Abstract
In the brain, both HIV-1 and methamphetamine (meth) use result in increases in oxidative and nitrosative stress. This redox stress is thought to contribute to the pathogenesis of HIV-associated neurocognitive disorder (HAND) and further worsening cognitive activity in the setting of drug abuse. One consequence of such redox stress is aberrant protein S-nitrosylation, derived from nitric oxide which may disrupt protein activity. Here, we report an improved, mass spectrometry-based technique to assess S-nitrosylated protein in human postmortem brains using selective enrichment of S-nitrosocysteine residues with an organomercury resin. The data show increasing S-nitrosylation of tricarboxylic acid (TCA) enzymes in the setting of HIV and HIV/meth use compared to control brains without CNS pathology. The consequence is systematic inhibition of multiple TCA cycle enzymes, resulting in energy collapse that can contribute to the neuronal and synaptic damage observed in HAND and meth use.
Keywords: HIV-1, Methamphetamine, Protein S-nitrosylation, TCA cycle, Mitochondrial dysfunction
Introduction
Emerging evidence suggests the heuristic model that both HIV/AIDS and several forms of drug abuse affect the brain via free radical damage involving the generation of reactive oxygen species (ROS, producing oxidative stress) and reactive nitrogen species (RNS), including nitric oxide (NO, resulting in nitrosative stress). For example, both HIV-associated neurocognitive disorder (HAND) and methamphetamine use (meth) appear to produce neuronal damage via oxidative and nitrosative redox stress (Cadet and Krasnova 2007; Cotto et al. 2019; Vazquez-Santiago et al. 2014). MR spectroscopy (MRS) studies have shown that the combination of HIV/meth contributes to both glial activation and neuronal injury (Chang et al. 2005), and oxidative stress and microglial inflammation have been proposed to contribute to the neuronal damage (Reiner et al. 2009). However, the molecular targets of nitrosative stress, which results in HIV/meth effects on brain and behavior, remain largely unknown (Soontornniyomkij et al. 2016). Clinically, these unknown targets represent a major challenge for the field and need to be identified in order both to develop biomarkers of disease and to allow screening for new therapies to prevent corruption of these targets by free radical stress. Here, we employed an innovative approach using newly emerging mass spectrometry (MS) techniques to identify in a systematic, selective fashion the posttranslational modifications (PTMs) of proteins resulting from such redox stress, specifically protein S-nitrosylation (Dedon and Tannenbaum 2004; Doulias et al. 2010; Doulias et al. 2013a; Forrester et al. 2009; Hess et al. 2005; Keshive et al. 1996; Keszler et al. 2010; Madej et al. 2008; Martinez-Ruiz et al. 2011; Mitchell and Marletta 2005; Nakamura et al. 2015; Raju et al. 2015; Seth and Stamler 2011; Smith and Marletta 2012) and hence identify new protein targets affected by AIDS and drug abuse. Heretofore, it has not been possible to identify the full range of proteins undergoing PTMs due to nitrosative and oxidative stress, but various new MS techniques allow this kind of identification (Doulias et al. 2013b; Seneviratne et al. 2016). The known role of RNS/ROS on HAND/meth incentivized us to look for molecular targets affected by nitrosative and oxidative stress because these are not generally known. We find protein S-nitrosylation of enzymes involved in transporter-mediated glutamate clearance and oxidative phosphorylation (OXPHOS)-related metabolic pathways in accord with previously reported disruption of excitatory amino acid (EAA) reuptake and energy metabolism in HIV or meth-associated synaptic toxicity and neuronal damage (Chang et al. 2007; Cotto et al. 2019; Kaul et al. 2001; Sanchez et al. 2016; Vazquez-Santiago et al. 2014). This approach thus yields a variety of new targets of potential therapeutic importance in patients in the setting of HAND and meth use.
Materials and methods
Chemicals and reagents
Reagents were obtained from Sigma-Aldrich. All chemicals and reagents used were of analytical grade.
Postmortem human brain samples
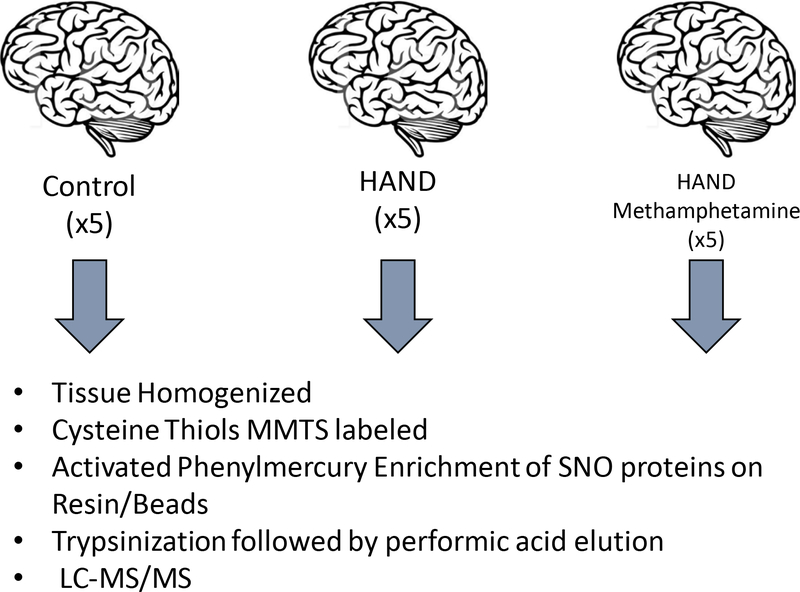
The use of all human postmortem brain samples was reviewed and approved by the Institutional Review Board of The Scripps Research Institute and the University of California, San Diego (UCSD), School of Medicine. At postmortem examination, brains were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until used by the UCSD Neuropathology Core HIV Brain Bank. Three groups of age-matched brains were collected, with 5 samples from the frontal lobe in each case (Table S1): HIV+ with evidence of HAND, HIV+ with evidence of HAND and meth use; control brains without CNS pathology. Note that predominantly middle-aged male brains were available from this HIV Brain Bank. The tissues were subsequently homogenized into 3 ml of lysis buffer (250 mM Hepes-NaOH at pH 7.7, containing 1 mM diethylenetriamine pentaacetic acid, 0.1 mM neocuproine, 1% Triton X-100, and protease inhibitors) with a Teflon pestle on ice. Homogenates were centrifuged at 13,000g for 30 min at 4 °C. Soluble protein fractions were collected, and protein concentration determined by Bradford assay. Sample preparation and negative control samples were generated as described (Doulias et al. 2010; Doulias et al. 2013a).
