Abstract
Allergic rhinitis (AR) is an IgE-mediated upper airway disease with a high worldwide prevalence. MicroRNA (miR)-205-5p upregulation has been observed in AR; however, its role is poorly understood. The aim of the present study was to investigate the effect of miR-205-5p on AR-associated inflammation. To establish an AR model, BALB/c mice were sensitized using an intraperitoneal injection of ovalbumin (OVA) on days 0, 7 and 14, followed by intranasal challenge with OVA on days 21–27. A lentiviral sponge for miR-205-5p was used to downregulate miR-205-5p in vivo via intranasal administration on days 20–26. Reverse transcription-quantitative PCR revealed that miR-205-5p was upregulated in AR mice. Notably, miR-205-5p knockdown reduced the frequency of nose-rubbing and sneezing, and attenuated pathological alterations in the nasal mucosa. The levels of total and OVA-specific IgE, cytokines IL-4, IL-5 and IL-13, and inflammatory cells, were decreased by miR-205-5p knockdown in AR mice. In addition, miR-205-5p knockdown inhibited nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activation by reducing the expression levels of NLRP3, apoptosisassociated specklike protein containing a CARD, cleaved caspase-1 and IL-1β by western blot analysis. B-cell lymphoma 6 (BCL6) was confirmed as a target of miR-205-5p by luciferase reporter assay. In conclusion, the present findings suggested that miR-205-5p knockdown may attenuate the inflammatory response in AR by targeting BCL6, which may be a potential therapeutic target for AR.
Keywords: allergic rhinitis, B-cell lymphoma 6, inflammatory response, microRNA-205-5p
Introduction
Allergic rhinitis (AR) is a chronic inflammatory disease of the upper airway with a high worldwide prevalence (10–40%). AR is characterized by sneezing, pruritus, rhinorrhea, retronasal drainage and nasal congestion (1–3). AR is triggered by allergen-specific IgE-mediated reactions, and type 2 helper T (Th2) cells further drive this inflammatory response against inhaled allergens in the environment (4). These inhaled allergens include pollens, dust mites, animal dander and mold (5). Multiple inflammatory cells, such as B and T cells, basophils and mast cells, are involved in this inflammatory process (6). AR has a severe impact on the quality of life of patients. Children or adults with AR may suffer from learning impairments, and sleep or emotional disturbances (7). In recent years, the prevalence of AR has increased at an alarming rate in adults and children worldwide (8). In addition, the presence of AR could significantly increase the risk of asthma (9); according to previous studies, ≤40% of individuals with AR either have or will get asthma (5,9–11). Furthermore, patients with AR usually suffer from allergic conjunctivitis (12). AR presents a great challenge to humans, due to its high prevalence and other complications. Pharmacotherapy is one of the major approaches to the management of AR, and nasal corticosteroids and antihistamines are the most common medications. Effective pharmacotherapy requires regular use of medications so that the symptoms do not recur following discontinuation (13–15).
MicroRNAs (miRNAs/miRs) are single-stranded non-coding RNA molecules ~22 nucleotides in length. miRNAs mediate silencing of target genes by binding to target mRNA (16). Mature miRNAs bind to the 3′-untranslated region (3′-UTR) of their target mRNA transcripts with the help of the RNA-induced silencing complex, and then induce the degradation of target mRNA or inhibit protein translation (17,18). miRNAs serve an important role in a variety of physiological and pathological processes. Abnormal miRNA expression has been reported to be involved in the occurrence and development of various human diseases, including AR (19,20). Suojalehto et al (21) reported that miR-205-5p was upregulated in the nasal mucosa of symptomatic patients with AR. The significance of miR-205-5p in regulating inflammation has also been demonstrated. Notably, IL-32α has been shown to suppress vascular inflammation and atherosclerosis by inhibiting miR-205-5p biogenesis in mice (22). Furthermore, miR-205-5p has been demonstrated to alleviate hip fracture-induced rat lung injury by decreasing inflammatory response through inhibition of the expression of high mobility group box 1 (23). These studies suggested that miR-205-5p may have different functions in different pathological environments, which can be explained by its target gene or regulatory mechanism. However, to the best of our knowledge, the role of miR-205-5p in AR remains unknown and requires further investigation.
B-cell lymphoma 6 (BCL6) is a sequence-specific transcriptional regulator that inhibits the transcription of target genes by binding to a specific DNA sequence in the promoter region (24,25). BCL6 serves an important role in the development of B cells, follicular Th cells and T regulatory cells, indicating its potential function in regulation of the immune response (26–28). BCL6 has also been reported to be downregulated in the nasal mucosa of patients with AR (29). Hiromura et al (30) reported that IL-21 upregulated BCL6 and relieved AR. In addition, BCL6 may inhibit the inflammatory response in human renal tubular epithelial cells and attenuate renal inflammation by negatively regulating the transcription of nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3) (31). These findings suggested the importance of BCL6 in the regulation of inflammation in human diseases. Therefore, it was hypothesized that BCL6 could also be involved in AR-related inflammation.
With the rising prevalence of AR, it is necessary to develop novel and effective approaches to AR treatment. In the present study, the functions of miR-205-5p and its underlying regulatory mechanism in AR were explored. The present study aimed to improve understanding of AR pathogenesis and a potential therapeutic target for AR.
Materials and methods
Animal model
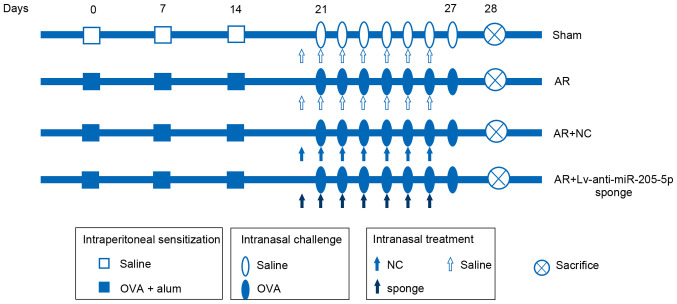
Female wild-type (WT) BALB/c mice (age, 4 weeks; weight, 18–22 g) were purchased from Liaoning Changsheng Biotechnology Co., Ltd. A total of 168 mice were used in the present study. All animal experiments were approved by the Ethics Committee of The Second Affiliated Hospital of Shenyang Medical College (approval no. 2020005; Shenyang, China). Mice were maintained under a 12-h dark/light cycle, at 45–55% humidity and 22±1°C room temperature with free access to food and water. After a 1-week acclimation period, all mice were randomly divided into the Sham, AR, AR + negative control (NC) and AR + Lv-anti-miR-205-5p sponge groups. A mouse model of AR was established according to a previous study (32). As shown in Fig. 1, in the AR group, the mice were sensitized via an intraperitoneal (i.p.) injection of 200 µl saline containing 25 µg ovalbumin (OVA; Shanghai Aladdin Biochemical Technology Co., Ltd.) and 2 mg aluminum hydroxide on days 0, 7 and 14. The mice were then subjected to continuous intranasal challenges with 100 µg OVA (50 µl; 2 mg/ml) on days 21–27. The sham group was administered an equivalent amount of saline without OVA or aluminum hydroxide on days 0, 7 and 14, and on days 21–27. A total of 24 h before each OVA challenge on days 21–27, a recombinant lentiviral vector carrying an anti-miR-205-5p sponge or NC (empty vector) was constructed and intranasally administered to the mice (20 µl; 1×108 TU/ml).
Figure 1.
