Abstract
There is a need to develop novel approaches to improve the balance between efficacy and toxicity for transcription factor t therapies. In this study, we exploit context dependent differences in RNAPII processivity as an approach to improve the activity and limit the toxicity of the EWS-FLI1 targeted small molecule, mithramycin, for Ewing sarcoma. The clinical activity of mithramycin for Ewing sarcoma is limited by off-target liver toxicity that restricts the serum concentration to levels insufficient to inhibit EWS-FLI1. In this study, we perform an siRNA screen of the druggable genome followed by a matrix drug screen to identify mithramycin potentiators and a synergistic “class” effect with CDK9 inhibitors. These CDK9 inhibitors enhanced the mithramycin-mediated suppression of the EWS-FLI1 transcriptional program leading to a shift in the IC50 and striking regressions of Ewing sarcoma xenografts. In order to determine if these compounds may also be liver protective, we performed a qPCR screen of all known liver toxicity genes in HepG2 cells to identify mithramycin-driven transcriptional changes that contribute to the liver toxicity. Mithramycin induces expression of the BTG2 gene in HepG2 but not Ewing sarcoma cells which leads to a liver-specific accumulation of reactive oxygen species (ROS). siRNA silencing of BTG2 rescues the induction of ROS and the cytotoxicity of mithramycin in these cells. Furthermore, CDK9 inhibition blocked the induction of BTG2 to limit cytotoxicity in HepG2, but not Ewing sarcoma cells. These studies provide the basis for a synergistic and less toxic EWS-FLI1 targeted combination therapy for Ewing sarcoma.
Keywords: Ewing sarcoma, EWS-FLI1, Mithramycin, Cyclin Dependent Kinase 9, BTG2
Introduction
Ewing sarcoma (ES) requires the sustained activity of the oncogenic transcription factor EWS-FLI1 for cell survival (1). EWS-FLI1 dysregulates transcription at hundreds of genes, promoting the expression of pro-survival genes while limiting the expression of tumor-suppressor genes (2). Furthermore, elimination of EWS-FLI1 reverses the tumorigenic phenotype in ES cells (3). We previously identified mithramycin as a potent and specific inhibitor of EWS-FLI1 in a high-throughput cell-based screen of EWS-FLI1 driven NR0B1 promoter luciferase activity (4). The screen was successful because NR0B1 promoter activity is highly specific for EWS-FLI1 activity (5). Indeed, both FLI1 and EWS-FLI1 can bind the promoter GGAA EWS-FLI1 response element but only EWS-FLI1 can transactivate (6). Furthermore, deletion of the GGAA microsatellite within the NR0B1 promoter using CRISPR/Cas9 leads to a loss of expression of NR0B1 (7). We showed that 100 nM mithramycin blocked promoter activity, reduced expression of NR0B1 mRNA and protein, and reversed the gene signature of EWS-FLI1 (4). Importantly, mithramycin decreased both Ewing sarcoma cell viability in vitro and tumor size in orthotopic xenografts.
We subsequently opened a phase I/II clinical trial in patients with Ewing sarcoma using single-agent mithramycin (8). In the modern era of supportive care, mithramycin treatment was well-tolerated. However, reversible grade 3/4 liver toxicity limited the serum concentration of mithramycin to levels that were lower than that needed to inhibit EWS-FLI1. Therefore, the trial was closed. We have subsequently reported a second-generation mithramycin analog called EC8042 with an improved toxicity profile that may be able to achieve the therapeutic suppression of EWS-FLI1. However, the clinical translation of EC8042 is challenged by the rarity of the disease and the time it takes to translate novel compounds to the clinic. Therefore, as a complementary approach, in this study, we sought to identify a compound or class of compounds that is able to amplify the mithramycin mediated suppression of EWS-FLI1 with non-overlapping toxicity to realize an EWS-FLI1 targeted therapy.
In order to develop this combination therapy, we performed an siRNA screen of the druggable genome followed by a matrix drug screen to identify agents that potentiate the mithramycin-mediated block of EWS-FLI1. We identified a focus of synergy at the 20 nM concentration of mithramycin that we achieved in our published clinical trial with cyclin-dependent kinase 9 (CDK9) inhibitors (8). CDK9 is the catalytic subunit of positive transcription elongation factor b (P-TEFb) that promotes the shift from abortive to productive elongation by phosphorylating the catalytic subunit of RNA polymerase II (RNAPII), negative elongation factor (NELF) and DRB sensitivity inducing factor (DSIF) both releasing paused RNAPII and suppressing negative regulators of productive transcription (9–11). Because mithramycin appears to block transcription initiation, we reasoned that CDK9 downstream targeting might provide an opportunity to capitalize on context-dependent transcriptional changes to amplify “on target” activity while limiting toxicity.
CDK9 inhibition amplified the mithramycin-mediated suppression of EWS-FLI1 activity to suppress the expression of the EWS-FLI1 transcriptional program. These effects were cell line selective and “on target” leading to a marked shift in the IC50 of mithramycin and striking regressions of a Ewing sarcoma xenograft in combination with mithramycin at a fraction of the dose used in previous preclinical studies. Importantly, the context dependence did indeed provide cell line selectivity as CDK9 blockade did not cooperate with mithramycin to suppress the proliferation of HepG2 liver-derived cells. In addition, CDK9 inhibition blocked the induction of the BTG2 gene and the accumulation of reactive oxygen species (ROS) which has been previously described with BTG2 and doxorubicin (12). Together, these data provide the basis for the clinical realization of EWS-FLI1 suppression by mithramycin in combination with CDK9 blockade, a novel approach to identify transcription targeted therapies and insight into the therapeutic targeting of EWS-FLI1.
Materials and Methods
Cell Culture
All cell lines were maintained in RPMI 1640 growth media (Invitrogen) with 10% fetal bovine serum (Atlanta Biologicals), 100 U/mL penicillin,100 μg/mL streptomycin and 2 mM L-glutamine (Invitrogen) at 37°C in an atmosphere of 5% CO2. Ewing sarcoma cell lines TC32, TC71, TC252, and CHLA9 were a gift from Tim Triche (Children’s Hospital of Los Angeles) and authenticated using short tandem repeat genotyping and screened bi-annually for mycoplasma. For live cell imaging, TC32 cells were transfected with IncuCyte NucLight Green Lentivirus Reagent (EF1a, Puro) (Essen BioScience) at a multiplicity of infection of 6 in 2 μg/mL Polybrene (MilliporeSigma) and batch selected in media containing 1 μg/mL puromycin (Thermo-Fischer Scientific Incorporated).
Compounds
Mithramycin (Tocris Bioscience) was dissolved as a 1 mg/mL stock solution in PBS, and frozen in aliquots. PHA-767491 and other small molecule inhibitors (Selleck Chemicals) were dissolved in DMSO as stock solutions. Small molecule stocks were stored at −80 °C for storage or −20 °C for experimental use, minimizing freeze-thaw cycles. For in vivo experiments, a 30 mg/mL stock solution of PHA-767491 (Adooq Bioscience) was made by dissolving the powder in a solution of 1 mg/10mL D-α-Tocopherol polyethylene 1000 succinate (MilliporeSigma) for oral gavage.
siRNA screen for Mithramycin Sensitizers
The Ambion Silencer Select Druggable Genome siRNA Library (Thermo-Fisher Scientific) was used to silence ~9000 genes with three different siRNAs per gene in TC32 cells. Briefly, siRNAs were spotted to 384 well plates (Corning 3570) and 0.07 μL of Lipofectamine RNAiMAX (Thermo-Fischer Scientific) was added to each well in 20 μL of serum free media for 30 minutes followed by the addition of 1000 cells per well in 2x serum containing media. This yielded final transfection mixtures with 20nM siRNA. 48 hours later, mithramycin was added in 5 μL of media to a final concentration of 40 nM. The same volume of fresh media was added to a replicate set of screening plates. 48 hours after the addition of mithramycin, cell viability was measured using CellTiter Glo Assay Reagent (Promega). Luminescence was measured on a Perkin Elmer Envision Reader (PerkinElmer Incorporated). Viability measurements were normalized to the median of negative control wells on each plate (Ambion Silencer Select Negative Controls #2, n=16 per plate) and the log2 fold-change between mithramycin and vehicle treated samples were calculated for each siRNA. Hits were selected from genes with more than one siRNA exhibiting a greater than 2-fold change in viability.
Matrix Drug Screening
7 Serial dilution of mithramycin (ranging from 1.28 μM to 10 nM), a transcription inhibitor, control compound (Table S1)(dilutions centered on the IC50 of the individual compound) or medium controls were plated in quadruplicate in 10 μL total volume 384-well plates using an epMotion 5075 (Eppendorf) and stored at −80 °C until needed. Plates were thawed, allowed to come to room temperature and 2000 cells in a volume of 30 μL were added to the matrix with a VIAFLO384 electronic 384 channel pipette (INTEGRA Biosciences). The mixture was incubated at 37 °C for 60 hours after which, 8 μL of CellTiter Glo Assay Reagent was added and the plate incubated for 1 hour. Additional confirmation of synergy between mithramycin and a CDK9 inhibitor was performed in triplicate using the same approach in 96-well plates with serial dilution of compounds and additional cell lines TC32 (5000 cells per well), TC252 (10000 cells per well), or CHLA9 (10000 cells per well). Drug synergy was calculated using Bliss independence via the synergy finder package in R v 3.5.2 (https://cran.r-project.org/).
