Abstract
The transcription factor CHOP (C/EBP homologous protein 10) is a bZIP protein induced by a variety of stimuli that evoke cellular stress responses and has been shown to arrest cell growth and to promote programmed cell death. CHOP cannot form homodimers but forms stable heterodimers with the C/EBP family of activating transcription factors. Although initially characterized as a dominant negative inhibitor of C/EBPs in the activation of gene transcription, CHOP-C/EBP can activate certain target genes. Here we show that CHOP interacts with members of the immediate-early response, growth-promoting AP-1 transcription factor family, JunD, c-Jun, and c-Fos, to activate promoter elements in the somatostatin, JunD, and collagenase genes. The leucine zipper dimerization domain is required for interactions with AP-1 proteins and transactivation of transcription. Analyses by electrophoretic mobility shift assays and by an in vivo mammalian two-hybrid system for protein-protein interactions indicate that CHOP interacts with AP-1 proteins inside cells and suggest that it is recruited to the AP-1 complex by a tethering mechanism rather than by direct binding of DNA. Thus, CHOP not only is a negative or a positive regulator of C/EBP target genes but also, when tethered to AP-1 factors, can activate AP-1 target genes. These findings establish the existence of a new mechanism by which CHOP regulates gene expression when cells are exposed to cellular stress.
The transcription factor CHOP (C/EBP homologous protein 10, also known as GADD153) (17, 38) was initially isolated based on its inducibility by genotoxic stress (17). Subsequently it was shown that the expression of the chop gene is induced by a variety of environmental signals that provoke cellular stress, such as nutrient deprivation (10, 11), hypoxia (35), and protein misfolding in the endoplasmic reticulum (ER) (6, 13, 23, 48). The expression of CHOP appears to play a role in the control of the cell division cycle since its forced overexpression in cells results in growth arrest (5, 52) and since an altered form of CHOP, TLS-CHOP, formed by a reciprocal translocation of the chop gene and a gene encoding an RNA-binding protein, is associated with human myxoid liposarcoma (14, 36). CHOP has also been implicated in the inhibition of adipocyte differentiation (7) and the programmed cell death pathway (54). Embryonic fibroblasts obtained from mice with a targeted disruption of the chop gene are partially resistant to apoptosis-inducing stimuli (54).
CHOP is a member of a large family of transcription factors known as bZIP proteins because they have structurally similar DNA-binding and dimerization domains (reviewed in reference 30). These domains consist of a sequence of basic amino acids followed by a sequence of leucine heptad repeats that constitute a surface for protein dimerization, a so-called leucine zipper. Broadly defined, the bZIP proteins are composed of at least three subfamilies of proteins: the cyclic AMP (cAMP)-responsive CREB/ATF factors, the immediate-early response Jun- and Fos-related proteins that collectively make up the activator protein 1 (AP-1) complex and are involved in cell proliferation, and the C/EBPs, of which CHOP is a member. The functions of bZIP proteins in the regulation of gene transcription require that they form dimers, either homo- or heterodimers. The proteins within a given bZIP subfamily readily form homo- and heterodimers and thereby provide a diversity of transcription factor complexes to regulate transcription by interactions with specific DNA control sequences. Dimerization of bZIP factors of a subfamily with those of another subfamily is much more selective and is somewhat unusual. Although CHOP was initially determined to function as a dominant negative inhibitor of gene transcription by forming stable heterodimers with C/EBPs and preventing them from binding to DNA (38), later studies showed that CHOP-C/EBP heterodimers are capable of recognizing novel DNA target sequences and thereby of activating gene transcription (45).
The AP-1 complex is composed of both homo- and heterodimers of the Jun and Fos families of transcription factors. There are three distinct Jun proteins, i.e., c-Jun, JunB, and JunD, and four Fos members, i.e., c-Fos, FosB, Fra1, and Fra2 (reviewed in reference 4). Jun/Fos heterodimers are transactivators of transcription, whereas Jun homodimers may be activators or repressors depending on the context of the promoter context, the sequence of the cognate DNA-binding site, the cell phenotype, and the environmental milieu of the cell, e.g., conditions that favor quiescence or proliferation. The Fos proteins do not form stable homodimers. JunB, c-Jun, and the Fos proteins are rapidly and markedly induced when serum-deprived quiescent cells are induced to proliferate by repletion of serum; this is the immediate-early response. The behavior of JunD is quite different from that of c-Jun and JunB. Serum repletion of serum-starved cells does not induce JunD. Rather, the levels of JunD are increased in quiescent compared to growing cells and the overexpression of JunD slows cell growth. Moreover, the cellular distribution of the expression of JunD differs from that of the c-Jun and the Fos proteins.
In an earlier report we described the activation of the promoter of the somatostatin gene by the interactions of C/EBPβ with the cAMP response element (CRE) of the promoter (46). We also showed by DNA-protein-binding assays that multiple proteins bound to the somatostatin CRE (46). We now show that one of these proteins is JunD and, unexpectedly, that CHOP directly interacts with JunD and other members of the AP-1 family of transcription factors, c-Jun and c-Fos, to activate the transcription of certain genes. Thus a cross talk exists between CHOP, a member of the C/EBP family, and the AP-1 family of bZIP proteins. These findings are somewhat surprising because the bZIP regions of the Jun/Fos subfamily of transcription factors have only modest similarities to the corresponding bZIP regions of the C/EBP proteins (30) and would not necessarily be expected to interact with them. These findings further emphasize that the transcription factor CHOP can serve either as a dominant negative inhibitor of the binding of C/EBPs to certain regulatory elements in the promoter of genes or as an activator of gene transcription when tethered to members of the Jun/Fos proteins that comprise the family of AP-1 complex factors.
MATERIALS AND METHODS
Reagents.
DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) or Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Radioactive compounds were obtained from Du Pont-New England Nuclear (Boston, Mass.). Nucleotides were purchased from Pharmacia-LKB (Piscataway, N.J.). Tissue culture media and reagents were obtained from Gibco-BRL (Grand Island, N.Y.). All other molecular biology grade reagents were obtained from Sigma Chemical Co. (St. Louis, Mo.). Oligonucleotides were synthesized at the Molecular Biology Core Facility of the Massachusetts General Hospital.
Cell lines.
Rat islet somatostatin-producing RIN-1027-B2 (33), HeLa, and human choriocarcinoma JEG-3 (ATCC 36-HTB) cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. NIH 3T3 fibroblasts (ATCC 1658-CRL) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum. All cells were cultured in the presence of the antibiotics penicillin (100 U/ml) and streptomycin (100 μg/ml).
Plasmid constructs.
The transcriptional reporter construct containing the coding sequence for the chloramphenicol acetyltransferase (CAT) driven by the somatostatin CRE in front of a minimal promoter (SCRE TK CAT) has been described previously (47), as has the reporter driven by the human JunD promoter (−219-CAT) (9). The CAT reporter driven by the collagenase promoter was created by inserting a double-stranded oligonucleotide corresponding to the collagenase 12-O-tetradecanoylphorbol-13-acetate response element (TRE) and its flanking sequence in front of the minimal promoter (−41TK CAT). The oligonucleotide sequence was 5′CCAAGAGGATGTTATAAAGCATGAGTCAGACACCTCTGGCTTTCT3′. Expression plasmids for CHOP, CHOP LZ− (38), CHOPΔBR (5), CREB (46), and c-Jun, c-Fos, and JunD (25) have been described previously. To express a JunD-Gal4 DNA-binding domain fusion protein, the coding region of the mouse JunD cDNA was amplified by PCR and inserted into the pM plasmid (Clontech, Palo Alto, Calif.) already containing the sequence for the DNA binding domain of Gal4. The resulting construct, pM-JunD, was sequenced and tested for appropriate expression of the fusion protein. A luciferase reporter vector containing three binding sites for Gal4 was described previously (48).
