What is currently known on the topic
The gut microbiome, i.e., the community of bacteria and other microorganisms living in the human gut, has been implicated both directly and indirectly (mediating the effects of diet) on human health [1, 2]. The associations between gut microbiome composition and disease status have been widely reported, while recent studies have demonstrated a role for the gut microbiome in influencing remote organs, mucosal, and immune function [3, 4]. Considerable effort is currently focused on understanding the natural history of microbiome development in humans in the context of health outcomes, in parallel with improving our knowledge of microbiome–host molecular interactions. These efforts ultimately aim to develop effective approaches to rehabilitate perturbed human microbial ecosystems as a means to restore health and prevent disease. This review details the role of the gut microbiome in chronic diseases (Fig. 1) and ways it can be modulated for the management or prevention of chronic conditions.
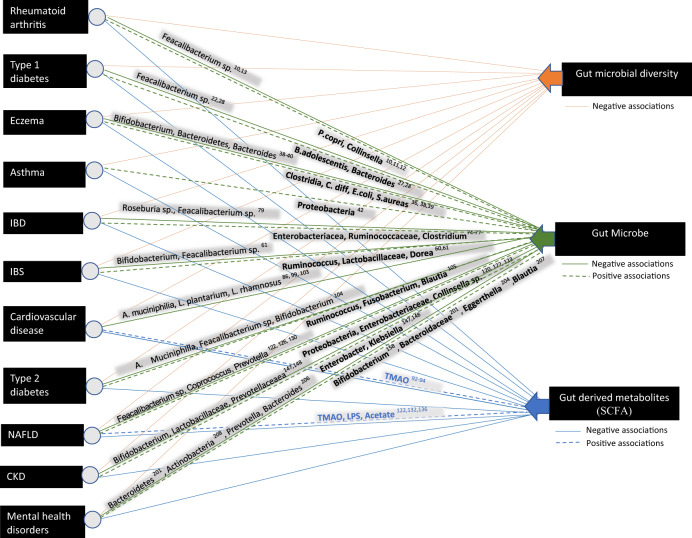
Fig. 1. Schematic representation of the association of the composition of the gut microbiome and gut-derived metabolites with chronic diseases.
The solid lines represent negative associations and the dashed lines represent positive associations of the disease phenotype with gut microbes and metabolites.
Differences in gut microbiome composition and function have been associated with a variety of chronic diseases ranging from gastrointestinal inflammatory and metabolic conditions to neurological, cardiovascular, and respiratory illnesses.
The aim of this narrative review is to describe the associations between gut microbiome composition and various types of chronic diseases and to discuss the links to habitual diet and dietary components (Table 1).
Table 1.
Summary of key findings outlining the role of the gut microbiome in chronic disease.
| Key messages |
|---|
| Gut microbiome composition is significantly different in healthy individuals compared to affected individuals with a broad range of chronic diseases. Lower microbiome diversity appears to be a common theme across many of the diseases |
| Auto-immune diseases, in common with cardiometabolic diseases and irritable bowel syndrome, show low abundances of SCFA producing bacteria such as Bifidobacterium sp, Faecalibacterium sp, Roseburia sp, or Coprococcus eutactus |
| High abundances of pathogenic bacteria (such as E. coli, S. aureus, and C. difficile) are common in some of these diseases |
| SCFAs but also other microbial metabolites are involved in the health effects of gut microbiome composition and microbial metabolites can be better predictors of health outcomes than microbiome diversity |
| Gut microbiome composition is modifiable by various therapeutic strategies. Dietary interventions can result in robust changes in both gut microbiome composition and function and in the corresponding health effects |
Gut microbiome and autoimmune disease
The pathogenesis of autoimmune diseases (AIDs) is not only attributed to genetic susceptibilities but also environmental factors, among which, a disturbed gut microbiota has attracted increasing attention. Compositional and functional changes of gut microbiota have been reported in various autoimmune diseases, and increasing evidence suggests that disturbed gut microbiota contributes to their immunopathogenesis.
Rheumatoid arthritis (RA)
Rheumatoid arthritis (RA) is a systemic autoimmune inflammatory condition that manifests in joint damage. It was recently demonstrated that different environmental factors are involved in the development of both intestinal/oral dysbiosis and arthritis onset and outcome, among which the most relevant are diet, smoking, and infections [5, 6]. The observation that germ-free mice are protected from development of experimental arthritis [7] suggests a possible role for the microbiome in the pathogenesis of this disease. The composition of the gut microbiota in RA patients free of therapy is severely altered compared to healthy controls. Chen et al. reported that compared with healthy controls, patients with RA show decreased gut microbial diversity, which correlates with autoantibody levels and disease duration [8]. In addition, at the compositional level, patients with RA show an increased abundance of Prevotella species, including Prevotella copri [8, 9], and recent preclinical phase studies on RA patients in European countries were shown to harbor a high abundance of this species in the intestine, suggesting that dysbiosis precedes the development of arthritis [10]. In contrast, Faecalibacterium, which is generally recognized as a beneficial microbe, is decreased in RA patients. Furthermore, the relative abundance of Collinsella was found to be increased in RA patients [11]. Interestingly, inoculation of Collinsella into collagen-induced arthritis (CIA)-susceptible mice induces severe arthritis. In vitro experiments showed that Collinsella Aerofaciens increases gut permeability and induces IL-17A expression, a key cytokine involved in the pathogenesis of RA, suggesting that Collinsella is a candidate arthritogenic bacterium in the human intestine [11]. In summary, Prevotella copri [12] and Collinsella are the dominant gut microbiota in patients with early RA and may be involved in its pathogenesis.
Recent links have been made between dietary intake of short-chain fatty acids (SCFAs) and autoimmune arthritis in mice, wherein SCFAs play an important role in the suppression of inflammation in RA [13, 14]. Mice deficient for SCFA receptors showed exacerbated inflammation in modes of RA [14]. Butyrate, one of the most abundant SCFAs, acts as an endogenous histone deacetylase (HDAC) inhibitor and has been shown to decrease inflammation in animal models of RA and other inflammatory diseases [15]. A recent study has revealed a role for intestinal barrier function, and specifically for zonulin, a peptide that controls epithelial tight junction permeability, in regulating the onset of joint disease in mice with collagen-induced arthritis (CIA) and potentially also in patients with RA [16]. Increased levels of zonulin have been associated with leaky intestinal barrier, dysbiosis, and inflammation. Restoration of the intestinal barrier in the period before clinical arthritis, either by dietary supplementation with the SCFA butyrate or pharmacological agents such as a zonulin antagonist may help delay disease onset and reduce the severity of RA.
Type 1 diabetes (T1D)
In humans, intestinal microbiota alterations, including loss of bacterial diversity, preceded the onset of metabolic symptoms associated with T1D [17]. Previous studies have linked several facets of gut health with the onset of T1D in humans and animal models [18–20]. In animal models, antibiotic-induced dysbiosis, altered microbial lipid metabolism, and suppressed enteric Th17 and T-reg cell populations have led to increased incidence of T1DM-like disease in mice [21]. Various human-based studies have also reported altered intestinal microbiota in connection to T1D from several ethnic groups [17,20, 22–24]. The common findings from these studies include increased numbers of Bacteroides species, and deficiency of bacteria that produce SCFAs [25, 26] in cases of T1D. Specifically, the butyrate producer Faecalibacterium prausnitzii has been found to be decreased in abundance in children with diabetes-related autoantibodies [26].
In addition, increased intestinal permeability [22] and decreased microbial diversity [17] before T1D diagnosis have been reported. A multicenter case–control study of 783 children, showed that the microbiomes of healthy children contained more genes that were related to the fermentation and the biosynthesis of SCFAs, but these were not consistently associated with the abundance of particular taxa across geographically diverse clinical centers, suggesting that microbial function rather than composition was associated more with T1D [27].
The protective effects of SCFAs in T1D have been elucidated in animal studies that have looked at nonobese diabetic (NOD) mouse models [28]. For example, NOD mice fed specialized diets resulting in high bacterial release of acetate and butyrate were almost completely protected from T1D [28] mainly via the immune-modulating effects of SCFAs. In animal models of autoimmune diabetes, increased gut permeability precedes the development of diabetes, and environmental factors that modulate the permeability thereby modulate the incidence of the disease. Gut permeability appears to be an important factor in the relationship between the intestinal microbiota and the development of T1D. Both gut permeability and inflammation have been linked to the development of T1D in humans [26, 29].
Atopic eczema
Atopic eczema, a chronic inflammatory skin disorder, is most prevalent in early childhood. The pathogenesis of eczema has been attributed to skin-barrier dysfunctions, immune dysregulation as well as environmental–host–microbial interactions [30]. Environmental factors and modern lifestyle trends have been shown to indirectly contribute to pathogenesis of the disease through modulation of the gut microbiome [31].
