Abstract
Since the outset of the coronavirus disease 2019 (COVID-19) pandemic, the gut microbiome in COVID-19 has garnered substantial interest, given its significant roles in human health and pathophysiology. Accumulating evidence is unveiling that the gut microbiome is broadly altered in COVID-19, including the bacterial microbiome, mycobiome, and virome. Overall, the gut microbial ecological network is significantly weakened and becomes sparse in patients with COVID-19, together with a decrease in gut microbiome diversity. Beyond the existence of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), the gut microbiome of patients with COVID-19 is also characterized by enrichment of opportunistic bacteria, fungi, and eukaryotic viruses, which are also associated with disease severity and presentation. Meanwhile, a multitude of symbiotic bacteria and bacteriophages are decreased in abundance in patients with COVID-19. Such gut microbiome features persist in a significant subset of patients with COVID-19 even after disease resolution, coinciding with ‘long COVID’ (also known as post-acute sequelae of COVID-19). The broadly-altered gut microbiome is largely a consequence of SARS-CoV-2infection and its downstream detrimental effects on the systemic host immunity and the gut milieu. The impaired host immunity and distorted gut microbial ecology, particularly loss of low-abundance beneficial bacteria and blooms of opportunistic fungi including Candida, may hinder the reassembly of the gut microbiome post COVID-19. Future investigation is necessary to fully understand the role of the gut microbiome in host immunity against SARS-CoV-2 infection, as well as the long-term effect of COVID-19 on the gut microbiome in relation to the host health after the pandemic.
Keywords: COVID-19, Gut, Microbiome, Immunity, Infection
Introduction
The ongoing global pandemic of coronavirus disease 2019 (COVID-19) is a disease caused by the RNA virus severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), primarily infecting the respiratory tract and resulting in various symptoms at various severity levels in patients after infection [1]. Around 5%–33% COVID-19 patients had gastrointestinal (GI) symptoms, including diarrhea, nausea, and vomiting [2], [3], [4]. Several studies have detected SARS-CoV-2 in stool samples and anal swabs [5], [6], suggesting that the digestive tract might be an extra-pulmonary site for SARS-CoV-2 infection. Although most cases of COVID-19 are mild, disease can be severe and result in hospitalization, respiratory failure, or death [1]. Such remarkable differences in individual’s presentations and symptoms of COVID-19 arise from the heterogeneous immune statuses and responses against SARS-CoV-2 infection [7], [8], [9]. The GI tract is the largest immune organ in humans, playing critical roles in combating infections of pathogens [10]. Living inside the gut of humans are trillions of microorganisms — bacteria, fungi, viruses, and other life forms that are collectively known as the microbiome — regulating host immunity [11]. As of now, accumulating evidence suggests that the gut microbiome ecology is broadly altered in patients with COVID-19 and that the gut microbiome configurations are associated with immune responses and disease presentations in COVID-19 [12], [13], [14], [15]. The SARS-CoV-2 infection course is crucial for the alterations in the ecology and dynamics of human gut microbiome, in both the short term and long term, which in return influence the human host’s health. Moreover, the presence of active SARS-CoV-2 virus in the gut and altered ecology of the gut microbiome may lead to an unfavorable gut milieu, which facilitates opportunistic bloom of certain fungi and pathogenic bacteria, further hindering the community assembly and function of the gut microbiome, as well as weakening the host immunity [9], [12], [13], [14], [15], [16]. Herein, we summarize the impact of COVID-19 on the human gut microbiome in association with disease phenotypes, from the perspective of the gut microbial ecology, including that of bacteria, fungi, and viruses.
The gut bacterial microbiome in COVID-19
Compositional changes of the gut bacterial microbiome
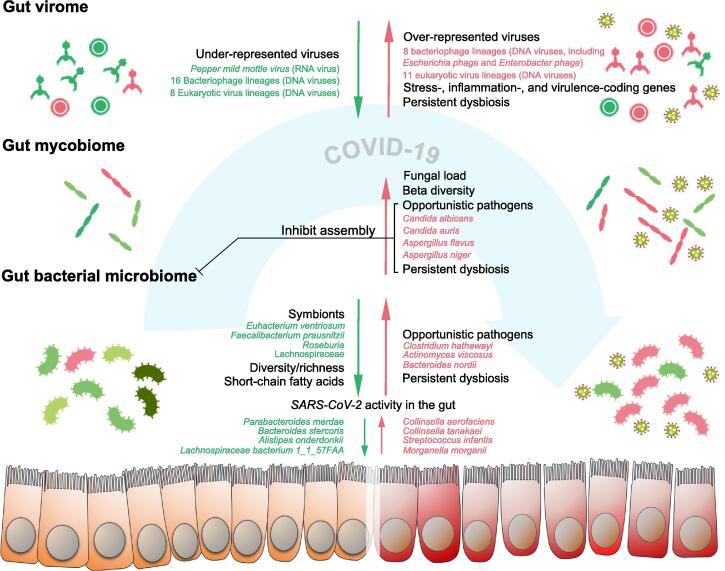
Our early study showed that the gut bacterial microbiome of patients with COVID-19 was significantly altered compared with healthy controls, characterized by depletion of beneficial commensals and enrichment of opportunistic pathogens in the gut [15]. The feces of COVID-19 patients was enriched for opportunistic pathogens known to cause bacteremia, including Clostridium hathewayi, Actinomyces viscosus, and Bacteroides nordii (Figure 1; Table 1) [15], as a secondary infection/bloom post onset of COVID-19 due to disrupted gut microbial ecology and colonization resistance [17], [18]. The patients treated with antibiotics at hospitalization displayed a further depletion of bacterial species, particularly symbionts beneficial to host immunity including Faecalibacterium prausnitzii, Lachnospiraceae bacterium 5_1_63FAA, Eubacterium rectale, Ruminococcus obeum, and Dorea formicigenerans [15]. Such alterations in the bacterial microbiome ecology persisted over the disease course of COVID-19 and even after clearance of SARS-CoV-2 from the respiratory tract [15]. Consistently, another study also showed a similar pattern of gut microbiome dysbiosis in COVID-19 patients [19]. The abundance of butyrate-producing bacteria, such as Faecalibacterium prausnitzii, Clostridium butyricum, Clostridium leptum, and Eubacterium rectale, was significantly decreased in patients with COVID-19 compared to controls [19]. In contrast, the abundance of the common opportunistic pathogens Enterobacteriaceae and Enterococcus was significantly increased in patients with COVID-19 compared to controls [19]. At the genus level, the genera Streptococcus, Rothia, Veillonella, and Actinomyces (all opportunistic pathogens) were enriched in the feces of COVID-19 patients, whereas the genera Romboutsia, Faecalibacterium, and Fusicatenibacter were enriched in the feces of healthy controls [20]. An ecological network analysis revealed significant positive correlations across COVID-19-enriched genera [20], indicating co-expansion of opportunistic bacteria dominating the ecological network of the gut microbiome due to SARS-CoV-2 infection. A high baseline abundance of opportunistic bacteria Coprobacillus, Clostridium ramosum, and Clostridium hathewayi in patients’ feces at hospitalization was associated with a more severe COVID-19 disease course, whereas the anti-inflammatory bacterium Faecalibacterium prausnitzii showed an inverse correlation [15], suggesting baseline gut microbiome calibration of host immunity, thereby affecting disease response upon SARS-CoV-2 infection.
