the statsmodel.api package in Python (v3.7).
Results
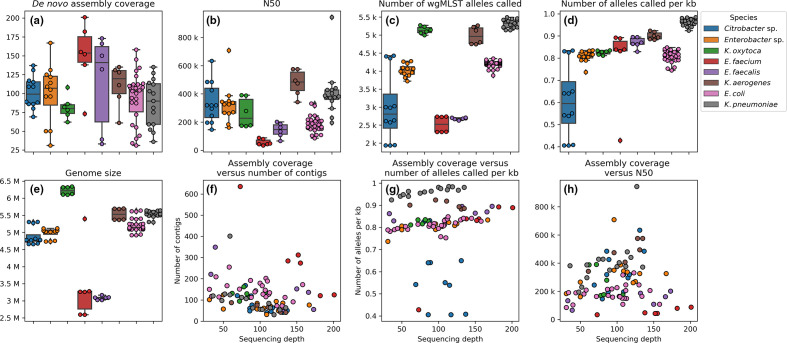
The assembly coverage, or the depth of coverage, of all isolates ranged from 30 to 203 (Fig. 1a). The N50 score, indicative for how fragmented a de novo assembly is, ranged from 33712 bp (E. faecium) to 942715 bp (K. pneumoniae) and showed clear species dependence (Fig. 1b). Assemblies of E. coli, E. faecalis and E. faecium showed a lower N50 score, indicating the difficulties of assembling such genomes (Fig. 1b). The number of contigs also varied per species, and overall had a significant negative correlation with the sequencing depth (P<0.01, Spearman rank correlation, Fig. 1f).
Fig. 1.
Distribution of various quality parameters pre and post de novo assembly. Subplots (a–e) show boxplots with interquartile (IQ) range. Whiskers range up to 1.5 times the IQ range. All single datapoints are represented as single dots. Subplots (f-h) show scatterplots of relations between two quality metrics.
The number of wgMLST alleles called ranged from 1933 (Citrobacter sp.) alleles to 5493 (K. pneumoniae, Fig. 1c). Furthermore, the average number of alleles per kilobase (kb) ranged from 0.41 to 0.98. A significant positive correlation between the normalized allele count and sequencing depth was observed (P<0.05, Spearman rank correlation Fig. 1g). Surprisingly, the Citrobacter sp. datasets seemed to showed a low coding density (range 0.41 to 0.65) compared to the median of the entire dataset (0.83). Further inspection of the Citrobacter sp. genome assemblies using blast webservice (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed 1 April 2020), showed low homology (~85% DNA identity score) to known Citrobacter sp. isolates available in the NCBI database (accessed on 1 Aril 2020, data not shown).
One dataset of E. faecium-1 had an unusually large genome size of 5.4 Mb (Fig. 1e). This dataset also had a higher number of contigs; (636, median of 274 for E. faecium, Fig. 1f ), and showed a lower number of alleles per kb (0.43, median of 0.84. Fig. 1d) compared to the other E. faecium datasets. This indicates contamination in the NGS dataset of a non- E. faecium microbe. Manual inspection of the assembly, using blast webservice, showed the presence of contigs belonging to Cutibacterium (formerly known as Propionibacterium), a skin commensal and previously described as a common contaminant of NGS datasets [31–33].
