ABSTRACT
Streptococcus intermedius, an oral commensal bacterium, is found at various sites, including subgingival dental plaque, purulent infections, and cystic fibrosis lungs. Oral streptococci utilize proteins on their surface to adhere to tissues and/or surfaces localizing the bacteria, which subsequently leads to the development of biofilms, colonization, and infection. Among the 19 genomically annotated cell wall-attached surface proteins on S. intermedius, Pas is an adhesin that belongs to the antigen I/II (AgI/II) family. Here, we have structurally and functionally characterized Pas, particularly focusing on its microbial-host as well as microbial-microbial interactions. The crystal structures of VPas and C123Pas show high similarity with AgI/II of Streptococcus mutans. VPas hosts a conserved metal binding site, and likewise, the C123Pas structure retains its conserved metal binding sites and isopeptide bonds within its three DEv-IgG domains. Pas interacts with nanomolar affinity to lung alveolar glycoprotein 340 (Gp340), its scavenger receptor cysteine-rich domains (SRCRs), and with fibrinogen. Both Candida albicans and Pseudomonas aeruginosa, the opportunistic pathogens that cohabitate with S. intermedius in the lungs of CFTR patients were studied in dual-species biofilm studies. The Pas-deficient mutant (Δpas) displayed significant reduction in dual-biofilm formation with C. albicans. In similar studies with P. aeruginosa, Pas did not mediate the biofilm formation with either the acute isolate (PAO1) or the chronic isolate (FRD1). However, the sortase A-deficient mutant (ΔsrtA) displayed reduced biofilm formation with both C. albicans and P. aeruginosa FRD1. Taken together, our findings highlight the role of Pas in both microbial-host and interkingdom interactions and expose its potential role in disease outcomes.
IMPORTANCEStreptococcus intermedius, an oral commensal bacterium, has been clinically observed in subgingival dental plaque, purulent infections, and cystic fibrosis lungs. In this study, we have (i) determined the crystal structure of the V and C regions of Pas; (ii) shown that its surface protein Pas adheres to fibrinogen, which could potentially ferry the microbe through the bloodstream from the oral cavity; (iii) characterized Pas’s high-affinity adherence to lung alveolar protein Gp340 that could fixate the microbe on lung epithelial cells; and (iv) most importantly, shown that these surface proteins on the oral commensal S. intermedius enhance biofilms of known pathogens Candida albicans and Pseudomonas aeruginosa.
KEYWORDS: Candida albicans, Streptococcus intermedius, protein structure-function, surface antigens
INTRODUCTION
Members of the anginosus/milleri group of streptococci, which includes Streptococcus intermedius, S. constellatus, and S. anginosus, are commensal residents of the oral cavity. However, in recent years, they have been observed in various sites of purulent infection, including the brain, liver, and spleen (1–3). Apart from the oral cavity, S. intermedius specifically has been found in gastrointestinal and urogenital tracts (4) and infective endocarditis (2) and has been detected in the airways of cystic fibrosis (CF) patients with chronic lung disease (5–7).
Oral streptococci have evolved multiple mechanisms to colonize the oral cavity (8–14). The first contact with the host seems to occur through proteins on the bacterial surface, which aid the microbe in adhering/attaching to surfaces as well as tissues that they encounter. These proteins on Gram-positive bacteria are anchored onto the peptidoglycan layer through the signature “LPXTG” motif that is present on their C-terminal hydrophobic tail (15). In oral streptococci, the sortase A (srtA) protein, in a penicillin-like reaction, covalently attaches these surface proteins to the peptidoglycan layer (16–18). In general, these surface proteins have a ligand binding region that is projected away from the cell wall through a set of repeat sequences/domains that either form elongated domain architectures and/or secondary structures that are fibrillar (19).
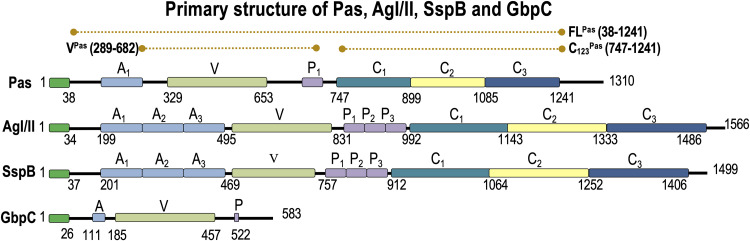
Among the oral streptococcal surface proteins, the antigen I/II (AgI/II, also known as SpaP and P1) has been well characterized (20). AgI/II has an apical lectin-like V domain that is projected away from the cell surface through an elongated fibrillar architecture, which is fused together from two distinct regions that are well separated in the primary structure (Fig. 1) (21). These are the alanine-rich A region and the proline-rich P region, which adopt an alpha-helix (right-handed) and a PP-II-like helix (left-handed), respectively. These two helices intertwine in a novel manner, giving rise to elongated fibrils (21). At the C-terminal region, there are three domains that adopt the DEv-IgG fold, which is universally observed in Gram-positive bacteria (22). These individual DEv-IgG domains (C domains) contain a conserved Ca2+ binding site and are stabilized by an isopeptide bond displaying different versions to adhere to ligands (23).
FIG 1.
Primary structures of Pas, AgI/II, SspB, and GbpC. Each of these proteins has very similar domain arrangements except for GbpC, which only has the V region. The extents of the Pas constructs FLPas, VPas, and C123Pas used in this study are shown in dashed lines on the top.
AgI/II’s adherence to the tooth surface is mediated through the secreted glycoprotein 340 (Gp340), also known as salivary agglutinin (24, 25). Gp340 is an innate immune receptor, and apart from saliva, Gp340 exists as a normal secreted component in tears and breast milk. It also exists on the surfaces of vaginal, gastrointestinal, lung alveolar, and pancreatic epithelial cells (26–32). The primary structure of Gp340 (Fig. S1 in the supplemental material) consists of 14 scavenger receptor cysteine-rich domains (SRCR), 2 C1s/Uegf binding domains (CUB), and 1 zona pellucida domain (ZP) (33). Each SRCR domain contains about 110 amino acids, and there are 13 of these in tandem that are linked through 30- to 32-amino-acid SID domains, which stands for SRCR interspersed domains that host the O-glycosylation sites. Previously, we had described the adherence of Streptococcus mutans AgI/II and its homolog on Streptococcus gordonii SspB to recombinant SRCR1 and SRCR123 domains and reported that a single SRCR domain contains distinct sites with which the V and C domains interact with high affinity (34, 35). Apart from its adherence to Gp340, we discovered that S. mutans AgI/II promotes interkingdom interactions with the opportunistic fungus Candida albicans (36). In the same study, we reported that AgI/II promoted acid production, one of the known factors that result in the erosion of tooth enamel (36).
In addition to AgI/II, we characterized the crystal structure of GbpC (37), the glucan binding protein of S. mutans. This structure resembled the apical V domain of AgI/II, and based on this observation, we showed that it adheres to Gp340 and its SRCR domains in addition to its adherence to dextran (37). While dextran did not impede the binding affinity to SRCR in competition experiments, it certainly produced steric hindrance signals, indicating that the binding sites for dextran and SRCR are in close proximity. These examinations revealed a key loop region on the structure of GbpC that drives the adhesion to dextran, which is missing in both AgI/II and SspB (37), thus structurally explaining the exclusive affinity of GbpC to dextran.
In this study, we characterize the role of Pas (AgI/II-like protein) on S. intermedius (38). The pas gene is much shorter, and while it does retain the apical V- and C-terminal domains, it contains only a single A-P repeat compared to other AgI/II-like proteins, which often carry 3 to 5 repeats (Fig. 1). We present the crystal structures of the V (VPas) and the C domains (C123Pas) of Pas and their adherence characteristics to SRCR domains of Gp340. Their binding affinities are comparable to AgI/II and SspB and are in line with the structural similarity observed. Pas also interacts with nanomolar affinity to fibrinogen. In monospecies biofilm studies, Pas exerts significant influence, and in dual-species biofilm studies with C. albicans (CA), the Pas mutant demonstrates reduced biofilm formation. In dual-biofilm studies with P. aeruginosa, the S. intermedius Pas mutant did not affect biofilm formation with acute P. aeruginosa (PAO1) or the chronic P. aeruginosa (FRD1). Interestingly, the S. intermedius sortase A mutant significantly inhibited the dual biofilm formation with FRD1, suggesting cell wall-anchored proteins other than Pas are involved in the dual-species biofilm formation with P. aeruginosa.
RESULTS
The VPas structure.