Organomercury enrichment and identification of S-nitrosylation (SNO)-sites in the proteome
Experimental details for preparation and activation of columns, and reaction of homogenate with organomercury resin for S-nitrosocysteine enrichment have been previously presented (Doulias et al. 2010; Doulias et al. 2013b). For each group (HIV HIV/meth, control), five samples were analyzed with technical duplicates. As determined previously for this method in our laboratory (Doulias et al. 2013a), the false identification rate was <6% for brain samples. For column washes, we started with 50 bed volumes of 50 mM tris-HCl (pH 7.4) containing 300 mM NaCl and 0.5% SDS. This was followed by 50 bed volumes of the same buffer containing 0.05% SDS. Columns were then washed with 50 bed volumes of 50 mM tris-HCl containing 300 mM NaCl (pH 7.4), 1% Triton X-100, and 1 M urea. Columns were next washed with 50 bed volumes of the same buffer containing 0.1% Triton X-100 and 0.1 M urea, and then 200 bed volumes of water. This was followed by on-column digestion of proteins into peptides for which the columns were initially washed with 10 bed volumes of 0.1 M ammonium bicarbonate.
Next, the bound proteins were subjected to digestion with Trypsin Gold (1 μg/mL) (Promega) added in one bed volume of 0.1 M ammonium bicarbonate in the dark at room temperature for 16 hours. This was followed by washing the resin with 40 bed volumes of 1 M ammonium bicarbonate (pH 7.4) containing 300 mM NaCl, followed by 40 volumes of the same buffer without salt, 40 volumes of 0.1 M ammonium bicarbonate, and 200 volumes of deionized water. To elute the peptides bound to the resin, the columns were then incubated with one bed volume of performic acid in water synthesized as described (Doulias et al. 2010; Doulias et al. 2013a). To ensure recovery of eluted peptides, the column was then washed with one volume of deionized water. Eluates were stored at −80°C overnight, and then lyophilized and resuspended in 300 μL of 0.1% formic acid. Following resuspension, peptides placed into low-retention tubes (Axygen) and reduced to a volume of 30 μL by speed vacuum. Following, peptide suspensions were desalted using Stage Tips (ThermoFisher Scientific) and weretransferred to a high-performance liquid chromatography vial and submitted for LC-MS/MS analysis. For SNO-peptide identification, 20 μL of the peptide suspension were submitted for high-performance liquid chromatography (LC)–MS/MS analysis using a Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer (ThermoFisher Scientific). Subsequent analysis, as described (Doulias et al. 2013a), determined the S-nitrosocysteine proteome and identified S-nitrosylated peptides and proteins for each sample group (Tables S2 to S4). Following S-nitrosoproteome identification, functional analysis was performed to characterize the biological pathways affected using gene ontology (GO) knowledgebase (2020–12-08 release) enrichment, pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG release 96.0, October 1, 2020 ) and STRING protein-protein interaction network functional enrichment (Mi et al. 2013).
Human induced pluripotent stem cell (hiPSC)-derived neurons
We differentiated hiPSCs used a standard protocol for generating cerebrocortical neurons (Talantova et al. 2013). Briefly, feeder-free hiPSCs were cultured on Matrigel in mTeSR 1 medium (StemCell Technologies) and induced to differentiate by a one week exposure to the following: A83–01, Dorsomorphin, and PNU74654 (each at 2 μM) in DMEM/F12 medium supplemented with 20% Knock Out Serum Replacement (Invitrogen). Next, PAX6+ neurospheres were formed by manually scraping cells and then maintained for 2 weeks in DMEM/F12 medium supplemented with N2 and B27 (Invitrogen) and 20 ng ml−1 of basic FGF (R&D Systems). Finally, the neurospheres were seeded on polyornithine/laminin-coated dishes to form rosettes and human neural progenitor cells (hNPCs) were manually picked and expanded. Terminal neuronal differentiation was accomplished by seeding at a 1:1 ratio with neonatal mouse astrocytes onto polyornithine/laminin-coated glass coverslips in DMEM/F12 medium supplemented with B27, N2, GDNF (20 ng ml−1) and BDNF (20 ng ml−1)(Peprotech), and 0.5% FBS (Invitrogen). Experiments were conducted after a total of 5–6 weeks of differentiation.
Tricarboxylic acid (TCA) cycle enzymatic assays
In cell-based assays, isocitrate dehydrogenase (IDH) activity was determined using a colorimetric IDH Assay Kit (Abcam, ab102528). Aconitase activity was determined using an enzyme activity assay kit (Abcam, ab109712).
In in vitro enzymatic assays using recombinant protein, dihydrolipoyl dehydrogense (DLD) activity was measured in the forward direction in which DLD catalyzes the conversion of dihydrolipoic acid and NAD+ to lipoic acid and NADH (Carothers et al. 1989; Patel et al. 1995). In brief, recombinant human DLD protein (R&D Systems 8646DH) was incubated with 200 μM SNOC in the dark for 20 min at room temperature to induce SNO-DLD formation. Recombinant DLD was then added to assay buffer (100 mM potassium phosphate buffer, pH 8.0, 1mM EDTA) containing 3mM dihydrolipoic acid and 3 mM NAD+. The NADH production is monitored by following the increase in absorbance at 340 nm at room temperature. As a blank control, assay solution without recombinant protein was used. For the calculation of enzymatic activity, an extinction coefficient of 6.22 mM−1 cm−1 for NADH was used (Carothers et al. 1989; Patel et al. 1995). The dehydrogenase activity was expressed as 1 μmol of NAD+ reduced per min per amount of DLD (μg).
Statistical analysis
Normally distributed data were displayed as mean ± SEM. Data were analyzed with GraphPad Prism software. Statistical difference between a pair of values was determined by a two-tailed Student’s t test, and multiple groups were analyzed by two-way ANOVA with an appropriate post-hoc test.
