Abstract
The ability for biologics to access intracellular targets hinges on the translocation of active, unmodified protein. This is often achieved using nanoscale formulations, which enter cells through endocytosis. This uptake mechanism often limits the therapeutic potential of the biologics, as the propensity of the nanocarrier to escape the endosome becomes the key determinant. To appropriately evaluate and compare competing delivery systems of disparate compositions, it is therefore critical to assess endosomal escape efficiencies. Unfortunately, quantitative tools to assess endosomal escape are lacking and standard approaches often lead to erroneous interpretation of cytosolic localization. In this study, we use a split-complementation endosomal escape (SEE) assay to evaluate levels of cytosolic caspase-3 following delivery by polymer nanogels and mesoporous silica nanoparticles. In particular, we use SEE as a means to enable systematic investigation of the effect of polymer composition, polymer architecture (random vs. block), hydrophobicity, and surface functionality. Although polymer structure had little influence on endosomal escape, nanogel functionalization with cationic and pH-sensitive peptides significantly enhanced endosomal escape levels and further, significantly increased the amount of nanogel per endosome. This work serves as a guide for developing an optimal caspase-3 delivery system, as this caspase-3 variant can be easily substituted for a therapeutic caspase-3 cargo in any system that results in cytosolic accumulation and cargo release. In addition, these data provide a framework that can be readily applied to a wide variety of protein cargos to assess the independent contributions of both uptake and endosomal escape of a wide range of protein delivery vehicles.
Keywords: intracellular protein delivery, casp-3, endosomal escape, split GFP, stimuli responsive materials, nanogel, apoptosis
Graphical Abstract
Introduction:
Protein biologics are an important emergent class of pharmaceuticals which have immense therapeutic potential due to their specificity. Nevertheless, using these biologics is challenging because of their degradation in the bloodstream, short half-lives and the potential for off-target action.1,2 Thus, biologics often require conjugation within a safe delivery system, utilizing native residues or engineered reactive handles, to facilitate intracellular translocation and improved pharmacokinetics.3,4 Substantial efforts have focused on developing efficient vehicles to deliver biologics via covalent or supramolecular formulation using polymer,5 dendrimer,6 lipid and inorganic materials.7-10 These vehicles provide benefits beyond cellular entry, including cargo protection and cell targeting. Additional requirements include sufficient cytosolic localization of an unmodified biologic while maintaining intrinsic structure and function.9,11 To better understand the factors that impact delivery efficiencies, it is essential to distinguish cellular uptake followed by cytosolic localization from a simple cellular uptake, which often results in endosomal entrapment.
The overarching hurdle in evaluating intracellular biologic delivery is the dependence on endosomal escape, for which robust and quantitative evaluation tools are lacking.12-14 The field of delivery instead relies heavily on labeled-cargo imaging or endpoint therapeutic assays to imply intracellular delivery. Microscopy can be used to visualize endosomal entrapment or escape and flow cytometry or total cell lysate analyses can elucidate internalization. Nevertheless, microscopy and flow studies rely on dye-functionalization of delivery vehicles or cargo, affording obstructions from dye-induced hydrophobicity and membrane-association, complicating interpretation.15-18 For example, our group has observed mitochondrial targeting upon cyanine dye (Cy3) functionalization to anionic polymers.19 Moreover, the process of cell fixation allows redistribution of endocytotic vesicles, often compromising membrane integrity.20-22 On the other hand, flow cytometry or total cell lysate analysis solely reveal total internalization, regardless of cytosolic or endosomal localization. Therefore, alternative methods, such as the use of split GFP, have recently been investigated to reveal true cytosolic translocation of protein cargos and antibodies.23-28
Significant endosomal escape is related to cargo concentration in each endosome as well as the number of endosomes containing the delivered agent.12,29 Many efforts have focused on enhancing these parameters by incorporating moieties within delivery systems that induce endosomal membrane fusion, membrane destabilization, or osmotic rupture through the proton sponge effect.12,14,30 Delivery systems must rely on a balance of membrane influence with cytotoxicity as there is typically a correlated relationship. The field of delivery would significantly benefit from routine use of assays that directly evaluate and quantify endosomal escape, despite the challenges and increased complexity of these assessment tools.12,16,31 In this work, we present an approach using split GFP to directly assess cargo localization. This approach facilitates optimization of nanogel (NG) polymer composition and surface functionalization for endosomal escape (Scheme 1).28 While the approach holds promise for many protein cargos, we focused this project on caspases because of their potential for targeted cell killing, in the context of proliferative diseases including cancer, rheumatoid arthritis and scleroderma. Furthermore, caspase-3 has been delivered by many materials32-38 in addition to our redox-responsive systems,39,40 underlying merit for this development. The caspase-3 cargo for split GFP developed herein can be utilized to evaluate endosomal escape or simply, cytosolic concentration, for any system so long as they allow release of the cargo protein in the cytosol. Caspases are cysteine aspartate proteases that play a vital role in the initiation and propagation of apoptosis, a form of programmed cell death that allows removal of cells without collateral damage in adjacent cells.41,42 In contrast to small-molecule cytotoxins which have dangerous implications upon excretion into the environment, as human proteins, caspases are biodegradable, environmentally friendly, and non-immunogenic. In addition, because caspases are inherently catalytic, they can function at sub-stoichiometric levels.
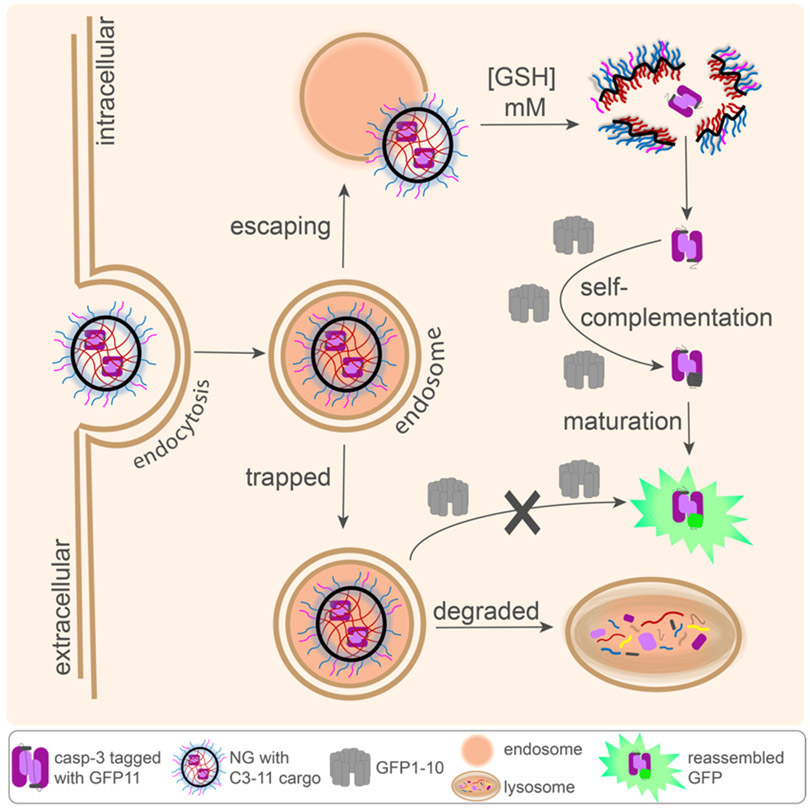
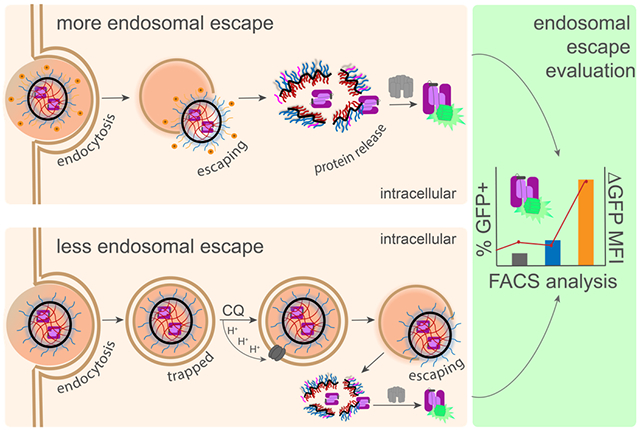
Scheme 1. Split-Complementation Endosomal Escape (SEE) pathways for NG delivering tagged casp-3 (C3-11).
Only once nanogels (NG) escape the endosome into the cytosol will C3-11 be released by cytosolic glutathione, leading to reassembly with cytosolically expressed GFP1-10 and GFP fluorescence.
The principal goal of the work described here has been to develop a method that would allow quantitation of endosomal escape that could guide development of effective cytosolic delivery and enable the direct comparison of various delivery vehicles. Using split GFP to trace cytosolic delivery of casp-3, we have demonstrated herein that endosomal escape: i) is independent of extent of protein cargo loaded in the nanogels; ii) is independent of the carrier architecture, i.e. whether it is based on block copolymers or random copolymers, iii) is enhanced with cationic and pH-responsive peptides on the surface of the nanocarriers and iv) that is assessed using this methodology can be translated to other delivery systems capable of delivering casp-3.
Results and Discussion:
Redox-responsive polymers provide many promising characteristics for caspase delivery including stealth, post-modification capability, efficient protein encapsulation, reductant-induced traceless protein release and cargo-induced cell death.39,43 We previously generated and characterized a casp-3 variant capable of cytosolic tracking using split GFP complementation. Specifically, casp-3 was tagged with the 11th strand of GFP containing three solubilizing mutations, which we refer to as C3-11 (also called C3-11M3 but refereed to here as C3-11 for simplicity). C3-11 maintains properties of native casp-3 and can report on cytosolic localization upon reassembly with GFP1-10, which is constitutively expressed cytosolically (Scheme 1).28 We often used a catalytically dead variant generated by knocking out (KO) the active site cysteine residue generating C3KO-11.28 Using this catalytically dead variant allowed us to track cytosolic levels of casp-3 without inducing apoptosis. Nevertheless C3KO, but should mimic an active casp-3 therapeutic cargo in polymer-protein formulation, nanoparticle uptake and resultant translocation.