Transfections and transactivation assays.
JEG-3 cells and NIH 3T3 fibroblasts were transfected by the calcium phosphate precipitation method (20). Cells were harvested 48 h after transfection. Initially, CAT activity was measured by a simple phase extraction assay (41), and then it was measured by a nonradioactive fluorimetric assay (Fast CAT; Molecular Probes, Eugene, Oreg.). When the simple phase extraction assay was used, CAT values were expressed as percent conversion. CAT values obtained from the fluorimetric assay were expressed in arbitrary fluorimetric units per milligram of protein. All values are expressed as mean ± standard error of the mean (SEM) of at least three experiments carried out in duplicate. Experiments carried out to confirm previous observations were performed in duplicate, and the data from one representative experiment is shown. Unless otherwise specified, all cotransfections were performed with 10 μg of reporter plasmid and 0.5 μg of expression plasmid or the corresponding empty vectors.
In vivo mammalian two-site hybrid system (protein-protein interaction assay).
An in vivo protein-protein interaction assay system, the mammalian two-hybrid system, was constructed as described previously (2). A luciferase reporter construct driven by three Gal4-binding sites (GBS) was cotransfected with pM-JunD to express a fusion protein comprising JunD and the DNA-binding domain of Gal4 (Gal4-JunD). Enhancement of the transcriptional activity of the reporter construct by CHOP and not by the empty vector or deletion mutants was interpreted as evidence of in vivo physical and functional interaction. To increase the sensitivity of the assay without increasing the nonspecific background of the assay, transfections were performed in HeLa cells by using the high-efficiency transfection reagent GenePorter (Gene Therapy Systems, San Diego, Calif.).
Electrophoretic mobility shift assays (EMSA).
DNA-protein binding assays were carried out with nuclear extracts prepared as described previously (39), in the presence of the protease inhibitors pepstatin A (1 mg/ml), leupeptin (10 mg/ml), aprotinin (10 mg/ml), and p-aminobenzamidine (0.1 mM). Protein concentrations were determined by the Bio-Rad protein assay with bovine serum albumin as a standard. Synthetic complementary oligonucleotides with 5′ GATC overhangs were annealed and labeled by a fill-in reaction with [32P]dATP and Klenow enzyme. The sequences of the oligonucleotides used are 5′CCGGCGCCTCCTTGGCTGACGTCAGAGAGAGAG 3′ for CRE and 5′ CCAAGAGGATGTTATAAAGCATGAGTCAGACACCTCTGGCTTTCT 3′ for TRE.
Binding reactions were carried out in the presence of 2 μg of poly(dI-dC), using nuclear extracts (10 μg of protein) or recombinant bacterially produced c-Fos (wb-Fos) and c-Jun (wb-Jun), provided by T. Curran (1). Incubations were performed in the presence of 20,000 cpm of radiolabeled probe (approximately 6 to 10 fmol) in a total volume of 20 μl containing 20 mM potassium phosphate (pH 7.9), 70 mM KCl, 1 mM dithiothreitol, 0.3 mM EDTA, and 10% glycerol.
In vitro translation and coimmunoprecipitation assays.
In vitro translation reactions were carried out in rabbit reticulocyte lysates (Promega, Madison, Wis.), with mRNAs encoding the different proteins, in the presence of [35S]methionine as specified by the manufacturer. mRNAs were prepared by transcribing the linearized plasmids bearing the corresponding cDNAs with T7 or SP6 DNA polymerases. Immunoprecipitations were carried out as previously described (38) in a buffer containing 200 mM NaCl, 50 mM HEPES (pH 7.9), 0.1% Nonidet P-40, 5 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride.
Protein zipper blot assays.
To examine for a direct protein-protein interaction between CHOP and the components of the AP1 complex, a zipper blot assay was used (38). CHOP bZIP was produced in Escherichia coli as a glutathione S-transferase (GST) fusion protein and labeled with 32P by using protein kinase A (38). This probe was reacted for 1 h with the other leucine zipper-containing proteins, which had been previously immobilized on a nitrocellulose membrane, in buffer DZ (20 mM potassium phosphate [pH 7.9], 250 mM KCl, 5 mM NaF, 1 mM dithiothreitol, 0.2 mM EDTA, 10% nonfat dry milk). This buffer was also used for extensive washing prior to exposure for autoradiography.
GST pull-down assays.
In vitro-translated c-Fos, c-Jun, and JunD proteins were incubated with recombinant GST-CHOP and its deletion mutant control GST-CHOP LZ− at 4°C for 1 h in a buffer containing 20 mM sodium phosphate (pH 7.3), 150 mM NaCl, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, and 0.5 μg of leupeptin per ml. Protein complexes were precipitated by centrifugation with glutathione-Sepharose beads (Pharmacia-LKB). After extensive washing, bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected by autoradiography.
RESULTS
The somatostatin gene CRE is a DNA regulatory element that binds multiple transcription factors.
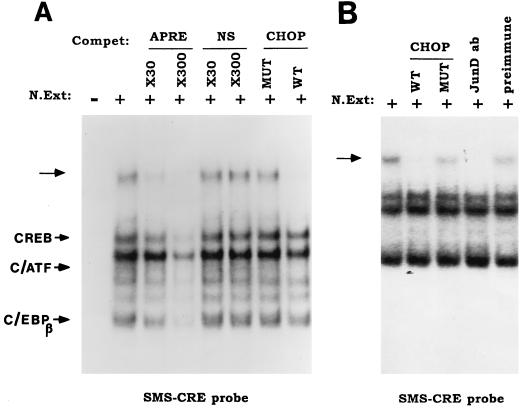
Nuclear extracts prepared from the RIN1027-B2 insulinoma cell line contain several proteins that bind to the CRE of the rat somatostatin promoter. In earlier studies (46), we showed in EMSA that the two most prominent protein-DNA complexes consisted of CREB and C/ATF and that a faster-migrating complex is formed by C/EBPβ (Fig. 1A). An upper, more slowly migrating complex was also identified. Although the identity of the slowly migrating complex was not defined at that time, it appeared to be related to the C/EBP family of transcription factors because preincubation of the nuclear extracts with a higher molar ratio of CHOP, a dominant negative inhibitor of the binding of C/EBPs to DNA, prevented the formation of this more slowly migrating complex (Fig. 1A). However, none of the known C/EBP proteins were identified as being part of this complex (46). This complex has now been identified as JunD, as shown by inhibition of the formation of this specific complex by an antiserum specific to JunD (Fig. 1B).
FIG. 1.