Previous studies have found that the gut microbiome in early life is associated with age of onset, severity, and remission of atopic eczema [32, 33]. However, the association between gut microbiome diversity and atopic eczema development remains contradictory with few studies reporting an inverse relationship between gut diversity and the severity of atopic eczema [34, 35]. The development of atopic eczema may instead be driven by the interactions between specific gut microbiome signatures, the immune system, and the host. Specifically, in patients with atopic eczema, the proportion of Clostridia, Clostridium difficile, Escherichia coli, and Staphylococcus aureus in the gut microbiome is higher than in healthy controls, whereas that of Bifidobacteria, Bacteroidetes, and Bacteroides is decreased [34, 36–38]. The abundance of Clostridia and Escherichia coli in the intestine may be associated with atopic eczema via eosinophilic inflammation [36]. Butyrate-producing bacteria, such as Coprococcus eutactus, are increased in infants with milder atopic eczema or healthy infants than in those with severe atopic eczema [34]. Furthermore, fecal samples from patients with atopic eczema showed decreased levels of butyrate and propionate, which have anti-inflammatory effects. This is likely a consequence of a dysbiosis in the SCFA producer F prausnitzii that reduces the number of high butyrate and propionate strains [38].
It is evident that the compositional and proportional differences in the gut microbiome are associated with the development of AID via an immunomodulatory effect of the gut microbiome. The gut microbiome may contribute to the development, persistence, and severity of AID via immunologic, metabolic, and neuroendocrine pathways. It is however still unclear whether compositional changes in the gut microbiome precede the development of atopic eczema, and thereby shift the immune system and disrupt the gut epithelial barrier, making it amenable to the development of atopic eczema.
Atopic asthma
The rapid increase in asthma prevalence in industrialized nations over the past several decades cannot be explained by genetic risk factors alone and is thought to be related to altered environmental exposures associated with Western lifestyles. Early life is the most important period during which microbiota dysbiosis in the gut may lead to the development of many respiratory diseases, as the gut microbiota has a significant influence on immune-cell maturation and resistance to pathogens [39]. A number of validated epidemiological observations have implicated early-life environmental exposures in increased risk for childhood asthma. Many of these exposures are known to shape the nascent gut microbiome, including cesarean birth [40], antibiotic use [41], formula feeding, and other environmental factors including airborne toxins [42]. Further evidence for an intricate relationship between environmental exposure, the gut microbiome, and allergic airway disease comes from an expanding body of work, particularly those utilizing experimental animal models. For example, treating neonatal mice with antibiotics was shown to diminish gut microbial diversity, alter metabolite profiles, exacerbate immune cell response, and increase susceptibility to allergic lung inflammation [43, 44]. Furthermore, supplementation with SCFAs ameliorated airway inflammation in these mice, with the mechanism attributed to decreased activity of immune-modulating markers such as T cells, IL-4-producing CD4+ T cells, and reduced levels of circulating IgE [44].
Observational studies in humans have identified Proteobacteria to be the most dominant phylum overrepresented in patients with asthma compared with nonasthmatic volunteers across several studies [39]. The Proteobacteria phylum is represented by potentially pathogenic bacteria, including those that belong to the genera Haemophilus, Moraxella, and Neisseria [39].
Asthma pathogenesis and severity are linked with pro-inflammatory mechanisms. The effects of the gut microbiota on asthma are at least partially mediated by bacterial metabolites, which may influence immune responses in distal parts of the body. The most known metabolites with demonstrated protective properties in human airway inflammation are SCFAs. Children with high amounts of butyrate and propionate in feces at 1 year of age have significantly less atopic sensitization and are less likely to have asthma between 3 and 6 years [45]. In addition, soluble fiber may ameliorate the effects by exerting anti-inflammatory action via SCFAs binding to associated G-protein-coupled receptors (GPCRs) [46, 47].
Of recent interest are studies showing that gut bacteria in humans are able to produce other metabolites with pro- and anti-inflammatory potential, such as biogenic amines (including histamine) [48] and oxylipins such as 12,13-diHOME [49]. The number of histamine-secreting bacteria is significantly higher in fecal samples of asthma patients compared with nonasthmatic volunteers [50]. Furthermore, the number of histamine-secreting bacteria correlates with disease severity.
Although the mechanisms mediating communication between the gut and lungs are still unclear, it has been suggested that epithelial cells, other structural cells, and immune cells absorb signals from the gut endothelium to form a local cytokine microenvironment, which leads to changes in immune responses at distal sites [51]. Specifically, SCFAs derived from gut bacteria have inhibitory effects on proinflammatory responses in the lungs.
Gut microbiome–gut inflammation/bowel disorders
Irritable bowel syndrome (IBS)
IBS is generally characterized by abdominal pain, discomfort, and altered bowel habits. Although the etiology is multifactorial, recent understanding of the pathophysiology of IBS has revealed that variations in the normal gut microbiota may have a role to play in the low-grade intestinal inflammation associated with the syndrome [52]. Microbial dysbiosis in the gut is thought to be involved in IBS pathogenesis [53] and a recent study revealed a clear separation between the gut microbiota of patients with IBS and that of the controls. IBS was characterized by an increase in Firmicutes and, more specifically, in the numbers of Ruminococcus, Clostridium, and Dorea, in addition to a marked reduction of beneficial microbes such as Bifidobacterium and Faecalibacterium spp. [54]. Furthermore, systematic reviews have demonstrated the potentially harmful microbiota in patients with IBS, including phylum Proteobacteria, family Enterobacteriaceae (phylum Proteobacteria), family Lactobacillaceae, and genus Bacteroides (phylum Bacteroidetes). The Enterobacteriaceae family contains several pathogenic bacteria; for instance, Escherichia, Shigella, Campylobacter, and Salmonella [55]. These could either suggest previous intestinal infection in such patients or a change in the intestinal environment. By-products from these potentially harmful bacteria have been associated with some of the classical symptoms of IBS, including abdominal pain, bloating, and diarrhea [55].
The most consistent finding of a potentially “protective” bacterial group in IBS patients was found in the group of uncultured Clostridiales. Although the association is not causal and the mechanism of a protective effect in IBS symptoms is unclear. The genus Faecalibacterium, especially Faecalibacterium prausnitzii, which belongs to the similar order as the uncultured Clostridia, has been associated with maintaining gut mucosal health. This bacterium was considered as the main butyrate-producing and anti-inflammatory organism [56] and reduced IBS symptoms via mediation of IL-17 expression in a rat model [57, 58], as well as maintained gut-barrier integrity [59]. Furthermore, the genus Bifidobacterium was decreased significantly in IBS patients regardless of IBS subtype. Therefore, it was another promising potential genus in ameliorating IBS symptoms. A systematic review of probiotics in IBS has highlighted that Bifidobacterium-containing interventions reduce IBS symptoms, which are not seen in products that contain Lactobacillus alone [60].
Management of IBS symptoms can be achieved by the restriction of fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP). Clinical studies have now shown the clinical efficacy of the low FODMAP diet in reducing IBS symptoms [61]. One consequence of this dietary intervention is its impact on gut microbiome composition as FODMAPs can modulate microbial composition and microbial metabolite production [62] and not all IBS subjects respond and there may be challenges with implementing a low FODMAP diet. Given the role of the microbiome in metabolizing poorly absorbed carbohydrates that depend on a person’s microbiome composition, that there has been increasing attention on the potential role of the gut microbiome in predicting low FODMAP diet efficacy [62] and recent data suggest that there are potential baseline differences in microbiome activity and composition, which can distinguish between low FODMAP diet responders and nonresponders [63].
Inflammatory bowel disease (IBD)
Long-lasting inflammation and ulceration of the colon are predominantly the main characteristics of IBD, which are features of Crohn’s disease (CD) and ulcerative colitis (UC). IBD, encompassing both ulcerative colitis and Crohn’s disease, is characterized by chronic and relapsing inflammation of the GI tract. The onset of both conditions is, in general, not thought to be due to a single causal organism but by a general microbial dysbiosis in the gut [64]. A role for gut microbes in the manifestation of IBD has been indicated by a number of studies and the gut microbiota are thought to be essential components in the development of mucosal lesions [65]. The alterations in the composition and functionality of the gut microbiota in IBD compared with non-IBD controls have been shown previously. In general, microbial dysbiosis in IBD is characterized by a decrease in diversity and stability of the microbiota [66–69]. Specifically, a decrease in Firmicutes and an increase in Proteobacteria taxa is the most consistent outcome from IBD microbiome studies. Furthermore, a common signature of microbial dysbiosis among IBD patients, especially in (active) CD, is the decreased abundance of Firmicutes bacteria belonging to the families Ruminococcaceae and Lachnospiraceae as opposed to healthy control samples [70–73]. Both families are important functional members of the human gut microbiota since most butyrate-producing bacteria from the human gut belong to them. Therefore, depletion of these bacterial families in IBD can be linked to the observed disturbance on a functional level, including a lower butyrate-producing capacity of the IBD microbiota [74]. In addition, a metagenomic and proteomics study in ileal CD microbiota demonstrated an underrepresentation of genes for SCFA production and a decrease in metagenomic reads and proteins of important butyrate producers Faecalibacterium prausnitzii and Roseburia sp. [75].
Butyrate has therapeutic potential in IBD because it serves as the main energy source for colonocytes, enhances epithelial barrier integrity, and inhibits inflammation. Recent observational and interventional studies have explored an alternative approach of probiotics, which involves the consumption of butyrate-producing bacteria to increase in situ butyrate production [76–78]. This may suggest that targeting microbial dysbiosis by supplementing butyrate-producing bacteria could restore gut homeostasis and health in IBD.