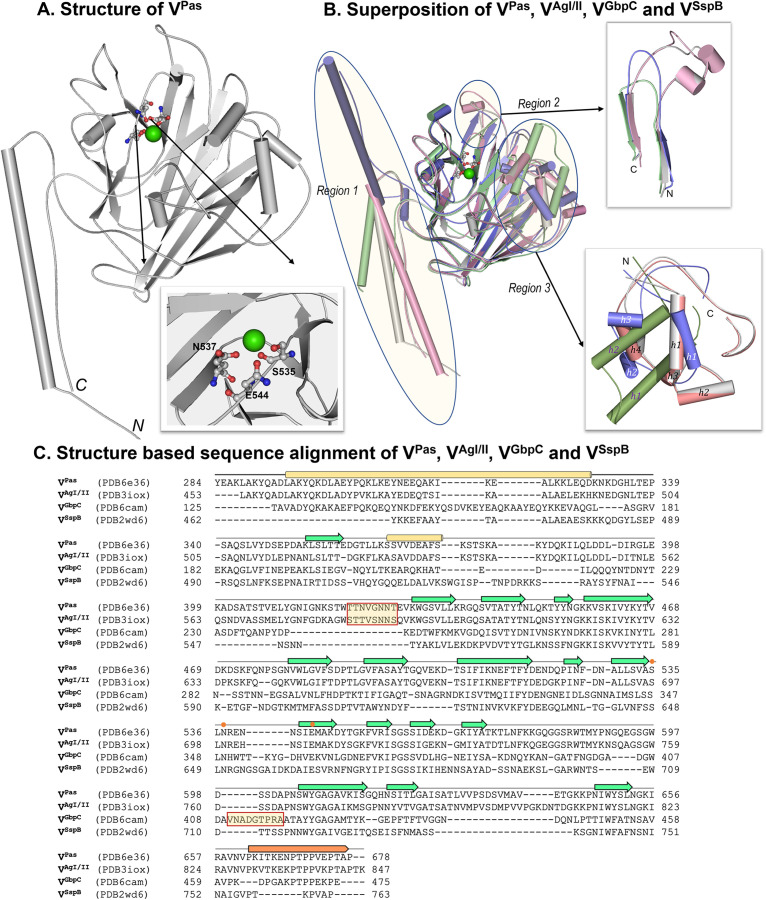
The cloning primers and primary structure for VPas and the extents of the cloning are shown in Table 1 and Fig. 1. The crystal structure of VPas adopts a similar fold as the V domain of AgI/II (VAgI/II) and superposes with a root mean square deviation (RMSD) of 0.55 Å for 2,285 (of 2,846) aligned atoms (1.57 Å for all 388 C-α atom pairs), thus indicating a high similarity in their secondary structures. X-ray diffraction data collection and refinement statistics for VPas are listed in Table 2. Like the other V domains of VAgI/II, VGbpC, and VSspB, VPas has a conserved metal binding site, which is coordinated by Ser535, Asn537, and Glu544. VPas was crystallized using higher concentrations of MgCl2 (250 mM), and therefore, this structure contains an Mg2+ ion instead of Ca2+ present in other structures. The coordination geometry in the structure of VPas with shorter Mg to O distances (2.1 to 2.2 Å) compared to Ca to O distances (2.4 to 2.5 Å) is notable (Table S1). While the putative binding pockets in VAgI/II, VGbpC, VSspB, and VPas are highly homologous and well conserved, three distinct regions are notable where we observe structural differences. Region 1 (Fig. 2) forms a fibrillar architecture between the alpha-helical alanine-rich A region at the N terminus and the PPII-like helix with the proline-rich P region in VPas (residues 295 to 329; 662 to 677), VAgI/II (residues 460 to 494; 829 to 844), VGbpC (residues 125 to 175; 460 to 475), and VSspB (residues 459 to 493; 752 to 764). The largest difference in region 1 is seen in VGbpC, as it projects this segment in an opposite direction from VPas, VAgI/II, and VSspB (Fig. 2), while VPas and VAgI/II show high structural similarity, although there appears to be an inherent flexibility in projecting this region. Region 2, consisting of a strand-loop-strand secondary structural motif (VPas, residues 618 to 653; VAgI/II, residues 780 to 820; VSspB, residues 730 to 748; and VGbpC, residues 432 to 455) (Fig. 2), is present in all four V domains, where VPas and VAgI/II distinctly include two short helices, whereas the same loop in VSspB and VGbpC is much shorter (∼10 residues). The third region (region 3) (Fig. 2) in VPas (residues 362 to 429) contains a helix-turn-helix (HTH) motif and is similar to VAgI/II (residues 527 to 593) with 65% sequence identity; it superposes with an RMSD of 0.7 Å, but it is very different to the much shorter (∼25 residues less) stretches in VGbpC (residues 203 to 244) and VSspB (residues 730 to 748). Region 3 of VPas reveals substantial structural homology to HTH-type transcriptional regulators such as RutR (RMSD, ∼3 Å for an alignment length of 48 residues that encompass this motif), albeit low sequence identity (6 to 8%). Finally, VAgI/II (residues 584 to 590) and VPas (residues 420 to 427) contain a loop region that hovers over the metal binding site (Fig. 2) that is involved in biofilm formation and dextran-dependent aggregation (23). This loop is absent in both VSspB and VGbpC; however, VGbpC uniquely displays another loop region (residues 410 to 418) that resides on the opposite side atop the metal binding site compared to VAgI/II and VPas (23). In VGbpC, the metal site hosts the dextran (glucan) binding site, and this flexible loop may open and close to accommodate the glucose molecules, thereby locking them in place (23).
TABLE 1.
Protein constructs and primers used in this study
| Construct or primer | Residue (range) | Sequence(s) (5′–3′)b | MW (Da) | |
|---|---|---|---|---|
| Protein constructs | ||||
| FLa | 1220 (38–1241) | Forward, GCGACCATGGGAAGAAACTACAAAATCA; reverse, AATCCTCGAGGCCCTGAAAATACAGGTTTTCAGACGCATAGGCAACTTT | 133,767 | |
| Vhel | 417 (289–682) | Forward, GCTCCATGGGCTGATTACGAAGCAAAA; reverse, | 46,235 | |
| GCGTCTCGAGGCCCTGAAAATACAGGTTTTCAGTAGGAGCTTGTGG | ||||
| C123 | 512 (747–1241) | Forward, GCTACCATGGCATTTTCGTTACTATAAACTA; reverse, AATCCTCGAGGCCCTGAAAATACAGGTTTTCAAGACGCATAGGCAACTTT | 56,906 | |
| Primers used in the construction of Pas mutant in S. intermedius | ||||
| Pas flanking up 5′ | AACGCATTTGCTGACTACACTGAAGT | |||
| Pas flanking down 3′ | GCACGATTATTGGCTACAGCAGTTACATA | |||
| Pas up 3′ overlap | GAGTGTTATTGTTGCTCGGGTAAATTGGTCGCTGGATTTCCAGT | |||
| Pas down 5′ overlap | GGTATACTACTGACAGCTTCGCTATCATCAGCACCTGCTACCCTT | |||
| Pas_Com-F | GCTCGTCGACATGAAGAAAAGAAAAGAAGTTTTTGGTTTTC | |||
| Pas_Com-R | GCTCGGTACCCTATTTTTCGTTACGTTTTAATTGACCTA | |||
| Primers used in the construction of sortase A mutant in S. intermedius | ||||
| Sortase A flanking up 5′ | CTGTTCTAGATGCTGCCGA | |||
| Sortase A flanking down 3′ | ATTAGTCTCTACCAGCACACACG | |||
| Sortase A up 3′ overlap | GAGTGTTATTGTTGCTCGGGCTAACTTGATACCTATTGGTATGC | |||
| Sortase A down 5′ overlap | GGTATACTACTGACAGCTTCAGACGCAGCAGCAACATC |
FL, full length.
Boldfacing indicates restriction enzyme sites.
TABLE 2.
Crystallographic data and refinement statistics
| Characteristica | Data for: |
|
|---|---|---|
| VPas (PDB ID 6E36) | C123Pas (PDB ID 6E3F) | |
| Space group | P21 | I4122 |
| a (Å) | 71.04 | 99.59 |
| b (Å) | 91.31 | 99.59 |
| c (Å) | 71.89 | 514.88 |
| α (°) | 90.0 | 90.0 |
| β (°) | 116.8 | 90.0 |
| γ (°) | 90.0 | 90.0 |
| Resolution (Å) | 1.7 | 2.7 |
| Rmerge (%) | 0.074 (0.927) | 0.124 |
| Rpim (%) | 0.051 (0.648) | 0.041 (0.443) |
| CC1/2 (%) | 0.993 (0.583) | 0.997 (0.737) |
| I/σI | 8.5 (1.6) | 25.4 (2.1) |
| Completeness (%) | 98.9 (99.8) | 100 (100) |
| Redundancy | 3.0 (3.0) | 14.4 (14.8) |
| Refinement | ||
| Resolution (Å) | 1.7 | 2.7 |
| No. of reflections | 88,935 (6,629) | 36,440 (2,642) |
| Rwork/Rfree | 21.8/24.7 | 21.5/24.4 |
| No. of atoms | ||
| Protein | 6,135 | 3,890 |
| Ion | 2 (Mg2+) | 2 (Ca2+) |
| Water | 411 | 73 |
| B-factors (Å2) | ||
| Wilson B | 33.8 | 55.1 |
| Protein | 39.8 | 75.2 |
| Ions | 30.9 | 93.6 |
| Water | 43.4 | 47.7 |
| RMD deviation | ||
| Bond lengths (Å) | 0.012 | 0.013 |
| Bond angles (°) | 1.52 | 1.75 |
| Ramachandran statistics | ||
| Favored (%) | 97.7 | 94.8 |
| Allowed (%) | 2.3 | 4.8 |
| Outliers (%) | 0 | 0.4 |
| MolProbity score | 0.84 | 1.77 |
| Clash score | 0.90 | 4.16 |
CC1/2, half-set correlation coefficient; I/σI, intensity-to-noise ratio; Rwork, refinement R-factor of work set; Rfree, refinement R-factor of test set; RMS, root mean square.
FIG 2.
V domain. (A) The crystal structure of VPas has a conserved metal binding site coordinated by S535, N537, and E544. This site is occupied by a Mg2+, as opposed to Ca2+ ion observed in crystal structures of VAgI/II, VGpbC, and VSspB. (B) The superposition of VPas (gray), VAgI/II (pink), VGpbC (blue), and VSspB (green) is shown, where three regions (1, 2, and 3) show maximal variations. (C) Structure-based sequence alignment of VPas, VAgI/II, VGpbC, and VSspB where the metal binding site is indicated by orange dots, and the boxed regions are the loop regions that hover over the putative binding site on VPas, VAgI/II and VGpbC.
The C123Pas structure.