Resistance genes and plasmid ORIs
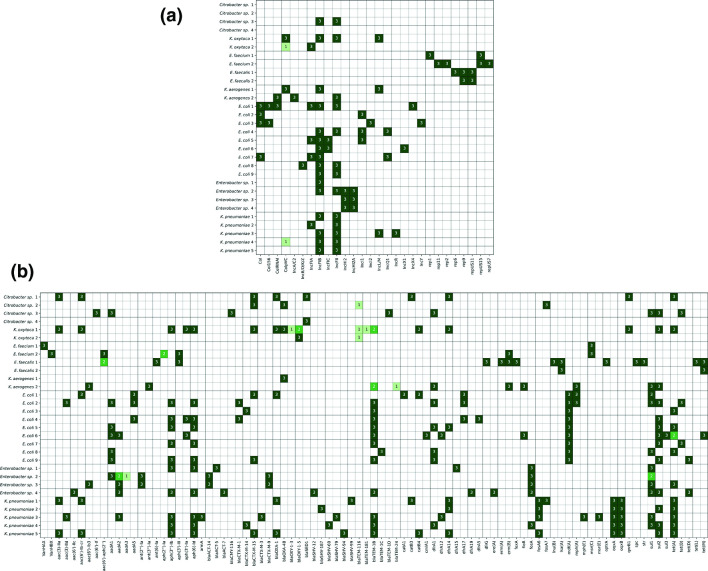
Overall, a good consensus was obtained for the genotyping of plasmid ORIs and AMR genes (Fig. 2a, b). A total of 973 AMR genes and ORIs were called with a precision of 99.0% and sensitivity of 99.2%. For four isolates, a genotype was not called in one of the datasets. The missed genotypes were for E. cloacae-2 a sul1 gene, for E. coli-6 a tet(A) gene, for E. faecalis-1 an aac(6')-aph(2') gene, and for E. faecium-2 an aph(2'')-Ia gene. For Citrobacter sp.-2 and K. oxytoca-2, a false discovery of a blaTEM-116 was observed, as this genotype was not called in either of the other two datasets of these isolates. For four isolates, a discrepant genotype was called. These discrepancies were observed for K. aerogenes-2 (blaTEM), for E. cloacae-2 (aadA), and for K. oxytoca-1 (blaOXY and blaTEM). Twice, an unexpected ColpVC was found in a K. oxytoca-2 and K. pneumoniae-4 dataset, which were from two different centres, indicating either loss of this plasmid in the other dataset of this isolate, or contamination during DNA isolation or library preparation (Fig. 2a).
Fig. 2.
(a) Heatmap of the number of genotype calls for various origins of replication among the isolates subjected to WGS. Genotype calls per locus was summed up for each centre’s isolate if this locus was detected in their dataset. (b) Heatmap of the number of genotype calls for various AMR genes, among the isolates subjected to WGS. Genotype calls per locus was summed up for each centre’s isolate if this locus was detected in their dataset.
Inter-laboratory variation in cgMLST profiles
To assess the baseline genetic variation of identical isolates when these isolates were sequenced in different sequencing institutes, we compared the cgMLST and wgMLST profiles among the isolates from the three participating institutes. Only for E. coli and K. pneumoniae cgMLST schemes were available for use in BioNumerics. On average, 2441 (97.1%) and 615 (97.1%) core-genome alleles were called for E. coli and K. pneumoniae respectively (Fig. S1, available in the online version of this article). In total, 27 and 15 pairwise allelic distances were calculated among the nine E. coli and five K. pneumoniae isolates. In 25/27 (93%) and 12/15 (80%) comparisons, a perfect concordance of cgMLST profiles was observed. If no concordance in cgMLST profiles was observed, only one allele was differently called (Fig. S2).
Inter-laboratory variation in wgMLST profiles
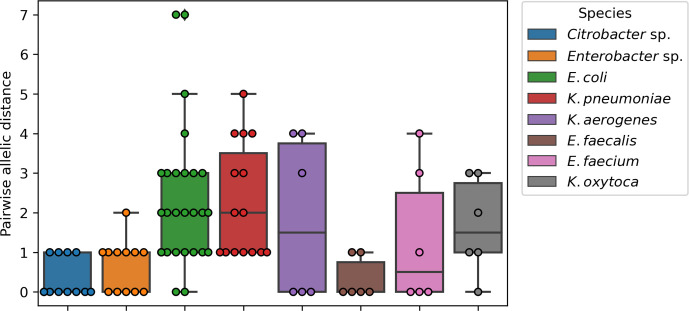
In total 90 pairwise comparisons were made for K. oxytoca (6), Citrobacter sp. (12), E. coli (27), K. pneumoniae (15), E. cloacae (12), K. aerogenes (6), E. faecalis (6) and E. faecium (6). Perfect concordance in wgMLST profiles was obtained in 26/90 (29%) comparisons (Fig. 3). In 44/90 (49%) pairwise comparisons, one or two discrepant alleles were observed. Only 23/90 (22%) comparisons showed more than two discrepant alleles, with a maximum of seven alleles different for an E. coli. For E. faecium-1 with the contamination of Cutibacterium had a perfect concordance of wgMLST profiles was observed (data not shown). This indicates the robustness of allele-based typing despite contamination with bacterial DNA from different species. For all species, an average allelic distance of 1.6 alleles (standard deviation 1.6) was observed.