Experimental protocol for sensitization with OVA and treatment with a lentivirus-mediated sponge for miR-205-5p. The AR mice were sensitized via an intraperitoneal injection of saline containing 25 µg OVA and 2 mg aluminum hydroxide on days 0, 7 and 14. The mice were then subjected to continuous intranasal challenges with 100 µg OVA (50 µl; 2 mg/ml) on days 21–27. The sham group was administered an equivalent amount of saline without OVA or aluminum hydroxide on days 0, 7 and 14, and on days 21–27. A total of 24 h before each OVA challenge on days 21–27, a recombinant lentiviral vector carrying a anti-miR-205-5p sponge or NC (20 µl; 1×108 TU/ml) was intranasally administered to the mice in the AR + NC and AR + Lv-anti-miR-205-5p sponge groups, respectively. AR, allergic rhinitis; miR, microRNA; NC, negative control; OVA, ovalbumin.
The frequency of nose-rubbing or sneezing within 15 min from the final OVA challenge was recorded. A total of 24 h after the final OVA challenge, mice were euthanized with an overdose of sodium pentobarbital (150 mg/kg, i.p.). Blood (~1 ml) was collected from the retrobulbar vein immediately and serum was isolated for use (33,34). Nasal lavage fluid (NLF) was harvested by cannulating the upper part of the trachea into nasal cavity and lavaging with 1 ml PBS, followed by centrifugation at 277 g for 10 min at 4°C. The collected serum, NLF and some nasal mucosa tissues were frozen in liquid nitrogen and then stored at −70°C for Gimesa staining, ELISA, reverse transcription-quantitative (RT-q)PCR and western blot analysis. Some nasal mucosa tissues were fixed in 4% paraformaldehyde at room temperature for 24 h for histological analysis.
RT-qPCR
Total RNA was isolated from the nasal mucosa using TRIpure (BioTeke Corporation) and reverse transcribed into first-strand cDNA at 25°C for 10 min, 42°C for 50 min and 80°C for 10 min using Super M-MLV Reverse Transcriptase (BioTeke Corporation). qPCR was then performed using an Exicycler™ 96 Real-time PCR system (Bioneer Corporation) and SYBR Green kit (Merck KGaA) according to the manufacturer's instructions. The reaction conditions were as follows: 94°C for 5 min, 94°C for 15 sec, 60°C for 25 sec and 72°C for 30 sec; followed by 40 cycles of 72°C for 5 min 30 sec, 40°C for 5 min 30 sec; melting at 60–94°C for 34 sec and final extension for 2 min at 25°C.
The transcript levels of genes were analyzed quantitatively using the 2−ΔΔCq method and normalized to 5S or β-actin (35). The primer sequences used were as follows: mmu-miR-205-5p, forward, 5′-TCCTTCATTCCACCGGAGTCTG-3′ and reverse, 5′-GCAGGGTCCGAGGTATTC-3′; 5S, forward, 5′-CTAAAGATTTCCGTGGAGAG-3′ and reverse, 5′-TGGTGCAGGGTCCGAGGTAT-3′; BCL6, forward, 5′-CGGAAGGGTCTGGTAGT-3′ and reverse, 5′-CATTCTGATTGAGGCTGTTG-3′; β-actin, forward, 5′-CTGTGCCCATCTACGAGGGCTAT-3′ and reverse, 5′-TTTGATGTCACGCACGATTTCC-3′.
Histological analysis
H&E staining was performed to assess histopathological alterations in nasal mucosa tissues. Briefly, mouse nasal mucosa tissues fixed with 4% paraformaldehyde at room temperature for 24 h were dehydrated in increasing ethanol gradients and embedded in paraffin. The tissues were cut into 5-µm slices and stained with 0.2% hematoxylin (Beijing Solarbio Science & Technology Co., Ltd.) for 5 min and 0.35% eosin (Sangon Biotech Co., Ltd.) for 3 min at room temperature for histopathological examination. Histopathological changes were observed under a BX53 light microscope (Olympus Corporation) at a magnification of ×200. The replicates of histological analysis came from seven different mice in each group and only the representative image was displayed.
Semi-quantification of inflammatory cells in NLF
Inflammatory cells in NLF were measured by cytochemical staining with Wright's-Giemsa. Mouse NLF was collected and centrifuged at 300 × g for 10 min at the room temperature. Nasal lavage smears were stained with Wright's-Giemsa (cat. no. D010; Nanjing Jiancheng Bioengineering Institute) at room temperature according to the manufacturer's instructions, and inflammatory cells were identified as leukocytes, eosinophils, neutrophils, lymphocytes and macrophages, according to their morphology. NLF cell differentials were counted under a light microscope at a magnification of ×200. In total, five random fields of each sample were captured for average counting.
Western blot analysis
The nasal mucosa tissue of each mouse was ground in liquid nitrogen with protein lysis buffer (Beyotime Institute of Biotechnology) containing 1 mM PMSF and placed on ice for 5 min. After centrifugation at 10,000 × g and 4°C for 5 min, the supernatant was collected and quantified using a BCA kit (Beyotime Institute of Biotechnology). The mean protein concentration was 3.46 µg/µl (Sham group), 3.31 µg/µl (AR group), 3.81 µg/µl (AR + NC group) and 3.88 µg/µl (AR + Lv-anti-miR-205-5p sponge group). The amount of protein loaded for BCL6 detection was 20 µg per sample, and that for the detection of IL-1β, caspase-1, ASC or NLRP3 was 40 µg per sample. The proteins were separated by SDS-PAGE on 10% gels, transferred to polyvinylidene difluoride membranes (MilliporeSigma), and then blocked with 5% skimmed milk solution for 1 h at room temperature. The membranes were incubated with primary antibodies (Table I) overnight at 4°C. After washing with TBS-Tween-20 (1.5% m/v) buffer, the membranes were incubated with goat anti-rabbit IgG-HRP (dilution, 1:5,000; cat. no. A0208; Beyotime Institute of Biotechnology) or goat anti-mouse IgG-HRP secondary antibodies (dilution, 1:5,000; cat. no. A0216; Beyotime Institute of Biotechnology) for 45 min at 37°C. β-actin was used as an internal reference. All antibodies used for western blot analysis were polyclonal antibodies. Protein bands were visualized and analyzed using enhanced chemiluminescence reagent (Beyotime Institute of Biotechnology) under a gel imaging Gel-Pro-Analyzer system (WD-9413B; Beijing Liuyi Biotechnology Co., Ltd.). Image-Pro Plus software 6.0 (Media Cybernetics, Inc.) was used for semi-quantification. The western blot analysis replicates were from seven different mice in each group.
Table I.
Primary antibodies used in western blot analysis.
| Primary antibody | Dilution | Supplier | Cat. no. |
|---|---|---|---|
| BCL6 | 1:500 | ProteinTech Group, Inc. | 21187-1-AP |
| IL-1βa | 1:400 | Affinity Biosciences, Ltd. | AF5103 |
| Caspase-1b | 1:1,000 | Affinity Biosciences, Ltd. | AF5418 |
| ASC | 1:500 | ABclonal Biotech Co., Ltd. | A1170 |
| NLRP3 | 1:1,000 | ABclonal Biotech Co., Ltd. | A5652 |
| β-actin | 1:1,000 | Santa Cruz Biotechnology Co., Ltd. | sc-47778 |
IL-1β antibody recognizes both pro-IL-1β (31 kDa) and mature IL-1β (17 kDa)
caspase-1 antibody recognizes both pro-caspase-1 (45 kDa) and cleaved caspase-1 (20 kDa). ASC, apoptosisassociated specklike protein containing a CARD; BCL6, B-cell lymphoma 6; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3.