CHOP interacts with JunD, when bound to the CRE of the rat somatostatin promoter in nuclear extracts of RIN1027-B2 cells. Data shown are EMSA demonstrating RIN1027-B2 nuclear proteins binding to 32P-labeled CRE oligonucleotide probe. (A) The oligonucleotide (CRE31) (46), consisting of the somatostatin promoter containing a CRE (SMS-CRE probe). Binding of the 32P-labeled probe is fully competed with a 10-fold excess of unlabeled APRE. NS is a nonspecific oligonucleotide control. Note that addition of wild-type CHOP (WT) to the nuclear extract eliminates binding of the C/EBPs whereas addition of the mutant CHOP (MUT) lacking the leucine zipper dimerization domain (CHOP-LZ−) does not interfere with the binding of the proteins. The unknown slowest-migrating complex (arrow) is eliminated by incubation of the nuclear extract with CHOP. (B) The slower-migrating complex is eliminated by preincubation of the nuclear extract with recombinant CHOP and with an antiserum specific for the detection of JunD.
JunD activates transcription of the CRE in the somatostatin gene promoter.
The potential relevance of the binding of JunD to the CRE of the somatostatin promoter was examined in a functional transactivation assay with the placental choriocarcinoma cell line JEG-3, which is particularly conducive for activation of cAMP-responsive genes due to low levels of the inducible cAMP early repressor, ICER (28). Transfection of a transcriptional reporter plasmid containing the somatostatin CRE with expression plasmids for JunD, or CREB as a positive control, showed a strong activation in the presence of the cAMP agonist 8-bromo cAMP (Fig. 2A). The activation by JunD in response to 8-bromo cAMP (7.5-fold) was higher than the corresponding activation by CREB (4.2-fold). Furthermore, cotransfection and expression of CHOP with JunD resulted in enhanced transcription from the somatostatin promoter (Fig. 2B). Notably, the mutant CHOP with a deleted leucine zipper, CHOP-LZ− (38), did not augment activation by JunD (Fig. 2B).
FIG. 2.
CHOP enhances the transcriptional activation of JunD on the somatostatin promoter CRE, and activation depends on the presence of the leucine zipper dimerization domain of CHOP. (A) JunD activates the transcription of a reporter plasmid containing the somatostatin CRE (CRETK-CAT). The reporter plasmid was cotransfected to JEG-3 cells with expression plasmids for either JunD or CREB, in the presence or absence of 8-bromo cAMP (8Br cAMP). EV (empty vector) is the plasmid vector used for expression of JunD and CREB. The data shown are representative of two experiments with similar results. (B) CHOP augments JunD activation of the somatostatin promoter. The mutant CHOP without the leucine zipper dimerization domain (CHOP-LZ−) does not affect transactivation by JunD. The data are the mean and SEM of three independent experiments.
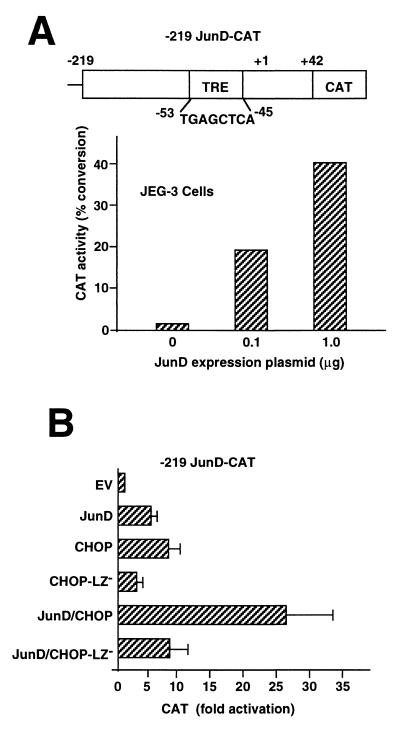
CHOP augments the activation of gene transcription from the JunD promoter.
Because of the somewhat paradoxical findings of gene activation by CHOP on the somatostatin promoter, we examined yet another promoter known to be strongly activated by JunD. The promoter of the JunD gene itself is strongly positively autoregulated (9, 15), as confirmed by coexpression of JunD and the −219 JunD promoter sequence in NIH 3T3 cells (Fig. 3A). The JunD promoter contains a DNA control sequence similar to a TRE that is present in the promoters of a large number of genes that bind and transcriptionally respond to AP-1 complexes of Jun and Fos proteins. It has been established that the activation of the JunD promoter requires the composite TRE sequence (9, 15). The effects of CHOP expression on the JunD promoter were examined and found to stimulate transcription equivalent to or greater than the autostimulation by JunD (Fig. 3B). Notably, the coexpression of JunD plus CHOP synergistically activates the JunD promoter by 25-fold, an effect not seen by expression of the mutant CHOP-LZ− protein (Fig. 3B). These findings indicate that CHOP not only functions as a dominant negative repressor of gene transcription, as originally described for the activation of genes by C/EBPs (38), but also can serve as an activator of transcription in the circumstances of the JunD gene promoter and that CHOP and JunD cooperate to further enhance gene transcription. Since JunD is constitutively expressed in most if not all cell types, the positive transcriptional effects of CHOP observed (Fig. 3) may represent an interaction with endogenous JunD.
FIG. 3.
CHOP activates transcription of the JunD promoter as efficiently as JunD does itself. (A) Activation of the −219 promoter of the human JunD gene by a JunD expression plasmid. The autoactivation of the JunD promoter occurs via the phorbol response element −53 to −45 (9). The JunD promoter-CAT reporter and JunD expression plasmids were transfected to NIH 3T3 cells. The experiment was performed twice with similar results. (B) CHOP also activates the JunD promoter and augments the activation by JunD (0.5 μg of expression plasmid), and the activation by CHOP is attenuated by deletion of the leucine zipper dimerization domain (CHOP-LZ−). Data are the mean of four experiments and SEM.
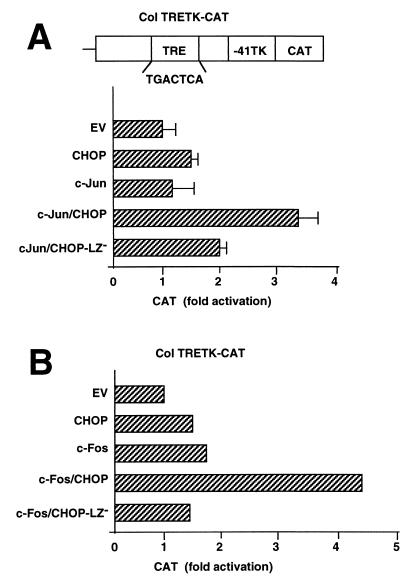
CHOP enhances the transcriptional activation of the human collagenase gene promoter in concert with c-Jun and c-Fos.
Because c-Jun and AP-1 complexes of c-Jun and c-Fos are best known to activate the transcription of genes by interactions with TREs, we examined the interactions of CHOP and AP-1 factors on the TRE sequence of the human collagenase promoter, one of the most thoroughly studied promoters regulated by AP-1 (4). To favor the identification of possible interactions of CHOP with AP-1 factors, we used NIH 3T3 cells, a well-studied cell line known to readily express AP-1 factors in response to growth-promoting signals. The transcriptional reporter vector Col-TRE TK-CAT was cotransfected with expression vectors for c-Jun and/or CHOP. After transfection, cells were placed in medium containing only 0.5% serum. Under these conditions of low serum concentration, NIH 3T3 cells are quiescent and the expression of the endogenous immediate-early Jun and Fos proteins is low (c-Jun, JunB, and the Fos and Fra proteins). The overexpression of c-Jun alone had little effect in stimulating transcription from the collagenase TRE, but CHOP expression significantly activated transcription. Notably, the coexpression of c-Jun and CHOP markedly stimulated gene transcription (Fig. 4A), an effect that is attenuated with the mutant CHOP, CHOP-LZ−, lacking the dimerization domain. The results obtained were consistent with the transcriptional activation of the JunD promoter by CHOP. Similar experiments were carried out with a c-Fos expression plasmid instead of a c-Jun expression plasmid. The results were similar to those of c-Jun expression: c-Fos or CHOP alone modestly activated the collagenase TRE reporter, whereas coexpression of c-Fos and CHOP strongly stimulated the reporter (Fig. 4B). The inactive mutant CHOP, CHOP-LZ−, gave no such stimulation when coexpressed with c-Fos.
