Abstract
Background
The aim is to study common etiological pathways for 3 major cardiovascular diseases (CVD), as reflected in multiple proteins.
Methods and Results
Eighty‐four proteins were measured using the proximity extension technique in 870 participants in the PIVUS (Prospective Investigation of Uppsala Seniors Study) cohort on 3 occasions (age 70, 75, and 80 years). The sample was followed for incident myocardial infarction, ischemic stroke or heart failure. The same proteins were measured in an independent validation sample, the ULSAM (Uppsala Longitudinal Study of Adult Men) cohort in 595 participants at age 77. During a follow‐up of up to 15 years in PIVUS and 9 years in ULSAM, 222 and 167 individuals experienced a CVD. Examining associations with the 3 outcomes separately in a meta‐analysis of the 2 cohorts, 6 proteins were related to incident myocardial infarction, 25 to heart failure, and 8 proteins to ischemic stroke following adjustment for traditional risk factors. Growth differentiation factor 15 and tumor necrosis factor‐related apoptosis‐inducing ligand receptor 2 were related to all 3 CVDs. Including estimated glomerular filtration rate in the models attenuated some of these relationships. Fifteen proteins were related to a composite of all 3 CVDs using a discovery/validation approach when adjusting for traditional risk factors. A selection of 7 proteins by lasso in PIVUS improved discrimination of incident CVD by 7.3% compared with traditional risk factors in ULSAM.
Conclusions
We discovered and validated associations of multiple proteins with incident CVD. Only a few proteins were associated with all 3 diseases: myocardial infarction, ischemic stroke, and heart failure.
Keywords: epidemiology, heart failure, myocardial infarction, protein, stroke
Subject Categories: Epidemiology, Cardiovascular Disease, Proteomics
Nonstandard Abbreviations and Acronyms
- CASP‐8
caspase‐8
- CCL3
C‐C motif chemokine 3
- EN‐RAGE
protein S100‐A12
- FABP‐4
fatty acid binding protein‐4
- FGF‐23
fibroblast growth factor 23
- GDF‐15
growth/differentiation factor 15
- MMP‐12
matrix metalloproteinase‐12
- PAPPA
pappalysin‐1
- PIVUS
the Prospective Investigation of Uppsala Seniors Study
- TIM‐1
T‐cell immunoglobulin and mucin domain 1
- TNFR‐1
TNF receptor‐1
- TRAIL‐R2
TNF‐related apoptosis‐inducing ligand receptor 2
- U‐PAR
urokinase plasminogen activator surface receptor
- ULSAM
Uppsala Longitudinal Study of Adult Men
Clinical Perspective
What Is New?
We related 86 proteins to incident cardiovascular disease (myocardial infarction, stroke and heart failure) in 2 cohorts.
GDF‐15 (growth differentiation factor 15) and TRAIL‐R2 (TNF‐related apoptosis‐inducing ligand receptor 2) were related to all 3 CVDs.
What Are the Clinical Implications?
A selection of 7 proteins improved discrimination of incident cardiovascular disease by 7.3% compared with traditional risk factors, suggesting the use of proteomics in risk prediction.
Myocardial infarction, stroke, and heart failure are 3 major cardiovascular diseases, that share common etiological factors, but that may also have separate causes. For example, atherosclerosis is a common denominator for myocardial infarction and ischemic stroke, and hypertension and left ventricular hypertrophy are common features of both myocardial infarction and heart failure. However, details at the molecular level linking some of the pathophysiology for these disorders are little understood.
Using the proximity extension assay technique to measure proteins in plasma,1 it is possible to evaluate a great number of proteins in a multiplex fashion. Using this technique, we have described several novel associations of proteins with specific cardiovascular outcomes such as ischemic stroke2 and heart failure,3 as well as with risk factors such as obesity, hypertension, dyslipidemia, diabetes mellitus, renal function, atherosclerosis,4, 5 and impaired left ventricular function.3 In order to understand the commonalities of the pathophysiology of the clinical cardiovascular diseases at the molecular level, one must compare if proteins are linked to 2 or more of these cardiovascular diseases (CVDs), or if proteins are uniquely linked to a certain CVD.
With that in mind, we first performed an analysis of associations of 84 proteins versus incident myocardial infarction, ischemic stroke and heart failure in separate analyses for each disease. Thereafter, we evaluated the protein profile versus a combined end point of the 3 CVDs, and also evaluated if some of the proteins could improve discrimination regarding incident CVD on top of traditional risk factors. We used 2 population‐based cohorts, PIVUS (Prospective Investigation of Uppsala Seniors study)6 and ULSAM (the Uppsala Longitudinal Study of Adult men),7 in which the proteins have been measured in the same fashion. As compared with our previous studies,2, 3 we have now updated the length of the follow‐up period, as well as analyzed the protein panel at 3 occasions in the PIVUS study, actions that will increase the power compared with previous reports.
Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Samples
Uppsala Longitudinal Study of Adult Men
In 1970 to 74, 2322 men all aged 50 years living in the city of Uppsala, Sweden, were investigated as part of the ULSAM (Uppsala Longitudinal Study of Adult Men) (https://www.pubcare.uu.se/ulsam/).7, 8 Of the invited individuals, 82% accepted to participate.
This cohort has been reinvestigated at ages 60, 70, 77, 82, 88, and 93 years of age. This study uses data from the 77‐year investigation, at which 839 subjects participated. Of those, we have proteomic data in 761 individuals. A total of 166 subjects were excluded from the analyses due to prevalent CVD at the baseline examination at age 77.
Prospective Investigation of Uppsala Seniors
In 2001 to 2004, 1016 men and women all aged 70 years living in the city of Uppsala, Sweden were investigated as part of the PIVUS (Prospective Investigation of Uppsala Seniors) study.6 Of the invited individuals, 50% accepted to participate. This cohort has been reinvestigated at 75 (n=826) and 80 years (n=604) of age. Proteomics analysis was performed at all 3 occasions, and was present at 98% of the investigations. A total of 146 subjects were excluded from the analyses due to prevalent CVD at age 70.
Both studies were approved by the Ethics Committee of Uppsala University and all study participants have given their informed consent to participate.
Traditional Risk Factors in Both Cohorts
Fasting blood samples were drawn in the morning after an overnight fast. Serum levels of cholesterol and triglycerides, and high‐density lipoprotein were assayed by enzymatic techniques. Friedewald's formula was used to calculate low‐density lipoprotein‐cholesterol. Blood glucose was measured using an oxidase method. Supine systolic and diastolic blood pressures were measured twice in the right arm after 10 minutes rest, and means were calculated. Data on smoking status and medications at baseline were based on a questionnaire.