Figure 1.
Alterations in the gut bacterial, fungal, and viral microbiome in patients with COVID-19
The gut bacterial microbiome in COVID-19 is characterized by decreased diversity and richness, and persistent bacterial microbiome dysbiosis even after disease resolution. The gut mycobiome in COVID-19 is characterized by increased fecal fungal load and increased beta-diversity (more heterogeneous), and it is unstable over time and also persistently altered after disease resolution. SARS-CoV-2 shows infectivity in the gut. Delayed SARS-CoV-2 viral shedding and persistent gut virome dysbiosis are both present after disease resolution. The gastrointestinal tract epithelial barrier is impaired in a subset of COVID-19 patients. The figure is created with BioRender.com.
Table 1.
Gut microbes with altered abundance in COVID-19
| Kingdom | Species | Change | Feature | Refs. |
|---|---|---|---|---|
| Bacteria | Actinomyces viscosus | ↑ | Opportunistic pathogens known to cause bacteremia | [15] |
| Clostridium hathewayi | ↑ | Associated with human infection and bacteremia | [15] | |
| Bacteroides nordii | ↑ | Opportunistic pathogens to cause bacteremia | [15] | |
| Eubacterium ventriosum | ↓ | Butyrate-producing | [15] | |
| Dorea formicigenerans | ↓ | Butyrate-producing | [15] | |
| Faecalibacterium prausnitzii | ↓ | Butyrate-producing | [15], [19] | |
| Eubacterium rectale | ↓ | Butyrate-producing | [15], [19] | |
| Ruminococcus obeum | ↓ | Preventing increases in intestinal permeability | [15] | |
| Lachnospiraceae bacterium 5_1_63FAA | ↓ | Butyrate-producing | [15] | |
| Enterococcus faecium | ↓ | Opportunistic pathogens | [15] | |
| Clostridium ramosum | ↓ | Immune-regulatory characteristics | [15] | |
| Fungi | Candida albicans | ↑ | Opportunistic infection and impairing microbiome assembly | [15] |
| Candida auris | ↑ | Healthcare-associated invasive infection | [14] | |
| Aspergillus flavus | ↑ | Associated with respiratory symptoms | [14] | |
| Escheruchia virus | ↑ | Bacteriophage | [14] | |
| Virus/phage | Edafosvirus | ↑ | Unknown host from the environment | [13] |
| Solumvirus | ↑ | Unknown host from the environment | [13] | |
| Bandra megavirus | ↑ | Unknown host from the environment | [13] | |
| Streptococcus virus | ↑ | Bacteriophage | [13] | |
| Paguma larvata circovirus | ↑ | Animal host | [13] | |
| Microcystis virus | ↑ | Bacteriophage | [13] | |
| Ralstonia phage | ↑ | Bacteriophage | [13] | |
| EBPR podovirus | ↑ | Bacteriophage | [13] | |
| Dishui lake phycodnavirus | ↑ | Algae host | [13] | |
| Homavirus | ↑ | Unknown host from the environment | [13] | |
| Cannes 8 virus | ↑ | Amoeba host | [13] | |
| Pigeonpox virus | ↑ | Columba host | [13] | |
| Sucra jujuba nucleopolyhedrovirus | ↑ | Geometrid host | [13] | |
| Phormidium phage | ↑ | Bacteriophage | [13] | |
| Lactococcus phage | ↑ | Bacteriophage | [13] | |
| Enterobacter phage | ↑ | Bacteriophage | [13] | |
| Columbid alphaherpesvirus | ↑ | Columba host | [13] | |
| Faustovirus | ↑ | Amoeba host | [13] | |
| Burkholderia virus | ↓ | Bacteriophage | [13] | |
| Lysinibacillus phage | ↓ | Bacteriophage | [13] | |
| Bdellovibrio phage | ↓ | Bacteriophage | [13] | |
| Citrobacter phage | ↓ | Bacteriophage | [13] | |
| Ranid herpesvirus | ↓ | Animal host | [13] | |
| Dunaliella viridis virus | ↓ | Algae host | [13] | |
| Marseillevirus | ↓ | Amoeba host | [13] | |
| Raccoonpox virus | ↓ | Animal host | [13] | |
| Kaumoebavirus | ↓ | Amoeba host | [13] | |
| Acinetobacter virus | ↓ | Bacteriophage | [13] | |
| Halomonas phage | ↓ | Bacteriophage | [13] | |
| Mycobacterium phage BGlluviae | ↓ | Bacteriophage | [13] | |
| Golden marseillevirus | ↓ | Amoeba host | [13] | |
| Silicibacter phage | ↓ | Bacteriophage | [13] | |
| Tokyovirus | ↓ | Amoeba host | [13] | |
| Sulfolobales virus YNP2 | ↓ | Archaea host | [13] | |
| Nocardia phage | ↓ | Bacteriophage | [13] | |
| Alteromonas phage | ↓ | Bacteriophage | [13] | |
| Hyperthermophilic archaeal virus 2 | ↓ | Archaea host | [13] | |
| Crassphage | ↓ | Bacteriophage | [13] | |
| Nodularia phage | ↓ | Bacteriophage | [13] | |
| Hamiltonella virus | ↓ | Bacteriophage | [13] | |
| Hudisavirus | ↓ | Human host | [13] | |
| Mesorhizobium phage | ↓ | Bacteriophage | [13] | |
| Uncultured crAssphage | ↓ | Bacteriophage | [13] | |
| Halocynthia phage | ↓ | Bacteriophage | [13] |
Note: “↑”, abundance increased; “↓”, abundance decreased.