FIG. 4.
CHOP cooperates with c-Jun or c-Fos to activate the human collagenase promoter containing a TRE. CHOP enhances the activation of the human collagenase transcriptional reporter, ColTRE-41TK-CAT, by c-Jun (A) or by c-Fos (B). The enhancement of transcription is attenuated when the leucine zipper of CHOP is deleted (CHOP-LZ−). Transcriptional reporter and transcription factor expression plasmids were transfected to NIH 3T3 cells. At 24 h after transfection, the cells were placed in medium containing only 0.5% serum for an additional 24 h to decrease the expression of endogenous AP1 proteins before harvesting.
These findings further demonstrate that on certain promoters and cell phenotypes, CHOP can serve as an activator of gene transcription, in addition to a dominant negative repressor of transcription on other promoters and cell phenotypes, and that CHOP cooperates positively with AP-1 factors in the activation of gene transcription.
CHOP forms protein-protein interactions with AP-1 complex proteins.
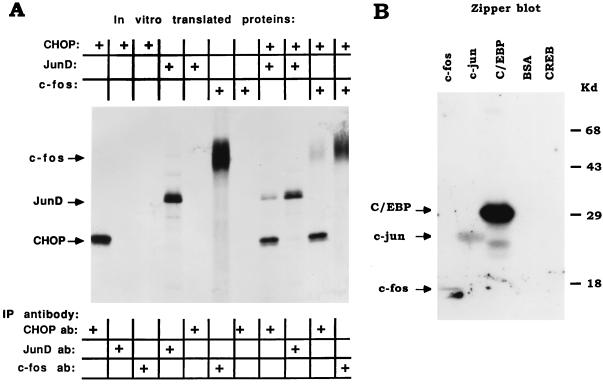
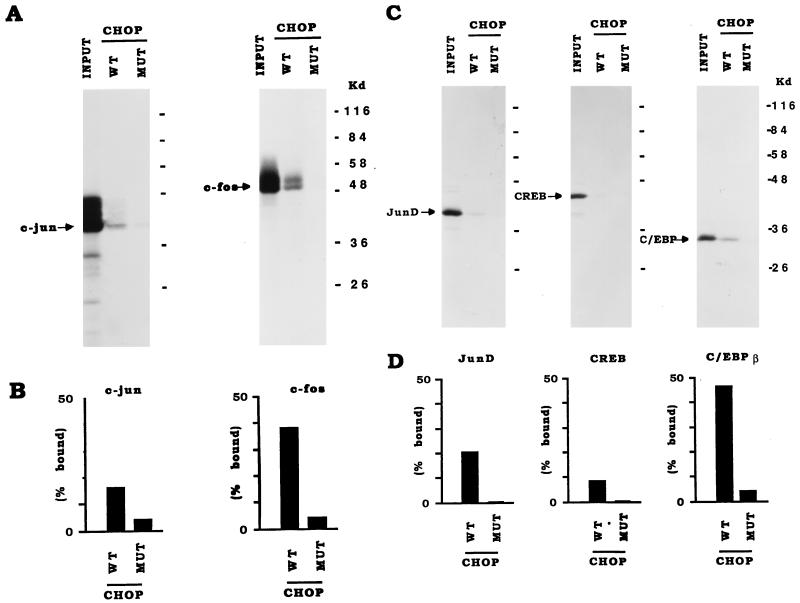
The above studies demonstrating positive transcriptional interactions of CHOP with AP-1 factors to stimulate gene transcription imply that CHOP may directly interact with JunD, c-Jun, and c-Fos. To examine the possibility of protein-protein interactions between CHOP and AP-1 factors, several different experimental approaches were used. To determine whether CHOP interacts directly with JunD and c-Fos, coimmunoprecipitation experiments were carried out with in vitro-translated proteins and antisera specific to CHOP, JunD, and c-Fos. The antiserum to CHOP coimmunoprecipitates both JunD and c-Fos (Fig. 5A), indicating that CHOP interacts directly with these two AP-1 complex proteins. That the interactions occur via the leucine zippers of these bZIP proteins was determined by “Zipper” blot experiments (38). A fusion protein, consisting of the phosphorylation domain of CREB (P-box, KID) and the leucine zipper dimerization domain of CHOP, was labeled with 32P catalyzed by protein kinase A and used to probe a nitrocellulose filter on which equivalent amounts of the truncated proteins (1) containing only their bZip domain of c-Jun, c-Fos, C/EBPβ (as a positive control), and CREB (negative control), were transferred from an SDS-PAGE gel. The 32P-labeled CHOP dimerized to both c-Jun and c-Fos, as well as to the C/EBP control, and not to CREB (Fig. 5B). The results of these studies indicate that the interactions of CHOP with c-Jun and c-Fos occur via their leucine zipper dimerization domains. Interactions of CHOP with c-Jun, c-Fos, and JunD were further substantiated by GST pull-down experiments with a GST-CHOP fusion protein as the hook (Fig. 6). GST-CHOP interacted nearly as efficiently with c-Fos as with the positive control C/EBPβ. It remains uncertain whether the seeming interaction of the negative control CREB is a legitimate interaction or represents the background of the GST pull-down experiments (Fig. 6).
FIG. 5.
CHOP directly interacts with JunD, c-Jun, and c-Fos, as shown by coimmunoprecipitation and zipper blot studies. (A) Coimmunoprecipitation studies. CHOP, JunD, and c-Fos were translated or cotranslated in a reticulocyte lysate system in the presence of [35S]methionine (top). The proteins were immunoprecipitated (IP) with antisera (antibody [ab]) to CHOP, JunD, and c-Fos (bottom). The immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography. Note that the antiserum to CHOP coimmunoprecipitates both JunD (lane 8) and c-Fos (lane 10). (B) Zipper blot. The proteins designated at the top were bacterially produced recombinant truncated proteins encompassing their respective bZIP DNA-binding and dimerization domain. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was then incubated with a truncated form of CHOP consisting of the bZIP domain that had been labeled with 32P (see Materials and Methods) (38). An autoradiogram of the SDS-PAGE gel shows that CHOP binds to c-Fos and c-Jun, as well as to C/EBPβ (positive control), and not to CREB or bovine serum albumin (BSA) (negative controls).
FIG. 6.
GST pulldown experiments show that CHOP binds c-Jun, c-Fos, and JunD. Either wild-type CHOP (WT) or CHOP-LZ− mutant (MUT) fused to GST was incubated with 35S-labeled c-Jun or c-Fos (A and B) or JunD, CREB, or C/EBPβ (C and D), and bound proteins were isolated by capture with a glutathione affinity resin. The proteins were analyzed by SDS-PAGE and autoradiography. Autoradiograms of GST-captured proteins compared to input of proteins (INPUT) are shown in panels A and C. Densitometric quantitation of the film images of panels A and C are shown in panels B and D, respectively. The experiments shown were done twice with similar results.
