ABSTRACT
Human microbiota influence the response of malignancies to treatment with immune checkpoint blockade; however, their impact on other forms of immunotherapy is poorly understood. This study explored the effect of blood microbiota on clinical efficacy, represented by progression-free survival (PFS) and overall survival (OS), of combined chemotherapy and adoptive cellular therapy (ACT) in advanced colon cancer patients.
Plasma was collected from colorectal cancer patients (CRC) treated with either chemotherapy alone (oxaliplatin and capecitabine) (XELOX CT alone group, n = 19), or ACT with a mixed dendritic cell/cytokine-induced killer cell product (DC-CIK) + XELOX (ICT group, n = 20). Circulating microbiota analysis was performed by PCR amplification and next-generation sequencing of variable regions V3~V4 of bacterial 16S rRNA genes. The association of the blood microbial diversity with clinical response to the therapy as measured by RECIST1.1 and OS was evaluated.
The baseline Chao index of blood microbial diversity predicted prolonged PFS and OS of DC/CIK immunotherapy. More diverse blood microbiota that included Bifidobacterium, Lactobacillus, and Enterococcus were identified among responders to DC/CIK compared with non-responders. The plasma bacterial DNA copy number is inversely correlated with the CD3−/CD16+/CD56+ NK cells in circulation and decreased following DC-CIK; however, the Chao index of plasma microbiota significantly increased after administration of the DC-CIK product and this subsequent change was correlated with the number of CD3−/CD16+/CD56+ and CD8+/CD28+ cells infused.
The diversity of the blood microbiome is a promising predictive marker for clinical responses to chemotherapy combined with DC-CIK. Cellular immunotherapy can affect the plasma microbiota’s diversity in a manner favorable to clinical responses.
KEYWORDS: Blood microbiome, adoptive cellular immunotherapy, colorectal cancer
Introduction
The role of the microbiota of the gastrointestinal tract, mouth, skin, and vagina in health and disease has been well established, but more recently, a role for the gut microbiota in modulating the anti-tumor efficacy and toxicity of immune checkpoint inhibitors (ICIs) has been proposed.1–4 For example, high diversity and abundance of particular bacteria in the stool are associated with clinical response to ICI in animal models and humans.5,6 Proposed mechanisms by which the microbiota influence immune responses include microbial metabolites, bacterial molecules that activate pattern-recognition receptors (PRRs), and cross-reactivity between bacterial and tumor antigens.7–10 In addition to the efforts to harness the gut microbiota to enhance ICI, it is critical to understand the effects of microbiota on other immunotherapeutic modalities and the impact of microbiota from other body sites on immunotherapy.
From among the numerous immunotherapy strategies, we have focused on adoptive cell therapy with DC-CIK, a combination of ex vivo generated, autologous dendritic cells (DC) and cytokine-induced killer cells (CIK) which has demonstrated activity alone and in combination with chemotherapy across malignancies.11,12 Pre-clinical models suggest that gut microbiota can modulate tumor responses to adoptive T cell therapy;13 however, there has been limited study of the effect of microbiota on DC-CIK therapy in humans.
Although the gut microbiome would be attractive to study, we reasoned that because the adoptively transferred cells traffic through the vasculature to sites of malignancy, they would interact with the microbiome more directly in the bloodstream. Although the bloodstream is often thought to be “sterile,” recent data suggest the presence of a bloodstream microbiome as well.14 In colorectal cancer (CRC) patients, dysfunctional integrity of subepithelial lamina propria (SBLP) macrophages may allow commensal gut bacteria to invade into the bloodstream15,16 as a source for the blood microbiome. Therefore we have assured that blood microbiome might impact the clinical responses of immunotherapies.17 We investigated whether there was an association between the bloodstream microbiota diversity and efficacy of DC-CIK therapy in CRC patients.
Materials and methods
Setting and participants
This was a retrospective cohort study based on Clinical Trial NCT01906632. The study was approved by the Ethics Committee of Beijing Shijitan Hospital (2019KYLS(51)). All patients gave written informed consent for participation. From March 1, 2014 to January 31, 2019, 39 colorectal cancer patients hospitalized at the Beijing Shijitan Hospital, Capital Medical University Cancer Center, who met the following criteria were enrolled in this study: treated with 2 cycles of standard capecitabine/oxaliplatin chemotherapy (CT group, n = 19) or 2 cycles of capecitabine/oxaliplatin chemotherapy combined with 2 cycles of DC-CIK therapy (Immunochemotherapy (ICT) group, n = 20) (as previously described12 (Figure Suppl 1). There was no DC-CIK alone group after March 1, 2014 when our Ethics Committee deemed it inappropriate to offer only DC-CIK in a setting where there were established survival benefits for chemotherapy. Patients who had received prior immunotherapy or chemotherapy within 8 weeks of the first cycle of XELOX were not included.
Clinical and laboratory data were extracted from the hospital electronic medical record and included age, sex, colorectal cancer stage, history of diabetes or hyperlipidemia (which was defined as cholesterol and/or triglycerides higher than normal level before the first cycle of XELOX); results of CEA, CA19-9, CA-153, and CA-125 before therapy, phenotypes of lymphocytes before therapy and after therapy, results of clinical assessment after 2 cycles of XELOX or XELOX combined with 2 cycles of DC-CIK.
All PFS and OS data were collected based on the hospital electronic database or by telephone interview until January 31, 2020. The loss to follow-up rates of PFS and OS were 10.2% and 7.6%, respectively. Clinical efficacy was evaluated according to Response Evaluation Criteria in Solid Tumors (RECIST)1.1 after 2 cycles of treatment.
Patients were deemed to have clinical benefit and were considered responders (R) if they had a CR (complete response), PR (partial response), or SD (stable disease). They were considered non-responders (NR) if they had progressive disease (PD). Characteristics of all patients before therapy are detailed in Table 1, Table S2, and Table S4. Study flow is presented in Figure S1.
Table 1.