Gut microbiome composition and cardiometabolic diseases
Cardiovascular disease
The alterations in gut microbiota composition and the metabolic potential of gut microbiota have been identified as a contributing factor in Cardiovascular Disease (CVD) development [79]. Recently, mechanistic links between the gut microbiota and the severity of myocardial infarction have been reported in rats [80, 81]. Hypertension is the most prevalent modifiable risk factor for CVD. However, results from human studies remain scarce and conflicting. In the TwinsUK cohort [82], self‐reported hypertension was not related to 68 various microbiota markers. Furthermore, in the CARDIA study of 529 individuals an increase in alpha diversity resulted in a modest decrease in objectively measured systolic blood pressure (BP) [83]. More recently, the association between the gut microbiome and hypertension was studied on the well-phenotyped population of 6953 individuals as part of the FINRISK study [84] using standardized BP measurements. Although the associations between overall gut taxonomic composition and BP were weak, individuals with hypertension demonstrated changes in several microbiota genera, with most of these genera belonging to the Firmicutes phylum. Interestingly, most of the negative associations with BP belonged to the Lactobacillus species, specifically the greater abundance of Lactobacillus paracasei, a known probiotic that was associated with lower mean arterial pressure and lower dietary sodium intake which has also been reported previously by Wilck et al. [85].
Metabolism of certain dietary components such as choline and carnitine, which comprise a large component of a Western diet, has been shown to increase risk of cardiovascular disease mainly via the metabolism of these compounds to produce trimethylamine oxide (TMAO) [86, 87]. Gut-derived metabolites such as SCFAs have been recently shown to modulate blood pressure [88]. The suggested mechanism of SCFAs to modulate blood pressure is via the host G-protein coupled receptor (GPR) pathways that impact renin secretion and blood pressure regulation [88]. A series of studies using GPR41 knockout mice further supports involvement of these receptors in blood pressure control. This suggests that the gut microbiota are potentially intertwined functionally to control blood pressure, and their dysfunctions could be associated with hypertension. A recent study found that a Mediterranean diet which typically encompasses a high fiber diet followed by fasting was shown to reduce systolic blood pressure, need for antihypertensive medications, body-mass index at three months post intervention compared to following a Mediterranean diet alone [89]. These data highlight fasting followed by a shift to a health-promoting diet as a promising non-pharmacological intervention for patients with hypertension and other cardiometabolic diseases.
In rodent model studies, the administration of probiotics such as Lactobacillus plantarum and Lactobacillus rhamnosus was associated with improved ventricular function and attenuated heart failure after myocardial infarction [80, 90]. Furthermore, a meta-analysis demonstrated a significant decrease in blood pressure in patients treated with probiotics [91]. Lastly, supplementation of atherosclerosis-prone mice with A. muciniphila, a mucus colonizer protected against atherosclerosis development induced by feeding a Western diet [92]. These observations may suggest that probiotics use, in combination with standard medication, could offer additional benefits in the prevention and management of cardiovascular events.
Type 2 diabetes
Adults with T2DM have an altered gut microbiota composition compared to healthy controls. However, it is unclear whether these changes represent a cause or an effect and required further investigation. Among the commonly reported findings, the genera of Bifidobacterium, Bacteroides, Faecalibacterium, Akkermansia, and Roseburia were negatively associated with T2D, while the genera of Ruminococcus, Fusobacterium, and Blautia were positively associated with T2D [93, 94]. A large metagenome-wide association study found a moderate degree of gut dysbiosis in patients with T2DM [95]. Of the identifiable bacterial species, the control samples were enriched in various butyrate-producing bacteria and increased abundance of Lactobacillus spp., while patients with T2DM were characterized by an increase in certain opportunistic pathogens, such as a number of Clostridium spp. [95]. Overall, T2DM was associated with a reduced abundance of butyrate-producing bacteria and SCFAs, in particular butyrate which has been linked with insulin sensitivity [9, 96, 97]. The link between SCFAs and insulin sensitivity lies in the ability of SCFAs to trigger the secretion of GLP-1 by intestinal L-cells via specific G protein receptors (GPR41, GPR43) which has a substantial impact on pancreatic function and insulin release, as well as central effects regulating appetite [98, 99].
In addition to SCFAs, many other metabolites have been implicated in the role of gut microbiome composition and risk to T2DM. We have in fact reported reproducible associations between serum metabolites of microbial origin and gut microbiome alpha diversity in 1018 individuals from the Twins UK cohort [100]. The circulating levels of these metabolites were combined into a microbiome-metabolite score using a linear combination. Importantly the association of the microbiome metabolite score with cardiometabolic traits was stronger than the association between gut microbiome diversity and the same clinical traits. Specifically, the microbiome metabolite score was associated with prevalence and incidence of T2DM in the discovery cohort and this result was replicated in a multi-ethnic independent US cohort (n = 1522) showing the relevance of microbiome-derived metabolites in predicting risk and supporting their role as the mechanistic link between microbiome composition and health [100].
Non-alcoholic fatty liver disease (NAFLD)
Non-alcoholic fatty liver disease (NAFLD) and the more advanced stage non-alcoholic steatohepatitis (NASH) are common comorbidities of obesity and T2DM with an increasing burden for society [101]. There is an increasing body of evidence linking the gut-liver axis to the development of NAFLD. Gut dysbiosis is directly related with increased intestinal permeability as a consequence of epithelial barrier deterioration, tight junctions’ alteration and bacterial translocation causing endotoxemia, which might reach and damage the liver through the portal vein [102–104]. Several studies have revealed alterations in the gut microbiome in people with NAFLD compared with healthy controls. We and others have reported a decrease in both alpha and beta diversities in NAFLD and its more advanced stages compared to healthy controls [105, 106]. Case control studies have reported consistent altered microbial signatures at phylum level such as increased Proteobacteria [107–110], at family level such as increased Enterobacteriaceae and genera such as an increase in the abundance of Collinsella sp, Escherichia, Dorea and a decrease in the abundance of Coprococcus, Eubacterium, Faecalibacterium and Prevotella [107, 111]. Similar microbial signatures were found in NASH as outlined in this recent review [112]. Although these initial results suggest a measurable contrast in microbial signatures between individuals with hepatic steatosis and controls, there are, however, large discrepancies found across studies with varying results in the literature. However, a common microbial signature that has been associated with other metabolic diseases is the reduction of levels of the butyrate producer F. prausnitzii in the more advanced forms of NAFLD [106, 113].
Several hypotheses have provided mechanistic insights into the pathways of how the gut microbiota might contribute to NAFLD development and its disease progression. Specific microbial species such as Collinsella sp. have been shown to metabolize bile acids to oxo-bile acid intermediates which may increase intestinal permeability and contribute to the development of NAFLD [114, 115]. Increased intestinal permeability may lead to lipopolysaccharide (LPS) release to the host, which can trigger tissue and systemic inflammation. Furthermore, the action of microbially-produced metabolites (including TMAO, choline or ethanol) and bile acid signaling can also affect host immunity [108, 116, 117]. Interestingly, unlike other diseases, SCFAs appear to prevent but also promote the development of NAFLD and NASH depending on the signaling pathway or mechanism they activate. Amongst the SCFAs, acetate was found to be enriched in fecal samples from adults with more advanced stages of NAFLD (i.e. fibrosis) [110], whereas butyrate and propionate were enriched in fecal samples from adults with mild or moderate NAFLD. This may suggest that levels and subtypes of SCFAs correlate with the severity of fatty liver disease. Since each SCFA exerts specific and somehow different metabolic effects, assessing their balance both at the fecal and systemic level in patients and after a dietary intervention using different substrates may help provide more information on their overall role in NAFLD development, exacerbation or improvement.
On the basis of the above, advances in discovering gut microbiota or microbiota-related metabolite signatures could be used as noninvasive diagnostic tools in NAFLD and its etiology.
Chronic kidney disease (CKD)
There has been a growing interest in studying the composition of the gut microbiota in patients with CKD as well as the mechanisms by which gut dysbiosis contributes to CKD progression. The influence of the gut microbiota on the gut–kidney axis acts in a reciprocal way with either CKD significantly modifying the composition and functions of the gut microbiota. Alternatively, the gut microbiota is able to manipulate the processes leading to CKD onset and progression through inflammatory, endocrine, and neurologic pathways. Therefore, understanding the complex interaction between these two organs may provide novel interventions to prevent the progression of CKD by targeting the gut microbiota.
The existence of intestinal microbiota alterations such as decrease of microbial richness, diversity, and uniformity has been related to CKD [118]. Patients with CDK show a lower colonization of Bifidobacterium sp, Lactobacillaceae, Bacteroidaceae, Akkermansia, and Prevotellaceae genera, and higher intestinal levels of Enterobacteriaceae, particularly Enterobacter, Klebsiella, and Escherichia, and also increased levels of Enterococci and Clostridium perfringes [119, 120]. The decrease in the abundance of Akkermansia muciniphilla, an important probiotic, in patients with CKD negatively correlated with plasma IL-10 levels, suggesting that an altered microbiota in CKD may promote chronic systemic inflammation [121]. This chronic systemic inflammation state represents a major risk factor for CKD progression.
CDK patients are characterized by decreased consumption of dietary fibers that are required for SCFA production. Lack of dietary fibers leads to increased amino nitrogen, which can be transformed into uremic toxins by the gut microbiota [122]. Patients with CDK are characterized by an imbalance between fermentative and proteolytic microbiota in favor of the latter. The imbalance in favor of proteolytic species is related to detrimental effects and also has a fundamental role in the progression of CKD [123]. Moreover, the main SCFAs especially butyrate, were found to be reduced in the feces and serum of patients during CKD development [123]. Further research is however needed to determine whether increasing levels of circulating SCFAs would provide any direct clinical benefit in patients with CDK.