Live Cell Imaging
Live cell imaging was performed using standard tissue culture procedures in clear bottom plates and imaged every 2 hours for cell confluence measurements and/or green object count (for GFP expressing cell lines) using the IncuCyte ZOOM. Cells were allowed to reach ~15% confluence before compound addition.
Reverse Transcription Quantitative Polymerase Chain Reaction
400,000 cells were plated in 6-well plates, allowed to recover overnight and incubated with 100 nM mithramycin, 20 nM mithramycin, 2 μM PHA-767491, or 20 nM mithramycin and 2 μM PHA-767491 for 18 hours. RNA was collected using a RNeasy kit (Qiagen) and quantified using a NanoDrop 2000 spectrophotometer (Thermo-Fischer). 1800 ng of RNA was reverse transcribed into cDNA using MultiScribe Reverse Transcriptase (Applied Biosystems), 10X RT Buffer (Applied Biosystems), 25X dNTPs (Applied Biosystems), and oligo-dT (Life Technologies). A 96-well thermocycler (Applied Biosystems) was used to run the following program: 25 °C for 10 minutes, 37 °C for two hours, and 85 °C for 5 minutes. 50 ng of cDNA was added to a mixture of SYBR Green PCR Master Mix (Thermo-Fischer) and gene specific primers. A CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories Incorporated) was used to acquire Ct values under the following program: 95°C for 10 minutes, 40 cycles of [95 °C for 30 seconds, 55 °C for 30 seconds, 72 °C for 30 seconds], 65 °C for 5 seconds, and melt curve analysis at 0.5 °C per step up to 95 °C. Fold change was determined using the ΔΔCT method with GAPDH used for normalization.
RNA Sequencing
RNA was collected as described as for qPCR after 12 hours of incubation with compound. Libraries were prepared from 500 ng of total RNA using the KAPA RNA HyperPrep Kit with RiboseErase (v1.16) (Kapa Biosystems). RNA was sheared to 300–400 bp. Prior to PCR amplification, cDNA fragments were ligated to IDT for Illumina Unique Dual Indexes (Integrated DNA Technologies). Quality and quantity of the finished libraries were assessed using a combination of Agilent DNA High Sensitivity chip (Agilent Technologies) and QuantiFluor® dsDNA System (Promega). Individually indexed libraries were pooled and 50 bp, paired end sequencing was performed on an Illumina NovaSeq6000 sequencer using a 150 bp S1 sequencing kit (Illumina). Base calling was done by Illumina RTA3 and output of NCS was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v1.9.0. Adapters and low-quality sequences were then removed with Trimgalore. Trimmed data was quality controlled with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then mapped with STAR to the mm10 genome (13). Raw gene counts from STAR were imported into R. Genes with less than 10 counts in a minimum of two samples were removed and counts were then normalized for library size and composition biases using the weighted trimmed mean of M-values as implemented in edgeR (14). Heatmaps were then generated for the Hancock targets using the logFC of each treatment sample to its group-matched solvent control (15).
Immunoblotting
1.5 million cells were plated in 10 cm2 dishes, allowed to recover overnight and incubated with 100 nM mithramycin, 20 nM mithramycin, 2 μM PHA-767491, or 20 nM mithramycin and 2 μM PHA-76741 for 18 hours. The cells were washed, collected in Dulbecco’s phosphate-buffered saline (DPBS), lysed in 4% LDS buffer and boiled for 10 minutes. The detergent was diluted with water and quantitated relative to BSA standard using the bicinchoninic acid (BCA) kit (Thermo-Fischer) and a Synergy NEO Microplate reading system (BioTek). 30 μg of each sample was resolved on a NuPAGE 4–12% Bis-Tris protein gels or NuPAGE 8% Bis-Tris protein gels (Invitrogen) in 1X NuPAGE MOPS SDS Running Buffer (Invitrogen). The protein was transferred overnight to nitrocellulose, blocked with TBST containing 5% milk and incubated at 4 °C with primary antibody (or 1 hour at room temperature for GAPDH) overnight. The membrane was then incubated with anti-mouse or anti-rabbit HRP-conjugated antibodies at room temperature for 1 hour in TBST with 5% dry milk. The antibodies used in this study: EZH2 (D2C9) XP rabbit monoclonal antibody (1:1000 dilution; Cell Signaling), FLI1 (ab133485) rabbit monoclonal antibody (1:1000 dilution; Abcam), NR0B1/Dax1 (ab196649) rabbit monoclonal antibody (1:1000 dilution; Abcam), GAPDH (ab8245) mouse monoclonal antibody (1:500 dilution; Abcam), RNAPII CTD repeat YSPTPS (phospho S2) rabbit polyclonal antibody (1:1000 dilution, Abcam), RPB1 CTD (4H8) mouse monoclonal antibody (1:1000 dilution Cell Signaling). Bands were visualized using an enhanced chemiluminescence (ECL) Western blotting analysis system (Amersham) and HyBlot CL autoradiography film (Denville Scientific).
RNAPII Processivity Assay
4*106 TC32 cells were plated on 15 cm tissue-culture treated dishes (Corning) in RPMI growth media for 48 hours. Cells were placed on ice for 5 minutes, washed in ice cold DPBS and collected by manual disruption. Cells were centrifuged at 300g at 4 °C for 10 minutes, washed in 10 mL ice cold DPBS. Cells were swelled in a solution of 10 mM Tris-HCl (pH 7.5), 200 μM MgCl2, and 300 μM CaCl2 then centrifuged at 400g at 4 °C for 10 minutes. Cells were first resuspended in a solution of 10% glycerol then lysed with a solution of 10% glycerol and 1% NP-40. After centrifugation at 600g for 5 minutes, nuclei where resuspended in a solution of 40% glycerol, 50 mM Tris-HCl (pH 8), 5 mM MgCl2, and 50 μM EDTA at a concentration of 107 nuclei per 100 μL. The nuclear run-on reaction occurred at 30 °C for 5 minutes and 850 rpm in a thermomixer (Eppendorf). Reaction volume was 200 μL and consisted of 100 μL of nuclei and 100 μL of a solution containing 500 μM N-laurylsarcosine, 10 mM Tris-HCl (pH 8), 5 mM MgCl2, 300 mM KCl, 1 mM DTT, 500 μM ATP, 500 μM GTP, 500 μM 5’-BrUTP, 2 μM CTP, and 200 U/mL SUPERase• In RNase Inhibitor (Invitrogen). Trizol Reagent (Invitrogen) and the Direct-zol RNA Kit (Zymo Research) were used to isolate RNA and then qRT-PCR was performed. The quantitation was performed by fitting the data to a standard curve for the EZH2 proximal and EZH2 distal amplicons using cDNA generated from solvent control cells. Nuclear run-on cDNA was fitted to the curve to obtain absolute cDNA quantity. Amplicon amount for treated nuclei was normalized to solvent control nuclei.
Xenografts for Combination Therapy
Athymic nude mice (Crl; Nu-Foxn1Nu/Nu) were implanted with 2 million TC32 cells in the left gastrocnemius muscle in a volume of 100 μL Hank’s Balanced Salt Solution (Thermo-Fischer Scientific). Once tumor diameter reached 0.5 cm, mice were randomly assigned to either the control, mithramycin, PHA-767491, or combination groups. Mithramycin was administered via an Alzet 1003D osmotic pump (Durect) placed within the peritoneum which delivered a dose continuous dose in a vehicle of DPBS with MgCl2 and CaCl2 (MilliporeSigma) and was removed after treatment. PHA-767491 was administered via oral gavage in 200 μL of its vehicle, 1 mg/10 mL D-α-Tocopherol polyethylene 1000 succinate (MilliporeSigma). Mice were euthanized by CO2 asphyxiation when tumor diameter reached greater than 2 cm3. Tumor size was determined by the equation( ), where D is the maximum diameter and d is the minimum diameter. All animal work was approved by the Van Andel Institute Animal Care and Use Committee.
Reactive Oxygen Species Analysis
2.5 million HepG2 cells were plated in 8 mL of media into 10 cm tissue-culture treated culture dishes and incubated for 24 hours at 37 °C in media or media containing siRNA for BTG2. Gene silencing was confirmed using RT-qPCR. Cells were then incubated with either solvent control media, media containing 20 nM mithramycin, or media containing 50 nM mithramycin. After 24 hours of treatment, cells were incubated for 30 minutes with spin probe 1-hydroxy-4-methoxy-2,2,6,6-tetramethylpiperidine (TMH, Enzo Life Sciences) (16) at 37 °C before collection into 0.6 mL Krebs-HEPES buffer (pH 7.4). The mixtures were then frozen in liquid nitrogen and the electron paramagnetic resonance (EPR) spectra measured. Cellular EPR signals were calculated by subtracting the EPR signal of a cell-free blank from the total EPR signal of cellular sample and expressed as nmol/liter based on TEMPOL calibration curve. The cellular signal was normalized to protein content (mg/mL) which was determined by Bradford method (BioRad).