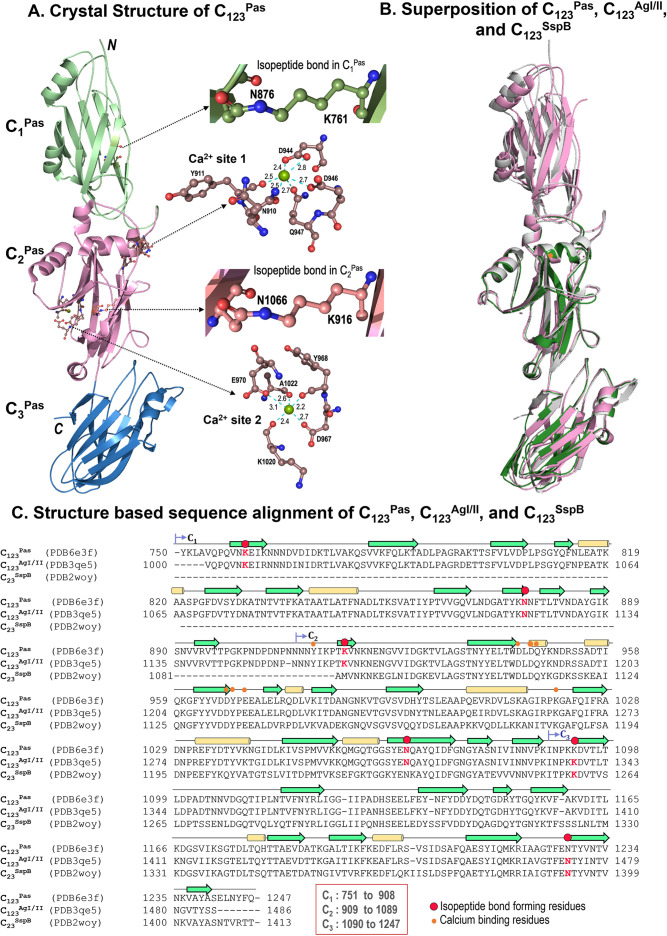
The cloning primers and the primary structure for C123Pas are shown in Table 1 and Fig. 1. X-ray diffraction data collection and refinement statistics for C123Pas are listed in Table 2. C123Pas and C123AgI/II present 92% identity and 96% homology in their primary structure. Consequently, structural superposition of C123Pas with C123AgI/II shows an RMSD of 0.87 Å for 395 (pruned) atom pairs, with an RMSD of 1.56 Å for all Cα atoms (487) and 1.81 Å for all matching atoms (3,744), thus indicating a high degree of similarity between the two structures. The overall superposition displays slightly altered orientations of the C1, C2, and C3 domains, where C1 has the most altered orientation (Fig. 3). However, superposing the individual domains of CPas and CAgI/II shows a much better structural similarity (∼0.4 Å RMSD for ∼80% of all matching atoms) and agrees well with the observed high sequence identity/homology. Comparison of the structures of C23Pas and C23SspB displays an RMSD of 0.77 Å for 305 (pruned) atom pairs and 1.81 Å for all matching 329 atom pairs; whereas C23AgI/II and C23SspB show an RMSD of 0.78 Å for 309 (pruned) atom pairs and 0.97 Å for all matching 323 atom pairs; all these numbers indicate a high degree of similarity between C123Pas, C123AgI/II, and C23SspB. While the superpositions do not identify too many differences, the surface charge plots clearly reveal that the makeup of each of these proteins differs due to sequence differences as well as structural differences that include a different orientation of individual domains (C1, C2, and C3) with respect to each other, which is especially relevant in the case of the highly homologous C123AgI/II and C123Pas (see Fig. S2 in the supplemental material).
FIG 3.
C domain. (A) Crystal structure of C123Pas with each domain highlighted. The insets show the two isopeptide bonds in C1Pas and C2Pas and the two calcium binding sites in C123Pas. (B) Superposition of C123Pas (gray), C123AgI/II (pink), and C123SspB (green) shows that they are very similar; however, C1Pas shows large deviations. (C) Structure-based protein sequence alignment of C123Pas, C123AgI/II, and C123SspB is shown.
Isopeptide bonds stabilize C123Pas.
The DEv-IgG fold has an isopeptide bond that stabilizes this domain. In the crystal structure of C123Pas (PDB ID 6E3F), only the C1 (K761-N876) and C2 (K916-N1066) domains display the isopeptide bond, whereas C3 does not (Fig. 3). In the C3 domain, K1093 makes instead a hydrogen bond with the backbone carbonyl oxygen of N1228 (2.7 Å) and the amide oxygen atom of N1243 (3.6 Å), while the side chain of N1228 points away for salt bridge interactions with D1142 (2.6 and 2.8 Å, respectively). This is similar to SspB (PDB ID 2WOY), where the C2 domain does not show the isopeptide bond, while it resides in the C3 domain (K1259-N1393). On the other hand, C123AgI/II (PDB IDs 3QE5) displays all three isopeptide bonds in the C1 (K1006-N1121), the C2 (K1161-N1311), and the C3 (K1338-N1473) domains.
The calcium binding sites on C123Pas.
Two calcium ions are observed in the structure of C123Pas, with one between the C1 and C2 domains and one in the C2 domain (Fig. 3; Table S1). The first calcium ion adopts bipyramidal geometry and is coordinated by side chain oxygens of N910, D944, D946, and Q947 and the main chain oxygen of Y911. The second calcium ion is coordinated by the side chain oxygens of D967 and E970 and through the main chain oxygen atoms of Y968, K1020, and A1022. These calcium binding sites are very similar to those observed in AgI/II and SspB (Table S1; Fig. 3).
The sugar binding sites on C123Pas.
Although no glucose/sugars are observed in the structure of C123Pas, superposition with C123AgI/II highlights the conservation of residues in C123Pas that interact with the glucose molecules in C123AgI/II (PDB IDs EQE5) (Table S2).
Models of A123VP123C123AgI/II and A1VP1C123Pas.
As previously reported, the AgI/II model for residues 202 to 1486 (V domain, 3 A-P repeats, and C domains) has an extended structure whose length is approximately 65 nm or 650 Å from top to bottom (Fig. S3). Modeling of Pas for residues 197 to 1241 (V domain, 1 A-P repeat, and C domains) results in a length of approximately 35 nm or 350 Å, where each A-P repeat extends to ∼15 nm (Fig. S3). The presence of a single A-P repeat in Pas compared to three A-P repeats in AgI/II thus has reduced the elongation of the structure by nearly half the size of AgI/II.
Adherence of FLPas, VPas, and C123Pas to SRCR.
The affinity coefficients of Pas constructs were determined using surface plasmon resonance studies (BIAcore 2000). Each construct of Pas (FLPas, VPas, and C123Pas) interacted with nanomolar affinity to SRCR domains of Gp340 (Table 3; Fig. S4).
TABLE 3.
Surface plasmon resonance studies on the adherence of Pas with SRCR /Gp340 and fibrinogen
| Analyte | Ligand | ka (1/Ms)a | kd (1/s) | KA (1/M) | KD (M) | Chi-square |
|---|---|---|---|---|---|---|
| VPas | SRCR1 | 2.51 × 104 | 4.27 × 10−4 | 5.88 × 107 | 1.70 × 10−8 | 1.54 |
| SRCR123 | 9.85 × 104 | 1.23 × 10−3 | 8.01 × 107 | 1.25 × 10−8 | 2.08 | |
| Gp340 | 2.70 × 104 | 4.94 × 10−4 | 5.47 × 107 | 1.83 × 10−8 | 0.81 | |
| C123Pas | SRCR1 | 7.46 × 105 | 1.61 × 10−3 | 4.64 × 108 | 2.16 × 10−9 | 4.05 |
| SRCR123 | 2.45 × 106 | 4.25 × 10−3 | 5.77 × 108 | 1.73 × 10−9 | 3.65 | |
| Gp340 | 4.98 × 104 | 1.18 × 10−3 | 4.23 × 107 | 2.36 × 10−8 | 3.75 | |
| FLPas | SRCR1 | 1.88 × 104 | 2.45 × 10−4 | 7.69 × 107 | 1.30 × 10−8 | 18 |
| SRCR123 | 3.49 × 103 | 1.38 × 10−4 | 2.53 × 107 | 3.96 × 10−8 | 19.7 | |
| Gp340 | 3.09 × 105 | 1.22 × 10−3 | 2.53 × 108 | 3.95 × 10−9 | 5.37 | |
| Fibrinogen | 1.15 × 105 | 3.18 × 10−4 | 3.61 × 108 | 2.77 × 10−9 | 14.3 |
ka, association rate constant; kd, dissociation constant; KA, equilibrium binding constant; KD, equilibrium dissociation constant.
Adherence of FLPas to fibrinogen.
The affinity coefficients for the full-length Pas construct to fibrinogen were determined to have nanomolar adherence using surface plasmon resonance (Table 3; Fig. S5).
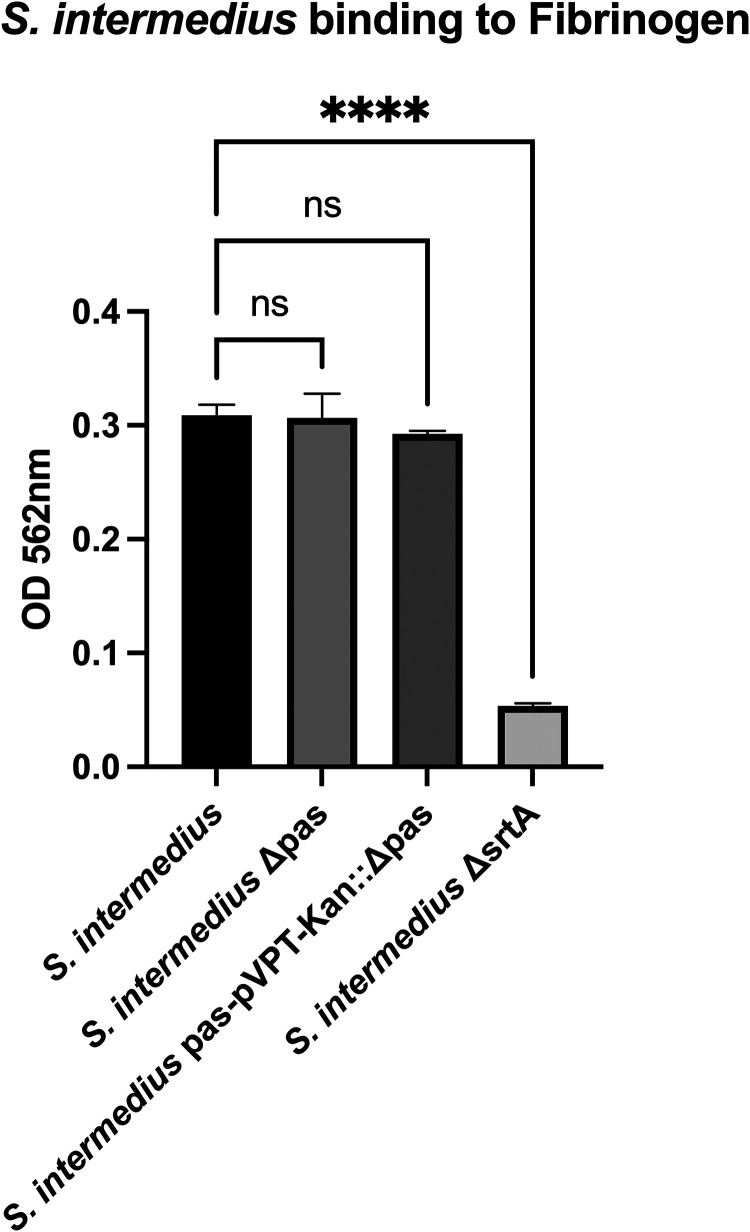
S. intermedius interaction with fibrinogen.
S. intermedius wild type (WT) adhered to 96-well plates coated with fibrinogen. The Δpas mutant did not affect the adherence to fibrinogen (Fig. 4); however, the sortase A mutant (ΔsrtA) significantly reduced S. intermedius binding. When the reverse experiments were conducted where the plates were coated with bacteria and probed with fibrinogen-conjugated Alexa Fluor 488, there was a notable reduction in the binding of S. intermedius Δpas compared to the WT (Fig. S6). When complemented with Pas, fibrinogen binding was restored.