Several experimental and clinical studies have shown the beneficial effects of prebiotic, probiotic, and synbiotic supplementation on gut microbiota–renal axis [124]. These have emerged as a potential therapeutic intervention to restore the imbalance of the gut microbiota, reduce inflammation or oxidative stress markers and modulate gut-derived uremic toxins, such as P‐cresyl sulfate (PCS), indoxyl sulfate (IS), and trimethylamine N‐oxide (TMAO), which have been implicated in the progression of CKD [125–127].
Mental health disorders
Gut microbes have been shown to also influence neurological functions and these links have been termed the “gut−brain axis” [128]. The gut microbiota communicates with the brain through three major pathways—the neural pathway (vagus nerve, enteric nervous system), the immune pathway (cytokines), and the endocrine pathway (HPA axis, gut hormones). Impaired functioning of this connection can lead to manifestation of mental disorders. Common gut microbial species belonging to the phylum Firmicutes and Actinobacteria, and the genera Bacteroides and Bifidobacterium, may be contributing to mental health disorders [129]. Different mental disorders are characterized by the differential gut microbial community that is generally predominated by a bacterial genus or a family as summarized in Table 2. Notably, in some of the disorders, there is an over-abundance of specific microbial species. For example, abundance of Lactobacillus sp. and Bifidobacterium sp. in disorders like schizophrenia (SCZ) [130] and attention-deficit hyperactivity disorder (ADHD) [131], respectively. The probable reason for this can be attributed to the complexity of the gut microbiota.
Table 2.
Studies (or reviews) on non-dietary interventions targeting the gut microbiome.
| Therapy | Indication | Type of study/Reference |
|---|---|---|
| FMT | T2D/metabolic syndrome | Interventional studies [163, 151] |
| FMT | or C difficile infection – | Systematic review of RCTs [164] |
| FMT | for ulcerative colitis – | Systematic review of RCTs [165] |
| FMT | T1D – | RCT in [166] |
| TMAO synthesis inhibitors | Lowering cholesterol and bile acid metabolism | Preclinical [167] |
| TMAO synthesis inhibitors | Chronic kidney disease | Preclinical [168] |
| TMAO synthesis inhibitors | Heart disease | Preclinical [169] |
Gut microbiota modulates the gut−brain axis via numerous direct and indirect ways. This includes maintaining gut permeability by modulating the integrity of tight junctions in the gut epithelium, producing a wide variety of metabolites including neurotransmitters, SCFAs, and amino acids. These gut-derived metabolites can affect the central nervous system by acting locally on the enteric nervous system or enter circulation and affect the brain. Furthermore, alteration in the levels of gut microbial metabolites, such as SCFAs, ammonia, tryptophan, and histamine have shown to be associated, directly or by breaking down into neuroactive catabolites with various neurological conditions like Parkinson’s disease (PD) [132], anorexia nervosa (AN) [133], Alzheimer’s disease (AD) [134], autism spectrum disorder (ASD) [135], and chronic stress and depression [136]. However, whether this disruption of homeostasis in the mental health disorders is the cause or effect of the alterations in gut microbiota and its functions is not emphatically clear and requires more studies.
Further, considering the evidence, several studies have been undertaken to specifically target the gut microbiota through different therapeutic interventions including administration of pre- and probiotics (psychobiotics) to treat mental health disorders and/or their symptoms [137, 138]. Human-intervention studies with psychotics such as the probiotic combinations of lactobacilli and Bifidobacteria showed significant reduction in psychological distress [139], improved cognition and communication among patients with AD [140] and ASD [141], and improving symptoms among PD patients [142]. Based on the promising effects of psychobiotics on modulating the gut−brain axis, more clinical trials are currently being conducted to identify the bacterial strains as potential candidates for treatment of mental disorders.
The gut microbiome as a therapeutic target in chronic diseases
Given the tight interplay between enteric microbial symbionts and host immunity (Fig. 1), efforts have focused on implementing various strategies targeting the gut microbiota to manage or prevent chronic diseases. Clinical approaches to modify gut microbiota generally focus on depleting overabundant members or the overall microbial load using antibiotics or antifungal agents, modulation through diet, or supplementation with live microbes (single or mixed species). There are a variety of proposed therapeutic approaches, such as dietary modifications, prebiotics, probiotics, and TMAO-synthesis inhibitors, to target the gut microbiome. More recently, fecal microbial transplantation (FMT) has been used in a range of infectious, neurological, and GI conditions, with promising outcomes (Table 3).
Table 3.
Summary table of key findings.
| Disease group | Specific disease | Gut microbes | Main findings |
|---|---|---|---|
| Autoimmune diseases | Rheumatoid arthritis | Prevotella copri., | Increased in abundance [12, 13] |
| Collinsella sp., | Increased in abundance [13] | ||
| Faecalibacterium sp. | Decreased in abundance, links to SCFA production [25] | ||
| Type-1 diabetes | Dialister invisus., Gemella sanguinis., Bifidobacterium longum | Increased in abundance (children), gut permeability [25] | |
| F. prausnitzii | Decreased in abundance (children), butyrate (SCFA) production [28] | ||
| Atopic eczema | Clostridium difficile | Increased in abundance [37] | |
| Escherichia coli | Increased in abundance, eosinophilic inflammation [37] | ||
| Staphylococcus aureus | Increased in abundance [37] | ||
| Bifidobacteria spp. | Decreased in abundance [37] | ||
| Bacteroidetes spp. | Decreased in abundance [38, 39] | ||
| Coprococcus eutactus | Decreased (in children), linked to butyrate production [39] | ||
| F. prausnitizii | Increased in abundance, SCFA production [41] | ||
| Atopic asthma | Various—Clostridium, Pediococcus, Escherichia, Klebsiella, Morganella, and Proteus spp. | Increased in abundance, increased bioamine (histamine) production [55] | |
| Enterococcus faecalis, Streptococcus spp., Bifidobacterium bifidum., Lactobacillus spp. | Increased in abundance, increased bioamine (histamine) levels, increased epoxide hydrolase production of oxylipins (12,13-diHOME) [55, 56] | ||
| Gut inflammation disorders | Irritable bowel syndrome | Ruminococcus spp., Clostridium spp., Dorea spp. | Increased in abundance [62] |
| Bifidobacterium spp. | Decreased significantly in abundance (all IBS subtypes) [62, 68] | ||
| Faecalibacterium spp. | Decreased in abundance, anti-inflammatory, butyrate production [62, 64, 65] | ||
| Enterobacteriaceae spp. | Increased, links to previous intestinal infection and pathogen byproducts [63] | ||
| Lactobacillaceae spp., Bacteroides spp. | Increased in abundance [63] | ||
| Inflammatory bowel disease | Ruminococcaceae spp., Lachnospiraceae spp. | Decreased in microbiome, butyrate production [78–81] | |
| Faecalibacterium prausnitzii, Roseburia sp. | Decreased in microbiome, butyrate production [83] | ||
| Cardiometabolic diseases | Cardiovascular disease (including hypertension) | Lactobacillus plantarum, Lactobacillus rhamnosus | Improved ventricular function and attenuated heart failure after myocardial infarction [90, 107]. Improved blood pressure [110] |
| Akkermansia muciniphila | Protected against atherosclerosis in mice [111] | ||
| Type-2 diabetes | Bifidobacterium, Bacteroides, Faecalibacterium Akkermansia and Roseburia spp. | Negative association [112, 113] | |
| Ruminococcus, Fusobacterium, and Blautia | Positive association [112, 113] | ||
| Lactobacillus spp. | Enriched in control microbiome, butyrate production [112, 114, 115], insulin sensitivity through GLP-1 [116–120] | ||
| Clostridium spp. | Increased, opportunistic pathogen | ||
| NAFLD | Collinsell spp. | Increased in abundance [128]. Increased intestinal permeability and lipopolysaccharide release through metabolism of bile-acids to oxo-bile [144, 145] | |
| Escherichia spp., Dorea spp. | Increased in abundance [131, 135, 136] | ||
|
Coprococcus spp., Eubacterium spp., Faecalibacterium spp., Prevotella spp. |
Decreased in abundance [131, 135, 137] | ||
| Chronic kidney disease | Bifidobacterium, Lactobacillaceae, Prevotellaceae | Decreased in abundance, SCFA producers associated with anti-inflammatory cytokines [148, 149] | |
| Enterobacter | Increased in abundance [149] | ||
| Klebsiella | Increased in abundance, pathogen associated with inflammatory disease states including Crohn’s [148, 149] | ||
| Clostridium perfringes | Increased in abundance, pathogen associated with intestinal diseases [148] | ||
| Mental health disorders | SCZ, ADHD | Lactobacillus spp, Bifidobacterium | Increased in abundance in specific disorders such as SCZ and ADHD [157, 158] |
| General anxiety disorder (GAD) |
Bacteriodetes, Ruminococcus gnavus, and Fusobacterium Bacteroidaceae, Enterobacteriaceae, and Burkholderiaceae |
Decreased in abundance in general anxiety disorder (GAD) [170]. Increased in abundance in GAD [171, 172] | |
| Post- traumatic stress disorder (PTSD) | Actinobacteria, Lentisphaerae, and Verrucomicrobia | Decreased in abundance in post-traumatic stress disorder (PTSD) [173] | |
| Depression |
Eggerthella., Holdemania, Turicibacter, Paraprevotella Prevotella, Dialister |
Increased in abundance amongst individuals with depression [174, 175]. Decreased in abundance amongst individuals with depression [176] |
|
| Dementia | Escherichia, Blautia, Bifidobacterium, Streptococcus, Lactobacillus, Dorea spp Actinobacteria, Bacteroides | Increased in abundance amongst individuals with dementia [177]. Decreased in abundance amongst individuals with dementia [177] |
More recently, the composition of the gut microbiome has been implicated in predicting the severity of COVID-19 possibly via its immune-modulatory properties. More specifically, gut commensals with known immunomodulatory potential, such as Faecalibacterium prausnitzii, Eubacterium rectale, and Bifidobacterium adolescentis, were found to be significantly under-represented in patients with COVID-19 compared with healthy controls and were also associated with disease severity after taking account of antibiotic use and patient age [143]. Furthermore, the microbial imbalance found in the COVID patients was also associated with raised levels of inflammatory cytokines and blood markers of tissue damage, such as C-reactive protein and certain enzymes. Based on these findings, increasing the levels of beneficial gut species depleted in COVID-19 could serve as a novel avenue to mitigate severe disease, highlighting the importance of the gut microbiota in the pathophysiology of COVID-19.