Statistical Methods
All analyses, unless explicitly stated, were conducted in Prism 5.0 (GraphPad Software Inc). Data are presented as mean values with standard deviation or 95% confidence intervals. 1-way ANOVA was performed to determine the statistical significance of differences between control and treatment groups with Dunnett’s post-hoc test. Assumptions of variance in ANOVA were verified using Bartlett’s test. Cell growth and tumor volume comparison was performed using 2-way ANOVA and a Bonferroni post-hoc test. All statistical tests were two-sided.
Results
CDK9 Inhibition Leads to a Synergistic Loss of Ewing Sarcoma Cell Viability When Combined with Mithramycin
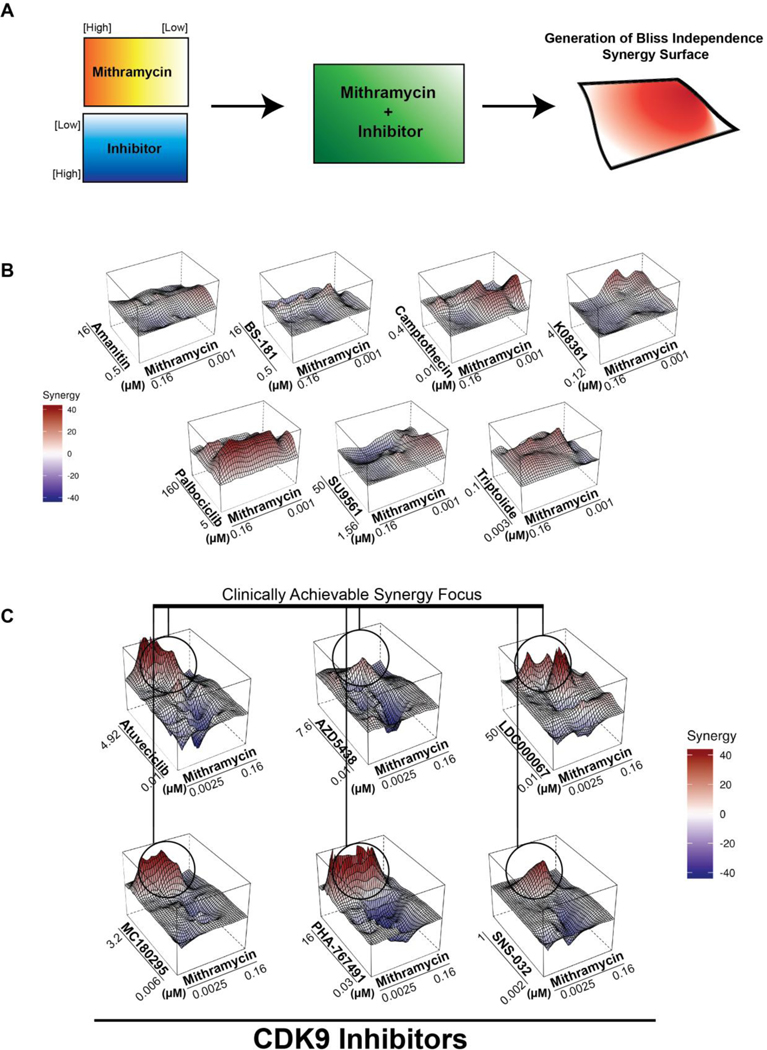
In order to identify agents that potentiate the mithramycin-mediated suppression of EWS-FLI1 activity, we screened a siRNA library of the druggable genome in the presence or absence of 40 nM mithramycin (Fig. S1A, Supplementary Table 1A). We focused on those hits that decreased ES cell viability by at least two-fold over the observed gene silencing alone with two or three independent siRNA (Fig. S1B, Supplementary Table 1B). We identified several hits that are crucial for eukaryotic transcription including proteins required for RNA polymerase II assembly or activity (e.g., POLR2A/E/F/H, GPN1)(Fig. S1C). The importance of directly targeting transcription was reinforced by Gene Ontology analysis (Supplementary Table 1C). A general blockade in transcription would likely potentiate toxicity as well as activity of mithramycin. However, we reasoned that eukaryotic transcription is both context dependent and highly regulated consisting of discrete steps including polymerase promoter binding (17), transcriptional initiation (18), productive elongation (19), and termination (20). Therefore, it might be possible to target the EWS-FLI1 transcriptome in series to increase the specificity for EWS-FLI1 while limiting the toxicity imposed by a general blockade in transcription. Since, we and others have found that mithramycin likely impacts transcription initiation, we assembled a panel of inhibitors that work downstream of this step and screened for synergy at 48 different concentration combinations in quadruplicate using a matrix approach (Table S2)(21–34). As an addition control, we included several cell-cycle inhibitors to control for general effects on proliferation (Fig. 1A).
Figure 1.
Mithramycin synergizes with cyclin-dependent kinase 9 inhibitors. A, Schematic of the drug matrix screen. Plates with serial dilutions of mithramycin and different transcriptional inhibitors were stamped onto 384-well plates to test 48 different drug combinations in quadruplicate with at least two independent experiments. TC32 cells were added to the drug combination and viability was determined at 60 hours. The degree of synergy is determined by Bliss independence and represented by the height of the peak and color (red = synergy; blue = antagonism). B, A panel of transcription inhibitors demonstrate limited synergy (amanitin, BS-181, SU9561), diffuse synergy (palbociclib, triptolide, camptothecin) or require high doses of compound (K08361). C, A focus of synergy between mithramycin and CDK9 inhibitors at the clinically achievable concentration of mithramycin with 6 different CDK9 inhibitors.
We identified several compounds that showed marked synergy with mithramycin at various concentrations (Fig. 1B-C). However, we focused on CDK9 inhibitors because the compounds provided a highly-focused area of synergy at the clinically achievable concentration of mithramycin as measured by Bliss Independence. We recently completed a clinical study of mithramycin and found that the drug accumulates to a maximal concentration (Cmax) of just under 20 nM (8). We found that several CDK9 inhibitors produced a synergy peak at this 20 nM concentration that induced a substantial loss of Ewing sarcoma viability (Fig. 1C). Importantly, this synergy was seen with highly selective CDK9 inhibitors including LDC000067 and MC180295 (35). Furthermore, the synergy observed in TC32 cells was seen in two other cell lines, TC252 and CHLA9 (Fig. S2). It is notable that there was some variability in the synergy observed particularly in the TC252 cells. However, these cells were substantially more sensitive to the single agent CDK9 inhibitors MC180295, atuveciclib and SNS-032 by themselves which masked the synergy. Importantly, the exquisite sensitivity to these drugs indicates likely clinical utility (Fig. S3). Together, these data suggest that the impairment of Ewing sarcoma cell viability by mithramycin can be sustained or improved in combination with CDK9 inhibitors.
Mithramycin Combined with CDK9 Inhibition Blocks EWS-FLI1 Activity
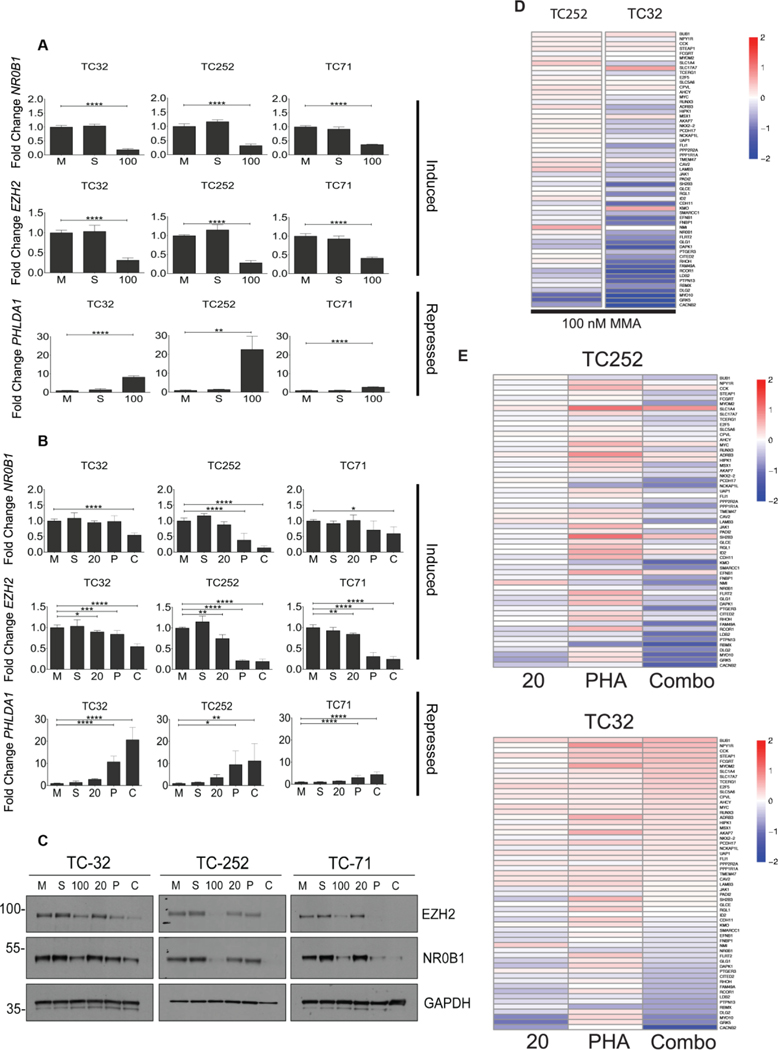
In order to determine if the combined effects of mithramycin plus a CDK9 inhibitor are “on-target”, we examined the impact of treatment on the EWS-FLI1 transcriptome. We chose PHA-767491 as a tool compound to further evaluate the combination because there was very similar synergy among all the CDK9 inhibitors indicative of a “class” effect and the synergistic micromolar concentrations of PHA-767491 are achievable in preclinical xenograft models (36). We evaluated the impact of exposure of TC32 cells to mithramycin and/or 2 μM PHA-767491 on the expression levels of two well-defined EWS-FLI1 induced targets (NR0B1 and EZH2)(2,5) and a well-established EWS-FLI1 repressed target PHLDA1 (37) using RT-qPCR. 100 nM mithramycin served as the positive control and consistent with previous observations decreased the mRNA expression of the EWS-FLI1 induced targets NR0B1 and EZH2, while inducing expression of the EWS-FLI1 repressed target, PHLDA1 (Fig. 2A). In contrast, the 20 nM concentration of mithramycin which represents the concentration achieved in patients did not have much of effect on the expression of these downstream targets. 2 μM was chosen as the concentration to evaluate PHA-767491 as an attempt to increase specificity for CDK9 as it was at the lower end of the broad synergy peak (see Fig 1C, S2) and had a limited effect by itself on EWS-FLI1 downstream target expression in TC32 cells. However, when 20 nM mithramycin and 2 μM PHA-767491 were combined, EWS-FLI1 was suppressed leading to decreased expression of induced targets (NR0B1 and EZH2) and induction of the EWS-FLI1 repressed target, PHLDA1 (Fig. 2B). Importantly, these are the identical concentrations that showed a synergistic reduction in cell viability.
