Summary
How lifespan and body weight vary as a function of diet and genetic differences is not well understood. Here we quantify the impact of differences in diet in a genetically diverse family of female mice, split into matched isogenic cohorts fed a low-fat chow (CD, n = 663) or a high-fat diet (HFD, n = 685). We further generate key metabolic data in a parallel cohort sacrificed at four time points. HFD feeding shortens lifespan by 12%— equivalent to a decade in humans. Initial body weight and early weight gains account for longevity differences of ~4–6 days/g. At 500 days, animals on a HFD typically gain 4× as much weight as control, but variation in weight gain does not correlate with lifespan. Classic serum metabolites, often regarded as health biomarkers, are not necessarily strong predictors of longevity. Our data indicate that responses to a high fat diet are substantially modulated by gene-by-environmental interactions, highlighting the importance of genetic variation in making accurate individualized dietary recommendations.
Keywords: Lifespan, recombinant inbred strains, high fat diet, body weight, genetic reference population, gene-by-environment interaction (GXE), genometype, polyphenome
Introduction
Lifespan is among the most heterogeneous and complex of traits. Differences are dependent on innumerable gene-by-environmental (GXE) interactions 1–4. Nutrition, in particular, has a profound influence on health and lifespan 5. Relative to an ad libitum diet, a high fat diet (HFD) generally shortens lifespan and increases morbidities 6, but in one extreme case—a murine progeria model—a HFD increases lifespan ~75% 7. In comparison, caloric restriction, intermittent fasting, or a ketogenic diet generally improve lifespan and health 8–11 These dietary effects are not solely dependent on patterns of caloric intake, but are modulated by dietary macro- and micronutrient composition, the amount of time spent in different metabolic states, age of onset, periodicity of access to food, sex, and of greatest importance to us in this study—differences in genometype (strain) and gene-by-dietary interactions 12,13.
While the effects of differences in dietary composition and caloric restriction on lifespan have been studied extensively, key results remain controversial 14–16. Caloric restriction increases lifespan on average, but there is good evidence that this effect is not universal, and certain individuals and genotypes do not benefit in all environments 12,17–19. Surprisingly, there are no corresponding large-scale studies of the range of effects of a high fat diet on lifespan in any large and diverse population of mice.
Mice share most protein-coding genes with humans 20,21. Their short lifespan enables longitudinal studies in highly controlled environments and under multiple experimental and dietary conditions 22–24. Unfortunately, with the important exception of the NIA Interventional Testing Program 22, most studies using mice fail to incorporate a level of genetic complexity that matches that of human populations 25–27. In fact, effects of diets, drugs, and environmental perturbations are often studied on a single genome—most often that of the inbred C57BL/6 strain. This reductionist single-genome focus compromises generality and translational utility of discoveries 26,28.
To address this problem, we rely on a large family of BXD strains of mice that segregate for over 6 million variants 29–31. Collectively, this family also segregates for an impressive level of variation in phenotypes related to aging, metabolism, and mRNA and protein expression in liver, muscle, brain, and many other tissues and cell types 32–37. Previous studies of about 30 BXD strains have demonstrated two-fold variation in lifespan on a standard diet—from 12–15 months for the shortest-lived strains up to about 30 months for the longest-lived strains 34,38. Heritability of lifespan is in the range of 25–45%, but the effective heritability (h2RI)—a value that accounts for the depth of resampling of genetically identical individuals (n = 8 to 12/strain)—is as high as 80% 1,39. The BXD family is particularly well suited to study gene-by-environmental (GXE) interactions because diverse but balanced cohorts can be treated in parallel with different diets 19,32,40–42. This exchangeability property makes it possible to test causal models 43,44 and to greatly improve the precision of measured effects of diets and other interventions on lifespan and weight gain as a function of genometype. It is also practical in the case of the BXDs to accumulate extensive multiscalar phenomes across multiple environments—what we refer to as a family polyphenome.
In this study, we have measured lifespan, body weight, weight gain, and selected metabolites across balanced cohorts of up to 73 fully sequenced BXD family members and their progenitors—C57BL/6J, DBA/2J, and their F1 progeny D2B6F1. We studied cases and controls on diets that differed greatly in fat content—controls on a standard low fat chow diet (CD, 17% of calories from fat) and cases on a very high fat diet (HFD, 60% calories from fat). To the best of our knowledge this is the largest and most rigorously controlled GXE experiment on the effects of diet on lifespan and weight change.
Here we address a set of questions about the impact of diet on lifespan across highly diverse genometypes of mice. How does a HFD modulate variability in lifespan within and between strains? Put more simply: What is the strength of evidence in favor of GXE effects on lifespan? We ask if youthful adult body weight (~120 days) predicts lifespan. Is the change in body weight in adults in response to a HFD a causal predictor of lifespan? Finally, we ask whether levels of classic serum metabolites or metabolic hormones measured in middle-age or old-age predict variation in lifespan? Our focus is both on overall effects and on strain-specific difference in effect of diet on lifespan and weight gain, rather than on specific genetic modifiers or loci of lifespan.
Results
High fat diet shortens lifespan with wide strain variation
Balanced sets of females from 73 BXD strains and their parents were assigned to either CD (n = 663) or HFD (n = 685) at an average of 120 days of age, weighed every other month, and followed until natural death or extreme morbidity (Figure 1A). The HFD has a strong average effect on lifespan across the family. Mean lifespan decreased significantly (p = 6.6E–21, paired t-test; CD:HFD r = 0.55) from 690 ± 8 SE (±199 SD) days on the control diet to 605 ± 6 SE (±169 SD) days on the HFD. Median longevity decreased 77 days—from 703 to 626 (Figures 1B, box plot inset). The 75% quantile change in longevity decreased from 796 days to 699 days. Assuming linear scaling and that a dietary change was imposed in humans at about 20 years-of-age, this 77- to 97-day difference would compare roughly to a 8–10 year loss of longevity in humans 45.
Figure 1.
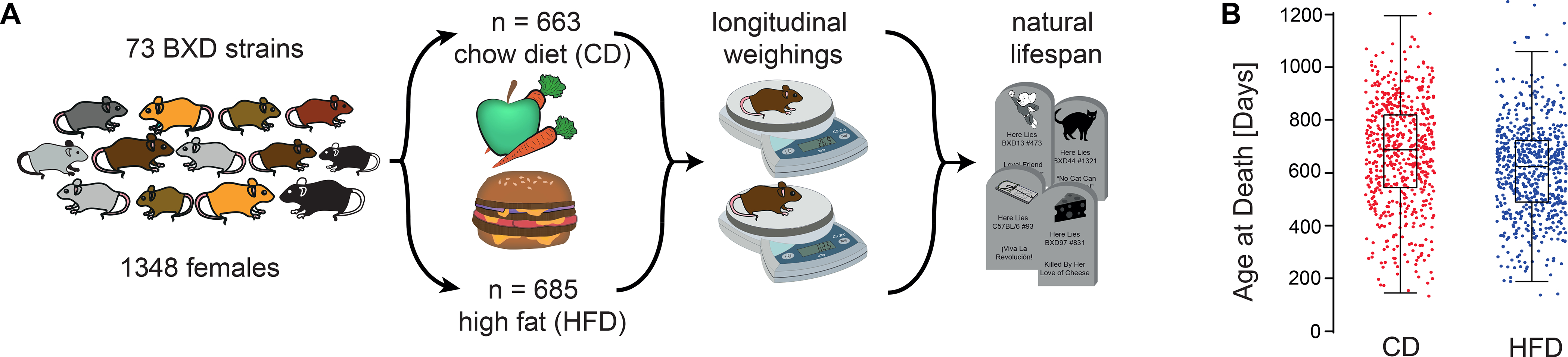
Study overview. (A) Balanced sets of females from 73 BXD strains and their parents were assigned to the low-fat chow diet (CD) or the high fat (HFD) diet and weighed every other month. (B) High fat diet modulates lifespan. When grouped by diet—irrespective of genetic background—median lifespan of CD (n = 663) cohort exceeded by 77 days that of the HFD (n = 685) cohort (see inset for box plot which spans from 25th to 75th percentile, with centerline at median, whiskers extend to maximum and minimum data points which is no more than 1.5 times the length of the box away from the box). Red and blue dots represent individuals on CD and HFD, respectively. For strain averages of lifespan in the two cohorts see Extended figure E1, panels A and B.
Using a mixed-effects Cox model with diet as a fixed effect and strain as random effect, we estimated a hazard ratio of 2.0, indicating that animals on HFD have a two-fold higher age-adjusted risk of death compared to their matched CD-fed controls (Figure 2A). The hazard ratio is relatively constant throughout the study and there is no crossing of the cumulative hazard curves (Figure 2B).
Figure 2.
