Abstract
Background
A severe form of acute hemorrhagic diarrhea syndrome (AHDS) occurred in dogs in the Oslo region of Norway during autumn 2019.
Objectives
To characterize the fecal microbiota of dogs with AHDS during the outbreak and compare it to that of healthy dogs from the same period and before the outbreak.
Animals
Dogs with AHDS (n = 50), dogs with nonhemorrhagic diarrhea (n = 3), and healthy dogs (n = 11) were sampled during the outbreak. In addition, 78 healthy dogs from the same region were sampled before the outbreak between 2017 and 2018.
Methods
Retrospective case‐control study. The fecal microbiotas were characterized using 16S rRNA gene amplicon sequencing.
Results
Dogs with AHDS had significantly different microbiota composition (R 2 = .07, P < .001) and decreased intestinal diversity relative to healthy dogs from the outbreak period (median, 2.7; range, 0.9‐3.5 vs median, 3.2; range, 2.6‐4.0; P < .001). The microbiota in dogs with AHDS was characterized by a decrease of Firmicutes and an outgrowth of Proteobacteria, with increased numbers of Clostridium perfringens and Providencia spp. Among the Providencia spp., 1 showed 100% sequence identity with a Providencia alcalifaciens strain that was cultivated and isolated from the same outbreak. No Providencia spp. was found in healthy dogs sampled before the outbreak.
Conclusions and Clinical Importance
Dogs with AHDS had marked changes in fecal microbiota including increased numbers of Providencia spp. and C. perfringens, which may have contributed to the severity of this illness.
Keywords: canine, dysbiosis, hemorrhagic diarrhea, intestinal microbiota, Norway
Abbreviations
- AHDS
acute hemorrhagic diarrhea syndrome
- ASV
amplicon sequence variants
- GI
gastrointestinal
- NMBU
Norwegian University of Life Sciences
- NSC
Norwegian Sequencing Centre
1. INTRODUCTION
Idiopathic acute hemorrhagic diarrhea syndrome (AHDS) in dogs is characterized by acute onset of hemorrhagic diarrhea, often associated with vomiting, leading to severe hemoconcentration and lethargy.1, 2 In contrast to humans, where syndromes are clearly described, the syndrome of AHDS in dogs is poorly defined. This severe form of necrotizing enteritis is of unknown etiology, but dietary components and bacterial toxins are considered important contributors.3 The peracute clinical signs in conjunction with Clostridium perfringens proliferation have led to the assumption that toxins, presumably produced by these bacteria, are involved.4 However, these toxins are not detected in feces in all cases, and they also are found in healthy dogs. These observations cast doubt on the importance of these toxins as a direct cause of AHDS in dogs.5, 6, 7 Susceptibility to AHDS appears to differ among dogs, including dogs that live in similar environments. Thus, it is likely that other factors, such as properties of the intestinal microbiota, influence disease development.8, 9, 10 A few reports, based on bacterial culture, have described the presence of Providencia alcalifaciens in dogs with diarrhea.11, 12 Previous studies using targeted quantitative polymerase chain reaction (qPCR) methods have shown that a dysbiotic intestinal microbiota in dogs with acute diarrhea and AHDS is characterized by changes such as higher abundance of Proteobacteria, C. perfringens, Fusobacterium, and Turibacter and lower abundance of Ruminococcaceae, Faecalibacterium, Prevotella, Blautia, Eubacterium, Lachnospiraceae, Sutterella, and Bifidobacterium spp.8, 9, 10 These studies, however, did not find increased abundance of Providencia spp. in dogs with AHDS.
Between August and November 2019, an unusually high number of dogs with acute hemorrhagic diarrhea was observed in the Oslo region of Norway. Dogs presented with acute clinical signs including vomiting, lethargy, anorexia, and profuse watery, hemorrhagic diarrhea. Because of the severity of clinical signs, a comprehensive outbreak investigation was undertaken to identify underlying causes.13 Clinical examinations and diagnostic testing identified no obvious underlying cause, leading to the presumptive diagnosis of idiopathic AHDS. However, a main finding was positive fecal culture results for P. alcalifaciens.
We used 16S rRNA gene amplicon sequencing to characterize the fecal microbiota in dogs with AHDS during an outbreak and compared it to the fecal microbiota of healthy dogs from the same time period (2019) and healthy dogs before the outbreak (2017‐2018). We hypothesized that dogs with AHDS would have fecal dysbiosis and a distinct profile of gut microbes, including increased abundance of Providencia spp., which would not be apparent in healthy dogs.
2. MATERIALS AND METHODS
2.1. Animals and samples
Ours was a retrospective case control study. Dogs with AHDS (n = 50) were client‐owned dogs that presented to the University Hospital at the Norwegian University of Life Sciences (NMBU) between August 1, 2019 and October 31, 2019. Inclusion criteria were dogs with acute hemorrhagic diarrhea for which no underlying cause based on history and clinical investigations was found. The clinical investigation was determined based on the veterinarian's discretion and included tests such as routine hematology and biochemistry, diagnostic imaging (radiology or ultrasound examination of abdomen or both), coagulation tests, fecal examination for parasites, fecal culture for Salmonellae and Campylobacter, fecal PCR tests for C. perfringens A/B and NetE/NetF enterotoxin genes with or without fecal PCR testing for parvovirus. The diagnostic evaluation of these dogs was conducted in collaboration with the National Veterinary Institute and the Norwegian Food Safety Authority, in part to determine the cause of the outbreak. Fecal samples were collected from dogs to characterize the microbiota as described here. Written consent was given by dog owners for using collected fecal samples for investigative and research purposes.
In addition to the dogs with AHDS, 3 dogs with nonhemorrhagic diarrhea were included. These dogs were examined and treated during the same period as the dogs with AHDS. In these dogs, history and clinical investigation did not indicate AHDS (Figure S1).
Healthy dogs (n = 11) were staff‐owned dogs without evidence of gastrointestinal (GI) diseases, no prior history of previous GI diseases, and had not been given any medications. Samples from these dogs were collected during the outbreak.
Fecal samples were collected after natural defecation from dogs as soon as possible, and no later than 2 days after they presented to NMBU. Samples were frozen at −80°C before processing.