Fig. 3.
Boxplot of the allelic distance based on wgMLST between the triplicates that were selected for WGS. Boxes show IQ range and whiskers range up to 1.5 times the IQ range. All single pairwise observations were plotted as dots.
For the four Citrobacter sp., a highly diverse number of wgMLST alleles were called, ranging from 1933 to 4426. The genome size did not vary strongly (mean 4.88 Mb, range 4.66 Mb to 5.31 Mb). The normalized allele counts were lower for Citrobacter sp. (mean 0.61, range 0.41 to 0.83) than in other species in this study (mean 0.84, range 0.43 to 0.98). Therefore, the variation in the number of alleles in the wgMLST scheme for Citrobacter sp. cannot be determined in this study, as an incomplete set of alleles were called.
Reference free wgSNP
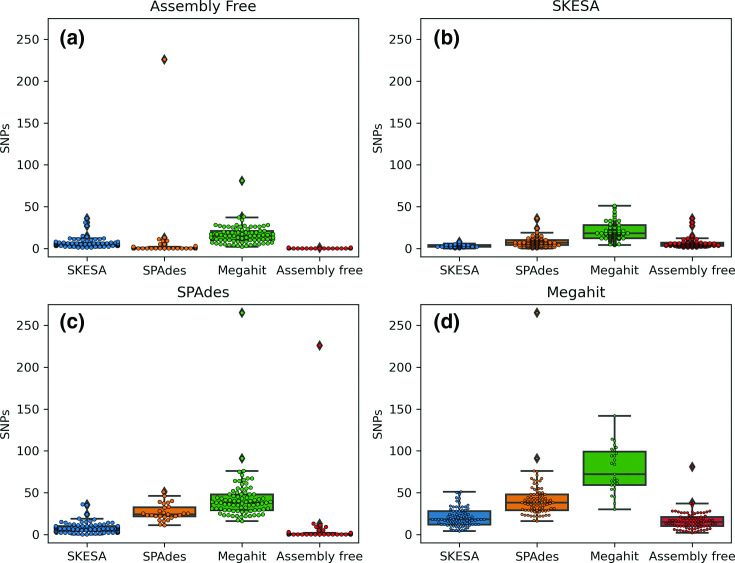
As mutations in the genome can also arise in intergenic regions (which are not taken into account in MLST-based methods), all assemblies of each isolate were screened using pairwise SNPs. First, the most optimal assembler for this task was chosen. For this, we determined the inter- and intra-assembler variation introduced on the number of pairwise SNP between two de novo assemblies. The best assembler was chosen based on the one that introduced the least number of pairwise SNPs in the datasets from the same isolates with the intra-assembler comparison. To reduce complexity, only the E. coli dataset was used. Secondly, the number of pairwise SNPs was determined for the entire dataset using the best suited assembler. Additionally, we also used the assembly-free method for determining SNPs, as in implemented by SKA.
The mean intra-assembly variation was 0.2 SNPs (assembly free), 2.7 SNPs (SKESA), 26.6 SNPs (SPAdes) and 77.8 SNPs (Megahit) (Fig. 4a–d). The mean inter-assembler variation ranged from 3.9 (assembly free compared to SPAdes) up to 43.0 SNPs (‘SPAdes to megahit’). All combinations, except the ‘assembly-free to assembly-free’ and ‘SKESA to SKESA’, revealed pairwise comparisons with over 20 SNPs for the E. coli dataset. Therefore, only these two methods were used to analyse the complete dataset.