ELISA
ELISA was used to measure the serum levels of total and OVA-specific IgE, and those of IL-4, IL-5 and IL-13 in nasal mucosa tissues. The serum levels of total and OVA-specific IgE were detected using a Mouse IgE ELISA kit [cat. no. EK275; Hangzhou Multisciences (Lianke) Biotech Co., Ltd.] and Mouse OVA-sIgE ELISA kit (cat. no. JL46328; Jianglaibio Biology), respectively, according to the manufacturer's instructions. To detect the levels of IL-4, IL-5 and IL-13, nasal mucosa tissues were homogenized in saline and centrifuged at 430 × g for 10 min at the room temperature. The concentration of proteins in the supernatant was quantified using a BCA kit (Beyotime Institute of Biotechnology). The concentration of IL-4 (cat. no. EK0405), IL-5 (cat. no. EK0408) or IL-13 (cat. no. EK0425) was assessed using the corresponding ELISA kit (Boster Biological Technology). The levels of IL-4, IL-5 or IL-13 in nasal mucosa tissues were expressed as the ratio of cytokine concentration to isolated protein concentration (pg/mg protein). Replicates were from seven different mice in each group.
Luciferase reporter assay
Luciferase reporter assay was performed to verify the binding of miR-205-5p to its target mRNA. The putative target was predicted according to the prediction website (TargetScanHuman; targetscan.org/vert_72/); BCL6 was revealed to be a potential target of miR-205-5p. The wild-type (WT) 3′-UTR of BCL6 and its mutant (MT) sequence, synthesized by GenScript (Nanjing) Co., Ltd., were cloned into the pmirGLO vector [GenScript (Nanjing) Co., Ltd.]. 293T cells (Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd.) at the density of 70% were co-transfected with the recombinant vector (1.5 µg, 0.5 µg/ml) containing the WT or MT 3′-UTR of BCL6 plus miR-205 or NC mimics (75 pmol, 25 pmol/ml) at 37°C for 4 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The sequences, synthesized by JTS scientific, were as follows: miR-205-5p mimics, 5′-UCCUUCAUUCCACCGGAGUCUG-3′ and 5′-GACUCCGGUGGAAUGAAGGAUU-3′; NC mimics, 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. Cells were grown in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Cytiva) and collected 48 h after transfection to determine luciferase signals using a dual-luciferase assay reporter kit (Promega Corporation). The relative luciferase activities represented the firefly/Renilla luciferase ratios. The experiment was repeated at least three times.
Immunohistochemistry
The fixed and paraffin-embedded nasal mucosa tissue slides were deparaffinized, rehydrated and subjected to boiling antigen retrieval for 10 min using 10% citrate buffer solution. Subsequently, the slides were exposed to 3% H2O2 (Sinopharm Chemical Reagent Co., Ltd.) for 15 min and then blocked with 100% goat serum (Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at room temperature. These slides were stained with a polyclonal anti-BCL6 primary antibody (dilution, 1:100; cat. no. Bs-2734R; Beijing Biosynthesis Biotechnology Co., Ltd.) overnight at 4°C. The slides were then washed with PBS three times, followed by incubation with polyclonal HRP-conjugated secondary antibody (dilution, 1:500; cat. no. 31460; Thermo Fisher Scientific, Inc.) at 37°C for 60 min. Chromogenic detection was performed using a DAB Substrate kit (Beijing Solarbio Science & Technology Co., Ltd.), followed by counterstaining with hematoxylin. Immunohistochemistry images were captured at a magnification of ×400 under a light microscope. The replicates came from seven different mice in each group and only the representative image was displayed. For semi-quantification of immunohistochemistry, three random fields were captured, and integrated optical density and area of each image were measured by Image-Pro Plus software 6.0 (Media Cybernetics, Inc.). The mean density represented BCL6 expression in the nasal mucosa.
Immunofluorescence staining
After dewaxing and rehydration, the fixed and paraffin-embedded nasal mucosa tissue slides underwent antigen retrieval as aforementioned, followed by blocking with 100% goat serum (Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at room temperature. The slides were then incubated with a mixture of polyclonal anti-BCL6 (dilution, 1:100; cat. no. Bs-2734R; Beijing Biosynthesis Biotechnology Co., Ltd.) and monoclonal anti-CD4 (dilution, 1:50; cat. no. Sc-20079; Santa Cruz Biotechnology, Inc.) or anti-F4/80 (dilution, 1:50; cat. no. Sc-377009; Santa Cruz Biotechnology, Inc.) antibodies overnight at 4°C. Subsequently, the sections were incubated with a mixture of Cy3-conjugated goat anti-rabbit IgG (dilution, 1:200; cat. no. A0516; Beyotime Institute of Biotechnology) and fluorescein isothiocyanate-labelled goat anti-mouse IgG (dilution, 1:200; cat. no. A0568; Beyotime Institute of Biotechnology) antibodies at room temperature for 90 min. Nuclei were counterstained with DAPI (Shanghai Aladdin Biochemical Technology Co., Ltd.). The slides were visually examined under a BX53 immunofluorescence microscope (Olympus Corporation) at a magnification of ×400. The replicates came from seven different mice in each group, and only the representative image was displayed.
Statistical analysis
All experiments were performed with seven biological replicates. Data are presented as the mean ± SD. Statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc.). One-way ANOVA followed by Tukey's post-hoc test was used for multiple comparisons among groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-205-5p knockdown alleviates OVA-induced AR
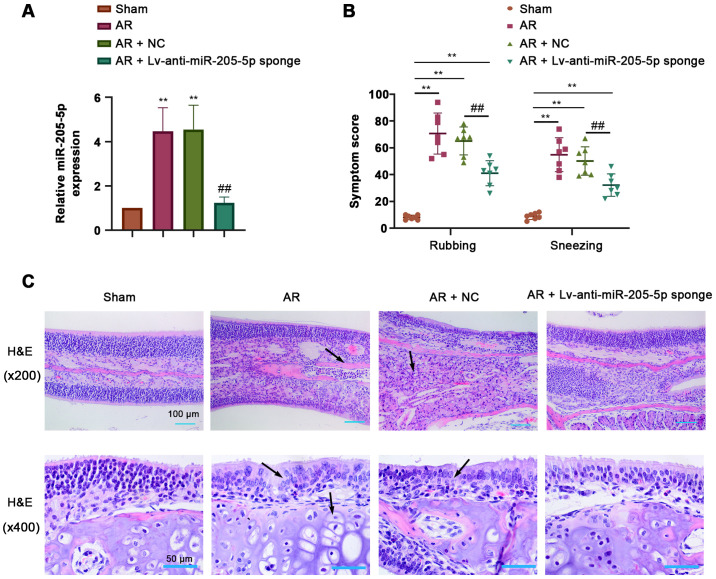
To determine the role of miR-205-5p in AR, the expression levels of miR-205-5p were detected by RT-qPCR in nasal mucosa tissues from AR mice sensitized by OVA. miR-205-5p expression was significantly increased in response to OVA-induced AR compared with that in the sham group, and was efficiently downregulated by the intranasal administration of a lentiviral sponge for miR-205-5p (Fig. 2A). The frequency of nose-rubbing or sneezing within 15 min of the final OVA challenge was recorded to evaluate the OVA-triggered AR response. The frequency of nose-rubbing or sneezing of AR mice was markedly increased compared with that of sham mice but was significantly alleviated by miR-205-5p knockdown (Fig. 2B). H&E staining showed apparent pathological alterations in the nasal mucosa of AR mice, including disarrangement of the epidermis, capillary edema and inflammatory cell infiltration. miR-205-5p knockdown attenuated hyperemia and inflammatory cell infiltration in the nasal mucosa of AR mice (Fig. 2C). These results suggested that miR-205-5p knockdown may alleviate OVA-induced AR.