Figure 2.
The combination of mithramycin and PHA-767491 reverses the activity of EWS-FLI1. A and B, 100 nM mithramycin for 18 hours blocks the expression of the EWS-FLI1 induced targets EZH2 and NR0B1 while inducing the expression of the repressed target PHLDA. Lower concentration (20 nM for 18 hours) of mithramycin had a minimal impact on expression of EWS-FLI1 induced (NR0B1, EZH2) or repressed targets (PHLDA1) unless combined with 2 μM PHA-767491. Data represents fold change (2ΔΔCT) in expression relative to GAPDH as measured by RT-qPCR in TC32 (n=6), TC252 (n=6), and TC71 (n=3) treated with either media (M), solvent (S), 100 nM mithramycin (100), 20 nM mithramycin (20), 2 μM PHA-767491 (P), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (C) for 18 hours. Each biological replicate had 3 or 4 technical replicates. * = P < 0.05, ** = P < 0.01, *** = P <0.001, **** = P < 0.0001, error bars show standard deviation. C, EWS-FLI1 downstream target proteins are suppressed with high dose mithramycin or the combination of mithramycin and PHA-767491. Immunoblot showing expression of the EWS-FLI1 downstream targets (NR0B1, EZH2) relative to loading control (GAPDH) following 18-hour exposure to medium (M), solvent (S), 100 nM mithramycin (100), 20 nM mithramycin (20), 2 μM PHA-767491 (P), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (C). Immunoblots shown are representative of three independent experiments per cell line. D and E, Reversal of the EWS-FLI1 gene signature requires either high dose mithramycin or combination treatment as demonstrated by RNA sequencing following treatment with medium (M), solvent (S), or 100 nM mithramycin (100 MMA), 20 nM mithramycin (20), 2 μM PHA-767491 (PHA), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (Combo) for 12 hours in TC32 (n=3) and TC252 (n=3) cell lines.
Similar effects on the protein expression of downstream targets were observed with this drug combination (Fig. 2C). 100 nM mithramycin markedly suppressed expression of NR0B1 and EZH2 in the Ewing sarcoma cell lines TC32, TC252, and TC71. In contrast, neither 20 nM mithramycin or 2 μM PHA-767491 had much of an impact on EWS-FLI1 downstream target expression as single agents but repressed NR0B1 and EZH2 when combined in all three Ewing sarcoma cell lines (Fig. 2C). It is important to note that the synergistic suppression of EWS-FLI1 target genes appears to be greater than the sum of the effect of both individual agents particularly in the TC252 cells (Fig. 2C, E). Finally, we showed that the effect of this combination on EWS-FLI1 downstream target expression extends beyond these well-established targets to a previously described “gene-signature” of EWS-FLI1 targets. Again, 100 nM mithramycin markedly suppressed the expression of this gene signature while neither 20 nM mithramycin or 2 μM PHA-767491 led to reduction in expression of these target genes (Fig. 2D, 2E see also Supplementary Fig. S4). In fact, 2 μM PHA-767491 led to striking induction of expression of most of these genes. Nevertheless, the combination of 20 nM mithramycin and 2 μM PHA-767491 phenocopied the effect of high-dose (100 nM) mithramycin and reversed the expression of this gene signature of EWS-FLI1 (Fig. 2E). Importantly, PHA-767491 suppressed EWS-FLI1 protein expression which may contribute to the dramatic effect observed on the EWS-FLI1 transcriptome when combined with mithramycin (Fig. S5). Loss of EWS-FLI1 expression has been observed before with the bromodomain inhibitor JQ1 which also decreases expression of EWS-FLI1 in some, but not all, Ewing sarcoma cell lines (38).
Both Mithramycin and CDK9 Inhibition Decrease RNAPII Processivity
In order to determine if mithramycin synergized with PHA-767491 due to inhibition of CDK9, we evaluated the impact of 20 nM mithramycin and 2 μM PHA-767491 on CDK9 signaling and RNAPII processivity. CDK9 is critical to the switch from abortive to productive elongation by phosphorylating S2 serine residues of the heptapeptide repeats of the carboxy-terminal repeat domain (CTD) of RNAPII (39,40). 2 μM PHA-767491 caused a reduction of phospho-serine 2 levels in three Ewing sarcoma cell lines (Fig. 3A). To confirm CDK9 blockade with an additional assay, we evaluated the effect of drug treatment on the expression of endogenous retroviruses (ERVs) in TC32 Ewing sarcoma cells (Fig. 3B). Zhang and colleagues have reported that CDK9 inhibition increases the expression of ERV mRNA (35). Indeed, we found marked induction in expression of ERV mRNA with 2 μM PHA-767491. To show that this blockade of CDK9 does indeed impair RNAPII processivity at the concentrations employed throughout the study, we performed a nuclear run-on assay at the EWS-FLI1 target gene EZH2. We chose EZH2 because it is a large gene spanning 20 exons and over 75,000 base pairs and a well-established EWS-FLI1 target gene (41). We designed qPCR primer pairs to both a proximal and distal region of the EZH2 gene and compared the nascent transcription of proximal and distal amplicons (Fig. 3C). Consistent with the proposed mechanism, 2 μM PHA-767491 inhibited the progression of RNAPII to productive elongation at this target gene. As expected, 20 nM mithramycin poisoned initiation when added to the run-on reaction or after an 18-hour pretreatment (Fig. 3D). These data are consistent with a model where this combination targets the EWS-FLI1 transcriptome in series with mithramycin poisoning initiation and CDK9 inhibition poisoning productive elongation: further supporting a relationship between EWS-FLI1 and CDK9 as described by Gorthi & Bishop et al. (42). Silencing of EWS-FLI1 therefore releases this repression of EWSR1 leading to a further reduction of serine-2 phosphorylation and impairment of transcription. It follows that incomplete blockade of EWS-FLI1 with low-dose mithramycin might not reverse this process leading to incomplete blockade of serine 2 phosphorylation but could be rescued by synergistic downstream direct targeting of serine-2 phosphorylation with CDK9 blockade.
Figure 3.
2 μM PHA-767491 inhibits CDK9. A, 2 μM PHA-767491 blocks serine-2 phosphorylation independently or in combination with mithramycin. Immunoblot showing RNAPII and RNAPII CTD phosphoserine-2 relative to GAPDH loading control in TC32, TC252, and TC71 cell lines following exposure to medium (M), solvent (S), 100 nM mithramycin (100), 20 nM mithramycin (20), 2 μM PHA-767491 (P), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (C) for 18 hours. Data representative of three independent experiments. B, 2 μM PHA-767491 induces the expression of endogenous retroviral RNA (ERV). Data represents fold change in expression (2ΔΔCT) of ERV-F and ER9–1 relative to GAPDH in TC32 (n=3), TC252 (n=3), and TC71 (n=3) cells following exposure to medium (M), solvent (S), 100 nM mithramycin (100), 20 nM mithramycin (20), 2 μM PHA-767491 (P), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (C) for 18 hours. C, Schematic of nuclear run on assay used to measure RNA processivity. Primer pairs to both a proximal and distal region on the EZH2 locus were used for RT-qPCR. D, Processivity of RNA as measured by qPCR enrichment of mRNA from the proximal (start) vs. distal (end) amplicon of EZH2 relative to solvent after a TC32 nuclear run-on assay. Nuclei were exposed to 2 μM PHA-767491 during the run-on reaction (PHA), 20 nM mithramycin during the run-on reaction (MMA Run on), or cells were pretreated with 20 nM mithramycin for 18 hours before the run-on reaction (MMA Pre). Each biological replicate in the figure had three technical replicates. * = P < 0.05, ** = P < 0.01, *** = P <0.001, **** = P < 0.0001, error bars show standard deviation.