Despite encouraging evidence from animal models in which inflammatory conditions were successfully treated via gut microbiota manipulation, data from human trials is less conclusive. Findings from recent work showed that the ability of an introduced microbe to successfully colonize the gut depends on the structure of the resident gut microbiota [144], a factor that has so far been overlooked in microbial intervention-based clinical trials and that might explain discrepancies in animal models and human trials. Nonetheless, intervention with a multispecies consortium of bacteria has shown to be effective in the maintenance of remission in patients with UC but not in those with CD [145]. Meta-analysis of intervention trials in which various combinations of bacterial strains were administered to adults with T2DM showed moderate improvements in hyperglycemia [146]. Moreover, probiotic supplementation during the first 27 days of life reduced the risk of islet autoimmunity in a large multicenter prospective cohort study of children at high genetic risk for T1DM, compared with no supplementation or supplementation later in infancy [147].
Intervention studies involving live microbial supplementation have also shown encouraging results, although more attention to microbial strain selection based on functional attributes, defined timing or duration of supplementation, and/or tailoring of the supplemented organisms to the endogenous gut microbiome of the recipient may significantly improve the efficacy in future studies. Ongoing studies are focused on understanding the basis of microbe-microbe interactions in order to identify discrete gut microbiomes that more readily respond to specific microbial interventions.
There is also a great need for tailored interventions that consider the microbial individuality of the recipient in order to prevent or manage chronic diseases. Such approaches are likely to result in a transition away from historically used probiotic strains, which are frequently poorly adapted to the enteric microenvironment, toward personalized multispecies microbial consortia sourced from healthy human enteric ecosystems. To further enhance the efficacy of microbial supplementation, nutritional support in the form of targeted dietary modifications, tailored to the specific substrate requirements [148], should also be considered as discussed in the section below. Such an integrated approach to microbiome-based therapeutics, built on independent observations in the field of gut microbiome research, may offer more effective, predictable, and sustainable microbial restitution in cases of chronic disease in which microbiome perturbation and functional gene loss are prominent features.
FMT has proven effective in >90% of patients with recurrent antibiotic-resistant C. difficile infection [149]. The success rate of FMT administration for other chronic intestinal inflammatory conditions such as IBD, however, is more modest, with clinical remission being less predictable [150]. More recently, FMT has been applied to chronic inflammatory conditions affecting organs distal to the GI tract. FMT from lean donors improved insulin sensitivity in men with obesity and metabolic syndrome, and this improvement was linked to changes in plasma metabolites, in particular increased gamma-aminobutyric acid (GABA), tryptophan, and phenylalanine [151].
Effects of a healthy diet are mediated via the gut microbiome
The most obvious way of targeting the gut microbiome is through dietary modifications. Already, several controlled clinical dietary intervention studies targeting the human gut microbiota have been reported [2, 152]. For example, diets rich in fiber were shown to significantly improve glucose control and promote a healthier metabolic profile in T2DM patients as well as reduce the risk of coronary heart disease [153]. Other studies showed marked interindividual variation in postprandial glycemic responses after consumption of identical meals [154, 155]. The consideration of microbial composition alongside known disease-risk factors (i.e., body mass index, fasting glucose) enabled accurate glucose-response prediction, thus allowing the design of more effective, personalized diets for improved glycemic control [155] in these studies.
Some of these same studies have recently investigated the links between gut microbiome structure, habitual diet, and blood cardiometabolic markers [156] in a cohort of 1100 healthy individuals from the United States and the United Kingdom. This study reported that microbial diversity and composition were highly correlated with both fasting and postprandial biomarkers. The strongest associations were with high-density lipoprotein cholesterol (HDLC), which positively correlated with microbial species richness. Cardiometabolic markers, which have in the recent past been shown to be strongly predictive of incident diabetes and heart disease, such as glycoprotein acetyl (GlycA) were also associated (positively or negatively) with microbiome richness. One of the most striking findings of the study was that gut microbiome composition is a better predictor of postprandial triglyceride and insulin concentrations than of glucose levels. The authors also reported that the diversity of healthy plant-based foods in habitual diet shapes gut microbiome composition. Approximately 10% of the variance in gut microbial composition could be explained by variation in healthy eating patterns [156].
Bacterial species were found to segregate into groups associated with more and less healthy plant- and animal- based foods. In addition, the authors reported a microbial signature of cardiometabolic health levels consistent across diet, obesity indicators, and cardiometabolic risks. The authors reported a consistent set of microbial species that strongly linked to all of the following: healthy eating patterns, indicators of obesity and cardiometabolic health, and fasting and postprandial circulating metabolites connected with cardiometabolic risk [156]. Other much smaller studies have also reported that dietary habits are strongly correlated with gut microbiome composition, disease status, and SCFA production [129].
Although a key weakness of the above study is its cross-sectional observational nature, consistent changes in gut microbiome composition and health outcomes have been recently reported by an international consortium investigating the effect of 12 months of Mediterranean diet on 612 elderly individuals from five countries [157]. Shankar Ghosh et al. [157] reported significant differences in the association patterns of bacterial taxa that increased (DietPositive) or decreased (DietNegative) in response to the Mediterranean diet intervention. The DietPositive bacteria had consistent negative associations (significantly lower than the DietNegative bacteria) with the pro-inflammatory circulating markers hsCRP and IL-17 levels as well as with functional measures associated with increased frailty. In contrast, DietPostive bacterial taxa showed positive correlations with measures of improved cognitive function, hand-grip strength, and two of the anti-inflammatory cytokines. DietPositive species included butyrogenic taxa such as F. prausnitzii, Eubacterium and Roseburia, whereas DietNegative bacteria included Ruminococcus torques, Collinsella aerofaciens, Coprococcus comes, Dorea formicigenerans, and Clostridium ramosum. Interestingly, there is no overlap between the “DietPositive” bacteria whose abundance increased after 12 months of a Med Diet reported by Shankar Ghosh et al. [157] and the taxa associated with “healthy diet indices” reported by Asnicar et al. On the other hand, the “DietPositive” and “DietNegative” taxa have substantial overlap with results from our own recent interventional study. We compared the changes in gut microbiome composition after 6 weeks of 500 mg of omega-3 fatty acids or 20 g of soluble fiber (inulin) in 70 middle-age and elderly individuals who had low fiber intake at baseline [158]. We found that, consistent with what is known of prebiotics such as inulin, that Bifidobacterium sp was remarkably increased and this correlated with butyrate production. On the other hand, omega-3 supplementation resulted in a significant decrease in Collinsella sp, both of which are in line with the findings from the multicenter Mediterranean diet intervention [157]. Importantly, many of the changes in bacterial abundances and SCFA concentrations were accompanied by decreases in inflammatory or atherogenic molecular mediators such as VLDL or IL4. Unlike the effects seen through a complex dietary intervention, such as the Mediterranean diet, we showed that it is possible to see significant effects both on microbiome composition and cardiometabolic health mediators.
Other recent studies have highlighted the role of the type of dietary fat on gut microbiome composition. In a randomized trial, 38 overweight and obese subjects were assigned to consume an excess of 1000 kcal/day of diets rich in saturated fat, unsaturated fat, or simple sugars for 3 weeks [159]. The relative abundances of individual taxa were altered in a diet-specific manner, wherein the overfeeding of saturated fat increased Proteobacteria, while unsaturated fat increased butyrate producers.
It is indeed possible to target the increase of specific taxa and the production of specific metabolites and the extent to which different carbohydrate sources (digestible by gut bacteria but not by humans) alter gut microbiome composition and function was recently tested by Deehan et al. [148] in a randomized controlled trial using a four-arm design. They gave increasing doses of three forms of resistant starch from maize, potato, or tapioca and a human-digestible corn starch (placebo) to 10 healthy volunteers per arm. They compared the effect of these carbohydrate substrates on gut microbiome composition in humans and found that chemically modified resistant starches with small structural differences induced highly specific effects on the gut microbiome that direct changes in the output of either propionate or butyrate. Dominant effects were remarkably consistent within treatment groups and dose-dependent with a plateau at a dose of 35 g/day. For example, maize-derived resistant starch resulted in increases in B. adolescentis, E. rectale, Oscillibacter, and Ruminococcus-related taxa and this correlated with increases in butyrate production. Tapioca-resistant starch on the other hand, which induced shifts in propionate proportions, as positively correlated with increases in P. distasonis an important succinate-producing bacterium. Succinate is then converted to propionate by other commensal bacteria in the gut. This very elegant design also clearly demonstrated the dose dependency between carbohydrate sources and the production of specific SCFAs. Similar strong and specific effects on gut microbiome composition have been shown by many interventional studies, both in healthy [160, 161] and diseased populations [162].