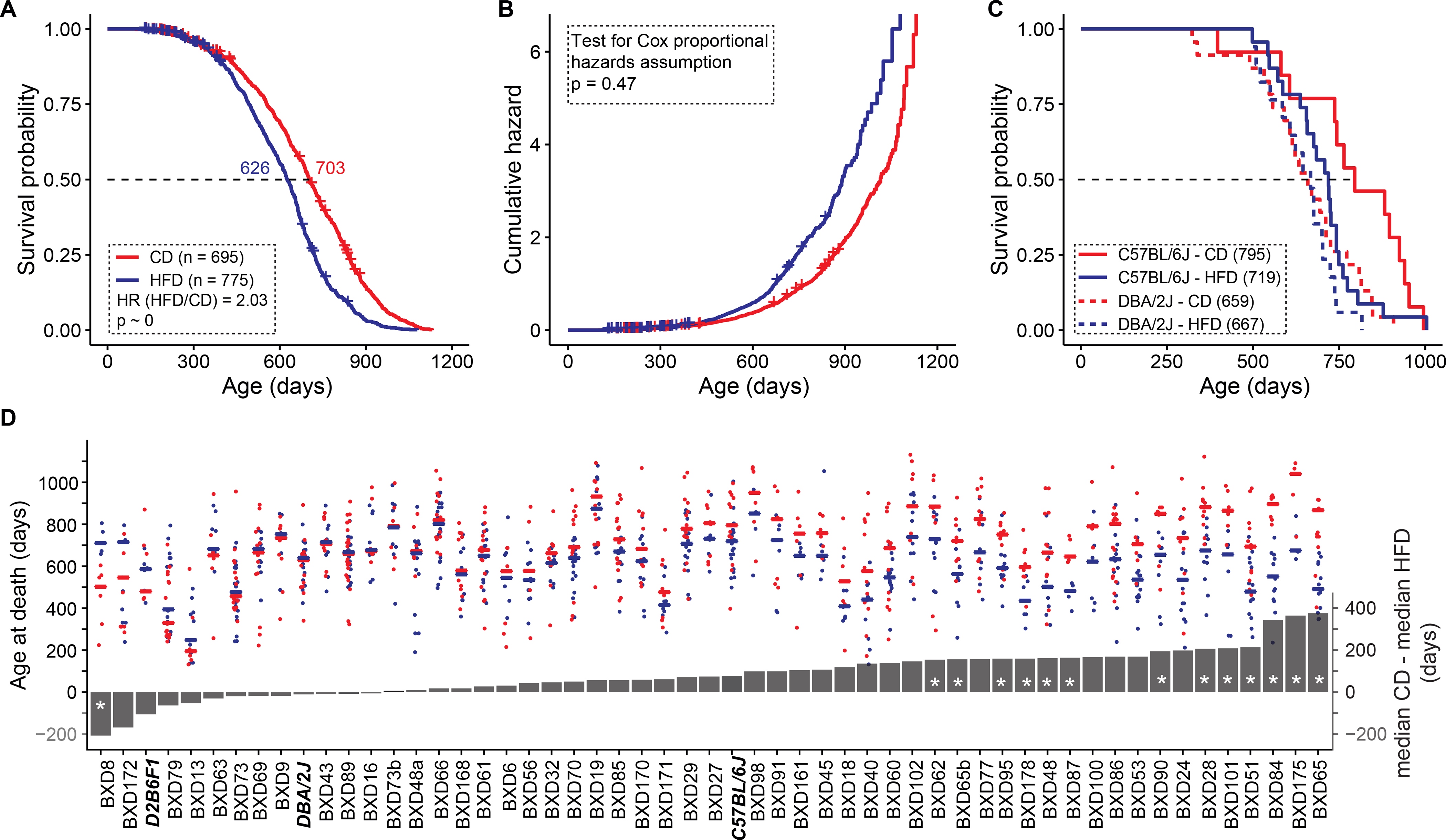
Diet influence on lifespan in female mice (A) Median lifespan decreased by 77 days on HFD. Cases fed HFD have a two-fold higher risk of death compared to those fed CD. (B) Cumulative hazard curves by diet do not cross and the hazards ratio of 2.0 is relatively constant throughout the study. (C) The lifespan of C57BL/6J, but not DBA/2J, is influenced by diet. Numbers in parentheses are median lifespans in days. (D) Lifespan on a high fat diet depends strongly on strain. Red points represent lifespans of cases on CD and blue points those on HFD. Lines represent median survival (left y axis). Grey bars represent the difference in median survival on the diets (right y axis). Negative values to the left indicate higher survival on HFD, positive values indicate higher survival on CD. Parental strains and F1 are denoted by bold italics font. Asterisks in bars denote significant FDR scores at a q value of 0.1. Censored cases in A–B are still alive and are marked by + signs.
While HFD decreased lifespan at the family level, individual strains differed greatly in how they reacted to diet (Extended Figure 1 A–B). The parents exemplified this marked difference—the lifespan of the DBA/2J paternal strain was unaffected by diet, whereas the C57BL/6J maternal strain lost 76 days on the HFD (Figure 2C). In fact, some progeny lived even longer on the HFD, demonstrating that the longevity-diet relation was modulated by a forceful GXE component (Figure 2D). Overall, lifespan in 21 out of 67 strains (those with four or more natural deaths in both cohorts) was significantly affected by the HFD at a nominal p threshold of 0.05. To correct for multiple testing (n = 67), we computed q values (Figure 2D), and with this correction only 15 strains had significantly different lifespans on the two diets. Interestingly, BXD8 had a significantly longer lifespan on the HFD, with a median increase of 208 days (t = 4.0, p = 0.0052, q <0.05, two-tailed). One other strain also trended in the same direction—BXD172 (146 day increase on HFD, p = 0.073), but with a high q value of 0.85. As expected, the HFD reduced longevity for most strains (Fig 2D, right side)—most prominently for BXD65, with a decrease in median longevity of nearly a year (345 days, t = 9.3 p = 9.0E–7, q = 6.0E–7). Of interest, this strain ages most rapidly based on its epigenetic clock on a HFD 46. Adjusting for strain differences, mean lifespan is decreased by 89 ± 14 SE days on the HFD (p<0.0001). Genetic variation explained 30% of the total variance in lifespan whereas diet accounted for 5%, and genetics-by-dietary interaction accounted for 6%.
Modulation of variability in lifespan by high fat diet
We computed coefficients of variation (CV, SD/mean) for all strains in the two cohorts with sample sizes of greater than 6 cases (GN traits BXD_21533 and BXD_21534). The mean CV on the control diet is 0.27 ± 0.015 SE (n = 52) compared to 0.25 ± 0.013 SE on HFD (n = 50)—a small drop in CV on the HFD, but not significant. Using a random effects model, we estimated the lnCV ratio between CD and HFD to be 0.0757 (SE 0.0653); the estimate of between-strain heterogeneity (I2) was 48.2%. The CVs of the inbred strains are slightly higher on average than those of the outbred female mice of about 0.21 (Miller et al., 2007, see GN2 Trait ITP_10001).
Youthful adult body weight is a strong predictor of lifespan
Individuals were weighed at bimonthly intervals throughout their lifespan. As expected, those on HFD typically gained much more weight than those on CD (Figure 3A). Initial weights recorded at point of entry into the aging colony at about 120 days-of-age had a significant influence on lifespan after adjusting for differences in age at start of the HFD (p = 0.0006, r = –0.5) (Figure 3B). A one-gram increase in weight at this stage is associated with a 6-day loss of lifespan. By design there was no appreciable difference between cases assigned to the HFD (23.11 ± 0.22 SE g, n = 685) versus those continuing on the CD for the remainder of their lives (23.26 ± 0.22 g, n = 659). Of interest, the slope of –6 days/g at ~120 days was not affected by subsequent diet. This is highlighted by the parallel red and blue lines in Figure 3B, and indicates that the young adult body size-lifespan relation was relatively insensitive to GXE.
Figure 3.
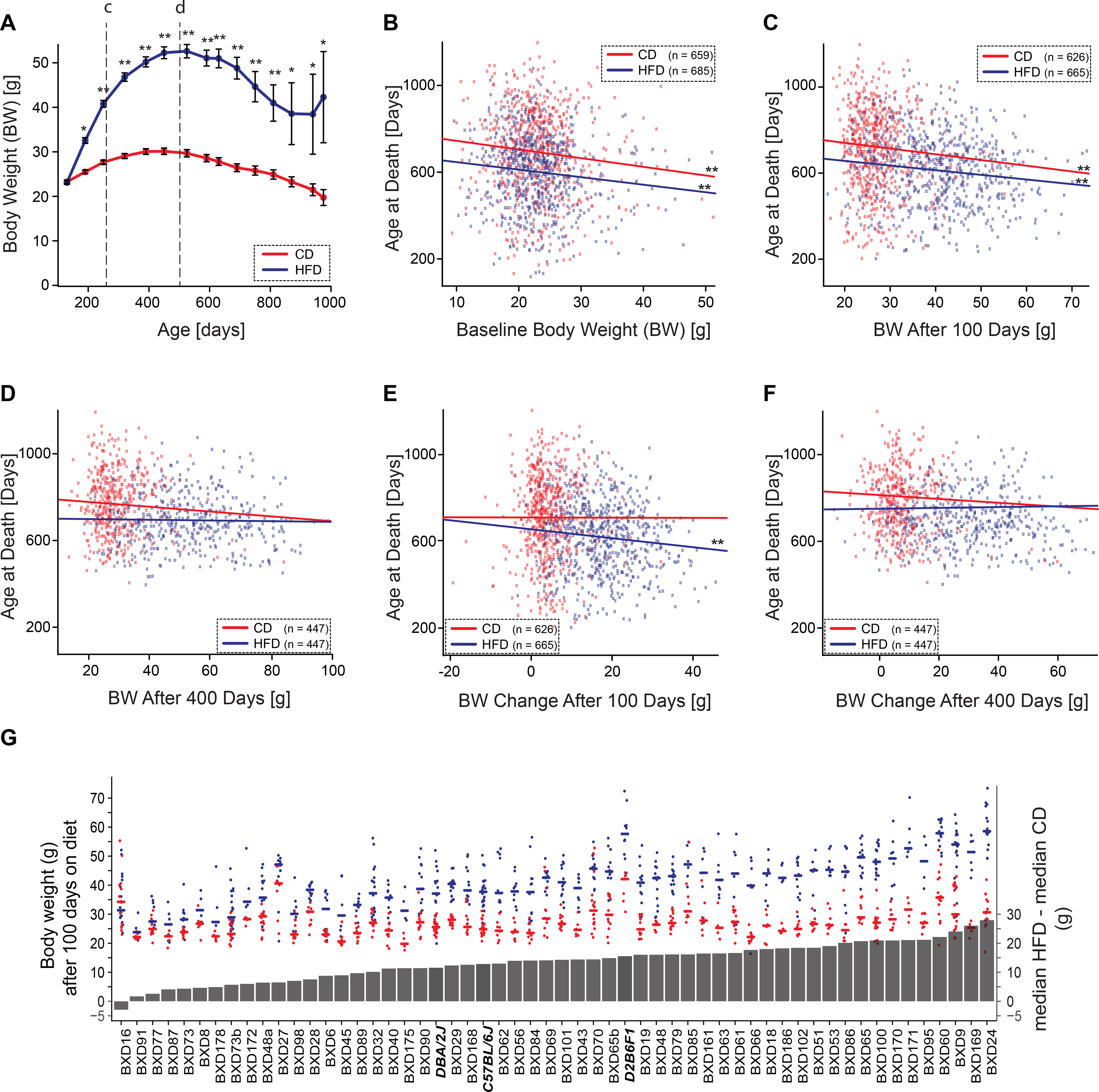
Effect of body weight on lifespan. Effects of body weight on lifespan across all strains were analyzed using fixed-effects linear model in R. Correlations are Pearson’s r. (A) Body weight by diets and age (CD, red line and HFD, blue line). Data are presented as mean ± SEM. Single and double asterisks denote significance at p <0.05 and <0.001. Body weight declines on both diets after about 500 days of age. (B) Initial body weight—the weight taken at entry into colony prior to the point of any animal being shifted to the HFD—has a modest but consistent negative slope with lifespan (–6 days/g, p = 0.0006, r = 0.1) that is not exacerbated by the HFD (n = 659 on CD, 685 on HFD). (C) Body weight after 100 days on both diets (~260 days age) correlates negatively with lifespan (–4 days/g, p <0.001, r = 0.3, see line labeled c in Panel A) (n = 626 on CD, 665 on HFD). (D) Early weight change in response to HFD (blue line)—the difference from baseline after 100 days on diet—was negatively related with lifespan (–4 days/g, p = 0.004, r = 0.1), but this is not true of cases remaining on CD. (E) After 400 days on diet (~500 days age), body weight does not predict variance in lifespan (see line labeled d in Panel A) (p = 0.63, r = 0.01) (n = 447 on CD and HFD). (F) Substantial weight change after prolonged HFD feeding—difference from baseline to 400 days on diet (blue line)—does not predict lifespan (p = 0.26, r = 0.02). (G) Strain-wise changes in median weight after 100 days on diets. Red points represent lifespans of cases on CD and blue points those on HFD. Lines represent median body weight (left y axis). Grey bars represent differences in median body weight on the diets (right y axis). Parental strains and F1 hybrids are denoted by bold italic font. Strain averages of body weight at 500 days of age in the two cohorts are shown in Extended figure E1, panels C and D.