We also obtained fecal samples from 78 healthy female and male dogs from a range of breeds and ages, collected during the period from October 2017 to August 2018 (healthy dogs preoutbreak). These samples were obtained by consenting dog owners who collected approximately 2 g of fresh feces into collection tubes containing 3 mL 96% ethanol to stop any further bacterial replication. Samples subsequently were sent by mail to the University of Oslo, where they were stored at −20°C pending processing. These samples were collected as part of another study and are included here only for comparison with the outbreak cohort. Sequence data can be made available upon request. A flow diagram shows the different groups of dogs included in the study (Figure 1).
FIGURE 1.
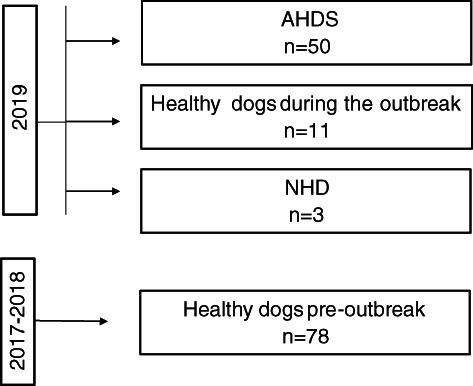
The flow diagram shows the different groups of dogs included in this study. AHDS, acute hemorrhagic diarrhea syndrome; NHD, nonhemorrhagic diarrhea
2.2. 16S rRNA sequencing
Fecal DNA was extracted using the MagAttract PowerSoil DNA KF kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Library preparation for DNA sequencing was carried out as previously described,14 targeting the V4 region of the 16S rRNA gene with the 515f‐805r primer pair. Sequencing of the 2 × 300 pair‐end reads was performed using the MiSeq platform at the Norwegian Sequencing Centre (NSC).
Sequence read demultiplexing was carried out using a custom routine developed at NSC (https://github.com/nsc-norway/triple_index-demultiplexing). Primer sequences were trimmed using the “fastx_truncate” function in the USEARCH software suite.15 Further sequence data processing was performed using the Divisive Amplicon Denoising Algorithm as implemented in the dada2 v1.16 R‐package.16 Taxonomic classification of amplicon sequence variants (ASVs) was done using the Ribosomal Database Project v16 training set.17
2.3. Sequence comparison among ASVs classified as Providencia with P. alcalifaciens outbreak isolates
For a comparative study as part of the outbreak investigation,18 8 P. alcalifaciens isolates from dogs with AHDS (Figure S2) were genome sequenced using a combination of the Nanopore and Illumina sequencing platforms. These DNA sequences were used to generate hybrid assemblies for each isolate. Then, all 16S rRNA gene sequences from the 8 isolate assemblies were extracted using CBS Feature extractor tool (http://www.cbs.dtu.dk/services/FeatureExtract/, October 2020). Each of the 8 isolates had 7 copies of the 16S rRNA gene.
All 56 rRNA gene V4 sequences from these 8 strains were aligned, with default settings, using an online implementation of the MUSCLE alignment algorithm (https://www.ebi.ac.uk/Tools/msa/muscle/, October 2020). We identified and included all 7 V4 16S rRNA regions of P. alcalifaciens, as found in 3 annotated isolates (strains FDAARGOS_408, NCTC10286, 1701003) using Geneious Prime (version 11.0.3 + 7). In Geneious, we aligned these 21 sequences with the sequences from the 8 isolates, and the 6 ASV 16S rRNA amplicon sequences from our study. For those isolates that had ≥2 16S rRNA sequences with a pairwise 100% sequence similarity, we used a reference sequence in the alignment to minimize redundancy. The resulting alignment was used to create an unrooted phylogenetic tree of the V4 region using 21 16S rRNA genes from outbreak isolates, reference strains, and 6 amplicon sequences. The tree was generated in Geneious Prime using the Neighbor‐joining algorithm with the Jukes‐Cantor distance model and 1000 replicates. This alignment was used to compare the ASVs with the corresponding region from the isolated strains.
2.4. Statistical analysis of data
Tests for ASV enrichment among groups were done using the test for differential expression based on the negative binomial distribution, as implemented in the DESeq2 v1.28.1 R‐package.19 All P values reported from these tests were subjected to Benjamini‐Hochberg correction for multiple hypothesis testing. Multivariate analysis of variance with permutation (PERMANOVA) was carried out with the “adonis” function in the vegan v.2.5.6 R‐package (https://CRAN.R-project.org/package=vegan). Two‐sided, unpaired Wilcoxon rank sum tests were carried out using the “wilcox.test” function. The logistic regression model used to test for any relationship between health condition (dogs with AHDS vs healthy dogs during the outbreak) and sex was done using the “glm” function with binomial errors. All statistical tests were carried out in R v.4.0.2.
2.4.1. Data acessibility
The sequence data have been deposited in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with accession number PRJNA725169.
3. RESULTS
The 50 dogs with AHDS were between 4 months and 14 years with a median age of 6.5 years. Various breeds were represented, with 30% (17/50) of the dogs being of a miniature breed (<10 kg). Healthy dogs during the outbreak (n = 11) were between 4 months and 15 years of age with a median of 4 years, and none of the dogs in this group was of a miniature dog breed (Figure S1). No specific breed was overrepresented in the cohort. No significant difference in age (P > .30, Wilcoxon rank sum test) or sex (P > .15, logistic regression model) distribution was found between the dogs with AHDS and the healthy dogs during the outbreak. The healthy dogs pre‐outbreak consisted of various breeds and ages of both sexes.
3.1. Microbial analysis
3.1.1. Sequencing analysis
After quality filtering, pair merging and chimera removal, the outbreak sequence data consisted of 2 697 402 reads, with a per sample mean of 42 147 (±13 030 SD). The total number of observed ASVs was 790. To account for differences in sampling depth, we used common scaling20 to the lowest sample read number (13 056 reads). This read number appears to capture the main diversity in the fecal samples (Figure S3).
3.1.2. Fecal microbiota composition—alpha and beta diversity in dogs with AHDS vs healthy dogs during the outbreak
The overall fecal microbiota composition in dogs with AHDS was significantly different from that of healthy dogs (R 2 = .07, P < .001, PERMANOVA; Figure 2).
FIGURE 2.