Where is this field moving toward?
Over the past decade, considerable evidence from both animal and human studies has accumulated showing a clear link between the gut microbiome in chronic diseases, including inflammatory autoimmune disorders, gut inflammation-related disorders, and cardiometabolic diseases (Fig. 1). It is increasingly clear that bacterial metabolites are at least in part, the key agents involved in the role of the gut microbiome on human health and among such metabolites, SCFAs appear to be the most important ones. Butyrate-producing bacteria are associated with lower risk of inflammatory autoimmune and cardiometabolic diseases, and also irritable bowel syndrome (Table 1). A number of therapeutic strategies to target the gut microbiome are possible, but nutritional changes appear to be the most obvious, noninvasive, and immediate way of altering the gut microbiome composition and function. Recent randomized controlled trials have shown that both composition and function respond in consistent ways to specific dietary interventions. Dietary fiber and unsaturated fat, separately or in a healthy diet such as the Mediterranean diet, result in higher relative abundances of butyrate-producing bacteria and these bacteria and the SCFAs produced in turn result in improved health outcomes. Different types of dietary fiber result in different bacterial changes and different SCFAs. The possibility of designing dietary interventions targeted specifically at increasing certain bacterial metabolites to improve cardiometabolic and inflammatory health outcomes appears well within reach within the next half decade.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1038/s41430-024-01467-z
Change history
4/4/2022
The super title Challenges in Nutrition was added.
Change history
6/26/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41430-024-01467-z
References
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerreiro CS, Calado Â, Sousa J, Fonseca JE. Diet microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front Med. 2018;5:349. doi: 10.3389/fmed.2018.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017;152:1–12. doi: 10.1111/imm.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells PM, Adebayo AS, Bowyer RCE, Freidin MB, Finckh A, Strowig T, et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: a cross-sectional study. Lancet Rheumatol. 2020;2:e418–e427. doi: 10.1016/S2665-9913(20)30064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78:590–3. doi: 10.1136/annrheumdis-2018-214514. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z, Andreev D, Oeser K, Krljanac B, Hueber A, Kleyer A, et al. Th2 and eosinophil responses suppress inflammatory arthritis. Nat Commun. 2016;7:11596. doi: 10.1038/ncomms11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Disco. 2009;8:969–81. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. 2020;11:1995. doi: 10.1038/s41467-020-15831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyötyläinen T, Hämäläinen A-M, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12:154–67. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 19.Paun A, Yau C, Danska JS. The influence of the microbiome on type 1 diabetes. J Immunol. 2017;198:590–5. doi: 10.4049/jimmunol.1601519. [DOI] [PubMed] [Google Scholar]
- 20.Needell JC, Zipris D. The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr Diab Rep. 2016;16:89. doi: 10.1007/s11892-016-0781-z. [DOI] [PubMed] [Google Scholar]
- 21.Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. 2016;1:16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maffeis C, Martina A, Corradi M, Quarella S, Nori N, Torriani S, et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev. 2016;32:700–9. doi: 10.1002/dmrr.2790. [DOI] [PubMed] [Google Scholar]
- 23.Mejía-León ME, Petrosino JF, Ajami NJ, Domínguez-Bello MG, de la Barca AMC. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64:3510–20. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–77. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 26.de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–44. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562:589–94. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18:552–62. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 29.Vaarala O, Atkinson MA, Neu J. The ‘perfect storm’ for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–62. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung DYM, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment approaches. J Allergy Clin Immunol. 2014;134:769–79. doi: 10.1016/j.jaci.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ta LDH, Chan JCY, Yap GC, Purbojati RW, Drautz-Moses DI, Koh YM, et al. A compromised developmental trajectory of the infant gut microbiome and metabolome in atopic eczema. Gut Microbes. 2020;12:1–22. doi: 10.1080/19490976.2020.1801964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pothmann A, Illing T, Wiegand C, Hartmann AA, Elsner P. The microbiome and atopic dermatitis: a review. Am J Clin Dermatol. 2019;20:749–61. doi: 10.1007/s40257-019-00467-1. [DOI] [PubMed] [Google Scholar]
- 33.Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the gut-skin axis in atopic dermatitis. Allergy Asthma Immunol Res. 2018;10:354–62. doi: 10.4168/aair.2018.10.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70:241–4. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 35.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–40. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 36.Lee E, Lee S-Y, Kang M-J, Kim K, Won S, Kim B-J, et al. Clostridia in the gut and onset of atopic dermatitis via eosinophilic inflammation. Ann Allergy Asthma Immunol. 2016;117:91–92.e1. doi: 10.1016/j.anai.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 37.Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut. 2002;51:51–55. doi: 10.1136/gut.51.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song H, Yoo Y, Hwang J, Na Y-C, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137:852–60. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 39.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. 2020;52:241–55. doi: 10.1016/j.immuni.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–26. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in finnish pre-school children. Nat Commun. 2016;7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. 2016;6:31775. doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018;11:785–95. doi: 10.1038/mi.2017.75. [DOI] [PubMed] [Google Scholar]
- 45.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74:799–809. doi: 10.1111/all.13660. [DOI] [PubMed] [Google Scholar]
- 46.Williams LM, Scott HA, Wood LG. Soluble fibre as a treatment for inflammation in asthma. J Nutr Intermed Metab. 2019;18:100108. [Google Scholar]
- 47.McLoughlin R, Berthon BS, Rogers GB, Baines KJ, Leong LEX, Gibson PG, et al. Soluble fibre supplementation with and without a probiotic in adults with asthma: A 7-day randomised, double blind, three way cross-over trial. EBioMedicine. 2019;46:473–85. doi: 10.1016/j.ebiom.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pugin B, Barcik W, Westermann P, Heider A, Wawrzyniak M, Hellings P, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Micro Ecol Health Dis. 2017;28:1353881. doi: 10.1080/16512235.2017.1353881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart CJ. Homing in on 12,13-diHOME in asthma. Nat Microbiol. 2019;4:1774–5. doi: 10.1038/s41564-019-0599-y. [DOI] [PubMed] [Google Scholar]
- 50.Barcik W, Pugin B, Brescó MS, Westermann P, Rinaldi A, Groeger D, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74:899–909. doi: 10.1111/all.13709. [DOI] [PubMed] [Google Scholar]
- 51.Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 52.Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EMM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329–36. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- 53.Ghoshal UC, Shukla R, Ghoshal U, Gwee K-A, Ng SC, Quigley EMM. The gut microbiota and irritable bowel syndrome: friend or foe? Int J Inflam. 2012;2012:151085. doi: 10.1155/2012/151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajilić-Stojanović M, Biagi E, Heilig HGHJ, Kajander K, Kekkonen RA, Tims S, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 55.Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 56.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–52. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M, Qiu X, Zhang H, Yang X, Hong N, Yang Y, et al. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS One. 2014;9:e109146. doi: 10.1371/journal.pone.0109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Gong J, Wang W, Long Y, Fu X, Fu Y, et al. Are there any different effects of bifidobacterium, lactobacillus and streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS One. 2014;9:e90153. doi: 10.1371/journal.pone.0090153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ford AC, Harris LA, Lacy BE, Quigley EMM, Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharm Ther. 2018;48:1044–60. doi: 10.1111/apt.15001. [DOI] [PubMed] [Google Scholar]
- 61.Dionne J, Ford AC, Yuan Y, Chey WD, Lacy BE, Saito YA, et al. A systematic review and meta-analysis evaluating the efficacy of a gluten-free diet and a low fodmaps diet in treating symptoms of irritable bowel syndrome. Am J Gastroenterol. 2018;113:1290–1300. doi: 10.1038/s41395-018-0195-4. [DOI] [PubMed] [Google Scholar]
- 62.Chumpitazi BP. The gut microbiome as a predictor of low fermentable oligosaccharides disaccharides monosaccharides and polyols diet efficacy in functional bowel disorders. Curr Opin Gastroenterol. 2020;36:147–54. doi: 10.1097/MOG.0000000000000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leshem A, Segal E, Elinav E. The gut microbiome and individual-specific responses to diet. mSystems. 2020;5. 10.1128/mSystems.00665-20. [DOI] [PMC free article] [PubMed]