Fig. 4.
Boxplots of the inter- and intra-assembly difference in de novo assemblies based on SNPs, using SKA for the E. coli dataset. De novo assembly method compared to is indicated above each box. (a) Assembly free, (b) SKESA, (c) SPAdes and (d) Megahit. Boxes show IQ range. Whiskers range up to 1.5 times the IQ range. All single pairwise observations were plotted as dots.
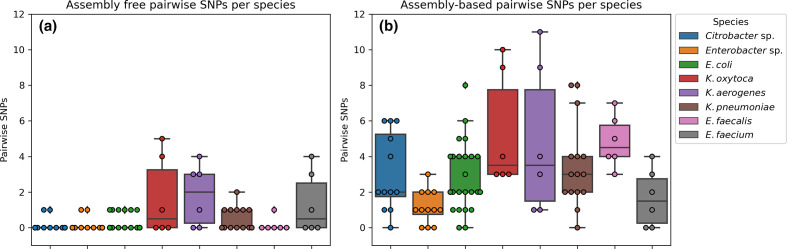
Using the assembly-free approach, 63/90 (70%) and 21/90 (21%) comparisons show zero or one pairwise SNP, respectively (Fig. 5a). Only for K. pneumoniae, E. faecium, K. oxytoca and K. aerogenes was more than one pairwise SNP observed, with a maximum of five SNPs for K. oxytoca. Using the assembly-based approach zero SNPs were observed among assemblies in 10/90 (10%) comparisons (Fig. 5b). Less than five pairwise SNPs were observed in 72/90 (80%) of the comparisons. Interestingly, in the K. aerogenes and K. oxytoca datasets, more than eight pairwise SNPs were observed. However, on wgMLST no more than four alleles' difference was observed. On average, 3.4 (standard deviation 2.6) pairwise SNPs were observed between assemblies of the same isolates (but sequenced in different institutes). Overall, more pairwise SNPs were observed when assemblies were used for SNP analysis compared to screening raw reads for SNPs. The difference in number of k-mers between the assemblyfree and assembly-based methods ranged from −2.1–1.2% (Fig. S3), indicating that a similar amount of k-mers were compared in both methods.
Fig. 5.
Boxplot of the SNP distance between the triplicates that were selected for WGS. Boxes show IQ range and whiskers range up to 1.5 times the IQ range. All single pairwise observations were plotted as dots. (a) SNP distances using the raw reads as input for SKA. (b) SNP distances based on the de novo assembly using SKESA.
Discussion
Using an inter-laboratory ring trial we evaluated the reproducibility of whole-genome sequencing for outbreak surveillance purposes. Participating institutes subjected the same set of 30 bacterial isolates of various Enterobacteriaceae and Enterococci species to whole-genome sequencing. As a first step, we assessed various QC measures and observed a slight positive trend of the sequencing depth on the normalized number of alleles called in the sequencing depth range of 30 to 207-fold. It remains unclear what sequencing depth is needed to correctly reconstruct the maximum possible number of correct alleles in the genome. Kluytmans-Van Den Bergh et al. [21] demonstrated an increase in resolution for phylogenetic reconstruction of Enterobacteriaceae if wgMLST is implemented compared to cgMLST. This would indicate that making more alleles available for comparison will improve the surveillance of outbreaks by cgMLST or wgMLST methods. Therefore, it is advisable to generate WGS data of sufficient depth to maximize the number of loci in the de novo assembly. On the other hand, deeper sequencing after a certain depth may not improve the phylogenetic signal any further, and does increase the run-time of subsequent de novo assembly.
Prokaryotes show a coding density of 1 CDS per 1 kb [34], however we observed a lower allele density. The majority of our datasets showed a lower number allele density (0.83 per kb, Fig. 1d), which could be caused by the quality-filtering step in allele calling. However, the low number of called alleles for most Citrobacter sp. may be explained by incomplete allele schemes, which do not contain the complete diversity of alleles. This indicates that the diversity of Citrobacter sp. genome assemblies present in public databases is incomplete, and our data may represent the discovery of a new antibiotic-resistant Citrobacter sp. in The Netherlands.