CVD Diagnosis
Data on causes of death and hospitalizations were retrieved from the Swedish Cause of Death Register and the Swedish Hospital Discharge Register, respectively. The 3 major cardiovascular diseases were defined as: myocardial infarction (International Classification of Diseases, Eighth, Ninth, and Tenth Revisions [ICD]‐8 code 410, ICD‐9 code 410, or ICD‐10 code I20), ischemic stroke (ICD‐8 codes 431, 433–436, ICD‐9 code 431, 433–436, ICD‐10 code I63–I66), and heart failure (ICD‐8 codes 427.00, 427.10, 428.99, ICD‐9, 428 and ICD‐10 code I50 and I11.0). The accuracy of these diagnoses in the Swedish registers have been deemed high quality,9 but since the heart failure diagnosis is less precise, we performed additional chart review based validation of heart failure events, as previously described.10
Protein Analysis
We used the Olink Proseek Multiplex Cardiovascular I 96×96 kit to simultaneously measure proteins in plasma by real‐time PCR using the Fluidigm BioMark HD (Olink, Uppsala, Sweden).1 Ninety‐two proteins were measured, and 84 of these proteins with a call‐rate >75% were further evaluated in the analyses. Mean intra‐assay and inter‐assay variation were 8% and 12%, respectively. Further details regarding levels of detection, reproducibility, and validations are given at Olink´s webpage (https://www.olink.com/resources‐support/document‐download‐center/). Values below levels of detection were replaced by levels of detection/20.5.
Estimated Glomerular Filtration Rate
In both cohorts, plasma creatinine and cystatin C were measured by a standard enzymatic method and by an enhanced turbidimetric method, respectively. A validated formula to calculate estimated glomerular filtration rate (eGFR) using both of these markers was used.11
Statistical Analysis
All protein values were log2‐transformed to achieve a normal distribution and thereafter transformed to the semantic differential scale in order to obtain comparable estimates.
Generally, for each protein 2 Cox proportional hazards regression models were investigated for each outcome. The first model adjusted for age and sex (only age in ULSAM as it is an all‐male cohort), and the second model also adjusted for the traditional risk factors systolic blood pressure, diabetes mellitus, high‐density lipoprotein and low‐density lipoprotein‐cholesterol, body mass index, and smoking. A third model also included eGFR (ln‐transformed due to a skewed distribution) as an independent variable in addition to the traditional risk factors.
The proportional hazard assumption for the Cox analysis was evaluated by visual inspection of the ‐ln[‐ln(S(t))] versus ln(t) version of the Kaplan‐Meier plot, using a low and a high group of the proteins created by a split of the distribution by the median.
Since we have measurements of proteins and covariates at 3 occasions in PIVUS (at age 70, 75, and 80 years), we updated all of them at each examination for that sample, implying that the follow‐up period of 15 years was split into three 5‐year periods with data from each examination serving as the baseline for that 5‐year period, so called time‐dependent Cox proportional hazards regression. The analyses in ULSAM only used one baseline (at age 77).
First, in the analyses of the separate outcomes (myocardial infarction, ischemic stroke, and heart failure), we fit the models to the PIVUS and ULSAM cohorts separately and meta‐analyzed the results using the fixed‐effects inverse variance‐weighted method. In these analyses, prevalent cases at baseline of the respective outcome were excluded before analysis. Since we did not have a replication step using this approach, we applied Bonferroni adjustment for 84 tests (P<0.000625) for the crude model together with P<0.05 for the multivariable‐adjusted model for significance.
Second, we evaluated a composite end point of CVD (myocardial infarction or ischemic stroke or heart failure). For those analyses, we used the PIVUS cohort for discovery and the ULSAM cohort for validation. We considered associations statistically significant that passed a false discovery rate of <0.05 in the age‐ and sex‐adjusted analyses in both samples, and a nominal multivariable‐adjusted P value of <0.05 in the validation sample. Subjects with prevalent CVD at baseline in the 2 samples were excluded in these analyses.
To evaluate if some of the proteins could improve discrimination for the composite end point of CVD (myocardial infarction or ischemic stroke or heart failure) compared with the traditional risk factors, we first used lasso for logistic regression in the PIVUS cohort with the split sample technique using all 84 proteins (forcing age and sex into the model) to select proteins included in the model with the best fit. Thereafter, we compared the discrimination (C‐statistic) for a logistic regression model in the ULSAM cohort with the traditional risk factors (systolic blood pressure, diabetes mellitus, high‐density lipoprotein and low‐density lipoprotein‐cholesterol, body mass index, and smoking) to a model further adding the proteins identified using lasso in the PIVUS cohort.
STATA16 (Stata inc, College Station, TX, USA) was used for the analyses.
Results
Baseline characteristics of the 2 cohorts are given in Table 1.
Table 1.
Baseline Characteristics in PIVUS at Age 70 (n=870) and ULSAM at Age 77 (n=595)
| PIVUS | ULSAM | |
|---|---|---|
| Variable | Mean (SD) | Mean (SD) |
| Age, y | 70.1 (0.1) | 77.5 (0.7) |
| Sex (% female) | 51 | 0 |
| Systolic blood pressure, mm Hg | 149 (22) | 150 (20) |
| Smoker, % | 11 | 8.2 |
| HDL‐cholesterol, mmol/L | 1.52 (0.42) | 1.32 (0.33) |
| LDL‐cholesterol, mmol/L | 3.40 (0.88) | 3.48 (0.87) |
| Body mass index, kg/m2 | 26.9 (4.3) | 26.2 (3.5) |
| Diabetes mellitus, % | 11 | 14 |
| eGFR, mL/min/BSA | 86 (16) | 75 (20) |
BSA indicates body surface area; eGFR, estimated glomerular filtration rate; HDL, high‐density lipoprotein; LDL, low‐density lipoprotein; PIVUS, the Prospective Investigation of Uppsala Seniors Study; and ULSAM, Uppsala Longitudinal Study of Adult Men.
Incidence of Cardiovascular Events
Over 15 years of follow‐up in the discovery cohort (PIVUS), 222 CVD events (83, 89, and 132 cases of myocardial infarction, stroke or heart failure, respectively, of which some had more than one outcome each) occurred during 10 666 person years at risk. In the validation cohort (ULSAM), 167 CVD events (59, 81, and 112 cases of myocardial infarction, stroke or heart failure occurred, respectively) occurred during 5777 person years at risk.