Early body weight gain associated with reduction in lifespan
Body weight measured after 100 days on both diets also correlates negatively with lifespan, after adjusting for strain differences (Figure 3C), a one-gram increase now corresponding to a decrease of 4 days (p<0.0001, r = –0.22). Looking at change in body weight after 100 days on diet, early body weight gain in response to HFD, but not CD, trended to be negatively correlated with lifespan, with a one-gram gain corresponding to a decrease of ~1.5 days (p = 0.08, r = –0.06) (Figure 3D).
Diet significantly alters lifespan, not weight gain per se
We chose to focus on two time points for body weight analyses—100 days on diet as a point to evaluate early weight gain on HFD, and 400 days on diet, a stage that is close to the maximal weight on both diets. The mean weight of the population plateaus around 500 days of age and declines thereafter on both diets. By 500 days of age, cases had been on HFD for 400 ± 44 days and gained an average of 29.5 g. Those on the CD gained only 6.2 g (mean weight on CD = 29.7 ± 0.35 SE g, n = 447; mean weight on HFD = 52.6 ± 0.63 SE g, n = 447).
However, the substantial increase in body weight on the HFD for 400 days is not significantly associated with decrease in lifespan (Figure 3E). Only 10% of the effect of diet on lifespan is mediated through body weight gain after adjusting for strain-specific differences in lifespan. Mirroring this interesting observation, sustained weight gain after prolonged feeding of HFD (Figure 3F) has no predictive value, emphasizing that the diet itself, rather than weight gain per se, modulates lifespan. Adiposity in harvested females on both diets (n CD = 309, n HFD = 292, 47 BXD strains), calculated from adipose tissue weights (subcutaneous, perirenal, and perigonadal fat pads) as a percentage of body weight, was significantly increased upon high fat feeding (mean CD 4.4 ± 0.22, HFD 14.4 ± 0.41, p<2.2E–16 Welch’s t-test). The increased adiposity in HFD fed animals showed a moderate negative association with lifespan (p = 0.03, r = –0.39). Further, we examined the difference between subcutaneous and visceral adiposity ( calculated from perirenal and perigonadal adipose tissue weights), which are known to exhibit different intrinsic properties which make visceral fat a more pathogenic depot with greater impact on metabolic health than subcutaneous fat 47,48. In our HFD cohort, visceral adiposity trended to have a negative impact on lifespan (p = 0.09). Overall, 53 out of 67 strains had significant weight gain on the HFD after 100 days (p <0.05, q <0.1, t values ranging from –3.03 to –13.43) (Figure 3G). At 500 days of age, 45 out of 57 strains gained significant weight on the HFD over 400 days (p <0.05, q <0.1, t values ranging from –3.03 to –13.14) (Extended Figure 1 C–D). Diet had a substantial impact on body weight accounting for 45% of variance in body weight versus 19% explained by genometype, 10% by gene-diet interaction, and 25% as unexplained residual.
Association of lifespan with serum metabolites and hormones
In a separate subsample of animals on both diets harvested at approximately 6, 12, 18, and 24 months-of-age, the HFD elevated circulating levels of glucose, cholesterol, and triglycerides (n = 255 CD, 254 HFD, ~30 strains). Glucose was elevated from 10.34 ± 0.89 on the CD to 11.54 ± 0.22 mmol/L on the HFD (p <4.77E–5 Welch’s t-test, see GN traits BXD_21607 and BXD_21608). Total cholesterol climbed from 1.7 ± 0.46 to 2.4 ± 0.06 mmol/L (p <2.2E–16 Welch’s t-test, traits BXD_21609 and BXD_21610). Triglyceride levels increased more modestly—from 0.47 ± 0.83 to 0.54 ± 0.03 mmol/L (p = 0.04 Welch’s t-test, BXD_24377 and BXD_24378).
In contrast, levels of free fatty acids did not change significantly. The mean on the CD was 0.57 ± 0.51 while that on the HFD was 0.59 ± 0.02 mmol/L (p = 0.53 Welch’s t-test, traits BXD_24379 and BXD_24380, Figure 4 A–D).
Figure 4.
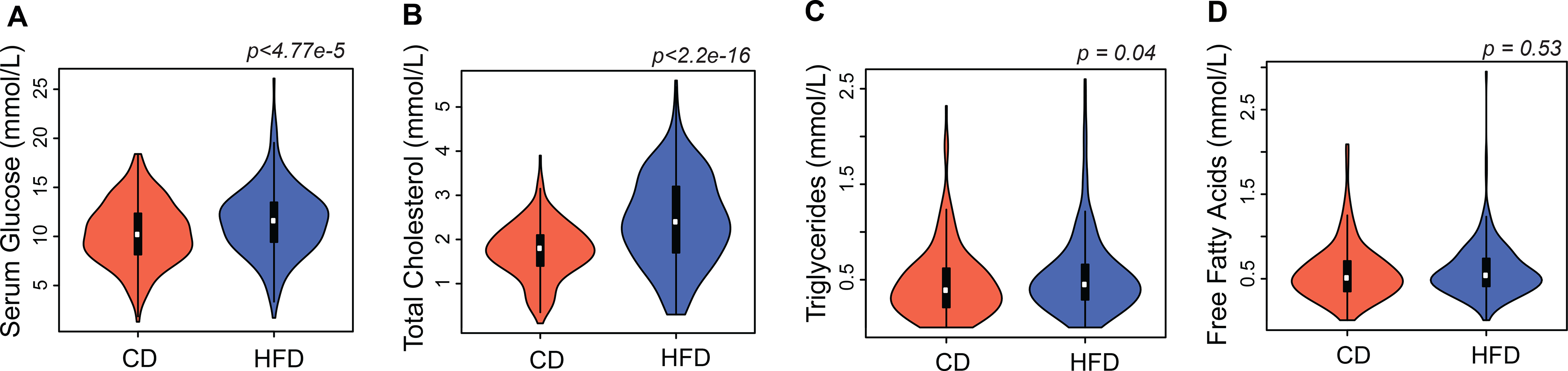
Diet effect on serum metabolites. Violin plots of (A) Glucose (B) Total cholesterol (C) Triglycerides (D) Free fatty acids on CD (n = 255) and HFD (n = 254), across ~30 BXD strains. See inset for box plot which spans from 25th to 75th percentile, with centerline at median, whiskers extend to maximum and minimum data points which is no more than 1.5 times the length of the box away from the box. HFD significantly elevated circulating levels of serum glucose (p <4.77E-5), total cholesterol (p <2.2E-16), triglycerides (p = 0.04) while free fatty acid levels did not change significantly with diet (p = 0.53). Statistical significance determined by two-sided Welch’s t-test.
Glucose (p <0.0001, r = –0.28) and total cholesterol (p = 0.0077, r = –0.17) showed a weak negative association with age at death, on the high fat diet. Almost all strains trended towards a shorter lifespan as serum glucose and cholesterol levels increased. Glucose (p = 0.03, r = 0.29) and total cholesterol (p = 0.07, r = 0.44) showed a moderate positive association with final body weight.
In an expanded set of cases and controls on both diets (n on CD = 312, n on HFD = 302, ~50 BXD strains), harvested at 6, 12, 18, and 24 months, serum insulin levels were significantly higher on a high fat diet with mean values of CD 7.03 ± 0.044 log pg/mL, HFD value of 7.56 ± 0.051 log pg/mL, p = 1.051E-14 Welch’s t-test (GN trait BDL_10055). Homeostatic model assessment-insulin resistance (HOMA-IR), a measure of insulin sensitivity, is also significantly higher in HFD-fed mice compared to CD (2.195 vs 2.839 log units, p <2.2E-16 Welch’s t-test). Among other metabolites analyzed, the HFD significantly elevated the circulating levels of leptin with a mean CD value of 7.52 ± 0.083 log pg/mL, a HFD value of 8.82 ± 0.067 log pg/mL, p <2.2E-16 Welch’s t-test (GN trait BDL_10059), and C-peptide with a mean CD value of 5.94 ± 0.059 log pg/mL, a HFD value of 6.56 ± 0.061 log pg/mL, p = 9.1E-14 Welch’s t-test (GN trait BDL_10056) (Figure 5 A–D).
Figure 5.
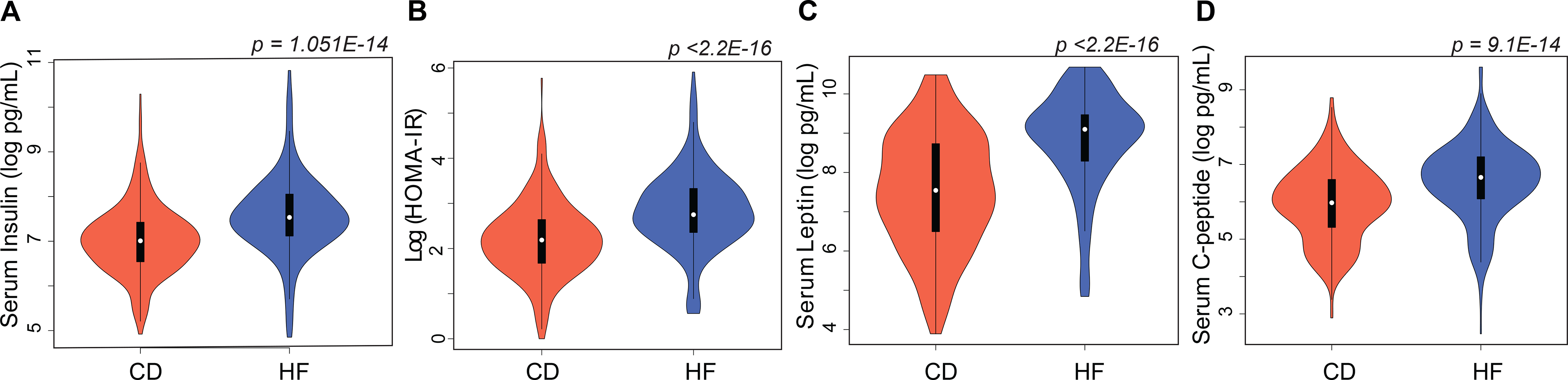
Diet effect on serum metabolic hormones. Violin plots of (A) insulin (B) HOMA-IR (C) leptin (D) C-peptide on CD (n = 312) and HFD (n = 302), across ~50 BXD strains. HOMA-IR was calculated as fasting serum insulin (uIU/mL) multiplied by fasting glucose (mg/dL). See inset for box plot which spans from 25th to 75th percentile, with centerline at median, whiskers extend to maximum and minimum data points which is no more than 1.5 times the length of the box away from the box. HFD significantly elevated circulating levels of serum insulin (p = 1.051E-14), HOMA-IR (p <2.2E-16), leptin (p <2.2E-16), and C-peptide (p = 9.1E-14). Statistical significance determined by two-sided Welch’s t-test.