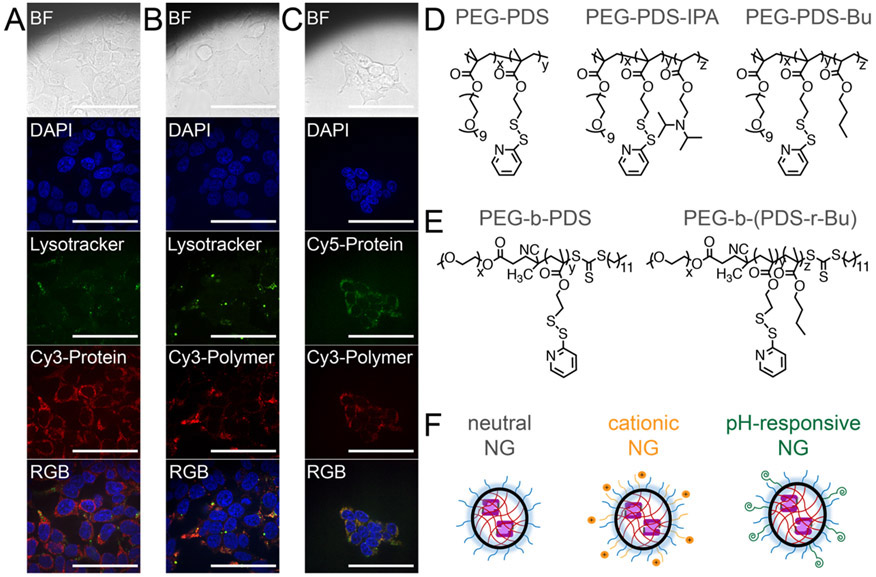
Casp-3 was delivered by random copolymers composed of poly(ethylene glycol) (PEG) and pyridyl disulfide (PDS), termed PEG-PDS.39 Crosslinked PEG-PDS NG demonstrate excellent stability of non-covalently and covalently encapsulated guests, small molecule or protein, in both the absence44-46 and presence47 of serum. A significantly punctate distribution39 was visualized upon microscopy, which is often taken to be consistent with endosomal entrapment. This was true independent of whether the protein (Figure 1A) or polymer (Figure 1B) is fluorescently labeled. Accordingly, imaging of dual-labeled NG (Cy3-polymer, Cy5-protein) demonstrates punctate colocalization of the cargo and delivery vehicle (Figure 1C), implying great complex internalization. Despite this endosomal entrapment, it is clear that the amount of casp-3 that does escape the endosome is sufficient to induce cell death,28,39 but enhancing endosomal escape is a warranted investigation to optimize efficacy.
Figure 1.
Confocal fluorescence microscopy of (A) Cy3-C3KO-11 protein delivered by unlabeled NG. (B) Cy3-PEG-PDS NG delivering unlabeled C3KO-11. (C) Cy3-PEG-PDS NG delivering Cy5-C3KO-11. HEK293T cells, 60X objective, BF (bright-field), DAPI (nucleus stain, 405 nm), Lysotracker (acidic endosomal stain, 488 nm), Cy3 (532 nm), Cy5 (632 nm). Scale bars indicate 80 microns. (D) Random copolymer structures of poly(ethylene glycol) and pyridyl disulfide (PEG-PDS) derivatives. (E) Block copolymer structures. (F) Representation of nanogels (NGs) with varying surface functionalities.
To determine if polymer structure could significantly improve endosomal escape, we generated a series of linear polymer derivatives with varying compositions and architectures. All of the polymers evaluated herein maintained the PDS group necessary for casp-3 encapsulation and traceless release (Figure S1) but have alterations in particle charge or hydrophobicity. Our group has previously synthesized PEG-PDS with random incorporation of diisopropylamino moieties (IPA), PEG-PDS-IPA.45 We hypothesized that PEG-PDS-IPA may afford better endosomal escape than PEG-PDS, as PEG-PDS-IPA demonstrates an increase in zeta potential upon decreased pH.45 To understand the effect of increasing polymer hydrophobicity, PEG-PDS with butyl (Bu) moieties, PEG-PDS-Bu, were also synthesized (Figure 1B). As changes in particle size, rigidity and hydrophobics have been shown to influence cellular internalization,48-50 we also aimed to change the hydrophilic-hydrophobic positioning by using block copolymers of PEG-PDS and PEG-PDS-Bu, termed PEG-b-PDS and PEG-b-(PDS-r-Bu) respectively (Figure 1C). To assess the importance of hydrophobic component, Bu composing 25% of the hydrophobic block (PEG-b-PDS-Bu25) or 40% (PEG-b-PDS-Bu40) were compared. In addition, the effect of NG surface functionalization by cationic or pH-sensitive moieties, was also explored (Figure 1D). PEG-PDS NGs can easily be functionalized via PDS substitution with any ligand containing an available thiol (Figure S1).39
To track endosomal escape of casp-3 we developed a split-complementation endosomal escape (SEE) assay. Casp-3 tagged with GFP11 (C3KO-11)28 was delivered by NG into HEKs1-10, which are HEK cells stably expressing the other ten beta strands of GFP (GFP1-10).51 Following endocytosis and subsequent endosomal escape, high cytosolic glutathione (1-10 mM)52,53 mediates NG-disassembly, liberating C3KO-11 from the NG to reassemble with GFP1-10.28 PEG-PDS-IPA and PEG-PDS-Bu polymers maintained the ability to encapsulate and release C3KO-11 from NG, although the encapsulation efficiency by PEG-PDS was consistently higher (Figure S2). Furthermore, the three polymers delivered comparable levels of C3KO-11 overall (Figure S2). Nevertheless, upon first assessment of NG delivered C3KO-11, no GFP signal was observed, regardless of the polymer used (Figure 2A). As endosomal escape is an extremely inefficient process, a lack of GFP signal can be attributed to low levels of escape, however, these data may hinge on sensitivity limitations. It has recently been reported that biological concentrations above 10 μM are required to observe split GFP mediated endosomal escape in cells.54
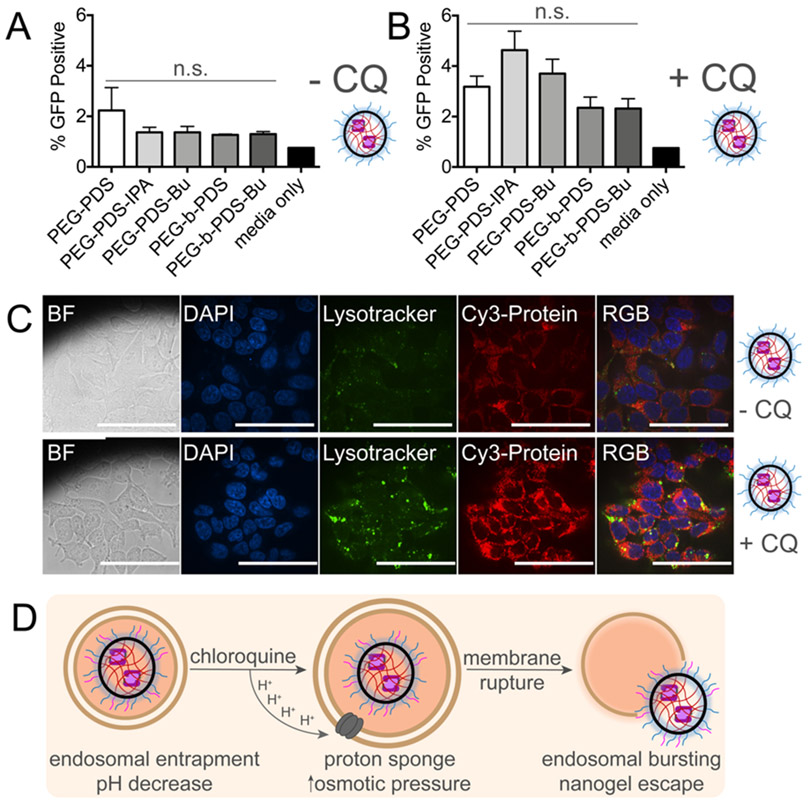
Figure 2. Incorporation of isopropyl amine or butyl functionalities within PEG-PDS polymers has little influence on C3KO-11 SEE.