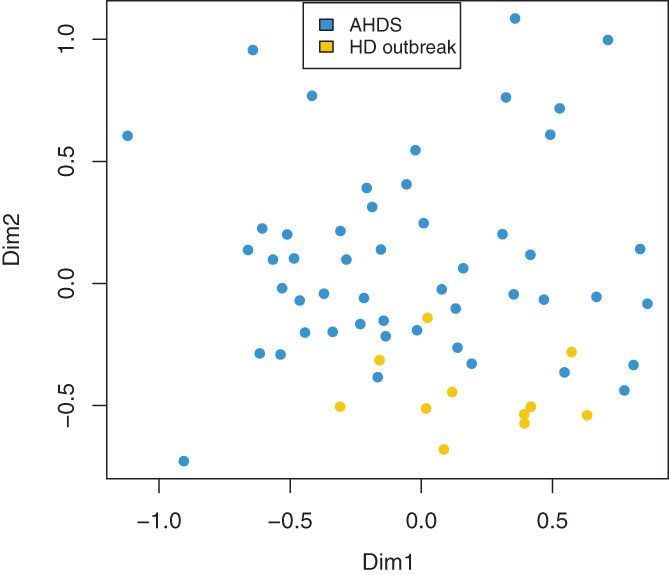
Nonmetric multidimensional scaling plot based on Bray‐Curtis distances for the acute hemorrhagic diarrhea syndrome (AHDS) and healthy dogs (HD) from the outbreak. The microbiota composition in the dogs with AHDS (n = 50) was significantly different from the healthy dogs (n = 11; PERMANOVA, R 2 = 0.07, P < .001)
Dogs with AHDS had significantly decreased Shannon index (median, 2.7; range, 0.9‐3.5 vs median, 3.2; range, 2.6‐4.0; P < .001, Wilcoxon rank sum test; Figure 3A) and species richness (median, 67; range, 32‐91 vs median, 80; range, 69‐141; P < .001, Wilcoxon rank sum test; Figure 3B) relative to the healthy dogs.
FIGURE 3.
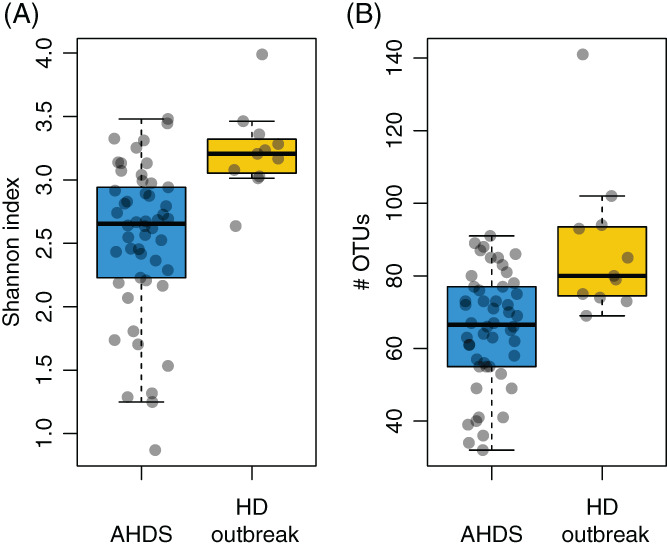
A,B, Comparison of gut bacterial diversity between AHDS and healthy dogs (HD) from the outbreak. A, Diversity as represented by the Shannon index was significantly reduced in dogs with acute hemorrhagic diarrhea syndrome (AHDS) (P < .001, Wilcoxon rank sum test; median, 2.7; range, 0.9–3.5) relative to healthy dogs (median, 3.2; range 2.6‐4.0). B, Dogs with AHDS had reduced observed species richness (P < .001; median, 67, median range 32‐91) relative to healthy dogs (median, 80; range, 69‐141)
3.1.3. Differences in microbiota populations in dogs with AHDS vs dogs with nonhemorrhagic diarrhea and healthy dogs sampled during the outbreak
Figure 4A,B shows the most abundant phyla and families in dogs with AHDS and healthy dogs during the outbreak, respectively.
FIGURE 4.
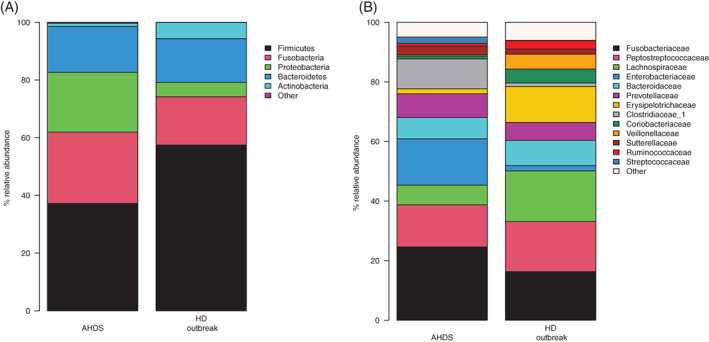
A,B, Mean relative abundances of microbiota composition at the phylum and family level in dogs with acute hemorrhagic diarrhea syndrome (AHDS) (n = 50) and healthy dogs (HD) from the outbreak (n = 11) dogs
Significant phylum‐level differences were identified in the microbiota composition of dogs with AHDS compared to healthy dogs. The dogs with AHDS showed, on average, a 4‐fold increase in relative abundance of Proteobacteria (P < .001, Wilcoxon rank sum test), whereas Firmicutes was decreased by 20% (P = .017) and Actinobacteria was decreased by a factor of 5 (P < .001). On the family level, dogs with AHDS were depleted for Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae and Veillonellaceae (P < .001 for all comparisons, Wilcoxon rank sum test), whereas they showed significantly increased abundance of Entrobacteriaceae (P < .001) and Clostridiaceae_1 (P = .03).