Results
S-Nitrosoproteome in HIV/ HIV/meth human brains vs. normal controls
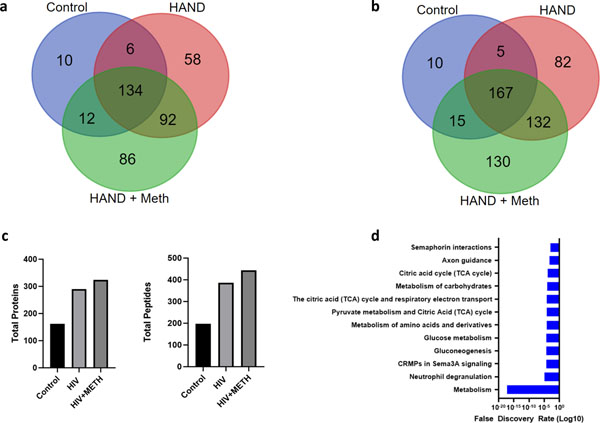
To determine the S-nitrosoproteome in brains of HIV-1 infected individuals with HAND (abbreviated HIV brains), HAND plus meth use (abbreviated HIV/meth), and controls, we analyzed human brain tissues from the UCSD Neuropathology Core Brain Bank (schema of procedures shown in Fig. 1). In brief, we prepared tissue homogenates from cortical brain samples from the two affected populations and age- and sex-matched non-affected controls. S-nitrosylated proteins were captured on activated phenylmercury resin. The captured S-nitrosylated proteins we digested on-column with trypsin, the bound peptides were eluted using performic acid and the peptides were analyzed by LC-MS/MS. Fig. 2a presents a Venn diagram indicating that 58 S-nitrosylated proteins were found exclusively in the HIV group and 86 exclusively in the HIV/meth group. An additional 92 S-nitrosylated proteins were shared in the HIV and HIV/meth groups and not found in the control samples. Fig. 2b depicts the same kind of information for the number of S-nitrosocysteine peptides. The total number of proteins and peptides identified is shown in Fig. 2c. The HIV+ meth group contained the largest number of S-nitrosylated proteins (324), followed by HIV (290) and non-disease controls (162). The majority of the proteins had a single S-nitrosylated cysteine-containing peptide, but proteins with up to 7 SNO-sites were detected. A gene ontology (GO) enrichment analysis of all S-nitrosylated proteins identified in all samples showed a large proportion of the modified proteins is functionally involved in metabolism, immunity and neuroinflammation (Fig. 2d).
Fig. 1.
Schema of human postmortem brain analysis for S-nitrosylated proteins
Abbreviation: MMTS, methyl methanethiosulfonate.
Fig. 2.
Venn diagram of S-nitrosylated (SNO)-proteins (a) Venn diagram of proteins found in the three groups: HIV, HIV/meth, and normal (control). (b) Venn diagram of peptides found in the three groups. (c) Comparison of total proteins (left) and peptides (right) identified in organomercury enrichment. (d) Gene ontology (GO) enrichment analysis of proteins found in all samples.
S-Nitrosoproteome of normal human brain
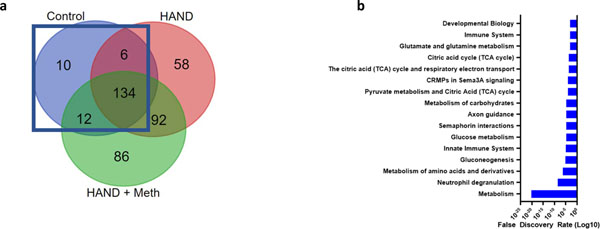
GO analysis on the 162 proteins found to be S-nitrosylated in normal brain (Fig. 3a, b) indicated enrichment of proteins participating in the TCA cycle. This suggests that several of the proteins in this metabolic pathway are S-nitrosylated to some extent under normal physiological conditions in the human brain, as had previously been reported for normal mouse brain (Doulias et al. 2013a).
Fig. 3.
Normal brain S-nitrosoproteome (a) Venn diagram outlining the 162 SNO-proteins found in normal brain. (b) GO enrichment analysis showed that the TCA cycle was enriched with an FDR if 1E-5.
S-Nitrosoproteome of HIV human brain
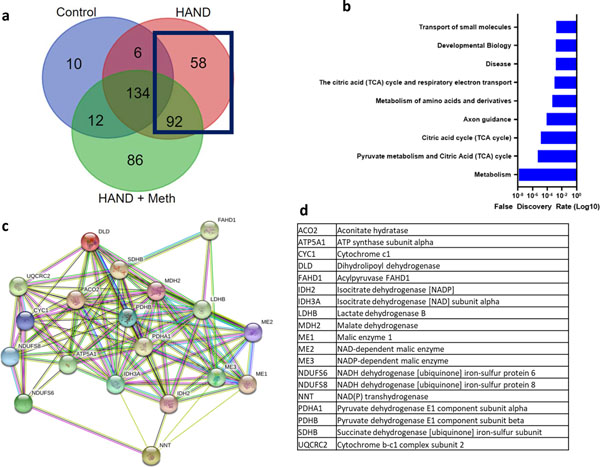
We next performed a gene ontology analysis of the 147 proteins that were found to be S-nitrosylated in HIV brains but not in normal human brains (Fig. 4a). Among the GO enrichment terms, transport of small molecules was significant, as evidenced by the FDR (Fig. 4b). In the list of individual S-nitrosylated proteins, the excitatory amino acid transporter (EAAT) 2 was found in HIV brain but not in normal brain (Tables S2 vs. S3). Prior results from our laboratory and others had shown that EAAT2 function was compromised in models of HAND, thus increasing glutamate levels and contributing to neurotoxicity (Dreyer and Lipton 1995; Kaul et al. 2001; Vazquez-Santiago et al. 2014; Wang et al. 2003). Moreover, oxidation of critical thiol groups could account for this inhibition of EAAT2 activity (Trotti et al. 1997). Because protein S-nitrosylation represents one such oxidation reaction, our new finding of S-nitrosylated -EAAT2 in HIV brains could therefore account, at least in part, for the loss of transporter activity.
Fig. 4.