Evidence has been accumulating that a substantial number of COVID-19 patients experienced systemic and/or organ-specific afflictions during follow-up after disease resolution, including fatigue, muscle weakness, sleep difficulties, anxiety, depression, diarrhea, and poor glycemic controls [21], [22], [23], [24], a phenomenon known as ‘long COVID’. Interestingly, the GI tract is also affected in a long term in COVID-19, as demonstrated by a prolonged shedding of viral RNA in stool specimens up to 42 days and the presence of SARS-CoV-2 virus in the gut epithelium up to 90 days after disease resolution in some patients [25], [26]. Concordantly, long-lasting gut microbiome dysbiosis is also consistently observed in subjects recovered from COVID-19 [12], [15], [27], [28], implying that gut microbiome is closely linked to host health in a post-COVID-19 age.
Relationship between gut microbiome changes, SARS-CoV-2 infection, and host immunity
In a six-month follow-up study on the gut microbiome of patients with COVID-19, significant decreases in the richness (Chao1 index) of gut microbiome were observed across the acute, convalescence, and post-convalescence phases of COVID-19 [27]. In addition, COVID-19 patients had a significantly reduced gut bacterial diversity [20], [29]. Microbial diversity is a critical determinant of microbial ecosystem stability [30]. Stable ecosystems provide colonization resistance to opportunistic pathogens [31]. Therefore, the reduction in gut microbiota diversity and richness may somewhat contribute to the expansion of opportunistic bacteria and have long-term impact in patients with COVID-19 [32]. Concordantly, a critical proportion of patients with COVID-19 also experienced persistent symptoms following disease resolution and hospital discharge, known as ‘long COVID-19’ [21], [24]. Patients with lower post-convalescence bacterial microbiome richness had higher levels of COVID-19 severity (worse pulmonary functions) and blood C-reactive protein (CRP) during the acute phase [27], suggesting a relationship between gut dysbiosis and hyper-inflammatory response in COVID-19. A more recent study also showed that the gut microbiome ecology was stratified well with COVID-19 severity, as demonstrated in the principal component analysis (PCA) visualization that the gut microbiome communities followed a continuum along the mild, moderate, severe, and critical gradients of COVID-19 severity [12]. Moreover, the gut microbiota composition was correlated with plasma concentrations of inflammatory cytokines and blood parameters, such as CRP, lactate dehydrogenase, aspartate aminotransferase, and gamma-glutamyl transferase [12]. These data together suggest that SARS-CoV-2 infection may cause immune-pathophysiological changes in the human host, including the gut, resulting in gradual changes in the gut microbial ecology in relation to illness severity. In favor of this hypothesis, a recent proof-of-principle study in a mouse model of COVID-19 demonstrated that SARS-CoV-2 infection elicited immune/infection-related gene expression in the gut epithelial cells, leading to a change in the gut milieu where the microbiota were affected [33]. Following that, we found that the SARS-CoV-2 activity in the gut might be a prominent factor in shaping the gut microbiome composition [34]. Patients with high SARS-CoV-2 infectivity in the gut displayed a high abundance of the bacterial species Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, and Morganella morganii (Figure 1; Table 1), as well as a high functional capacity for nucleotide de novo biosynthesis, amino acid biosynthesis, and glycolysis [34]. However, patients with low-to-none SARS-CoV-2 infectivity in the gut displayed a high abundance of short-chain fatty acid (SCFA)-producing bacteria, Alistipes onderdonkii, Parabacteroides merdae, Bacteroides stercoris, and Lachnospiraceae bacterium 1_1_57FAA [34]. Among them, Alistipes onderdonkii was a bacterial species, the abundance of which also showed a negative correlation with COVID-19 severity [15]. Interestingly, Alistipes species are indole-positive, involved in the serotonin precursor tryptophan metabolism and in maintaining gut immune homeostasis [35], [36]. This is later validated in animals that tryptophan metabolism in the gut was altered as a result of SARS-CoV-2 infection [28]. In addition, the hyper-inflammatory response of COVID-19 patients was associated with disrupted gut permeability and microbial translocation [16], [37]. The amount of fecal calprotectin, a marker of intestinal inflammation as a consequence of translocation of granulocytes and monocytes/macrophages into the gut lumen, was elevated in the feces of patients with COVID-19 [38], indicating immune dysfunction of the gut and altered gut niche in COVID-19 patients. Taken together, the compositional alterations in the gut microbiome of COVID-19 patients are likely the result of host immune responses and altered gut milieu during SARS-CoV-2 infection.