- 64.Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–36. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 65.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 66.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One. 2013;8:e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez C, Antolin M, Santos J, Torrejon A, Casellas F, Borruel N, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–8. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 70.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–99. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halfvarson J, Brislawn CJ, Lamendella R, Vázquez-Baeza Y, Walters WA, Bramer LM, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mottawea W, Chiang C-K, Mühlbauer M, Starr AE, Butcher J, Abujamel T, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat Commun. 2016;7:13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–51. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 75.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Immerseel F, Ducatelle R, De Vos M, Boon N, Van De Wiele T, Verbeke K, et al. Butyric acid-producing anaerobic bacteria as a novel probiotic treatment approach for inflammatory bowel disease. J Med Microbiol. 2010;59:141–3. doi: 10.1099/jmm.0.017541-0. [DOI] [PubMed] [Google Scholar]
- 77.Miquel S, Martín R, Rossi O, Bermúdez-Humarán LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–61. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 78.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–70. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 79.Tang WHW, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120:1183–96. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lam V, Su J, Koprowski S, Hsu A, Tweddell JS, Rafiee P, et al. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. 2012;26:1727–35. doi: 10.1096/fj.11-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lam V, Su J, Hsu A, Gross GJ, Salzman NH, Baker JE. Intestinal microbial metabolites are linked to severity of myocardial infarction in rats. PLoS One. 2016;11:e0160840. doi: 10.1371/journal.pone.0160840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson MA, Verdi S, Maxan M-E, Shin CM, Zierer J, Bowyer RCE, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9:2655. doi: 10.1038/s41467-018-05184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun S, Lulla A, Sioda M, Winglee K, Wu MC, Jacobs DR, Jr, et al. Gut microbiota composition and blood pressure. Hypertension. 2019;73:998–1006. doi: 10.1161/HYPERTENSIONAHA.118.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palmu J, Salosensaari A, Havulinna AS, Cheng S, Inouye M, Jain M, et al. Association between the gut microbiota and blood pressure in a population cohort of 6953 Individuals. J Am Heart Assoc. 2020;9:e016641. doi: 10.1161/JAHA.120.016641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–9. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker JL, Havas KA, Miller LA, Lacy WA, Schlanser J. Gunshot wounds in military working dogs in operation enduring freedom and operation Iraqi freedom: 29 cases (2003-9) J Vet Emerg Crit Care. 2013;23:47–52. doi: 10.1111/j.1476-4431.2012.00823.x. [DOI] [PubMed] [Google Scholar]
- 88.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA. 2013;110:4410–5. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maifeld A, Bartolomaeus H, Löber U, Avery EG, Steckhan N, Markó L, et al. Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat Commun. 2021;12:1970. doi: 10.1038/s41467-021-22097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gan XT, Ettinger G, Huang CX, Burton JP, Haist JV, Rajapurohitam V, et al. Probiotic administration attenuates myocardial hypertrophy and heart failure after myocardial infarction in the rat. Circ Heart Fail. 2014;7:491–9. doi: 10.1161/CIRCHEARTFAILURE.113.000978. [DOI] [PubMed] [Google Scholar]
- 91.Khalesi S, Sun J, Buys N, Jayasinghe R. Effect of probiotics on blood pressure: a systematic review and meta-analysis of randomized, controlled trials. Hypertension. 2014;64:897–903. doi: 10.1161/HYPERTENSIONAHA.114.03469. [DOI] [PubMed] [Google Scholar]
- 92.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in apoe-/- mice. Circulation. 2016;133:2434–46. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 93.Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 96.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824–31. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 97.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–26. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–71. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fava S. Glucagon-like peptide 1 and the cardiovascular system. Curr Diabetes Rev. 2014;10:302–10. doi: 10.2174/1573399810666141030125830. [DOI] [PubMed] [Google Scholar]
- 100.Menni C, Zhu J, Le Roy CI, Mompeo O, Young K, Rebholz CM, et al. Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes. 2020;11:1632–42. doi: 10.1080/19490976.2020.1778261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123–33. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Gottardi A, McCoy KD. Evaluation of the gut barrier to intestinal bacteria in non-alcoholic fatty liver disease. J Hepatol. 2011;55:1181–3. doi: 10.1016/j.jhep.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Sabaté J-M, Jouët P, Harnois F, Mechler C, Msika S, Grossin M, et al. High prevalence of small intestinal bacterial overgrowth in patients with morbid obesity: a contributor to severe hepatic steatosis. Obes Surg. 2008;18:371–7. doi: 10.1007/s11695-007-9398-2. [DOI] [PubMed] [Google Scholar]
- 104.Moreira APB, Texeira TFS, Ferreira AB, Peluzio M, do CG, Alfenas R, de CG. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br J Nutr. 2012;108:801–9. doi: 10.1017/S0007114512001213. [DOI] [PubMed] [Google Scholar]
- 105.Astbury S, Atallah E, Vijay A, Aithal GP, Grove JI, Valdes AM. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes. 2020;11:569–80. doi: 10.1080/19490976.2019.1681861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun. 2019;10:1406. doi: 10.1038/s41467-019-09455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoyles L, Fernández-Real J-M, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24:1070–80. doi: 10.1038/s41591-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen F, Zheng R-D, Sun X-Q, Ding W-J, Wang X-Y, Fan J-G. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16:375–81. doi: 10.1016/S1499-3872(17)60019-5. [DOI] [PubMed] [Google Scholar]
- 109.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868–75. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 110.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25:1054. doi: 10.1016/j.cmet.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–75. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, et al. Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat Rev Gastroenterol Hepatol. 2020;17:279–97. doi: 10.1038/s41575-020-0269-9. [DOI] [PubMed] [Google Scholar]
- 113.Iebba V, Guerrieri F, Di Gregorio V, Levrero M, Gagliardi A, Santangelo F, et al. Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci Rep. 2018;8:8210. doi: 10.1038/s41598-018-26509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, et al. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol. 2018;84. 10.1128/AEM.00235-18. [DOI] [PMC free article] [PubMed]
- 115.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:G227–34. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 116.Aron-Wisnewsky J, Gaborit B, Dutour A, Clement K. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect. 2013;19:338–48. doi: 10.1111/1469-0691.12140. [DOI] [PubMed] [Google Scholar]
- 117.Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33:128–33. doi: 10.1097/MOG.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Onal EM, Afsar B, Covic A, Vaziri ND, Kanbay M. Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens Res. 2019;42:123–40. doi: 10.1038/s41440-018-0144-z. [DOI] [PubMed] [Google Scholar]
- 119.Jazani NH, Savoj J, Lustgarten M, Lau WL, Vaziri ND. Impact of gut dysbiosis on neurohormonal pathways in chronic kidney disease. Diseases. 2019;7. 10.3390/diseases7010021. [DOI] [PMC free article] [PubMed]
- 120.Sircana A, De Michieli F, Parente R, Framarin L, Leone N, Berrutti M, et al. Gut microbiota, hypertension and chronic kidney disease: recent advances. Pharm Res. 2019;144:390–408. doi: 10.1016/j.phrs.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 121.Li F, Wang M, Wang J, Li R, Zhang Y. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front Cell Infect Microbiol. 2019;9:206. doi: 10.3389/fcimb.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–15. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 123.Yang T, Richards EM, Pepine CJ, Raizada MK. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat Rev Nephrol. 2018;14:442–56. doi: 10.1038/s41581-018-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mafra D, Borges N, Alvarenga L, Esgalhado M, Cardozo L, Lindholm B, et al. Dietary components that may influence the disturbed gut microbiota in chronic kidney disease. Nutrients. 2019;11. 10.3390/nu11030496. [DOI] [PMC free article] [PubMed]
- 125.Tayebi-Khosroshahi H, Habibzadeh A, Niknafs B, Ghotaslou R, Yeganeh Sefidan F, Ghojazadeh M, et al. The effect of lactulose supplementation on fecal microflora of patients with chronic kidney disease; a randomized clinical trial. J Ren Inj Prev. 2016;5:162–7. doi: 10.15171/jrip.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wanchai K, Yasom S, Tunapong W, Chunchai T, Thiennimitr P, Chaiyasut C, et al. Prebiotic prevents impaired kidney and renal Oat3 functions in obese rats. J Endocrinol. 2018;237:29–42. doi: 10.1530/JOE-17-0471. [DOI] [PubMed] [Google Scholar]
- 127.Van Hung T, Suzuki T. Dietary fermentable fibers attenuate chronic kidney disease in mice by protecting the intestinal barrier. J Nutr. 2018;148:552–61. doi: 10.1093/jn/nxy008. [DOI] [PubMed] [Google Scholar]
- 128.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–38. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maldonado-Contreras A, Noel SE, Ward DV, Velez M, Mangano KM. Associations between diet, the gut microbiome, and short-chain fatty acid production among older Caribbean Latino adults. J Acad Nutr Diet. 2020;120:2047–.e6. doi: 10.1016/j.jand.2020.04.018. [DOI] [PubMed] [Google Scholar]