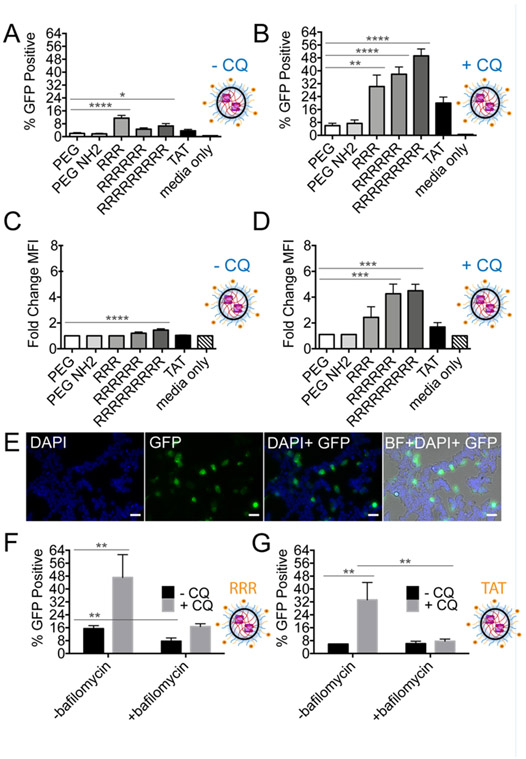
(A) SEE after 24-hour incubation with nanogel (NG) complexes at 0.75 mg/mL polymer concentration in HEKs1-10 cells demonstrates little influence of polymer structure on endosomal escape. Measured by split GFP, endosomal escape is quantified as GFP positive cells (Ex. 488 nm). Untreated, single-cell HEKs1-10 population are gated for baseline GFP fluorescence levels and any cell beyond the gate is categorized as GFP positive. SEM error bars pertain to individual biological replicates from independent NG batches, analyzed on different days. (B) NG complexes, in the presence of 80 μM chloroquine diphosphate (CQ), show an increased GFP positive population. (C) Confocal microscopy of NG (Cy3-C3KO-11 protein) in HEK293T cells. 60X objective, BF (bright-field), DAPI (nucleus stain, 405 nm), Lysotracker (acidic endosomal stain, 488 nm), Cy3 (532 nm), scale bars indicate 80 microns. (D) Hypothesized proton sponge mechanism of chloroquine mediated endosomal disruption, liberating NG entrapment in endosomes. SEM error bars pertain to four or more individual biological replicates from independent NG batches analyzed on different days. For (A,B), one-way ANOVA was performed against PEG-PDS, no significant (n.s.) differences found.
Alternative strategies to improve NG-mediated endosomal escape were undertaken as the next step of development of an optimal casp-3 delivery vehicle. NG doses were titrated up to 2 mg/mL and the time required for both NG incubation and cellular reassembly of the GFP fragments were monitored (8 – 48 hrs). As expected, GFP signal increased with increased incubation time, but otherwise, no significant improvements were observed. Endosomal-buffering agents, such as chloroquine diphosphate (CQ), can liberate NG trapped in endosomes.28 Notably, CQ has been shown to mitigate challenges beyond endosomal escape such as decreasing nanoparticle immunological clearance and increasing cytotoxic therapeutic efficacy in cancer tissues.55 Accordingly, the use of CQ in the evaluation of delivery systems has become increasingly prevalent as CQ largely increases delivery system efficiency via the proton sponge effect.56-59 Upon protonation, CQ neutralizes the acidification of endosomes, preventing endosomal-lysosomal fusion, and inducing osmotic swelling, leading to vesicle rupture.60,61 In these studies, CQ is a useful chemical tool, because when combined with data analyzing total delivered protein content, it can allow calculation of the ratio of uptake to endosomal escape. Upon the addition of CQ (80 μM), we observed a slight increase in the total percentage of GFP positive cells yet, the new polymer structures did not outperform PEG-PDS (Figure 2B). CQ induced changes in vesicle number and volume were visualized using an endosomal marker, consistent with the expected CQ-mediated vesicle swelling (Figure 2C). We ensured that the addition of CQ did not meaningfully favor the total amount of protein delivered (Figure S3) or the total amount of polymer delivered (Figure S4). These data strongly indicating that CQ herein is acting on endosomal escape, not uptake, through the proton sponge mechanism (Figure 2D).
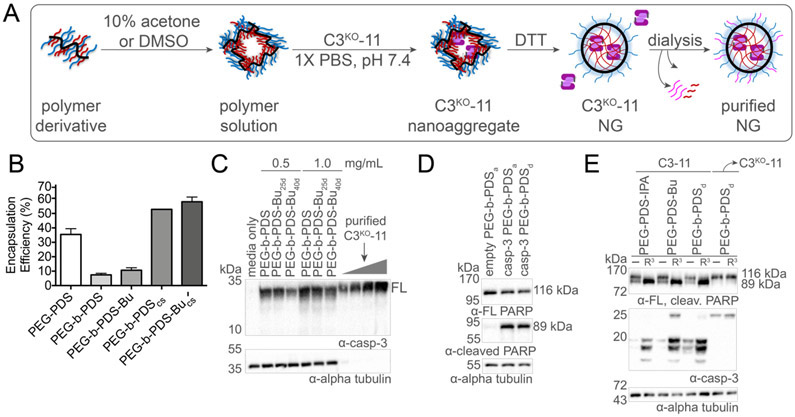
To determine whether the total amount of cytosolic casp-3 could be enhanced by increasing protein encapsulation within the NG, we altered the protein-polymer assembly strategy for the block copolymers. Traditionally, PEG-PDS random copolymer NG assemblies demonstrated encapsulation efficiencies of approximately 30 wt% (Figure S2). Comparatively, block copolymer casp-3 assemblies demonstrated notably lower amounts of encapsulation efficiencies and with minimal cellular uptake observed upon immunoblot analysis (Figure S5). The lower encapsulation efficiencies of block copolymer may be due to a pre-formed dense and difficult-to-penetrate polymeric core. To increase dynamics within the block copolymer aggregates and enhance protein-polymer reactivity, we explored co-solvent formulations.49 We selected dimethyl sulfoxide (DMSO), which is known to be compatible with casp-3 activity at low concentrations (>8%), and the volatile solvent acetone as two co-solvents for block copolymer-protein assemblies, denoted with a subscript for the co-solvent: PEG-b-PDSd and PEG-b-PDSa respectively. Co-solvent C3KO-11 NG formation begins by dissolving the polymer in the organic solvent. Casp-3 in aqueous buffer is then added protein dropwise, followed by normal NG crosslinking and purification procedures (Figure 3A). Protein release, monitored by SDS-PAGE analysis (Figure S6), demonstrated an increase in block copolymer encapsulation efficiency from approximately 10% to 40% using the co-solvent methodology, compared to the aqueous formulation strategy (Figure 3B). Fittingly, effective intracellular delivery of casp-3 was demonstrated by immunoblot analysis of total cell lysates (Figure 3C) at levels of 0.48 ± 0.21 μg/mL (0.5 mg/mL NG dose) and 0.69 ± 0.11 μg/mL (1.0 mg/mL NG dose). Thus, it is clear that increasing casp-3 cargo loaded into NG delivery vehicles increases the intracellular casp-3 titer that can be achieved.
Figure 3. Co-solvent block copolymer NG formation.
(A) Co-solvent block copolymer nanogel (NG) formation begins by dissolving the polymer in 10% solvent followed by addition of an aqueous solution containing C3KO-11 dropwise with stirring. The general NG encapsulation, crosslinking and purification procedure is then followed. (B) Encapsulation efficiencies of the block copolymers is enhanced by the co-solvent methodology (cs subscript). (C) Visualization of PEG-b-PDS NG mediated delivery of C3KO-11 to MCF7 cells at two different doses (1.0 and 0.5 mg/mL polymer concentration). Purified C3KO-11 was added at 0.2, 0.4, 0.8 and 1.6 μg/mL. (D) PEG-b-PDS NG prepared using co-solvents acetone (a subscript) or DMSO (d subscript) mediated delivery of WT casp-3 demonstrated cleavage of poly (ADP-ribose) polymerase 1 (PARP) indicating delivery of active casp-3 WT and apoptosis with an empty NG control prepared using the co-solvent methodology. (E) Delivery of active C3-11 by PEG-PDS NG derivatives, unfunctionalized (−) and Arg3 (R3) functionalized, and resultant PARP cleavage. These data demonstrate C3KO-11’s ability to maintain activity against apoptotic substrates in cells and that R3 functionalization increased total protein delivered and subsequently increased the resultant PARP cleavage for various NG. Delivery of inactive C3KO-11 used as control to demonstrate lack of apoptosis from an inactive casp-3 cargo.
The co-solvent-assembled block copolymers can be therapeutically impactful, only if casp-3 retains activity upon encapsulation. To ensure that co-solvents did not prevent the ability of C3KO-11 to generate GFP fluorescence for SEE, we tested NG-released reassembly. Released C3KO-11 demonstrated robust fluorescence in the presence of GFP1-10 (data not shown), demonstrating that DMSO-mediated encapsulation did not negatively impact split GFP reassembly. Furthermore, to ensure that co-solvent encapsulation did not irreversibly denature or otherwise inactivate casp-3 , we encapsulated active WT casp-3 in block copolymer NGs which should activate apoptosis and result in poly (ADP-ribose) polymerase 1 (PARP) cleavage62,63 if co-solvent encapsulated WT casp-3 remained active. Co-solvent block copolymer casp-3 NG delivery resulted in significant disappearance of full-length PARP and appearance of cleaved PARP, indicating that active casp-3 was effectively delivered in both accounts. In contrast, no significant apoptosis-induction was induced from empty NG prepared using co-solvent methodologies (Figure 3D). Likewise, delivering the active casp-3 variant capable of split GFP (C3-11)28 in aqueous random or co-solvent block formulations again displayed significant PARP cleavage (Figure 3E). Together these results suggest that both aqueous and co-solvent formulations allow delivery of active casp-3.
Despite the increased casp-3 encapsulation efficiencies in the block copolymer assemblies, levels of SEE-mediated fluorescence were identical to aqueous formulations. These data suggest that endosomal escape rates are better for the aqueously formulated block co-polymer NG, since the same number of casp-3 molecules would be contained in a greater number of aqueous NG. We reasoned that addition of surface functionalization that could increase uptake or endosomal escape could increase cytosolic delivery of casp-3. In line with that prediction, NGs functionalized on the surface with a tri-arginine peptide (R3) demonstrated higher levels of both casp-3 delivery and subsequent PARP cleavage (Figure 3E). Given this result and we next investigated the effect of cationic surface functionalization on SEE.