Commonality in Associations of Proteins With Incidence of Separate Cardiovascular Events
Six proteins were associated with incident myocardial infarction (Table 2), 25 with incident heart failure (Table 3), and 8 proteins with incident ischemic stroke (Table 4), in Bonferroni‐adjusted meta‐analyses of the 2 cohorts. Following further adjustment for eGFR on top of traditional risk factors, 2 of the 6 proteins still showed P<0.05 for myocardial infarction, 18 of the 25 for heart failure and all 8 proteins regarding stroke. Those proteins with P<0.05 also following adjustment for eGFR are indicated with a * following the traditional risk factor adjusted P value in Tables 2, 3, 4. The proportional hazard assumption was fulfilled for all the proteins given in Tables 2, 3, 4.
Table 2.
Proteins Associated With Incident Myocardial Infarction in a Meta‐Analysis of ULSAM and PIVUS
| Protein | Age Adjusted | Adjustment for Traditional Risk Factors | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI Lower | 95% CI Higher | P Value | HR | 95% CI Lower | 95% CI Higher | P Value | |
| MMP‐12 | 1.61 | 1.37 | 1.89 | 6.30e‐09 | 1.31 | 1.08 | 1.57 | 0.00041* |
| GDF‐15 | 1.56 | 1.32 | 1.83 | 9.75e‐08 | 1.37 | 1.12 | 1.67 | 0.0018* |
| TRAIL‐R2 | 1.41 | 1.22 | 1.62 | 1.90e‐06 | 1.35 | 1.12 | 1.62 | 0.0012 |
| U‐PAR | 1.49 | 1.23 | 1.80 | 0.000042 | 1.32 | 1.07 | 1.64 | 0.0087 |
| VEGF‐A | 1.33 | 1.13 | 1.56 | 0.00043 | 1.21 | 1.01 | 1.45 | 0.033 |
| IL‐6 | 1.28 | 1.11 | 1.48 | 0.00055 | 1.18 | 1.004 | 1.39 | 0.044 |
Hazard ratios (HRs) are given for a 1‐SD increase in the proteins. Only proteins with age‐ or sex‐adjusted P value <0.000625 and a multi‐adjusted P value <0.05 are shown. GDF‐15 indicates growth/differentiation factor 15; IL‐6, interleukin‐6; MMP‐12, matrix metalloproteinase‐12; PIVUS, the Prospective Investigation of Uppsala Seniors Study; TRAIL‐R2, TNF‐related apoptosis‐inducing ligand receptor 2; ULSAM, Uppsala Longitudinal Study of Adult Men; U‐PAR, urokinase plasminogen activator surface receptor; and VEGF‐A, vascular endothelial growth factor A.
P<0.05 also after adjustment for estimated glomerular filtration rate.
Table 3.
Proteins Associated With Incident Heart Failure in a Meta‐Analysis of ULSAM and PIVUS
| Protein | Age Adjusted | Adjustment for Traditional Risk Factors | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI Lower | 95% CI Higher | P Value | HR | 95% CI Lower | 95% CI Higher | P Value | |
| NTproBNP | 2.14 | 1.84 | 2.49 | 4.77e‐23 | 2.11 | 1.79 | 2.48 | 3.17e‐19* |
| TRAIL‐R2 | 1.49 | 1.36 | 1.63 | 3.15e‐19 | 1.40 | 1.25 | 1.56 | 1.25e‐09* |
| GDF‐15 | 1.58 | 1.39 | 1.79 | 6.62e‐13 | 1.44 | 1.24 | 1.67 | 1.65e‐06* |
| FGF‐23 | 1.50 | 1.34 | 1.69 | 6.39e‐12 | 1.39 | 1.21 | 1.59 | 2.49e‐06* |
| TIM‐1 | 1.51 | 1.34 | 1.70 | 7.41e‐12 | 1.39 | 1.21 | 1.59 | 1.26e‐06* |
| FABP4 | 1.50 | 1.32 | 1.71 | 9.40e‐10 | 1.31 | 1.11 | 1.55 | 0.0012* |
| MMP‐12 | 1.48 | 1.30 | 1.68 | 1.87e‐09 | 1.42 | 1.23 | 1.63 | 1.12e‐06* |
| SPON1 | 1.57 | 1.35 | 1.83 | 2.62e‐09 | 1.51 | 1.29 | 1.77 | 2.72e‐07* |
| CSF‐1 | 1.45 | 1.26 | 1.67 | 2.20e‐07 | 1.31 | 1.11 | 1.53 | 0.00077* |
| TNF‐R1 | 1.41 | 1.24 | 1.61 | 2.25e‐07 | 1.24 | 1.05 | 1.47 | 0.0094 |
| U‐PAR | 1.48 | 1.27 | 1.73 | 5.51e‐07 | 1.33 | 1.12 | 1.58 | 0.00090* |
| CCL20 | 1.32 | 1.18 | 1.48 | 5.96e‐07 | 1.28 | 1.14 | 1.45 | 0.000027* |
| HGF | 1.37 | 1.20 | 1.55 | 1.49e‐06 | 1.23 | 1.07 | 1.43 | 0.0040* |
| PlGF | 1.37 | 1.20 | 1.57 | 2.24e‐06 | 1.28 | 1.09 | 1.49 | 0.0014 |
| IL‐6 | 1.30 | 1.16 | 1.45 | 2.57e‐06 | 1.20 | 1.06 | 1.36 | 0.0027* |
| Follistatin | 1.35 | 1.19 | 1.54 | 4.43e‐06 | 1.26 | 1.10 | 1.45 | 0.00080* |
| hK11 | 1.32 | 1.16 | 1.49 | 0.000011 | 1.24 | 1.07 | 1.43 | 0.0031 |
| TNF‐R2 | 1.31 | 1.15 | 1.48 | 0.000028 | 1.17 | 1.01 | 1.37 | 0.035 |
| CD40 | 1.31 | 1.15 | 1.50 | 0.000053 | 1.18 | 1.02 | 1.37 | 0.024 |
| ST2 | 1.31 | 1.14 | 1.50 | 0.000076 | 1.30 | 1.13 | 1.50 | 0.00014* |
| EN‐RAGE | 1.27 | 1.12 | 1.44 | 0.00012 | 1.25 | 1.10 | 1.43 | 0.00038* |
| AGRP | 1.30 | 1.13 | 1.49 | 0.00020 | 1.25 | 1.08 | 1.45 | 0.0026* |
| CHI3L1 | 1.25 | 1.11 | 1.41 | 0.00022 | 1.14 | 1.01 | 1.30 | 0.031 |
| LOX‐1 | 1.27 | 1.11 | 1.45 | 0.00027 | 1.23 | 1.07 | 1.42 | 0.0026* |
| CCL3 | 1.20 | 1.08 | 1.32 | 0.00033 | 1.14 | 1.02 | 1.29 | 0.027 |
Hazard ratios (HRs) are given for a 1‐SD increase in the proteins. Only proteins with age‐ or sex‐adjusted P value <0.000625 and a multi‐adjusted P value <0.05 are shown. AGRP indicates agouti‐related protein; CCL3, C‐C motif chemokine 3; CCL20, C‐C motif chemokine 20; CD40, CD40L receptor; CHI3L1, chitinase‐3‐like protein 1; CSF‐1, macrophage colony‐stimulating factor; EN‐RAGE, protein S100‐A12; FABP4, fatty acid‐binding protein 4; FGF‐23, fibroblast growth factor 23; GDF‐15, growth/differentiation factor 15; HGF, hepatocyte growth factor; hk11, kallikrein‐11; IL‐6, interleukin‐6; LOX‐1, lectin‐like oxidized LDL receptor 1; MMP‐12, matrix metalloproteinase‐12; NTproBNP, N‐terminal pro‐B‐type natriuretic peptide; PIGF, placenta growth factor; PIVUS, the Prospective Investigation of Uppsala Seniors Study; SPON1, spondin‐1; ST2, ST2 protein; TIM‐1, T‐cell immunoglobulin and mucin domain 1; TNF‐R1, tumor necrosis factor receptor 1; TRAIL‐R2, TNF‐related apoptosis‐inducing ligand receptor 2; ULSAM, Uppsala Longitudinal Study of Adult Men; and U‐PAR, urokinase plasminogen activator surface receptor.