Genotyping AMR genes and ORIs
We next performed identification of AMR genes and plasmid ORIs. Overall, an excellent reproducibility was achieved, as a precision of 99% was obtained. Most discrepancies could be explained by the variation in the variant calling of a specific resistance gene. There was an unexplained absence of a resistance gene four times in 973 genotype calls. Although the DNA isolation method used here showed good results for the application of WGS [35], some loci could still be missed due to inefficient isolation of plasmid DNA, where these AMR genes can be located. Only twice, and in different institutes, an unexpected ColpVC ORI was found in one of the sequencing datasets, which may indicate contamination during DNA isolation or library preparation. Strauß and co-authors reported a 1.7% discordance between WGS and micro-array for the detection of resistance and virulence genes [36]. In this study, a similar reproducibility in typing resistance genes and ORIs was obtained and previously described by Kozyreva et al., which found a reproducibility rate of 99.97% [37].
Genetic variation
It is of great importance that the genetic distance between technical duplicates does not surpass commonly used thresholds to classify isolates into clusters. In this study some variation among the wgMLST allelic profiles was observed, translating to an average of 0.49 discrepant allele per 1000 alleles. Kluytmans-Van Den Bergh et al. [21] reported a variation in genetic distance based on wgMLST in a range of 0 to 0.001 (which translates to five alleles difference, based on 5000 alleles compared) for five E. coli and three K. pneumoniae, which were sequenced in duplicate [21]. This is in concordance with our study, where 88/90 comparisons differed by no more than five alleles. Additionally, clonal thresholds reported by these authors were roughly 26 and 2 alleles difference for E. coli and K. pneumoniae on cgMLST, respectively. For wgMLST this was 29, 23, 8 and 14 alleles difference for E. coli, K. pneumoniae, Citrobacter sp. and Enterobacter sp., respectively. These clonal thresholds are higher by a safe margin than the variation between any of the replicates in our study presented here. Although variation on a genetic level was observed, the level of disparity remained below other thresholds commonly employed for hospital outbreak surveillance purposes [9]. Previous work suggested a cut-off of 10 alleles for MDR E. coli and K. pneumoniae based on cgMLST [38, 39]. Therefore, it is safe to assume that if harmonized protocols are used, the technical genetic variation will remain within these previously described thresholds.
In the wgSNP analysis, all methods except for the ‘assembly-free to assembly-free’ and ‘SKESA to SKESA’ showed pairwise comparisons with more than 20 SNPs. This indicates that using SPAdes or Megahit in combination with a SNP-based method is unsuitable for outbreak surveillance, as datasets from identical isolates have more SNPs than commonly used outbreak thresholds [9], indicating that these isolates would be considered not clonally related, thus not belonging to the same outbreak. Furthermore, this also held true when comparing two assembly methods, which implies that comparing bacterial assemblies should be avoided at all costs if centres employ different methodologies to generate de novo assemblies for WGS outbreak surveillance. Potential outbreaks could be missed due to the large number of SNPs detected, resulting in identical isolates not being flagged as clonally related. This would subsequently have implications for infection prevention and control. For the assembly-free method, we observed most replicates to have no SNPs between each other (70%), which is in line with the GenomeTrakr proficiency-test study, which found a similar fraction of datasets showed having no SNPs (73%) [40].