The Combination of Mithramycin and CDK9 Inhibition Stops Ewing Sarcoma Cell Proliferation and Induces Tumor Lysis in Orthotopic Xenografts
To determine how the combination of EWS-FLI1 and CDK9 blockade impacts cell viability, we looked at the impact of drug treatment on proliferation both in vitro and in vivo. The addition of 2 μM PHA-767491 to mithramycin shifted the half maximal growth inhibitory concentration (IC50) of mithramycin in TC32 cells by almost an order of magnitude from 15 nM (95% CI = 15–16 nM) to 2.4 nM (95% CI = 2.3–2.6 nM)(Fig. 4A). Similar results were seen in two other cell lines leading to a shift in the IC50 of mithramycin from 17 nM (95 % CI = 16 −19 nM) to 5 nM (95 % CI = 4.4–5.7 nM) in TC252 cells and from 37 nM (95% CI = 34–41 nM) to 14 nM (95% CI = 12–18 nM) in CHLA9 cells (Fig. 4A). It is notable that these effects were sustained in vitro and less total drug exposure was required to suppress proliferation. The cells required a full 48 hour of exposure to either 20 nM mithramycin or 2 μM PHA-767491 to suppress continued proliferation (Fig. 4B). In contrast only 24 hours of exposure to the combination of 20 nM mithramycin and 2 μM PHA-767491 led to sustained effects on proliferation for up to 110 hours or more than 96 hours after drug removal from the medium (Fig. 4B).
Figure 4.
The combination of 20 nM mithramycin and 2 μM PHA-767491 decreases Ewing sarcoma cell viability. A, Mithramycin (MMA) concentration response curves alone or in combination 2 μM PHA-767491 (MMA + PHA). Data represents percent viability normalized to medium (MMA) or 2 μM PHA-767491 (MMA + PHA) in TC32, TC252, and CHLA9 Ewing sarcoma cell lines as measured by MTS assay. Best fit lines represent 3rd order polynomial with variable slope and error bars represent standard deviation. B, Proliferation as measured by cell count of GFP labeled nuclei every two hours of TC32 cells incubated with solvent (48 hours), 20 nM mithramycin (48 hours), 2 μM PHA-767491 (48 hours), or 20 nM mithramycin and 2 μM PHA-767491 (combo, 24 hours). *** = P <0.001, shading represents standard deviation. C, Spaghetti plot showing TC32 xenograft growth of individual mice as a function of treatment with vehicle, 1.5 mg/kg mithramycin intraperitoneal as a continuous infusion over 72 hours, 50 mg/kg PHA-767491 oral gavage twice daily for three days or the combination of both on the same schedule. 2 million TC32 cells were implanted into the gastrocnemius muscle of nude athymic female mice (n=11–12) and allowed to establish to a minimum diameter of 0.5 mm prior to starting treatment. Three mice were sacrificed for weight loss of unknown etiology (see Fig. S9)((PHA group, n =1 of 12; combination group (n =2 of 12)). D, Waterfall plot displaying best response as measured as the largest reduction in tumor volume following treatment. Each bar represents an individual animal. E, Same experiment as in C but with 3 mg/kg mithramycin eluted over 72 hours (n=6). Three mice in combination had end-of-infusion toxicity and were found dead in the combination cohort (see Fig. S9).
Next, to examine the effect of combining mithramycin and PHA-767491 on tumor volume, we tested the combination in an orthotopic mouse model. We have recently determined that mithramycin is more effective in Ewing sarcoma when administered as a continuous infusion (Flores, Boguslawski et al. in preparation). Mice were split into four groups (n = 11–12) and treated with 1.5 mg/kg of mithramycin eluted from an implanted intraperitoneal pump over 72 hours. It is notable that this total dose is <20% of the total dose that we have previously shown was effective in Ewing sarcoma in this xenograft (4). The PHA-767491 group received a twice daily oral gavage of 50 mg/kg PHA-767491 for three days. Animals in the combination group received both mithramycin infusion and twice daily PHA-767491 by oral gavage for three days. All animals showed suppression of tumor growth with the combination of mithramycin and PHA-767491 despite the exceptionally low total dose of mithramycin and the brief 3-day treatment (Fig. 4C, 4D). Six of the 12 mice showed striking regressions that were not seen with treatment with the individual agents (Fig. 4D). Indeed, one combination animal had a regression of tumor volume from 590 mm3 to 88 mm3 that remained below 340 mm3 for more than 40 days post initial treatment (Fig. 4C, expanded graph in Fig. S6). At these doses, there was limited toxicity and there was minimal weight loss in the cohorts (Fig. S8). One animal was found dead in the combination cohort (n =12) and two had to be sacrificed for appearance (1 from the PHA cohort and 1 from the coxmbination cohort). All of these animals were responding to therapy and both that were sacrificed had elevated blood potassium (see below and Supplementary Table S3 for blood chemistries). Unfortunately, it was not possible to retreat the animals because of IACUC limitations on second surgeries and the tumors eventually returned.
In an attempt to increase the efficacy of our combination therapy, we repeated the above experiment (n=6) but doubled the dose of mithramycin so that 3 mg/kg would be eluted from an intraperitoneal pump over 72 hours alone and in combination with 3-day treatment with PHA-767491 (Fig. 4E). In this experiment, 1 animal in the PHA-767491 cohort responded to treatment as did the mice treated with mithramycin alone (expanded graph in Fig. S7). All of the mice treated with the combination showed marked and sustained regressions of tumor growth including one animal with a tumor volume of 1000 mm3 at the beginning of treatment that regressed to less than 62 mm3 for 46 days. Unfortunately, mithramycin was toxic at the 3 mg/kg dose. All animals experienced weight loss with mithramycin treatment alone and in combination with PHA-767491. Some animals recovered in both cohorts (Fig. S9) but there was limited transient weight loss at the lower 1.5 mg/kg dose level of mithramycin (Fig. S8).
To better understand the toxicity of mithramycin in animals, we analyzed the blood of a euthanized combination treatment animal that appeared sick from the 1.5 mg/kg mithramycin and PHA-767491 group above. The serum ALT of this animal was significantly elevated at 853 U/L (>60 U/L) consistent with previously observed liver toxicity (8). Though an analysis of the liver by H&E did not show evidence of toxicity or necrosis (Fig. S10), there was significant elevation in serum potassium (>8.5 mmol/L) and serum phosphorous (9.8 mg/dL) (Supplementary Table S3). These metabolic abnormalities are consistent with tumor lysis syndrome caused by widespread tumor cell death (43). The potassium level was so elevated that it was out of range of detection, a level that would be cardiotoxic, lead to arrhythmias, and potentially explain the death of the animals who received combination treatment. Tumor lysis may explain both the effectiveness and toxicity of higher dose mithramycin in Ewing sarcoma xenografts. While challenging to manage preclinically in mouse models, tumor lysis syndrome is routinely managed in the clinic in patients with acute lymphoblastic leukemia (43,44). In addition, the unexpected alterations in serum glucose in more than one mouse would be easily managed in the clinic.