CHOP enhancement of AP-1 transcriptional activation depends on tethering to the AP-1 complex and not to formation of new DNA-binding complexes.
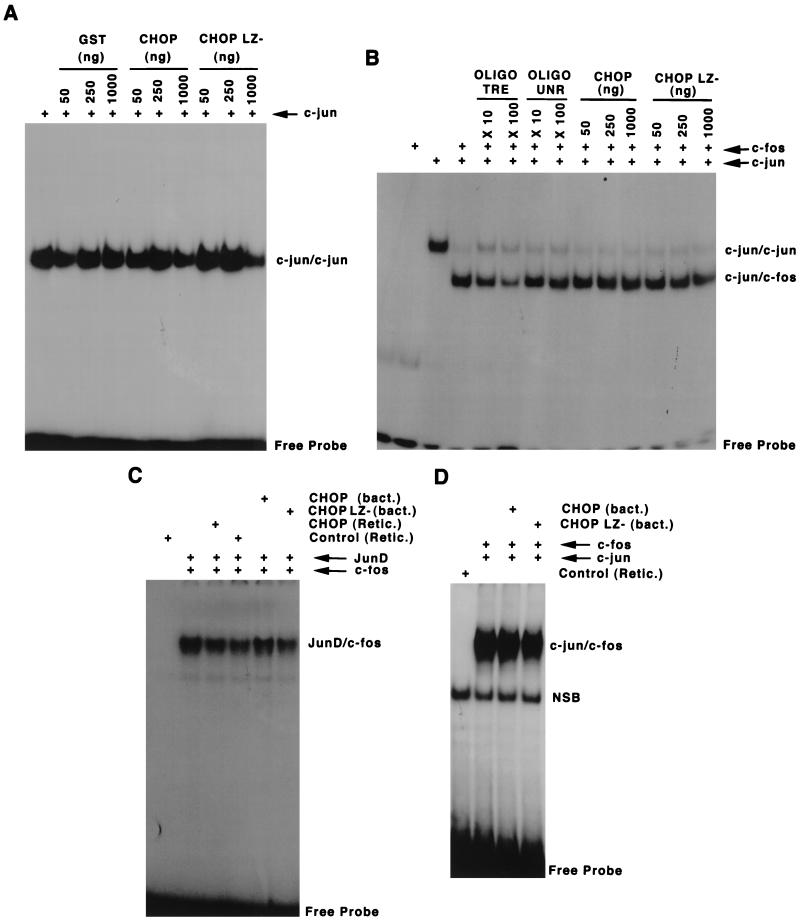
Our data indicate the existence of a protein-protein interaction between CHOP and the AP-1 family members, JunD, c-Jun, and c-Fos. Since the interaction maps to their corresponding leucine zipper dimerization domains, we considered the possibility that the enhancement of AP-1 transactivation induced by CHOP was a consequence of the formation of new heterodimers between CHOP and AP1 proteins. These new complexes could have a higher affinity than Jun/Jun homodimers and Jun/c-Fos heterodimers for the AP-1-responsive element. To test this hypothesis, we studied the electrophoretic shifting pattern of a TRE in the presence of various amounts of both recombinant (Fig. 7A and B) and in vitro-translated (Fig. 7C and D) proteins. Under these conditions, no new or lower-mobility complexes were observed in the presence of CHOP (Fig. 7). Additional gel retardation experiments preformed with nuclear extracts from different cell types and different treatments also failed to show evidence for the formation of heterodimers between CHOP and AP-1 proteins on the TRE element (data not shown). These observations, along with the finding that the interaction between CHOP and AP-1 proteins appears to be somewhat weaker than that between CHOP and C/EBPs (Fig. 5B and 6), suggested the possibility that rather than direct binding to DNA, CHOP is tethering to the AP-1 complex. This mechanism of tethering has been demonstrated recently for other bZIP proteins such as ATF6 (see Discussion).
FIG. 7.
CHOP fails to form new stable complexes with AP-1 factors on a TRE. EMSA with the oligonucleotide containing TRE as a gel shift probe and recombinant or in vitro-translated proteins, c-Jun, c-Fos, JunD, and CHOP or its corresponding deletion mutants were used to test the possibility of direct DNA binding of CHOP–AP-1 complexes. (A) A constant amount of recombinant c-Jun was incubated with increasing amounts of recombinant CHOP, its leucine zipper-minus mutant, and the control GST protein. No additional shifting bands were observed in the presence of CHOP. (B) The sifting pattern produced by c-Jun and c-Fos heterodimers was not changed by the presence of increasing amounts of CHOP. A competitive nonlabeled TRE oligonucleotide, but not an unrelated oligonucleotide (UNR), decreased the formation of c-Jun/c-Fos heterodimers in a dose-dependent manner. (C and D) No new retardation complexes, nor changes in binding affinity, were observed when in vitro-translated JunD and c-Fos (C), or c-Jun and c-Fos (D) full-length proteins were used.
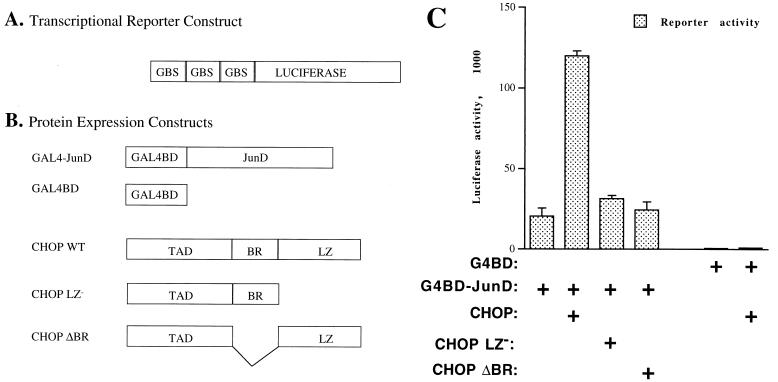
To examine the possibility that the interaction between CHOP and AP1 proteins is tethering and occurs in vivo, a previously described modification of the mammalian two-hybrid system was used (54). One of the proteins, JunD, was fused to the DNA-binding domain of Gal4 (Fig. 8B) to study its effect on a heterologous promoter driven by three Gal4 binding sites (GBS) (Fig. 8A). CHOP, the second partner of the interaction, was not fused to a heterologous transactivation domain, because it contains a strong transactivation domain (45). CHOP exerted a strong enhancement of the Gal4-JunD transcriptional activity, acting on a reporter driven by three Gal4-binding sites (Fig. 8C). This effect was exerted through an in vivo interaction with JunD, because CHOP did not have any effect on the reporter activity when a control Gal4-binding domain construct was used instead of Gal4-JunD (Fig. 8C). A deletion mutant of CHOP, lacking the leucine zipper dimerization domain, that fails to interact in vitro with JunD also failed to enhance in vivo the transcriptional activity of the Gal4-JunD fusion protein (Fig. 8C). These findings indicate that the enhancement by CHOP of AP-1-mediated transcription is specific and that it depends on an in vivo interaction between the two proteins. To further exclude the possibility that the interaction is mediated through a leucine zipper interaction with an unknown inhibitor of JunD, we examined the effect of a basic-region deletion mutant of CHOP (CHOP ΔBR) that retains an intact leucine zipper dimerization motif. This mutant CHOP ΔBR failed to enhance the activity of the Gal4-JunD construct (Fig. 8C), thus excluding this possibility.
FIG. 8.