FIG 4.
S. intermedius adherence to fibrinogen. S. intermedius binding to fibrinogen coated on a plate is not mediated by Pas but is greatly impacted by a knockout of sortase A.
Pas impacts single, as well as dual, species biofilms.
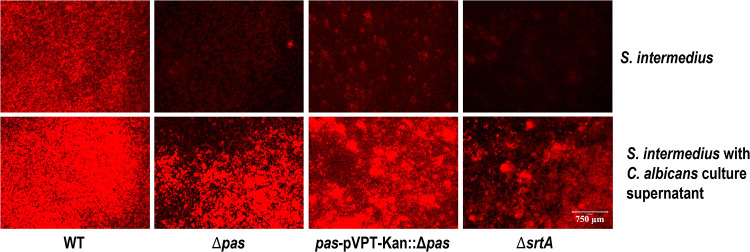
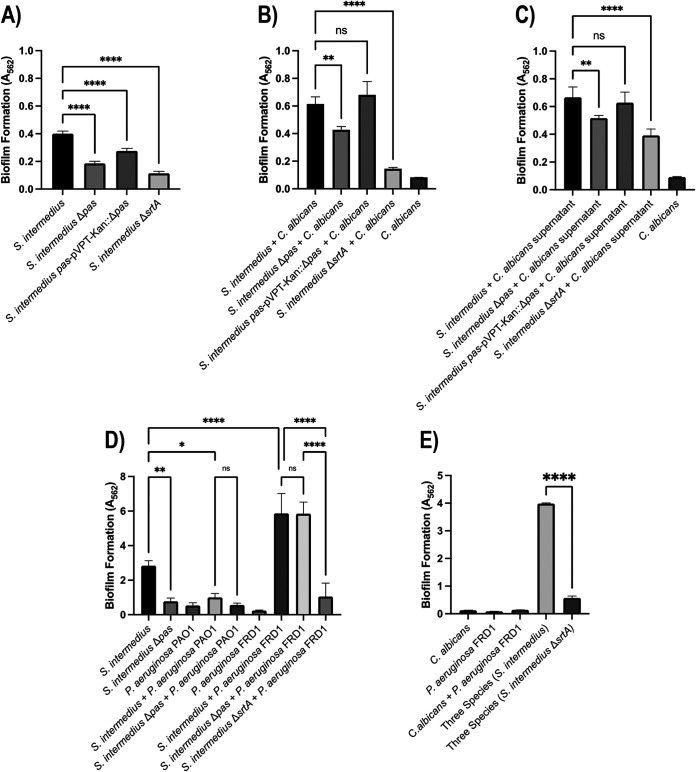
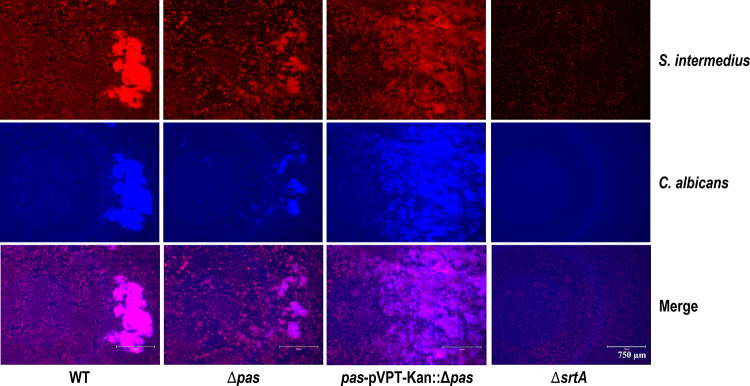
In single-species biofilm assays, there was a significant reduction (54%) in biofilm formation with Δpas, and when complemented, this phenotype was mostly (79%) restored (Fig. 5 and 6). The sortase A mutant, which does not allow the surface proteins to be anchored onto the peptidoglycan layer, displayed the largest reduction (71%) in biofilm formation. In the dual-species study with C. albicans, the impact of the pas knockout was reduced but still significant (31%) and was fully recovered by the complemented strain (see Fig. 6 and 7). The sortase A mutant again had a greater impact in the dual-species biofilm with C. albicans than the pas knockout (Fig. 6B). To determine the role of components released by S. intermedius and C. albicans, their respective supernatants were used to study the biofilm formation. The culture supernatants from S. intermedius wild-type and mutant strains did not alter C. albicans monospecies biofilm level (data not shown), suggesting that C. albicans needs to physically interact with the streptococci to form biofilms. However, in reverse experiments, the addition of C. albicans culture supernatant significantly increased the biofilm level of all tested S. intermedius strains (Fig. 5), indicating that extracellular components from C. albicans facilitate S. intermedius biofilm formation.
FIG 5.
S. intermedius monospecies biofilm. The bottom panels for S. intermedius, which is grown with C. albicans culture supernatant, exhibited more robust biofilm than the top panels with the same strain (hexidium iodide red stain). S. intermedius strains were grown in THB, diluted at a ratio of 1:2,000 into 1 ml fresh TSB medium, and grown for 24 h at 5% CO2 and 37°C under static conditions.
FIG 6.
Biofilm studies, crystal violet staining. (A) In the monospecies biofilm, there was a significant reduction (54%) in biofilm formation with Δpas, and when complemented, this phenotype was mostly (79%) restored. The sortase A mutant also displayed significant reduction (71%) in biofilm formation. (B) The dual-species study showed less reduction with the Pas knockout (31%) and was fully recovered by the complemented strain. (C) The S. intermedius with C. albicans supernatant has similar reduction to the dual species biofilm. (D) Pas and sortase mediate different interactions with acute and chronic strains of P. aeruginosa. PAO1, acute P. aeruginosa; FRD1, chronic P. aeruginosa. Biofilms were grown in Todd Hewitt with 20 mM glucose for 16 h and incubated at 37°C with 5% CO2. (E) Sortase A knockout of S. intermedius also reduces biofilm with three species biofilm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All statistics were analyzed by one-way ANOVA using GraphPad Prism 9.
FIG 7.
Dual-species biofilm. S. intermedius and C. albicans biofilm was assessed using hexidium iodide (S. intermedius) or calcofluor white (C. albicans). S. intermedius strains with C. albicans were grown in THB and were diluted each at a ratio of 1:2,000 into 1 ml fresh TSB medium and grown for 24 h at 5% CO2 and 37°C under static conditions.
Since S. intermedius has been found to colonize the cystic fibrosis (CF) lung, we questioned whether this bacterium interacts with the major CF pathogen, P. aeruginosa, in a Pas-dependent manner. Using acute (PAO1) and chronic CF (FRD1) isolates of P. aeruginosa, we discovered that in dual-species studies, PAO1 produced a smaller amount of biofilm than the S. intermedius single species, and more importantly, the loss of Pas did not significantly reduce the dual PAO1 and S. intermedius biofilm (Fig. 6). The S. intermedius-FRD1 biofilm formation was robust and larger than PAO1; however, this promotion was again not mediated by Pas. Finally, the loss of sortase A (ΔsrtA) reduced the FRD1 and S. intermedius dual biofilm, as well as the three-species biofilm with FRD1 and C. albicans (Fig. 6), firmly suggesting that a surface protein(s) other than Pas mediates the interaction between S. intermedius and this chronic P. aeruginosa isolate. Overall, these data suggest that S. intermedius utilizes different surface proteins to interact with specific microbes.
DISCUSSION
Over the last 2 decades, the surface proteins on microbes have garnered attention, and their roles in specific host interactions have been elucidated (39, 40). In Gram-positive bacteria, the enzyme sortase anchors these surface proteins on the peptide-glycan layer through a penicillin-like reaction (18). Both the commensal and pathogenic oral streptococcal strains display a variety of surface proteins, among which the antigen I/II family (AgI/II) homologs have been well studied (20, 41–47). AgI/II facilitates various interactions with the host as well as with other microorganisms in the biofilm matrix (48–51). In group A streptococci, the AgI/II-homolog AspA was shown to be an important factor in infecting the epithelial cells of the respiratory tract (52), and the homolog in group B streptococci, BspC, interacts with vimentin in the progression of group B Streptococcus (GBS) meningitis disease (53). AgI/II is also essential for the interkingdom interactions between S. mutans and C. albicans (36).
While S. intermedius is identified to be an oral commensal, its multifaceted role as an opportunistic pathogen is still emerging. In the oral cavity, it is observed in subgingival purulent infections, and elsewhere in the human body, it is often associated with purulent infections. Most importantly, in cystic fibrosis lungs, S. intermedius (milleri group) is known to display dual characters, both exacerbating the condition of the patient, as well as providing clinical stability through enhancing microbial diversity and reducing inflammation of the CF lung (54).
In this study, we attempted to characterize the structure/function attributes of Pas, an AgI/II family protein of S. intermedius. The X-ray crystal structures for VPas and C123Pas of the S. intermedius AgI/II homolog Pas (Fig. 2 and 3; Table 2) were resolved at 1.7 and 2.7 Å resolution, respectively. The VPas is highly similar to equivalent structures in VAgI/II (RMSD, 0.55 Å) and VGbpC (RMSD, 1.45 Å) of S. mutans, and similarly, the C123Pas display structural homology to S. mutans C123AgI/II (RMSD, 0.40 Å) and S. gordonii C123SspB (RMSD, 0.77 Å). The overall elongated fibrillar structure is much shorter (at 350 Å about half of AgI/II or SspB) (see Fig. S3 in the supplemental material) where Pas contains only one A-P repeat, whereas AgI/II homologs have 3 to 5 A-P repeats, contributing much to their length.