CDK9 Inhibition Reverses Mithramycin-Induced BTG2 Expression and Hepatocyte Free Radical Accumulation
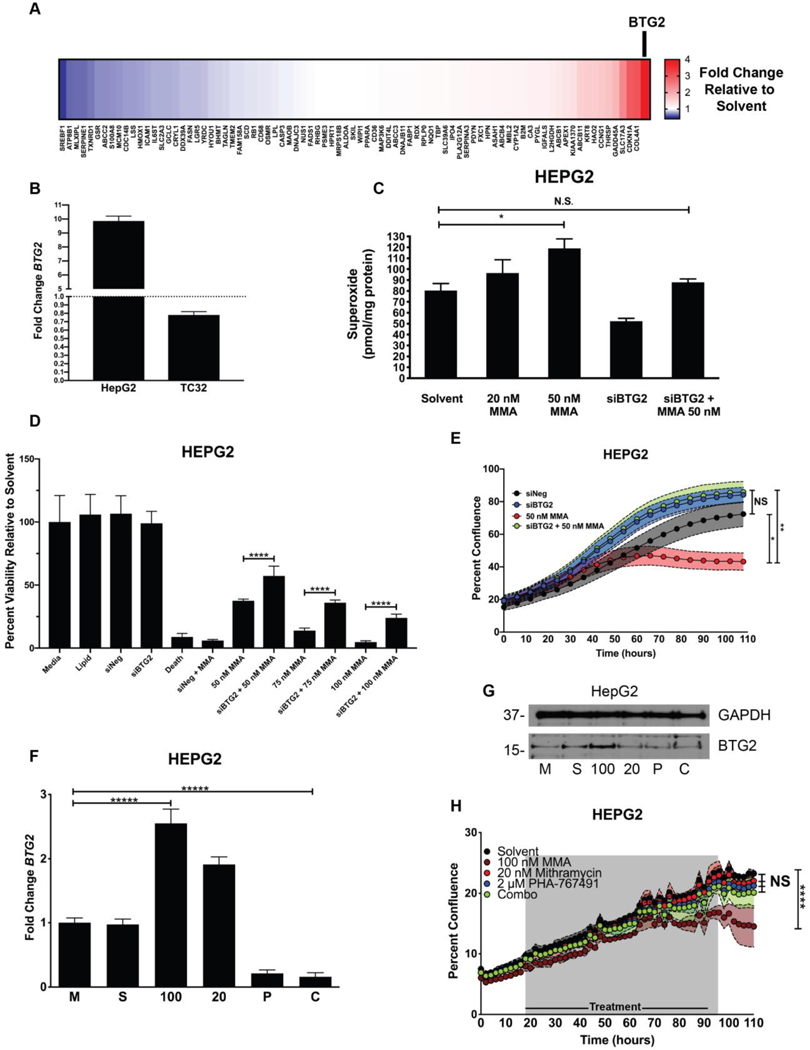
Next, we hypothesized that the observed liver toxicity seen in patients may be due to context-dependent changes in gene expression. It is known that transcription is context-dependent and tightly coordinated by tissue-specific chromatin structure and by expression of cell-specific transcription factors (45). The toxicity of mithramycin that we have observed is also context-dependent favoring the liver relative to other tissues and different across species. It is, therefore, likely that alterations in gene expression induced by mithramycin specific to the liver contribute to the selective toxicity of the drug for this organ. To test this hypothesis, we used a RT-qPCR screen in HepG2 cells to determine the impact of mithramycin exposure on the expression of the majority of genes implicated in liver toxicity (84 total) (Fig. 5A). Although HepG2 cells lack cytochromes and do not model all types of hepatotoxicity, they are an acceptable model for many types of hepatotoxicity and certainly a relevant model to evaluate context-dependent gene expression changes (46). Consistent with the specificity of MMA for specific transcription factors in distinct cellular contexts, the majority of hepatotoxicity genes remained unchanged in expression after mithramycin treatment. However, the gene BTG2 was significantly upregulated in liver cells after treatment with MMA. In contrast, the expression of BTG2 in TC32 Ewing sarcoma cells actually decreased with mithramycin treatment consistent with the hypothesis (Fig. 5B). BTG2 is a complicated protein with pleiotropic effects that promotes cell survival and differentiation in some cellular contexts while driving apoptosis in others (47). Importantly, BTG2 has been shown to exacerbate the cytotoxicity of doxorubicin by significantly increasing the intracellular concentration of reactive oxygen species (ROS) (12). Since the core structure of MMA and doxorubicin is similar, we reasoned that mithramycin may induce BTG2 in an analogous fashion in liver cells to generate increased ROS that makes these cells hypersensitive to mithramycin.
Figure 5.
Mithramycin induces BTG2 expression and ROS production in liver cells which is reversed by PHA-767491. A, Heat map showing fold change increase (red) or decrease (blue) in expression of 84 different liver toxicity changes as measured by qPCR (2ΔΔCT) for each gene relative to GAPDH control in HepG2 cells following exposure to 50 nM mithramycin for 6 hours. BTG2 was the gene induced to the greatest degree with treatment (see BTG2 and arrow). B, Expression of BTG2 in HepG2 liver cells and TC32 Ewing sarcoma cells. Fold change was calculated using 2ΔΔCT method using GAPDH as a control. C, Mithramycin induces a dose dependent increase in superoxide that is rescued with siRNA silencing of BTG2. The data represents the amount of superoxide radicals per unit mass of protein as measured by electron paramagnetic resonance following exposure to 20 nM or 50 nM mithramycin. Silencing of BTG2 with siRNA for 30 hours reduces the accumulation of superoxide and rescues the induction caused by 50 nM mithramycin. * = P < 0.05, error bars represent standard error. MMA = mithramycin D, Silencing of BTG2 with siRNA has no impact on viability but mitigates mithramycin cytotoxicity at 3 different concentrations of drug. Data represents viability as determined by MTS assay following 30 hours of silencing for a total of 60 hours in HepG2 cells relative to medium, solvent, a non-targeting siRNA (siNEG), siRNA targeting of BTG2 (siBTG2), a positive control (siDeath) or 50 nM, 75 nM or 100 nM mithramycin (MMA) alone or in combination with silencing of BTG2 (siBTG2 + 50, 75, 100 nM MMA). E, Cellular proliferation of HepG2 cells (n=3) following silencing of BTG2 (siBTG2) alone or in combination with 50 nM mithramycin (MMA) relative to a non-targeting siRNA control (siNeg) as measured by percent confluence on an IncuCyte ZOOM system. * = P < 0.05, ** = P < 0.01, error bars represent standard deviation. F and G, 2 μM PHA-767491 blocks the induction of BTG2 (F) mRNA or protein (G) expression. Data represents qPCR fold change ( 2ΔΔCT) in expression of BTG2 relative to GAPDH as a control following exposure of TC32 cells to medium (M), solvent (S), 100 nM mithramycin (100), 20 nM mithramycin (20), 2 μM PHA-767491 (P), or a combination of 20 nM mithramycin and 2 μM PHA-767491 (C) for 6 hours or immunoblot following identical exposure for 18 hours. Each biological replicate had 3 technical replicates. **** = P < 0.0001, error bars represent standard deviation. Immunoblots shown are representative of three replicates per cell line. GAPDH used as loading control. H, HepG2 growth over time as measured by percent confluence on an IncuCyte ZOOM with exposure to solvent, 100 nM mithramycin (100 nM MMA), 20 nM mithramycin (20 nM MMA), 2 μM PHA-767491, or 20 nM mithramycin and 2 μM PHA-767491 (Combo) for 72 hours and allowed to grow for 110 hours. **** = P < 0.0001, shaded regions represent standard deviation.
Superoxide is a major physiological ROS and also the precursor for hydrogen peroxide and hydroxyl radicals (48). To investigate whether BTG2 induction leads to ROS generation in liver cells, we used electron paramagnetic resonance spectroscopy to measure the level of superoxide radicals in HepG2 cells after 24-hour mithramycin administration (Fig. 5C). We observed a concentration-dependent and significant increase in superoxide levels after mithramycin administration. This effect was rescued with siRNA silencing of BTG2 in the same cells with the same exposure to drug (Fig. 5C). This rescue of superoxide production mitigated the toxicity of mithramycin in these cells at several concentrations of drug (Fig. 5D). In addition, when viewed over time as percent confluence, silencing of BTG2 eliminated the cytotoxic effects of 50 nM mithramycin (Fig. 5E). These effects were phenocopied by the CDK9 inhibitor, PHA-767491 which suppressed the expression of BTG2 as a single agent (Fig. 5F). Strikingly, PHA-767491 prevented induction of BTG2 mRNA and protein even after exposure to a higher 100 nM concentration of mithramycin (Fig. 5F, G). These effects antagonized the cytotoxic effects of mithramycin and protected HepG2 cells from mithramycin induced cytotoxicity (Fig. 5H). The identical combination of 20 nM mithramycin and 2 μM PHA-767491 that synergistically eliminated Ewing sarcoma cellular proliferation with just 24 hours of drug exposure (Fig. 4B) had no effect on HepG2 proliferation even with 72 hours of exposure (Fig. 5H). Together the data suggest that CDK9 blockade may shift the IC50 of mithramycin enough in Ewing sarcoma cells to achieve the therapeutic suppression of EWS-FLI1 at clinically achievable concentrations of mithramycin. Furthermore, the exposure of Ewing sarcoma cells to mithramycin may actually increase in patients in combination with PHA-767491 due to a context-dependent loss of BTG2 induction in hepatocytes leading to a more effective and less hepatotoxic therapy.
Discussion
It has been known for more than 20 years that Ewing sarcoma cells depend on the EWS-FLI1 transcription factor for survival. While a number of promising studies that seek to target EWS-FLI1 are currently entering the clinic including shRNA targeting of EWS-FLI1, TK216, seclidemstat, and trabectedin in combination with irinotecan. Unfortunately, the therapeutic suppression of EWS-FLI1 has not yet been achieved (3,49,50). Previous clinical studies of cytarabine and mithramycin failed to show evidence of activity or EWS-FLI1 suppression (8,51). In the case of mithramycin, we believe that this failure was due to the off-target liver toxicity that limited serum concentrations of drug to levels that are insufficient to inhibit EWS-FLI1 activity. To overcome this, we have reported a second-generation analog of mithramycin, EC8042, with an improved toxicity profile that should achieve high enough serum concentration to inhibit the target (52). However, it is challenging to develop novel compounds for rare tumors such as Ewing sarcoma. Therefore, a complementary approach employed in this study was to identify a compound that potentiates the suppression of EWS-FLI1, does not have overlapping toxicity and could allow for the clinical realization of a mithramycin-based EWS-FLI1 targeted therapy.