To validate that SARS-CoV-2 infection is the driving force of the alterations in the gut microbiome ecology, Sokol et al. used a non-human primate model (rhesus macaques and cynomolgus macaques) challenged with SARS-CoV-2 and subsequently analyzed the impact of SARS-CoV-2 infection on dynamic changes of the gut microbiome [28]. Strikingly, the gut microbiome gradually changed from day 0 until day 13 post infection, at which time point the gut microbiome was most different from the baseline microbiome in terms of fecal microbial community structure [28]. This result indicates that SARS-CoV-2 infection virtually induces alterations in the gut microbiome ecology in COVID-19. To delineate the dynamic changes caused by SARS-CoV-2 infection, the authors compared the composition at each time point post infection with time points before infection. Consistent with the findings in humans [15], [20], the abundance of opportunistic bacteria from the Proteobacteria phylum was increased, whilst the abundance of beneficial members from the Firmicutes phylum (especially those from the Ruminococcaceae and Lachnospiraceae families) was decreased after SARS-CoV-2 infection [28]. Although some alterations in the gut microbiome were resolved at later time points, certain perturbations persisted even after disease resolution [28], analogous to the observations in humans [15], [20]. This finding further addresses that SARS-CoV-2 infection may have long-lasting impact on the gut microbiome ecology. Ecological network analysis of the bacterial–bacterial interactions in the gut microbiome of macaques before vs. after SARS-CoV-2 infection revealed a sparse, atrophied bacterial microbiome ecological network after SARS-CoV-2 infection compared to a dense, interconnected network before SARS-CoV-2 infection [28]. The gut microbiome ecological network reflects the complex interplay of microbial communities [39]. In a steady state, the gut microbiome exhibits a dense, intricate microbial ecological network, whereas under gut inflammation conditions, such as inflammatory bowel disease (IBD) and Clostridioides difficile infection (CDI), it manifests a significant sparse one [11], [40], [41]. The significantly weakened ecological microbial network after SARS-CoV-2 infection both in humans and macaques implies that SARS-CoV-2 infection may induce host inflammatory responses resulting in disrupted gut microbiome ecology.
Studies have shown that intestinal microbiota can affect viral replication and systemic pathogenesis [42], [43], [44], [45]. Depletion of the intestinal microbiota in mice by antibiotics rendered the mice less susceptible to poliovirus disease and supported minimal viral replication in the intestines of mice [43]. Exposure of poliovirus to bacteria enhanced host–cell association and infection, since poliovirus binds to lipopolysaccharide [43]. The pathology of reovirus (an unrelated enteric virus) infection was also more severe in the presence of intestinal microbes [43]. In addition, antibiotics prevented persistent murine norovirus infection, which was reversed by replenishment of the bacterial microbiota [45]. In parallel, enteric bacteria were also found to promote human and mouse norovirus infection of B cells [44]. These studies together suggest that gut microbes influence virus infection and that viruses may exploit intestinal microbes for replication and transmission. That being said, the gut microbiome may play a role in SARS-CoV-2 susceptibility and infectivity, which remains to be verified in future studies.
COVID-19 is essentially a lung disease, and it has been established that gut can affect lung through the gut–lung axis [46], [47]. Beyond the local immune regulation by the gut microbiota, the far-reaching immune impact of gut microbiota is also well recognized, especially on the pulmonary immune system [48]. SCFAs, a group of prototypic metabolites produced by gut bacteria, translocate across the intestinal barrier, reach the systemic circulation, and modulate the lung immune response [49], [50]. They are mainly produced by bacterial degradation and fermentation of dietary fibers, acting as signaling molecules in the lungs on resident antigen-presenting cells to attenuate the inflammatory and allergic responses [49], [51], [52]. Decreases in the abundance of SCFA-producing bacteria observed in the gut microbiota of patients with COVID-19 [12], [15], [34] may represent one of the critical mechanisms contributing to the gut–lung crosstalk and thereby disease severity in COVID-19.
Angiotensin-converting enzyme 2 and the gut microbiome
Studies have provided direct evidence that angiotensin-converting enzyme 2 (ACE2) is the binding site of SARS-CoV-2 for host entry [53], [54]. ACE2 is highly expressed in the respiratory tract and the intestines, especially in nasal epithelial cells and colonocytes of humans [55]. ACE2 has also been demonstrated to regulate amino acid transport, expression of antimicrobial peptides, microbial ecology, and inflammation in the gut [56]. These lines of evidence underscore an interplay between ACE2 expression, SARS-CoV-2 infection, and the gut microbiome in the host. Bacterial species from the Bacteroidetes phylum were shown to down-regulate ACE2 expression, while species from the Firmicutes phylum displayed variable effects in modulating ACE2 expression in the murine colon [57]. Interestingly, our study in the gut microbiome of COVID-19 patients showed that the fecal abundance of the Bacteroidetes species, Alistipes onderdonkii and Bacteroides ovatus, was inversely correlated with COVID-19 severity, and the abundance of 4 Bacteroidetes species, Bacteroides dorei, Bacteroides thetaiotaomicron, Bacteroides massiliensis, and Bacteroides ovatus, showed inverse correlation with the fecal viral load of SARS-CoV-2 [15], [34]. Amongst these Bacteroidetes species, Bacteroides dorei was previously shown to inhibit colonic ACE2 expression [57] and to calibrate host immune response [58], [59]. Intriguingly, subjects with pre-existing chronic diseases (such as diabetes mellitus, hypertension, obesity, and coronary artery disease) were characterized by a low abundance of Bacteroides species and had the highest COVID-19 mortality and morbidity [60], [61], [62]. Collectively, these data imply a sophisticated quaternary relationship between SARS-CoV-2, gut microbiome, ACE2 expression, and host immunity, underlying the varying anti-SARS-CoV-2 immune responses and thereby disease severity in the host.