Baseline characteristics of patients treated with ICT and separated into clinical responders (R) and non-responders (NR)
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Variables | ICT-R (n = 13) | ICT-NR (n = 7) | P-value | OR | 95% CI | P-value |
| Gender (F:M) | 8:5 | 4:3 | ||||
| Age (years) median (range) | 60 (36–78) | 60 (47–86) | ||||
| BMI (kg/m2) median (range) | 22.7 (17.7–27.0) | 20.7 (13.7–26.0) | ||||
| Hyperlipemia (n) | 0 | 0 | ||||
| Diabetes (n) | 4 | 0 | ||||
| TNM staging | ||||||
| III(n) | 3 | 0 | ||||
| IV(n) | 10 | 7 | ||||
| Primary surgery for CRC (n) | 8 | 7 | ||||
| Interval between primary surgery and the treatment (month) | 10(1.9–33) | 8(1.9–84) | ||||
| CA-199 (U/ml) median (range) | 31.1 (<2–119.9) | 111.1 (<2–3515.5) | ||||
| CA-125 (U/ml) median (range) | 26.2(2.7–994.1) | 75.8 (9.4–473.3) | ||||
| CA-153 (U/ml) median (range) | 10.1 (6.4–129.2) | 8.4 (6.7–11.8) | ||||
| CEA (ng/ml) median (range) | 8.6 (1.0–>15000) | 21.3 (2.8–898.0) | ||||
| T lymphocyte subgroup | ||||||
| CD3+ (%)median (range) | 74.5(51.7–82.6) | 71.2(61.1–80.5) | ||||
| CD3+/CD4+ (%)median (range) | 36.2(30.0–62.0) | 36.8(32.2–55.3) | ||||
| CD3+/CD8+ (%)median (range) | 28.9(15.9–50.6) | 26.7(22.0–31.4) | ||||
| CD3–/CD16+/CD56+ (%)median (range) | 14.4(9.4–30.3) | 17.2(12.9–29.3) | ||||
| CD3+/CD16+/CD56+ (%)median (range) | 3.1(1.3–11.3) | 1.6(0.9–11.6) | ||||
| CD19+ (%)median (range) | 7.4(3.3–16.3) | 9.7(4.1–10.8) | ||||
| CD4+/CD25+ (%)median (range) | 1.3(0.3–3.3) | 2.6(1.1–3.0) | ||||
| CD8+/CD28– (%)median (range) | 21.7(8.1–39.8) | 18.3(9.7–26.6) | ||||
| CD8+/CD28+ (%)median (range) | 11.3(7.6–18.1) | 10.3(6.9–15.3) | ||||
| CIK cell infused median (range), 1st-cycle | 7.9(3.3–11.1)×109 | 4.7(2.6–6.8)×109 | ||||
| CIK cell infused median (range), 2nd-cycle | 5.7(1.21–9.6)×109 | 4.7(2.2–5.4)×109 | ||||
| Quantification of total bacterial 16S DNA gene copy number (copies/ml): pre-therapy | 70930(39377–111409) | 68610(32377–91399) | ||||
| Bacterial diversity: pre-therapy | ||||||
| Chao | 354.59(189.33–537.94) | 211.50(153.20–321.67) | 0.011 | 1.019 | 1.001–1.037 | 0.042 |
| Shannon | 3.73(2.57–4.54) | 3.05(2.11–4.04) | 0.039 | |||
| Simpson | 0.07(0.03–0.26) | 0.11(0.05–0.19) | ||||
| Median for relative abundance of microbe at genus level(%) | ||||||
| Bifidobacterium | 2.76(0.19–36.68) | 0.33(0.00–1.33) | 0.014 | |||
| Lactobacillus | 2.76(0.12–7.19) | 0.37(0.09–3.34) | 0.014 | |||
| Enterococcus | 0.77(0.02–22.42) | 0.14(0.00–0.53) | 0.030 | |||
| Pseudomonas | 25.43(4.96–44.35) | 39.78(27.85–45.39) | 0.037 | |||
| The P-value is >0.05 when it is not indicated. | ||||||
| ICT: DC-CIK therapy combined with chemotherapy; R, response; NR, no-response. | ||||||
Patients treated with ICT were separated into responders (R) and non-responders (NR) according to the clinical assessment. The gender, age, BMI, number of patients with primary surgery, interval time between primary surgery and the treatment, tumor markers (CA-199, CA-125, CA-153, and CEA), proportion of T lymphocyte subgroup and total number of CIK cells infused was not significantly different between subgroups ICT-R (n = 13) and ICT-NR (n = 7). The total bacterial 16S DNA gene copy number of plasma between sub-group ICT-R and ICT-NR was also not statistically different. The bacterial diversity index (Chao and Shannon) and the relative abundance of Bifidobacterium, Lactobacillus, Enterococcus, Pseudomonas (at genus level) was statistically different before therapy. The multivariate analysis revealed that the Chao index was associated with response to ICT therapy.
Sample collection and preservation
Time points of the sample collection are shown in Figure S1. Fasting venous blood was collected in K2 EDTA tubes (BD Vacutainer) and centrifuged at 1500 rpm for 20 minutes to isolate the plasma fraction, which was placed in DNA-free, sterile collecting tubes and stored frozen at −80°C until further analysis.
DNA extraction and quantification of total bacterial 16S DNA gene copy number
All microbial DNA isolation procedures were performed in a biological flow cabinet, and all consumables used were sterile. QIAamp UCP Pathogen Mini Kit (Cat No. 50214, Qiagen Inc, Venlo, the Netherlands) was used to isolate microbial DNA from plasma following the manufacturer’s instructions. DNA was quantified with a Qubit Fluorometer by using the Qubit® dsDNA BR Assay kit (Invitrogen, USA) and the quality was checked by running aliquots on 1% agarose gels.
Total bacterial 16S rRNA gene copy number in plasma was quantified using TaqMan qPCR as described previously.18 Extraction blanks (DNA-free water) served as environmental DNA contamination controls.
Bacterial 16S rRNA sequencing
The plasma microbial composition and diversity were assessed by real-time full-length 16S rRNA gene sequencing (BGI Genomics, Shenzhen, China). All samples were coded before they were sent to BGI so the technicians performing the assays would be blinded to the study group with which a particular sample was associated.
Library construction
Variable regions V3~V4 of bacterial 16S rRNA gene were amplified with degenerate PCR primers, 341 F (5ʹ-ACTCCTACGGGAGGCAGCAG-3ʹ) and 806 R (5ʹ-GGACTACHVGGGTWTCTAAT-3’). Forward and reverse primers were both tagged with Illumina pad, adapter, and linker sequences. PCR enrichment was performed in 50 μL reactions that contained a fusion PCR primer, a PCR master mix, and 30 ng template. PCR cycling conditions were as follows: 94°C for 3 minutes, 30 cycles of 94°C for 30 seconds, 56°C for 45 seconds, 72°C for 45 seconds, and final extension for 10 minutes at 72°C for 10 minutes. AmpureXP beads were used for PCR product purification, and purified product was eluted in the Elution buffer. Libraries were qualified with an Agilent 2100 bioanalyzer (Agilent, USA). The validated libraries were sequenced on an Illumina MiSeq platform (BGI, Shenzhen, China) following standard pipelines of Illumina, and generating 2 × 300 bp paired-end reads.