P<0.05 also after adjustment for estimated glomerular filtration rate.
Table 4.
Proteins Being Associated With Incident Ischemic Stroke in a Meta‐Analysis of ULSAM and PIVUS
| Protein | Age Adjusted | Adjustment for Traditional Risk Factors | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI Lower | 95% CI Higher | P Value | HR | 95% CI Lower | 95% CI Higher | P Value | |
| NTproBNP | 1.69 | 1.41 | 2.01 | 6.25e‐09 | 1.70 | 1.41 | 2.06 | 2.61e‐08* |
| GDF‐15 | 1.41 | 1.21 | 1.65 | 0.000011 | 1.35 | 1.13 | 1.61 | 0.00068* |
| Osteoprotegerin | 1.42 | 1.21 | 1.67 | 0.000017 | 1.36 | 1.15 | 1.61 | 0.00025* |
| TRAIL‐R2 | 1.30 | 1.15 | 1.48 | 0.000031 | 1.27 | 1.11 | 1.46 | 0.00062* |
| CSF‐1 | 1.38 | 1.18 | 1.63 | 0.000073 | 1.34 | 1.12 | 1.60 | 0.0010* |
| Adrenomedullin | 1.43 | 1.19 | 1.71 | 0.00010 | 1.41 | 1.17 | 1.71 | 0.00032* |
| Follistatin | 1.32 | 1.14 | 1.54 | 0.00023 | 1.28 | 1.10 | 1.50 | 0.0010* |
| TIM‐1 | 1.29 | 1.11 | 1.50 | 0.00055 | 1.21 | 1.03 | 1.42 | 0.018* |
Hazard ratios (HRs) are given for a 1‐SD increase in the proteins. Only proteins with age‐ or sex‐adjusted P value <0.000625 and a multi‐adjusted P value <0.05 are shown. CSF‐1 indicates macrophage colony‐stimulating factor 1; GDF‐15, growth/differentiation factor 15; NTproBNP, N‐terminal pro‐B‐type natriuretic peptide; PIVUS, the Prospective Investigation of Uppsala Seniors Study; TIM‐1, T‐cell immunoglobulin and mucin domain 1; TRAIL‐R2, TNF‐related apoptosis‐inducing ligand receptor 2; and ULSAM, Uppsala Longitudinal Study of Adult Men.
P<0.05 also after adjustment for estimated glomerular filtration rate.
GDF‐15 (growth/differentiation factor 15) and TRAIL‐R2 (TNF‐related apoptosis‐inducing ligand receptor 2) were related to each of the 3 CVDs. Another 7 proteins were related to 2 out of 3 CVDs (follistatin, IL‐6 [interleukin‐6], CSF‐1 [macrophage colony‐stimulating factor 1], MMP‐12 [matrix metalloproteinase‐12], NT‐proBNP [N‐terminal pro‐B‐type natriuretic peptide], TIM‐1 [T‐cell immunoglobulin and mucin domain 1] and U‐PAR [urokinase plasminogen activator surface receptor]). The overlap is illustrated in Figure.
Figure 1. Associations of proteins with incidence of myocardial infarction, ischemic stroke, and heart failure.
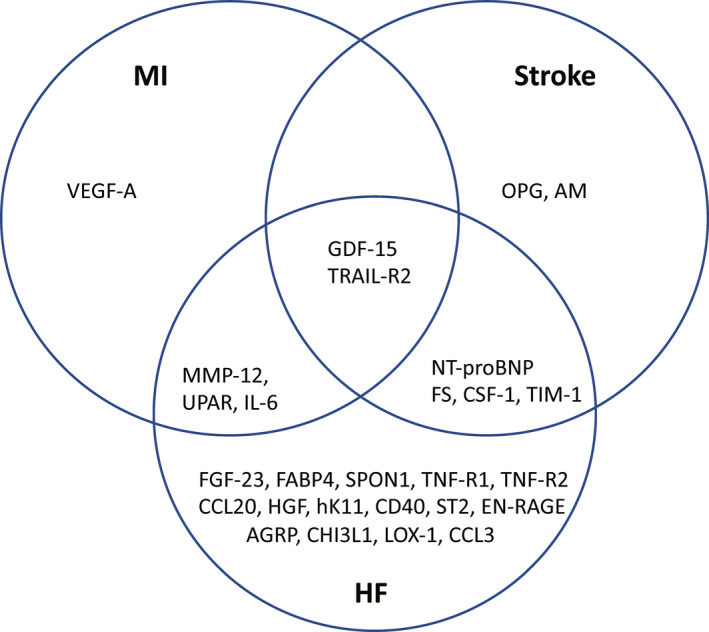
Venn diagram. Only false discovery rate‐adjusted statistically significant associations are shown. AGRP indicates Agouti‐related protein; AM, adrenomedullin; CCL3, C‐C motif chemokine 3; CCL20, C‐C motif chemokine 20; CD40, CD40L receptor; CHI3L1, chitinase‐3‐like protein 1; CSF‐1, macrophage colony‐stimulating factor 1; EN‐RAGE, protein S100‐A12; FABP4, fatty acid binding protein‐4; FGF‐23, fibroblast growth factor 23; FS, follistatin; GDF‐15, growth/differentiation factor 15; HF, heart failure; HGF, hepatocyte growth factor; hK11, kallikrein‐11; IL‐6, interleukin‐6; LOX‐1, lectin‐like oxidized LDL receptor 1; MI, myocardial infarction; MMP‐12, matrix metalloproteinase‐12; NT‐proBNP, N‐terminal pro‐B‐type natriuretic peptide; OPG, osteoprotegerin; SPON1, spondin‐1; ST2, ST2 protein; TIM‐1, T‐cell immunoglobulin and mucin domain 1; TNF‐R1, TNF receptor‐1; TRAIL‐R2, TNF‐related apoptosis‐inducing ligand receptor 2; UPAR, urokinase plasminogen activator surface receptor; and VEGF‐A, vascular endothelial growth factor A.