Analysis using DESeq2 identified 32 ASVs that had a significant differential abundance in dogs with AHDS compared with healthy dogs (DESeq2, Table 1). Of these, 24 ASVs were more abundant in dogs with AHDS, including Escherichia spp./Shigella spp. (P < .001), Haemophilus haemoglobinophilus (P < .001), Sutterella spp. (P < .01), as well as 3 ASVs affiliated with Providencia spp., of which 1 (ASV number 6) was particularly prevalent (P < .001;Figure 5A and DESeq2 Table 1). Furthermore, of the 25 ASVs classified as Clostridium sensu stricto, 1 ASV (ASV number 3) showed 100% sequence identity to C. perfringens (Figures S4 and S5). This ASV was highly enriched in dogs with AHDS (P < .001, DESeq2, Table 1 and Figure 5B), and we henceforth refer to it as C. perfringens.
TABLE 1.
The ratio of log2 fold change of bacterial taxa (ASVs) in healthy dogs from the outbreak period and AHDS dogs as detected and filtered by DESeq2
| Genus/Species | ASV no. | log2 fold change | P value |
|---|---|---|---|
| Providencia | Asv39 | 23,90 | <.001 |
| Escherichia/Shigella | Asv38 | 23,42 | <.001 |
| Alloprevotella | Asv59 | 23,12 | <.001 |
| Clostridium_sensu_stricto | Asv40 | 22,79 | <.001 |
| Escherichia/Shigella | Asv64 | 22,71 | <.001 |
| Providencia | Asv71 | 22,64 | <.001 |
| Enterobacteriaceaea | Asv61 | 22,40 | <.001 |
| Fusobacterium | Asv66 | 22,29 | <.001 |
| Clostridium_XlVa | Asv94 | 22,23 | <.001 |
| Clostridium_XlVb | Asv79 | 21,82 | <.001 |
| Streptococcus | Asv37 | 21,72 | <.001 |
| Paraprevotella | Asv47 | 21,62 | <.001 |
| Clostridium_XI | Asv85 | 21,13 | <.001 |
| Fusobacterium | Asv52 | 21,00 | <.001 |
| Clostridium_XI | Asv57 | 20,61 | <.001 |
| Clostridium_XI | Asv119 | 20,50 | <.001 |
| Haemophilus haemoglobinophilus | Asv95 | 20,19 | <.001 |
| Bacteroides | Asv121 | 18,97 | <.001 |
| Sutterella | Asv138 | 18,96 | <.001 |
| Sutterella | Asv68 | 6,80 | <.01 |
| Streptococcus | Asv18 | 4,98 | <.01 |
| Escherichia/Shigella | Asv5 | 4,59 | <.001 |
| Providencia | Asv6 | 3,49 | <.001 |
| Clostridium_sensu_strictob | Asv3 | 2,64 | <.01 |
| Lachnospiraceaea | Asv14 | −2,91 | <.001 |
| Blautia | Asv32 | −3,02 | <.01 |
| Faecalibacterium prausnitzii | Asv25 | −3,35 | <.001 |
| Catenibacterium mitsuokai | Asv20 | −5,97 | <.001 |
| Coprobacillus | Asv271 | −11,28 | <.01 |
| Fusobacterium | Asv169 | −24,39 | <.001 |
| Cetobacterium | Asv157 | −25,84 | <.001 |
| Collinsella | Asv284 | −27,54 | <.001 |
Note: A log2 fold change with a positive value indicates increased presence of the given bacterial taxa in AHDS dogs relative to the healthy dogs, whereas a negative value indicates reduced presence of the given bacterial taxa.
Abbreviations: AHDS, acute hemorrhagic diarrhea syndrome; ASV, amplicon sequence variants.
Classified to family as lowest level.
According to BLAST, this ASV was classified with 100% identify to Clostridium perfringens. The other ASVs classified as Clostridium_sensu_stricto were also implemented in a BLAST search, but did not reveal similar high identity.
FIGURE 5.
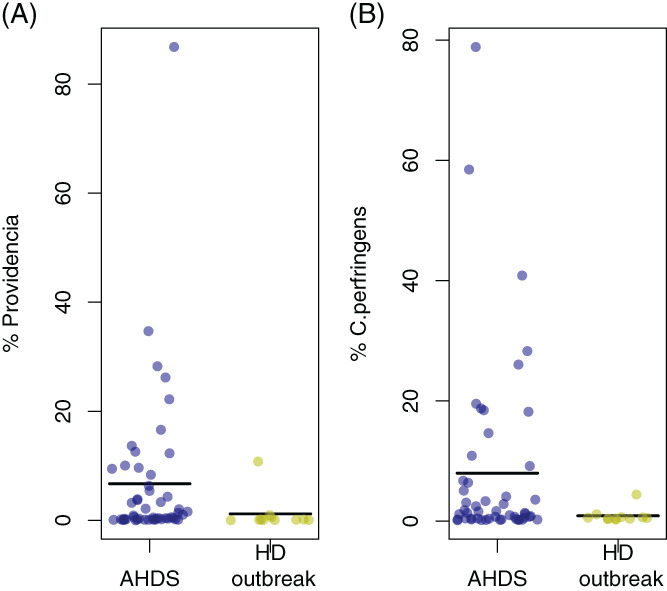
A,B. Mean relative abundances of Providencia spp. and Clostridium perfringens in AHDS dogs 6 ASVs) were significantly more abundant in dogs with AHDS. The figure shows the combined relative abundance of 6 ASVs classified as this genus. Three of these were significantly enriched in dogs with AHDS (DESeq2, Table 1) B. Clostridium perfringens in dogs with AHDS and healthy dogs. The figure shows data for a single ASV (ASV no. 3) that showed 100% identity to known C. perfringens isolates, and that was significantly more abundant in dogs with AHDS (DESeq2, Table 1). AHDS, acute hemorrhagic diarrhea syndrome; ASV, amplicon sequence variants
Eight ASVs, including Faecalibacterium prausnitzii, Blautia spp. and Collinsella, were significantly more abundant in healthy dogs (P < .01, DESeq2, Table 1). The 3 dogs with nonhemorrhagic diarrhea also had evidence of fecal dysbiosis with lower abundance of Actinobacteria and an increase in C. perfringens, but with levels of Providencia similar to the healthy dogs during the outbreak (Figures S6 and S7). Because of the small number of samples in this category, these are anecdotal observations without statistical significance.