Variation in SNPs among isolates showed a lower number of SNPs based on the assembly-free method compared to the assembly-based method. It is unlikely that this is caused by different numbers of k-mers that were compared for SNPs, as there was only a modest difference for the number of k-mers compared between the assembly-free and the assembly-based SNP analysis, which ranged from −2.1–1.2% difference in compared k-mers (Fig. S3). Therefore, it is more likely that de novo assembly introduces phylogenetic noise in regions difficult to assemble, like regions such as mobile elements (transposons and plasmids). Previously described work employing SNP-based methodologies to infer phylogeny among bacterial isolates often mask regions in the genome that are sensitive for non-informative SNPs for phylogenetic reconstruction, such as mobile genetic elements (MGE). Masking of these regions requires specialized tools such as Gubbins [41] that are able to recognize regions with elevated numbers of base substitutions in the genome. Unfortunately, using this reference-free methodology makes this masking impossible to perform in an unbiased and automated fashion like in the Gubbins pipeline. Therefore, we must assume the possibility of overestimation of SNPs among isolates in our study.
Study limitations
For this study, only three centres participated in this ring-trial, all part of the i-4-1-Health study group. Here, ESBL-producing and ciprofloxacin-resistant Enterobacteriaceae and vancomycin-resistant Enterococcus were defined of primary interest, however other important nosocomial bacterial pathogens such as Pseudomonas sp., Staphylococci and Acinetobacter sp. were not included in the study. Furthermore, all three centres used the same protocols for the extraction and library preparation for sequencing on the Illumina MiSeq. Recommendations for future research would therefore be to determine if these harmonized wet-lab protocols and subsequent bioinformatic data processing are indeed required for the reconstruction of outbreak clusters.
Conclusion
Overall, the work presented here demonstrated that whole-genome sequencing generates reproducible results when comparing results across laboratories that use identical wet-lab and dry-lab methodologies for WGS. Furthermore, to make multi-centre outbreak surveillance feasible in the future, we recommend that laboratories share raw sequencing reads, because systematic errors were introduced in the de novo assemblies by different assemblers. Finally, work presented here lays the foundation for routine proficiency testing in clinical microbiology laboratories.
Supplementary Data
Funding information
The i-4-1-Health project was financed by the Interreg V Flanders-The Netherlands program, the cross-border cooperation program with financial support from the European Regional Development Fund (ERDF). Additional financial support was received from the Dutch Ministry of Health, Welfare and Sport, the Dutch Ministry of Economic Affairs, the Province of Noord-Brabant, the Belgian Department of Agriculture and Fisheries, the Province of Antwerp and the Province of East-Flanders.’ The authors are free to publish the results from the project without interference from the funding bodies. Selective and non-selective agar plates, ETEST stips and VITEK 2 AST cards were provided by bioMérieux (Marcy l’Etoile); FecalSwabs and tryptic soy broths were provided by Copan Italy (Brescia, Italy). The authors are free to publish the results from the project without interference from bioMérieux or Copan Italy.
Acknowledgements