Following further adjustment for eGFR, only GDF‐15 was significantly related to all 3 CVDs, while NT‐proBNP, TIM‐1, CSF‐1, and follistatin were related to 2 of the CVDs.
Associations of Proteins With Incidence of Combined Cardiovascular Events
Fifteen proteins were discovered and validated at the false discovery rate <0.05 level to be associated with incident CVD. The top 3 associations were NT‐proBNP, FGF‐23 (Fibroblast growth factor 23), and adrenomedullin. These top 3 proteins were confirmed using a Bootstrap calculation with 1000 repetitions.
Details are given in Table 5. Following further adjustment for eGFR, only 9 of the 15 proteins still showed P<0.05 in the validation step in ULSAM. Those 9 proteins are indicated with a * following the multi‐adjusted P value in Table 2.
Table 5.
Validation of Associations of Proteins With Incidence of Combined Cardiovascular Events (Myocardial Infarction, Stroke, or Heart Failure) in the ULSAM Cohort
| Protein | Age‐ and Sex‐Adjusted Models | Adjustment for Traditional Risk Factors | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI Lower | 95% CI Higher | P Value | HR | 95% CI Lower | 95% CI Higher | P Value | |
| NT‐proBNP | 1.85 | 1.54 | 2.23 | 5.78e‐11 | 1.81 | 1.51 | 2.18 | 2.69e‐10* |
| FGF‐23 | 1.44 | 1.25 | 1.66 | 5.00e‐07 | 1.36 | 1.18 | 1.57 | 0.000029* |
| AM | 1.55 | 1.31 | 1.84 | 5.17e‐07 | 1.48 | 1.24 | 1.76 | 9.65e‐06* |
| GDF‐15 | 1.46 | 1.24 | 1.72 | 6.64e‐06 | 1.36 | 1.14 | 1.61 | 0.00058* |
| PlGF | 1.37 | 1.18 | 1.59 | 0.000037 | 1.28 | 1.1 | 1.49 | 0.0012 |
| TIM‐1 | 1.35 | 1.17 | 1.57 | 0.000050 | 1.26 | 1.08 | 1.47 | 0.0037* |
| TRAIL‐R2 | 1.39 | 1.18 | 1.63 | 0.000074 | 1.31 | 1.11 | 1.55 | 0.0012* |
| Follistatin | 1.36 | 1.16 | 1.59 | 0.00016 | 1.33 | 1.13 | 1.56 | 0.00054* |
| SPON1 | 1.33 | 1.13 | 1.57 | 0.00066 | 1.32 | 1.12 | 1.56 | 0.0011* |
| TNF‐R1 | 1.32 | 1.12 | 1.56 | 0.0011 | 1.23 | 1.04 | 1.45 | 0.014 |
| FABP4 | 1.30 | 1.10 | 1.53 | 0.0023 | 1.21 | 0.99 | 1.46 | 0.046 |
| MMP‐12 | 1.28 | 1.09 | 1.5 | 0.0024 | 1.19 | 1.01 | 1.41 | 0.043 |
| CSF‐1 | 1.29 | 1.10 | 1.53 | 0.0025 | 1.23 | 1.04 | 1.45 | 0.012 |
| Osteoprotegerin | 1.25 | 1.07 | 1.46 | 0.0050 | 1.23 | 1.05 | 1.44 | 0.010* |
| Interleukin‐6 | 1.22 | 1.06 | 1.42 | 0.0070 | 1.19 | 1.02 | 1.39 | 0.029 |
Hazard ratios (HRs) are given for a 1 ‐SD increase in the proteins. Only associations with false discovery rate‐adjusted P value <0.05 in age and sex‐adjusted models, and multivariable‐adjusted P value <0.05 in both ULSAM and PIVUS are shown. CSF‐1 indicates macrophage colony‐stimulating factor; FAB4, fatty acid‐binding protein 4; FGF‐23, fibroblast growth factor 23; GDF‐15, growth/differentiation factor 15; IL‐6, interleukin‐6; MMP‐12, matrix metalloproteinase‐12; NTproBNP, N‐terminal pro‐B‐type natriuretic peptide; PIGF, placenta growth factor; PIVUS, the Prospective Investigation of Uppsala Seniors Study; SPON1, spondin‐1; TIM‐1, T‐cell immunoglobulin and mucin domain 1; TNF‐R1, Tumor necrosis factor receptor 1; TRAIL‐R2, TNF‐related apoptosis‐inducing ligand receptor 2; and ULSAM, Uppsala Longitudinal Study of Adult Men.
P<0.05 also after adjustment for estimated glomerular filtration rate.
The proportional hazard assumption was fulfilled for all 15 proteins in Table 5 except for FABP4 (fatty acid binding protein‐4). In this case, logistic regression analyses resulted in almost identical estimates as when Cox proportional hazard models were used.
Risk Prediction of Combined Cardiovascular Events
In a model with all 84 proteins in the PIVUS cohort, seven proteins were selected using lasso to produce the best fitting model for incident CVD ([lambda 0.014], NT‐proBNP, CSF‐1, PAPPA [Pappalysin‐1], CCL3 [C‐C motif chemokine 3], EN‐RAGE [Protein S100‐A12], CASP‐8 [Caspase‐8], and FABP‐4). Performing a correlation matrix of the 7 chosen proteins, the correlation coefficients versus NT‐proBNP ranges from 0.27 at most to 0.06. In general, the correlations between the other 6 proteins were in the 0.10 to 0.20 range, with some exceptions; CSF‐1 versus FABP4 with r=0.47, CSF‐1 versus CCL3 with r=0.45, and EN‐RAGE versus CASP‐8 with r=0.61. When a model with these 7 proteins and the traditional risk factors was fit to the ULSAM cohort, it improved discrimination (C‐statistic) from 0.64 (95% CI, 0.59–0.69) to 0.72 (95% CI, 0.67–0.76; P=0.002 for difference) over a model with only the traditional risk factors. The major part of this improvement was however due to NT‐proBNP alone (adding NT‐proBNP to a model with traditional risk factors improved C‐statistic to 0.71 [95% CI, 0.66–0.76; P=0.004]).