3.1.4. Providencia spp. in dogs with AHDS vs healthy dogs from the outbreak period
We found 6 ASVs classified as the genus Providencia, of which 3 were significantly enriched in the dogs with AHDS (ASV numbers 6, 39 and 70; DESeq2, Table 1). In dogs with AHDS, ASV number 6 was found in all but 1 sample and had a mean relative abundance of 5.6%. In contrast, its abundance was 1.2% in healthy dogs from the outbreak period. On the other hand, ASV number 39 and ASV number 71 were present at lower abundances, representing 0.49% and 0.31% of the mean relative abundances in dogs with AHDS, respectively. These 2 ASVs were not observed in healthy dogs.
We compared the 16S rRNA V4 regions from the 6 Providencia spp. ASVs with the corresponding sequences from 8 P. alcalifaciens isolates obtained from dogs suffering from AHDS during the outbreak, as well as 3 reference strains (Figure 6). We found that only ASV number 6 matched these isolates with 100% sequence identity, demonstrating that ASV number 6 is closely related to the P. alcalifaciens strains implicated in the AHDS outbreak.
FIGURE 6.
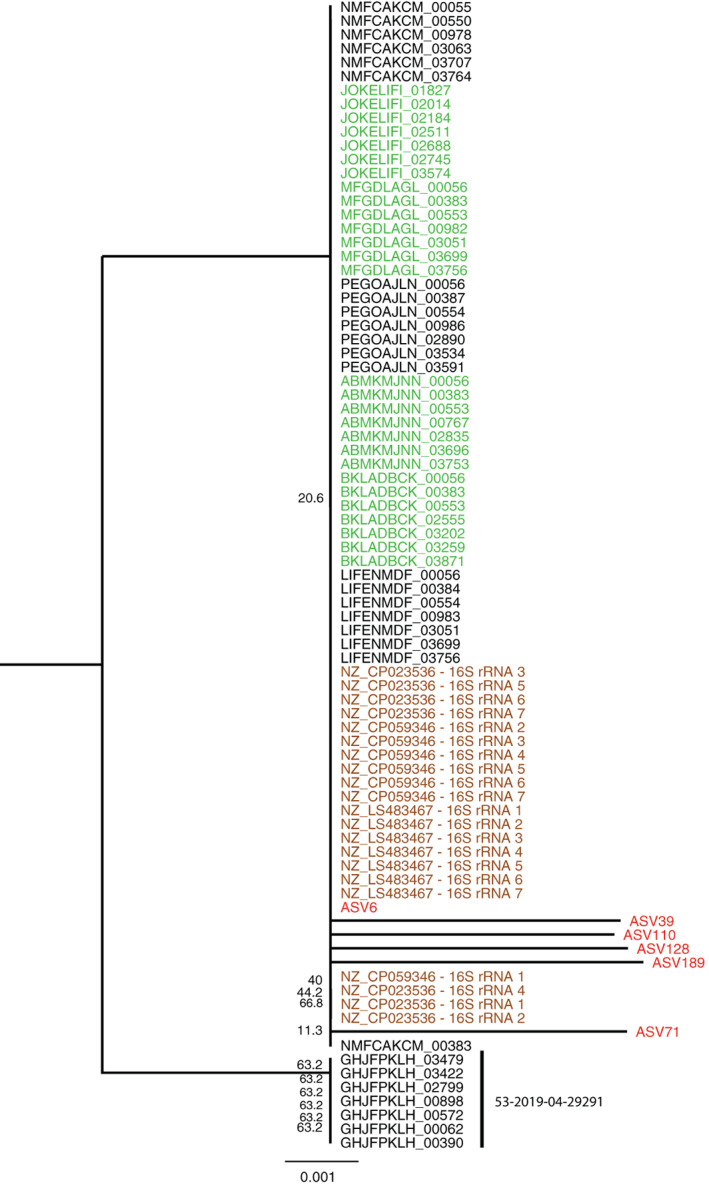
Neighbor‐joining tree of the V4 region of 83 16S rRNA genes from outbreak isolates, reference strains and 6 amplicon sequences. Sequences from each isolate with a pairwise 100% similarity of the V4 16S rRNA region were replaced by a representative sequence to reduce the redundancy. The number of combined sequences is indicated in brackets for each sequence. The amplicon sequence variants (ASVs) are marked in red, the reference strain sequences in brown, and the outbreak strains in green. In black are sequences from nonoutbreak isolates. The phylogeny shows that all Providencia alcalifaciens sequences cluster at the same node together with ASV6, except for the sequences of isolate 53‐2019‐04‐29291, which is the outgroup here. The numbers at the nodes indicate bootstrap values higher than 20
The other 5 ASVs classified as Providencia showed nucleotide differences with all of the 16S rRNA gene V4 region sequences to which they were compared. This finding indicates that these represent different P. alcalifaciens strains, or possibly other species.
3.1.5. Providencia in healthy dogs sampled before the outbreak
To determine if Providencia is part of the normal microbiota of dogs in the region, we compared our sequencing results from dogs with AHDS (n = 50) and healthy dogs during the outbreak (n = 11) with sequencing results from healthy dogs before the outbreak (n = 78). In healthy dogs before the outbreak, 5 818 558 16S rRNA gene amplicon sequences were generated, from which we failed to identify a single ASV classified as Providencia. For comparison, an ASV identical to C. perfringens (ASV number 3) was found in 77 of the 78 samples at a mean relative abundance of 0.7% (range, 0%‐11%). Small but significant differences (R 2 = .05, P = .01, PERMANOVA) were found between healthy dogs before and during the outbreak. When comparing dogs with AHDS and healthy dogs preoutbreak, a significant difference was found in microbiota composition (PERMANOVA, R 2 = .06, P < .01; Figure S8).
4. DISCUSSION
We report increased abundance of Providencia spp. and C. perfringens in dogs with AHDS compared to healthy dogs. Furthermore, we found widespread fecal dysbiosis in dogs with AHDS, with a general overgrowth of Proteobacteria and depletion of putatively beneficial species. One ASV classified as Providencia (ASV number 6) showed 100% sequence identity to 16S rRNA gene V4 sequences of P. alcalifaciens isolates from dogs with AHDS during the outbreak. We also report that Providencia spp. were not part of the normal dog microbiota in the time before the AHDS outbreak in 2019. However, the small but significant difference between healthy dogs before and during the outbreak may indicate that Providencia spp. entered the population in the time before the outbreak. However, whether this bacterium was the actual cause of the outbreak, or whether its presence was favored by the altered microbiota, remains unknown, and the question is beyond the scope of our study.