Functionality changes of the gut microbiome in COVID-19
Beyond the compositional changes, the functionality of the gut microbiome is also changed in COVID-19. The fecal microbiome of patients with high SARS-CoV-2 infectivity in the gut was enriched for functional pathways involved in amino acid biosynthesis (L-lysine biosynthesis II and superpathway of L-serine and glycine biogenesis), carbohydrate metabolism (glycolysis II from fructose-6-phosphate), and nucleotide metabolism (superpathway of adenosine nucleotide de novo biosynthesis II, superpathway of adenosine nucleotide de novo biosynthesis I, superpathway of guanosine deoxyribonucleotide de novo biosynthesis II, purine ribonucleoside degradation, adenosine deoxyribonucleotide de novo biosynthesis II, and guanosine nucleotide de novo biosynthesis II) compared to samples with low-to-none SARS-CoV-2 infectivity [34]. The augmented functionality of amino acid and nucleotide biosynthesis and carbohydrate metabolism in the gut microbiome suggests an enhanced production of building blocks for macromolecules and energy extraction in bacterial cells. In the macaque model of COVID-19, a decrease in the concentration of SCFAs and alterations in the concentration of several bile acids and tryptophan metabolites in the feces of infected animals were observed, as revealed by targeted quantitative metabolomics [28]. The decreases in the concentration of SCFAs were in agreement with the microbiome compositional changes in COVID-19 patients, where the abundance of SCFA-producing bacteria was significantly decreased compared with controls [15], [34]. Reductions in SCFA production were also observed in animals during influenza infection, which contributed to further gut microbial dysbiosis and pulmonary pneumococcal superinfection [63]. Moreover, SARS-CoV-2 infection led to an impairment of the fecal bile acid pool, where the primary-to-secondary bile acid ratio (as a function of bile acid transformation by the gut microbiota) was changed and positively correlated with serum levels of chemokines such as C-X-C motif chemokine ligand 13 (CXCL13) [28]. Overall, an increase in the fecal bile acid levels was seen in the macaques infected with SARS-CoV-2, suggesting that the infection leads to accelerated transit and/or impaired bile acid reabsorption in the ileum [28]. Incidentally, the amount of tryptamine, a tryptophan metabolite of microbiota known to accelerate bowel transit [64], [65], was also increased in the feces of SARS-CoV-2-infected macaques [28]. Two end-products (quinolinic and picolinic acids) of tryptophan metabolism by the host indoleamine 2,3-dioxygenase (IDO) pathway was detected in the feces, indicating the presence of intestinal inflammation [64]. Collectively, the functional alterations in the gut microbiome of COVID-19 patients are likely the result of both compositional alterations of the gut microbiome and host immune responses against SARS-CoV-2 infection, which are intertwined with host pathophysiology.
The gut mycobiome in COVID-19
The human gut also harbors a large number of fungi, known as the gut mycobiome. The gut fungi have been demonstrated to be causally implicated in microbiome assembly, ecology, and immune development [66], [67]. Our study in patients with COVID-19 also showed alterations in the gut mycobiome, characterized by enrichment of Candida albicans and highly heterogeneous mycobiome configurations [14]. The abundance of opportunistic fungal pathogens, Candida albicans, Candida auris, and Aspergillus flavus was increased in the feces of COVID-19 patients during the disease course (Figure 1; Table 1) [14]. Fungal pathogens associated with pneumonia and respiratory symptoms, Aspergillus flavus and Aspergillus niger, were detected in the fecal samples from a subset of patients with COVID-19, even after disease resolution [14]. Unstable gut mycobiomes and prolonged dysbiosis persisted in a significant proportion (∼ 30%) of COVID-19 patients [14]. Another study investigated the gut mycobiota in both COVID-19 and H1N1-infected patients and found increased fungal load and enrichment of fungi, including Candida species, in both groups of patients [68]. Presence of Aspergillus niger was positively correlated with diarrhea, while the abundance of Penicillium citrinum was inversely correlated with blood levels of CRP [68]. Aspergillus infections were recently reported in respiratory tract secretions and tracheal aspirates in patients with COVID-19 [69], [70]. Aspergillus is a genus of ubiquitous fungi that cause a variety of pulmonary and respiratory symptoms [71]. Aspergillus may harness the host with immune dysfunction and affect the clinical features and disease course [71]. Cough was found to be more frequent in COVID-19 subjects with Aspergillus infections than those who were not infected [72]. COVID-19 patients who had Aspergillus flavus presence in feces also presented with cough during hospitalization, suggestive of a link of gut mycobiome in the gut–lung axis. These data suggest a gut mycobiome dysbiosis in COVID-19 and its relationship with a systemic dysregulation of host immunity.
Overall, such fungal bloom in the gut of patients with COVID-19 is likely a result of SARS-CoV-2 infection. Secondary fungal infection or co-infection in patients with COVID-19 during the pandemic was frequently observed [73], [74], [75]. Candida and Aspergillus lineages were amongst the specific opportunistic fungal pathogens enriched in patients with COVID-19 during the disease course, particularly Candida albicans [14], [68]. Candida albicans has been shown to impair gut microbiome assembly in both humans and mice, including gut microbiome reassembly after disruption by antibiotics and inflammation [76], [77]. A recent multi-kingdom microbiome study in preterm infants to elucidate the ecological drivers of gut microbiota assembly and dynamics found that between-kingdom interactions have a key role in community dynamics and that the single fungal species, Candida albicans, inhibits multiple dominant genera of gut bacteria (including Klebsiella and Escherichia) [78]. Our prior fecal microbiota transplantation (FMT) study in CDI also demonstrated that presence of Candida albicans in donors or recipients impairs colonization of donor bacteria into recipients, therefore nullifying FMT efficacy in clearing CDI, in both humans and mice [76]. Surprisingly, such inhibitory effect of a single fungus on the restoration of the gut bacterial microbiome can be extended to other fungi, including Aspergillus penicillioides and Penicillium brocae [76]. These studies suggest a crucial role of gut fungi in the gut microbiome ecology, revealing the centrality of simple microbial–microbial interactions in shaping host-associated microbiota. Moreover, gut colonization by Candida albicans can aggravate inflammation in the gut and non-gut tissues [79], [80]. Therefore, the opportunistic expansion of certain fungi in COVID-19 patients potentially has a deleterious role on gut microbiome assembly, where a persistent gut microbiome dysbiosis is consistently seen even after disease resolution and hospital discharge. The long-term effect of gut fungi on the gut microbiome and host health remains to be further investigated.
The gut virome in COVID-19
In addition to the bacteria and fungi, the human gut also harbors an immense diversity of viruses collectively known as the gut virome [81], [82]. Virome consists of both RNA and DNA viruses that chronically infect their eukaryotic (humans, animals, and plants) and prokaryotic hosts (bacteria) [81]. The gut virome serves to modulate the ecology of the co-resident gut bacterial microbiota as well as the immunity of the mammalian host [82].