In order to accomplish this goal, we performed a siRNA screen of the druggable genome to identify targets that would sensitize Ewing sarcoma cells to mithramycin by enhancing the suppression of EWS-FLI1. Consistent with the goal of the project, the top hits from the screen included several subunits of RNAPII and other important components of RNAPII assembly and activity. Therefore, we assembled a panel of relatively non-specific transcription inhibitors and used a matrix drug screening approach to identify synergistic combinations that had a focus of synergy at the 20 nM clinically achievable concentration of mithramycin. We found a “class effect” with CDK9 inhibitors that drove a dramatic shift in the IC50 of mithramycin, amplified the suppression of the EWS-FLI1 transcriptome and even led to a loss of expression of EWS-FLI1 itself. Importantly, the loss of cell viability due to the mithramycin-CDK9 inhibitor combination was selective for Ewing sarcoma cells relative to a liver-derived cell line and the identical concentration that eliminated proliferation of Ewing sarcoma cells had no effect on HepG2 proliferation (Fig. 6). We linked this cytotoxic selectivity to the context-dependent induction of BTG2 expression which led to the accumulation of ROS in liver cells that could be rescued with siRNA silencing of BTG2. PHA-767491 also blocked the induction of BTG2 potentially providing a more effective and less toxic therapy. These studies provide proof of principle of this combination in Ewing sarcoma. However, it is notable that this does appear to be a “class effect” with CDK9 inhibitors. Therefore, any of the CDK9 inhibitors could be selected for clinical development with limited validation based on practical consideration such as pharmacokinetics, toxicity profile and availability of compound.
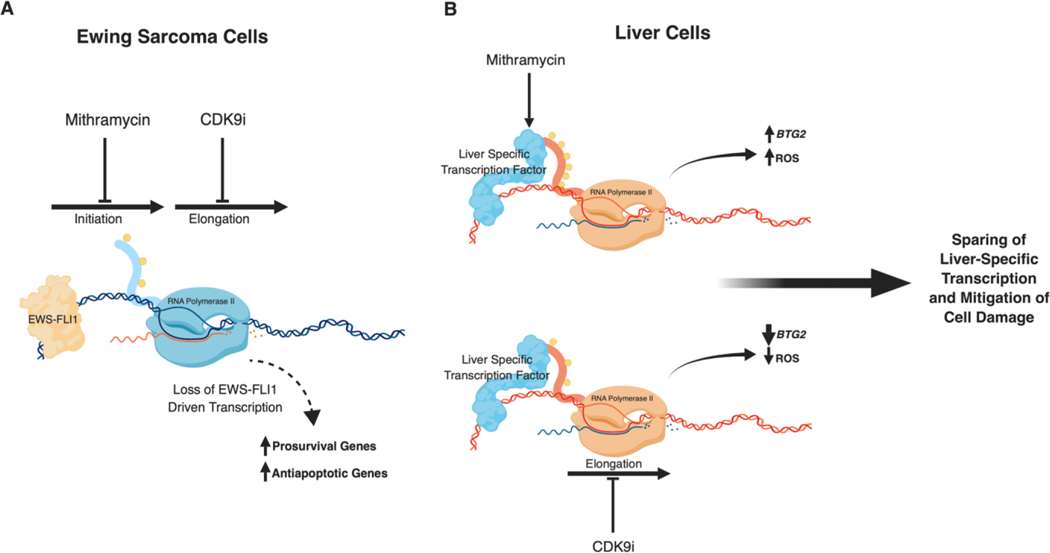
Figure 6.
Summary of therapeutic strategy. Created with BioRender.com. A, EWS-FLI1 suppression by mithramycin requires a clinically unachievable concentration of 50–100 nM. A clinically achievable concentration of drug can be used if combined with sequential targeting of the transcription process. B, Since only Ewing sarcoma cells express EWS-FLI1, this strategy should have minimal effect on non-Ewing sarcoma cells. Furthermore, mithramycin induced liver damage is also transcriptionally regulated as BTG2 induction leads to ROS accumulation and a loss of liver cell viability. CDK9 inhibition abolishes the induction of BTG2 thus mitigating mithramycin induced damage in liver cells.
Interestingly, CDK9 was not identified as a top candidate in our initial siRNA screen for mithramycin sensitizers even though it was contained in the library. This is most likely due to the fact that there is a reserve pool of CDK9 that is associated with the long non-coding RNA 7SK and the protein HEXIM1 (53). In our initial siRNA screen, we would not have silenced CDK9 long enough to exhaust the available pool. The approach of sequential screening allowed us to identify a non-subtle class effect with CDK9 inhibitors which we validated was “on-target” using complementary western blots of serine-2 phosphorylation, qPCR of ERV induction, and nuclear run-on assays. It has been suggested that the induction of ERVs may render tumors more sensitive to immunotherapy (54). An intriguing possibility is that this combination may render Ewing sarcoma more immunogenic.
This study has a number of important implications for therapies that target specific transcription factors. The therapeutic targeting of specific transcription factors remains a challenge and there are few examples of highly specific agents that have successfully targeted specific transcription factors with a favorable toxicity profile. It is likely that this limited specificity is due to the biology of transcription which requires multiple proteins and multi-subunit complexes to direct the expression of a given transcriptional program. Indeed, even RNAPII itself is a multi-subunit complex. Therefore, the realization of transcription factor targeting may require combination therapies that exploit context-dependent changes in gene expression to improve the balance between efficacy and toxicity. Here, we provide a road map for identifying combination therapies of this sort and a new approach to target oncogenic transcription factors that we call context dependent sequential targeting. Importantly, transcriptional cyclin dependent kinases such as CDK7 have shown activity in other transcription factor driven diseases such as MYCN-amplified neuroblastoma (55). In addition, CDK12 inhibitors synergize with PARP in Ewing sarcoma and show evidence of synthetic lethality (56). Here we show, another use of these compounds is to directly target discrete steps in the transcription process as a means to capitalize on context-dependent transcription to lead to more effective target suppression. Since EWS-FLI1 is only found in Ewing sarcoma cells, by combining an EWS-FLI1 inhibitor and a general transcriptionally directed agent, cooperative suppression would only occur at EWS-FLI1 target genes. The reason this effect is so prominent in the EWS-FLI1 transcriptional program may be due to releasing the EWS-FLI1 mediated block of EWSR1 that has been previously described leading to a further reduction in gene expression (42). Our data show that CDK9 blockade by itself minimally impacts EWS-FLI1 target gene expression and even induces many of the targets. However, in combination with a clinically achievable concentration of mithramycin, PHA-767491 greatly enhances suppression to eliminate expression of several downstream targets. We show that this results from effects on RNAPII processivity at the EWS-FLI1 target gene EZH2. In addition, context dependent transcription changes also may lead to less combined toxicity based on a blockade of induction of BTG2. The net result is reversal of the gene signature, a dramatic reduction in tumor size, and evidence of tumor lysis in mice which is extremely uncommon in solid tumors. While the management of tumor lysis in our mouse model was difficult, in the clinic this would be routine as it is frequently needed for standard management for B-cell leukemia (44). In summary, these data provide a novel EWS-FLI1 targeted combination therapy and a unique approach to improve the therapeutic window of transcription directed cancer therapies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karsten Melcher, Stefan Jovinge, and David Kirsch for insight and critiques on this project and Kristen Gehlhaus (Genetics Branch, CCR, NCI) for assistance with optimizing the siRNA screening conditions.
FUNDING
The work was supported by Alex’s Lemonade Stand, the National Cancer Institute (PJG: R01CA188314), internal funds at the Van Andel Institute (PJG) as well as by Intramural Research Program of the National Cancer Institute (NCI), Center for Cancer Research (CCR) (NJC: ZIA BC 011704 and Z01 BC 010939) and the National Center for Advancing Translational Sciences.
Footnotes
Grant Support:
Disclosure of Potential Conflicts of Interest: None to disclose.