Comparison of the crystal structures of VPas and C123Pas to AgI/II, SspB, and GbpC clearly illustrates that they are more related to AgI/II of S. mutans and, to a lesser degree, to SspB of S. gordonii. On closer examination of the V domains, we find subtle but significant differences in three distinct regions, which may contribute to yet unknown functional differences. In region 3 of VPas, we observe a helix-turn-helix motif. The structural homology search indicated a substantial structural match (RMSD, ∼3 Å) with DNA-binding HTH-like transcriptional regulators despite low sequence identity for the observed helix-turn-helix motif in the V domains of the AgI/II homologs. It would not be surprising if these motifs possibly adhered to extracellular genomic DNA (eDNA), which is a known structural component of biofilm (55–57). In the VAgI/II homolog, the metal binding site is often occupied by a Ca2+ ion, whereas in VPas, we confirmed the presence of Mg2+, which indicates flexibility within this metal binding site to accommodate various group IIA metals, and this might be relevant, as altered levels of magnesium in serum have been observed in both purulent infections and chronic obstructive pulmonary disease (58).
In the case of the structures of C123 domains among the AgI/II homologs, all the C domains adopt the DEv-IgG fold, and we observed that the orientations of these C-terminal domains vary considerably with respect to each other. Particularly, C1 had shifted by as much as 2.0 Å at some sites. In addition, there is an apparent rotation of this domain by about 5 to 10 degrees, and comparison of the charge distribution between C123Pas and C123AgI/II shows that these are very different (Fig. S2 in the supplemental material). The distinct differences in the C1 domains are highlighted with ovals in Fig. S2. More importantly, the two arrows highlight the charge distribution in the interface between the C1-C2 and C2-C3 domains that are known to host the sites of adherence to both carbohydrates and/or Gp340/SRCR domains (Fig. S2). Such differences in charge distribution are perhaps evolved adaptations by S. intermedius to its local environment.
The adherence of Staphylococcus aureus DEv-IgG domains has been well characterized with clumping factors ClfA, ClfB, and Staphylococcus epidermis SdrG, where they use the “dock, lock, and latch” mechanism to adhere to various regions of fibrinogen (59–61). These interactions are facilitated by two DEv-IgG domains (60, 62), where the first domain has an inherent gap between the “D” and “E” strands into which the fibrinopeptides dock, and thereafter, it is latched and locked by an extended strand from the second DEv-IgG domain. In the case of the C domains of AgI/II-like proteins, such a mechanism would not be possible, as these domains do not carry the inherent gap between the “DE” strands (22). The C-terminal regions that carry the DEv-IgG domains in oral streptococci must have developed their own unique adherence mechanism, which has yet to be discovered. Our future efforts will focus on structurally characterizing the oral streptococcal DEv-IgG domains with their binding partners in order to enumerate the specific binding motif/mechanism.
Previously, we had shown that AgI/II and its homologs (34, 37) adhere with nanomolar affinity to Gp340 and its SRCR domains. Hence, we examined the binding of FLPas, VPas, and C123Pas to SRCR, and similar to the AgI/II-family, they display nanomolar affinity (Table 3). The shortened length of Pas did not affect the binding to SRCR/Gp340, further confirming that the AP regions are perhaps just the projection mechanism used by oral streptococci to initially adhere through the V domain and thereafter adhere with high affinity amid laminar flow, thus establishing a firmer contact with the host. Among the bacterial cell wall-anchored surface proteins, these gene duplications are often observed as a predominant feature. For example, in Staphylococcus aureus, (i) the B domains of the collagen binding adhesin Cna, (ii) the fibronectin binding repeats of FnBPA , and (iii) the G5-E domains project the ligand binding domain away from the microbial cell surface through these repeat domains (63). It is quite plausible that S. intermedius acquired the pas gene through lateral gene transfer, and the resulting single A-P repeat could have been evolutionarily and environmentally driven. These studies conclude that all AgI/II homologs adhere with very high affinity to SRCR/Gp340.
Biofilm studies show that Pas does influence the single-species biofilm formation, and this result is different from that of AgI/II of S. mutans (64). Pas also contributes to interkingdom interactions, where its absence results in lower dual-species biofilm with C. albicans. Previously, we had shown that AgI/II is an important factor in this dual-species biofilm formation (36), and it appears from the present study that Pas exerts a similar role. Given these results, it now establishes that the AgI/II-like family of proteins are directly involved in such interkingdom interactions with other microbes in the oral cavity. The existing paradigm on biofilm formation of diseased sites in dental caries is mostly attributed to the glucosyl transferases (GTFs). These GTFs, which are secreted into the milieu surrounding the microbe, produce soluble/insoluble glucans from ingested sucrose, and they are supposed to provide the bed layer/base, where other microbes will bind and form biofilms. However, our current and the previous study on AgI/II family of proteins now directly implicate a role for its surface antigens in biofilm formation, particularly for dual-species biofilm with C. albicans.
Pas belongs to the AgI/II family of proteins, which have previously been shown to interact with C. albicans. In particular, the SspB of S. gordonii was shown to interact with Als3 of C. albicans (65). In studies conducted in our lab, AgI/II of S. mutans and S. intermedius Pas do not appear to interact with Als3 (data not shown). Our results now confirm that secreted components in the C. albicans supernatant promote dual-species biofilm formation (Fig. 5). While Pas mediates direct attachment to Gp340 and fibrinogen, it interacts indirectly with C. albicans. Future studies will be aimed at identifying the biofilm-enhancing components secreted by C. albicans.
In addition to interactions with C. albicans, we also studied the role of P. aeruginosa with S. intermedius, another known pathogen in CF lung microbiota (66, 67). Our biofilm studies with a Δpas mutant demonstrated that Pas does not mediate interactions with the acute cystic fibrosis isolate (PAO1) or the chronic isolate (FRD1) of P. aeruginosa. However, there was a significant increase in the S. intermedius and FRD1 dual biofilm, and loss of sortase A resulted in a severe reduction in the dual-species biofilm with the chronic CF, as well as the three-species biofilm containing C. albicans, P. aeruginosa (FRD1), and S. intermedius, which are species relevant to CF lung disease. These results indicate there are other surface proteins on S. intermedius that are involved in such interactions. Genomic annotations reveal 18 other putative cell wall-anchored surface proteins on S. intermedius (Table S3), and further studies are necessary to identify the factors that are involved in the S. intermedius-P. aeruginosa interface, which might provide clues for targeted microbial therapies that could specifically attenuate the biofilm.
Physical interactions between surface proteins of S. intermedius and C. albicans suggest a potential synergistic role in CF lung disease. C. albicans is commonly coisolated from P. aeruginosa biofilms in CF lung infections (68). Additionally, anginosus group streptococci, like S. intermedius, are known to cause exacerbations in CF patients by promoting P. aeruginosa virulence (69). Taken together, our data demonstrate potential interplay between these three airway pathogens, which may be mediated by streptococcal surface proteins. Our future studies will focus on identifying additional factors that modulate multispecies interactions between S. intermedius, C. albicans, and P. aeruginosa.
Within the oral cavity, S. intermedius is associated with periodontitis and has been isolated from lesions in clinical reports (70). Outside the oral environment, S. intermedius and other S. anginosus group bacteria are able to infect the host, presenting themselves in endocarditis and/or purulent abscesses found in the brain, liver, and spleen (4). While knowledge on specific mechanisms that facilitate S. intermedius’s transmission and infection is currently very limited, various case studies provide evidence for hematogenous dissemination of S. intermedius within the same organ and, more importantly, simultaneous presentation of abscesses in multiple organs (71). S. intermedius’s ability to infect a variety of organs suggests it adapted for low shear forces found in venous blood flow. One study showed that the binding of anginosus streptococci to fibrinogen induced platelet aggregation, and this could perhaps be one of the dissemination mechanisms to various sites of abscesses and endocarditis (72). Therefore, we investigated the interaction of Pas with fibrinogen. The Pas knockout did not impact S. intermedius interaction to immobilized fibrinogen, whereas the sortase A (ΔsrtA) mutant significantly affected adherence (Fig. 4). In reverse experiments where S. intermedius was immobilized, we observe that the Δpas mutant displays reduced adherence (Fig. S6), and complementation restores the phenotype. These results confirm that, compared to Pas, other surface proteins on S. intermedius (Table S3) significantly contribute to fibrinogen binding (5). Using surface plasmon resonance, we further quantified that full-length Pas interacts with fibrinogen with nanomolar affinity (Table 3), which is higher than the micromolar affinity observed for staphylococcal surface adhesins (59, 61, 73–75). In staphylococcal infections, interaction with fibrinogen allows cross-linking and encasement of bacterial cells, which provides localization within the vasculature (76–78). Further investigations are needed to establish the role of S. intermedius’s binding to platelets, and whole-cell experiments assessing binding to various blood components would provide valuable insights into such interactions and could offer attractive targets for developing therapeutics.
It is our hypothesis that S. intermedius could gain access to the bloodstream by binding to fibrinogen (this study) and/or platelets (72) to reach the lungs. Once it reaches the lungs, the nanomolar adherence of Pas to membrane alveolar macrophage protein Gp340/SRCR would allow S. intermedius to localize on the lung alveoli in CF patients. Subsequently, the interaction of S. intermedius Pas with C. albicans and yet-to-be-identified surface adhesins with P. aeruginosa, the two other opportunistic CF pathogens, could promote biofilm formation and colonization. The biofilm formation, colonization, and its interkingdom interactions of S. intermedius in CF patients is known to exacerbate and/or reduce symptoms (54). Our studies conclude that the multifunctionality of S. intermedius’s surface antigen Pas could play a major role in the transmissibility, localization, and colonization of the microbe. Future studies could focus on determining the interacting partners on C. albicans and P. aeruginosa and enumerating the mechanisms of such interactions, which could lead to development of therapeutic interventions.
MATERIALS AND METHODS
Cloning of Pas constructs.