Discussion
The present study shows that a proteomic approach to the etiology of CVDs can yield new insights. Above and beyond traditional risk factors, NT‐proBNP added most to risk prediction for the combined cardiovascular events outcome, and GDF‐15 and TRAIL‐R2 were also separately associated with each of the specific cardiovascular outcomes.
This study confirms notions from our previous studies of the usefulness of the proteomic approach for understanding of combined cardiovascular events. In a study conducted in 1211 people with diabetes mellitus from 6 different cohorts, MMP‐12, IL‐27 subunit α (IL‐27a), KIM‐1 (kidney injury molecule‐1), FGF‐23, protein S100‐A12, TNFR‐1 (TNF receptor‐1), TNFR‐2, and TRAIL‐R2 were related to incident CVD.12 Of note, IL‐27a and protein S100‐A12 did not seem important in the present study, raising the possibility that those associations might be specific for people with diabetes mellitus. Another study in patients with acute coronary syndromes report associations between GDF‐15 and NT‐proBNP and several specific cardiovascular outcomes, such as myocardial infarction, heart failure, and sudden cardiac death.13
When looking at the analyses of proteins versus the 3 separate outcomes and compared those finding to the 15 proteins being related to the combined CVD end point, most proteins being related to the combined end point were also related to incident heart failure. Of the 3 CVDs, it was also heart failure that showed the largest number of significant protein associations. If this discrepancy versus the other 2 CVDs was due to the fact that the number of incident cases of heart failure was higher than for ischemic stroke and myocardial infarction, or if heart failure as a disease is linked to more pathophysiological pathways than ischemic stroke and myocardial infarction remains to be evaluated.
Associations of NT‐proBNP with incidence of cardiovascular events have been well documented previously, using both the PEA technique and conventional ELISAs. In an individual‐participant‐data meta‐analysis conducted in almost 100 000 individuals from 60 different samples, elevated levels of NT‐proBNP were associated with increased risk of the major CVDs.14 In the clinical setting, NT‐proBNP is mainly used to diagnose and follow heart failure patients, but our study along with previous data supports the view that NT‐proBNP might also have a clinically important role in risk prediction in the primary prevention setting.14 In the present study, we used the lasso approach to select proteins for prediction of incident CVD in the PIVUS cohort and validated the 7 identified proteins in the ULSAM cohort, and found a substantial improvement in discrimination. It should however be pointed out that almost all this improvement was due to NT‐proBNP alone. Higher NT‐proBNP is likely not causal for higher CVD risk, but a protective response to an increased risk. Genetic variants in or close to the BNP (NPPB) gene reported to be associated with NT‐proBNP levels were not associated with an increased risk of heart failure in a large population‐based cohort.15 In another genetic study, one single‐nucleatide variation (rs198389) was associated with increased NT‐proBNP levels, a reduced blood pressure, and decreased cardiovascular mortality.16 In another study using Mendelian randomization, genetically determined higher NT‐proBNP levels were causally related to lower blood pressure and a reduced risk of large‐artery stroke.17
GDF‐15 belongs to the transforming growth factor‐beta cytokine superfamily. It is normally produced by immunocompetent cells, but cardiac cells and tumor cells can secrete GDF‐15 during stress. Elevated levels of GDF‐15 have previously been related to mortality in both CVD and cancer,18 and GDF‐15 has been associated with abdominal obesity, other risk factors for CVDs, and markers of subclinical CVD such as endothelial dysfunction, atherosclerosis, left ventricular hypertrophy, and a reduced left ventricular ejection fraction.19 It is therefore unsurprising that this marker was linked to risk of all of the 3 major CVDs in the present study.
Also, TRAIL‐R2 levels were related to all 3 major CVDs. TRAIL‐R2 is one of the receptors for the TRAIL, a protein known to be involved in apoptosis. TRAIL has been shown to induce apoptosis and upregulation of inflammatory genes in endothelial cells.20 In patients with chronic renal failure, low levels of TRAIL were related to an accelerated plaque progression over 2 years,21 and in subjects with advanced atherosclerosis, TRAIL‐R2 in plaque was related to apoptosis and to symptomatic plaque. High circulating TRAIL‐R2 levels have been associated with later CVD events both in that high‐risk sample and in the general population.22, 23 Higher levels of TRAIL‐R2 have also been linked to atrial fibrillation, a major risk factor for stroke and heart failure.24
MMP‐12, U‐PAR, and IL‐6 were linked to both myocardial infarction and heart failure. Possible pathophysiological connections between those 2 CVDs are high blood pressure, arterial stiffness, left ventricular hypertrophy, and a poor left ventricular systolic function following a myocardial infarction. MMP‐12 and U‐PAR have previously been linked to a reduced left ventricular ejection fraction.3 MMP‐12 has furthermore been linked to arterial stiffness25 and left ventricular hypertrophy26 in experimental studies, suggesting that especially MMP‐12 is worthwhile to explore further regarding the connection between myocardial infarction and heart failure.
NT‐proBNP, follistatin, CSF‐1, and TIM‐1 were linked to both heart failure and stroke. Hypertension is the most likely common denominator of these 2 traits; atherosclerosis could be another. This is an example where mechanistic studies of proteins might provide additional insight of shared pathophysiological pathways.
Some proteins were linked only to subsequent heart failure. Of those, hK‐11 (Kallikrein‐11) was unique in being highly significantly related to heart failure, but not to myocardial infarction and stroke. All other proteins linked to only one of the 3 CVDs had some association (P<0.05) with the other 2 CVDs. Human kallikreins are a family of 15 highly conserved serine proteases. hK‐11 has mainly been evaluated as a marker for cancer,27 but we have previously found this protease to be linked to an impaired kidney function28 and albuminuria.29 Although few studies have linked hK‐11 to CVD, genetic studies in humans and mice have shown close links between the kallikrein/kinin system and cardiovascular structure and function, as reviewed recently.30 Given also the close connection between the kallikrein/kinin system and the renin‐angiotensin system, it is plausible that a protein in the kallikrein/kinin system could be linked to heart failure, and further studies on this protein are warranted.