Insulin (p = 0.0004, r = 0.49), leptin (p = 0.01, r = 0.36) and HOMA-IR (p = 0.03, r = 0.40) showed a moderate positive association with final body weight on the HFD. In contrast, only HOMA-IR (p = 0.07, r = –0.19) displayed a weak negative association with age of death on the HFD.
We learned a consensus Bayesian network model that links serum metabolites to both body weight at 500 days and to lifespan. We tested several versions of these causal models, grouping strains by diet or by the age at which serum metabolites were measured. Here we define two age groups— between 6 and 12 months of age (middle-aged) or between 18 and 24 months of age (old-aged). Our models included up to six metabolites (see Methods) as predictors of lifespan—1. serum glucose, 2. total cholesterol, 3. triglycerides, 4. high-density lipoprotein cholesterol, 5. low-density lipoprotein cholesterol, and 6. free fatty acids. The consensus model highlighted a potential causal effect of diet on peak body weight measured relatively late in life (500 days), acting through circulating levels of total and high-density lipoprotein cholesterol measured in the old-aged group (Extended Figure 2). The Bayesian network analysis, as we structured it, failed to show any causality between serum metabolites and variability in lifespan.
Association between lifespan and metabolic organ weights
We measured weight of certain metabolic organs and tissues of a subsample of cases on both diets at ~500 days of age. HFD mice (n = 63) had 84% greater fat mass, 25% greater heart mass, 19% greater liver mass, and 18% greater kidney mass at ~500 days compared to controls (n = 71). However, HFD did not influence brain mass (Supplemental Table). The correlation between lifespan and organ weights at 18 months is not significant, except for liver weight on the HFD which shows a weak negative association with lifespan (p = 0.03, r = –0.1).
Major morbidities contributing to death in the aging colony
We carried out gross necropsies, with or without histopathology, for a total of 155 individuals. Seventy-six CD cases from 45 strains had a single cause of morbidity; the remaining 7 had multiple likely causes of death. Similarly, 71 HFD cases from 43 strains had a single cause of morbidity; the remaining 8 had multiple causes. Likely cause of death was clear at necropsy or following histopathology in 87% of cases. Hematopoietic neoplasias—lymphomas and histiocytic sarcomas—were leading causes of death, accounting for ~35% on both diets. We also detected miscellaneous non-neoplastic conditions in 24% of CD- and 32% of HFD-fed animals.
Diet appeared to modulate causes of morbidity and mortality (Table 1). In the HFD cohort there was a higher prevalence and severity of cardiovascular disease and lesions, including atrial thrombosis, cardiomyopathy and cardiac dilation sometimes with hepatic steatosis and centrilobular atrophy than in the CD cohort. Nineteen percent of these cases (15 out of 79) had heart pathologies that we rated moderate to severe. In contrast, only a single CD cases had a heart lesion rated at least moderately severe (two-tail Fisher’s exact test p = 0.0003). Four HFD cases, but no CD cases, had systemic polyarteritis. Conversely, some pathologies were evidently more pervasive in the CD cohort. Thirty-six percent displayed non-hematopoietic malignant neoplasia (27 of 76 CD–11 sarcomas, 15 carcinomas, and 1 teratoma) compared to 13% HFD cases with non-hemopoietic malignancy (10 of 79 HFD, 6 sarcomas and 4 carcinomas). While this contrast is nominally significant (Fisher p = 0.0012), we have not corrected for multiple comparisons.
Table 1.
Major morbidities contributing to death in BXD family
| Morbidity Type | Diet | n with single morbidity | n with multiple morbidities | p value |
|---|---|---|---|---|
|
| ||||
| Unnatural causes | CD | 7 | 1 | 0.1257 |
| HFD | 3 | 0 | ||
|
| ||||
| Non-neoplastic conditions | CD | 16 | 2 | 0.3715 |
| HFD | 22 | 2 | ||
|
| ||||
| Heart/ cardiovascular | CD | 0 | 1 | 0.0003** |
| HFD | 8 | 7 | ||
|
| ||||
| Polyarteritis (autoimmune) | CD | 0 | 0 | 0.1204 |
| HFD | 3 | 1 | ||
|
| ||||
| Lymphoma | CD | 25 | 3 | 0.7402 |
| HFD | 26 | 1 | ||
|
| ||||
| Sarcoma | CD | 9 | 2 | 0.2042 |
| HFD | 4 | 2 | ||
|
| ||||
| Carcinoma | CD | 12 | 3 | 0.0065* |
| HFD | 4 | 0 | ||
|
| ||||
| Renal | CD | 0 | 1 | 0.3673 |
| HFD | 1 | 3 | ||
|
| ||||
| Teratoma | CD | 0 | 1 | 0.4903 |
| HFD | 0 | 0 | ||
Significant at nominal alpha of 0.05
Significant with Bonferroni correction for multiple tests
Unnatural causes = flooded cage, foot injury in wire lid, eye abrasion leading to euthanasia, and other.
Miscellaneous non- neoplastic conditions = inflammation, infection, amyloidosis, ulcerative dermatitis
Heart = major contribution of cardiovascular failure other than polyarteritis
Polyarteritis = systemic multifocal arteritis suggestive of autoimmune disease
Lymphoma = hematopoietic neoplasia including lymphoma and histiocytic sarcoma
Sarcoma = nonhistiocytic sarcoma, spindle cell, hemangiosacoma, leiomyosarcoma, sarcoma NOS
Carcinoma = carcinomas (hepatocellular, squamous cell)
Renal = renal failure due to severe nephropathy
Discussion
We have studied effects of a high fat diet on lifespan in a large and genetically diverse family of mice. In the introduction, we pose three sets of questions for which we now have good answers. A diet that is high in fat reduces longevity by an average of 12% in female members of the BXD family, and as in humans, the risk of cardiovascular disease is elevated on the HFD. However, this is not a universal response. Differences among genomes and GXE effects are strong for both lifespan and weight gain. Even after we correct for multiple comparisons, one strain lives significantly longer and another strain gains no weight on the HFD. We confirm that lower weight at an early age is linked to longer lifespan, and that this effect is also true on a HFD. There is at best only a modest association between weight gain after maturity and lifespan. Weight gain measured later in life—between ages of 12–18 months—accounts for minimal variation in lifespan. Diet modulates lifespan and has a stronger effect than weight gain per se. Key serum metabolites, including glucose and total cholesterol exhibit a modest negative association with lifespan. Similar effects are seen with key serum hormones like insulin and leptin. While these specific biochemical assays can be linked causally to body weight, they cannot be linked causally to lifespan. By far the strongest effect on lifespan and weight gain is genometype.
In the interest of avoiding overly broad generalization from this study of GXE effects, we highlight three limitations. First, we only studied females—the main reason being that unlike males, they can be housed without overt and detrimental aggression. Males generally do not live as long as females 49, and in the large NIA Interventional Testing Program the overall difference is 10%: 808 ± 2.88 days for males (n = 7147) and 894 ± 2.43 days for females (n = 6139) (see GeneNetwork Trait ITP_10001). Of even greater practical importance to us, variability of lifespan is much higher in males than females—a coefficient of variation (CV) of 0.30 versus 0.21. As a result, studies of lifespan in females are statistically twice as efficient as those in males—the ratio of the squares of CVs 22,50. At this point we can only presume that the same GXE effects of diet are likely to apply to males 32,41. Second, we have studied only one dietary contrast. Each diet has the potential to reveal novel GXE effects as a function of macro- and micro-nutrients 12. The dietary difference in this study unequivocally causes differences in weight gain and lifespan as a function of genotype (more correctly, genometype), but we are cautious in terms of attributing effects strictly to fat content. Differences in fiber content—150 versus 66 g/kg on CD and HFD, respectively—and other differences in cholesterol and micronutrients are also likely to contribute 51,52. Third, 75 of 76 genometypes of mice we have studied here—all except D2B6F1 hybrids—are fully inbred individuals of the type now used pervasively in biomedical research. One might expect this unusual genetic architecture to have both generally deleterious effects on lifespan, and possibly in higher variability within and between strains than in outbred stock such as used in the NIA ITP program. This last caveat is discussed in more detail below.
A high fat diet decreases lifespan by an average of almost three months across BXD females, roughly scaling to a decade decrease in humans. The high fat diet is associated with an average two-fold higher age-adjusted risk of death compared to the control chow diet. Lifespan under the two diets correlates moderately well (r = 0.55, GN traits BXD_18435 and 18441). However, the strains of mice display wide variation in responses to diet, and despite the strong effect, diet only accounts for 5% of the total variance in lifespan. In comparison, strain as a factor accounts for 30% of variance. Combined across the two diets, lifespan varies from a low of 307 ± 37 days in BXD13 (n = 21) to a high of 852 ± 33 days in BXD168 (n = 23). Some strains are fully resistant to the negative effects of HFD on body weight and lifespan while others are strongly affected. While mean lifespan is shortened by an average of 10% on a HFD, genetic factors account for roughly a two-fold range. At least one strain—BXD8—actually lives significantly longer on the HFD (+37%). Lifespans of other strains, including BXD16 and BXD73, are apparently unaffected by the HFD challenge. Of course, lifespan of most strains is adversely affected (n = 67), and in the case of BXD65 is cut in half. Weight gain is also characterized by a forceful GXE effect—at least four BXD family members are resistant to weight gain, including BXD16, BXD77, BXD87, and BXD91, gaining at most 5% over 100 days of the HFD. There is a mild positive correlation (r = 0.20, n = 63 strains) between weight change after 100 days on the HFD and median lifespan differences (control minus HFD). This again indicates that that weight gain accounts for only 4–5% of the change in lifespan.