The pas gene was custom synthesized (GeneArt), and subclones were developed from this full-length construct. Three constructs (primers used for cloning are listed in Table 1), full-length (FLPas) (residues 38 to 1241), VPas (289 to 682), and C123Pas (747 to 1241), were designed with a cleavable (TEV protease) C-terminal histidine tag (Fig. 1) utilizing the fast-digest restriction enzymes NcoI and XhoI (Thermo Fisher Inc.) for cloning into the pET23d vector (Novagen). PCR amplification of the fragments was done using Phusion DNA polymerase followed by digestion of both the PCR fragments and vector with appropriate enzymes. The products were ligated with T4 DNA ligase (NEB), transformed into Escherichia coli DH5α cells, and grown on LB agar plates supplemented with ampicillin (50 μg/ml). Single colonies were grown in fresh 5-ml LB cultures, and the plasmids were harvested using the miniprep kit (Zymo). DNA sequencing was carried out at the UAB Heflin center, which established the presence of the appropriate fragments in the pET23d vector. After confirmation, these plasmids were transformed into E. coli BL21(DE3) cells for protein expression.
Expression and purification.
E. coli BL21(DE3) cells harboring the plasmids for each Pas construct were inoculated into a 20-ml starter terrific broth (TB) culture overnight at 37°C. The next day, these starter cultures were transferred to shaker flasks containing 1 liter of TB, and cells were grown to an optical density at 600 nm (OD600) of 1.0, at which point they were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 30°C, followed by subsequent growth overnight at 18°C. The cells were harvested by centrifugation at 5,000 × g for 20 min in a Beckman Avanti JL-25 centrifuge, and the cell pellets were resuspended in nickel affinity column binding buffer (50 mM Tris [pH 8.0], 500 mM sodium chloride), which was augmented with a Complete EDTA-free protease inhibitor (Roche). Cells were ruptured by sonication (Fisherbrand sonicator) for a total of 5 min while maintaining a maximum temperature of 10°C. Lysed cells were centrifuged at 35,000 rpm for 1 h using a Ti70 rotor, and the supernatant was collected and filtered through a 0.22-μm filter before being loaded onto a 20-ml HisPrep nickel column (GE Healthcare, Inc.). Using a step gradient of 50 mM imidazole, the nonspecifically bound proteins were gently removed from the column, and then the bound protein was eluted with a 50 to 300 mM gradient. The protein fractions were selected based on purity as visualized on an SDS-PAGE gel and thereafter pooled and dialyzed overnight into MonoQ buffer (50 mM Tris [pH 8.0], 50 mM sodium chloride, and 1 mM EDTA). This sample was loaded onto a MonoQ column (GE Healthcare, Inc.), and the protein was eluted with a 0 to 400 mM NaCl gradient. The purest single-banded fractions as identified by SDS-PAGE gels were then pooled and concentrated under 55 lb/in2 nitrogen gas using an Amicon stirring concentrator. Protein concentration was measured using a modified method described elsewhere (79).
Crystallization and data collection.
FLPas, VPas, and C123Pas were subjected to crystallization trials using our Gryphon (Art Robbins Instruments) robot and commercial screens. Initial hits for VPas and C123Pas were obtained from the Hampton crystallization screen (CrystalScreen HT), with hit conditions being further refined to routinely obtain crystals. Briefly, 1 μl of reservoir solution was mixed with 1 μl of concentrated protein (VPas at 10.0 mg/ml and C123Pas at 12.0 mg/ml) in a hanging drop vapor diffusion setup using Linbro boxes. VPas crystals were flash frozen in buffer of the crystallization condition (100 mM Tris [pH 8.5], 25% polyethylene glycol 3350 [PEG 3350], and 250 mM magnesium chloride) supplemented by 20% ethylene glycol. Diffraction data for VPas were collected using a MarMosaic 300HS charge-coupled-device (CCD) detector at the SER-CAT 22-ID beamline of the advanced photon source (APS) at the Argonne National Laboratory (ANL) in Chicago. Similarly, C123Pas crystals were flash frozen in buffer of the crystallization condition (100 mM sodium citrate buffer [pH 5.8], 200 mM ammonium sulfate, and 900 mM lithium sulfate) supplemented by 20% glycerol. Diffraction data for C123Pas were collected using an ADSC quantum 315 CCD detector at the NE-CAT 24-ID-E beamline of APS at ANL in Chicago. Data for C123Pas were integrated, merged, and scaled with HKL2000 (80). Data for VPas were integrated, merged, and scaled with XDS (81) followed by Aimless (82) in CCP4 (83). Data collection parameters are listed in Table 2.
Crystal structure determination and refinement.
The structures were initially solved by molecular replacement using Phaser (84) with the V domain and the C domains of antigen I/II (PDB IDs 3IOX and 3QE5) as initial search models. After replacement with the residues from the Pas sequence of both the V domain and C domains, further refinements were performed using a combination of both Refmac5 (85) in CCP4 and Phenix (86). For both of these structures, the isotropic B-factor refinements were carried out using a translation-libration-screw model (87). Coot was used for all model building (88), and figures were created with PyMol (version 2.2.0; Schrödinger, LLC). For model quality assessment, we employed validations using Phenix, QualityCheck (https://smb.slac.stanford.edu/jcsg/QC/), and the wwPDB validation service (https://validate-rcsb-2.wwpdb.org/). The conserved Ca2+ sites (see Table S1 in the supplemental material) in C123Pas and the replacement of Mg2+ instead of Ca2+ in VPas were verified through the CheckMyMetal web server (https://csgid.org/csgid/metal_sites/). Refinement statistics are shown in Table 2.
Modeling of A123VP123C123AgI/II and AVPC123Pas.
The A123VP123C123 region for AgI/II (residues 202 to 1486) is based on the crystal structures of A3VP1AgI/II (PDB ID 3IOX) and C123AgI/II (PDB ID 3QE5). The other two A-P repeats (A2-P2; A1-P3) were modeled based on homology with the A3-P1 repeat in A3VP1AgI/II. For the AVPC123Pas model, we used the two structures (PDB IDs 6E36 and 6E3F) of VPas and C123Pas described here and modeled the single A-P repeat by homology with the A-P repeat in the A3VP1AgI/II structure (PDB ID 3IOX).
Structural homology search.
A structural homology search was performed using the CAME web server (https://topsearch.services.came.sbg.ac.at/) with the coordinates for region 3 of VPas (residue range, 362 to 429).
Surface plasmon resonance studies.
To determine binding affinities, surface plasmon resonance studies were performed using immobilized SRCR1 and SRCR123 that were recombinantly expressed and purified from insect cells as previously described (22). For binding affinity to fibrinogen, fibrinogen depleted of plasminogen, von Willebrand factor, and fibronectin was purchased from Enzyme Research Laboratories (catalog no. FIB 3). Briefly, ligands SRCR1, SRCR123, and fibrinogen were immobilized on a CM5 chip using ethanolamine chemistry, and the chip surface was prepared for binding experiments. Analytes FLPas, VPas, and C123Pas were injected over SRCR1 and SRCR123 at various concentrations (serial dilutions within 0.0625 to 8 μM), dissociation was measured for 600 s following injections, and the sensorgrams were recorded using a Biacore2000. The running buffer for the SRCR/Gp340 studies was 20 mM HEPES (pH 7.4), 150 mM NaCl, and 1 mM CaCl2, and regenerations were done with 10 mM HCl. Similarly, FLPas was injected over fibrinogen at various concentrations with a running buffer of 20 mM MES (morpholineethanesulfonic acid; pH 5.5), 50 mM NaCl, and 1 mM CaCl2. Each experiment was carried out in triplicate. The sensorgrams were fitted using BIAevaluation software, where both the residuals and chi-square values were refined to convergence. The results of these experiments are presented in Tables 3; corresponding sensorgrams are shown in Fig. S4 and S5.
Bacterial strains and culture conditions.
S. intermedius ATCC 27335, C. albicans SC5314, and Pseudomonas aeruginosa strains PAO1 and FRD1 were grown in tryptic soy broth (TSB) and yeast extract-peptone-dextrose (YPD) (2% dextrose, 2% Bacto peptone, and 1% yeast extract) in an atmosphere of 5% CO2 at 37°C. Erythromycin (10 μg/ml) and kanamycin (500 μg/ml) were used to grow S. intermedius strains with corresponding antibiotic resistance.
Construction of the Pas and sortase A mutants.
S. intermedius antigen I/II-like protein is encoded by the pas gene. PCR ligation mutagenesis was used to construct the Pas deletion mutant (Δpas) and the sortase A deletion mutant (ΔsrtA) in S. intermedius ATCC 27335. The S. intermedius JTH08 genome sequence was used to design primers for Δpas and ΔsrtA (Table 1). Two amplified flanking regions of pas/srtA were ligated in an IFDC2 cassette on 5′ and 3′ ends via overlapping PCR (89). The resulting PCR product was transformed into the S. intermedius wild-type strain. The transformation method was similar to the one used for S. mutans with some modifications (90). Overnight cultures of S. intermedius were diluted 1:20 and grown for 2 h at 5% CO2 and 37°C. Five hundred nanograms of PCR product was mixed with 200 μl of S. intermedius culture and incubated for 2 h in the same condition. The Pas mutant transformants were selected on 10 μg/ml erythromycin TSB agar. The deletion mutant Δpas was complemented with the shuttle vector pVPT-Kan bearing the full-length pas DNA fragment; the construct pas-pVPT-Kan was then transformed into Δpas and selected using kanamycin resistance TSB agar plates. Using overnight cultures of S. intermedius WT, Δpas, Pas complement (pas-pVPT-Kan::Δpas), and ΔsrtA, Western blots were probed with cross-reacting 3-8D antibody (a gift from L.J. Brady, UFL) (91), which confirmed the expression of Pas, in both the wild-type strain and the Pas-complemented strain, whereas no such expression was observed in the Δpas and ΔsrtA mutants (data not shown).
Biofilm formation assay.
Single- and dual-species biofilms of S. intermedius strains with C. albicans or P. aeruginosa were grown overnight in THB and YPD media, respectively. S. intermedius and C. albicans were diluted each at a ratio of 1:2,000 into 1 ml fresh TSB medium. These samples in triplicate were transferred to a Costar 24-well plate to grow for 24 h at 5% CO2 and 37°C under static conditions. The plate was washed three times with distilled water and stained with crystal violet as previously described (92) and/or fluorescent hexidium iodide (S. intermedius) with calcofluor white (C. albicans). In a separate set of experiments, the role of extracellular components released from either S. intermedius strains or C. albicans was determined. Supernatants were extracted using 0.2- filters from S. intermedius and C. albicans that were grown overnight in TSB medium. Subsequently, biofilm growth medium was prepared with fresh TSB medium with the supernatants in a 1:1 ratio. Specifically, the S. intermedius supernatant was used for growing C. albicans biofilm, and the C. albicans supernatant was used for growing S. intermedius biofilm, which were assessed using hexidium iodide (S. intermedius) or calcofluor white (C. albicans). Biofilm images were obtained using the Invitrogen EVOS 5000 microscope digital imaging system and collected with a 4× lens objective at room temperature. For two- and three-species biofilm studies involving (i) S. intermedius, (ii) PAO1, acute P. aeruginosa; and FRD1, chronic P. aeruginosa; and (iii) C. albicans, biofilms were grown in Todd Hewitt with 20 mM glucose for 16 h and incubated at 37°C with 5% CO2. One-way ANOVA was used in all statistical analyses using GraphPad Prism 9.