References
- 1.May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proceedings of the National Academy of Sciences of the United States of America 1993;90(12):5752–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer research 2008;68(7):2176–85 doi 10.1158/0008-5472.can-07-1761. [DOI] [PubMed] [Google Scholar]
- 3.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer research 2005;65(19):8984–92 doi 10.1158/0008-5472.can-05-0565. [DOI] [PubMed] [Google Scholar]
- 4.Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. Journal of the National Cancer Institute 2011;103(12):962–78 doi 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendiola M, Carrillo J, Garcia E, Lalli E, Hernandez T, de Alava E, et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int J Cancer 2006;118(6):1381–9 doi 10.1002/ijc.21578. [DOI] [PubMed] [Google Scholar]
- 6.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PloS one 2009;4(3):e4932 doi 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson KM, Taslim C, Saund RS, Lessnick SL. Identification of two types of GGAA-microsatellites and their roles in EWS/FLI binding and gene regulation in Ewing sarcoma. PloS one 2017;12(11):e0186275 doi 10.1371/journal.pone.0186275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grohar PJ, Glod J, Peer CJ, Sissung TM, Arnaldez FI, Long L, et al. A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS-FLI1 fusion transcript. Cancer chemotherapy and pharmacology 2017. doi 10.1007/s00280-017-3382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booth GT, Parua PK, Sanso M, Fisher RP, Lis JT. Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nature communications 2018;9(1):543 doi 10.1038/s41467-018-03006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, Murphy S. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nature structural & molecular biology 2015;22(5):396–403 doi 10.1038/nsmb.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vos SM, Farnung L, Urlaub H, Cramer P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature 2018;560(7720):601–6 doi 10.1038/s41586-018-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim YB, Park TJ, Lim IK. B cell translocation gene 2 enhances susceptibility of HeLa cells to doxorubicin-induced oxidative damage. The Journal of biological chemistry 2008;283(48):33110–8 doi 10.1074/jbc.M804255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–40 doi 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell cycle (Georgetown, Tex) 2008;7(2):250–6 doi 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 16.Dikalov SI, Kirilyuk IA, Voinov M, Grigor’ev IA. EPR detection of cellular and mitochondrial superoxide using cyclic hydroxylamines. Free radical research 2011;45(4):417–30 doi 10.3109/10715762.2010.540242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel AB, Louder RK, Greber BJ, Grunberg S, Luo J, Fang J, et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science (New York, NY) 2018;362(6421) doi 10.1126/science.aau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoll ER, Zhu ZI, Sarkar D, Landsman D, Morse RH. Role of the pre-initiation complex in Mediator recruitment and dynamics. eLife 2018;7 doi 10.7554/eLife.39633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuber PK, Hahn L, Reinl A, Schweimer K, Knauer SH, Gottesman ME, et al. Structure and nucleic acid binding properties of KOW domains 4 and 6–7 of human transcription elongation factor DSIF. Scientific reports 2018;8(1):11660 doi 10.1038/s41598-018-30042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miki TS, Carl SH, Grosshans H. Two distinct transcription termination modes dictated by promoters. Genes & development 2017;31(18):1870–9 doi 10.1101/gad.301093.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albertini V, Jain A, Vignati S, Napoli S, Rinaldi A, Kwee I, et al. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer Streptomyces argillaceus exhibits improved transcriptional repressor activity: implications for cancer therapy. Nucleic Acids Research 2006;34(6):1721–34 doi 10.1093/nar/gkl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest 1991;88(5):1613–21 doi 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray R, Thomas S, Miller DM. Mithramycin selectively inhibits the transcriptional activity of a transfected human c-myc gene. The American journal of the medical sciences 1990;300(4):203–8. [DOI] [PubMed] [Google Scholar]
- 24.Natoni A, Murillo LS, Kliszczak AE, Catherwood MA, Montagnoli A, Samali A, et al. Mechanisms of action of a dual Cdc7/Cdk9 kinase inhibitor against quiescent and proliferating CLL cells. Molecular cancer therapeutics 2011;10(9):1624–34 doi 10.1158/1535-7163.mct-10-1119. [DOI] [PubMed] [Google Scholar]
- 25.Ali MA, Choy H, Habib AA, Saha D. SNS-032 prevents tumor cell-induced angiogenesis by inhibiting vascular endothelial growth factor. Neoplasia (New York, NY) 2007;9(5):370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KS, Sack JS, Tokarski JS, Qian L, Chao ST, Leith L, et al. Thio- and oxoflavopiridols, cyclin-dependent kinase 1-selective inhibitors: synthesis and biological effects. Journal of medicinal chemistry 2000;43(22):4126–34. [DOI] [PubMed] [Google Scholar]
- 27.Albert TK, Rigault C, Eickhoff J, Baumgart K, Antrecht C, Klebl B, et al. Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. British journal of pharmacology 2014;171(1):55–68 doi 10.1111/bph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nature chemical biology 2011;7(3):182–8 doi 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane ME, Yu B, Rice A, Lipson KE, Liang C, Sun L, et al. A novel cdk2-selective inhibitor, SU9516, induces apoptosis in colon carcinoma cells. Cancer research 2001;61(16):6170–7. [PubMed] [Google Scholar]
- 30.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Molecular cancer therapeutics 2004;3(11):1427–38. [PubMed] [Google Scholar]
- 31.Alexander LT, Mobitz H, Drueckes P, Savitsky P, Fedorov O, Elkins JM, et al. Type II Inhibitors Targeting CDK2. ACS chemical biology 2015;10(9):2116–25 doi 10.1021/acschembio.5b00398. [DOI] [PubMed] [Google Scholar]
- 32.Luzzio MJ, Besterman JM, Emerson DL, Evans MG, Lackey K, Leitner PL, et al. Synthesis and antitumor activity of novel water soluble derivatives of camptothecin as specific inhibitors of topoisomerase I. Journal of medicinal chemistry 1995;38(3):395–401. [DOI] [PubMed] [Google Scholar]
- 33.Ali S, Heathcote DA, Kroll SH, Jogalekar AS, Scheiper B, Patel H, et al. The development of a selective cyclin-dependent kinase inhibitor that shows antitumor activity. Cancer research 2009;69(15):6208–15 doi 10.1158/0008-5472.can-09-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stirpe F, Fiume L. Effect of alpha-amanitin on ribonucleic acid synthesis and on ribonucleic acid polymerase in mouse liver. The Biochemical journal 1967;103(3):67p–8p. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Pandey S, Travers M, Sun H, Morton G, Madzo J, et al. Targeting CDK9 Reactivates Epigenetically Silenced Genes in Cancer. Cell 2018;175(5):1244–58.e26 doi 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nature chemical biology 2008;4(6):357–65 doi 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- 37.Boro A, Pretre K, Rechfeld F, Thalhammer V, Oesch S, Wachtel M, et al. Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing’s sarcoma. International journal of cancer 2012;131(9):2153–64 doi 10.1002/ijc.27472. [DOI] [PubMed] [Google Scholar]
- 38.Hensel T, Giorgi C, Schmidt O, Calzada-Wack J, Neff F, Buch T, et al. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget 2016;7(2):1451–63 doi 10.18632/oncotarget.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell cycle (Georgetown, Tex) 2016;15(4):519–27 doi 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeronimo C, Collin P, Robert F. The RNA Polymerase II CTD: The Increasing Complexity of a Low-Complexity Protein Domain. Journal of molecular biology 2016;428(12):2607–22 doi 10.1016/j.jmb.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Rossier C, Antonarakis SE. Cloning of a human homolog of the Drosophila enhancer of zeste gene (EZH2) that maps to chromosome 21q22.2. Genomics 1996;38(1):30–7 doi 10.1006/geno.1996.0588. [DOI] [PubMed] [Google Scholar]
- 42.Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018;555(7696):387–91 doi 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DP, Mahmoud H, Chesney RW. Tumor lysis syndrome: pathogenesis and management. Pediatric nephrology (Berlin, Germany) 1995;9(2):206–12. [DOI] [PubMed] [Google Scholar]
- 44.van der Hoven B, Thunnissen PL, Sizoo W. Tumour lysis syndrome in haematological malignancies. The Netherlands journal of medicine 1992;40(1–2):31–5. [PubMed] [Google Scholar]
- 45.Filtz TM, Vogel WK, Leid M. Regulation of transcription factor activity by interconnected post-translational modifications. Trends in pharmacological sciences 2014;35(2):76–85 doi 10.1016/j.tips.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Lechon MJ, Tolosa L, Donato MT. Upgrading HepG2 cells with adenoviral vectors that encode drug-metabolizing enzymes: application for drug hepatotoxicity testing. Expert opinion on drug metabolism & toxicology 2017;13(2):137–48 doi 10.1080/17425255.2017.1238459. [DOI] [PubMed] [Google Scholar]
- 47.Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN. Tumor suppressors BTG1 and BTG2: Beyond growth control. Journal of cellular physiology 2018. doi 10.1002/jcp.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radi R. Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proceedings of the National Academy of Sciences of the United States of America 2018;115(23):5839–48 doi 10.1073/pnas.1804932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao DD, Jay C, Wang Z, Luo X, Kumar P, Eysenbach H, et al. Preclinical Justification of pbi-shRNA EWS/FLI1 Lipoplex (LPX) Treatment for Ewing’s Sarcoma. Molecular therapy : the journal of the American Society of Gene Therapy 2016;24(8):1412–22 doi 10.1038/mt.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harlow ML, Chasse MH, Boguslawski EA, Sorensen KM, Gedminas JM, Kitchen-Goosen SM, et al. Trabectedin Inhibits EWS-FLI1 and Evicts SWI/SNF from Chromatin in a Schedule-dependent Manner. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25(11):3417–29 doi 10.1158/1078-0432.Ccr-18-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DuBois SG, Krailo MD, Lessnick SL, Smith R, Chen Z, Marina N, et al. Phase II study of intermediate-dose cytarabine in patients with relapsed or refractory Ewing sarcoma: a report from the Children’s Oncology Group. Pediatric blood & cancer 2009;52(3):324–7 doi 10.1002/pbc.21822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osgood CL, Maloney N, Kidd CG, Kitchen-Goosen S, Segars L, Gebregiorgis M, et al. Identification of Mithramycin Analogues with Improved Targeting of the EWS-FLI1 Transcription Factor. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(16):4105–18 doi 10.1158/1078-0432.ccr-15-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Molecular cell 2003;12(4):971–82. [DOI] [PubMed] [Google Scholar]
- 54.Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat Rev Cancer 2019;19(3):151–61 doi 10.1038/s41568-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 55.Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nature genetics 2018;50(9):1240–6 doi 10.1038/s41588-018-0191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iniguez AB, Stolte B, Wang EJ, Conway AS, Alexe G, Dharia NV, et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer cell 2018;33(2):202–16.e6 doi 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.