Evidence of in vivo physical and functional interactions between CHOP and AP-1 proteins. The mammalian two-hybrid system was used to demonstrate in vivo protein-protein interactions. To determine whether such a protein-protein interaction is also functional, the CHOP transactivation domain was retained instead of being replaced with a heterologous activation domain. (A) Map of the luciferase reporter construct used in these experiments. In this reporter, luciferase expression is driven by three GBS as previously described (49). (B) JunD transactivation of the Gal4 reporter was achieved by fusing the DNA-binding domain of Gal4 (GAL4BD) to JunD to create Gal4-JunD. The maps for the wild-type CHOP (CHOP WT) and the corresponding mutants without the leucine zipper (CHOP LZ−) or the basic region (CHOP ΔBR) are also shown. TAD, transactivation domain. (C) CHOP, but not CHOP LZ− or CHOP ΔBR mutants, enhances the transcriptional activity of Gal4-JunD by severalfold. No direct effect of CHOP was observed on the reporter activity, even when the Gal4 binding domain (G4BD) control was cotransfected along CHOP. These data not only demonstrate that the interaction between CHOP and JunD also occurs in vivo but also indicate that this interaction has a functional relevance.
DISCUSSION
The results of these studies show that CHOP, a member of the C/EBP family of bZIP transcription factors, can directly interact with Jun and Fos members of the AP-1 complex of transcription factors and does so to activate promoters of selected genes, i.e., the somatostatin, JunD, and collagenase promoters. The strong transactivation properties of CHOP in the context of association with Jun or Fos and selective promoters may have been predicted from earlier studies showing that the amino-terminal sequence of CHOP fused to a GAL4 DNA-binding domain strongly activated a transcriptional reporter containing GAL4-binding sites (45). The experimental findings reported in this study, prompted by the fortuitous observation that CHOP interacts with JunD on the somatostatin promoter, were unanticipated because CHOP was identified and defined initially as an interacting partner with C/EBP proteins that served as a dominant negative inhibitor of the actions of C/EBP on conventional C/EBP-binding sites of gene promoters (38). In at least two circumstances, CHOP has been shown to serve as an activator of gene transcription (43, 45). Similar to the Fos proteins, CHOP does not form stable homodimers and thereby depends on heterodimerization with other proteins to exert functions on the control of gene transcription (38).
Several examples of heterodimerization among members of different bZIP protein families have been reported. Members of the activating-transcription factor (ATF) family dimerize with Jun proteins (8, 22, 24, 27). ATF2 dimerizes with both c-Jun and JunD, and ATF4 dimerizes with JunD. C/ATF, a protein that closely resembles ATF4 (47), also forms stable heterodimers with C/EBPs, a not surprising finding because the bZIP domains of C/ATF and ATF4 are closely related to those of the C/EBPs (30, 47). Notably, CHOP has been implicated in interactions with ATF3, since CHOP appears to repress the transactivational activity of ATF3 in liver cells, possibly by direct interactions of the two proteins (12). Recently it was demonstrated that the tumor necrosis factor alpha gene is regulated by the interactions of c-Jun and C/EBPβ and that this interaction contributes to the expression of the tumor necrosis factor alpha gene in myelomonocytic cells (51). This interaction was unique in that it did not require the transactivation domain of c-Jun. DNA-binding assays suggested that C/EBPβ and c-Jun interact in vitro and that the interaction may be DNA dependent (51). The observations reported here represent the first evidence that CHOP can interact with proteins of the Fos/Jun AP-1 complex.
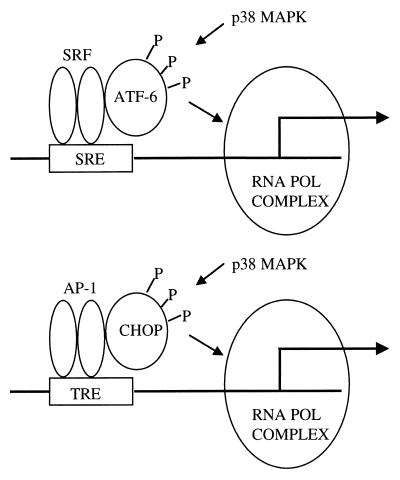
Although we show that CHOP can form protein-protein interactions with the AP-1 members JunD, c-Jun, and c-Fos, we did not initially fully understand the nature of these interactions. The zipper blot experiments, in conjunction with the consistent findings of an attenuation of transcriptional activation potential of CHOP with a mutated inactive leucine zipper (CHOP-LZ−), suggest that the protein-protein interactions occur via the leucine zipper domain of CHOP. When partnered with C/EBP, CHOP represses transcription from the acute-phase response element (APRE) of the angiotensinogen promoter and other C/EBP binding sites of promoters of several genes. CHOP acts as a dominant negative repressor of C/EBP-activated transcription from the APRE, because CHOP-C/EBP heterodimers cannot bind to the APRE (38) (Table 1). However, CHOP becomes an activator on a different set of genes, acting on a related DNA-binding element (43, 45). Through this mechanism, CHOP-C/EBP heterodimers activate transcription of the carbonic anhydrase VI gene (43) (Table 1). In this regard, it is worth noting that the consensus binding site for CHOP-C/EBP heterodimers, RRRTGCAAT (R = purine), differs considerably from that of C/EBP dimers, TTNNNGCAAT (45). In this paper, we report new findings indicating that CHOP can activate the transcription of additional genes by a completely different and unexpected tethering mechanism. By this mechanism, CHOP tethers to preexisting transcription factor complexes bound to DNA control elements. A similar mechanism has been described recently for at least another bZIP protein, ATF6 (44, 50, 53). Transcriptional activation mediated by ATF6 on the c-Fos promoter (53) and the atrial natriuretic factor (ANF) promoter (44) results from tethering with serum response factor (SRF) bound to serum response elements (SREs) present in both promoters. ATF6, however, does not bind directly to the SRE, although it contains a DNA-binding domain consisting of a basic region and a leucine zipper similar to those of CREB-RP, ATF1, CREB, and CREM. On the ANF promoter, ATF6 mediates the activation induced by the stress-dependent protein kinase p38 mitogen-activated protein kinase (MAPK) through direct phosphorylation (44). This effect does not appear to be an artifact resulting from overexpression of ATF6, because suppression of endogenous ATF6 by an antisense technique abolishes p38-mediated induction of ANF (44). ATF6 or a proteolysis product of ATF6 uses a tethering mechanism to regulate the expression of several genes encoding molecular chaperones, known as glucose-regulated proteins (GRPs), contained within the ER. When cells are exposed to agents that produce stress in the ER, GRP gene expression is induced (50). The induction of the GRP genes, GRP78 (BiP), GRP94, and calreticulin, comprises a cellular response known as the unfolded protein response. This response is conserved between mammals and yeasts, where it becomes orchestrated by another bZIP protein, Hac1. ATF6 in vertebrates seems to operate as the Hac1 homologs in yeast. We propose a model in which tethering to preexisting transcriptional complexes and phosphorylations mediated by the p38 stress-activated protein kinase on both CHOP and ATF6 results in changes of the expression of a set of genes involved in the cellular response to ER stress (Fig. 9).
TABLE 1.