1,2 Sequencing and bioinformatics analysis
Raw reads were filtered to remove adaptors, low-quality, and ambiguous bases. Then, paired-end reads were added to tags using Fast Length Adjustment of Short reads program (FLASH, v1.2.11)19 to get the tags. With a cutoff value of 97%, the tags were clustered into operational taxonomic units (OTUs) using the UPARSE software (v7 .0.1090).20 The chimera sequences were detected by comparing with the Gold database using UCHIME (v4.2.40).21 Using the Ribosomal Database Project (RDP) Classifier v.2.2 with a minimum confidence threshold of 0.6, the OTU representative sequences were taxonomically classified. Then, they were trained on the Greengenes database v201305 by QIIME v1.8.0.22 All the tags were compared back to OTU by USEARCH_global23 to get the OTU abundance statistics table of each sample. Alpha and beta diversity were estimated by QIIME (v1.8.0)22 and MOTHUR (v1.31.2)24 at the OTU level, respectively. GraPhlan map of species composition was created using GraPhlAn. Linear discriminant analysis effect size (LEfSe) cluster analysis was conducted using LEfSe. Barplot and heatmap and different classification levels were plotted with the R package v3.4.1 and Morpheus (https://software.broadinstitute.org/morpheus/), respectively.
Statistical analysis
Statistical analysis of patient characters
The non-normal continuous variables were expressed as median (range) and compared using Kruskal–Wallis tests. Categorical variables were compared using chi-square tests.
Statistical analysis for bacteria-related results
Bacterial 16S rRNA gene copy number was compared between different groups using Kruskal–Wallis tests. Alpha diversity (including Chao, Shannon, and Simpson) was compared between different groups using the Mann–Whitney test. Pairwise comparisons of taxonomic abundances based on the response to therapy were also conducted using the Mann–Whitney test. Variables associated with response to therapy in the univariate analysis (P ≤ 0.05) were included in a logistic regression analysis to identify those independently associated with response to therapy. Relationships between variables were determined by Spearman’s correlation analysis. The ability of significant bacterial variables to predict response to DC/CIK therapy was assessed by computing receiver operating characteristic (ROC) curves. The Kaplan–Meier survival method was used to present median PFS and OS between different groups. All statistical evaluations were carried out using SPSS software (Statistical Package for the Social Science, version 17.0, SPSS Inc.) and GraphPad Prism 5 (Version 5.01, GraphPad Software, Inc.). A value of p < .05 was considered to be statistically significant.
Results
Baseline diversity of microbiota is associated with clinical response to immunochemotherapy
Bacterial diversity at the baseline was assessed by subjecting full-length 16S rRNA gene libraries, generated from plasma obtained prior to initiation of therapy (pre-CT (n = 19), pre-ICT (n = 20)), to next-generation sequencing and microbiota statistical analysis. To better understand the impact of the baseline plasma microbiota on the efficacy of immune therapies, the blood microbial composition prior to therapy was compared between the responders (CR/PR/SD) and non-responders (PD). At the OTU level, for patients receiving ICT, the Chao and Shannon indices of diversity were higher for clinical responders than non-responders (Chao: p = .011; Shannon: p = .039) (Table 1, Figure 1a-c). The Simpson index did not differ statistically (p = .096). For patients receiving CT only, there were no differences between clinical responders and non-responders in any of the indices of bacterial diversity (Table S4, Figure S8A-C).
Figure 1.
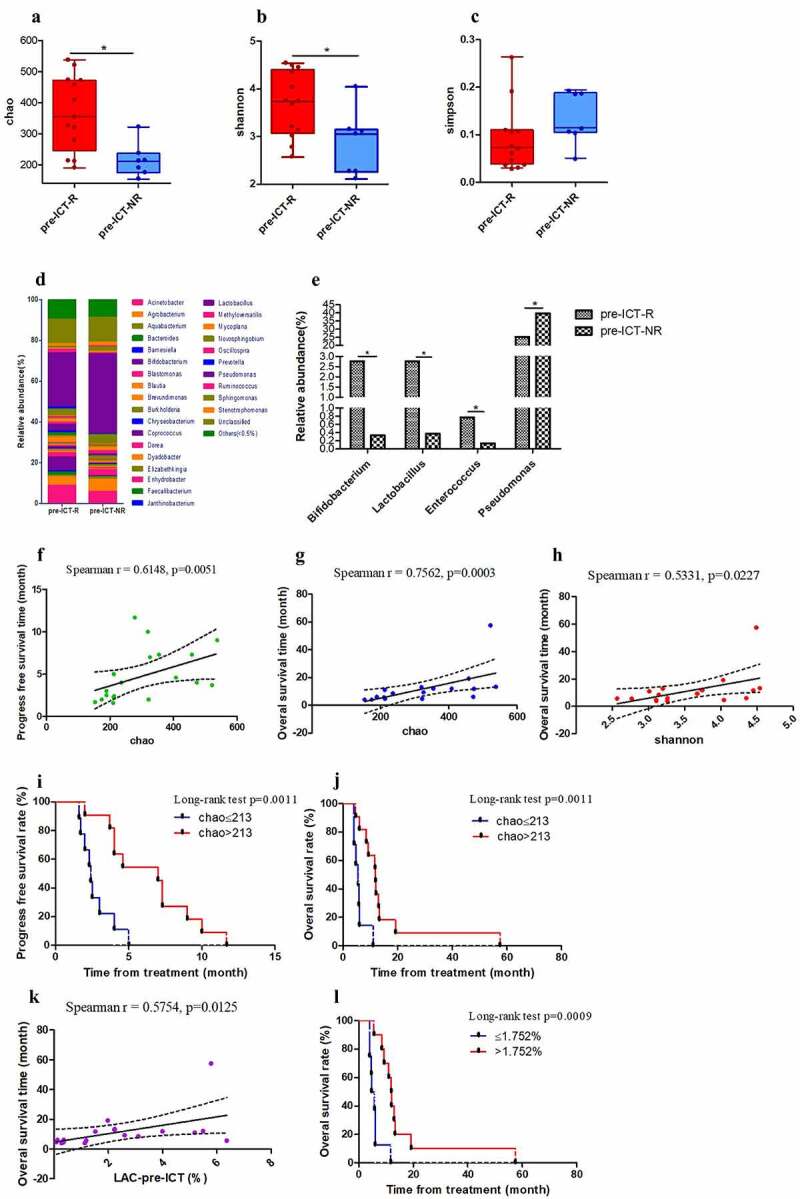
Microbial analysis of pre-ICT samples according to clinical assessment. Chao (a), Shannon (b) and Simpson (c) indices of pre-ICT samples at the operational taxonomic unit (OTU) level according to clinical assessment. Median for relative abundance of OTUs at genus level (d) is shown in R- and NR subgroups by bar plots. Median for relative abundance of Bifidobacterium, Lactobacillus, Enterococcus, Pseudomonas at genus level is displayed by R and NR sub-groups (e). The correlation ship between chao index and PFS (f) or OS (g). The correlation ship between Shannon index and OS (h). Effects of pre-ICT chao index on ICT-patients’ PFS (i) and OS (j). Correlation ship between the relative abundance of Lactobacillus at genus level and OS (k-l). (*p < .05, **p < .01 and ***p < .001). ICT, Immunotherapy combined with chemotherapy; R, response; NR, no-response; pre-, before therapy
In the ICT group, the PFS of responders (subgroup ICT-R) was statistically longer than the PFS of sub-group non-responders (ICT-NR) (p = .0008). OS of the sub-group ICT-R was also statistically different from that of the sub-group ICT-NR (p = .0004). The baseline Chao index of pre-ICT was related to PFS (p = .0051) and OS (p = .0003). The baseline Shannon index of pre-ICT was related to OS (p = .0227). The PFS was statistically different between patients with Chao index ≤213 and Chao index >213 in the ICT group (p = .0011). The OS was also significantly different (p = .0011) between patients with Chao index ≤213 and >213 (Figure 1f-j). In the CT group, the baseline Chao index of pre-CT was not related to PFS (p = .5477) or OS (p = .1043), respectively (Figure S8G-H). The PFS was statistically different between the sub-group CT-R and CT-NR (p = .0053), but OS was not significantly different between the sub-group CT-R and CT-NR (p = .7137). These data suggest that PFS and OS were routinely associated with indices of microbial diversity for the chemotherapy plus DC/CIK (ICT group) but not in the chemotherapy alone group, suggesting that bacterial diversity interacts with DC-CIK.