Our findings can be compared to strain variation and GXE effects in response to dietary restriction. Dietary restriction without malnutrition is regarded as having an almost universal benefit on longevity 53–55. One exception is a pair of studies on the impact of moderately intense restriction—a 40% reduction in caloric intake—across a large family of LXS strains of mice (n of up 44 strains with 10–20 replicates per strain) 17,19. Their most notable finding was high variation in strain-specific changes in lifespan—shortened in some strains by up to 671 days, but lengthened in others by up to 300 days (GN trait LXS_10164). Both the Liao and Rikke papers were controversial 56, but this puzzle is mirrored to some extent in similar studies of non-human primates 57. Given these contrasts in outcome it would be worthwhile extending the analysis of HFD to dietary restriction and other modifications in the expanded BXD family (now 150 strains, 31). Given what we now know of the precise quantitative effects of one dietary intervention on lifespan, with what level of accuracy can we predict the impact of other interventions?
Variations in lipid metabolism mediates differences in lifespan acting via different dietary, genetic, pharmacological and surgical interventions in model organisms 58. Age-related changes in glucose and fat metabolism have been quantified extensively under tightly-controlled experimental conditions using young (3 months) and old (22 months) C57BL/6J males on the chow diet 51. We have extended this work in the BXD family using high fat-fed females and by evaluating causal links between serum metabolites and metabolic hormones and lifespan and body weight. The HFD cohort had increased adiposity and higher concentrations of serum metabolites including serum glucose, total cholesterol, and triglycerides, with weak negative associations to age at death. All three metabolites are linked to cardiovascular disease 59 and diabetic cardiomyopathy in mice 60,61. The HFD cohort had significantly higher serum insulin levels coupled with increased insulin resistance as compared to the CD cohort, with moderate positive associations to final body weight but a much weaker negative association with age at death. Previous work has shown that mice with reduced circulating insulin levels have improved age-dependent insulin sensitivity and metabolic homeostasis, extending lifespan 62,63. While the role of insulin levels and resistance on health is experimentally well supported and evolutionarily conserved, our results suggest that the linkage to lifespan is complex and surprisingly modest in this family of female mice. Interestingly, there is also evidence that insulin sensitivity and longevity may belong to different causal pathways that are not interconnected64.
We used Bayesian causal modeling to formulate and test networks that link the diet intervention to key serum metabolites and to two outcome measures—body weight at 500 days and lifespan. The best model indicates that circulating levels of HDL and total cholesterol measured at the older ages (18 to 24 months) mediate variation in peak body weight measured at 500 days of age. In contrast, the Bayesian causal analysis does not define any similar mediating effects on lifespan for any combination of the six metabolites. We therefore cannot find any support that these common metabolites predict lifespan alone or in combination. A possible reason could be that the Bayesian networks that we explored did not incorporate strain genometypes.
Recent observational studies using Mendelian randomization emphasize the difficulty in evaluating causal links between lipid metabolites and human cardiometabolic diseases 65. In the cases of low-density lipoprotein cholesterol and triglycerides, the link to coronary heart disease risk was reconfirmed, whereas in the case of high-density lipoprotein cholesterol, a causal link could not be established 66. Even in elderly populations there is no compelling causality linking triglyceride levels to longevity 67. Similarly, there is no causal link between C-reactive protein levels and risk of coronary artery disease despite evidence that inflammation plays a key role in the pathogenesis of coronary heart disease 68.
Major morbidities and likely causes of death among different members of the BXD family are influenced by diet. Females on the HFD have an increased prevalence and severity of cardiovascular disease and lesions. However, the effects of a HFD on cardiovascular disease incidence is modest in mice, unlike that observed in human populations 69,70. Interestingly, the incidence of sarcomas and carcinomas in the BXD females are higher on the CD than HFD. Large prospective studies of humans have failed to detect strong associations between dietary fat and cancer risk 71. The impact of dietary fat on increasing cancer risk depends on tumor type, location and context, making it difficult to glean any conclusive data in the setting of large trials. Combining data from prospective cohort trials with mechanistic studies in mice reveal that certain tumors like prostate cancer respond directly to dietary fat, while breast cancer is affected indirectly by the resulting obesity and related complications such as inflammation and insulin resistance 72. Evaluating a causal role of diet-induced obesity in the etiology of several chronic diseases and cancers has been difficult due to correlations with numerous lifestyle factors and resulting confounding biases. Obesity dramatically modifies the adipose tissue microenvironment which may present a hospitable environment to adjacent developing tumors and facilitate metastatic progression 73. Mendelian randomization provides a practical way to assess causal links between risk factors and diseases 74. Well-controlled animal experiments can similarly provide additional understanding of causes and mediators underlying these complex relations, while also enabling well controlled interventions of specific replicable genotypes.
Chronically high levels of fat consumption lead to substantial weight gain, associated metabolic disorders, and shortened lifespan in humans and mice, but causal interactions among these critical variables remain controversial. In general, a diet high in fat leads to obesity and reduced lifespan in diverse species including Drosophila, C. elegans, and mice 75–77. In our study, higher youthful body weight—between 100 to 200 days-of-age—is associated with a reduction in lifespan of ~4 days per gram. Higher body weight in young adulthood has been associated with accelerated epigenetic aging in a separate subsample of BXDs on both diets 46. This corroborates much previous work that demonstrates that larger body size within a species is generally associated with a shorter lifespan. For example, in outcrossed mice, for each gram increase in weight at 180 days, lifespan is reduced by 10 to 15 days78 (their Table 1). Among breeds of dog, for each kilogram increase in weight, lifespan is reduced by ~15 days 79. In young adult and middle-aged humans, for each kilogram increase in body weight, lifespan is reduced by 80 to 146 days 80 (their figure 6). In these three species, a 5% gain in young adult body weight is associated with a 1–3% loss in lifespan. In both mice and dogs, the relation between early weight gain and longevity is in part a well-known function of growth hormone (GH)–insulin-like growth factor 1 (IGF1) activity, although other mechanisms and gene variants are likely to contribute. In humans the causes of this relation are probably a result of an interplay of many factors, especially nutritional composition, smoking, mean activity levels, and health care systems. For example, tall stature protects against cardiovascular disease due to lower levels of adiposity, lipid fractions, blood pressure, and better lung function 81. The linkage of longevity to IGF1 levels is also more tenuous in humans than in either mice or dogs 82, although short stature and lower GH/IGF1 signaling is associated with resistance to most types of cancers 83,84
In humans an increase in body mass index from 27 to 42 kg/m2 increases all-cause mortality hazard ratio 1.6-fold 85. We see an even greater effect in the BXD family—the HFD diet increases weight 1.8-fold and the mortality hazard ratio 2-fold. But these averages mask impressive modulation by genetic differences. While 45 of 57 family members gain significant weight, another 12 gain only modest, and statistically insignificant weight. Even after 400 days on the HFD, BXD16 is only 1.05-fold heavier than control, whereas BXD24 is 2.1-fold heavier. Remarkably, only 10% of the effect of diet on lifespan is mediated through weight gain. HFD itself exerts a stronger direct effect on lifespan in BXDs than weight gain per se.
Future Directions
Impressive GXE differences among BXD family members emphasize the complexity of interactions among diet, weight gain, and lifespan. These effects need to be teased apart and then also reassembled and explained at genetic, epigenetic, phenome, and even polyphenome levels—work that is now in progress using the same genometypes and cohorts 32,36,41,42,46,86–88. Substantial diversity in outcomes among members the BXD family highlight the fact that population averages can obscure major GXE effects. Not much can be said with certainty from a single genometype or strain of mouse or even from a small number of diverse strains. In our case, detecting strong GXE interactions required data from 10 isogenic individuals from each of 76 genometypes under both conditions. Data of this type can be a foundation for specific predictions and for the design a second wave of experiments using diallel cross progeny and the NIA Interventions Testing Program intended to extend lifespan and vigor 22,23. Furthermore, these data can be used to map genetic modifiers of lifespan; work that is now in progress.
When extrapolated to humans, our results indicate that conventional dietary recommendations will often be too general to provide genuinely individualized guidance for improved health or lifespan. While general dietary recommendations are well intentioned, and even effective at the population level, there is much to be gained by achieving more refined and accurate recommendations that account for the unique genomes and environmental exposures of each individual.
Methods
All experimental procedures were in accordance with the Guidelines for the Care and Use of Laboratory Animals published by the National Institutes of Health and were approved by the UTHSC institutional Animal Care and Use Committee.
Animals
Animals were raised and housed in a specific pathogen-free (SPF) facility at UTHSC (Memphis, TN), at 20–24 °C on a 12-hour light cycle. During the course of this study, serum samples from sentinel mice were tested quarterly for the following 12 pathogens: ectromelia virus, epizootic diarrhea of infant mice (EDIM), lymphocytic choriomeningitis (LCM), Mycoplasma pulmonis, mouse hepatitis virus (MHV), murine norovirus (MNV), mouse parvovirus (MPV), minute virus of mice (MVM), pneumonia virus of mice (PVM), respiratory enteric virus III (REO3), Sendai, and Theiler’s murine encephalomyelitis (TMEV GDVII). We tested twice a year for endoparasites in intestinal contents, and ectoparasites by direct pelt microscopy. All sentinel tests were negative.
From October 2011 through to December 2018, both parental strains, C57BL/6J and DBA/2J, their F1 progeny D2B6F1, and 73 BXD strains were followed from their entry into the aging colony from a large breeding colony —typically around 120 ± 66 days of age but with a wide range, from 26 days to 358 days—until their death (details below on entry age effects). Animals were inspected daily, and deaths were recorded for each animal with a precision of one day. Moribund animals (~10%) were euthanized, and those above the age of 200 days were included in lifespan calculations. Criteria for euthanasia were based on an assessment by our veterinary staff following AAALAC guidelines. All animals were initially raised by dams on the standard chow diet in the breeding colony. Upon entry into the aging colony, females were aged in groups of up to 10 individuals in polypropylene cages (935 cm2) provisioned with Envigo Teklad 7087 soft cob bedding. We provided all cages with enrichment and nesting materials: Bed-r’Nest (www.andersonslabbedding.com/irradiated/bed-rnest), Bio-Huts (www.bio-serv.com/product/Bio_Huts), and torn autoclaved paper towels.