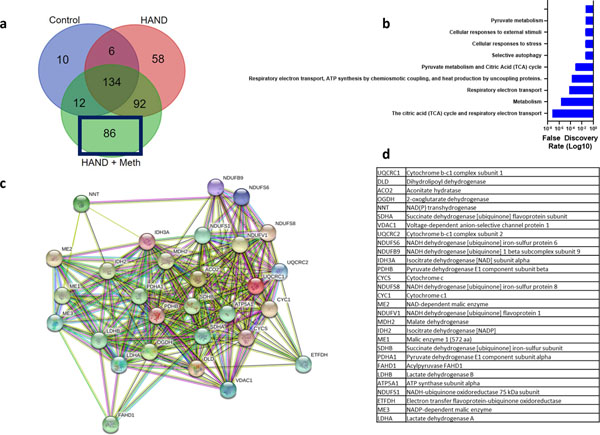
S-Nitrosylated proteins found in HIV brain but not in normal. (a) Venn diagram shows that the SNO-proteins in HIV brain but not in normal included 150 proteins. (b) GO enrichment analysis again showed that metabolism and specifically the TCA cycle were among the most affected pathways. (c) STRING network analysis of 9 proteins found in normal plus 10 found in HIV brain or HIV/meth brain (see also Fig. 5) show that multiple components of the TCA cycle and enzymes leading into the TCA cycle (e.g., pyruvate dehydrogenase) were S-nitrosylated. (d) List of protein abbreviations shown in the network analysis.
Many TCA cycle proteins were found to be S-nitrosylated in non-disease control human brains and even moreso in HIV brains (Fig. 4b). We performed a STRING network analysis of the 9 TCA cycle proteins S-nitrosylated encountered in normal brains and HIV brains plus the10 additional TCA cycle proteins found exclusively in HIV brains to show their interconnectedness (Fig. 4c, d). The widespread presence of S-nitrosylated enzymes in the TCA cycle in HIV brain indicates the potential for aberrant regulation and disruption of energy production via this metabolic pathway, which is known to be critical for basal neuronal function (Belanger et al. 2011; Demetrius et al. 2014; Machler et al. 2016). Notably, the TCA cycle and the respiratory electron transport chain (ETC), also listed in the GO pathway analysis, overlap at succinate dehydrogenase, which is not only an enzyme in the TCA cycle but also represents complex II of the ETC. Indeed, among the individual proteins S-nitrosylated in HIV brain but not in normal brain, we found succinate dehydrogenase (Fig. 4d).
S-Nitrosoproteome of HIV/meth human brain
Next, we performed a gene ontology analysis of the 84 proteins exclusively found to be S-nitrosylated in the HIV/meth brains and not found in HIV brains or normal control brains (Fig. 5a, b). Again, S-nitrosylated TCA cycle enzymes were significantly enriched in this group. Moreover, additional members of the TCA cycle were found to be S-nitrosylated, such as 2-oxoglutarate dehydrogenase (also known as α-ketoglutarate dehydrogenase, αKGDH) that were not found in normal or HIV brains, suggesting that this energy-generating system is further targeted with methamphetamine exposure. STRING network analysis of the 29 tabulated TCA cycle enzyme (Fig. 5c, d) found in the HIV/meth or HIV brains shows that nearly all members of the TCA cycle were affected, suggesting potential severe impact of this energy system that largely contributes to the baseline health of neurons (Belanger et al. 2011).
Fig. 5.
SNO-proteins found in HIV/meth brain. (a) Venn diagram outlining the 86 proteins involved. (b) GO enrichment annotation of these proteins again showing the predominant involvement of metabolism including the TCA cycle and associated electron transport chain (ETC). (c). Network analysis of all proteins found involved in the TCA cycle showing a clear enrichment of S-nitros(yl)ation in HIV/meth and HIV brain tissue. (d) List of protein abbreviations shown in the network analysis.
Functional impact of S-nitrosylation on key TCA cycle enzymes
Many enzymes in the TCA cycle have been shown to be inhibited by oxidation (S-nitrosylation is in fact a form of oxidation) (Nakamura and Lipton 2017; Sun et al. 2007), and often the cysteine residue affected by these reactions is at or near the active site, resulting in enzymatic inhibition (Chouchani et al. 2010; Foster and Stamler 2004; Gibson et al. 2010; Scheving et al. 2012; Smith et al. 1991; Yan et al. 2012). Therefore, we tested the functional consequences of specific S-nitrosylation reactions on a subset of TCA enzymes. We took two approaches to this experiment, one in a cell-based assay using human induced pluripotent stem cell (hiPSC)-derived neurons so we could test enzyme function in a human context, and a second comprising an in vitro enzymatic assay on purified human protein. We tested the first three enzymes of the TCA cycle for their ability to be inhibited by S-nitrosylation. These enzymes are comprised of aconitate hydratase (also known as aconitase, AC), which catalyzes the conversion of citrate to isocitrate; IDH, with the IDH3 subtype generally considered the rate-limiting step in the forward TCA cycle, which converts isocitrate to α-ketoglutarate; and αKGDH complex, which catalyzes α-ketoglutarate to succinyl-CoA and, importantly, produces NADH equivalents for the ETC and hence fuels the production of ATP. To induce S-nitrosylation, we used the well-known physiological nitric oxide (NO) donor, S-nitrosocysteine (SNOC), as previously described (Lei et al. 1992; Lipton et al. 1993).