Guanidine- and other positively-charged moieties on the surface of delivery vehicles increase membrane-association, permit cell entry through endocytosis and aid in endosomal escape.64-67 Our group has utilized varying cationic moieties, such as arginine-repeat peptides39,44 ,68 or TAT69, to functionalize the NG surface allowing for the addition of surface-exposed guanidine groups. To assess the impact of surface functionalization on casp-3 delivery, C3KO-11 PEG-PDS NGs were functionalized via thiol exchange (Figure S1) with PEG-amine, arginine peptides of various lengths (R3, R6, R9) or TAT (Figure S7).39 Fortunately, in addition to spectroscopic assessment, reductant-mediated release of higher degree oligopeptides can be observed by SDS-PAGE analysis (Figure S7). To ensure identical protein encapsulation and crosslinking, NGs were always prepared as a large batch prior to functionalization which resulted in similarly-sized NGs for post-functionalization (Figure S7).
Following treatment of cells with Arg-functionalized NG, we observed a 3- to 6-fold increase in the number of GFP-positive cells due to SEE (Figure 4A) relative to unfunctionalized NG, strongly suggesting that cationic NGs promote endosomal escape. Upon the addition of CQ, cationic functionalized NGs demonstrated an additional 4- to 13-fold increase in GFP signal compared to non-functionalized NGs in the presence of CQ (Figure 4B). In the absence of CQ, we observed 1.2-1.4 fold increase in the GFP mean fluorescence intensity (MFI) with CQ present (Figure 4C). Although minimal, this increase was evident as we previously observed no significant GFP positive population for the non-functionalized NG and correspondingly, no change in MFI. However, the addition of CQ significantly increased the GFP MFI further, 2.4-4.5 fold. Due to the effects of CQ, we hypothesize that this increase could be attributed Arg-mediated increases in the concentration of NGs per endosome. However, we cannot rule out the possibility there may also be more endosomes to accommodate increased numbers of NG.29 CQ-liberated endosomally entrapped R3-NG were also readily visualized by microscopy (Figure 4E, supplemental images Figure S8). In summary, arginine-functionalized caspase-containing NG demonstrated enhanced endosomal escape, although the results with CQ imply that a significant population of NGs still remain trapped in endosomes. Therefore, as noted for other delivery systems,24,25 arginine moieties enhance general uptake but these data demonstrate that arginine can also directly enhance endosomal escape, although not necessarily to 100% of the NG particles uptaken. To further probe the uptake mechanism, bafilomycin, an ATPase inhibitor that prevents endosomal acidification, was applied to NG treated cells.52,70,71 SEE was significantly decreased in the presence of bafilomycin for R3-NG (Figure 4F), in the absence of presence of CQ. SEE was also decreased for TAT-NG (Figure 4G), underscoring our assertion that arginine protonation and CQ influences endosomal escape through the proton sponge effect.64
Figure 4. NG functionalization with cationic peptides enhances endosomal escape.
(A) SEE after 24-hour incubation with surface modified PEG-PDS NG complexes at 0.75 mg/mL polymer concentration in HEKs1-10 cells indicates that Arg-functionalization aids in endosomal escape. SEM error bars pertain to individual biological replicates from independent NG batches, analyzed on different days. (B) In the presence of 80 μM CQ, a significant increase in fluorescence was observed suggesting that Arg-functionalization may also increase number of NG within endosomes. (C) SEE represented as change in GFP mean fluorescent intensity (MFI) of cells in CQ-free media. The change in MFI is generated after normalizing GFP MFI to the untreated, viable single-cell HEKs1-10 population. (D) In the presence of 80 μM CQ. (E) Visualization of CQ-liberated endosomes after 24-hour incubation with R3-PEG-PDS NG (20X objective: BF (bright-field), DAPI (nucleus stain, Ex. 405 nm), GFP (reassembly of C3KO-11 and GFP1-10, Ex. 488 nm), scale bar indicates approximately 50 microns). (F) SEE of R3-PEG-PDS NG and (G) TAT-PEG-PDS NG with and without CQ and bafilomycin, an endosomal acidification inhibitor. For (A-D), one-way ANOVA performed against PEG-PDS where *p < 0.05; **p < 0.01; ***p <0.001; ****p < 0.0001. If no label present, no significant (n.s.) differences were found. For (F and G), individual unpaired t-tests were performed.
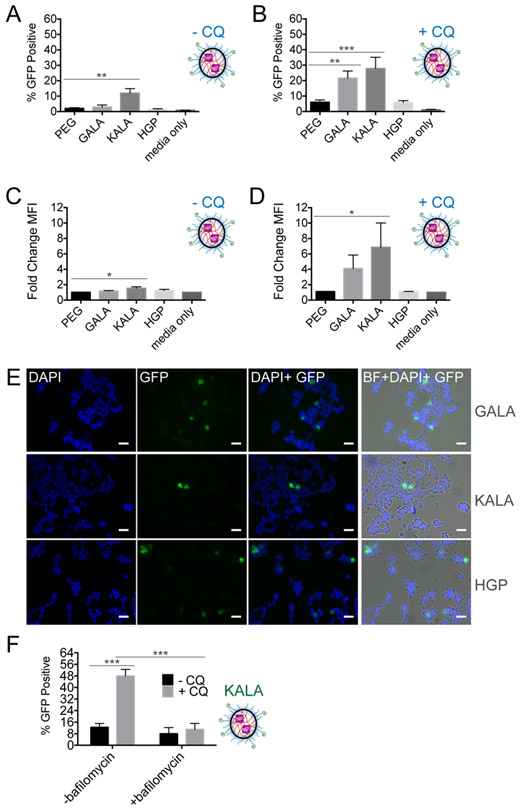
Having shown that NG surface functionalization influences translocation, we reasoned that improved delivery may be achievable by exploiting the inherent pH decrease upon early-to-late-endosomal processing. To date, surface modifications of polymeric NG with peptides capable of pH-responsive conformational changes has yet to be investigated. Importantly, pH-responsive functionalities are hypothesized to permit endosomal escape through mechanisms beyond the proton sponge alone, i.e., via a combination of rapid osmotic swelling and membrane destabilization leading to endosomal rupture.72-74 Inspired by the N-terminal anionic domain of an Influenza virus protein, synthetic pH-sensitive peptides composed of varying glutamic and aspartic acid residues have been generated to explore their membrane-disrupting properties.13,75 GALA, a 30-mer glutamic acid-alanine-leucine-alanine repeat, and KALA, a cationic derivative composed of lysine-alanine-leucine-alanine repeats, have been applied to enhance endosomal escape of cationic liposomes, cationic polymers and antibodies.13,76 Due to their length and hydrophobicity, truncated versions of these peptides,77 as well as another peptide based on the endodomain of the HIV enveloped glycoprotein gp41, termed HGP,78,79 can be more easily used. We incorporated GALA, KALA and HGP onto the surface of our nanogels by adding a C-terminal cysteine to the peptides,75,79-82 for reaction with PEG-PDS (Figure S9). SEE analysis demonstrated that only functionalization with KALA resulted in significant GFP signal, independent of CQ (Figure 5A). These data suggest that the combination of pH-responsive and cationic properties by KALA affords more endosomal escape than the pH-responsive properties alone in GALA and HGP. As aforementioned, often the enhancement of endosomal escape is compromised due to the toxicity associated with these peptides, which was observed for KALA at high degrees of functionalization (Figure S10). Halving the dose of KALA-NG decreased the induced cellular debris (cell death), but correspondingly decreased the subsequent amount of GFP fluorescence observed (17% GFP positive at 1 mg/mL dose vs 5% GFP positive at 0.5 mg/mL dose). These data imply that NG dose influenced the amount of NG per endosome. GALA and HGP NG appear to remained trapped until endosome acidification. We observed a significant increase in GFP positive cells (Figure 5B) and fold change in GFP fluorescence (Figure 5C,5D) upon the addition of CQ which could subsequently be visualized by microscopy (Figure 5E, supplemental images S11). As pH-responsive moieties may be facilitating endosomal escape through a proton sponge effect, we assayed KALA-NG in the presence of bafilomycin. We found statistical significance between KALA-NG, with and without CQ, as well as KALA-NG with CQ, with and without bafilomycin but did not find significance for KALA-NG with and without bafilomycin (Figure 5F). Interestingly, GALA and HGP demonstrated a clear functionalization-dependent response with CQ (Figure S12), confirming that there is still significant endosomal entrapment. These data may suggest that the extent of GALA and HGP functionalization influences the amount of NG per endosome or that higher amounts of these functionalities may work additively with CQ.
Figure 5. NG functionalization with pH-sensitive peptides influences endosomal escape.
(A) SEE after 24-hour incubation with surface modified PEG-PDS NG complexes at 0.75 mg/mL polymer concentration in HEKs1-10 cells and (B) In the presence of 80 μM CQ. SEM error bars pertain to individual biological replicates from independent NG batches, analyzed on different days. (C) SEE represented as change in GFP mean fluorescent intensity (MFI) of cells in CQ-free media. The change in MFI is generated after normalizing GFP MFI to the untreated, viable single-cell HEKs1-10 population. (D) In the presence of 80 μM CQ. (E) Visualized CQ-liberated endosomes after 24-hour incubation with the different pH-responsive-NG (20X fluorescent microscope objective: BF (bright-field), DAPI (nucleus stain, 405 nm), GFP (reassembly of C3KO-11 and GFP1-10, 488 nm)). SEM error bars pertain to four individual biological replicates from independent NG batches analyzed on different days. (F) SEE of KALA-PEG-PDS NG, with and without CQ and bafilomycin, an endosomal acidification inhibitor. For (A-D), one-way ANOVA performed against PEG-PDS where *p < 0.05; **p < 0.01; ***p <0.001; ****p < 0.0001. If no label present, no significant (n.s.) differences were found. For (F), individual unpaired t-tests were performed.