While the aging colony at UTHSC is still in operation, for this analysis we only consider individuals with deaths between April 2012 and November 2018. The colony was moved to a new vivarium in the Translational Science Research Building in April 2016 from the Nash Annex Building, both at UTHSC. Approximately 60% of the individuals lived and died in the original vivarium, ~35% were born in the Nash Annex but lived in both vivaria, and ~5% were born and spent their entire lives in the new facility. We evaluated birth and death data over all seasons for both vivaria and have ruled out any site-specific or seasonal effect on lifespan.
Diet
Controls were provisioned with a single standard low fat chow diet (CD, Envigo Teklad LM-485 7912, 17% calories from fat (fatty acids comprise of 0.8% saturated, 1.3% monounsaturated, and 2.9% polyunsaturated fats), 25% from protein, 58% from carbohydrates, caloric content of 3.1 kcal/g). Cases were provisioned with a high fat diet (HFD, Envigo Teklad TD06414, 60.3% calories from fat (fatty acid profile comprises of 37% saturated, 47% monounsaturated, 16% polyunsaturated fats), 18.4% from protein, 21.3% from carbohydrates, caloric content of 5.1 kcal/g). All individuals had ad libitum access to food and aquifer-sourced autoclaved municipal tap water. We used a conventional CD to align our new data with over 30 years of previous data on conventional diets, and to conform to recent metabolic genetic studies by Auwerx and colleagues using the same two diets 41,42,86. The use of a conventional diet does raise an important question regarding specific macro- and micronutrients likely to be responsible for effects on lifespan or weight. Almeida-Suhett and colleagues demonstrated that conventional non-formulated CD (18% calories from fat) and LFD (10% calories from fat) have similar effects on weight gain, glucose tolerance, and behavioral outcomes. Only cholesterol levels differ significantly89.
Lifespan cohort
We studied a total of 1348 female individuals (n = 663 on CD, n = 685 on HFD) from 76 strains (Supplemental Table; Lifespan cohort). Animals were labeled using ear tags and were randomly assigned to a diet. Baseline weight was measured at age of entry into the study. Seventy-seven percent of animals (n = 527) started on HFD at ages between 50–185 days, but some started on the diet as early as 26 days or as late as 358 days. Only 12 cases were placed on HFD at an age greater than 365 days, and these have been excluded from the Results. There was no appreciable correlation between age at which cases were started on the HFD and lifespan (p = 0.22, r = 0.008, n = 685), and this is also true when the analysis was restricted only to those cases that started on the HFD between 26 and 150 days-of-age (p = 0.60, r = 0.0006, n = 487). Fewer than 20% of animals were retired breeders that entered the study at more than 180 days-of-age and as shown in Results, this variable also does not covary with lifespan or weight gain. Individuals were weighed to the nearest 0.1 gram every other month until their death (Supplemental Table; Body weight data).
Biochemical and body composition cohort
A separate subpopulation of 662 animals (n = 334 on CD, n = 328 on HFD) from ~50 matching strains were sacrificed for tissue collection and biochemical evaluation at time-points centered on 6, 12, 18 and 24 months (Supplemental Table; Biochemical cohort). We generated data on selected 17 serum metabolites (Supplemental Table; Serum metabolites e.g., glucose, total cholesterol, triglycerides, free fatty acids) and major metabolic organ weights (liver, heart, kidneys). Cases and controls were removed from the aging colony and fasted overnight prior to sacrifice. Sacrifices started at approximately 9 am using the anesthetic Avertin (0.2 mL per 10 g of body weight), followed by complete blood draw from the vena cava (~1 mL), and then by perfusion with ice-cold phosphate buffered saline. Blood was collected in lithium-heparin tubes, shaken, and stored on ice. Samples were centrifuged at 4500 rpm for 10 min at 4 °C before being flash-frozen in liquid nitrogen. Blood serum was stored at –80 °C. Serum metabolite measurements and analysis was performed as described in Williams and colleagues 41.
An additional 11 metabolites (Supplemental Table; Serum hormones. eg., insulin, c-peptide, leptin) were analyzed using the Milliplex MAP Mouse Metabolic Hormone Extended Panel (Cat # MMHE-44K; Millipore, MA) according to manufacturer’s instructions. Samples were analyzed on a MagPix System (Luminex Corp. TX). Data analysis was performed using the xPONENT software. A curve fit was applied to the standards and the sample concentrations extrapolated from the standard curve using five-parameter logistic method. HOMA-IR was calculated as fasting serum insulin (uIU/mL) multiplied by fasting glucose (mg/dL).
Adiposity was calculated using summed weights of subcutaneous, perirenal, and perigonadal fat pads as a percentage of body weight.
Bayesian network analysis
We evaluated causal models linking diet through the subset of metabolites that we measured to the key outcome measures—lifespan and body weight at 500 days of age. We used Bayesian network modeling software (BNW, compbio.uthsc.edu/BNW) 90,91. The 1000 highest scoring networks were averaged, and those directed edges—unidirectional connections between phenotype nodes—that had posterior probabilities greater than 0.5 were retained in final consensus models. For computational efficiency we constrained nodes to have at most four parents. We assigned nodes into one of three tiers: 1. diet in the primary causal intervention tier; 2. serum metabolites in the mediator tier, and 3. the outcome measures, lifespan and body weight at 500 days of age (close to maximum body weight for cases and controls). During model learning, we constrained edges to be between diet and the mediator tier, and between the mediator tier and outcomes.
Pathology
Most individuals were fixed by immersion in 10% neutral-buffered formalin within 24 hours of death. The body cavity was opened to improve preservation. Evenly balanced but randomly selected cases and controls (45 strains and 76 CD controls; 43 strains and 71 HFD cases) were selected based on fixation quality for necropsy with histopathology of tissues. A board-certified veterinary pathologist (RWR) performed necropsies and judged probable cause of death and other morbidities.
Data availabiliity
All data conform fully to FAIR standards (findable, accessible, interoperable and reusable;92) and Supplementary table S1 provides Research Resource identifiers (RRIDs, www.rrids.org) for all strains.
Mean, median, and 75% quantile lifespan data from cases and controls are available at GeneNetwork (www.genenetwork.org, GN) under the headings Species: Mouse; Group: BXD Family; Type: Traits and Cofactors, and Dataset: BXD Published Phenotypes (e.g., GN traits BXD_18435, 18441, 19451, 19452, 21302, 21450). Body weight data at 6, 12, 18 and 24 months is also available in GN (e.g., traits BXD_19126, 19130, 19131, 19167, 19168, 19169, 19170, and 19171). For example, the following uniform resource locator (URL) with query string parameters will retrieve mean lifespan data for HFD cases: www.genenetwork.org/show_trait?trait_id=18435&dataset=BXDPublish where the number in bold can be replaced with other ID numbers to obtain and download any data from this work.
Organ weight data for a large subset of cases and controls that were sacrificed between 6 and 24 months-of-age (liver, heart, kidneys, and brain) are available but these data are only covered briefly here (e.g. GN traits BXD_20156, 20157, 20158, 20159, 20353, 20354, 20148, 20149, 20150, 20151, 20146, 20147).
Individual data are also available for all cases, both in the Supplementary table (the precise data used in all analyses here) and in GN under the headings Species: Mouse; Group: BXD NIA Longevity Study; Type: Traits and Cofactors; and Dataset: BXD-NIA-Longevity Phenotypes. For example, individual data for lifespan, irrespective of diet, is accessible here: www.genenetwork.org/show_trait?trait_id=10002&dataset=BXD-HarvestedPublish.
Note that lifespan data sets in GeneNetwork are part of a long-term, and still active genetic study of lifespan in the BXD family, and some datasets will therefore include additional strains as well as outliers excluded from the fixed Supplementary table.
Code availability
Source code and raw data used for the fixed-effects linear model, random effects meta-analysis model, and survival analysis in R are available at https://github.com/genenetwork/bxd_gxelongevity_2020. We have generated a Jupyter notebook as well, detailing our source code with computational and statistical output.
Statistics
While this study has most of the hallmarks of a classic prospective and interventional randomized controlled trial, in some respects our design is also similar to an observational prospective study. The main complicating factor is that to jump-start this lifespan study in 2013 we accepted females, including retired breeders, into the HFD limb over a broad age range (see Results for details). We used statistical methods common to observational analyses to adjust for variability in age-of-entry and to test and eliminate this variable as a confounder. On a positive note, heterogenization of this type can improve robustness of key findings 93.
Lifespan and body weight data were stratified by diet and by strain. Effects of diet, and body weight on lifespan across all strains were analyzed using fixed-effects linear model in R (version 4.0.0). Strain-level (genetic) effects were analyzed by a random-effects model in R using the metafor package 2.4–0 94. Hazard ratios were calculated using a mixed-effects Cox proportional hazard model using Therneau’s coxme R package 2.2.−10 (cran.r-project.org/web/packages/coxme/index.html) 95. Survival analyses were performed using the survival package for R and the data were right-censored (e.g. Fig. 2A–C, censored cases CD, n = 32; HFD, n = 80). Survival curves were computed by ANOVA and regression analyses were performed using R. The variance explained by strain, diet, GXE, and unexplained variance (non-diet, non-genetic) was calculated by fixed-effects two-way ANOVA. Two-sided Welch’s t-test was used to determine statistical significance in the effects of diet on serum metabolites and metabolic hormones. Correlations are Pearson’s r. All graphs were generated in R, and final figures were all prepared with Adobe Illustrator.
Extended Data
Extended Data Fig. 1. Diet effect on lifespan and body weight at 500 days of age.