S. intermedius binding to fibrinogen.
One hundred microliters of 100 μg/ml fibrinogen from human plasma was added to each well and incubated at 4°C overnight. Overnight cultures of S. intermedius (WT, Δpas, pas-pVPT-Kan::Δ pas and ΔsrtA) 1 ml were pelleted, washed with 1 ml phosphate-buffered saline (PBS) buffer, and resuspended with 1 ml binding buffer (10 mM HEPES [pH 7.5], 5 mM CalCl2, 5 mM MgCl2, and 50 mM NaCl). Experiments were conducted in triplicate, where 100 μl was added to each fibrinogen-coated well and incubated at 37°C for 1 h in a 5% CO2 incubator. Wells were washed three times with PBS buffer with 0.1% Tween 20 and stained with crystal violet. Bacterial binding to the plate was measured at optical density at 562 nm. One-way analysis of variance (ANOVA) was used in all statistical analyses using GraphPad Prism 9.
Overnight cultures of S. intermedius (WT, Δpas, pas-pVPT-Kan::Δ pas, and ΔsrtA) 1 ml were pelleted and resuspended in 1 ml of bicarbonate buffer, pH 9.6, 100 μl was aliquoted into each well (96-well Corning Costar EIA/RIA), and the plate was air dried overnight. Subsequently, the plate was washed three times with distilled water followed by 100 μl of binding buffer (10 mM HEPES [pH 7.4], 200 mM NaCl, and 5 mM CaCl2). Thereafter, the plate was blocked with 1% bovine serum albumin (BSA) for 1 h, washed, and incubated with 20 μg/ml of fibrinogen-conjugated Alexa Fluor 488 (Invitrogen) for 2 h with gentle swirling. Finally, after washing the plate three times with PBS buffer, images were obtained using the Invitrogen EVOS 5000 microscope digital imaging system and collected with a 40× lens objective at room temperature.
Data availability.
All data needed to evaluate the conclusions in the manuscript are present in the main text or the supporting information. The coordinates for VPas (PDB ID 6E36) and C123Pas (PDB ID 6E3F) have been deposited in the Protein Data Bank (RCSB).
ACKNOWLEDGMENTS
The X-Ray Crystallography Core Facility at UAB, which is supported by the O’Neal Comprehensive Cancer Center, is gratefully acknowledged for providing the resources to carry out X-ray Crystallography. High-resolution data were collected at the South Eastern Regional Collaborative Access Team (SERCAT) at APS, ANL, Chicago. The Heflin-Genomic Core at UAB is acknowledged for the DNA sequencing.
J.L.M. conducted SPR experiments, analyzed results, and cowrote the manuscript; N.S. refined the structures, deposited the structures to PDB, and cowrote the manuscript; R.W. carried out mutational analysis and biofilm studies; M.P. expressed and purified Pas fragments and crystallized the proteins; S.P. also contributed to SPR experiments; H.W. contributed to biological studies and revised the manuscript; J.S. carried out P. aeruginosa biofilm studies and cowrote the manuscript; and C.D. conceived ideas, collected X-ray data, solved the structures, and prepared the manuscript for publication.
J.L.M., N.S., R.W., H.W., and C.D. are supported by R01 DE029007. J.L.M. was previously a T-90 fellow supported by T-90DE022736-06. H.W. was also supported by R01 DE017954 and R01DE022350. J.S. is supported by R00DE025913.
Footnotes
Supplemental material is available online only.
Contributor Information
Champion Deivanayagam, Email: champy@uab.edu.
Ann M. Stock, Rutgers University-Robert Wood Johnson Medical School
REFERENCES
- 1.Mishra AK, Fournier PE. 2013. The role of Streptococcus intermedius in brain abscess. Eur J Clin Microbiol Infect Dis 32:477–483. 10.1007/s10096-012-1782-8. [DOI] [PubMed] [Google Scholar]
- 2.Tran MP, Caldwell-McMillan M, Khalife W, Young VB. 2008. Streptococcus intermedius causing infective endocarditis and abscesses: a report of three cases and review of the literature. BMC Infect Dis 8:154. 10.1186/1471-2334-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalya S, Komal B, Tulpule S, Raoof N, Sen S. 2017. Isolated Streptococcus intermedius pulmonary nodules. IDCases 8:48–49. 10.1016/j.idcr.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy S, Singh K, Hughes S. 2018. Liver abscesses caused by Streptococcus intermedius in an immunocompromised patient. Cureus 10:e2107. 10.7759/cureus.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson AB, Kent H, Sibley CD, Grinwis ME, Mabon P, Ouellette C, Tyson S, Graham M, Tyler SD, Van Domselaar G, Surette MG, Corbett CR. 2013. Phylogenetic relationship and virulence inference of Streptococcus Anginosus Group: curated annotation and whole-genome comparative analysis support distinct species designation. BMC Genomics 14:895. 10.1186/1471-2164-14-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. 2008. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA 105:15070–15075. 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibley CD, Sibley KA, Leong TA, Grinwis ME, Parkins MD, Rabin HR, Surette MG. 2010. The Streptococcus milleri population of a cystic fibrosis clinic reveals patient specificity and intraspecies diversity. J Clin Microbiol 48:2592–2594. 10.1128/JCM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolenbrander PE, Egland PG, Diaz PI, PalmerRJ, Jr.. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol 13:11–15. 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, PalmerRJ, Jr.. 2002. Communication among oral bacteria. Microbiol Mol Biol Rev 66:486–505. 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 54:413–437. 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 11.Kolenbrander PE, Andersen RN, Kazmerzak K, Wu R, PalmerRJ, Jr.. 1999. Spatial organization of oral bacteria in biofilms. Methods Enzymol 310:322–332. 10.1016/s0076-6879(99)10026-0. [DOI] [PubMed] [Google Scholar]
- 12.Kolenbrander PE. 1997. Biofilm developmental biology. Trends Microbiol 5:475. 10.1016/S0966-842X(97)88501-0. [DOI] [PubMed] [Google Scholar]
- 13.Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral streptococci. Microbiol Spectr 6. 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav P, Verma S, Bauer R, Kumari M, Dua M, Johri AK, Yadav V, Spellerberg B. 2020. Deciphering streptococcal biofilms. Microorganisms 8:1835. 10.3390/microorganisms8111835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA 99:14434–14439. 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson GK, Mitchell TJ. 2004. The biology of Gram-positive sortase enzymes. Trends Microbiol 12:89–95. 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H, Marraffini LA, Schneewind O. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta 1694:269–278. 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi T, Asaga E, Goto N. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol Immunol 18:266–269. 10.1034/j.1399-302x.2003.00076.x. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Urano-Tashiro Y, Konishi K. 2013. Adhesins of oral streptococci. Nihon Saikingaku Zasshi 68:283–293. (In Japanese.) 10.3412/jsb.68.283. [DOI] [PubMed] [Google Scholar]
- 20.Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. 2010. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol 77:276–286. 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, Brady LJ, Deivanayagam C. 2010. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of alpha- and PPII-helices. Proc Natl Acad Sci USA 107:5983–5988. 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson MR, Rajashankar KR, Crowley PJ, Kelly C, Mitchell TJ, Brady LJ, Deivanayagam C. 2011. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. J Biol Chem 286:21657–21666. 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker EN, Squire CJ, Young PG. 2015. Self-generated covalent cross-links in the cell-surface adhesins of Gram-positive bacteria. Biochem Soc Trans 43:787–794. 10.1042/BST20150066. [DOI] [PubMed] [Google Scholar]
- 24.Reichhardt MP, Holmskov U, Meri S. 2017. SALSA-A dance on a slippery floor with changing partners. Mol Immunol 89:100–110. 10.1016/j.molimm.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 25.Madsen J, Mollenhauer J, Holmskov U. 2010. Review: Gp-340/DMBT1 in mucosal innate immunity. Innate Immun 16:160–167. 10.1177/1753425910368447. [DOI] [PubMed] [Google Scholar]
- 26.Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus KK, von Deimling A, Poustka A. 1997. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3-26.1 is deleted in malignant brain tumours. Nat Genet 17:32–39. 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 27.Jumblatt MM, Imbert Y, YoungWW, Jr., Foulks GN, Steele PS, Demuth DR. 2006. Glycoprotein 340 in normal human ocular surface tissues and tear film. Infect Immun 74:4058–4063. 10.1128/IAI.01951-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz BL, Oxley D, Packer NH, Karlsson NG. 2002. Identification of two highly sialylated human tear-fluid DMBT1 isoforms: the major high-molecular-mass glycoproteins in human tears. Biochem J 366:511–520. 10.1042/BJ20011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronellenfitsch S, Weiß C, Frommhold D, Koch L, Mollenhauer J, Poeschl J, Müller H. 2012. High DMBT1 concentrations in breast milk correlate with increased risk of infection in preterm and term neonates. BMC Pediatr 12:157. 10.1186/1471-2431-12-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diegelmann J, Czamara D, Le Bras E, Zimmermann E, Olszak T, Bedynek A, Göke B, Franke A, Glas J, Brand S. 2013. Intestinal DMBT1 expression is modulated by Crohn's disease-associated IL23R variants and by a DMBT1 variant which influences binding of the transcription factors CREB1 and ATF-2. PLoS One 8:e77773. 10.1371/journal.pone.0077773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoddard E, Cannon G, Ni H, Karikó K, Capodici J, Malamud D, Weissman D. 2007. gp340 expressed on human genital epithelia binds HIV-1 envelope protein and facilitates viral transmission. J Immunol 179:3126–3132. 10.4049/jimmunol.179.5.3126. [DOI] [PubMed] [Google Scholar]
- 32.Holmskov U, Lawson P, Teisner B, Tornoe I, Willis AC, Morgan C, Koch C, Reid KB. 1997. Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule. J Biol Chem 272:13743–13749. 10.1074/jbc.272.21.13743. [DOI] [PubMed] [Google Scholar]