We are grateful to David Barnett for proofreading this manuscript and the collaborators in the participating laboratories for their contribution to the collection of whole-genome-sequence data. These collaborators are part of the i-4-1-Health Study Group: Lieke van Alphen (Maastricht University Medical Centre+, Maastricht, the Netherlands), Nicole van den Braak (Avans University of Applied Sciences, Breda, the Netherlands), Caroline Broucke (Agency for proofreading); Care and Health, Brussels, Belgium), Anton Buiting (Elisabeth-TweeSteden Hospital, Tilburg, the Netherlands), Liselotte Coorevits (Ghent University Hospital, Ghent, Belgium), Sara Dequeker (Agency for Care and Health, Brussels, Belgium and Sciensano, Brussels, Belgium), Jeroen Dewulf (Ghent University, Ghent, Belgium), Wouter Dhaeze (Agency for Care and Health, Brussels, Belgium), Bram Diederen (ZorgSaam Hospital, Terneuzen, the Netherlands), Helen Ewalts (Regional Public Health Service Hart voor Brabant, Tilburg, the Netherlands), Herman Goossens (University of Antwerp, Antwerpen, Belgium and Antwerp University Hospital, Antwerp, Belgium), Inge Gyssens (Hasselt University, Hasselt, Belgium), Casper den Heijer (Regional Public Health Service Zuid- Limburg, Heerlen, the Netherlands), Christian Hoebe (Maastricht University Medical Centre+, Maastricht, the Netherlands and Regional Public Health Service Zuid-Limburg, Heerlen, the Netherlands), Casper Jamin (Maastricht University Medical Centre+, Maastricht, the Netherlands), Patricia Jansingh (Regional Public Health Service Limburg Noord, Venlo, the Netherlands), Jan Kluytmans (Amphia Hospital, Breda, the Netherlands and University Medical Centre Utrecht, Utrecht University, Utrecht, the Netherlands), Marjolein Kluytmans-van den Bergh (Amphia Hospital, Breda, the Netherlands and University Medical Centre Utrecht, Utrecht University, Utrecht, the Netherlands), Stefanie van Koeveringe (Antwerp University Hospital, Antwerp, Belgium), Sien De Koster (University of Antwerp, Antwerp, Belgium), Christine Lammens (University of Antwerp, Antwerp, Belgium), Isabel Leroux-Roels (Ghent University Hospital, Ghent, Belgium), Hanna Masson (Agency for Care and Health, Brussel, Belgium), Ellen Nieuwkoop (Elisabeth-TweeSteden Hospital, Tilburg, the Netherlands), Anita van Oosten (Admiraal De Ruyter Hospital, Goes, the Netherlands), Natascha Perales Selva (Antwerp University Hospital, Antwerp, Belgium), Merel Postma (Ghent University, Ghent, Belgium), Stijn Raven (Regional Public Health Service West-Brabant, Breda, the Netherlands), Veroniek Saegeman (University Hospitals Leuven, Leuven, Belgium), Paul Savelkoul (Maastricht University Medical Centre+, Maastricht, the Netherlands), Annette Schuermans (University Hospitals Leuven, Leuven, Belgium), Nathalie Sleeckx (Experimental Poultry Centre, Geel, Belgium), Arjan Stegeman (Utrecht University, Utrecht, the Netherlands), Tijs Tobias (Utrecht University, Utrecht, the Netherlands), Paulien Tolsma (Regional Public Health Service Brabant Zuid-Oost, Eindhoven, the Netherlands), Jacobien Veenemans (Admiraal De Ruyter Hospital, Goes, the Netherlands), Dewi van der Vegt (PAMM Laboratory for Pathology and Medical Microbiology, Veldhoven, the Netherlands), Martine Verelst (University Hospitals Leuven, Leuven, Belgium), Carlo Verhulst (Amphia Hospital, Breda, the Netherlands), Pascal De Waegemaeker (Ghent University Hospital, Ghent, Belgium), Veronica Weterings (Amphia Hospital, Breda, the Netherlands), Clementine Wijkmans (Regional Public Health Service Hart voor Brabant, Tilburg, the Netherlands), Patricia Willemse-Smits (Elkerliek Hospital, Helmond, the Netherlands), Ina Willemsen (Amphia Hospital, Breda, the Netherlands).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; CDS, coding sequence; cgMLST, core genome multi locus sequence typing; ESBL, extended spectrum beta-lactamase; MDR, multi-drug-resistant; MLVA, multi-locus variable-number tandem repeat analysis; MRSA, methicillin-resistant Staphylococcus aureus; ORI, origin of replication; PFGE, pulsed field gel electrophoresis; SNP, single nucleotide polymorphism; wgMLST, whole genome multi locus sequence typing; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures are available with the online version of this article.
References
- 1.WHO Global action plan on antimicrobial resistance. [Jul 02;2021 ]; https://www.who.int/publications/i/item/9789241509763 accessed.