By shotgun RNA sequencing of the fecal RNA virome, an active presence of SARS-CoV-2 was found in 47% of patients with COVID-19, even in the absence of GI symptoms and after respiratory clearance of SARS-CoV-2 [34]. Meanwhile, COVID-19 patients also had underrepresentation of pepper mild mottle virus (RNA virus), which may originate from diet [13]. By shotgun DNA sequencing of the fecal DNA virome, 19 virus species were identified to be enriched in COVID-19 patients, whereas 26 virus species were enriched in non-COVID-19 controls [13]. Among them, a majority (18 out of 26 virus species) of the DNA viruses enriched in the feces of non-COVID-19 controls were prokaryotic viruses, particularly bacteriophages (16 of 18) (Figure 1; Table 1) [13]. In contrast, more eukaryotic viruses (11 out of 19 virus species) were enriched in the feces of COVID-19 patients [13], which may be a result of SARS-CoV-2 infection. The eukaryotic viruses may harness the immune dysfunction of the host after SARS-CoV-2 infection to expand [81]. The gut virome in COVIID-19 showed more stress-, inflammation-, and virulence-associated gene coding capacities [13]. At patient baseline, the fecal abundance of the RNA virus, pepper chlorotic spot virus, and multiple bacteriophage species was inversely correlated with COVID-19 severity [13]. The abundance of these viruses was also inversely associated with blood levels of pro-inflammatory proteins, white cell count, and neutrophil count [13], indicating that gut resident viruses might tune host immune response to SARS-CoV-2 infection [81], [83]. These data highlight that the gut virome may contribute to immunological and physiological changes in the host during COVID-19. Administration of the antiviral medication lopinavir-ritonavir was associated with the decreased abundance of Listeria phage in COVID-19, suggesting that use of antivirals may tune host bacteriophage–bacteria ecology in the gut, also likely a result of its role in modulating host immune defense against SARS-CoV-2.
Among the COVID-19-enriched viruses, Escherichia phage and Enterobacter phage were prominent [13]. Expansion of these phages has been causally implicated in gut inflammation and host interferon response in mice and humans [41], [84]. In addition, the abundance of their host bacteria was also increased in the gut after SARS-CoV-2 infection [19], [28]. The co-expansion of Escherichia phage and Escherichia was also reported in gut inflammation, and the bloom of Escherichia phage is potentially triggered by lysis of its bacterial host Escherichia under inflammatory conditions [41], [85]. Gut inflammation per se is able to boost bacteriophage transfer between bacteria [86]. Therefore, the alterations in the ecology of gut virome, particularly in the bacteriophage community, are at least partly caused by the alterations of the bacterial microbiome under the influence of SARS-CoV-2 infection and the subsequent immune dysfunction. Similarly, the gut virome dysbiosis persisted along with the dysbiosis of the gut bacterial microbiome, even after disease resolution of COVID-19 [13]. A strong correlation between the composition of virome and bacterial microbiome in COVID-19 patients was observed [33]. Combined ecological network analysis of the virome and bacterial microbiome in COVID-19 revealed that three bacterial species, Faecalibacterium prausnitzii, Bacteroides vulgatus, and Ruminococcus gnavus (the abundance of these bacterial species was also associated with COVID-19 and/or disease severity [12], [15], [19], [28], [34]), and Microviridae bacteriophages constitute central network nodes [33]. These bacterial and viral species may be keystone species that play prominent roles in mediating microbial–microbial interactions in the gut microbial ecology.
Concluding remarks and perspectives
SARS-CoV-2 infection leads to complicated immunologic and pathophysiologic responses in the host. Along with the phenotypic changes in the host, the gut microbiome is broadly altered in COVID-19, including the bacterial microbiome, mycobiome, and virome. Moreover, subsequent blooms of opportunistic bacteria, fungi, and viruses under circumstances of SARS-CoV-2 infection and quiescent/overt gut inflammation in COVID-19 pose further threats to host health and gut microbiome restoration. Such expansions in certain microbial species and decreases in microbiome diversity in conjunction with the impaired host immunity may hinder reassembly of the gut microbiome post COVID-19. Consequently, the altered gut microbiome ecology persists even after disease resolution. Overall, the intricate microbiome ecological network in a steady state is significantly weakened in COVID-19, shifting to one predominated by COVID-19-enriched microbes.
It is well-known that confounding factors such as treatment and diet can significantly affect the gut microbiome composition. However, due to the acute nature of COVID-19, controlling for these confounding factors or including treatment-naïve COVID-19 patients seems infeasible. Therefore, some of the differences between the microbiomes of COVID-19 patients and controls, and of those between disease stages (i.e., mild vs. severe COVID-19 cases), could be attributed to treatment regimens and/or diet. Albeit, we observed consistent microbiome changes across studies, including decreases in the abundance of Eubaterium and SCFA-producing bacteria [12], [15], [19], [33], [34]. In addition, we observed that SARS-CoV-2 infection predominated over medications and diet in affecting the gut virome alterations in patients with COVID-19 [13]. These results together suggest that SARS-CoV-2 infection might be a crucial contributor to the gut microbiome dysbiosis in patients with COVID-19. Although studies have demonstrated that the infection of SARS-CoV-2 would lead to the altered gut microbiome, the causal relationships among the baseline gut microbiome (before infection) that regulates ACE2 expression and host immune status, infectivity/severity of SARS-CoV-2, and altered gut microbiome after infection are complicated. Potential causal loops may also exist, for example, compositions of the gut microbiome (baseline) favor the infection of SARS-CoV-2, and subsequent infection of SARS-CoV-2 induces the change of gut microbial ecology. Little is known about the relative contribution of the baseline status of the gut microbiome to the later-on infection and the dynamics of the altered gut microbiome. Beyond that, it is also paramount to further understand how the gut microbiome regulates host immunity against SARS-CoV-2 infection, therefore disease severity, as well as the long-term impact of COVID-19 on the gut microbiome reassembly in relation to host health after the pandemic.