Figure 2.
miR-205-5p knockdown alleviates symptoms of OVA-induced AR. (A) miR-205-5p expression levels in the nasal mucosa. (B) Frequency of nose rubbing and sneezing within 15 min of the final challenge with OVA. (C) Pathological alterations in the nasal mucosa of AR mice (magnification, ×200 and ×400; scale bars, 100 and 50 µm). Arrows indicate the necrotic cells of the nasal mucosa. Data are presented as the mean ± SD (n=7). **P<0.01 vs. Sham; ##P<0.01 vs. AR + NC. AR, allergic rhinitis; miR, microRNA; NC, negative control; OVA, ovalbumin.
miR-205-5p knockdown attenuates OVA-induced inflammatory response
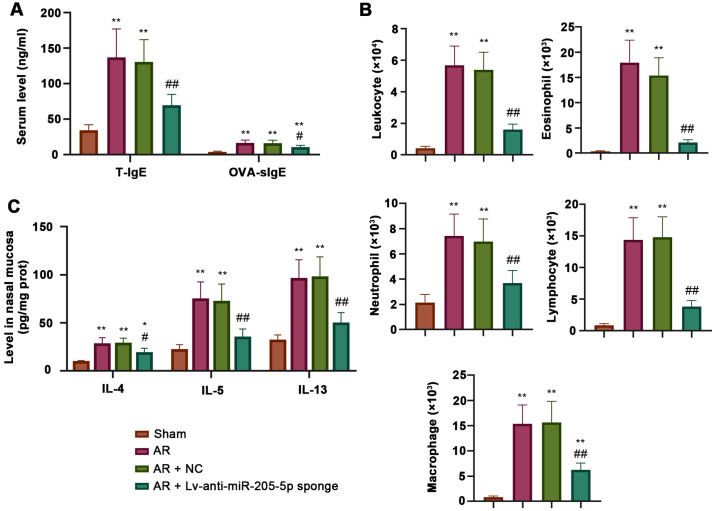
ELISA was performed to measure the levels of total and OVA-specific IgE, IL-4, IL-5 and IL-13 during AR. The concentrations of both total and OVA-specific IgE in the serum were increased in AR mice compared with those detected in sham mice, and were decreased by miR-205-5p knockdown (Fig. 3A). Wright's-Giemsa staining showed that the number of inflammatory cells, including leukocytes, eosinophils, neutrophils, lymphocytes and macrophages, was significantly higher in AR mice than that in sham mice. Conversely, the AR-induced increase in the number of inflammatory cells was significantly reduced by miR-205-5p knockdown (Fig. 3B). In addition, the levels of IL-4, IL-5 and IL-13 in the nasal mucosa were increased in AR mice compared with those in sham mice, but were decreased following miR-205-5p knockdown, as determined by ELISA (Fig. 3C). These data indicated that miR-205-5p knockdown may attenuate OVA-induced inflammatory reaction by reducing the levels of inflammatory cells and the production of proinflammatory cytokines.
Figure 3.
miR-205-5p knockdown decreases OVA-induced inflammatory response. (A) Levels of T-IgE and OVA-specific IgE in serum. (B) Number of inflammatory cells, including leukocytes, eosinophils, neutrophils, lymphocytes and macrophages in nasal lavage fluid. (C) Levels of IL-4, IL-5 and IL-13 in the nasal mucosa. Data are presented as the mean ± SD (n=7). *P<0.05, **P<0.01 vs. Sham; #P<0.05, ##P<0.01 vs. AR + NC. AR, allergic rhinitis; miR, microRNA; NC, negative control; OVA, ovalbumin; T-IgE, total IgE; OVA-sIgE, ovalbumin-specific serum immunoglobulin E.
miR-205-5p knockdown inhibits NLRP3 inflammasome activation in AR
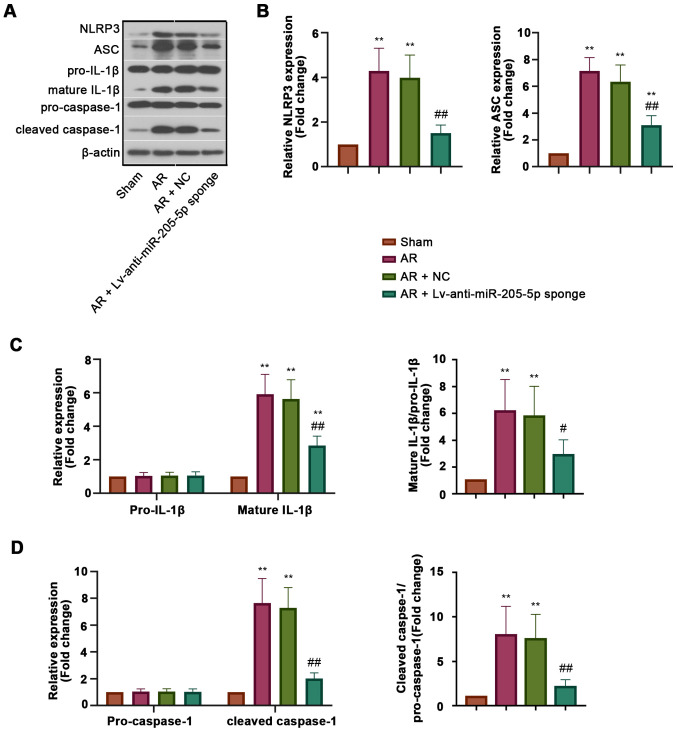
To explore the effect of miR-205-5p on NLRP3 inflammasome activation during AR, the expression levels of proteins associated with NLRP3 inflammasome activation, including NLRP3, apoptosisassociated specklike protein containing a CARD (ASC), cleaved caspase-1 and mature IL-1β, were detected in the nasal mucosa (Fig. 4A). The results of western blot analysis revealed that the protein expression levels of NLRP3 and ASC were increased in AR mice compared with those in sham mice, but were inhibited by miR-205-5p knockdown (Fig. 4B). The expression levels of pro-caspase-1 and pro-IL-1β in all groups exhibited no significant difference, whereas those of cleaved caspase-1 and mature IL-1β, which were recruited and matured by activated NLRP3 inflammasome, were decreased by miR-205-5p knockdown in AR mice (Fig. 4C and D). These results indicated that miR-205-5p knockdown may inhibit NLRP3 inflammasome activation in AR, which could help to explain the regulatory effect of miR-205-5p on inflammation.
Figure 4.
miR-205-5p knockdown inhibits NLRP3 inflammasome activation. (A) Western blot analysis of NLRP3 inflammasome components in the nasal mucosa. (B) Expression analysis of NLRP3 and ASC. (C) Relative expression levels of pro-IL-1β and mature IL-1β, and the mature IL-1β/pro-IL-1β ratio. (D) Relative expression levels of pro-caspase-1 and cleaved caspase-1, and the cleaved caspase-1/pro-caspase-1 ratio. Data are presented as the mean ± SD (n=7). **P<0.01 vs. Sham; ##P<0.01 vs. AR + NC. AR, allergic rhinitis; ASC, apoptosisassociated specklike protein containing a CARD; miR, microRNA; NC, negative control; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3.