At the genus level, there were distinct differences in abundance of various bacterial genera between responders and non-responders to ICT (Figure 1d, S8), while in the chemotherapy group, there was no clear distinctive profile based on clinical response (Figure S9 A-E). We found that the relative abundance of Bifidobacterium, Lactobacillus, and Enterococcus at the genus level was significantly higher in pre-ICT-R than in pre-ICT-NR and relative abundance of Pseudomonas was statistically lower in pre-ICT-R than in pre-ICT-NR (Table 1, Figure 1e), but these variables were not independently associated with response to ICT therapy.
The relative abundance of Lactobacillus of pre-ICT at the genus level was related to OS (p = .0125), but was not related to PFS (p = .2114). OS was statistically different between patients with relative abundance of Lactobacillus ≤1.752% and >1.752% in group ICT (p = .0009) (Figure 1k-l). The relative abundance of Bifidobacterium (PFS:p = .5394, OS:p = .1803) and Enterococcus (PFS:p = .7019, OS:p = .3212) of pre-ICT at the genus level was not related to PFS and OS, respectively. Receiver operating characteristic curves (ROC) were established for microbial variables in predicting response to ICT (Figure S10). Areas under the curve (AUCs) were 0.835 for Bifidobacterium (p = .016), 0.846 for Lactobacillus (p = .013) and 0.802 for Enterococcus (p = .029). There was no significant difference in the abundance of these three bacterial subgroups between pre-CT-R and pre-CT-NR (Table S5, Figure S9 D-F). The relative abundance of Lactobacillus of pre-CT at the genus level was not related to PFS (p = .9957) or OS (p = .5973), respectively. These data suggest that the relative abundance of Lactobacillus of pre-therapy may potentially be used to predict the prognosis of patients treated with DC-CIK.
Changes in bacterial diversity during ICT are associated with the number of DC/CIK cells infused
After 1 cycle of ICT, there were significant differences for Shannon (p = .025) and Simpson (p = .014) indices at the operational taxonomic unit (OUT) level (Figure 2e-f). Specifically, the Shannon and Simpson indices were greater following a cycle of ICT compared with the baseline indices. In contrast, there were no statistically significant differences in any of the indices following a cycle of CT compared with the baseline indices (Figure 2a-c).
Figure 2.
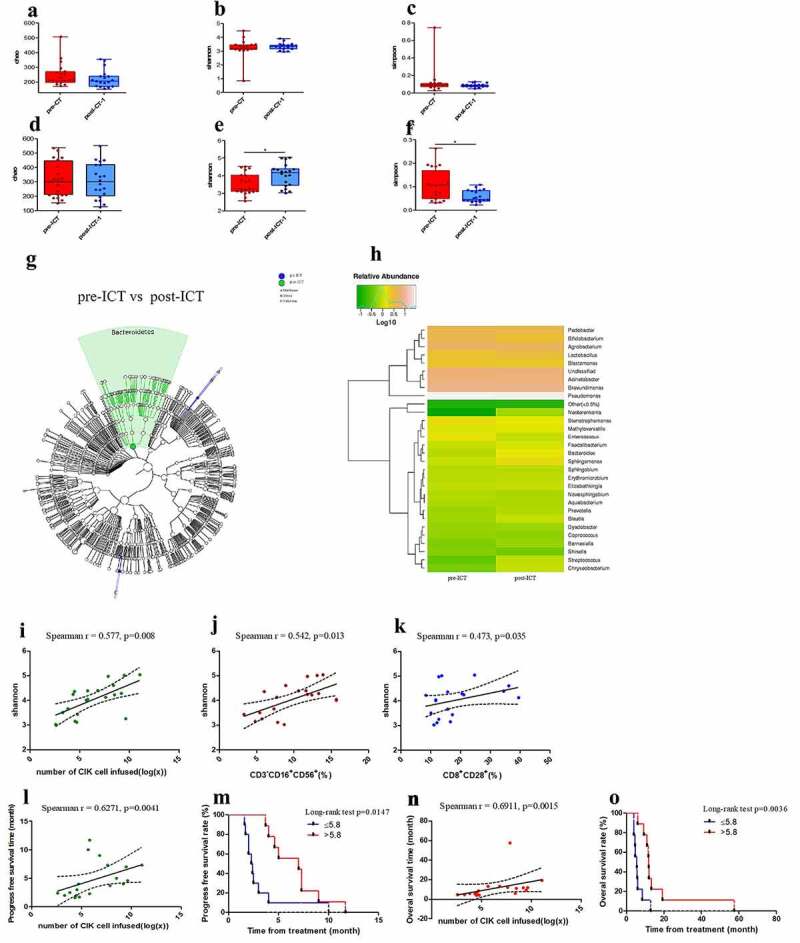
Bacterial diversity is increased by ICT and is associated with the number of CIK cells infused. Pairwise comparison of Chao (a, d), Shannon (b, e) and Simpson (c, f) indices at the operational taxonomic unit (OTU) level between pre- and post- therapy in the CT and ICT groups. Shannon (p = .025) and Simpson (p = .014) at OTU level were statistically different respectively between pre- and post- ICT; however they were not significantly different in the CT group. GraPhlAn output representing linear discriminant analysis effect size results which identified main features that are differentially abundant between pre- and post-therapy in the ICT group (g). Median for relative abundance of OTUs at genus level was shown in different pre- and post-ICT groups by bar plots (h). Spearman’s correlation analysis on Shannon (post-ICT-1) and the number of CIK cell infused, CD3−/CD16+/CD56+ and CD8+/CD28+ T-cell proportion in CIK infused (i-k), Spearman’s correlation analysis on the number of CIK cell infused and PFS (l) and OS (n), displaying linear fit curve (solid line) and 95% CI (dashed line). Effects of the CIK cell infused on ICT-patients’ PFS (m) and OS (o). (*p < .05, **p < .01 and ***p < .001). CT, chemotherapy; ICT, Immunotherapy combined with chemotherapy. pre-, before therapy; post-, after therapy; post-therapy-1, after 1 cycle of treatment
At the OTU level, the Chao, Shannon, and Simpson diversity were not significantly different between the group CT and ICT before therapy, respectively, but the Chao (p = .027), Shannon (p = .001), and the Simpson (p = .003) indices were statistically different between the CT and ICT groups after the therapy (Figure S6).