CRediT author statement
Tao Zuo: Conceptualization, Methodology, Writing - original draft. Xiaojian Wu: Writing - review & editing, Supervision. Weiping Wen: Writing - review & editing, Supervision. Ping Lan: Conceptualization, Supervision, Project administration. All authors have read and approved the final manuscript.
Competing interests
The authors have declared no conflict of interest.
Acknowledgments
TZ and PL are supported by the National Natural Science Foundation of China (Grant Nos. 32100134, 82172323, and 81970452), and a joint seed fund from the Sixth Affiliated Hospital of Sun Yat-sen University and Sun Yat-sen University, China. We thank Mingyue Cheng for his assistance in improving figure visualization.
Handled by Fangqing Zhao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Contributor Information
Tao Zuo, Email: zuot@mail.sysu.edu.cn.
Xiaojian Wu, Email: wuxjian@mail.sysu.edu.cn.
Weiping Wen, Email: wenwp@mail.sysu.edu.cn.
Ping Lan, Email: lanping@mail.sysu.edu.cn.
References
- 1.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z., et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69:1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- 5.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y.i., Li X., Zhu B., Liang H., Fang C., Gong Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson AC, Humbert M, Buggert M. The known unknowns of T cell immunity to COVID-19. Sci Immunol 2020;5:eabe8063. [DOI] [PubMed]
- 9.Ren X, Wen W, Fan X, Hou W, Su B, Cai P, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 2021;184:1895–913.e19. [DOI] [PMC free article] [PubMed]
- 10.Chassaing B., Kumar M., Baker M.T., Singh V., Vijay-Kumar M. Mammalian gut immunity. Biomed J. 2014;37:246–258. doi: 10.4103/2319-4170.130922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo T., Kamm M.A., Colombel J.F., Ng S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2018;15:440–452. doi: 10.1038/s41575-018-0003-z. [DOI] [PubMed] [Google Scholar]
- 12.Yeoh Y.K., Zuo T., Lui G.C.Y., Zhang F., Liu Q., Li A.Y., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo T., Liu Q., Zhang F., Yeoh Y.K., Wan Y., Zhan H., et al. Temporal landscape of human gut RNA and DNA virome in SARS-CoV-2 infection and severity. Microbiome. 2021;9:91. doi: 10.1186/s40168-021-01008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo T., Zhan H., Zhang F., Liu Q., Tso E.Y., Lui G.C., et al. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastroenterology. 2020;159:1302–1310.e5. doi: 10.1053/j.gastro.2020.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front Immunol 2021;12:686240. [DOI] [PMC free article] [PubMed]
- 17.Elsayed S., Zhang K. Human infection caused by Clostridium hathewayi. Emerg Infect Dis. 2004;10:1950–1952. doi: 10.3201/eid1011.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.tamilselvi R., Dakshinamoorthy M., Venkatesh A., Arumugam K. A literature review on dental caries vaccine-a prevention strategy. Indian J Public Health Res Dev. 2019;10:3041–3043. [Google Scholar]
- 19.Tang L., Gu S., Gong Y., Li B., Lu H., Li Q., et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6:1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis 2020;71:2669–78. [DOI] [PMC free article] [PubMed]
- 21.Huang C., Huang L., Wang Y., Li X., Ren L., Gu X., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montefusco L., Ben Nasr M., D’Addio F., Loretelli C., Rossi A., Pastore I., et al. Acute and long-term disruption of glycometabolic control after SARS-CoV-2 infection. Nat Metab. 2021;3:774–785. doi: 10.1038/s42255-021-00407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 24.Weng J., Li Y., Li J., Shen L., Zhu L., Liang Y., et al. Gastrointestinal sequelae 90 days after discharge for COVID-19. Lancet Gastroenterol Hepatol. 2021;6:344–346. doi: 10.1016/S2468-1253(21)00076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 2020;130:5235–44. [DOI] [PMC free article] [PubMed]
- 26.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222–225. doi: 10.1136/gutjnl-2021-324090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokol H., Contreras V., Maisonnasse P., Desmons A., Delache B., Sencio V., et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes. 2021;13:1–19. doi: 10.1080/19490976.2021.1893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Z., Wang H., Cui G., Lu H., Wang L., Luo H., et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut. 2021;70:1253–1265. doi: 10.1136/gutjnl-2020-323826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahti L., Salojärvi J., Salonen A., Scheffer M., de Vos W.M. Tipping elements in the human intestinal ecosystem. Nat Commun. 2014;5:4344. doi: 10.1038/ncomms5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buffie C.G., Pamer E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finlay B.B., Amato K.R., Azad M., Blaser M.J., Bosch T.C.G., Chu H., et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2010217118. e2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J., Wang C., Zhang Y., Lei G., Xu K., Zhao N., et al. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1887722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo T., Liu Q., Zhang F., Lui G.C.Y., Tso E.Y., Yeoh Y.K., et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276–284. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdu E.F., Hayes C.L., O’Mahony S.M. In: The gut-brain axis: dietary, probiotic, and prebiotic interventions on the microbiota. Hyland H., Stanton C., editors. Elsevier Inc.; Academic Press: 2016. Importance of the microbiota in early life and influence on future health; pp. 159–184. [Google Scholar]
- 37.Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe COVID-19 patients. Front Immunol. 2021;12:686240. doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Effenberger M., Grabherr F., Mayr L., Schwaerzler J., Nairz M., Seifert M., et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faust K., Sathirapongsasuti J.F., Izard J., Segata N., Gevers D., Raes J., et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo T., Wong S.H., Lam K., Lui R., Cheung K., Tang W., et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuo T., Lu X.J., Zhang Y., Cheung C.P., Lam S., Zhang F., et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domínguez-Díaz C., García-Orozco A., Riera-Leal A., Padilla-Arellano J.R., Fafutis-Morris M. Microbiota and its role on viral evasion: is it with us or against us? Front Cell Infect Microbiol. 2019;9:256. doi: 10.3389/fcimb.2019.