Mechanisms of transcriptional regulation by CHOP after ER stress
| Mechanism | Element | Target gene(s) | Effect |
|---|---|---|---|
| Dominant inhibition | CCAAT | C/EBPα (?), angiotensinogen | Inhibition of adipocyte differentiation; acute-phase response |
| DNA binding and transactivation | TGCAAT | Carbonic anhydrase VI | Intracellular pH control(?), apoptosis(?) |
| Tethering to AP-1 and CRE control elements | TGACTCA, TGACGTCA | JunD(?), somatostatin(?), collagenase(?), others(?) | Apoptosis(?), defense responses to stress(?), cell cycle control(?) |
FIG. 9.
CHOP and ATF6 are capable of gene expression regulation by tethering to a preexisting complex. A proposed model for the interaction between CHOP and the AP-1 complex is shown in parallel to the one previously described for ATF6 and SRF (44, 53). Tethering of bZIP proteins to preexisting DNA-protein complexes is a new mechanism by which bZIP transcription factors regulate gene expression. In this case, both CHOP and ATF6 are downstream effectors of the stress-induced p38 MAPK pathway. The tethering mechanism described may facilitate the regulation of a large number of genes by a limited number of transcription factors. This circumstance may have an important significance when cells are exposed to damaging or stressful conditions.
Studies of ATF6 have not described a role of the bZIP domain in the tethering function. In this report, we show that the basic region of the bZIP domain of CHOP is required for tethering to the AP1 proteins in vivo and serves a coactivation function. The requirement of the basic region for AP-1-mediated transactivation by CHOP suggests that this region may be involved in interactions with downstream effectors. That CHOP does contact DNA cannot be excluded, but if this is so, the contacts must be relatively weak, and under the conditions of the DNA-binding EMSA, the complexes dissociate from the DNA. The dilution of transcription factors in EMSA is known to produce false-negative results compared to nuclear offload and protein-DNA cross-linking experiments (31).
The importance of the base context of the DNA motif in the binding of proteins has been recognized for a long time. For example, the 5 to 10 bp flanking the core octamer sequence of the CRE (TGACGTCA) determine whether CREB will activate transcription from a CRE (16). It has been shown that the binding of thyroid hormone receptor or the yeast transactivator pheromone receptor transcription factor to two different DNA elements impart two different conformations to the proteins and that upon binding to DNA the transcription factors ATF2, ETS, and the glucocorticoid receptor (GR) undergo distinct changes in their intramolecular folding (26). The nature of the protein-protein interactions, whether dimerization or tethering in which the associated protein does not contact DNA, depends on the cell phenotype and the particular repertoire of proteins that are expressed or can be induced to express in that particular cell type. The environmental signaling milieu most often involves the state of phosphorylation of the DNA-binding and/or associated proteins. It is worth noting that AP-1 regulation of the collagenase gene was one of the first-described examples of tethering interactions in which the GR represses AP-1-mediated activation of the gene independent of its binding to DNA (3, 18, 40). Remarkably, mice carrying a mutant GR defective in dimerization and DNA binding are viable, whereas GR−/− mice die at birth. These circumstances suggest that tethering cross talk of the GR with other transcription factors maintains transcriptional functions of the GR in the absence of binding to DNA and is likely to be responsible for the survival of the mice with a DNA-binding defective GR (37).
The available evidence strongly indicates that CHOP is antiproliferative (5, 52), proapoptotic (54), and induced by activation of stress-associated signal transduction pathways (49). For example, microinjection of CHOP into cells induces growth arrest at the G1/S checkpoint (5), and a variety of agents that activate cell stress responses induce the expression of CHOP (6, 11, 13, 23, 35, 48). In particular, agents such as tunicamycin that result in the misfolding of proteins in the ER, so-called endoplasmic stress, strongly induce CHOP expression and are believed to involve the stress-activated p38/HOG kinase pathway (49). A role for CHOP in apoptosis is provided by studies of chop−/− mice in which cultured embryonic fibroblasts are relatively resistant to apoptosis (54). The chop−/− mice are also somewhat protected against tunicamycin-induced renal tubular necrosis, a process that involves programmed cell death of epithelial cells (54).
In marked contrast to CHOP, c-Jun and c-Fos are well recognized as immediate-early response proteins that are rapidly induced when serum-deprived quiescent cells are stimulated to proliferate by serum repletion (4). Although JunD is expressed at its highest levels in quiescent cells (32), the expression of c-Jun and c-Fos is undetectable in resting (G0) cells (4). It is worth noting that JunD, in contrast to c-Jun, suppresses the transformation of cells by an activated ras gene, suggesting that the two closely related transcription factors JunD and c-Jun can function in an opposing manner in the control of gene transcription and cell growth (32). The finding that CHOP can interact with both JunD and c-Jun raises interesting conjectural questions regarding the potential roles that CHOP may play in the control of cell growth mediated by the opposing actions of JunD and c-Jun. It is possible that the functions of CHOP are exerted at the time when proliferating cells, in which levels of c-Jun and c-Fos are high, are converted to a state of growth arrest in response to stressful stimuli that also induce CHOP. Thus, the tethering of CHOP to c-Jun/c-Fos may activate the transcription of genes that are typically expressed in quiescent cells and/or preapoptotic cells. Support for this notion comes from the observations that the AP-1 complex proteins, c-Fos and JunB, are implicated in apoptosis induced by a variety of stimuli (19, 21, 29, 34, 42). Exposure of WEH 17.2 thymoma cells to cAMP agonists activates a programmed cell death pathway that is preceded by an earlier activation of the expression of the c-fos and junB genes (21). Thus the role of the CHOP interaction with AP-1-complex proteins (JunD, c-Jun, and c-Fos) may be important in the regulation of a subset of genes expressed in the early phase following a stressful stimulation, when cells undergo growth arrest but have yet to decide whether to undergo repair of cellular damage and to subsequently reenter the cell division cycle or whether to commit suicide by programmed cell death because the damage is sensed to be beyond repair.
ACKNOWLEDGMENTS
We are grateful for the expert experimental assistance of L. Fucci and the helpful advice of M. Schmitt-Ney. We also thank T. Curran for purified c-Fos and c-Jun proteins and T. Budde for preparation of the manuscript.