LEfSe cluster analysis identified that the relative abundance of Bacteroidetes at the phylum level was different between pre- and post-ICT plasma samples. The relative abundance of Bacteroides at the genus level was increased after ICT compared with pre-ICT (Figure 2g-h), but it was decreased after CT compared with pre-CT levels (Figure S7).
The elevated Shannon index in the post-ICT group was related to the number of CIK cells infused as shown by a positive Spearman’s correlation (r = 0.577, p = .008) and was closely related to the number of CD3–/CD16+/CD56+ (r = 0.542, p = .013) and CD8+/CD28+ (r = 0.473, p = .035) cells infused, respectively (Figure 2i-k).
Although both the number of CD3–/CD16+/CD56+ (PFS: p = .1748, OS: p = .0799) and CD8+/CD28+ (PFS: p = .1748, OS: p = .9448) infused were not related to PFS and OS, respectively, we have found that PFS of the ICT group was related to the number of CIK cells infused (r = 0.6271, p = .0041), as well as OS (r = 0.6911, p = .0015). The PFS was statistically different between patients with CIK cells infused ≤5.8 × 109 and >5.8 × 109 (p = .0147), and OS was also different between these two groups (p = .0036) (Figure 2 l-o). These data suggest that infusion of CIK cells more than 5.8 × 109 will significantly improve the survival of patients.
Bacterial DNA quantity decreased following immunochemotherapy and negatively correlated with the CD3–/CD16+/CD56+ NK cell proportion in peripheral blood
All the pre- and post-therapy samples were tested for bacterial 16S rRNA gene copy number. Before therapy, the bacterial rRNA gene copies in the plasma of the CT and ICT groups were not significantly different (Figure 3a). After 1 and 2 cycles of standard chemotherapy, compared with pre-CT, bacterial DNA copies in the samples of post-CT-1 (after 1 cycle of chemotherapy) and post-CT-2 (after 2 cycles of chemotherapy) were not significantly different, respectively (Figure 3b). There were only 18 of 19 samples in the post-CT-2 group that were positive for 16S DNA. For the patients who were treated with 2 cycles of standard chemotherapy and 2 cycles of DC-CIK therapy, significantly lower bacterial DNA copies were found in the peripheral blood samples of the group post-ICT-2 (after 2 cycles of chemotherapy+DC-CIK: n = 16) compared with pre-ICT (p = .002) and post-ICT-1 (after 1 cycle of chemotherapy+DC-CIK; p = .020. Figure 3c).
Figure 3.
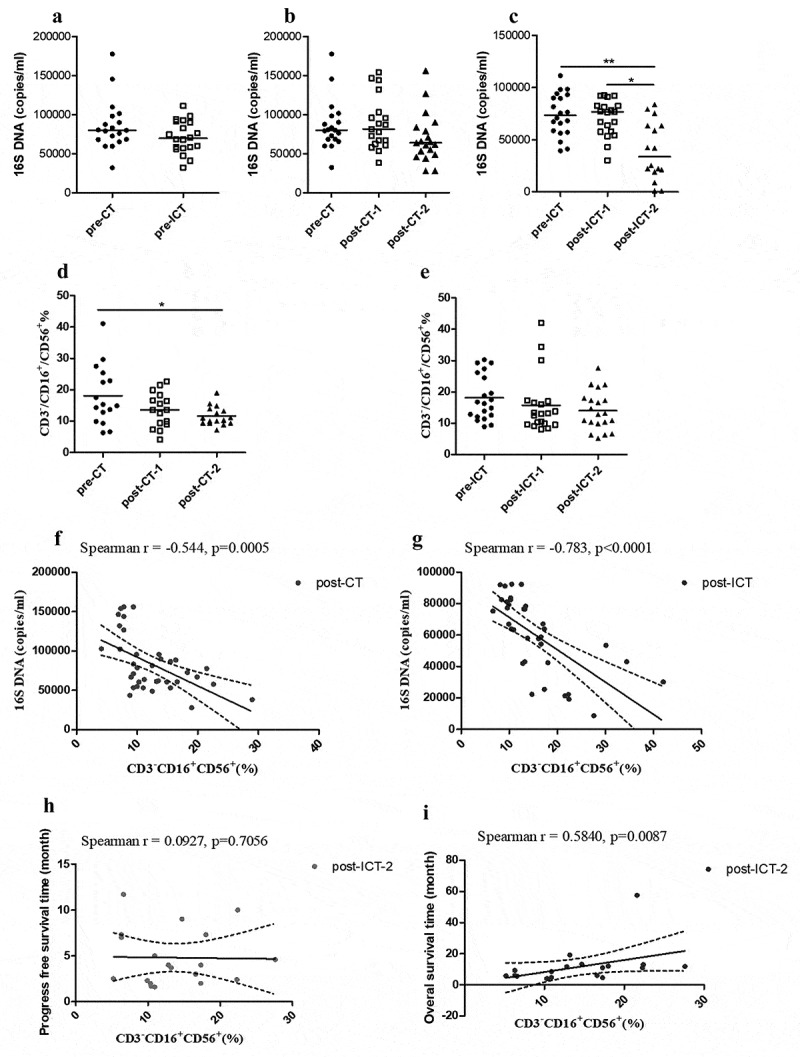
The bacterial DNA quantity of plasma was reduced by therapy in the ICT group. Bacterial 16S DNA baseline in pre-CT (n = 19) and pre-ICT (n = 20) samples (a). Comparison of plasma quantities of bacterial 16S DNA in paired pre-therapy and post-therapy CT and ICT patients (b, c) (post-CT-2: n = 18; post-ICT-2: n = 16). Median for 16S DNA copies is indicated per each group. Comparison of CD3−/CD16+/CD56+ T-cell proportion in peripheral blood before and after 2 cycle of therapy (d, e). Median for proportion of CD3−/CD16+/CD56+ cells is indicated per each group. Spearman’s correlation analysis on proportion of CD3−/CD16+/CD56+ cells and bacterial 16S DNA copy numbers (f, g), Spearman’s correlation analysis on proportion of post-ICT-2 CD3−/CD16+/CD56+ cells and PFS (h) and OS (i), displaying linear fit curve (solid line) and 95% CI (dashed line). (*p < .05, **p < .01 and ***p < .001). CT, chemotherapy; ICT, Immunotherapy combined with chemotherapy; pre-, before therapy; post-, after therapy; post-therapy-1, after 1 cycle of treatment; post-therapy-2, after 2 cycle of treatment
Lower amounts of plasma bacterial DNA were associated with higher proportions of CD3–/CD16+/CD56+ cells in peripheral blood, as shown by a negative Spearman’s correlation both in the CT group (r = −0.544, p = .0005) and in the ICT group (r = −0.783, p < .0001). The CD3–/CD16+/CD56+ T cell subset of peripheral blood significantly decreased in post-CT-2 compared with pre-CT (p = .045), but it did not decrease statistically after therapy in the ICT group (Figure 3d-g). These data suggest that peripheral blood CD3–/CD16+/CD56+ cells, which are maintained by DC-CIK therapy, may reduce bacteria in the bloodstream. In the ICT group, the CD3–/CD16+/CD56+ T cell subset of peripheral blood after 2 cycles of ICT was related to OS (r = 0.5840, p = .0087) but not related to PFS (r = 0.0927, p = .7056). In group CT, the peripheral blood CD3–/CD16+/CD56+ T cell subset of post-CT-2 was not related to PFS (r = 0.1596, p = .5548) and OS (r = 0.1378, p = .5857), respectively.
Discussion
Adoptive cellular immunotherapy (ACT) is an emerging strategy for cancer treatment, capable of direct cytotoxicity against cancer cells, modulation of immunosuppressive T cell populations, and enhancing recovery of exhausted T cells in vivo.25–27 We have previously reported the significant antitumor activity of DC/CIK therapy11,12,28 alone and in conjunction with chemotherapy.