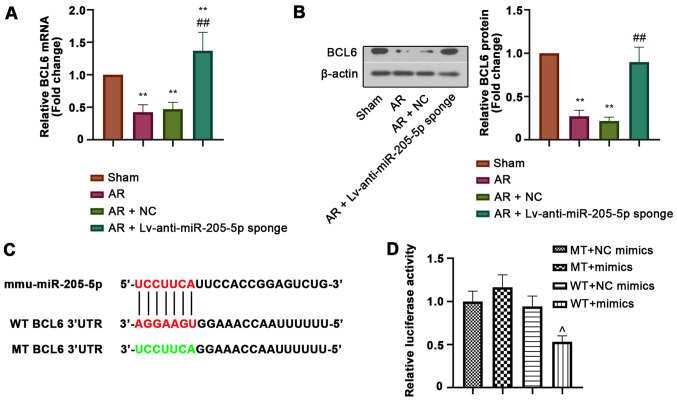
BCL6 is a target of miR-205-5p
BCL6 was predicted as a potential target gene of miR-205-5p. The relative mRNA and protein expression levels of BCL6 in the nasal mucosa were detected by RT-qPCR and western blot analysis, respectively. Both the mRNA and protein expression levels of BCL6 were downregulated in the nasal mucosa of AR mice compared with those in sham mice, but were markedly increased by miR-205-5p knockdown (Fig. 5A and B). In addition, luciferase reporter assay was performed to confirm the binding of miR-205-5p to BCL6. The WT or MT 3′-UTR sequence of BCL6 was cloned into the vector and then co-transfected into cells with miR-205-5p mimics or NC mimics (Fig. 5C). The decreased luciferase activity in WT cells transfected with miR-205-5p mimics indicated that miR-205-5p bound to the 3′-UTR of BCL6 (Fig. 5D). These results suggested that BCL6 was a target of miR-205-5p.
Figure 5.
BCL6 is a target gene of miR-205-5p. Relative (A) mRNA and (B) protein expression levels of BCL6. (C) Diagram of putative miR-205-5p binding sites in the WT or MT 3′UTR of BCL6. (D) Relative luciferase activity in cells co-transfected with the WT or MT 3′-UTR of BCL6 and miR-205-5p mimics or NC mimics. Data are presented as the mean ± SD (n=7). **P<0.01 vs. Sham; ##P<0.01 vs. AR + NC; ^P<0.01 vs. WT + NC mimics. 3′UTR, 3′-untranslated region; AR, allergic rhinitis; BCL6, B-cell lymphoma 6; miR, microRNA; MT, mutant; NC, negative control; WT, wild-type.
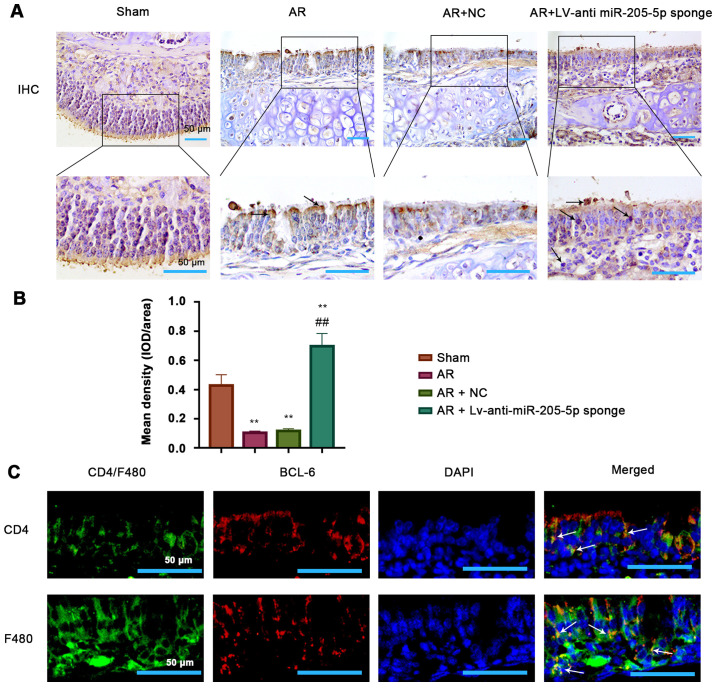
BCL6 is expressed in epithelial cells, T cells and macrophages within the nasal mucosa
The possible type of cells expressing BCL6 within the nasal mucosa tissue was investigated. Immunohistochemical images showed a clear epithelial structure and indicated that BCL6 was likely expressed in the epithelial cells of mouse nasal mucosa (Fig. 6A). The expression of BCL6 was clearly reduced in OVA-induced AR mice, but was recovered by miR-205-5p knockdown (Fig. 6B); this was consistent with the results of RT-qPCR and western blotting. Furthermore, double-immunofluorescence staining was performed for BCL6 and CD4 or F4/80 in the mouse nasal mucosa from the AR group (Fig. 6C). The results showed a co-localization of BCL6 and CD4+ T cells, suggesting that BCL6 was expressed in T cells of the nasal mucosa in the AR group. Moreover, BCL6 and F4/80 double-positive cells indicated that BCL6 was also expressed in macrophages from the nasal mucosa of mice in the AR group. Therefore, it was demonstrated that BCL6 was likely expressed in epithelial and immune cells, including T cells and macrophages, in the nasal mucosa in a mouse model of AR. These findings suggested that the function of BCL6 in OVA-induced inflammation may be linked to its expression in these cells.
Figure 6.
Presence of BCL6 in epithelial cells, T cells and macrophages. (A) Representative images (magnification, ×200 and ×400; scale bars, 100 and 50 µm) and (B) semi-quantification of immunohistochemical staining for BCL6 in the epidermis of nasal mucosa tissue. (C) Double-immunofluorescence staining of BCL6 (red) and CD4+ T cell (green) or F4/80 macrophage (green) in the nasal mucosa tissues in the AR group (magnification, ×400; scale bar, 50 µm). White arrows indicate double-positive cells (yellow). **P<0.01 vs. Sham; ##P<0.01 vs. AR + NC. AR, allergic rhinitis; BCL6, B-cell lymphoma 6; IHC, immunohistochemistry; miR, microRNA; NC, negative control.
Discussion
miRNA-based therapy is an emerging treatment for AR. Certain miRNAs have been reported to be differentially expressed in patients with AR and asthma, rendering them a diagnostic biomarker for these diseases (36). Several miRNAs have also been identified by RNA microarray analysis to be differentially expressed in patients with AR compared with in healthy participants (37). The therapeutic effects of miRNAs on AR have also been reported. miR-466a-3p has been shown to mitigate AR response by inhibiting Th2 cell priming (38). Furthermore, miR-133b has been demonstrated to alleviate allergic inflammation in AR by targeting NLRP3 (39). Overall, these studies supported the hypothesis that miRNAs are involved in the regulation of AR development; however, to the best of our knowledge, little research has been conducted on the effects of miR-205-5p on AR. The increased level of miR-205 in the nasal mucosa of symptomatic patients with AR was observed by Suojalehto et al (21). Consistent with this previous study, the present study reported an increased expression of miR-205-5p in mice with OVA-induced AR, accompanied by severe symptoms of AR. Notably, miR-205-5p knockdown alleviated the symptoms of AR by reducing the inflammatory response. The present study suggested that miR-205-5p may be involved in AR development and could be considered a novel therapeutic target for AR treatment.
miR-205-5p has been reported to stimulate MMP activity and inflammation in abdominal aortic aneurysm development (40). miR-205-5p has also been shown to suppress inflammatory responses in lipopolysaccharide-induced sepsis and lung injury following hip fracture (23,41). The contradictory function of miR-205 in inflammation may be attributed to different pathological environments. In the present study, an anti-inflammatory effect of miR-205-5p knockdown was observed in AR in vivo. Multiple inflammatory cells and cytokines are vital components of the inflammatory response in AR. Increased infiltration of inflammatory cells, such as leukocytes, eosinophils, neutrophils, lymphocytes and macrophages, is usually observed in AR (42,43). In the present study, it was revealed that the number of inflammatory cells in NLF was decreased by miR-205-5p knockdown following OVA challenge. The Th2 cytokines IL-4, IL-5 and IL-13 play major roles in allergic inflammation (44). In the present study, miR-205-5p knockdown effectively reduced the serum levels of total and OVA-specific IgE, as well as the production of IL-4, IL-5 and IL-13, in the nasal mucosa. These findings suggested that miR-205-5p knockdown may inhibit the inflammatory response in AR mice.