Although proliferation of C. perfringens and presence of enterotoxin genes have been associated with AHDS in dogs in several studies,5, 6, 21, 22 other factors may be involved in the pathogenesis. Providencia spp. also may have pathogenic properties. Providencia alcalifaciens previously has been linked with enteritis in dogs,11, 23 although others have claimed its presence is a consequence of the altered intestinal microenvironment in dogs with gastroenteritis.12 In humans, studies have found that Providencia spp., including P. alcalifaciens, are an important cause of traveler's diarrhea.24 Providencia alcalifaciens also has been associated with diarrhea in children, but other enteric pathogens also were present in these cases.25 In 2005, dogs from different areas in Oslo and nearby regions suffered from hemorrhagic diarrhea with unusually high numbers of P. alcalifaciens. Providencia alcalifaciens was isolated from 6 dogs, with 1 fatal outcome. Although it could not be determined whether or not this bacterium was the primary cause, the study demonstrated that it had the potential to invade mammalian epithelial cells in in vitro studies.26 Its potential to invade cells also has been demonstrated in other in vitro studies.27, 28 In addition to invasiveness, enterotoxins may be another contributing virulence mechanism, and cytholetal distending toxins produced by P. alcalifaciens and Providencia rettgeri have been associated with diarrhea in humans.29 Moreover, P. alcalifaciens may produce manganese superoxide dismutase which enhances protection against phagocytosis.30 Routine testing for enteropathogens usually does not include Providencia spp., because it is not recognized for being enteropathogenic and requires special culturing methods for identification.31 Thus, the presence of Providencia spp. in fecal samples from humans with diarrhea is likely not often detected. However, a study in humans found Providencia spp. in 7.5% of diarrhea samples.32 In dogs, the occurrence of Providencia spp. in diarrhea samples is unknown. Whether Providencia spp. should be regarded as enteropathogens and be investigated in dogs with diarrhea requires further study.
Members of the Firmicutes phylum such as Faecalibacterium prausnitzii and Blautia spp., as well as an ASV classified as family Lachnospiraceae, were depleted in dogs with AHDS, as has been reported in other studies.8, 9, 10 These bacteria belong to Clostridium clusters IV and XIVa, which contain efficient short chain fatty acid producers that are important for intestinal homeostasis.33 Thus, their depletion may contribute to development of gastroenteritis.34 Furthermore, these microbes contribute to the maintenance of mucosal integrity and protect against invasion.35 Dysbiosis characterized by the loss of these bacteria may further enhance mucosal damage caused by pathogens.36, 37 However, most knowledge about the effects of depletion of important bacterial groups in intestinal dysbiosis comes from studies in humans and mouse models of the human microbiome. The dog microbiome is very similar to that of humans, much more so than that of mice,38 emphasizing the relevance of microbiome studies in dogs. Nevertheless, more studies focusing on dogs, including in vitro experiments,39 are needed to determine the extent to which interactions among specific groups of intestinal microbes and the intestinal mucosa resemble those observed in humans.
In addition to depletion of putatively beneficial bacterial species, we also observed an enrichment of Proteobacteria, including Escherichia coli/Shigella and Sutterella. A previous study described a similar pattern of intestinal dysbiosis in dogs with diarrhea, including acute, hemorrhagic and chronic diarrhea, with dogs with ADHS experiencing the most marked microbiota alterations.10 Interestingly, we found that the 3 dogs with nonhemorrhagic diarrhea also had evidence of fecal dysbiosis with lower abundances of Actinobacteria and an increase in C. perfringens, yet with Providencia numbers similar to the healthy controls (Figures S6 and S7). Although this particular observation suggests a key role for Providencia in the etiology of AHDS, the small number of dogs with nonhemorrhagic diarrhea precludes any conclusion. However, the fact that we did not observe any Providencia sequences in a relatively large cohort sampled over 2 previous years indicates that these bacteria are not a normal part of the dog microbiota, at least not in the Oslo area. Thus, it is possible that they became established in the population during the period after the summer of 2018, eventually becoming widespread and highly abundant in many dogs. An epidemiological investigation of the outbreak has been performed,13 and further studies are underway that may provide an answer about the role of Providencia spp. in this outbreak.
Dogs with AHDS had increased relative abundances of C. perfringens compared to healthy dogs, which is consistent with previous observations.4 However, healthy dogs also may harbor relatively high numbers of C. perfringens,22 suggesting that this species is an opportunistic pathogen that can function as part of a healthy gut microbial system. Indeed, in our samples from 2017 to 2018 we found that this species is a normal and relatively abundant member of the healthy dog microbiota. A recent study found that the pore‐forming toxin genes NetE and NetF, commonly associated with certain strains of C. perfringens, were more prevalent in dogs with AHDS than in healthy controls.5 This finding indicates a potential role for C. perfringens in the etiology of AHDS, while also suggesting that it is possible to develop the disease in the absence of toxin‐producing strains, and that these strains can be found in healthy individuals. Although microbial factors likely influence the development of AHDS, their contribution may depend on the intestinal immune defenses of the individual dog, as reviewed previously.40
One limitation of our study was the low number of healthy dogs included in the control group. Ideally, we would have included a larger cohort of breed‐ and age‐matched control dogs to provide a more optimal comparison with dogs with AHDS. Furthermore, the 16S rRNA V4 gene fragment does not, in most cases, provide adequate information for accurate classification at the species level. Although the fact that ASV number 6 was identical in sequence to several outbreak isolates of P. alcalifaciens provides an indication that they are very closely related, we were not able to provide classification beyond the genus level for the other Providencia ASVs. Lastly, we do not know if Providencia spp. disappear from the host or remain members of the microbiome. This question is beyond the scope of our study, but follow‐up sampling of the dogs included in our study would provide valuable insight into potential effects on colonization dynamics. A previous study of dogs with AHDS suggested that changes in the intestinal microbiota might outlast GI inflammation.9 Thus, the environmental and ecological factors that led to the introduction and proliferation of Providencia in the population need to be further investigated.