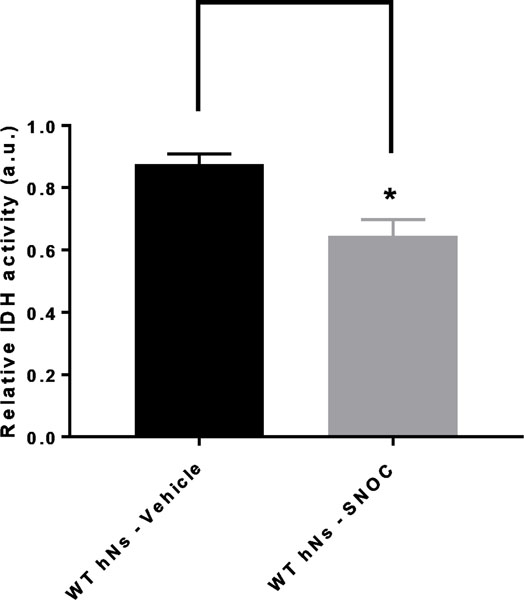
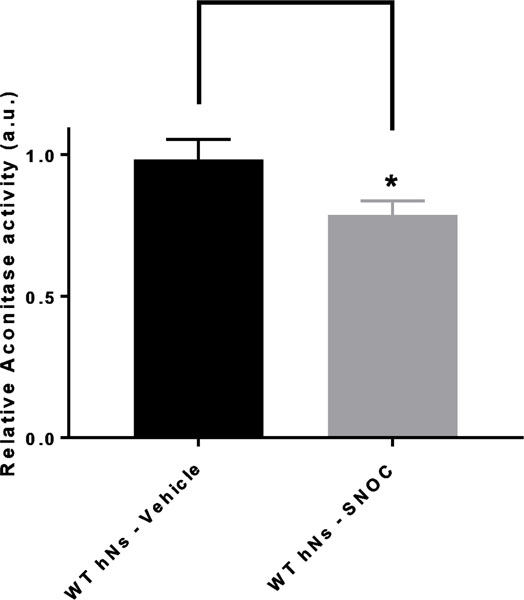
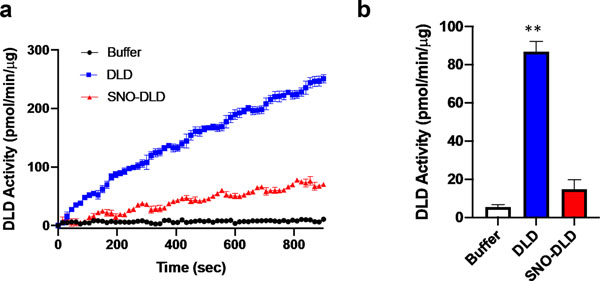
Indeed, our results strongly suggest that S-nitrosylation of IDH and aconitase inhibits their activity in hiPSC-derived neurons (Figs. 6 and 7). While these enzymes are nitrosylated even in normal brain (Table S2), affording physiological control of the TCA cycle via redox reactions (Doulias et al. 2013a), we found evidence for multiple subunits of the αKGDH complex being S-nitrsoylated only in HIV/meth brain (Fig. 5d and Table S4). Notably, there are three components of the αKGDH complex, and all three must function for enzyme activity. The first subunit (E1, or 2-oxoglutarate dehydrogenase) was found to be exclusively S-nitrosylated in the HIV/meth brains and not in HIV or control brains. In contrast, we found S-nitrosylation in all three groups of the E3 component of the αKGDH complex, termed DLD. The E3 component uses NAD+ and FAD+ as cofactors to produce NADH for the ETC. Hence, we wanted to test the effect of S-nitrosylation particularly on the E3 componet activity. In Fig. 8, we show by in vitro enzymatic assay, that S-nitrosylation of DLD inhibits its activity by approximately 65%, as occurs in the face of nitrosative stress in human HIV/meth brains.
Fig. 6.
IDH activity in hNs undergoing nitrosative stress hNs manifest inhibition of isocitrate dehydrogenase (IDH) enzyme activity in their mitochondria due to S-nitrosylation. Wild-type (WT) hiPSC-derived neurons (hNs) were exposed to 100 μM of the physiological NO donor S-nitrosocysteine (SNOC), and the lysates analyzed for IDH activity with isocitrate as substrate in the presence of NADPH with NADP+ being the assayed species reflective of IDH activity. A SpectraMax microplate instrument was used for the measurements. Values are mean + SEM. ∗p < 0.05 by Student’s t test; n = 3.
Fig. 7.
Aconitase activity in hNs undergoing nitrosative stress hNs manifest inhibition of aconitase enzyme activity in their mitochondria due to S-nitrosylation. Wild-type (WT) hiPSC-derived neurons (hNs) were exposed to 100 μM SNOC, and the lysates analyzed for aconitase activity with a commercial colorimetric assay kit (Abcam). This assay is based upon the rate of cis-aconitate production, measured spectrophotometrically, which is proportional to aconitase activity. Values are mean + SEM. ∗p < 0.05 by Student’s t test; n = 3.
Fig. 8.
Nitrosative stress inhibits DLD activity (measured as OD at 340 nm)
Dihydrolipoyl dehydrogenase (DLD) represents component #3 of the αKGDH complex, and is inhibited by S-nitrosylation by the NO donor S-nitrosocysteine (SNOC, 200 μM) in an in vitro activity assay. SNO-DLD: S-nitrosylated DLD. * p < 0.01 by two-tailed Student’s t-test.
Interestingly, the entry-level enzyme to the TCA cycle, the pyruvate dehyrogenase (PDH) complex, also has three components, and all three components (E1, E2, and E3) and their respective subunits were found to be S-nitrosylated in HIV/meth brain but not in HIV or normal brain (Tables S2–S5). Moreover, both the alpha and beta subunits of the E1 component of PDH were S-nitrosylated in HIV/meth and HIV brains but only the alpha subunit in normal control brains (Tables S2–S5). Additionally, more PDH alpha subunit peptides were S-nitrosylated in the HIV and HIV/meth brain than in controls. The fact that the E2 component of the mitochondrial PDH complex, designated dihydrolipoylysine-residue acetyltransferase (DLAT), and DLD (which comprises the E3 component of the PDH complex as well as the E3 component of the αKGDH complex) were both S-nitrosylated in HIV/meth brains, but not in HIV or normal brains, is consistent with the notion that the PDH complex was more inhibited in HIV/meth brains. Collectively, the PDH complex transforms pyruvate, NAD+ and coenzyme A into acetyl-CoA, CO2, and NADH (similar to the αKGDH complex), with NADH feeding the ETC for ATP production. Thus, inhibition of both the PDH and αKGDH complexes in HIV/meth brains would be expected to cause a much more severe compromise of energy production by the TCA cycle.
Furthermore, S-nitrosylation of pyruvate carboxylase, which catalyzes the converson of pyruvate to oxaloacetate was only found in HIV/meth brains and not in HIV or control brains. Other critical locations for energy production from the TCA cycle are also S-nitrosylated in HIV/meth and HIV brains compared to normal control brains; while all three groups of brains manifest some level of S-nitrosylation of malate dehdyrogenase (Tables S2–S4), another step that generates NADH for the ETC, only HIV/meth and HIV brains show S-nitrosylation of succinate dehyrogenase (SDH, Tables S3 and S4), which also represents complex II of the ETC. The step in the TCA cycle just prior to SDH, succinyl-CoA transferase (or succinyl-CoA ligase (SUCLG)) was also S-nitrosylated in HIV and HIV/meth brain to a greater extent than in normal controls, as judged from the number of peptides of this protein that we found to be S-nitrosylated on MS runs (Tables S2–S4).