The NLRP3 inflammasome is a signaling complex consisting of the inflammasome sensor molecule NLRP3, the adaptor ASC and the effector protease caspase-1. The NLRP3 inflammasome has been well characterized and is a key regulator in the maturation of pro-inflammatory cytokines IL-1β and IL-18 (45,46). The activated NLRP3 recruits the adaptor ASC, resulting in the recruitment and activation of pro-caspase-1 into its cleaved form, which cleaves and matures IL-1β and IL-18 (47,48). It has been reported that NLRP3 inflammasome activation may promote AR development in an IgE-independent manner (49). In a previous study, mice treated with MCC950, an NLRP3 inflammasome inhibitor, exhibited a reduced OVA-induced AR response (50). The NLRP3 inflammasome has been demonstrated to be a potential therapeutic target for AR (51). Therefore, the regulatory effect of miR-205-5p on NLRP3 inflammasome activation in AR was further investigated. NLRP3 inflammasome activation is represented by increased NLRP3 expression, ASC speck formation, and caspase-1 cleavage and therefore enhanced IL-1β and IL-18 secretion (52). Initiating the transcription of the NLRP3 gene to increase its expression is essential for activation of the NLRP3 inflammasome (48). Notably, AR has been found to contribute to the activation of the NLRP3 inflammasome by increasing the expression levels of its components, including NLRP3 and ASC, and via activation of caspase-1, led to the maturation of inflammatory cytokines IL-1β and IL-18 (49,53,54). In the present study, the levels of NLRP3, ASC and cleaved caspase-1 were decreased by miR-205-5p knockdown in the nasal mucosa, suggesting the involvement of miR-205-5p in NLRP3 inflammasome activation in AR. The deactivation of NLRP3 inflammasome by miR-205-5p knockdown resulted in the subsequent reduced production of proinflammatory cytokines, such as IL-1β, which critically contribute to AR development. In addition, NLRP3 has been reported to function as a Th2 transcription factor and promote a Th2-dependent allergic response (55). Accumulating evidence has indicated that caspase-1 and IL-1β are involved in Th2 immune response and asthma development (56,57). These results suggested that miR-205-5p knockdown may alleviate the inflammatory response in AR mice by inhibiting NLRP3 inflammasome activation.
To explore the underlying regulatory mechanism of miR-205-5p in the development of AR, BCL6 was identified as a candidate target gene of miR-205-p. A previous study demonstrated that BCL6 expression was significantly decreased in the nasal mucosa of patients with AR compared with that in healthy individuals (29). In the present study, the mRNA and protein expression levels of BCL6 were lower in the nasal mucosa of AR mice compared with those in sham mice, which was consistent with the previous finding. In addition, miR-205-5p inhibited BCL6 expression by binding to its 3′-UTR sequence, and miR-205-5p knockdown increased its expression. The expression of BCL6 was detected in both CD4+ T cells and F4/80 macrophages in the nasal mucosa of OVA-sensitized mice. Notably, BCL6 has been reported to suppress the expression of Th2 transcription factor GATA-binding protein 3 and Th2 genes including IL-4, IL-5 and IL-13 to regulate inflammation (58). BCL6 has been suggested to be a likely key regulator of the production of IL-1β and IL-18, which are produced by macrophages, inducing Th2 cell differentiation in allergic diseases. An increased percentage of macrophages producing IL-1β has been detected in asthmatic submucosa (54). In addition, BCL6 may serve a negative role in the regulation of key genes that predispose patients to allergies in a wide range of cells, including T cells, B cells, macrophages, mast cells and airway epithelial cells, resulting in the suppressed production of Th2 cytokines and IgE; therefore, BCL6 may act as a negative regulator in IgE-mediated allergic response (59,60). Furthermore, BCL6 has been demonstrated to be highly expressed in epithelial cells of the mouse nasal mucosa, and can suppress NLRP3 transcription by binding to the NLRP3 promoter, thus leading to attenuated inflammation in human renal tubular epithelial cells and in vivo (31). Therefore, the upregulation of BCL6 may have contributed to NLRP3 inflammasome deactivation following miR-205-5p knockdown in AR mice in the present study. The expression of NLRP3 and caspase-1 has been identified in human epithelial bronchus and cells, which are involved in asthma through the secretion of IL-1β (61,62). Whether BCL6 regulates the development of AR in an NLRP3 inflammasome-dependent pathway in the present study needs to be confirmed. The current study verified the likely presence of BCL6 in epithelial cells, T cells and macrophages in the nasal mucosa in a mouse model of AR. The function of BCL6 in AR inflammation may be linked to its expression in these cells. Overall, the results of the present study suggested that BCL6 may alleviate OVA-induced inflammation through the inhibition of Th2 cell differentiation, as indicated by decreased levels of Th2 cytokines IL-4, IL-5 and IL-13, as well as macrophage and NLRP3 inflammasome activation. However, the function and regulatory mechanism of BCL6 in a specific cell type in AR remain to be investigated.
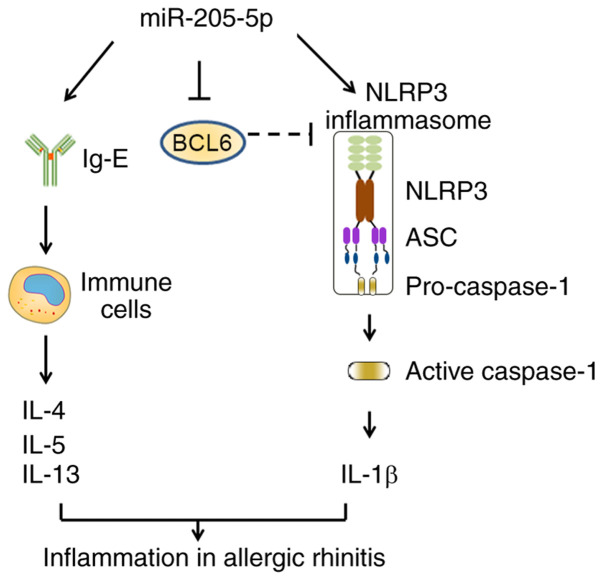
In conclusion, the present study demonstrated that miR-205-5p knockdown may ameliorate the inflammatory response in AR by suppressing the production of Th2 cytokines and activation of NLRP3 inflammasome (Fig. 7). As a target of miR-205-5p, BCL6 could serve a vital role in this process. The present study revealed the importance of the miR-205-5p/BCL6 axis in AR development, providing an improved understanding of AR pathogenesis and a potential therapeutic target for AR treatment.
Figure 7.
Schematic diagram of miR-205-5p regulatory mechanism in allergic rhinitis by targeting BCL6. ASC, apoptosisassociated specklike protein containing a CARD; BCL6, B-cell lymphoma 6; miR, microRNA; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the Science and Technology Project of Liaoning Province (grant no. 2019010183-JH8/103).