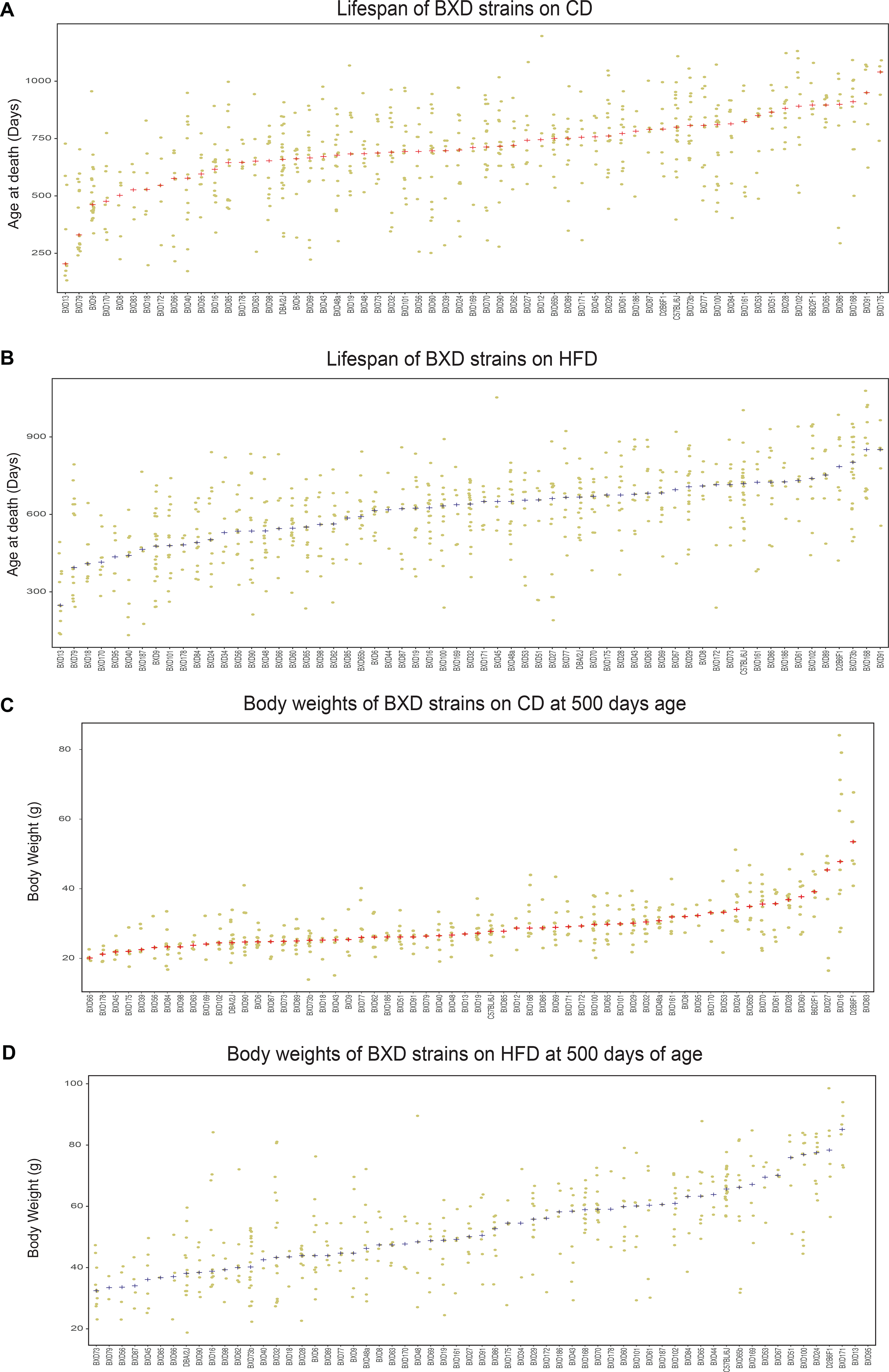
Related to Figure 1 and Figure 3. Diet effect on lifespan and body weight at 500 days of age (A) Data points represent lifespan of animals on low fat chow diet (CD) in BXDs with n ≥ 4 per strain. Red + denotes the strain median. (B) Data points represent lifespan on the high fat diet (HFD) in BXDs with n ≥ 4 per strain. Blue + denotes the strain median. (C) Data points represent body weight on CD at 500 days of age in BXDs with n ≥ 4 per strain. Red + denotes the strain median. (D) Data points represent body weight on HFD at 500 days of age in BXDs with n ≥ 4 per strain. Blue + denotes the strain median.
Extended Data Fig. 2. A Bayesian Network model of the impact of diet on serum metabolites and lifespan and peak body weight at 500 days age.
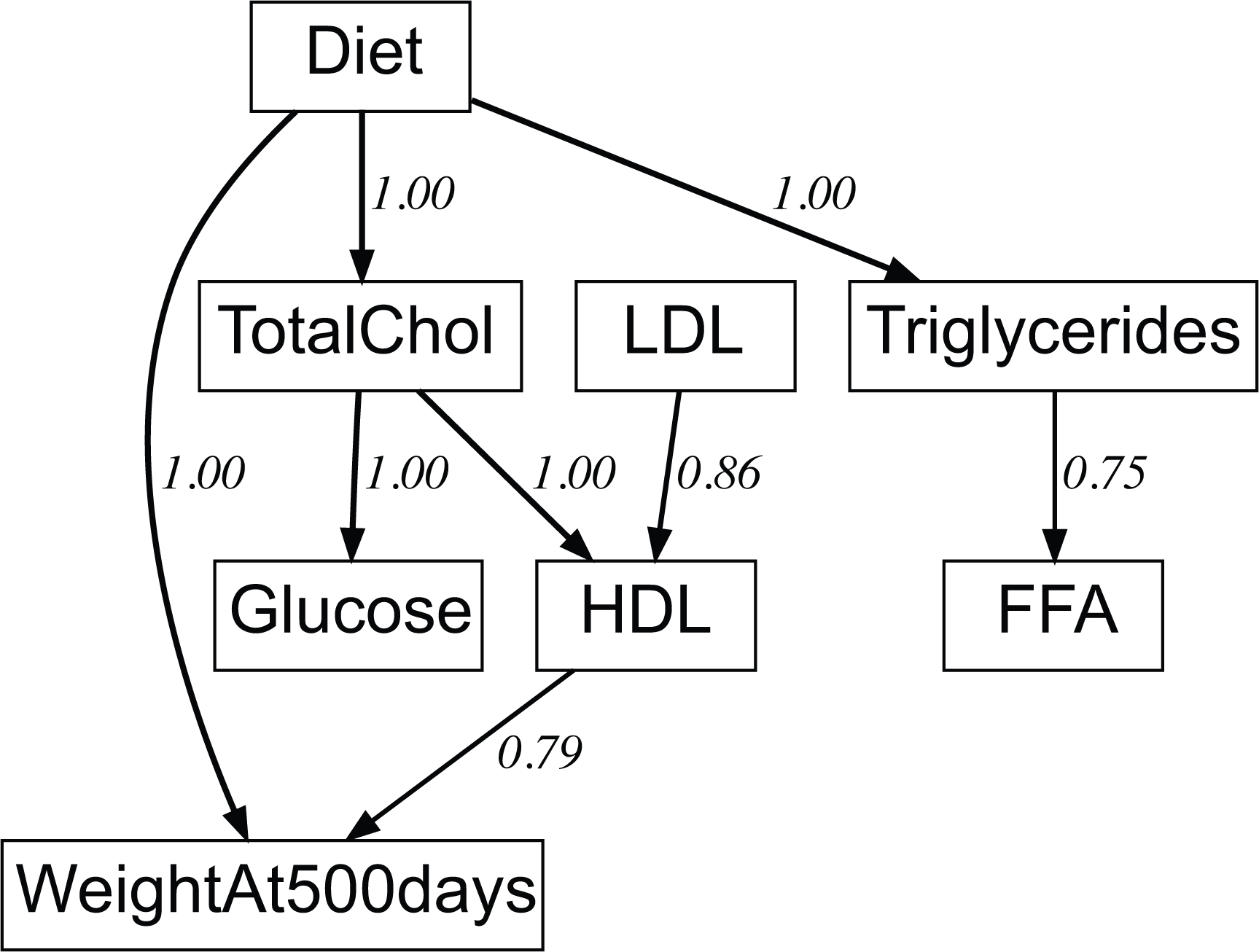
A Bayesian network model of the impact of diet on serum metabolites and lifespan and peak body weight at 500 days age. Edge weights in this network are the weighted fraction of best 1000 Bayesian models that have the same edge and polarity. A value of 1.0 means every one of 1000 “top-ranked models” has this edge. The upper 6 nodes are serum metabolites, and lifespan and body weight at 500 days age are the final outcomes.
Supplementary Material
Acknowledgements:
We thank Dr. James F. Nelson for helpful discussion on the LXS dietary restriction datasets. We thank Dr. Pjotr Prins, Zachary Sloan, and other members of the GeneNetwork team for superb informatics support. Finally, we thank Dr. Elizabeth A Fitzpatrick and team at the Regional Biocontainment Laboratory at UTHSC for generating serum hormone data.
Funding:
This work was supported by grants from the NIH R01AG043930 (RWW), NIH R01AG070913 (RWW), the University of Tennessee Center for Integrative and Translational Genomics (LL), the Ecole Polytechnique Fédérale de Lausanne, the European Research Council (AdG-787702) (JA), the Swiss National Science Foundation (310030B-160318) (JA), and the AgingX program of the Swiss Initiative for Systems Biology (RTD 2013/153) (JA). SS was supported by NIH P30 DA044223-04. EGW was supported by NIH F32 Ruth Kirchstein Fellowship (F32GM119190). KM was supported by NIH R21 AG055841. RWR was supported by TriMetis Life Sciences, Memphis TN. LM was supported by the American Heart Association and Methodist Mission Support Fund. CK was supported by NIH R01AG054180.
Footnotes
Competing interests: The authors declare no competing interests related to this work.