A major goal of this work has been to develop a method that allows robust comparison of delivery vehicles with disparate chemical properties. For this reason, we included in our analysis a delivery vehicle with dramatically different chemical characteristics, mesoporous silica (MSi) nanoparticles. We had previously developed MSi nanoparticles that effectively escape the endosome.83 To prepare casp-3-congugated MSi, lysine residues on C3KO-11 were functionalized with redox-responsive linkers capable of self-immolation (linker structure Figure S13), for traceless release of C3KO-11. These redox-responsive linkers contain a disulfide, PEG and a terminal aryl boronic acid moiety (Figure S13, linker structure 2). MSi-protein attachment is achieved through dative bond formation of the boronic acid functionalized protein and amine functionalized MSi.83 Efficient endosomal escape by MSi is hypothesized to be due to the proton sponge effect, as dissociation between the protein and MSi facilitates exposure of the amine group on MSi surface.30,64 Accordingly, MSi facilitated effective protein delivery of C3KO-11 to MCF7 cells, visible by immunoblot (Figure S13). In both HEK293T cells transfected with GFP1-10 and HEKs1-10, MSi also demonstrated effective casp-3-dependant GFP signal by SEE (Figure 6). MSi with smaller pore sizes demonstrated greater GFP positive populations (Figure 6A), with 7-fold change in GFP fluorescence (Figure 6B), compared to MSi with larger pore sizes. The evolution of the GFP signal can be observed in a dose-dependent manner (Figure 6C) and as a function of time (Figure 6D). Interestingly, it appears that removal of the PEG spacer from the redox-responsive linker (Figure S13, linker structure 1) comparably decreased GFP signal (Figure 6D), perhaps due to limiting GSH accessibility to the disulfide within the linker. Finally, MSi-mediated SEE signal doubles upon the addition of CQ (Figure 6E), indicating that some MSi remain trapped in endosomes. These data demonstrate that C3KO-11 delivery can be translated to other delivery systems with a range of assembly chemistries, permitting direct comparison of the resultant cytosolic protein levels by different nanomaterials. Hence, MSi mediated delivery of C3KO-11 resulted in higher cytosolic C3KO-11 levels than both unfunctionalized and functionalized polymeric NG, in the absence of CQ. Furthermore, we hypothesize that we can extrapolate the extent of “change” due to CQ as an indicator of the percentage of nanomaterial entrapment. The number of GFP positive cells from MSi C3KO-11 increased only 2-fold in the presence of CQ (Figure 6E), while functionalized polymeric NG delivering C3KO-11 increased over wider ranges, ~4-7-fold upon cationic functionalization (Figure 4B) and ~3-7-fold for pH-responsive functionalization (Figure 5B). These data may indicate that only half of MSi C3KO-11 complexes are entrapped, leaving half of the uptaken material to reach the cytosol for therapeutic effect. On the other hand, these calculations imply that only 25-33% of the polymeric materials may reach cytosol, leaving ~67-80% of the engulfed material endosomally entrapped. Correspondingly, we hypothesize that MSi mediated delivery of active casp-3 would likewise induce higher levels of cell death, a planned future investigation.
Figure 6. Detecting cytosolic delivery of C3KO-11 via mesoporous silica (MSi) nanoparticles by SEE.
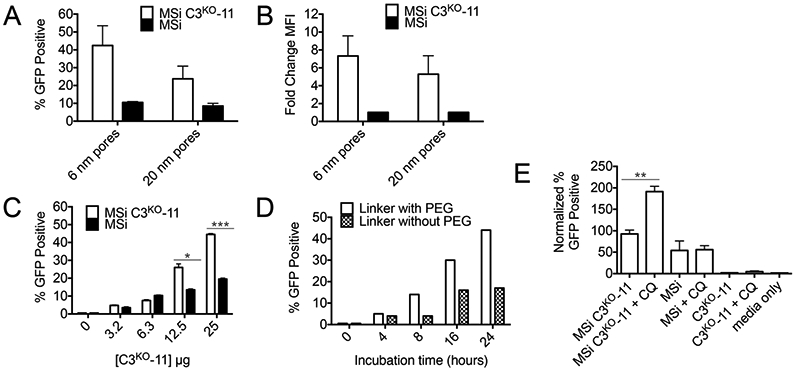
(A) Influence of MSi pore size on SEE after 24-hour incubation with MSi complexes in HEK293T cells transfected with GFP1-10 and (B) Change in MFI. (C) Dose dependence of SEE after 24-hour incubation with MSi complexes in HEKs1-10 cells. (D) Effect of incubation time on SEE. (E) SEE improvement of various MSi complexes in the presence of 100 μM CQ. For (C) two-way ANOVA performed and for (E) unpaired t-test (two-tailed) executed where *p < 0.05; **p < 0.01; ***p <0.001; ****p < 0.0001. If no label present, no significant (n.s.) differences were found. For (F), individual unpaired t-tests were performed.
Conclusions:
Known caspase properties, such as their catalytic ability to induce and propagate apoptosis through rapid cleavage of other procaspases and cellular substrates, suggest that low levels of cytosolic casp-3 would be sufficient to achieve effective cell death. Prior to this work, we lacked a robust method to estimate the amount of delivered casp-3 that reached the cytosol. Herein we utilized C3KO-11, capable of reassembly with GFP1-10, to generate fluorescence and report on casp-3 cytosolic localization in cells. SEE allowed effective monitoring of only cytosolic protein levels, after endosomal escape, and importantly lacked false positive results.
SEE was instrumental in evaluating the role of polymer composition and NG surface functionality in cytosolic delivery. In the field of cell death, the level of caspase activation required to induce apoptosis has not been quantified. Undertaking these studies, we anticipated that this approach would allow quantification of the threshold level of active casp-3 required for apoptosis. We were surprised to observe that the levels of casp-3 required to trigger apoptosis induction were so low that even a sensitive technique like SEE was not effective at quantification, indicating that the threshold casp-3 activation levels must be less than 10 μM.54 Nevertheless, we did observe that increasing protein cargo encapsulation efficiencies within NGs did not enhance SEE, while we know that changing the protein cargo’s sequence can influence SEE.28 Consistent with this effect, changing NG surface functionalization demonstrated significant improvements while altering delivery vehicle architecture did not. Thus, the low fluorescence signal from non-functionalized NG SEE contributes to the conclusion that i) SEE is not sensitive enough to detect very low levels of non-functionalized NG endosomal escape and ii) the amount of endosomal escape truly occurring without functionalization or lysosomotropic agents is low. Importantly this method allows direct comparison of commonly used approaches, such as cationic functionalization, to increase the amount of delivered protein that reaches the cytosol. Furthermore, we can compare the efficiency of different delivery vehicles, that vary by composition, surface functionalization and cargo tagging methodology, by signal generated solely from cytosolic protein cargo.
Materials and Methods:
C3-11 variant expression and purification –
Generation of C3KO-11 and C3-11 variants have been described previously.28 pET 23b plasmid encoding human WT casp-3, or variant, was transformed into BL21(DE3) E.coli cells via electroporation and plated on agar plates containing ampicillin. Single colony cultures were grown in 50 mL LB media with the corresponding antibiotic at 37 °C overnight. The following day 8L of LB was inoculated with ~5mL per L of the small seed culture and grown at 37 °C until an OD600 of ~0.6 was achieved. The incubation temperature was then reduced to 25 °C and cells were induced with a final concentration of 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and left to express for ~3 hours. Cells were then harvested via centrifugation for 10 minutes at 4700Xg and stored at −80 °C. Cell pellets were thawed and lysed using a microfluidizer (Microfluidics, Inc.) in a buffer containing 50 mM Na3PO4, 300 mM NaCl, 2 mM imidazole, pH 8. Lysed cells were centrifuged at 30,600xg for 55 minutes to remove cellular debris. The lysate supernatant was then loaded onto a pre-charged 5 mL HiTrap Ni-affinity column (GE Healthcare) and the column was subsequently washed with lysis buffer. Following the lysis wash, the column was further washed with an increased imidazole buffer, 50 mM, and the protein was finally eluted using a linear gradient to 300 mM imidazole. The eluted protein was diluted 7-fold in a buffer containing 20 mM Tris and 2 mM DTT, pH 8. This ~175 mL solution was then loaded onto a 5 mL HiTrap Q column (GE Healthcare) and finally eluted using a linear NaCl steep gradient in 20 mM Tris, 2 mM DTT, pH 8. The Q-fractions were analyzed for purity via SDS-PAGE and concentration concluded via A280 absorbance, using molar extinction coefficients ~25,900 M −1 cm−1 and subsequently stored at −80 °C.
Casp-3 variant random copolymer NG formation –
Casp-3 NG were formulated similarly to previous methods.39 20 mg of the appropriate random PEG-PDS derivative was added to a vial and dissolved in 1 mL 1X PBS, pH 7.4, and mixed via sonication for a final concentration of 20 mg/mL polymer. This polymer stock solution was stirred at 4 °C for ~1 hour before 0.5 mL was aliquoted into a new vial. To the 10 mg polymer aliquot, a casp-3 variant solution was added at a weight ratio of 25:1 polymer:protein with additional 1X PBS to achieve a final concentration of 10 mg/mL polymer(or a final volume of 1 mL). The polymer-protein solution was left to stir for ~3 hours at 4 °C. The conjugates were then crosslinked using an equivalent of dithiothreitol sufficient for 20-40% crosslinking (0.04 mg) and additional stirring for ~1 hour at 4 °C. Excess of a thiol containing ligand (two equivalents of the remaining PDS groups after crosslinking), such as a cysteine terminated arginine peptide used herein, was then added and stirred for two hours. The NG conjugates were then purified via dialysis against 1X PBS pH 7.4 using a 100kDa MWCO aqueous membrane for ~20 hours at 4 °C with multiple buffer changes. Dialysis against this large membrane allows removal of the crosslinking byproduct, excess ligand, DTT and unencapsulated protein simultaneously. Prior to dialysis, crosslinking and functionalization were quantified using 2 μL of the NG solution + 98 μL water ± excess DTT and recording the absorbance of the crosslinking byproduct at 343 nm using UV-Vis spectroscopy.