Funding
This work was supported by the Science and Technology Project of Liaoning Province (grant no. 2019010183-JH8/103).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZY and QT designed the experiments; SZ and SL performed the experiments and analyzed data; SZ wrote the paper; ZY and QT revised the paper. SZ and SL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal studies were approved by and performed in accordance with the Ethics Committee of the Second Affiliated Hospital of Shenyang Medical College (approval no. 2020005).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bousquet J, Van Cauwenberge P, Khaltaev N, Aria Workshop Group; World Health Organization Allergic rhinitis and its impact on asthma. J Allergy Clin Immunol. 2001;108(Suppl 1):S147–S334. doi: 10.1067/mai.2001.118891. [DOI] [PubMed] [Google Scholar]
- 2.Maurer M, Zuberbier T. Undertreatment of rhinitis symptoms in Europe: Findings from a cross-sectional questionnaire survey. Allergy. 2007;62:1057–1063. doi: 10.1111/j.1398-9995.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein DI, Schwartz G, Bernstein JA. Allergic rhinitis: Mechanisms and treatment. Immunol Allergy Clin North Am. 2016;36:261–278. doi: 10.1016/j.iac.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Wheatley LM, Togias A. Clinical practice. Allergic rhinitis. N Engl J Med. 2015;372:456–463. doi: 10.1056/NEJMcp1412282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378:2112–2122. doi: 10.1016/S0140-6736(11)60130-X. [DOI] [PubMed] [Google Scholar]
- 6.Han X, Krempski JW, Nadeau K. Advances and novel developments in mechanisms of allergic inflammation. Allergy. 2020;75:3100–3111. doi: 10.1111/all.14632. [DOI] [PubMed] [Google Scholar]
- 7.Meltzer EO. Quality of life in adults and children with allergic rhinitis. J Allergy Clin Immunol. 2001;108(Suppl 1):S45–S53. doi: 10.1067/mai.2001.115566. [DOI] [PubMed] [Google Scholar]
- 8.Schuler Iv CF, Montejo JM. Allergic rhinitis in children and adolescents. Pediatr Clin North Am. 2019;66:981–993. doi: 10.1016/j.pcl.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Guerra S, Sherrill DL, Martinez FD, Barbee RA. Rhinitis as an independent risk factor for adult-onset asthma. J Allergy Clin Immunol. 2002;109:419–425. doi: 10.1067/mai.2002.121701. [DOI] [PubMed] [Google Scholar]
- 10.Mastrorilli C, Posa D, Cipriani F, Caffarelli C. Asthma and allergic rhinitis in childhood: what's new. Pediatr Allergy Immunol. 2016;27:795–803. doi: 10.1111/pai.12681. [DOI] [PubMed] [Google Scholar]
- 11.Shaaban R, Zureik M, Soussan D, Neukirch C, Heinrich J, Sunyer J, Wjst M, Cerveri I, Pin I, Bousquet J, et al. Rhinitis and onset of asthma: A longitudinal population-based study. Lancet. 2008;372:1049–1057. doi: 10.1016/S0140-6736(08)61446-4. [DOI] [PubMed] [Google Scholar]
- 12.Bielory L. Allergic conjunctivitis and the impact of allergic rhinitis. Curr Allergy Asthma Rep. 2010;10:122–134. doi: 10.1007/s11882-010-0087-1. [DOI] [PubMed] [Google Scholar]
- 13.Cox L. Approach to Patients with Allergic Rhinitis: Testing and treatment. Med Clin North Am. 2020;104:77–94. doi: 10.1016/j.mcna.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Guilbert TW, Morgan WJ, Zeiger RS, Mauger DT, Boehmer SJ, Szefler SJ, Bacharier LB, Lemanske RF, Jr, Strunk RC, Allen DB, et al. Long-term inhaled corticosteroids in preschool children at high risk for asthma. N Engl J Med. 2006;354:1985–1997. doi: 10.1056/NEJMoa051378. [DOI] [PubMed] [Google Scholar]
- 15.Covar RA, Szefler SJ, Martin RJ, Sundstrom DA, Silkoff PE, Murphy J, Young DA, Spahn JD. Relations between exhaled nitric oxide and measures of disease activity among children with mild-to-moderate asthma. J Pediatr. 2003;142:469–475. doi: 10.1067/mpd.2003.187. [DOI] [PubMed] [Google Scholar]
- 16.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 19.Coskun M, Bjerrum JT, Seidelin JB, Nielsen OH. MicroRNAs in inflammatory bowel disease - pathogenesis, diagnostics and therapeutics. World J Gastroenterol. 2012;18:4629–4634. doi: 10.3748/wjg.v18.i34.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XH, Zhang YN, Liu Z. MicroRNA in chronic rhinosinusitis and allergic rhinitis. Curr Allergy Asthma Rep. 2014;14:415. doi: 10.1007/s11882-013-0415-3. [DOI] [PubMed] [Google Scholar]
- 21.Suojalehto H, Toskala E, Kilpeläinen M, Majuri ML, Mitts C, Lindström I, Puustinen A, Plosila T, Sipilä J, Wolff H, et al. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and asthma. Int Forum Allergy Rhinol. 2013;3:612–620. doi: 10.1002/alr.21179. [DOI] [PubMed] [Google Scholar]
- 22.Son DJ, Jung YY, Seo YS, Park H, Lee DH, Kim S, Roh YS, Han SB, Yoon DY, Hong JT. Interleukin-32α inhibits endothelial inflammation, vascular smooth muscle cell activation, and atherosclerosis by upregulating Timp3 and Reck through suppressing microRNA-205 biogenesis. Theranostics. 2017;7:2186–2203. doi: 10.7150/thno.18407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Chen X, Sun T. MicroRNA-205-5p Targets HMGB1 to suppress inflammatory responses during lung injury after hip fracture. BioMed Res Int. 2019;2019:7304895. doi: 10.1155/2019/7304895. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Li Q, Zhou L, Wang L, Li S, Xu G, Gu H, Li D, Liu M, Fang L, Wang Z, et al. Bcl6 modulates innate immunity by controlling macrophage activity and plays critical role in experimental autoimmune encephalomyelitis. Eur J Immunol. 2020;50:525–536. doi: 10.1002/eji.201948299. [DOI] [PubMed] [Google Scholar]
- 25.Béguelin W, Teater M, Gearhart MD, Calvo Fernández MT, Goldstein RL, Cárdenas MG, Hatzi K, Rosen M, Shen H, Corcoran CM, et al. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell. 2016;30:197–213. doi: 10.1016/j.ccell.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 27.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koh B, Ulrich BJ, Nelson AS, Panangipalli G, Kharwadkar R, Wu W, Xie MM, Fu Y, Turner MJ, Paczesny S, et al. Bcl6 and Blimp1 reciprocally regulate ST2+ Treg-cell development in the context of allergic airway inflammation. J Allergy Clin Immunol. 2020;146:1121–1136.e9. doi: 10.1016/j.jaci.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Y, Li XQ, Qiu QH. Detection of differentially expressed gene of allergic rhinitis based on RT2 profiler PCR array. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2017;31:869–872. doi: 10.13201/j.issn.1001-1781.2017.11.012. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 30.Hiromura Y, Kishida T, Nakano H, Hama T, Imanishi J, Hisa Y, Mazda O, et al. IL-21 administration into the nostril alleviates murine allergic rhinitis. J Immunol. 2007;179:7157–7165. doi: 10.4049/jimmunol.179.10.7157. [DOI] [PubMed] [Google Scholar]