References
- 1.Hook M et al. Genetic cartography of longevity in humans and mice: Current landscape and horizons. Biochim. Biophys. Acta 1864, 2718–2732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuningas M et al. Genes encoding longevity: from model organisms to humans. Aging Cell 7, 270–280 (2008). [DOI] [PubMed] [Google Scholar]
- 3.de Magalhães JP, Wuttke D, Wood SH, Plank M & Vora C Genome-environment interactions that modulate aging: Powerful targets for drug discovery. Pharmacol. Rev. 64, 88–101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDaid AF et al. Bayesian association scan reveals loci associated with human lifespan and linked biomarkers. Nat. Commun. 8, 15842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana L & Partridge L Promoting health and longevity through diet: From model organisms to humans. Cell 161, 106–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller AP, de Oliveira Dietrich M, Martimbianco de Assis A, Souza DO & Portela LV High saturated fat and low carbohydrate diet decreases lifespan independent of body weight in mice. Longev. Heal. 2, 10 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreienkamp R et al. Doubled lifespan and patient-like pathologies in progeria mice fed high-fat diet. Aging Cell 18, e12852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heilbronn LK & Ravussin E Calorie restriction and aging: review of the literature and implications for studies in humans. Am. J. Clin. Nutr. 78, 361–369 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Liang Y et al. Calorie restriction is the most reasonable anti-ageing intervention: a meta-analysis of survival curves. Sci. Rep. 8, 5779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts MN et al. A ketogenic diet extends longevity and healthspan in adult mice. Cell Metab. 26, 539–546.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speakman JR, Mitchell SE & Mazidi M Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp. Gerontol. 86, 28–38 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Barrington WT et al. Improving metabolic health through precision dietetics in mice. Genetics 208, 399–417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan KL et al. Caloric restriction study design limitations in rodent and nonhuman primate studies. J. Gerontol. Ser. A 73, 48–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel T & Holbrook NJ Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Keipert S, Voigt A & Klaus S Dietary effects on body composition, glucose metabolism, and longevity are modulated by skeletal muscle mitochondrial uncoupling in mice. Aging Cell 10, 122–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skorupa DA, Dervisefendic A, Zwiener J & Pletcher SD Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7, 478–490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao C-Y, Rikke BA, Johnson TE, Diaz V & Nelson JF Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell SJ et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rikke BA, Liao C-Y, McQueen MB, Nelson JF & Johnson TE Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp. Gerontol. 45, 691–701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azzu V & Valencak TG Energy metabolism and ageing in the mouse: A mini-review. Gerontology 63, 327–336 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Pennacchio LA & Rubin EM Comparative genomic tools and databases: providing insights into the human genome. J. Clin. Invest. 111, 1099–1106 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller RA et al. An Aging Interventions Testing Program: study design and interim report. Aging Cell 6, 565–575 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Strong R et al. Evaluation of resveratrol, green tea extract, curcumin, oxaloacetic acid, and medium-chain triglyceride oil on life span of genetically heterogeneous mice. J. Gerontol. A. Biol. Sci. Med. Sci. 68, 6–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan R, Peters LL & Paigen B Mice as a mammalian model for research on the genetics of aging. ILAR J. Natl. Res. Counc. Inst. Lab. Anim. Resour. 52, 4–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saul MC, Philip VM, Reinholdt LG & Chesler EJ High-diversity mouse populations for complex traits. Trends Genet. 35, 501–514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RW Animal models in biomedical research: ethics, challenges and opportunities. Princ. Mol. Med 2nd edition, 53–60 (2006). [Google Scholar]
- 27.Williams RW & Williams EG Resources for Systems Genetics. in Systems Genetics (eds. Schughart K & Williams RW) vol. 1488 3–29 (Springer; New York, 2017). [DOI] [PubMed] [Google Scholar]
- 28.Williams RW Herding cats: the sociology of data integration. Front. Neurosci. 3, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peirce JL, Lu L, Gu J, Silver LM & Williams RW A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5, 7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X et al. Joint mouse–human phenome-wide association to test gene function and disease risk. Nat. Commun 7, 10464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashbrook DG et al. A platform for experimental precision medicine: The extended BXD mouse family. Cell Syst. 12, 235–247.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreux PA et al. Systems genetics of metabolism: The use of the BXD murine reference panel for multiscalar integration of traits. Cell 150, 1287–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Haan G & Van Zant G Genetic analysis of hemopoietic cell cycling in mice suggests its involvement in organismal life span. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 13, 707–713 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Gelman R, Watson A, Bronson R & Yunis E Murine chromosomal regions correlated with longevity. Genetics 118, 693–704 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houtkooper RH et al. The metabolic footprint of aging in mice. Sci. Rep 1, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houtkooper RH et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams EG et al. An Evolutionarily conserved role for the aryl hydrocarbon receptor in the regulation of movement. PLOS Genet. 10, e1004673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang DH et al. Quantitative trait loci (QTL) analysis of longevity in C57BL/6J by DBA/2J (BXD) recombinant inbred mice. Aging Clin. Exp. Res. 22, 8–19 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Belknap JK Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav. Genet. 28, 29–38 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Hall RA et al. Systems genetics of liver fibrosis: Identification of fibrogenic and expression quantitative trait loci in the BXD murine reference population. PLoS ONE 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams EG et al. Systems proteomics of liver mitochondria function. Science 352, aad0189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158, 1415–1430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenland S & Robins JM Identifiability, exchangeability and confounding revisited. Epidemiol. Perspect. Innov. EPI 6, 4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearl J Causal inference in statistics: An overview. Stat. Surv. 3, 96–146 (2009). [Google Scholar]
- 45.Flurkey K, Currer JM & Harrison DE The Mouse in Biomedical Research - 2nd Edition. vol. 3 (2007). [Google Scholar]
- 46.Sandoval-Sierra JV et al. Body weight and high-fat diet are associated with epigenetic aging in female members of the BXD murine family. Aging Cell n/a, e13207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuster José J, Ouchi Noriyuki, Gokce Noyan, & Walsh Kenneth. Obesity-induced changes in adipose tissue microenvironment and their impact on cardiovascular disease. Circ. Res 118, 1786–1807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchkonia T et al. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 17, 644–656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Austad SN & Fischer KE Sex differences in lifespan. Cell Metab. 23, 1022–1033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng CJ, Gelfond JAL, Strong R & Nelson JF Genetically heterogeneous mice exhibit a female survival advantage that is age- and site-specific: Results from a large multi-site study. Aging Cell 18, e12905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houtkooper RH et al. The metabolic footprint of aging in mice. Sci. Rep. 1, 134 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrison KE, Jašarević E, Howard CD & Bale TL It’s the fiber, not the fat: significant effects of dietary challenge on the gut microbiome. Microbiome 8, 15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mair W & Dillin A Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Masoro EJ Caloric restriction-induced life extension of rats and mice: a critique of proposed mechanisms. Biochim. Biophys. Acta 1790, 1040–1048 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Weindruch R, Walford RL, Fligiel S & Guthrie D The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J. Nutr. 116, 641–654 (1986). [DOI] [PubMed] [Google Scholar]
- 56.Mattson MP Genes and behavior interact to determine mortality in mice when food is scarce and competition fierce. Aging Cell 9, 448–449 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Mattison JA et al. Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8, 14063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson AA & Stolzing A The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 18, e13048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura K, Fuster JJ & Walsh K Adipokines: A link between obesity and cardiovascular disease. J. Cardiol 63, 250–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calligaris SD et al. Mice long-term high-fat diet feeding recapitulates human cardiovascular alterations: An animal model to study the early phases of diabetic cardiomyopathy. PLoS ONE 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manrique C et al. Obesity and insulin resistance induce early development of diastolic dysfunction in young female mice fed a western diet. Endocrinology 154, 3632–3642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barzilai N, Huffman DM, Muzumdar RH & Bartke A The critical role of metabolic pathways in aging. Diabetes 61, 1315–1322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Templeman NM et al. Reduced circulating insulin enhances insulin sensitivity in old mice and extends lifespan. Cell Rep. 20, 451–463 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Barzilai N & Ferrucci L Insulin resistance and aging: A cause or a protective response? J. Gerontol. Ser. A 67, 1329–1331 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Holmes MV, Ala-Korpela M & Smith GD Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat. Rev. Cardiol. 14, 577–590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmes MV et al. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J. 36, 539–550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Z et al. Associations of triglyceride levels with longevity and frailty: A Mendelian randomization analysis. Sci. Rep 7, 41579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elliott P et al. Genetic Loci Influencing C-reactive protein levels and risk of coronary heart disease. JAMA J. Am. Med. Assoc. 302, 37–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menotti A & Puddu PE How the Seven Countries Study contributed to the definition and development of the Mediterranean diet concept: A 50-year journey. Nutr. Metab. Cardiovasc. Dis 25, 245–252 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Sacks Frank M et al. Dietary fats and cardiovascular Disease: A Presidential advisory from the American Heart Association. Circulation 136, e1–e23 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Willett WC Diet and Cancer. The Oncologist 5, 393–404 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Goncalves MD, Hopkins BD & Cantley LC Dietary fat and sugar in promoting cancer development and progression. 21 (2019). [Google Scholar]
- 73.Cozzo AJ, Fuller AM & Makowski L Contribution of adipose tissue to development of cancer. Compr. Physiol. 8, 237–282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao C et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int. J. Epidemiol 45, 896–908 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gáliková M & Klepsatel P Obesity and aging in the Drosophila model. Int. J. Mol. Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otabe S et al. Overexpression of human adiponectin in transgenic mice results in suppression of fat accumulation and prevention of premature death by high-calorie diet. Am. J. Physiol. - Endocrinol. Metab 293, E210–E218 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Yen CA & Curran SP Gene-diet interactions and aging in C. elegans. Exp. Gerontol. 86, 106–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller RA, Chrisp C & Atchley W Differential longevity in mouse stocks selected for early life growth trajectory. J. Gerontol. A. Biol. Sci. Med. Sci. 55, B455–461 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Kraus C, Pavard S & Promislow DEL The Size–life span trade-off decomposed: Why large dogs die young. Am. Nat. 181, 492–505 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Samaras TT, Storms LH & Elrick H Longevity, mortality and body weight. Ageing Res. Rev. 1, 673–691 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Nüesch E et al. Adult height, coronary heart disease and stroke: a multi-locus Mendelian randomization meta-analysis. Int. J. Epidemiol. 45, 1927–1937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vitale G, Pellegrino G, Vollery M & Hofland LJ Role of IGF-1 system in the modulation of longevity: Controversies and new insights from a centenarians’ perspective. Front. Endocrinol. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi YJ et al. Adult height in relation to risk of cancer in a cohort of 22,809,722 Korean adults. Br. J. Cancer 120, 668–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nunney L Size matters: height, cell number and a person’s risk of cancer. Proc. R. Soc. B Biol. Sci. 285, 20181743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wade KH, Carslake D, Sattar N, Davey Smith G & Timpson NJ BMI and mortality in UK Biobank: Revised estimates using Mendelian randomization. Obes. Silver Spring Md 26, 1796–1806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jha P et al. Systems analyses reveal physiological roles and genetic regulators of liver lipid species. Cell Syst. 6, 722–733.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jha P et al. Genetic regulation of plasma lipid species and their association with metabolic phenotypes. Cell Syst. 6, 709–721.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Williams EG et al. Multi-omic profiling of the liver across diets and age in a diverse mouse population. bioRxiv 2020.08.20.222968 (2021) (Cell Syst. in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Almeida-Suhett CP, Scott JM, Graham A, Chen Y & Deuster PA Control diet in a high-fat diet study in mice: Regular chow and purified low-fat diet have similar effects on phenotypic, metabolic, and behavioral outcomes. Nutr. Neurosci 22, 19–28 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Ziebarth JD & Cui Y Precise network modeling of systems genetics data using the Bayesian Network Webserver. Methods Mol. Biol. Clifton NJ 1488, 319–335 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Ziebarth JD, Bhattacharya A & Cui Y Bayesian Network Webserver: a comprehensive tool for biological network modeling. Bioinformatics 29, 2801–2803 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Voelkl B et al. Reproducibility of animal research in light of biological variation. Nat. Rev. Neurosci. 21, 384–393 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Viechtbauer W Conducting meta-analyses in R with the metafor package. J. Stat. Softw 36, 1–48 (2010). [Google Scholar]
- 95.Therneau TM & Grambsch PM Modeling Survival Data: Extending the Cox Model. (Springer-Verlag, 2000). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