Additionally, succinate-semialdehyde dehydrogenase (SSADH) was S-nitrosylated in HIV and HIV/meth brains but not in normal controls. Succinic semialdehyde is produced in inhibitory interneurons from gamma-aminobutyric acid (GABA), which is elevated when SSADH is inhibited, e.g., by S-nitrosylation. Succinic semialdehyde is normally converted to succinic acid which is shunted into the TCA cycle, but this pathway would be decreased with SNO-SSADH formation in HIV and HIV/meth brains compared to normal controls – again inhibiting the TCA cycle, in this case via substrate starvation.
Interestingly, in all three groups of brains we also found S-nitrosylation of ATP-citrate synthase, which catalyzes the reverse TCA cycle, with citrate synthase representing the forward first step of the TCA cycle catalyzing the conversion of oxaloacetate to citrate ((Verschueren et al. 2019). Notably, if this revserse TCA step is inhibited in conjunction with relative blockade of the forward TCA reactions from citrate through succinate, as occurs in HIV and HIV/meth brains to a greater degree than in controls, then flux through the TCA cycle in either direction would be expected to be seriously disrupted. Figure 9 schematically summarizes the relationships of these S-nitrosylated TCA-related enzymes.
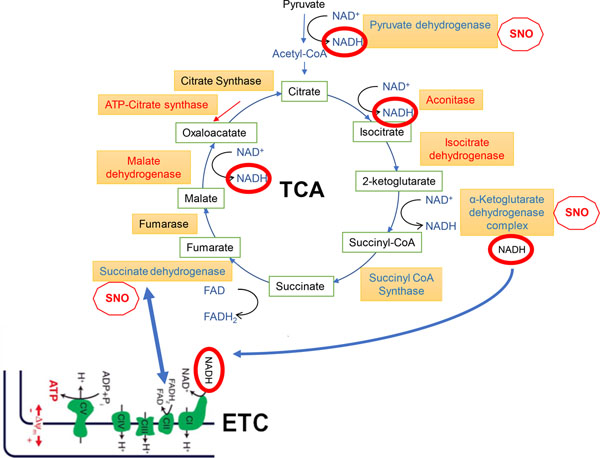
Fig. 9.
Schema of TCA cycle and ETC. Connections between the TCA cycle and ETC are shown by blue arrows. Red ‘stop SNO signs’ indicate significant inhibition of enzymatic activity by S-nitrosylation. Enzymes that are S-nitrosylated and thus inhibited to some degree are shown in red, while enzymes shown in blue have multiple components S-nitrosylated or are more heavily S-nitrosylated in HAND/meth and HAND brains compared to controls, as judged from the number of SNO-peptides recovered by MS (listed in Tables S2–S4); these enzymes are expected to be major sites of metabolic inhibition (termed SNO-STORM metablock) that occur in HAND and HAND/meth brains (see text for further explanation).
Discussion
To gain insight into the pathogenesis and possible treatment of HAND, particularly in the setting of methamphetamine use, we site-specifically identified by MS analysis 231 S-nitrosylated-proteins in HAND (designated as ‘HIV brains’ in the figures) and in HIV/meth brains that were not found in normal/control human brains. Pathway and network analysis revealed systems significantly affected by these aberrant redox reactions on proteins. Chief among these were small molecular transporters and metabolism, represented predominantly by enzymes involved in the TCA cycle.
Considering protein S-nitrosylation effects on transporters, the EAA2 transporter was found to be S-nitrosylated in HIV/meth > HIV >> control human brains (where no SNO-EAA2 was detected); the predominance in HIV/meth and HIV brains was reflected in the number of SNO-peptides from EAA2 recovered by MS (individually listed in Tables S2–S4). This membrane-bound protein, primarily on astrocytes, is the principal transporter that clears the excitatory neurotransmitter glutamate from the extracellular space at synapses in the central nervous system, and decrements in EAA2 activity are thought to contribute to neurotoxicity in HAND or HAND with meth use (Dreyer and Lipton 1995; Kaul et al. 2001; Vazquez-Santiago et al. 2014; Wang et al. 2003). Oxidative stress has also been reported to contribute to neuronal injury in both HAND and meth use (Reiner et al. 2009). Linking these processes, redox control via oxidation of EAAT2 is known to disrupt transporter function (Trotti et al. 1997), making it likely that this S-nitrosylation inhibits activity in the HIV and HIV/meth human brains that we studied, thus contributing to glutamate-related excitotoxicity.
Additionally, concerning protein S-nitrosylation-mediated disruption of metabolic pathways, we found increasing numbers of TCA enzymes to be S-nitrosylated in HIV/meth > HIV >> normal human brains. These findings are consistent with the notion that the oxidizing disease process leads to a concerted, systematic effect, inhibiting the entire metabolic pathway by S-nitrosylating enzymes at multiple sites within the pathway. We propose that such downregulation of the TCA cycle/OXPHOS could lead to metabolic compromise and contribute to neurotoxicity in HIV brains and to an even greater extent in HIV/meth brains. Confirmation of this concept comes from increasing evidence that metabolic disruption contributes to the pathogenesis of both HAND and meth-related neurological damage (Chang et al. 2007; Chang et al. 2005; Cotto et al. 2019; Sanchez et al. 2016).
By way of background, mature neurons (unlike neuronal precursor cells) are known to downregulate their use of glycolysis, and appear to rely on mitochondria via the TCA cycle/ETC for their basal activity energy production (Belanger et al. 2011; Demetrius et al. 2014; Machler et al. 2016; Zheng et al. 2016). That said, during intense stimulation, recent evidence has shown that neurons can use glucose for short bursts (Diaz-Garcia et al. 2017). However, long-term activity, learning and memory are all dependent on the TCA cycle ATP production to maintain neuronal health (Rangaraju et al. 2014; Rangaraju et al. 2019a; Rangaraju et al. 2019b). Thus, if the TCA cycle were shut down by inhibiting a series of its enzymes, e.g., IDH, AC, αKGDH, SUCLG, and SDH all in a row, neurons would be expected to become very vulnerable to synaptic loss and eventually die due to lack of ATP/energy proudction. In fact, this is exactly what we find, and the degree of S-nitrosylation occurs to a larger extent in HIV/meth brains than in HIV brains, and to a much lesser extent as part of physiological regulation of the TCA cycle in normal human and rodent brain (Fig. 9) (Doulias et al. 2013a).