Kidney function, as evaluated by eGFR, is well known to be related to CVDs, as well as to protein levels. In a recent study, in which we related the change in eGFR over 10 years to the changes in protein levels (the same ones as in the present study), we found that the changes in most proteins were negatively related to the change in eGFR, indicating that a reduction in GFR induces an increase in levels for many proteins.31 That complicates how to use GFR in the present study, since GFR is not likely to be a confounder, but rather that the protein level is in the causal pathway between GFR and incident CVD; GFR‐>protein level‐> incident CVD. In the present study we performed a secondary analysis adding eGFR to the models and found, as expected, that some of the previous findings just adjusting for traditional risk factors no longer showed P<0.05. It is however not easy to interpret these attenuations in some protein estimates given the above proposed chain of events, and they could possibly not be regarded as simple confounding.
One way to evaluate if a certain assay really measures the protein of interest is to perform genetic analysis and search for genetic loci in the gene coding for the protein of interest (so called cis‐pQTLs). We used preliminary updated results from the SCALLOP (Systematic and Combined AnaLysis of Olink Proteins) consortium,30 so far only deposited at BioRxive (https://doi.org/10.1101/2020.04.03.023804), and found significant cis‐pQTL for 75 of the 92 proteins on the CVD‐1 chip use in the present study, including the present top findings, NT‐proBNP, GDF‐15, MMP‐12, FGF‐23, TRAIL‐R2, adrenomedullin. Thus, from these preliminary data it is most likely that the PEA technique used in the present study are detecting and measuring the majority of the proteins evaluated in the present study in a valid way.
The strengths of the present study include the use of 2 samples from the same town, a discovery/validation approach, and repeated measurements of the proteins at 3 occasions in one cohort with updated covariate information. A limitation is that the use of 2 samples of elderly subjects from Sweden does not give generalizability to other ethnic and age groups or geographical locations, so our results have to be confirmed in other studies. In particular, the clinical utility of proteomics for improving cardiovascular risk prediction merits additional studies in large studies with a wider age range. Further, causality of the reported associations will have to be assessed using other study designs. Mendelian randomization studies of some of these questions are ongoing within the SCALLOP framework.32
In conclusion, in this discovery/validation study of 2 population‐based cohorts, NT‐proBNP added to risk prediction for cardiovascular events combined, and GDF‐15 and TRAIL‐R2 levels were associated with risks of incident myocardial infarction, stroke and heart failure separately, above and beyond traditional risk factors.
Sources of Funding
The PIVUS study was supported by Uppsala University Hospital.
Disclosures
None.
(J Am Heart Assoc. 2021;10:e017900. DOI: 10.1161/JAHA.120.017900.)
For Sources of Funding and Disclosures, see page 10.
References
- 1.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Bucht Thorsen S, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al. Homogenous 96‐Plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. DOI: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lind L, Siegbahn A, Lindahl B, Stenemo M, Sundström J, Ärnlöv J. Discovery of new risk markers for ischemic stroke using a novel targeted proteomics chip. Stroke. 2015;46:3340–3347. DOI: 10.1161/STROKEAHA.115.010829. [DOI] [PubMed] [Google Scholar]
- 3.Stenemo M, Nowak C, Byberg L, Sundström J, Giedraitis V, Lind L, Ingelsson E, Fall T, Ärnlöv J. Circulating proteins as predictors of incident heart failure in the elderly. Eur J Heart Fail. 2018;20:55–62. DOI: 10.1002/ejhf.980. [DOI] [PubMed] [Google Scholar]
- 4.Lind L, Ärnlöv J, Lindahl B, Siegbahn A, Sundström J, Ingelsson E. Use of a proximity extension assay proteomics chip to discover new biomarkers for human atherosclerosis. Atherosclerosis. 2015;242:205–210. DOI: 10.1016/j.atherosclerosis.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Lind L, Gigante B, Borne Y, Mälarstig A, Sundström J, Ärnlöv J, Ingelsson E, Baldassarre D, Tremoli E, Veglia F, et al. The plasma protein profile and cardiovascular risk differ between intima‐media thickness of the common carotid artery and the bulb: a meta‐analysis and a longitudinal evaluation. Atherosclerosis. 2020;295:25–30. DOI: 10.1016/j.atherosclerosis.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium‐dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol. 2005;25:2368–2375. DOI: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- 7.Hedstrand H. A study of middle‐aged men with particular reference to risk factors for cardiovascular disease. Ups J Med Sci Suppl. 1975;19:1–61. [PubMed] [Google Scholar]
- 8.Ärnlöv J, Ingelsson E, Sundström J, Lind L. Impact of body mass index and the metabolic syndrome on the risk of cardiovascular disease and death in middle‐aged men. Circulation. 2010;121:230–236. DOI: 10.1161/CIRCULATIONAHA.109.887521. [DOI] [PubMed] [Google Scholar]
- 9.Merlo J, Lindblad U, Pessah‐Rasmussen H, Hedblad B, Råstam J, Isacsson SO, Janzon L, Råstam L. Comparison of different procedures to identify probable cases of myocardial infarction and stroke in two Swedish prospective cohort studies using local and national routine registers. Eur J Epidemiol. 2000;16:235–243. DOI: 10.1023/a:1007634722658. [DOI] [PubMed] [Google Scholar]