A number of host and tumor factors modulate the efficacy of immunotherapy. The gut microbiome, for example, alters responsiveness to immune checkpoint inhibitors (ICI).5,29–31 Greater richness and diversity of gut microbiota are found in ICI responders. Pre-clinical mouse models suggest that gut microbiota may modulate tumor response to ACT;13,32 however, little is known about the interactions of peripheral blood bacteria, presumably present as a result of translocation of gut bacteria across the bowel epithelium, with ACT. In the current study, the bacterial richness, diversity, and composition (at the operational taxonomic unit (OTU) level) in the peripheral blood of colorectal cancer patients were analyzed before and after initiating immunochemotherapy (ICT) to explore the relationship between the blood microbiota and the clinical efficacy of ICT. Bacterial 16S rRNA gene copy number in plasma was also measured pre- and post-therapy in order to understand the effect of ICT on permeability of the gut barrier and bacteremia.
We observed that plasma microbiota before therapy in ICT responders was significantly different from that in ICT non-responders. Specifically, a highly rich and diverse plasma microbiota (as evidenced by higher Chao and Shannon indices) prior to ICT was found in responders. The baseline Chao index of ICT was related to PFS and OS. The Shannon index of pre-ICT was also related to OS. In the CT group, the richness and diversity of plasma microbiota prior to therapy was not significantly different between responders and non-responders. Although we do not have an ACT alone group, these findings suggest that the clinical benefits of DC/CIK infusions are affected by the blood microbial diversity.
Others have demonstrated that specific strains, including Lactobacillus rhamnosus, Bifidobacterium longum, and Enterococcus faecium, may influence the clinical response to ICI.31,33,34 When we analyzed the composition of plasma microbiota according to response to ICT, we found that before therapy, the relative abundance of Bifidobacterium, Lactobacillus, and Enterococcus in responders was significantly higher than that in non-responders, respectively, but there were no statistical differences in the abundance of these bacteria between responders and non-responders in the CT group. The relative abundance of Lactobacillus of pre-ICT was related to OS and when it was >1.752% (at the genus level), the ICT treatment was more effective. Although the relative abundance of Bifidobacterium and Enterococcus was not independently associated with the response to ICT therapy, the receiver operating characteristic curves (ROC) for these bacteria showed potential in predicting the response to ICT.
In addition to describing the impact of microbiota on the efficacy of ACT, we found that the diversity of plasma bacteria in CRC patients was affected by ICT such that greater diversity of plasma microbiota was found after ICT therapy compared with that before therapy. This may be due to direct effects of the DC-CIK on the plasma microbiome. Alternatively, because chemotherapy can cause intestinal wall disruption and sub-mucosal inflammation,35,36 a decrease in plasma bacterial 16S copy number in the ICT group (but not in the CT group) after 2 cycles of ACT administration, as we observed, may be due to positive effects on the mucosal barrier function of the gut, which would in turn enhance the efficacy of local and systemic anti-tumor immunity.
How the DC/CIK product interacts with the blood microbiome is still under evaluation. The DC-CIK product contains multiple cell types. We observed that the Shannon index of plasma bacterial diversity post-ICT was directly related to the number of CIK cells infused and the CD3–/CD16+/CD56+ and CD8+/CD28+ proportions within the infused CIK, suggesting a direct or indirect role of these cell types in sculpting the blood microbiome. For example, peripheral blood natural killer (NK) cells can localize to the gut and are essential for gut mucosal epithelial cell survival and barrier remodeling.37 Although the CT group experienced a decrease in CD3–/CD16+/CD56+ NK cells during therapy, there was no statistical decrease in the ICT group. Correlation analysis indicated that after therapy, the CD3–/CD16+/CD56+ cell subset in peripheral blood was negatively correlated with bacteria copy number, both in the CT and the ICT groups. The CD3–/CD16+/CD56+ cell subset in peripheral blood after 2 cycles of ICT was related to OS. These data suggest that the peripheral blood CD3–/CD16+/CD56+ cells might be maintained by ICT, and they in turn may have a role in repairing the gut barrier and enhancing bacterial diversity that benefited clinical outcomes.