Casp-3 variant block copolymer aqueous NG formation –
Casp-3 NG were formulated by dissolving 5 mg of the appropriate block PEG-PDS derivative in 0.5 mL 1X PBS, pH 7.4, via sonication for a final concentration of 5 mg/mL polymer, due to polymer solubility limitations. Using constant vortexing and sonication the polymers took several hours to dissolve in PBS. Next, a casp-3 variant solution was added at a weight ratio of 25:1 polymer:protein with additional 1X PBS to achieve a final concentration of 5 mg/mL polymer. The polymer-protein solution was left to stir for ~3 hours at 4 °C. The conjugates were then crosslinked, functionalized, dialyzed and characterized as described for the random copolymer NG.
Casp-3 variant block copolymer co-solvent NG formation –
Casp-3 NG were formulated by dissolving 10 mg of the appropriate block PEG-PDS derivative in 0.1 mL DMSO (10% of the final volume) via sonication and stirring at room temperature for ~ 15 minutes. To this solution, 0.9 mL of 1X PBS containing the appropriate amount of a casp-3 variant (ratio still 25:1) was added dropwise with stirring at room temperature. This vial was immediately moved to stir for ~3 hours at 4 °C. The conjugates were then crosslinked, functionalized, dialyzed and characterized as described for the random copolymer NG.
NG mediated protein encapsulation and release –
Purified nanogels were removed from dialysis and volumes normalized. To visualize protein release, 30 μL of the nanogel solution was incubated with either 10 μL of 1M DTT or autoclaved water and left for 15 minutes at RT. Next, 10 μL of SDS-PAGE 3X dye (with reductant) was added to the DTT-NG sample and reductant free SDS-PAGE 3X dye was added to the water-NG sample. The samples were immediately boiled at 95 °C for ~5 minutes and then added to a 16% SDS-PAGE and electrophoresis was executed at 175V for 60 minutes. Control protein samples were prepared using 30 μL of 10 μM protein and 10 μL of SDS-PAGE 3X dye, subsequently adding 10, 5, 2.5 and 1.25 μL to the gel, respectively. To compare encapsulation efficiencies, only when ran on the same gel, full-length C3KO-11 band intensities of different concentrations were made into a calibration curve and compared to NG-released C3KO-11 using Image Lab™ Software.
SEE Flow Cytometry experiments –
Two days prior to the assay, HEKs1-10 cells were plated at a density of ~5 x 104 in a 24-well plate and left to adhere for ~24 hours. Upon reaching confluence, cells were treated with purified nanomaterial diluted in a mixture of complete DMEM (Gibco #11965) and 10% v/v 1X PBS, pH 7.4. When CQ was used, a 100 mM CQ stock was freshly prepared weekly using milliQ water. On the day of the experiment, the 100 mM stock was freshly diluted into media (12.5-100 μM) and NG were diluted into these CQ-containing medias. If not specifically mentioned, 100 μM was used. After ~24 hours (unless otherwise noted), the cell incubation media was removed and cells were washed twice with 1X PBS (Gibco, Thermofisher) and incubated with 100 μL of 1X trypsin solution (prepared from 0.5% Trypsin-EDTA, Gibco #15400054) for ~5 minutes at 37 °C. 200 μL of FACS buffer (sterile filtered 0.5% BSA in 1X PBS, pH 7.4) was added to the wells and the entire solution was mixed then transferred to FACS compatible tubes. Samples were immediately assayed with 488 nm GFP laser line. Samples were analyzed first using a morphological gate (forward scatter-area vs. side scatter-area (FSC-A vs. SSC-A)) followed by a gate to include only single cells (FSC-A vs. forward scatter-height (FSC-H)). Finally, this single-cell population was gated against the media only (un-treated, cells only) control to have ~0.5-1% GFP positive cells (histogram of FITC-A). Percent GFP positive (GFP+) values were reported as quantified by the FlowJo, LLC software. For GFP MFI, samples were normalized to the media-only control. Data were analyzed by one-way ANOVA, with p-values calculated with GraphPad Prism 6.0. Differences were considered statistically significant when p≤ 0.05. P-values are illustrated with asterisks where *p < 0.05; **p < 0.01; ***p <0.001; ****p < 0.0001.
Bafilomycin Treatment –
Prior to NG addition to HEKs1-10 cells, cells were treated with bafilomycin (50 nM) containing media for forty-five minutes. Subsequently, bafilomycin media was removed and replaced with fresh bafilomycin-free media in the presence or absence of 80 μM chloroquine. NG were subsequently added and left to incubate for 24 hours before SEE analysis.
Protein modification with boronic acid linkers for inclusion in MSi NP –
120 μL C3KO-11 (123 μM, 0.46 mg), 200 μL H2O and 40 μL NaHCO3 solution (0.5 M) were dissolved in a 5 mL vial under stirring at room temperature. To the vial, 10 μL DMSO solution containing 1.5 mg linker 1 or 2 mg linker 2 was added (Figure S13). The reaction mixture was then stirred at room temperature for additional 6 h, followed by ultrafiltration purification with Amicon® Ultra Centrifugal Filters (MWCO = 3 kDa) for 5 times. The final modified protein was dissolved in 230 μL distilled deionized water (2 mg/mL) and stored at −20 °C.
Supplementary Material
Acknowledgment:
This work was supported by NIH R01 GM080532 and GM128181. Francesca Anson was supported by the UMASS BTP Program (NIH T32 GM108556). We thank the B. Huang lab at UCSF for the HEK293 cell line stably expressing GFP1-10. We likewise thank the Dr. Jim Chambers at the UMASS Light Microscopy Center as well as Dr. Amy Burnside at the UMASS Flow Cytometry center. We also thank Christopher Hango, Tew Lab at UMASS for productive scientific conversations.
Abbreviations:
- Casp-3
cysteine aspartate protease-3
- SEE
split-complementation endosomal escape
- WT
wild-type, native
- NG
nanogel
- NP
nanoparticle
- GFP1-10
superfolder GFP 1-10 optimized
- GFP11
superfolder GFP 11
- PEG
polyethylene glycol
- PDS
pyridyl disulfide
- PEG-PDS
polymer of PEG and PDS
- PP
PEG-PDS random copolymer
- PPI
PEG-PDS-IPA, isopropylamine, random copolymer
- PPB
PEG-PDS-Butyl random copolymer
- PbP
PEG-PDS block copolymer
- PbPB
PEG-b-(PDS-r-Bu) block copolymer
- GSH
glutathione
- DTT
dithiothreitol
- PBS
phosphate-buffered saline
- CQ
chloroquine diphosphate
- z-VAD-fmk
benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone
- DEVD-AMC
peptide N-acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin
- SDS-PAGE
SDS-polyacrylimide gel electrophoresis
- FCA
flow cytometry analysis
- TNG
20 mM Tris, 100 mM NaCl, 10% Glycerol, pH 7.5 buffer
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at https://pubs.acs.org. Materials & methods for synthesis and characterization of polymers as well as procedures for NG characterization (DLS, protein encapsulation and release, immunoblotting, additional microscopy photos) and silica synthesis and protein encapsulation are included. Supplemental Figures S1-S13 are present. The authors declare no competing financial interest.