- 2.Thaden JT, Li Y, Ruffin F. Increased costs associated with bloodstream infections caused by multidrug-resistant gram-negative bacteria are due primarily to patients with hospital-acquired infections. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01709-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Founou RC, Founou LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: A systematic review and meta-analysis. PLoS One. 2017;12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariappan S, Sekar U, Kamalanathan A. Carbapenemase-producing Enterobacteriaceae: Risk factors for infection and impact of resistance on outcomes. Int J Appl Basic Med Res. 2017;7:32–39. doi: 10.4103/2229-516x.198520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris SR, Feil EJ, Holden MTG. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pendleton S, Hanning I, Biswas D, Ricke SC. Evaluation of whole-genome sequencing as a genotyping tool for Campylobacter jejuni in comparison with pulsed-field gel electrophoresis and flaA typing. Poult Sci. 2013;92:573–580. doi: 10.3382/ps.2012-02695. [DOI] [PubMed] [Google Scholar]
- 7.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. Evaluation of whole genome sequencing for outbreak detection of salmonella enterica. PLoS One. 2014;9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, et al. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol. 2015;53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering R. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin Microbiol Infect. 2018;24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Silva M, Machado MP, Silva DN. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seemann T. Snippy. [May 18;2021 ]; https://github.com/tseemann/snippy accessed.
- 12.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris SR. SKA: Split KMER analysis toolkit for bacterial genomic epidemiology. bioRxiv. 2018 doi: 10.1101/453142. [DOI] [Google Scholar]
- 14.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling AE, Mau B, Perna NT. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 19.Harris SR, Cartwright EJP, Török ME. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: A descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rumore J, Tschetter L, Kearney A. Evaluation of whole-genome sequencing for outbreak detection of Verotoxigenic Escherichia coli O157:H7 from the Canadian perspective. BMC Genomics. 2018;19:870. doi: 10.1186/s12864-018-5243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluytmans-Van Den Bergh MFQ, Rossen JWA, Bruijning-Verhagen PCJ. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing enterobacteriaceae. J Clin Microbiol. 2016;54:2919–2927. doi: 10.1128/JCM.01648-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Den BM, Lammens C, Selva NP. Microbiological methods to detect intestinal carriage of highly-resistant microorganisms (HRMO) in humans and livestock in the i-4-1-health dutch- belgian cross-border project. Preprints. 2019:2019120216 [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Zankari E, Hasman H, Cosentino S. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, Garciá-Fernández A. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Souvorov A, Agarwala R, Lipman DJ. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19:153. doi: 10.1186/s13059-018-1540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 29.Köster J, Rahmann S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 30.Virtanen P, Gommers R, Oliphant TE. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tae H, Karunasena E, Bavarva JH, McIver LJ, Garner HR. Large scale comparison of non-human sequences in human sequencing data. Genomics. 2014;104:453–458. doi: 10.1016/j.ygeno.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangiovanni M, Granata I, Thind AS, Guarracino MR. From trash to treasure: Detecting unexpected contamination in unmapped NGS data. BMC Bioinformatics. 2019;20:168. doi: 10.1186/s12859-019-2684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016;8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin E, Wolf YI. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nouws S, Bogaerts B, Verhaegen B. Impact of DNA extraction on whole genome sequencing analysis for characterization and relatedness of Shiga toxin-producing Escherichia coli isolates. Sci Rep. 2020;10:14649. doi: 10.1038/s41598-020-71207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauß L, Ruffing U, Abdull S. Detecting Staphylococcus aureus virulence and resistance genes: A comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol. 2016 doi: 10.1128/JCM.03022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozyreva VK, Truong CL, Greninger AL, Crandall J, Mukhopadhyay R, et al. Validation and implementation of clinical laboratory improvements act-compliant whole-genome sequencing in the public health microbiology laboratory. J Clin Microbiol. 2017;55:2502–2520. doi: 10.1128/JCM.00361-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roer L, Hansen F, Thomsen MCF, Knudsen JD, Hansen DS, et al. WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J Antimicrob Chemother. 2017;72:1922–1929. doi: 10.1093/jac/dkx092. [DOI] [PubMed] [Google Scholar]
- 39.Zhou H, Liu W, Qin T, Liu C, Ren H. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae. Front Microbiol. 2017;8:371. doi: 10.3389/fmicb.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timme RE, Rand H, Leon MS. GenomeTrakr proficiency testing for foodborne pathogen surveillance: An exercise from 2015. Microb Genomics. 2018 doi: 10.1099/mgen.0.000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croucher NJ, Page AJ, Connor TR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.