CD3–/CD16+/CD56+ cells play a critical role in combating transformed and malignant cells.38,39 Increased peripheral blood CD8+/CD28+ T cells better predict early response to tumor therapy40 and are associated with length of progression-free survival (PFS) and overall survival (OS).41 As the number of CIK cells infused was associated with PFS and OS, it is possible that the DC-CIK product had direct antitumor effects but also may have enhanced the immune response by increasing the diversity of the blood microbiome. Therefore, modifications to increase the number of CIK cells infused and the proportion of CD3–/CD16+/CD56+ and CD8+/CD28+ T cell subtypes within the infused cellular product are warranted to enhance the efficacy of the ACT.
We also found that the difference in bacterial composition before and after ICT therapy was mainly manifest within the Bacteroides subgroup. The relative abundance of Bacteroides in plasma of CRC patients increased after ICT compared with pre-ICT, while it decreased after CT. Studies have provided evidence that Bacteroides are beneficial to human health and play key roles in interacting with host immunity.42 The relative abundance of Bacteroides may be increased by ICT, and this might contribute to a greater anti-tumor response.
In this study, we found that the baseline microbiome in peripheral blood prior to treatment of group CT and ICT was similar, while the relative abundance of Bifidobacterium was statistically different. Ohigashi et al.43 reported that obligate anaerobes such as Bacteroides and Bifidobacterium were diminished following colorectal cancer surgery. In contrast, the aerobe Pseudomonas was observed to increase after surgery. And this may be because of exposure to oxygen during colorectal surgery reported by Shogan et al.44 Although there was no significant difference in the proportion of primary colorectal cancer surgery between the CT and ICT groups, the interval time between primary surgery and treatment of group CT and ICT was statistically different. The interval time of the CT group was significantly less than that of the ICT group and the relative abundance of Bifidobacterium (p = .0046) and Lactobacillus (p = .0127) was related to interval time, respectively. This suggests that an appropriate interval time after primary surgery is needed to recover the damaged gut microbiome.
Serum levels of tumor marker molecules, carcinoembryonic antigen (CEA), carbohydrate antigen (CA-199), and CA-125 are routinely used in clinical practice to determine prognosis and monitor therapeutic responses in gastrointestinal cancers. CA-199 would decrease after chemotherapy combined with DC-CIK.12,45,46 In this study, CA-199 of patients who were not responders to combined therapy was increased significantly after ICT compared with before therapy. While in combined-responders, CA-199 was not increased statistically after therapy.
One limitation of this study is the lack of a DC-CIK therapy alone group that could not be included for ethical reasons. The use of antibiotics within 8 weeks before ICI therapy is associated with shorter OS;30 however, use of antibiotics within 8 weeks before DC-CIK therapy only occurred in two patients in this study.
In summary, mapping and modulating the microbiota to predict and improve therapeutic outcomes are areas of intense focus in cancer immunotherapy.47,48 In this study, the blood microbiota could predict the response to chemotherapy plus DC/CIK infusions. Follow-up clinical trials are needed to confirm these results. Further prospective testing of DC-CIK as a means to modulate the microbiome in animal models and larger clinical trials is needed.
Supplementary Material
Funding Statement
This work was supported by Enhancement Funding of Laboratory of Beijing Key Laboratory for Therapeutic Cancer Vaccines, Beijing Shijitan Hospital, Capital Medical University [2020-JS01]; Key Project of Beijing Municipal Committee of Science and Technology-Capital Clinical Featured Application Funding, Beijing [Z151100004015183].
Ethics approval and consent to participate
This was a retrospective cohort study based on Clinical Trial NCT01906632. The study was approved by the Ethics Committee of Beijing Shijitan Hospital (2019KYLS(51)). All patients gave written informed consent for participation.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure Statement
The authors declare that they have no competing interests.
Author contributions
Duo Yang, Xiaoli Wang, Xinna Zhou, Shuo Wang, Jing Zhao, Huabing Yang, Michael A. Morse, Amy Hobeika, Jun Ren, and Herbert Kim Lyerly conceived and designed the project and contributed to the interpretation of data.
Duo Yang, Jiangping Wu, Yanhua Yuan, Sha Li, and Jun Ren contributed to the acquisition and analysis of data.
Duo Yang, Michael A. Morse MD, Jun Ren, and Herbert Kim Lyerly drafted the manuscript.
Jun Ren and Herbert Kim Lyerly jointly supervised this work.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Wang Y, Wiesnoski DH, Helmink BA , et al.Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. NAT MED. 2018;24:1804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee KA, Shaw HM, Bataille V, Nathan P, Spector TD . Role of the gut microbiome for cancer patients receiving immunotherapy: Dietary and treatment implications. EUR J CANCER. 2020;138:149–55. [DOI] [PubMed] [Google Scholar]
- 3.Baruch EN, Youngster I, Ben-Betzalel G , et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. SCIENCE. 2021;371:602–9. [DOI] [PubMed] [Google Scholar]
- 4.Pezo RC, Wong M, Martin A . Impact of the gut microbiota on immune checkpoint inhibitor-associated toxicities. Therap Adv Gastroenterol. 2019;12:321907521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gopalakrishnan V, Spencer CN, Nezi L , et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. SCIENCE. 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Lv J, Guo F , et al. Gut Microbiome Influences the Efficacy of PD-1 Antibody Immunotherapy on MSS-Type Colorectal Cancer via Metabolic Pathway. FRONT MICROBIOL. 2020;11:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebbandi NR, Ronchi F, Wang J , et al. A Gut Microbial Mimic that Hijacks Diabetogenic Autoreactivity to Suppress Colitis. CELL. 2017;171:655–67. [DOI] [PubMed] [Google Scholar]
- 8.Keogh CE, Rude KM, Gareau MG . Role of pattern recognition receptors and the microbiota in neurological disorders. J Physiol. 2021;599:1379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith PM, Howitt MR, Panikov N , et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. SCIENCE. 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuo Q, Yu B, Zhou J , et al. Lysates of Lactobacillus acidophilus combined with CTLA-4-blocking antibodies enhance antitumor immunity in a mouse colon cancer model. Sci Rep. 2019;9:20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao G, Wang X, Zhou L , et al. Autologous Dendritic Cell-Cytokine Induced Killer Cell Immunotherapy Combined with S-1 Plus Cisplatin in Patients with Advanced Gastric Cancer: A Prospective Study. CLIN CANCER RES. 2019;25:1494–504. [DOI] [PubMed] [Google Scholar]