Notably, in our S-nitrosoproteome dataset of human brain, we found that the first five enzymes in the TCA cycle are S-nitrosylated in HIV/meth and HIV human brains, while only the first two (IDH and AC) are regulated by S-nitrosylation in normal brain. Critically, as reviewed by Tretter and Adam-Vizzi (Tretter and Adam-Vizi 2005), while AC and possibly IDH are more suceptible than αKGDH to oxidative/nitrosative stress, if αKGDH remains functional, then NADH generation and thus energy production is maintained by the TCA cycle (Fig. 9). What we have found to date is that in addition to oxidative stress, nitrosative stress is critical and inhibits all of these enzymes in functional activity assays – most importantly αKGDH is inhibited approximately 50% in the face of nitrosative stress, as occurs in human HIV/meth and HIV brains. Most importantly, these findings using this SNO-proteomics approach, have very substantial therapeutic implications. The data indicate that the HIV and HIV+meth groups impact the carboxylic acid metabolic processes in a broader biological function than just TCA, with pyruvate metabolic processes and respiration being affected as well. the impact on respiration and ATP production is likely to impact neurons in HIV and HIV +meth brains. Energy repletion, for example by rescuing TCA cycle intermediates, could potentially preserve neurons in the setting of HIV or HIV/meth insult. If this is the case, then new metabolic approaches could be identified for the treatment of HAND and to a greater extent HAND in the setting of meth use, where even more TCA enzymes are inhbited than in HAND alone.
Supplementary Material
Table S1. List of Human Brains Analyzed for S-Nitrosoproteome by Mass Spectrometry.
Table S3. S-Nitrosoproteome found in HIV+ Human Brains with Evidence of HAND.
Table S2. S-Nitrosoproteome found in Non-Diseased Control Human Brains.
Table S4. S-Nitrosoproteome found in HIV+ Human Brains with Evidence of HAND and Methamphetamine Use.
Table S5. S-Nitrosylation of the Various Pyruvate Dehyrogenase (PDH) Components.
Acknowledgements
We thank Eliezer Masliah (UC San Diego/NIA) for providing human brain tissues. This work was supported in part by NIH grants R01 NS086890, DP1 DA041722, RF1 AG057409, and R01 AG056259 (to S.A.L.), and R01 AG061845 (to T.N.) The NeuroAIDS tissue bank at UCSD was in part supported by the NIH/NIMH U24 MH100928.
Footnotes
Conflict of Interest The authors declare that they have no conflicts of interest.
References
- Belanger M, Allaman I, Magistretti PJ (2011) Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14: 724–38. [DOI] [PubMed] [Google Scholar]
- Cadet JL, Krasnova IN (2007) Interactions of HIV and methamphetamine: cellular and molecular mechanisms of toxicity potentiation. Neurotox Res 12: 181–204. [DOI] [PubMed] [Google Scholar]
- Carothers DJ, Pons G, Patel MS (1989) Dihydrolipoamide dehydrogenase: functional similarities and divergent evolution of the pyridine nucleotide-disulfide oxidoreductases. Arch Biochem Biophys 268: 409–25. [DOI] [PubMed] [Google Scholar]
- Chang L, Alicata D, Ernst T, Volkow N (2007) Structural and metabolic brain changes in the striatum associated with methamphetamine abuse. Addiction 102 Suppl 1: 16–32. [DOI] [PubMed] [Google Scholar]
- Chang L, Ernst T, Speck O, Grob CS (2005) Additive effects of HIV and chronic methamphetamine use on brain metabolite abnormalities. Am J Psychiatry 162: 361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RA, Murphy MP (2010) Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J 430: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto B, Natarajanseenivasan K, Langford D (2019) HIV-1 infection alters energy metabolism in the brain: Contributions to HIV-associated neurocognitive disorders. Prog Neurobiol 181: 101616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Tannenbaum SR (2004) Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys 423: 12–22. [DOI] [PubMed] [Google Scholar]
- Demetrius LA, Magistretti PJ, Pellerin L (2014) Alzheimer’s disease: the amyloid hypothesis and the Inverse Warburg effect. Front Physiol 5: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Garcia CM, Mongeon R, Lahmann C, Koveal D, Zucker H, Yellen G (2017) Neuronal Stimulation Triggers Neuronal Glycolysis and Not Lactate Uptake. Cell Metab 26: 361–374 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H (2010) Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci USA 107: 16958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias PT, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H (2013a) Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal 6: rs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulias PT, Tenopoulou M, Raju K, Spruce LA, Seeholzer SH, Ischiropoulos H (2013b) Site specific identification of endogenous S-nitrosocysteine proteomes. J Proteomics 92: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer EB, Lipton SA (1995) The coat protein gp120 of HIV-1 inhibits astrocyte uptake of excitatory amino acids via macrophage arachidonic acid. Eur J Neurosci 7: 2502–7. [DOI] [PubMed] [Google Scholar]
- Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol 27: 557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster MW, Stamler JS (2004) New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem 279: 25891–7. [DOI] [PubMed] [Google Scholar]
- Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF (2010) Cause and consequence: mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim Biophys Acta 1802: 122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–66. [DOI] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA (2001) Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410: 988–94. [DOI] [PubMed] [Google Scholar]
- Keshive M, Singh S, Wishnok JS, Tannenbaum SR, Deen WM (1996) Kinetics of S-nitrosation of thiols in nitric oxide solutions. Chem Res Toxicol 9: 988–93. [DOI] [PubMed] [Google Scholar]
- Keszler A, Zhang Y, Hogg N (2010) Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: How are S-nitrosothiols formed? Free Radic Biol Med 48: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA (1992) Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron 8: 1087–99. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS (1993) A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature 364: 626–32. [DOI] [PubMed] [Google Scholar]
- Machler P, Wyss MT, Elsayed M, Stobart J, Gutierrez R, von Faber-Castell A, Kaelin V, Zuend M, San Martin A, Romero-Gomez I, Baeza-Lehnert F, Lengacher S, Schneider BL, Aebischer P, Magistretti PJ, Barros LF, Weber B (2016) In Vivo Evidence for a Lactate Gradient from Astrocytes to Neurons. Cell Metab 23: 94–102. [DOI] [PubMed] [Google Scholar]