References:
- (1).Gu Z; Biswas A; Zhao M; Tang Y Tailoring Nanocarriers for Intracellular Protein Delivery. Chem. Soc. Rev 2011, 7, 3638–3655. [DOI] [PubMed] [Google Scholar]
- (2).Galliani M; Tremolanti C; Signore G Nanocarriers for Protein Delivery to the Cytosol: Assessing the Endosomal Escape of Poly(Lactide-Co-Glycolide)-Poly(Ethylene Imine) Nanoparticles. Nanomaterials 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Leader B; Baca QJ; Golan DE Protein Therapeutics: A Summary and Pharmacological Classification. Nat. Rev. Drug Discov 2008, 1, 21–39. [DOI] [PubMed] [Google Scholar]
- (4).Dipak SP; Kosloski MP; Balu-Iyer SV Delivery Of Therapeutic Proteins. J Pharm Sci. 2010, 6, 2557–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lv J; Fan Q; Wang H; Cheng Y Polymers for Cytosolic Protein Delivery. Biomaterials 2019, 119358. [DOI] [PubMed] [Google Scholar]
- (6).Ren L; Lv J; Wang H; Cheng Y A Coordinative Dendrimer Achieves Excellent Efficiency in Cytosolic Protein and Peptide Delivery. Angew. Chemie - Int. Ed 2020, 4711–4719. [DOI] [PubMed] [Google Scholar]
- (7).Zhang Y; Røise JJ; Lee K; Li J; Murthy N Recent Developments in Intracellular Protein Delivery. Curr. Opin. Biotechnol 2018, 52, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gu Z; Biswas A; Zhao M; Tang Y Tailoring Nanocarriers for Intracellular Protein Delivery. Chem. Soc. Rev 2011, 7, 3638. [DOI] [PubMed] [Google Scholar]
- (9).Lu Y; Sun W; Gu Z Stimuli-Responsive Nanomaterials for Therapeutic Protein Delivery. J. Control. Release 2014, 194, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fu A; Tang R; Hardie J; Farkas ME; Rotello VM Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjug. Chem 2014, 9, 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dutta K; Hu D; Zhao B; Ribbe AE; Zhuang J; Thayumanavan S Templated Self-Assembly of a Covalent Polymer Network for Intracellular Protein Delivery and Traceless Release. J. Am. Chem. Soc 2017, 139, 5676–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Smith SA; Selby LI; Johnston APR; Such GK The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem 2019, 30, 263–272. [DOI] [PubMed] [Google Scholar]
- (13).Shete HK; Prabhu RH; Patravale VB Endosomal Escape: A Bottleneck in Intracellular Delivery. J. Nanosci. Nanotechnol 2014, 14, 460–474. [DOI] [PubMed] [Google Scholar]
- (14).Leopold PL Endosomal Escape Pathways for Delivery of Biologics. Lysosomes Biol. Dis. Ther 2016, 3, 383–407. [Google Scholar]
- (15).Szeto HH; Schiller PW; Zhao K; Luo G Fluorescent Dyes Alter Intracellular Targeting and Function of Cell-Penetrating Tetrapeptides. FASEB J. 2005, 1, 118–120. [DOI] [PubMed] [Google Scholar]
- (16).Snipstad S; Hak S; Baghirov H; Sulheim E; Mørch Ý; Lélu S; von Haartman E; Bäck M; Nilsson KPR; Klymchenko AS; et al. Labeling Nanoparticles: Dye Leakage and Altered Cellular Uptake. Cytom. Part A 2017, 8, 760–766. [DOI] [PubMed] [Google Scholar]
- (17).Dougherty CA; Vaidyanathan S; Orr BG; Banaszak H Fluorophore:Dendrimer Ratio Impacts Cellular Uptake and Intracellular Fluorescence Lifetime. Bioconjug Chem 2015, 2, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zanetti-Domingues LC; Tynan CJ; Rolfe DJ; Clarke DT; Martin-Fernandez M Hydrophobic Fluorescent Probes Introduce Artifacts into Single Molecule Tracking Experiments Due to Non-Specific Binding. PLoS One 2013, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jiang Z; Liu H; He H; Yadava N; Chambers JJ; Thayumanavan S Anionic Polymers Promote Mitochondrial Targeting of Delocalized Lipophilic Cations. Bioconjug. Chem 2020, 5, 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Li Y; Almassalha LM; Chandler JE; Zhou X; Stypula-Cyrus YE; Hujask KA; Roth EW; Bleher R; Subramanian H; Szleifer I; et al. The Effects of Chemical Fixation on the Cellular Nanostructure. Exp Cell Res 2017, 2, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hobro AJ; Smith NI An Evaluation of Fixation Methods: Spatial and Compositional Cellular Changes Observed by Raman Imaging. Vib. Spectrosc 2017, 91, 31–45. [Google Scholar]
- (22).Stanly TA; Fritzsche M; Banerji S; Garcia E; De La Serna JB; Jackson DG; Eggeling C Critical Importance of Appropriate Fixation Conditions for Faithful Imaging of Receptor Microclusters. Biol. Open 2016, 9, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Choi D-K; Park S; Kim J; Bae J; Shin S-M; Kim D-M; Yoo TH; Kim Y-S Quantitative Assessment of Cellular Uptake and Cytosolic Access of Antibody in Living Cells by an Enhanced Split GFP Complementation Assay. Biochem. Biophys. Res. Commun 2015, 467, 771–777. [DOI] [PubMed] [Google Scholar]
- (24).Milech N; Longville BA; Cunningham PT; Scobie MN; Bogdawa HM; Winslow S; Anastasas M; Connor T; Ong F; Stone SR; et al. GFP-Complementation Assay to Detect Functional CPP and Protein Delivery into Living Cells. Sci. Rep 2015, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lönn P; Kacsinta AD; Cui XS; Hamil AS; Kaulich M; Gogoi K; Dowdy SF Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep 2016, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Schmidt S; Adjobo-Hermans MJW; Wallbrecher R; Verdurmen WPR; Bovée-Geurts PHM; Van Oostrum J; Milletti F; Enderle T; Brock R Detecting Cytosolic Peptide Delivery with the GFP Complementation Assay in the Low Micromolar Range. Angew. Chemie - Int. Ed 2015, 54, 15105–15108. [DOI] [PubMed] [Google Scholar]
- (27).Bale SS; Kwon SJ; Shah DA; Kane RS; Dordick JS A GFP Complementation System for Monitoring and Directing Nanomaterial Mediated Protein Delivery to Human Cellular Organelles. Biotechnol. Bioeng 2010, 6, 1040–1047. [DOI] [PubMed] [Google Scholar]
- (28).Anson F; Kanjilal P; Thayumanavan S; Hardy JA Tracking Exogenous Intracellular Casp-3 Using Split GFP. Protein Sci. 2021, 30, 366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Thompson David B. Villasenor Roberto Dorr Brent M Zerial M Liu DR Cellular Uptake Mechanisms and Endosomal Trafficking of Supercharged Proteins. Chem Biol 2012, 7, 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Freeman EC; Weiland LM; Meng WS Modeling the Proton Sponge Hypothesis: Examining Proton Sponge Effectiveness for Enhancing Intracellular Gene Delivery through Multiscale. J Biomater Sci Poly Ed 2013, 4, 398–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Jiang Z; He H; Liu H; Thayumanavan S Cellular Uptake Evaluation of Amphiphilic Polymer Assemblies: Importance of Interplay between Pharmacological and Genetic Approaches. Biomacromolecules 2019, 12, 4407–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Siprashvili Z; Reuter JA; Khavari PA Intracellular Delivery of Functional Proteins via Decoration with Transporter Peptides. Mol. Ther 2004, 5, 721–728. [DOI] [PubMed] [Google Scholar]
- (33).Zassler B; Blasig IE; Humpel C Protein Delivery of Caspase-3 Induces Cell Death in Malignant C6 Glioma, Primary Astrocytes and Immortalized and Primary Brain Capillary Endothelial Cells. J. Neurooncol 2005, 2, 127–134. [DOI] [PubMed] [Google Scholar]
- (34).Hentzen NB; Mogaki R; Otake S; Okuro K; Aida T Intracellular Photoactivation of Caspase-3 by Molecular Glues for Spatiotemporal Apoptosis Induction. J. Am. Chem. Soc 2020, 18, 8080–8084. [DOI] [PubMed] [Google Scholar]
- (35).Fu J; Yu C; Li L; Yao SQ Intracellular Delivery of Functional Proteins and Native Drugs by Cell-Penetrating Poly(Disulfide)s. J. Am. Chem. Soc 2015, 37, 12153–12160. [DOI] [PubMed] [Google Scholar]
- (36).Tang R; Kim CS; Solfiell DJ; Rana S; Mout R; Velázquez-Delgado EM; Chompoosor A; Jeong Y; Yan B; Zhu ZJ; et al. Direct Delivery of Functional Proteins and Enzymes to the Cytosol Using Nanoparticle-Stabilized Nanocapsules. ACS Nano 2013, 8, 6667–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kim CS; Mout R; Zhao Y; Yeh YC; Tang R; Jeong Y; Duncan B; Hardy JA; Rotello VM Co-Delivery of Protein and Small Molecule Therapeutics Using Nanoparticle-Stabilized Nanocapsules. Bioconjug. Chem 2015, 5, 950–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Esteban-Fernández De Ávila B; Ramírez-Herrera DE; Campuzano S; Angsantikul P; Zhang L; Wang J Nanomotor-Enabled PH-Responsive Intracellular Delivery of Caspase-3: Toward Rapid Cell Apoptosis. ACS Nano 2017, 6, 5367–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ventura J; Eron SJ; González-Toro DC; Raghupathi K; Wang F; Hardy JA; Thayumanavan S Reactive Self-Assembly of Polymers and Proteins to Reversibly Silence a Killer Protein. Biomacromolecules 2015, 10, 3161–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Raghupathi K; Eron SJ; Anson F; Hardy JA Utilizing Inverse Emulsion Polymerization to Generate Responsive Nanogels for Cytosolic Protein Delivery. Mol. Pharm 2017, 4515–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Riedl SJ; Shi Y Molecular Mechanisms of Caspase Regulation during Apoptosis. Nat. Rev. Mol. Cell Biol 2004, 11, 897–907. [DOI] [PubMed] [Google Scholar]