- 12.Jiang N, Qiao G, Wang X , et al. Dendritic Cell/Cytokine-Induced Killer Cell Immunotherapy Combined with S-1 in Patients with Advanced Pancreatic Cancer: A Prospective Study. CLIN CANCER RES. 2017;23: 5066–73. [DOI] [PubMed] [Google Scholar]
- 13.Uribe-Herranz M, Bittinger K, Rafail S , et al.Gut microbiota modulates adoptive cell therapy via CD8alpha dendritic cells and IL-12 JCI Insight. 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castillo DJ, Rifkin RF, Cowan DA, Potgieter M The Healthy Human Blood Microbiome: Fact or Fiction? Front Cell Infect Microbiol. 2019;9:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubio CA, Schmidt PT . Severe Defects in the Macrophage Barrier to Gut Microflora in Inflammatory Bowel Disease and Colon Cancer. ANTICANCER RES. 2018;38:3811–5. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Dong W, Wang S , et al. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. FOOD FUNCT. 2018;9:5588–97. [DOI] [PubMed] [Google Scholar]
- 17.Ouaknine KJ, Helly DTP, Dumenil C , et al. Role of antibiotic use, plasma citrulline and blood microbiome in advanced non-small cell lung cancer patients treated with nivolumab. J IMMUNOTHER CANCER. 2019;7:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Lin S, Kelen GD , et al. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J CLIN MICROBIOL. 2002;40:3449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magoc T, Salzberg SL , FLASH: fast length adjustment of short reads to improve genome assemblies. BIOINFORMATICS. 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC, UPARSE: highly accurate OTU sequences from microbial amplicon reads. NAT METHODS. 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . UCHIME improves sensitivity and speed of chimera detection. BIOINFORMATICS. 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J , et al. QIIME allows analysis of high-throughput community sequencing data. NAT METHODS. 2010;7:335–6. doi: 10.1093/bioinformatics/btq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC, et al. Search and clustering orders of magnitude faster than BLAST. BIOINFORMATICS. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- 24.Schloss PD, Westcott SL, Ryabin T , et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009; 75: 7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garofano F, Gonzalez-Carmona MA, Skowasch D , et al. Clinical Trials with Combination of Cytokine-Induced Killer Cells and Dendritic Cells for Cancer Therapy. INT J MOL SCI. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirjang S, Alizadeh N, Mansoori B , et al. Promising immunotherapy: Highlighting cytokine-induced killer cells. J CELL BIOCHEM. 2019;120:8863–83. [DOI] [PubMed] [Google Scholar]
- 27.Capellero S, Erriquez J, Melano C , et al. Preclinical immunotherapy with Cytokine-Induced Killer lymphocytes against epithelial ovarian cancer. Sci Rep. 2020;10:6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Wang X, Zhou X, Lyerly HK, Morse MA , Ren J. DC-CIK as a widely applicable cancer immunotherapy. Expert Opin Biol Ther. 2020;20:601–7. [DOI] [PubMed] [Google Scholar]
- 29.Abu-Sbeih H, Wang Y . Gut Microbiome and Immune Checkpoint Inhibitor-Induced Enterocolitis. Dig Dis Sci. 2020;65:797–9. [DOI] [PubMed] [Google Scholar]
- 30.Routy B, Le Chatelier E, Derosa L , et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. SCIENCE. 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 31.Matson V, Fessler J, Bao R , et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. SCIENCE. 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulos CM, Wrzesinski C, Kaiser A , et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J CLIN INVEST. 2007;117: 2197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivarelli S, Salemi R, Candido S , et al. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Gut Microbiota and Cancer: From Pathogenesis to Therapy. 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vetizou M, Pitt JM, Daillere R , et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. SCIENCE. 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalla RV, Ashbury FD . The MASCC/ISOO mucositis guidelines: dissemination and clinical impact. SUPPORT CARE CANCER. 2013;21:3161–3. doi: 10.3389/fimmu.2019.00961. [DOI] [PubMed] [Google Scholar]
- 36.Kozloff M, Yood MU, Berlin J , et al.Clinical outcomes associated with bevacizumab-containing treatment of metastatic colorectal cancer: the BRiTE observational cohort study. ONCOLOGIST. 2009;14:862–70. doi: 10.1172/JCI129338. [DOI] [PubMed] [Google Scholar]
- 37.Poggi A, Benelli R, Vene R , et al.Human Gut-Associated Natural Killer Cells in Health and Disease. FRONT IMMUNOL. 2019;10:961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodgins JJ, Khan ST, Park MM, Auer RC, Ardolino M . Killers 2.0: NK cell therapies at the forefront of cancer control. J CLIN INVEST. 2019;129:3499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morvan MG, Lanier LL . NK cells and cancer: you can teach innate cells new tricks. NAT REV CANCER. 2016;16:7–19. [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Hu Q, Hu K , et al. Increased CD8+CD28+ T cells independently predict better early response to stereotactic ablative radiotherapy in patients with lung metastases from non-small cell lung cancer. J TRANSL MED. 2019;17:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang L, Qiao G, Morse MA , et al. Predictive significance of T cell subset changes during ex vivo generation of adoptive cellular therapy products for the treatment of advanced non-small cell lung cancer. ONCOL LETT. 2019;18:5717–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibiino G, Lopetuso LR, Scaldaferri F, Rizzatti G, Binda C, Gasbarrini A . Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Dig Liver Dis. 2018;50:635–9. [DOI] [PubMed] [Google Scholar]
- 43.Ohigashi S, Sudo K, Kobayashi D, Takahashi T, Nomoto K, Onodera H . Significant changes in the intestinal environment after surgery in patients with colorectal cancer. J GASTROINTEST SURG. 2013;17:1657–64. [DOI] [PubMed] [Google Scholar]
- 44.Shogan BD, Smith DP, Christley S, Gilbert JA, Zaborina O, Alverdy JC . Intestinal anastomotic injury alters spatially defined microbiome composition and function. MICROBIOME. 2014;2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan X, Zhang AZ, Ren YL , et al. Cytokine-induced killer cells/dendritic cells and cytokine-induced killer cells immunotherapy for the treatment of esophageal cancer: A meta-analysis. Medicine (Baltimore). 2021;100:e24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao YJ, Sy ED, Hsu HP, Shan YS . Predictors for resectability and survival in locally advanced pancreatic cancer after gemcitabine-based neoadjuvant therapy. BMC SURG. 2014;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandi G, Frega G . Microbiota: Overview and Implication in Immunotherapy-Based Cancer Treatments. INT J MOL SCI. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz DJ, Rebeck ON, Dantas G. Complex interactions between the microbiome and cancer immune therapy . Crit Rev Clin Lab Sci. Medicine (Baltimore). 2019;56:567–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