- Madej E, Folkes LK, Wardman P, Czapski G, Goldstein S (2008) Thiyl radicals react with nitric oxide to form S-nitrosothiols with rate constants near the diffusion-controlled limit. Free Radic Biol Med 44: 2013–8. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Cadenas S, Lamas S (2011) Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic Biol Med 51: 17–29. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marletta MA (2005) Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol 1: 154–8. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Lipton SA (2017) ‘SNO’-Storms Compromise Protein Activity and Mitochondrial Metabolism in Neurodegenerative Disorders. Trends Endocrinol Metab 28: 879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Prikhodko OA, Pirie E, Nagar S, Akhtar MW, Oh CK, McKercher SR, Ambasudhan R, Okamoto S, Lipton SA (2015) Aberrant protein S-nitrosylation contributes to the pathophysiology of neurodegenerative diseases. Neurobiol Dis 84: 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Vettakkorumakankav NN, Liu TC (1995) Dihydrolipoamide dehydrogenase: activity assays. Methods Enzymol 252: 186–95. [DOI] [PubMed] [Google Scholar]
- Raju K, Doulias PT, Evans P, Krizman EN, Jackson JG, Horyn O, Daikhin Y, Nissim I, Yudkoff M, Nissim I, Sharp KA, Robinson MB, Ischiropoulos H (2015) Regulation of brain glutamate metabolism by nitric oxide and S-nitrosylation. Sci Signal 8: ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Calloway N, Ryan TA (2014) Activity-driven local ATP synthesis is required for synaptic function. Cell 156: 825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Lauterbach M, Schuman EM (2019a) Spatially Stable Mitochondrial Compartments Fuel Local Translation during Plasticity. Cell 176: 73–84 e15. [DOI] [PubMed] [Google Scholar]
- Rangaraju V, Lewis TL Jr., Hirabayashi Y, Bergami M, Motori E, Cartoni R, Kwon SK, Courchet J (2019b) Pleiotropic Mitochondria: The Influence of Mitochondria on Neuronal Development and Disease. J Neurosci 39: 8200–8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner BC, Keblesh JP, Xiong H (2009) Methamphetamine abuse, HIV infection, and neurotoxicity. Int J Physiol Pathophysiol Pharmacol 1: 162–179. [PMC free article] [PubMed] [Google Scholar]
- Sanchez AB, Varano GP, de Rozieres CM, Maung R, Catalan IC, Dowling CC, Sejbuk NE, Hoefer MM, Kaul M (2016) Antiretrovirals, Methamphetamine, and HIV-1 Envelope Protein gp120 Compromise Neuronal Energy Homeostasis in Association with Various Degrees of Synaptic and Neuritic Damage. Antimicrob Agents Chemother 60: 168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheving R, Wittig I, Heide H, Albuquerque B, Steger M, Brandt U, Tegeder I (2012) Protein S-nitrosylation and denitrosylation in the mouse spinal cord upon injury of the sciatic nerve. J Proteomics 75: 3987–4004. [DOI] [PubMed] [Google Scholar]
- Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai LH, Tannenbaum SR (2016) S-nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration. Proc Natl Acad Sci USA 113: 4152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Stamler JS (2011) The SNO-proteome: causation and classifications. Curr Opin Chem Biol 15: 129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Marletta MA (2012) Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr Opin Chem Biol 16: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Carney JM, Starke-Reed PE, Oliver CN, Stadtman ER, Floyd RA, Markesbery WR (1991) Excess brain protein oxidation and enzyme dysfunction in normal aging and in Alzheimer disease. Proc Natl Acad Sci U S A 88: 10540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontornniyomkij V, Kesby JP, Morgan EE, Bischoff-Grethe A, Minassian A, Brown GG, Grant I, Translational Methamphetamine ARCG (2016) Effects of HIV and Methamphetamine on Brain and Behavior: Evidence from Human Studies and Animal Models. J Neuroimmune Pharmacol 11: 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E (2007) Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res 101: 1155–63. [DOI] [PubMed] [Google Scholar]
- Talantova M, Sanz-Blasco S, Zhang X, Xia P, Akhtar MW, Okamoto S, Dziewczapolski G, Nakamura T, Cao G, Pratt AE, Kang YJ, Tu S, Molokanova E, McKercher SR, Hires SA, Sason H, Stouffer DG, Buczynski MW, Solomon JP, Michael S, Powers ET, Kelly JW, Roberts A, Tong G, Fang-Newmeyer T, Parker J, Holland EA, Zhang D, Nakanishi N, Chen HS, Wolosker H, Wang Y, Parsons LH, Ambasudhan R, Masliah E, Heinemann SF, Pina-Crespo JC, Lipton SA (2013) Aß induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proc Natl Acad Sci USA 110: E2518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter L, Adam-Vizi V (2005) Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos Trans R Soc Lond B Biol Sci 360: 2335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotti D, Rizzini BL, Rossi D, Haugeto O, Racagni G, Danbolt NC, Volterra A (1997) Neuronal and glial glutamate transporters possess an SH-based redox regulatory mechanism. Eur J Neurosci 9: 1236–43. [DOI] [PubMed] [Google Scholar]
- Vazquez-Santiago FJ, Noel RJ Jr., Porter JT, Rivera-Amill V (2014) Glutamate metabolism and HIV-associated neurocognitive disorders. J Neurovirol 20: 315–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, Van Beeumen J, Svergun D, Gutsche I, Savvides SN, Verstraete K (2019) Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature 568: 571–575. [DOI] [PubMed] [Google Scholar]
- Wang Z, Pekarskaya O, Bencheikh M, Chao W, Gelbard HA, Ghorpade A, Rothstein JD, Volsky DJ (2003) Reduced expression of glutamate transporter EAAT2 and impaired glutamate transport in human primary astrocytes exposed to HIV-1 or gp120. Virology 312: 60–73. [DOI] [PubMed] [Google Scholar]
- Yan LJ, Liu L, Forster MJ (2012) Reversible inactivation of dihydrolipoamide dehydrogenase by Angeli’s salt. Sheng Wu Wu Li Hsueh Bao 28: 341–350. [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Ma L, Hamm M, Gage FH, Hunter T (2016) Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5: e13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of Human Brains Analyzed for S-Nitrosoproteome by Mass Spectrometry.
Table S3. S-Nitrosoproteome found in HIV+ Human Brains with Evidence of HAND.
Table S2. S-Nitrosoproteome found in Non-Diseased Control Human Brains.
Table S4. S-Nitrosoproteome found in HIV+ Human Brains with Evidence of HAND and Methamphetamine Use.
Table S5. S-Nitrosylation of the Various Pyruvate Dehyrogenase (PDH) Components.