In conclusion, we found increased abundance of Providencia spp. in dogs with AHDS relative to healthy dogs, offering a plausible explanation for the unexpectedly high numbers of dogs suffering from a severe form of AHDS during this outbreak. Our results indicate the complexity of the AHDS outbreak, where Providencia spp. may have contributed to this severe outbreak of diarrhea. The condition could be compounded by concurrent outgrowths of C. perfringens in a dysbiotic background microbiota. Additional studies are needed to learn more about the population genomic structure of Providencia spp. and identify species and strains with pathogenic potential. Genomic and functional studies also are needed to elucidate the mechanisms of pathogenicity.
CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Authors declare no IACUC or other approval was needed.
HUMAN ETHICS APPROVAL DECLARATION
Authors declare human ethics approval was not needed for this study.
Supporting information
Figure S1 Demographic overview of the dogs
Figure S2 Overview isolates checked for the 16S rRNA
Figure S3 Rarefaction curves for the 61 outbreak samples. The dotted gray lines mark the number reads in the sample with the lowest read number (13 056).
Figure S4 List of 25 ASV sequences classified as Clostridium sensu stricto.
Figure S5 Neighbor joining tree showing the evolutionary relationship among the 25 ASVs classified as Clostridium sensu stricto, as well as the corresponding 16S rRNA gene region from Clostridium perfringens isolate ATCC 13124 downloaded from GenBank. The branch length scale is the number of nucleotide substitutions per site. The tree was made with MEGA X
Figure S6 Mean relative abundances of microbiota composition at the phylum level in dogs with AHDS (n = 50), healthy dogs (HD) during the outbreak (n = 11) dogs and dogs with nonhemorrhagic diarrhea (NHD, n = 3).
Figure S7 Mean relative abundances of Providencia spp. and Clostridium perfringens in dogs with AHDS, healthy dogs (HD) during the outbreak, and dogs with nonhemorrhagic diarrhea (NHD). Error bars are ±1 SE. A, Combined mean relative abundances of all 6 ASVs classified as Providencia. B, Mean relative abundances of ASV no. 3, which matched C. perfringens with 100% sequence identity.
Figure S8 Nonmetric multidimensional scaling plot based on Bray‐Curtis distances for the healthy dogs (HD) during the outbreak (n = 11) and healthy dogs (HD) preoutbreak (n = 78). There were small but significant differences (R 2 = .05, P = .01, PERMANOVA) between healthy dogs before and during the outbreak.
ACKNOWLEDGMENT
Funding provided by SPARK Norway and UiO:Life Science and we thank them for providing the funding for collection and analysis of the fecal samples from before the outbreak, as well as the volunteers that provided sample material.
Herstad KMV, Trosvik P, Haaland AH, Haverkamp THA, de Muinck EJ, Skancke E. Changes in the fecal microbiota in dogs with acute hemorrhagic diarrhea during an outbreak in Norway. J Vet Intern Med. 2021;35(5):2177–2186. 10.1111/jvim.16201
Kristin Herstad and Pål Trosvik contributed equally as first authors. Eric J. de Muinck and Ellen Skancke contributed equally as last authors.
Funding information SPARK Norway and UiO:Life Science
DATA AVAILABILITY STATEMENT
The sequence data have been deposited in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with accession number PRJNA725169.
REFERENCES
- 1.Mortier F, Strohmeyer K, Hartmann K, Unterer S. Acute haemorrhagic diarrhoea syndrome in dogs: 108 cases. Vet Rec. 2015;176(24):627. [DOI] [PubMed] [Google Scholar]
- 2.Leipig‐Rudolph M, Busch K, Prescott JF, et al. Intestinal lesions in dogs with acute hemorrhagic diarrhea syndrome associated with netF‐positive Clostridium perfringens type A. J Vet Diagn Invest. 2018;30(4):495‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall EJ, Day MJ. Diseases of the small intestine. In: Ettinger SJ, Feldman EC, Côte E, eds. Textbook of Veterinary Internal Medicine. 2.8th ed. Canada: Elsevier; 2017:1538‐1539. [Google Scholar]
- 4.Unterer S, Busch K, Leipig M, et al. Endoscopically visualized lesions, histologic findings, and bacterial invasion in the gastrointestinal mucosa of dogs with acute hemorrhagic diarrhea syndrome. J Vet Intern Med. 2014;28(1):52‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sindern N, Suchodolski JS, Leutenegger CM, et al. Prevalence of Clostridium perfringens netE and netF toxin genes in the feces of dogs with acute hemorrhagic diarrhea syndrome. J Vet Intern Med. 2019;33(1):100‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch K, Suchodolski JS, Kuhner KA, et al. Clostridium perfringens enterotoxin and Clostridium difficile toxin A/B do not play a role in acute haemorrhagic diarrhoea syndrome in dogs. Vet Rec. 2015;176(10):253. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein MR, Kruth SA, Bersenas AM, Holowaychuk MK, Weese JS. Detection and characterization of Clostridium perfringens in the feces of healthy and diarrheic dogs. Can J Vet Res. 2012;76(3):161‐165. [PMC free article] [PubMed] [Google Scholar]
- 8.Guard BC, Barr JW, Reddivari L, et al. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS One. 2015;10(5):e0127259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilmann RM, Guard MM, Steiner JM, Suchodolski JS, Unterer S. Fecal markers of inflammation, protein loss, and microbial changes in dogs with the acute hemorrhagic diarrhea syndrome (AHDS). J Vet Emerg Crit Care (San Antonio). 2017;27:586‐589. [DOI] [PubMed] [Google Scholar]
- 10.Suchodolski JS, Markel ME, Garcia‐Mazcorro JF, et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. 2012;7(12):e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Möhr AJ, van der Lugt JJ, Josling D, et al. Primary bacterial enteritis caused by Providencia alcalifaciens in three dogs. Vet Rec. 2002;150(2):52‐53. [DOI] [PubMed] [Google Scholar]
- 12.Tribe GW, Rood MJ. Providencia alcalifaciens in diarrhoeic dogs and cats. Vet Rec. 2002;150(12):386‐387. [PubMed] [Google Scholar]
- 13.Haaland AH, Herstad K, Nøstebø SF, et al. Outbreak of acute hemorrhagic diarrhea in dogs in Norway; is Providencia alcalifaciens involved? J Vet Intern Med. 2020;34:3058‐3166. 10.1111/jvim.15924. [DOI] [Google Scholar]
- 14.de Muinck EJ, Trosvik P, Gilfillan GD, Hov JR, Sundaram AYM. A novel ultra high‐throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform. Microbiome. 2017;5(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460‐2461. [DOI] [PubMed] [Google Scholar]
- 16.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261‐5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haverkamp T.H.A, et al. Complete genomes of eight Providencia alcalifaciens from diseased Norwegian dogs. 2021. Manuscript in preparation.