- 33.Osei KA, Deivanayagam C, Nichols JJ. 2018. Glycoprotein 340 in mucosal immunity and ocular surface. Ocul Surf 16:282–288. 10.1016/j.jtos.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Purushotham S, Deivanayagam C. 2014. The calcium-induced conformation and glycosylation of scavenger-rich cysteine repeat (SRCR) domains of glycoprotein 340 influence the high affinity interaction with antigen I/II homologs. J Biol Chem 289:21877–21887. 10.1074/jbc.M114.565507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purushotham S, Deivanayagam C. 2013. Cloning, expression and purification of the SRCR domains of glycoprotein 340. Protein Expr Purif 90:67–73. 10.1016/j.pep.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C, Scoffield J, Wu R, Deivanayagam C, Zou J, Wu H. 2018. Antigen I/II mediates interactions between Streptococcus mutans and Candida albicans. Mol Oral Microbiol 33:283–291. 10.1111/omi.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mieher JL, Larson MR, Schormann N, Purushotham S, Wu R, Rajashankar KR, Wu H, Deivanayagam C. 2018. Glucan binding protein C of Streptococcus mutans mediates both sucrose-independent and sucrose-dependent adherence. Infect Immun 86:e00146-18. 10.1128/IAI.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecharki D, Petersen FC, Assev S, Scheie AA. 2005. Involvement of antigen I/II surface proteins in Streptococcus mutans and Streptococcus intermedius biofilm formation. Oral Microbiol Immunol 20:366–371. 10.1111/j.1399-302X.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 39.Stones DH, Krachler AM. 2016. Against the tide: the role of bacterial adhesion in host colonization. Biochem Soc Trans 44:1571–1580. 10.1042/BST20160186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto-Shibayama K, Sato Y, Yamamoto Y, Ohta K, Kizaki H. 2006. Identification of a glucan-binding protein C gene homologue in Streptococcus macacae. Oral Microbiol Immunol 21:32–41. 10.1111/j.1399-302X.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 42.Lehner T, Russell MW, Caldwell J, Smith R. 1981. Immunization with purified protein antigens from Streptococcus mutans against dental caries in rhesus monkeys. Infect Immun 34:407–415. 10.1128/iai.34.2.407-415.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell MW, Childers NK, Michalek SM, Smith DJ, Taubman MA. 2004. A caries vaccine? The state of the science of immunization against dental caries. Caries Res 38:230–235. 10.1159/000077759. [DOI] [PubMed] [Google Scholar]
- 44.El-Sabaeny A, Demuth DR, Lamont RJ. 2001. Regulation of Streptococcus gordonii sspB by the sspA gene product. Infect Immun 69:6520–6522. 10.1128/IAI.69.10.6520-6522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demuth DR, Duan Y, Brooks W, Holmes AR, McNab R, Jenkinson HF. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol 20:403–413. 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 46.Chung WO, Demuth DR, Lamont RJ. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect Immun 68:6758–6762. 10.1128/IAI.68.12.6758-6762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407–450. 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hajishengallis G, Koga T, Russell MW. 1994. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res 73:1493–1502. 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 49.Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. 1990. Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect Immun 58:289–296. 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol Lett 268:158–165. 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakano K, Matsumura M, Kawaguchi M, Fujiwara T, Sobue S, Nakagawa I, Hamada S, Ooshima T. 2002. Attenuation of glucan-binding protein C reduces the cariogenicity of Streptococcus mutans: analysis of strains isolated from human blood. J Dent Res 81:376–379. 10.1177/0810376. [DOI] [PubMed] [Google Scholar]
- 52.Franklin L, Nobbs AH, Bricio-Moreno L, Wright CJ, Maddocks SE, Sahota JS, Ralph J, O'Connor M, Jenkinson HF, Kadioglu A. 2013. The AgI/II family adhesin AspA is required for respiratory infection by Streptococcus pyogenes. PLoS One 8:e62433. 10.1371/journal.pone.0062433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng L, Spencer BL, Holmes JA, Mu R, Rego S, Weston TA, Hu Y, Sanches GF, Yoon S, Park N, Nagao PE, Jenkinson HF, Thornton JA, Seo KS, Nobbs AH, Doran KS. 2019. The group B Streptococcal surface antigen I/II protein, BspC, interacts with host vimentin to promote adherence to brain endothelium and inflammation during the pathogenesis of meningitis. PLoS Pathog 15:e1007848. 10.1371/journal.ppat.1007848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott JE, O'Toole GA. 2019. The yin and yang of Streptococcus lung infections in cystic fibrosis: a model for studying polymicrobial interactions. J Bacteriol 201:e00115-19. 10.1128/JB.00115-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung CJ, Hsu RB, Shun CT, Hsu CC, Chia JS. 2017. AtlA mediates extracellular DNA release, which contributes to streptococcus mutans biofilm formation in an experimental rat model of infective endocarditis. Infect Immun 85:e00252-17. 10.1128/IAI.00252-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senpuku H, Nakamura T, Iwabuchi Y, Hirayama S, Nakao R, Ohnishi M. 2019. Effects of Complex DNA and MVs with GTF Extracted from Streptococcus mutans on the oral biofilm. Molecules 24:3131. 10.3390/molecules24173131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yumoto H, Hirota K, Hirao K, Ninomiya M, Murakami K, Fujii H, Miyake Y. 2019. The pathogenic factors from oral streptococci for systemic diseases. Int J Mol Sci 20:4571. 10.3390/ijms20184571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gumus A, Haziroglu M, Gunes Y. 2014. Association of serum magnesium levels with frequency of acute exacerbations in chronic obstructive pulmonary disease: a prospective study. Pulm Med 2014:1–5. 10.1155/2014/329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Höök M. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog 4:e1000226. 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, Gurusiddappa S, Narayana SV, Johnson AE, Höök M. 2008. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J Biol Chem 283:638–647. 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- 61.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217–228. 10.1016/S0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 62.Deivanayagam CC, Perkins S, Danthuluri S, Owens RT, Bice T, Nanavathy T, Foster TJ, Höök M, Narayana SV. 1999. Crystallization of ClfA and ClfB fragments: the fibrinogen-binding surface proteins of Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 55:554–556. 10.1107/s0907444998012426. [DOI] [PubMed] [Google Scholar]
- 63.Foster TJ. 2020. Surface proteins of Staphylococcus epidermidis. Front Microbiol 11:1829. 10.3389/fmicb.2020.01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. 2008. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect Immun 76:4259–4268. 10.1128/IAI.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun 78:4644–4652. 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scoffield JA, Wu H. 2015. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun 83:101–107. 10.1128/IAI.02396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marks-Austin KA, Fiel SB, Campbell PW, Stull TL. 1995. Infections in cystic fibrosis. Semin Pediatr Infect Dis 6:174–181. 10.1016/S1045-1870(05)80045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grainha T, Jorge P, Alves D, Lopes SP, Pereira MO. 2020. Unraveling Pseudomonas aeruginosa and Candida albicans communication in coinfection scenarios: insights through network analysis. Front Cell Infect Microbiol 10:550505. 10.3389/fcimb.2020.550505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill EM. 2017. The emergence of Streptococcus anginosus group as a cystic fibrosis pathogen. Clin Microbiol Newsl 39:143–147. 10.1016/j.clinmicnews.2017.09.002. [DOI] [Google Scholar]
- 70.Colombo AV, Silva CM, Haffajee A, Colombo AP. 2006. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol 55:609–615. 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 71.Giuliano S, Simone G, Rubini G, Conte A, Conti A, Goldoni P, Falcone M, Vena A, Venditti M, Morelli S. 2012. Streptococcus anginosus group disseminated infection: case report and review of literature. Infez Med 20:145–154. [PubMed] [Google Scholar]
- 72.Willcox MD. 1995. Potential pathogenic properties of members of the “Streptococcus milleri” group in relation to the production of endocarditis and abscesses. J Med Microbiol 43:405–410. 10.1099/00222615-43-6-405. [DOI] [PubMed] [Google Scholar]
- 73.Wann ER, Gurusiddappa S, Hook M. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J Biol Chem 275:13863–13871. 10.1074/jbc.275.18.13863. [DOI] [PubMed] [Google Scholar]
- 74.Hartford OM, Wann ER, Höök M, Foster TJ. 2001. Identification of residues in the Staphylococcus aureus fibrinogen-binding MSCRAMM clumping factor A (ClfA) that are important for ligand binding. J Biol Chem 276:2466–2473. 10.1074/jbc.M007979200. [DOI] [PubMed] [Google Scholar]
- 75.Vazquez V, Liang X, Horndahl JK, Ganesh VK, Smeds E, Foster TJ, Hook M. 2011. Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J Biol Chem 286:29797–29805. 10.1074/jbc.M110.214981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwiecinski JM, Horswill AR. 2020. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr Opin Microbiol 53:51–60. 10.1016/j.mib.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crosby HA, Kwiecinski J, Horswill AR. 2016. Staphylococcus aureus aggregation and coagulation mechanisms, and their function in host-pathogen interactions. Adv Appl Microbiol 96:1–41. 10.1016/bs.aambs.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182:319–326. 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 80.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 81.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 83.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 86.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winn MD, Isupov MN, Murshudov GN. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57:122–133. 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 88.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 89.Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. 2011. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol 77:8025–8033. 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peng X, Zhang Y, Bai G, Zhou X, Wu H. 2016. Cyclic di-AMP mediates biofilm formation. Mol Microbiol 99:945–959. 10.1111/mmi.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brady LJ, Cvitkovitch DG, Geric CM, Addison MN, Joyce JC, Crowley PJ, Bleiweis AS. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect Immun 66:4274–4282. 10.1128/IAI.66.9.4274-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu H, Zeng M, Fives-Taylor P. 2007. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun 75:2181–2188. 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6 and Tables S1 to S3. Download JB.00175-21-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Data Availability Statement
All data needed to evaluate the conclusions in the manuscript are present in the main text or the supporting information. The coordinates for VPas (PDB ID 6E36) and C123Pas (PDB ID 6E3F) have been deposited in the Protein Data Bank (RCSB).