The studies were supported in part by USPHS grant DK30457. J.F.H. is an Investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Abate C, Luk D, Gentz R, Rauscher F J I, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc Natl Acad Sci USA. 1990;87:1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal S K, Guru S C, Heppner C, Erdos M R, Collins R M, Park S Y, Saggar S, Chandrasekharappa S C, Collins C, Spiegel A M, Marx S J, Burns A L. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdorf H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 4.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 5.Barone M V, Crozat A Y, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, differ in their ability to induce G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett J, Luethy J, Carlson S, Sollott S, Holbrook N. Calcium ionophore A23187 induces expression of the growth arrest and DNA damage inducible CCAAT/enhancer-binding protein (C/EBP)-related gene, gadd153. J Biol Chem. 1992;267:20465–20470. [PubMed] [Google Scholar]
- 7.Batchvarova N, Wang X Z, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (gadd153) EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benbrook D M, Jones N C. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 9.Berger I, Shaul Y. The human junD gene is positively and selectively autoregulated. DNA Cell Biol. 1994;13:249–255. doi: 10.1089/dna.1994.13.249. [DOI] [PubMed] [Google Scholar]
- 10.Bruhat A, Jousse C, Wang X-Z, Ron D, Ferrara M, Fafournoux F. Amino acid limitation induces expression of chop, a CCAAT/enhancer binding protein related gene at both transcriptional and post-transcriptional levels. J Biol Chem. 1997;272:17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- 11.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Regulation of the C/EBP-related gene, gadd153, by glucose deprivation. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B P C, Wolfgang C D, Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol Cell Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q, Yu K, Holbrook N J, Stevens J L. Activation of the growth arrest and DNA damage-inducible gene gadd153 by nephrotoxic cysteine conjugates and dithiothreitol. J Biol Chem. 1992;267:8207–8212. [PubMed] [Google Scholar]
- 14.Crozat A Y, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma with t(12;16)(q13;p11) Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 15.de Groot R P, Karperien M, Pals C, Kruijer W. Characterization of the mouse junD promoter—high basal level activity due to an octamer motif. EMBO J. 1991;10:2523–2532. doi: 10.1002/j.1460-2075.1991.tb07792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch P J, Hoeffler J P, Jameson J L, Lin J C, Habener J F. Structural determinants for transcriptional activation by cAMP-responsive DNA elements. J Biol Chem. 1988;263:18466–18472. [PubMed] [Google Scholar]
- 17.Fornace A, Alamo I, Hollander M. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gack S, Vallon R, Schaper J, Ruther U, Angel P. Phenotypic alterations in fos-transgenic mice correlate with changes in Fos/Jun-dependent collagenase type I expression. Regulation of mouse metalloproteinases by carcinogens, tumor promoters, cAMP, and Fos oncoprotein. J Biol Chem. 1994;269:10363–10369. [PubMed] [Google Scholar]
- 19.Gillardon F, Eschenfelder C, Uhlmann E, Hartschuh W, Zimmerman M. Differential regulation of c-fos, fosB, c-jun, junB, bcl-2 and bax expression in rat skin following single or chronic ultraviolet irradiation and in vivo modulation by antisense oligodeoxynucleotide superfusion. Oncogene. 1994;9:3219–3225. [PubMed] [Google Scholar]
- 20.Gorman C M, Moffaat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grassilli E, de Prati A C, Monti D, Troiano L, Menegassi M, Barbieri D, Franceschi C, Suzuki H. Studies of the relationship between cell proliferation and cell death. II. Early gene expression during concanavalin A-induced proliferation or dexamethasone-induced apoptosis of rat thymocytes. Biochem Biophys Res Commun. 1992;188:1261–1266. doi: 10.1016/0006-291x(92)91367-y. [DOI] [PubMed] [Google Scholar]
- 22.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halleck M, Holbrook N, Skinner J, Liu H, Stevens J. The molecular response to reductive stress in LLC-PK1 renal epithelial cells: coordinate transcriptional regulation of gadd153 and grp78 genes by thiols. Cell Stress Chaperones. 1997;2:31–40. doi: 10.1379/1466-1268(1997)002<0031:tmrtrs>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeffler J P, Lustbader J W, Chen C-Y. Identification of multiple nuclear factors that interact with cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol Endocrinol. 1991;5:256–266. doi: 10.1210/mend-5-2-256. [DOI] [PubMed] [Google Scholar]
- 25.Kobierski L A, Chu H M, Tan Y, Comb M J. cAMP-dependent regulation of proenkephalin by JunD and JunB: positive and negative effects of AP-1 proteins. Proc Natl Acad Sci USA. 1991;88:10222–10226. doi: 10.1073/pnas.88.22.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefstin J A, Yamamoto K R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor P F, Abate C, Curran T. Direct cloning of leucine zipper proteins: Jun binds cooperatively to the CRE with CRE-BP1. Oncogene. 1990;5:451–458. [PubMed] [Google Scholar]
- 28.Mao D, Warner E A, Gurwitch S A, Dowd D R. Differential regulation and transcriptional control of immediate early gene expression in forskolin-treated WEH17.2 thymoma cells. Mol Endocrinol. 1998;12:492–503. doi: 10.1210/mend.12.4.0084. [DOI] [PubMed] [Google Scholar]
- 29.Marti A, Jehn B, Costello E, Keon N, Ke G, Martin F, Jaggi R. Protein kinase A and AP-1 (c-Fos/JunD) are induced during apoptosis of mouse mammary epithelial cells. Oncogene. 1994;9:1213–1223. [PubMed] [Google Scholar]
- 30.Meyer T E, Habener J F. Cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev. 1993;14:269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- 31.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfarr C M, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 33.Philippe J, Chick W L, Habener J F. Multipotential phenotypic expression of genes encoding peptide hormones in rat insulinoma cell lines. J Clin Investig. 1987;79:351–358. doi: 10.1172/JCI112819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preston G A, Lyon T T, Yin Y, Lang J E, Solomon G, Annab L, Srinivasan D G, Allcorta D A, Barrett J C. Induction of apoptosis by c-Fos protein. Mol Cell Biol. 1996;16:211–218. doi: 10.1128/mcb.16.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price B, Calderwood S. Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of glucose-regulated proteins. Cancer Res. 1992;52:3814–3817. [PubMed] [Google Scholar]
- 36.Rabbitts T H, Forster A, Larson R, Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 37.Reichardt H M, Kaestner K H, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schütz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 38.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ’mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber M, Baumann B, Cotten M, Angel P, Wagner E F. Fos is an essential component of the mammalian UV response. EMBO J. 1995;14:5338–5349. doi: 10.1002/j.1460-2075.1995.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seed B, Sheen J Y. A simple phase-extraction assay for chloramphenicol acetyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 42.Smeyne R J, Vendrell M, Hayward M, Baker S J, Miao G G, Schilling K, Robertson L M, Curran T, Morgan J I. Continuous c-fos expression precedes programmed cell death in vivo. Nature. 1993;363:166–169. doi: 10.1038/363166a0. [DOI] [PubMed] [Google Scholar]
- 43.Sok J, Wang X Z, Batchvarova N, Kuroda M, Harding H, Ron D. CHOP-dependent stress-inducible expression of a novel form of carbonic anhydrase IV. Mol Cell Biol. 1999;19:495–504. doi: 10.1128/mcb.19.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thuerauf D J, Arnold N D, Zechner D, Hanford D S, De Martin K M, McDonough P M, Prywes R, Clembotski C C. P38 mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum responsive element. J Biol Chem. 1998;273:20636–20643. doi: 10.1074/jbc.273.32.20636. [DOI] [PubMed] [Google Scholar]
- 45.Ubeda M, Wang X-Z, Zinszner H, Wu I, Habener J, Ron D. Stress-induced binding of the transcription factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallejo M, Gosse M E, Beckmann W, Habener J F. Impaired cyclic AMP-dependent phosphorylation renders CREB a repressor of C/EBP-induced transcription of the somatostatin gene in an insulinoma cell line. Mol Cell Biol. 1995;15:415–424. doi: 10.1128/mcb.15.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallejo M, Ron D, Miller C P, Habener J F. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad USA. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X-Z, Lawson B, Brewer J, Zinszner H, Sanjay A, Mi L, Boorstein R, Kreibich G, Hendershot L, Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X-Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP-kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of cis-acting endoplasmic reticulum stress response element for transcriptional induction of mammalian glucose-regulated proteins. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 51.Zagariya A, Mungre S, Lovis R, Birrer M, Ness S, Thimmapaya B, Pope R. Tumor necrosis factor alpha gene regulation: enhancement of C/EBPβ-induced activation by c-Jun. Mol Cell Biol. 1998;18:2815–2824. doi: 10.1128/mcb.18.5.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhan Q, Liebermann D A, Alamo I, Hollander M C, Ron D. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that cooperatively suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum responsive factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot R T, Remotti H, Stevens J L, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]