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks SL, Kather EJ, Kass PH, Melli AC. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med. 2002;16(5):533‐540. [DOI] [PubMed] [Google Scholar]
- 22.Weese JS, Staempfli HR, Prescott JF, Kruth SA, Greenwood SJ, Weese HE. The roles of Clostridium difficile and enterotoxigenic Clostridium perfringens in diarrhea in dogs. J Vet Intern Med. 2001;15(4):374‐378. [PubMed] [Google Scholar]
- 23.Król J, Janeczek M, Pliszczak‐Król A, Janeczek W, Florek M. Providencia alcalifaciens as the presumptive cause of diarrhoea in dog. Pol J Vet Sci. 2007;10(4):285‐287. [PubMed] [Google Scholar]
- 24.Yoh M, Matsuyama J, Ohnishi M, et al. Importance of Providencia species as a major cause of travellers' diarrhoea. J Med Microbiol. 2005;54(Pt 11):1077‐1082. [DOI] [PubMed] [Google Scholar]
- 25.Albert MJ, Faruque AS, Mahalanabis D. Association of Providencia alcalifaciens with diarrhea in children. J Clin Microbiol. 1998;36(5):1433‐1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fauske A, Hirsch M. Kan Providencia alcalifaciens være en primærpatogen bakterie ved diaré hos hund? [Bachelor Thesis]. Oslo, Norway: Norges Veterinærhøgskole Høgskolen i Oslo, Oslo; 2006.
- 27.Janda JM, Abbott SL, Woodward D, Khashe S. Invasion of HEp‐2 and other eukaryotic cell lines by Providenciae: further evidence supporting the role of Providencia alcalifaciens in bacterial gastroenteritis. Curr Microbiol. 1998;37(3):159‐165. [DOI] [PubMed] [Google Scholar]
- 28.Rahman M, Monira S, Nahar S, et al. TnphoA mutants of Providencia alcalifaciens with altered invasiveness of HEp‐2 cells. J Med Microbiol. 2002;51(8):682‐686. [DOI] [PubMed] [Google Scholar]
- 29.Shima A, Hinenoya A, Asakura M, et al. Molecular characterizations of cytolethal distending toxin produced by Providencia alcalifaciens strains isolated from patients with diarrhea. Infect Immun. 2012;80(4):1323‐1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Kodama T, Iida T, Honda T. Demonstration and characterization of manganese superoxide dismutase of Providencia alcalifaciens . Microbiol Immunol. 2007;51(10):951‐961. [DOI] [PubMed] [Google Scholar]
- 31.Shah MM, Odoyo E, Ichinose Y. Epidemiology and pathogenesis of Providencia alcalifaciens infections. Am J Trop Med Hyg. 2019;101(2):290‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shima A, Hinenoya A, Samosornsuk W, Samosornsuk S, Mungkornkaew N, Yamasaki S. Prevalence of Providencia strains among patients with diarrhea and in retail meats in Thailand. Jpn J Infect Dis. 2016;69(4):323‐325. [DOI] [PubMed] [Google Scholar]
- 33.Livanos AE, Snider EJ, Whittier S, et al. Rapid gastrointestinal loss of Clostridial clusters IV and XIVa in the ICU associates with an expansion of gut pathogens. PLoS One. 2018;13(8):e0200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minamoto Y, Otoni CC, Steelman SM, et al. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. 2015;6(1):33‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathogen. 2013;5(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep. 2019;7(1):3‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Huang Y‐J, Yoon JY, et al. Primary human colonic mucosal barrier crosstalk with super oxygen‐sensitive Faecalibacterium prausnitzii in continuous culture. Med. 2020;2(1):74‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coelho LP, Kultima JR, Costea PI, et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome. 2018;6(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandra L, Borcherding DC, Kingsbury D, et al. Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biol. 2019;17(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allenspach K. Clinical immunology and immunopathology of the canine and feline intestine. Vet Clin North Am Small Anim Pract. 2011;41(2):345‐360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Demographic overview of the dogs
Figure S2 Overview isolates checked for the 16S rRNA
Figure S3 Rarefaction curves for the 61 outbreak samples. The dotted gray lines mark the number reads in the sample with the lowest read number (13 056).
Figure S4 List of 25 ASV sequences classified as Clostridium sensu stricto.
Figure S5 Neighbor joining tree showing the evolutionary relationship among the 25 ASVs classified as Clostridium sensu stricto, as well as the corresponding 16S rRNA gene region from Clostridium perfringens isolate ATCC 13124 downloaded from GenBank. The branch length scale is the number of nucleotide substitutions per site. The tree was made with MEGA X
Figure S6 Mean relative abundances of microbiota composition at the phylum level in dogs with AHDS (n = 50), healthy dogs (HD) during the outbreak (n = 11) dogs and dogs with nonhemorrhagic diarrhea (NHD, n = 3).
Figure S7 Mean relative abundances of Providencia spp. and Clostridium perfringens in dogs with AHDS, healthy dogs (HD) during the outbreak, and dogs with nonhemorrhagic diarrhea (NHD). Error bars are ±1 SE. A, Combined mean relative abundances of all 6 ASVs classified as Providencia. B, Mean relative abundances of ASV no. 3, which matched C. perfringens with 100% sequence identity.
Figure S8 Nonmetric multidimensional scaling plot based on Bray‐Curtis distances for the healthy dogs (HD) during the outbreak (n = 11) and healthy dogs (HD) preoutbreak (n = 78). There were small but significant differences (R 2 = .05, P = .01, PERMANOVA) between healthy dogs before and during the outbreak.
Data Availability Statement
The sequence data have been deposited in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with accession number PRJNA725169.