- 10.Ingelsson E, Ärnlöv J, Sundström J, Lind L. The validity of a diagnosis of heart failure in a hospital discharge register. Eur J Heart Fail. 2005;7:787–791. DOI: 10.1016/j.ejheart.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, Kusek JW, Manzi J, Van Lente F, Zhang YL, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367:20–29. DOI: 10.1056/NEJMoa1114248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowak C, Carlsson AC, Östgren CJ, Nyström FH, Alam M, Feldreich T, Sundström J, Carrero JJ, Leppert J, Hedberg P, et al. Multiplex proteomics for prediction of major cardiovascular events in type 2 diabetes. Diabetologia. 2018;61:1748–1757. DOI: 10.1007/s00125-018-4641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindholm D, James SK, Gabrysch K, Storey RF, Himmelmann A, Cannon CP, Mahaffey KW, Steg PG, Held C, Siegbahn A, et al. Association of multiple biomarkers with risk of all‐cause and cause‐specific mortality after acute coronary syndromes: a secondary analysis of the PLATO biomarker study. JAMA Cardiol. 2018;3:1160–1166. DOI: 10.1001/jamacardio.2018.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natriuretic Peptides Studies Collaboration , Willeit P, Kaptoge S, Welsh P, Butterworth AS, Chowdhury R, Spackman SA, Pennells L, Gao P, Burgess S, et al. Natriuretic peptides and integrated risk assessment for cardiovascular disease: an individual‐participant‐data meta‐analysis. Lancet Diabetes Endocrinol. 2016;4:840–849. DOI: 10.1016/S2213-8587(16)30196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfister R, Luben RN, Khaw KT, Wareham NJ. Common genetic variants of the natriuretic peptide gene locus are not associated with heart failure risk in participants in the EPIC‐Norfolk study. Eur J Heart Fail. 2013;15:624–627. DOI: 10.1093/eurjhf/hft007. [DOI] [PubMed] [Google Scholar]
- 16.Seidelmann SB, Vardeny O, Claggett B, Yu B, Shah AM, Ballantyne CM, Selvin E, MacRae CA, Boerwinkle E, Solomon SD. An NPPB promoter polymorphism associated with elevated N‐terminal pro‐B‐type natriuretic peptide and lower blood pressure, hypertension, and mortality. J Am Heart Assoc. 2017;6:e005257. DOI: 10.1161/JAHA.116.005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thériault S, Sjaarda J, Chong M, Hess S, Gerstein H, Paré G. Identification of circulating proteins associated with blood pressure using mendelian randomization. Circ Genom Precis Med. 2020;13:e002605. DOI: 10.1161/CIRCGEN.119.002605. [DOI] [PubMed] [Google Scholar]
- 18.Wallentin L, Zethelius B, Berglund L, Eggers KM, Lind L, Lindahl B, Wollert KC, Siegbahn A. GDF‐15 for prognostication of cardiovascular and cancer morbidity and mortality in men. PLoS One. 2013;8:e78797. DOI: 10.1371/journal.pone.0078797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lind L, Wallentin L, Kempf T, Tapken H, Quint A, Lindahl B, Olofsson S, Venge P, Larsson A, Hulthe J, et al. Growth‐differentiation factor‐15 is an independent marker of cardiovascular dysfunction and disease in the elderly: results from the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) Study. Eur Heart J. 2009;30:2346–2353. DOI: 10.1093/eurheartj/ehp261. [DOI] [PubMed] [Google Scholar]
- 20.Li JH, Kirkiles‐Smith NC, McNiff JM, Pober JS. TRAIL induces apoptosis and inflammatory gene expression in human endothelial cells. J Immunol. 2003;171:1526–1533. DOI: 10.4049/jimmunol.171.3.1526. [DOI] [PubMed] [Google Scholar]
- 21.Arcidiacono MV, Rimondi E, Maietti E, Melloni E, Tisato V, Gallo S, Valdivielso JM, Fernández E, Betriu À, Voltan R, et al. Relationship between low levels of circulating TRAIL and atheromatosis progression in patients with chronic kidney disease. PLoS One. 2018;13:e0203716. DOI: 10.1371/journal.pone.0203716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonçalves I, Singh P, Tengryd C, Cavalera M, Yao Mattisson I, Nitulescu M, Flor Persson A, Volkov P, Engström G, Orho‐Melander M, et al. sTRAIL‐R2 (soluble TNF [tumor necrosis factor]‐related apoptosis‐inducing ligand receptor 2) a marker of plaque cell apoptosis and cardiovascular events. Stroke. 2019;50:1989–1996. DOI: 10.1161/STROKEAHA.119.024379. [DOI] [PubMed] [Google Scholar]
- 23.Mattisson IY, Björkbacka H, Wigren M, Edsfeldt A, Melander O, Fredrikson GN, Bengtsson E, Gonçalves I, Orho‐Melander M, Engström G, et al. Elevated markers of death receptor‐activated apoptosis are associated with increased risk for development of diabetes and cardiovascular disease. EBioMedicine. 2017;26:187–197. DOI: 10.1016/j.ebiom.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chua W, Purmah Y, Cardoso VR, Gkoutos GV, Tull SP, Neculau G, Thomas MR, Kotecha D, Lip GYH, Kirchhof P, et al. Data‐driven discovery and validation of circulating blood‐based biomarkers associated with prevalent atrial fibrillation. Eur Heart J. 2019;40:1268–1276. DOI: 10.1093/eurheartj/ehy815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu SL, Bae YH, Yu C, Monslow J, Hawthorne EA, Castagnino P, Branchetti E, Ferrari G, Damrauer SM, Puré E, et al. Matrix metalloproteinase‐12 is an essential mediator of acute and chronic arterial stiffening. Sci Rep. 2015;5:17189. DOI: 10.1038/srep17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L, Zhang Q, Pu LJ, Peng WH, Yan XX, Wang LJ, Chen QJ, Zhu ZB, Michel JB, Shen WF. Dysregulation of matrix metalloproteinases and their tissue inhibitors is related to abnormality of left ventricular geometry and function in streptozotocin‐induced diabetic minipigs. Int J Exp Pathol. 2008;89:125–137. DOI: 10.1111/j.1365-2613.2008.00579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unal D, Tasdemir A, Oguz A, Eroglu C, Cihan YB, Turak EE, Karaman H, Soyuer S. Is human kallikrein‐11 in gastric cancer treated with surgery and adjuvant chemoradiotherapy associated with survival? Pathol Res Pract. 2013;209:779–783. DOI: 10.1016/j.prp.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Carlsson AC, Ingelsson E, Sundström J, Carrero JJ, Gustafsson S, Feldreich T, Stenemo M, Larsson A, Lind L, Ärnlöv J. Use of proteomics to investigate kidney function decline over 5 years. Clin J Am Soc Nephrol. 2017;12:1226–1235. DOI: 10.2215/CJN.08780816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlsson AC, Sundström J, Carrero JJ, Gustafsson S, Stenemo M, Larsson A, Lind L, Ärnlöv J. Use of a proximity extension assay proteomics chip to discover new biomarkers associated with albuminuria. Eur J Prev Cardiol. 2017;24:340–348. DOI: 10.1177/2047487316676134. [DOI] [PubMed] [Google Scholar]
- 30.Alhenc‐Gelas F, Bouby N, Girolami JP. Kallikrein/K1, kinins, and ACE/Kininase II in homeostasis and in disease insight from human and experimental genetic studies, therapeutic implication. Front Med (Lausanne). 2019;6:136. DOI: 10.3389/fmed.2019.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lind L, Sundström J, Larsson A, Lampa E, Ärnlöv J, Ingelsson E. Longitudinal effects of aging on plasma proteins levels in older adults – associations with kidney function and hemoglobin levels. PLoS One. 2019;14:e0212060. DOI: 10.1371/journal.pone.0212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Folkersen L, Fauman E, Sabater‐Lleal M, Strawbridge RJ, Frånberg M, Sennblad B, Baldassarre D, Veglia F, Humphries SE, Rauramaa R, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. DOI: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]