- 130.Zhu F, Ju Y, Wang W, Wang Q, Guo R, Ma Q, et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat Commun. 2020;11:1612. doi: 10.1038/s41467-020-15457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prehn-Kristensen A, Zimmermann A, Tittmann L, Lieb W, Schreiber S, Baving L, et al. Reduced microbiome alpha diversity in young patients with ADHD. PLoS One. 2018;13:e0200728. doi: 10.1371/journal.pone.0200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Caspani G, Kennedy S, Foster JA, Swann J. Gut microbial metabolites in depression: understanding the biochemical mechanisms. Micro Cell Fact. 2019;6:454–81. doi: 10.15698/mic2019.10.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roubalová R, Procházková P, Papežová H, Smitka K, Bilej M, Tlaskalová-Hogenová H. Anorexia nervosa: gut microbiota-immune-brain interactions. Clin Nutr. 2020;39:676–84. doi: 10.1016/j.clnu.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 134.Zhang L, Wang Y, Xiayu X, Shi C, Chen W, Song N, et al. Altered gut microbiota in a mouse model of Alzheimer’s disease. J Alzheimers Dis. 2017;60:1241–57. doi: 10.3233/JAD-170020. [DOI] [PubMed] [Google Scholar]
- 135.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci. 2012;57:2096–102. doi: 10.1007/s10620-012-2167-7. [DOI] [PubMed] [Google Scholar]
- 136.Galland L. The gut microbiome and the brain. J Med Food. 2014;17:1261–72. doi: 10.1089/jmf.2014.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barbosa RSD, Vieira-Coelho MA. Probiotics and prebiotics: focus on psychiatric disorders – a systematic review. Nutr Rev. 2020;78:437–50. doi: 10.1093/nutrit/nuz080. [DOI] [PubMed] [Google Scholar]
- 138.Ansari F, Pourjafar H, Tabrizi A, Homayouni A. The effects of probiotics and prebiotics on mental disorders: a review on depression, anxiety, Alzheimer, and autism spectrum disorders. Curr Pharm Biotechnol. 2020;21:555–65. doi: 10.2174/1389201021666200107113812. [DOI] [PubMed] [Google Scholar]
- 139.Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105:755–64. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- 140.Akbari E, Asemi Z, Daneshvar Kakhaki R, Bahmani F, Kouchaki E, Tamtaji OR, et al. Effect of probiotic supplementation on cognitive function and metabolic status in Alzheimer’s disease: a randomized, double-blind and controlled trial. Front Aging Neurosci. 2016;8:256. doi: 10.3389/fnagi.2016.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nimgampalle M, Kuna Y. Anti-Alzheimer properties of probiotic, lactobacillus plantarum MTCC 1325 in Alzheimer’s disease induced albino rats. J Clin Diagn Res. 2017;11:KC01–KC05. doi: 10.7860/JCDR/2017/26106.10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tamtaji OR, Taghizadeh M, Daneshvar Kakhaki R, Kouchaki E, Bahmani F, Borzabadi S, et al. Clinical and metabolic response to probiotic administration in people with Parkinson’s disease: a randomized, double-blind, placebo-controlled trial. Clin Nutr. 2019;38:1031–5. doi: 10.1016/j.clnu.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 143.Yeoh YK, Zuo T, Lui GC-Y, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021. 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed]
- 144.Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 145.Abraham B, Quigley EMM. Antibiotics and probiotics in inflammatory bowel disease: when to use them? Frontline Gastroenterol. 2020;11:62–9. doi: 10.1136/flgastro-2018-101057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Samah S, Ramasamy K, Lim SM, Neoh CF. Probiotics for the management of type 2 diabetes mellitus: a systematic review and meta-analysis. Diabetes Res Clin Pr. 2016;118:172–82. doi: 10.1016/j.diabres.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 147.Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY Study. JAMA Pediatr. 2016;170:20–8. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador L, et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020;27:389–404.e6. doi: 10.1016/j.chom.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 149.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl J Med. 2013;368:407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 150.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8:1569–81. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kootte RS, Levin E, Salojärvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26:611–.e6. doi: 10.1016/j.cmet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 152.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–8. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 153.Pereira MA, O’Reilly E, Augustsson K, Fraser GE, Goldbourt U, Heitmann BL, et al. Dietary fiber and risk of coronary heart disease: a pooled analysis of cohort studies. Arch Intern Med. 2004;164:370–6. doi: 10.1001/archinte.164.4.370. [DOI] [PubMed] [Google Scholar]
- 154.Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf J, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26:964–73. doi: 10.1038/s41591-020-0934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–94. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 156.Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021. 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed]
- 157.Ghosh TS, Rampelli S, Jeffery IB, Santoro A, Neto M, Capri M, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut. 2020;69:1218–28. doi: 10.1136/gutjnl-2019-319654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vijay A, Astbury S, Le Roy C, Spector TD, Valdes AM. The prebiotic effects of omega-3 fatty acid supplementation: a six-week randomised intervention trial. Gut Microbes. 2021;13:1–11. doi: 10.1080/19490976.2020.1863133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jian C, Luukkonen P, Sädevirta S, Yki-Järvinen H, Salonen A. Impact of short-term overfeeding of saturated or unsaturated fat or sugars on the gut microbiota in relation to liver fat in obese and overweight adults. Clin Nutr. 2021;40:207–16. doi: 10.1016/j.clnu.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 160.So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. 2018;107:965–83. doi: 10.1093/ajcn/nqy041. [DOI] [PubMed] [Google Scholar]
- 161.Myhrstad MCW, Tunsjø H, Charnock C, Telle-Hansen VH. Dietary fiber, gut microbiota, and metabolic regulation-current status in human randomized trials. Nutrients. 2020;12. 10.3390/nu12030859. [DOI] [PMC free article] [PubMed]
- 162.Houghton D, Hardy T, Stewart C, Errington L, Day CP, Trenell MI, et al. Systematic review assessing the effectiveness of dietary intervention on gut microbiota in adults with type 2 diabetes. Diabetologia. 2018;61:1700–11. doi: 10.1007/s00125-018-4632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hartstra AV, Schüppel V, Imangaliyev S, Schrantee A, Prodan A, Collard D, et al. Infusion of donor feces affects the gut-brain axis in humans with metabolic syndrome. Mol Metab. 2020;42:101076. doi: 10.1016/j.molmet.2020.101076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hammeken LH, Baunwall SMD, Hvas CL, Ehlers LH. Health economic evaluations comparing faecal microbiota transplantation with antibiotics for treatment of recurrent Clostridioides difficile infection: a systematic review. Health Econ Rev. 2021;11:3. doi: 10.1186/s13561-021-00301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao HL, Chen SZ, Xu HM, Zhou YL, He J, Huang HL, et al. Efficacy and safety of fecal microbiota transplantation for treating patients with ulcerative colitis: A systematic review and meta-analysis. J Dig Dis. 2020;21:534–48. doi: 10.1111/1751-2980.12933. [DOI] [PubMed] [Google Scholar]
- 166.de Groot P, Nikolic T, Pellegrini S, Sordi V, Imangaliyev S, Rampanelli E, et al. Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut. 2021;70:92–105. doi: 10.1136/gutjnl-2020-322630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pathak P, Helsley RN, Brown AL, Buffa JA, Choucair I, Nemet I, et al. Small molecule inhibition of gut microbial choline trimethylamine lyase activity alters host cholesterol and bile acid metabolism. Am J Physiol Heart Circ Physiol. 2020;318:H1474–H1486. doi: 10.1152/ajpheart.00584.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gupta N, Buffa JA, Roberts AB, Sangwan N, Skye SM, Li L, et al. Targeted inhibition of gut microbial trimethylamine N-Oxide production reduces renal tubulointerstitial fibrosis and functional impairment in a murine model of chronic kidney disease. Arterioscler Thromb Vasc Biol. 2020;40:1239–55. doi: 10.1161/ATVBAHA.120.314139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Organ CL, Li Z, Sharp TE, 3rd, Polhemus DJ, Gupta N, Goodchild TT, et al. Nonlethal inhibition of gut microbial trimethylamine N-oxide production improves cardiac function and remodeling in a murine model of heart failure. J Am Heart Assoc. 2020;9:e016223. doi: 10.1161/JAHA.119.016223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jiang H-Y, Zhang X, Yu Z-H, Zhang Z, Deng M, Zhao J-H, et al. Altered gut microbiota profile in patients with generalized anxiety disorder. J Psychiatr Res. 2018;104:130–6. doi: 10.1016/j.jpsychires.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 171.Chen Y-H, Bai J, Wu D, Yu S-F, Qiang X-L, Bai H, et al. Association between fecal microbiota and generalized anxiety disorder: severity and early treatment response. J Affect Disord. 2019;259:56–66. doi: 10.1016/j.jad.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 172.Chen Y-H, Bai J, Wu D, Yu S-F, Qiang X-L, Bai H, et al. Corrigendum to ‘association between fecal microbiota and generalized anxiety disorder: severity and early treatment response’ [Journal of Affective Disorders 259(2019) 56–66] J Affect Disord. 2020;260:489. doi: 10.1016/j.jad.2019.09.033. [DOI] [PubMed] [Google Scholar]
- 173.Hemmings SMJ, Malan-Müller S, van den Heuvel LL, Demmitt BA, Stanislawski MA, Smith DG, et al. The microbiome in posttraumatic stress disorder and trauma-exposed controls: an exploratory study. Psychosom Med. 2017;79:936–46. doi: 10.1097/PSY.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stevens BR, Goel R, Seungbum K, Richards EM, Holbert RC, Pepine CJ, et al. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut. 2018;67:1555–7. doi: 10.1136/gutjnl-2017-314759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kelly JR, Borre Y, O’ Brien C, Patterson E, El Aidy S, Deane J, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–18. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 176.Lin P, Ding B, Feng C, Yin S, Zhang T, Qi X, et al. Prevotella and klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord. 2017;207:300–4. doi: 10.1016/j.jad.2016.09.051. [DOI] [PubMed] [Google Scholar]
- 177.Zhuang Z-Q, Shen L-L, Li W-W, Fu X, Zeng F, Gui L, et al. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis. 2018;63:1337–46. doi: 10.3233/JAD-180176. [DOI] [PubMed] [Google Scholar]