- 31.Chen D, Xiong XQ, Zang YH, Tong Y, Zhou B, Chen Q, Li YH, Gao XY, Kang YM, Zhu GQ. BCL6 attenuates renal inflammation via negative regulation of NLRP3 transcription. Cell Death Dis. 2017;8:e3156. doi: 10.1038/cddis.2017.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho SW, Zhang YL, Ko YK, Shin JM, Lee JH, Rhee CS, Kim DY. Intranasal Treatment With 1, 25-Dihydroxyvitamin D3 alleviates allergic rhinitis symptoms in a mouse model. Allergy Asthma Immunol Res. 2019;11:267–279. doi: 10.4168/aair.2019.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuntz C, Wunsch A, Rosch R, Autschbach F, Windeler J, Herfarth C. Short- and long-term results after laparoscopic vs conventional colon resection in a tumor-bearing small animal model. Surg Endosc. 2000;14:561–567. doi: 10.1007/s004640000130. [DOI] [PubMed] [Google Scholar]
- 34.Robert R, Nail S, Marot-Leblond A, Cottin J, Miegeville M, Quenouillere S, Mahaza C, Senet JM. Adherence of platelets to Candida species in vivo. Infect Immun. 2000;68:570–576. doi: 10.1128/IAI.68.2.570-576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A, Ishmael FT. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137:1423–1432. doi: 10.1016/j.jaci.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 37.Shaoqing Y, Ruxin Z, Guojun L, Zhiqiang Y, Hua H, Shudong Y, Jie Z. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. 2011;25:e242–e246. doi: 10.2500/ajra.2011.25.3682. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Deng Y, Li F, Xiao B, Zhou X, Tao Z. MicroRNA-466a-3p attenuates allergic nasal inflammation in mice by targeting GATA3. Clin Exp Immunol. 2019;197:366–375. doi: 10.1111/cei.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao L, Jiang L, Hu Q, Li Y. MicroRNA-133b Ameliorates Allergic Inflammation and Symptom in Murine Model of Allergic Rhinitis by Targeting Nlrp3. Cell Physiol Biochem. 2017;42:901–912. doi: 10.1159/000478645. [DOI] [PubMed] [Google Scholar]
- 40.Kim CW, Kumar S, Son DJ, Jang IH, Griendling KK, Jo H. Prevention of abdominal aortic aneurysm by anti-microRNA-712 or anti-microRNA-205 in angiotensin II-infused mice. Arterioscler Thromb Vasc Biol. 2014;34:1412–1421. doi: 10.1161/atvb.34.suppl_1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Wang J, Li Z, Li J, Sang M. MicroRNA-205 5b inhibits HMGB1 expression in LPS-induced sepsis. Int J Mol Med. 2016;38:312–318. doi: 10.3892/ijmm.2016.2613. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Wang B, Luo Y, Zhang Q, Bian Y, Wang R. Resveratrol-mediated SIRT1 activation attenuates ovalbumin-induced allergic rhinitis in mice. Mol Immunol. 2020;122:156–162. doi: 10.1016/j.molimm.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Benson M, Strannegård IL, Strannegård O, Wennergren G. Topical steroid treatment of allergic rhinitis decreases nasal fluid TH2 cytokines, eosinophils, eosinophil cationic protein, and IgE but has no significant effect on IFN-gamma, IL-1beta, TNF-alpha, or neutrophils. J Allergy Clin Immunol. 2000;106:307–312. doi: 10.1067/mai.2000.108111. [DOI] [PubMed] [Google Scholar]
- 44.Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev. 2011;242:31–50. doi: 10.1111/j.1600-065X.2011.01020.x. [DOI] [PubMed] [Google Scholar]
- 45.Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20:3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhen Y, Zhang H. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol. 2019;10:276. doi: 10.3389/fimmu.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo EK, Kim JK, Shin DM, Sasakawa C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell Mol Immunol. 2016;13:148–159. doi: 10.1038/cmi.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588–606. doi: 10.1038/nrd.2018.149. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z, Liang C, Wang T, Zou Q, Zhou M, Cheng Y, Peng H, Ji Z, Deng Y, Liao J, et al. NLRP3 inflammasome activation promotes the development of allergic rhinitis via epithelium pyroptosis. Biochem Biophys Res Commun. 2020;522:61–67. doi: 10.1016/j.bbrc.2019.11.031. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Ba G, Tang R, Li M, Lin H. Ameliorative effect of selective NLRP3 inflammasome inhibitor MCC950 in an ovalbumin-induced allergic rhinitis murine model. Int Immunopharmacol. 2020;83:106394. doi: 10.1016/j.intimp.2020.106394. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Y, Xu W, Su W. NLRP3 inflammasome: A likely target for the treatment of allergic diseases. Clin Exp Allergy. 2018;48:1080–1091. doi: 10.1111/cea.13190. [DOI] [PubMed] [Google Scholar]
- 52.Zhao W, Ma L, Cai C, Gong X. Caffeine inhibits NLRP3 inflammasome activation by suppressing MAPK/NF-κB and A2aR signaling in LPS-induced THP-1 macrophages. Int J Biol Sci. 2019;15:1571–1581. doi: 10.7150/ijbs.34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sousa AR, Lane SJ, Nakhosteen JA, Lee TH, Poston RN. Expression of interleukin-1 beta (IL-1beta) and interleukin-1 receptor antagonist (IL-1ra) on asthmatic bronchial epithelium. Am J Respir Crit Care Med. 1996;154:1061–1066. doi: 10.1164/ajrccm.154.4.8887608. [DOI] [PubMed] [Google Scholar]
- 55.Bruchard M, Rebé C, Derangère V, Togbé D, Ryffel B, Boidot R, Humblin E, Hamman A, Chalmin F, Berger H, et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol. 2015;16:859–870. doi: 10.1038/ni.3202. [DOI] [PubMed] [Google Scholar]
- 56.Iwata A, Nishio K, Winn RK, Chi EY, Henderson WR, Jr, Harlan JM. A broad-spectrum caspase inhibitor attenuates allergic airway inflammation in murine asthma model. J Immunol. 2003;170:3386–3391. doi: 10.4049/jimmunol.170.6.3386. [DOI] [PubMed] [Google Scholar]
- 57.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, Dinarello CA, Paul WE. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci USA. 2009;106:7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawant DV, Sehra S, Nguyen ET, Jadhav R, Englert K, Shinnakasu R, Hangoc G, Broxmeyer HE, Nakayama T, Perumal NB, et al. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol. 2012;189:4759–4769. doi: 10.4049/jimmunol.1201794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda N, Arima M, Tsuruoka N, Okada S, Hatano M, Sakamoto A, Kohno Y, Tokuhisa T. Bcl6 is a transcriptional repressor for the IL-18 gene. J Immunol. 2003;171:426–431. doi: 10.4049/jimmunol.171.1.426. [DOI] [PubMed] [Google Scholar]
- 60.Arima M, Fukuda T, Tokuhisa T. Role of the transcriptional repressor BCL6 in allergic response and inflammation. World Allergy Organ J. 2008;1:115–122. doi: 10.1097/WOX.0b013e31817dc522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirota JA, Hirota SA, Warner SM, Stefanowicz D, Shaheen F, Beck PL, Macdonald JA, Hackett TL, Sin DD, Van Eeden S, et al. The airway epithelium nucleotide-binding domain and leucine-rich repeat protein 3 inflammasome is activated by urban particulate matter. J Allergy Clin Immunol. 2012;129:1116–25.e6. doi: 10.1016/j.jaci.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 62.Birrell MA, Eltom S. The role of the NLRP3 inflammasome in the pathogenesis of airway disease. Pharmacol Ther. 2011;130:364–370. doi: 10.1016/j.pharmthera.2011.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.