- (42).Pop C; Salvesen GS Human Caspases: Activation, Specificity, and Regulation. J. Biol. Chem 2009, 284, 21777–21781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Raghupathi K; Thayumanavan S Nano-Armoring of Enzymes: Rational Design of Polymer-Wrapped Enzymes. Methods Enzymol. 2017, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ryu J; Bickerton S; Zhuang J; Thayumanavan S Ligand-Decorated Nanogels: Fast One-Pot Synthesis and Cellular Targeting †. Biomacromolecules 2012, 13, 1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Li L; Raghupathi K; Yuan C; Thayumanavan S Surface Charge Generation in Nanogels for Activated Cellular Uptake at Tumor-Relevant PH. Chem. Sci 2013, 4, 3654–3660. [Google Scholar]
- (46).Jiwpanich S; Ryu JH; Bickerton S; Thayumanavan S Noncovalent Encapsulation Stabilities in Supramolecular Nanoassemblies. J. Am. Chem. Soc 2010, 31, 10683–10685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Liu B; Thayumanavan S Importance of Evaluating Dynamic Encapsulation Stability of Amphiphilic Assemblies in Serum. Biomacromolecules 2017, 12, 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Xiong X-B; Binkhathlan Z; Molavi O; Lavasanifar A Amphiphilic Block Co-Polymers: Preparation and Application in Nanodrug and Gene Delivery. Acta Biomater. 2012, 6, 2017–2033. [DOI] [PubMed] [Google Scholar]
- (49).Kataoka K; Harada A; Nagasaki Y Block Copolymer Micelles for Drug Delivery: Design, Characterization and Biological Significance. Adv. Drug Deliv. Rev 2001, 47, 113–131. [DOI] [PubMed] [Google Scholar]
- (50).Moraes J; Peltier R; Gody G; Blum M; Recalcati S; Klok HA; Perrier S Influence of Block versus Random Monomer Distribution on the Cellular Uptake of Hydrophilic Copolymers. ACS Macro Lett. 2016, 12, 1416–1420. [DOI] [PubMed] [Google Scholar]
- (51).Kamiyama D; Sekine S; Barsi-Rhyne B; Hu J; Chen B; Gilbert LA; Ishikawa H; Leonetti MD; Marshall WF; Weissman JS; et al. Versatile Protein Tagging in Cells with Split Fluorescent Protein. Nat. Commun 2016, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jiang X; Yu Y; Chen J; Zhao M; Chen H; Song X; Matzuk AJ; Carroll SL; Tan X; Sizovs A; et al. Quantitative Imaging of Glutathione in Live Cells Using a Reversible Reaction-Based Ratiometric Fluorescent Probe. ACS Chem. Biol 2015, 3, 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Forman HJ; Zhang H; Rinna A Glutathione: Overview of Its Protective Roles, Measurement, and Biosynthesis. Mol Asp. Med 2009, 30, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Teo SLY; Rennick JJ; Yuen D; Al-Wassiti H; Johnston APR; Pouton CW Unravelling Cytosolic Delivery of Endosomal Escape Peptides with a Quantitative Endosomal Escape Assay (SLEEQ). bioRxiv 2020, 2020.08.20.258350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Pelt J; Busatto S; Ferrari M; Thompson EA; Wolfram J Chloroquine and Nanoparticle Drug Delivery: A Promising Combination. Pharmacol. Ther 2018, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Erbacher P; Roche AC; Monsigny M; Midoux P Putative Role of Chloroquine in Gene Transfer into a Human Hepatoma Cell Line by DNA/Lactosylated Polylysine Complexes. Exp. Cell Res 1996, 1, 186–194. [DOI] [PubMed] [Google Scholar]
- (57).Heath N; Osteikoetxea X; De Oliveria TM; Lázaro-Ibáñez E; Shatnyeva O; Schindler C; Tigue N; Mayr LM; Dekker N; Overman R; et al. Endosomal Escape Enhancing Compounds Facilitate Functional Delivery of Extracellular Vesicle Cargo. Nanomedicine 2019, 21, 2799–2814. [DOI] [PubMed] [Google Scholar]
- (58).Turner JJ; Ivanova GD; Verbeure B; Williams D; Arzumanov AA; Abes S; Lebleu B; Gait MJ Cell-Penetrating Peptide Conjugates of Peptide Nucleic Acids (PNA) as Inhibitors of HIV-1 Tat-Dependent Trans-Activation in Cells. Nucleic Acids Res. 2005, 21, 6837–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Du Rietz H; Hedlund H; Wilhelmson S; Nordenfelt P; Wittrup A Imaging Small Molecule-Induced Endosomal Escape of SiRNA. Nat. Commun 2020, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cervia LD; Chang CC; Wang L; Yuan F Distinct Effects of Endosomal Escape and Inhibition of Endosomal Trafficking on Gene Delivery via Electrotransfection. PLoS One 2017, 2, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Dallüge R; Haberland A; Zaitsev S; Schneider M; Zastrow H; Sukhorukov G; Böttger M Characterization of Structure and Mechanism of Transfection-Active Peptide-DNA Complexes. Biochim. Biophys. Acta - Gene Struct. Expr 2002, 1–2, 45–52. [DOI] [PubMed] [Google Scholar]
- (62).Walsh JG; Cullen SP; Sheridan C; Lüthi AU; Gerner C; Martin SJ Executioner Caspase-3 and Caspase-7 Are Functionally Distinct Proteases. Proc. Natl. Acad. Sci 2008, 35, 12815–12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).McComb S; Chan PK; Guinot A; Hartmannsdottir H; Jenni S; Dobay MP; Bourquin JP; Bornhauser BC Efficient Apoptosis Requires Feedback Amplification of Upstream Apoptotic Signals by Effector Caspase-3 or −7. Sci. Adv 2019, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Bus T; Traeger A; Schubert US The Great Escape: How Cationic Polyplexes Overcome the Endosomal Barrier. J. Mater. Chem. B 2018, 43, 6904–6918. [DOI] [PubMed] [Google Scholar]
- (65).El-Sayed A; Futaki S; Harashima H Delivery of Macromolecules Using Arginine-Rich Cell-Penetrating Peptides: Ways to Overcome Endosomal Entrapment. AAPS J. 2009, 1, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Fuchs SM; Raines RT Pathway for Polyarginine Entry into Mammalian Cells. Biochemistry 2004, 9, 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Najjar K; Erazo-Oliveras A; Mosior JW; Whitlock MJ; Rostane I; Cinclair JM; Pellois JP Unlocking Endosomal Entrapment with Supercharged Arginine-Rich Peptides. Bioconjug. Chem 2017, 12, 2932–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Gonza DC; Ryu J; Chacko RT; Zhuang J; Thayumanavan S Concurrent Binding and Delivery of Proteins and Lipophilic Small Molecules Using Polymeric Nanogels. J. Am. Chem. Soc 2012, 6964–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ryu J; Jiwpanich S; Chacko R; Bickerton S; Thayumanavan S Surface-Functionalizable Polymer Nanogels with Facile Hydrophobic Guest Encapsulation Capabilities. J. Am. Chem. Soc 2010, 132, 8246–8247. [DOI] [PubMed] [Google Scholar]
- (70).Brock DJ; Kustigian L; Jiang M; Graham K; Wang TY; Erazo-Oliveras A; Najjar K; Zhang J; Rye H; Pellois JP Efficient Cell Delivery Mediated by Lipid-Specific Endosomal Escape of Supercharged Branched Peptides. Traffic 2018, 6, 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Cervia LD; Chang CC; Wang L; Yuan F Distinct Effects of Endosomal Escape and Inhibition of Endosomal Trafficking on Gene Delivery via Electrotransfection. PLoS One 2017, 2, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Creusat G; Rinaldi AS; Weiss E; Elbaghdadi R; Remy JS; Mulherkar R; Zuber G Proton Sponge Trick for PH-Sensitive Disassembly of Polyethylenimine-Based Sirna Delivery Systems. Bioconjug. Chem 2010, 5, 994–1002. [DOI] [PubMed] [Google Scholar]
- (73).Peeler DJ; Sellers DL; Pun SH PH-Sensitive Polymers as Dynamic Mediators of Barriers to Nucleic Acid Delivery. Bioconjug. Chem 2019, 2, 350–365. [DOI] [PubMed] [Google Scholar]
- (74).Tang H; Zhao W; Yu J; Li Y; Zhao C Recent Development of PH-Responsive Polymers for Cancer Nanomedicine. Molecules 2019, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Li W; Nicol F; Szoka FC GALA: A Designed Synthetic PH-Responsive Amphipathic Peptide with Applications in Drug and Gene Delivery. Adv. Drug Deliv. Rev 2004, 7, 967–985. [DOI] [PubMed] [Google Scholar]
- (76).Morris MC; Chaloin L; Heitz F; Divita G Translocating Peptides and Proteins and Their Use for Gene Delivery. Curr. Opin. Biotechnol 2000, 5, 461–466. [DOI] [PubMed] [Google Scholar]
- (77).Turk MJ; Reddy JA; Chmielewski JA; Low PS Characterization of a Novel PH-Sensitive Peptide That Enhances Drug Release from Folate-Targeted Liposomes at Endosomal PHs. Biochim. Biophys. Acta - Biomembr 2002, 1, 56–68. [DOI] [PubMed] [Google Scholar]
- (78).Kwon EJ; Bergen JM; Pun SH Application of an HIV Gp41-Derived Peptide for Enhanced Intracellular Trafficking of Synthetic Gene and SiRNA Delivery Vehicles. Bioconjug. Chem 2008, 4, 920–927. [DOI] [PubMed] [Google Scholar]
- (79).Kwon EJ; Liong S; Pun SH A Truncated HGP Peptide Sequence Retains Endosomolytic Activity and Improves Gene Delivery Efficiencies. Mol. Pharm 2010, 3, 1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Ogino C; Ishii J; Nishimura Y; Ezawa R; Takeda K; Kondo A A Display of PH-Sensitive Fusogenic GALA Peptide Facilitates Endosomal Escape from a Bio-Nanocapsule via an Endocytic Uptake Pathway. J. Nanobiotechnology 2014, 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).De La Fuente-Herreruela D; Monnappa AK; Muñoz-Úbeda M; Morallón-Piña A; Enciso E; Sánchez L; Giusti F; Natale P; López-Montero I Lipid-Peptide Bioconjugation through Pyridyl Disulfide Reaction Chemistry and Its Application in Cell Targeting and Drug Delivery. J. Nanobiotechnology 2019, 1, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kuehne J; Murphy RM Synthesis and Characterization of Membrane-Active GALA-OKT9 Conjugates. Bioconjug. Chem 2001, 5, 742–749. [DOI] [PubMed] [Google Scholar]
- (83).Liu B; Ejaz W; Gong S; Kurbanov M; Canakci M; Anson F; Thayumanavan S Engineered Interactions with Mesoporous Silica Facilitate Intracellular Delivery of Proteins and Gene Editing. Nano Lett. 2020, 5, 4014–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.