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuss S.K., Best G.T., Etheredge C.A., Pruijssers A.J., Frierson J.M., Hooper L.V., et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones M.K., Watanabe M., Zhu S., Graves C.L., Keyes L.R., Grau K.R., et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldridge M.T., Nice T.J., McCune B.T., Yokoyama C.C., Kambal A., Wheadon M., et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enaud R., Prevel R., Ciarlo E., Beaufils F., Wieërs G., Guery B., et al. The gut–lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. doi: 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 48.Chiu L., Bazin T., Truchetet M.E., Schaeverbeke T., Delhaes L., Pradeu T. Protective microbiota: from localized to long-reaching co-immunity. Front Immunol. 2017;8:1678. doi: 10.3389/fimmu.2017.01678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trompette A., Gollwitzer E.S., Yadava K., Sichelstiel A.K., Sprenger N., Ngom-Bru C., et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 50.Bingula R., Filaire M., Radosevic-Robin N., Bey M., Berthon J.Y., Bernalier-Donadille A., et al. Desired turbulence? Gut–lung axis, immunity, and lung cancer. J Oncol. 2017;2017:1–15. doi: 10.1155/2017/5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anand S., Mande S.S. Diet, microbiota and gut–lung connection. Front Microbiol. 2018;9:2147. doi: 10.3389/fmicb.2018.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cait A., Hughes M.R., Antignano F., Cait J., Dimitriu P.A., Maas K.R., et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018;11:785–795. doi: 10.1038/mi.2017.75. [DOI] [PubMed] [Google Scholar]
- 53.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J., Zhao S., Liu M., Zhao Z., Xu Y., Wang P., et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020 2020.02.05.20020545. [Google Scholar]
- 56.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943.e11. doi: 10.1016/j.cell.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vatanen T., Kostic A.D., d’Hennezel E., Siljander H., Franzosa E.A., Yassour M., et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida N., Emoto T., Yamashita T., Watanabe H., Hayashi T., Tabata T., et al. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. 2018;138:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 60.Cai Q., Chen F., Wang T., Luo F., Liu X., Wu Q., et al. Obesity and COVID-19 severity in a designated hospital in Shenzhen, China. Diabetes Care. 2020;43:1392–1398. doi: 10.2337/dc20-0576. [DOI] [PubMed] [Google Scholar]
- 61.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hill M.A., Mantzoros C., Sowers J.R. Commentary: COVID-19 in patients with diabetes. Metabolism. 2020;107:154217. doi: 10.1016/j.metabol.2020.154217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sencio V., Barthelemy A., Tavares L.P., Machado M.G., Soulard D., Cuinat C., et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30:2934–2947.e6. doi: 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 64.Agus A., Planchais J., Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Tilburg Bernardes E., Pettersen V.K., Gutierrez M.W., Laforest-Lapointe I., Jendzjowsky N.G., Cavin J.B., et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun. 2020;11:2577. doi: 10.1038/s41467-020-16431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santus W., Devlin J.R., Behnsen J., Ottemann K.M. Crossing kingdoms: how the mycobiota and fungal–bacterial interactions impact host health and disease. Infect Immun. 2021;89 doi: 10.1128/IAI.00648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lv L., Gu S., Jiang H., Yan R., Chen Y., Luo R., et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 and associations with immune and gastrointestinal symptoms. Commun Biol. 2021;4:480. doi: 10.1038/s42003-021-02036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kosmidis C., Denning D.W. The clinical spectrum of pulmonary aspergillosis. Thorax. 2015;70:270–277. doi: 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- 72.Baba R., Takaoka H., Kamo T., Arai D., Takahashi H., Masaki K., et al. Clinical interpretations and therapeutic significance of isolating Aspergillus species from respiratory specimens. Am J Respir Crit Care Med. 2020;201:A2117. [Google Scholar]
- 73.Cox M.J., Loman N., Bogaert D., O’Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pemán J., Ruiz-Gaitán A., García-Vidal C., Salavert M., Ramírez P., Puchades F., et al. Fungal co-infection in COVID-19 patients: should we be concerned? Rev Iberoam Micol. 2020;37:41–46. doi: 10.1016/j.riam.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song G., Liang G., Liu W. Fungal co-infections associated with global COVID-19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020;185:599–606. doi: 10.1007/s11046-020-00462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuo T., Wong S.H., Cheung C.P., Lam K., Lui R., Cheung K., et al. Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun. 2018;9:3663. doi: 10.1038/s41467-018-06103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Downward J.R.E., Falkowski N.R., Mason K.L., Muraglia R., Huffnagle G.B. Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci Rep. 2013;3:2191. doi: 10.1038/srep02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao C., Coyte K.Z., Bainter W., Geha R.S., Martin C.R., Rakoff-Nahoum S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021;591:633–638. doi: 10.1038/s41586-021-03241-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sonoyama K., Miki A., Sugita R., Goto H., Nakata M., Yamaguchi N. Gut colonization by Candida albicans aggravates inflammation in the gut and extra-gut tissues in mice. Med Mycol. 2011;49:237–247. doi: 10.3109/13693786.2010.511284. [DOI] [PubMed] [Google Scholar]
- 80.Kumamoto C.A. Inflammation and gastrointestinal Candida colonization. Curr Opin Microbiol. 2011;14:386–391. doi: 10.1016/j.mib.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Virgin H. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neil J.A., Cadwell K. The intestinal virome and immunity. J Immunol. 2018;201:1615–1624. doi: 10.4049/jimmunol.1800631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liang G., Bushman F.D. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021:1–14. doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gogokhia L., Buhrke K., Bell R., Hoffman B., Brown D.G., Hanke-Gogokhia C., et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe. 2019;25:285–299.e8. doi: 10.1016/j.chom.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Norman J.M., Handley S.A., Baldridge M.T., Droit L., Liu C.Y., Keller B.C., et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diard M., Bakkeren E., Cornuault J.K., Moor K., Hausmann A., Sellin M.E., et al. Inflammation boosts bacteriophage transfer between Salmonella spp. Science. 2017;355:1211–1215. doi: 10.1126/science.aaf8451. [DOI] [PubMed] [Google Scholar]