Abstract
Tsetse flies (Glossina spp.) house a population-dependent assortment of microorganisms that can include pathogenic African trypanosomes and maternally transmitted endosymbiotic bacteria, the latter of which mediate numerous aspects of their host’s metabolic, reproductive, and immune physiologies. One of these endosymbionts, Spiroplasma, was recently discovered to reside within multiple tissues of field captured and laboratory colonized tsetse flies grouped in the Palpalis subgenera. In various arthropods, Spiroplasma induces reproductive abnormalities and pathogen protective phenotypes. In tsetse, Spiroplasma infections also induce a protective phenotype by enhancing the fly’s resistance to infection with trypanosomes. However, the potential impact of Spiroplasma on tsetse’s viviparous reproductive physiology remains unknown. Herein we employed high-throughput RNA sequencing and laboratory-based functional assays to better characterize the association between Spiroplasma and the metabolic and reproductive physiologies of G. fuscipes fuscipes (Gff), a prominent vector of human disease. Using field-captured Gff, we discovered that Spiroplasma infection induces changes of sex-biased gene expression in reproductive tissues that may be critical for tsetse’s reproductive fitness. Using a Gff lab line composed of individuals heterogeneously infected with Spiroplasma, we observed that the bacterium and tsetse host compete for finite nutrients, which negatively impact female fecundity by increasing the length of intrauterine larval development. Additionally, we found that when males are infected with Spiroplasma, the motility of their sperm is compromised following transfer to the female spermatheca. As such, Spiroplasma infections appear to adversely impact male reproductive fitness by decreasing the competitiveness of their sperm. Finally, we determined that the bacterium is maternally transmitted to intrauterine larva at a high frequency, while paternal transmission was also noted in a small number of matings. Taken together, our findings indicate that Spiroplasma exerts a negative impact on tsetse fecundity, an outcome that could be exploited for reducing tsetse population size and thus disease transmission.
Author summary
Endosymbiotic bacteria regulate numerous aspects of their host’s reproductive physiology. Natural populations of the tsetse fly, Glossina fuscipes fuscipes (Gff), house heterogeneous infections with the bacterium Spiroplasma glossinidia. Infection with the bacterium results in the presentation of several phenotypes in both male and female Gff that would put them at a significant reproductive disadvantage when compared to their counterparts that do not house the bacterium. These Spiroplasma induced phenotypes include changes in sex–biased gene expression in the reproductive organs, a depletion in the availability of metabolically critical lipids in pregnant females that results in delayed larval development, and compromised sperm fitness. These findings indicate that Spiroplasma exerts an overall negative impact on both male and female reproductive fitness and thus likely has a profound effect on fly population structure. This outcome, in conjunction with the fact that Spiroplasma infected tsetse are unusually refractory to infection with pathogenic African trypanosomes, indicates that the bacterium could be experimentally exploited to reduce disease transmission through the fly.
Introduction
Tsetse flies (Glossina spp.), which vector pathogenic African trypanosomes, reproduce via a process called adenotrophic viviparity. Following mating, female tsetse ovulate one oocyte per gonotrophic cycle (GC). The oocyte is fertilized in the maternal uterus by sperm that are released from the spermathecae. Unlike in most arthropods, tsetse embryos and larvae develop exclusively in utero, and larvae receive nourishment in the form of maternal milk secretions. Following the completion of larvigenesis, the mother gives birth to a fully developed 3rd instar larva that pupates within 30 minutes. This process repeats itself approximately every 10 days [1]. During the mating process, male tsetse transfer seminal fluid (SF), which contains sperm as well as numerous male accessory gland (MAG) derived proteins, into the reproductive tract of receptive females. Once in the female’s uterus, this mixture forms into a proteinaceous spermatophore that facilitates successful transfer of sperm into the spermathecae for long-term storage [2–6]. Because tsetse’s viviparous reproductive strategy results in the production of relatively few offspring, targeting reproduction can be a most effective approach to reduce tsetse populations. Methods to inhibit tsetse fecundity can be highly effective in reducing disease transmission given that the fly is an obligate vector for parasite transmission [1,7]. In fact, the application of sterile male programs has been very successful with tsetse [8] and is currently endorsed to eliminate tsetse populations on the African continent [8,9].
Tsetse flies have evolved long-term associations with several vertically transmitted endosymbiotic bacteria that impact numerous aspects of their host’s nutritional, developmental and reproductive physiology. All tsetse flies house the obligate endosymbiont Wigglesworthia, which provides nutrients absent from the fly’s vertebrate blood-specific diet [10–13]. In addition to Wigglesworthia, laboratory reared and natural tsetse populations can also harbor facultative Sodalis as well as parasitic Wolbachia and Spiroplasma [14,15]. To date Spiroplasma infections in tsetse have been found exclusively within tsetse flies of the subgenus Palpalis [14,15]. In both field captured and laboratory reared Glossina fuscipes fuscipes (Gff), the bacterium resides in reproductive and digestive tissues as well as hemolymph [14]. Spiroplasma induces an immune protective effect in laboratory reared Gff, as flies that house the bacterium are significantly more resistant to infection with trypanosomes than are flies that do not [15]. The mechanism that underlies Spiroplasma enhanced refractoriness to trypanosome infection in Gff is currently unknown. However, in other insects the bacterium confers pathogen (e.g., nematodes, fungi and parasitoid wasps) resistant phenotypes through the production of immune effector molecules and/or through nutrient scavenging that limits metabolically critical nutrients for other pathogens [16–19].
Infection with Spiroplasma can also impact host reproductive fitness and lead to reproductive abnormalities. In Drosophila, females infected with Spiroplasma are less fecund and produce fewer eggs, which may be a consequence of nutritional competition between the fly and bacterium for metabolically important free (hemolymph-borne) lipids [20]. In several insect taxa, the bacterium induces a male killing phenotype [21–23]. The potential functional role of Spiroplasma in tsetse reproductive physiology is currently unknown. Herein we performed a detailed investigation of the molecular and physiological associations that characterize reproductive aspects of the Gff-Spiroplasma symbiosis. Specifically, we performed RNA sequencing (RNA-seq) of reproductive tissues from field captured (Uganda) Spiroplasma infected and uninfected male and female Gff and report on genes and pathways that are differentially regulated in the presence of the bacterium. We also made use of a Gff lab line that carries a heterogenous infection with Spiroplasma to characterize the trans-generational transmission dynamics of this endosymbiont and to characterize Spiroplasma-induced metabolic and reproductive phenotypes in tsetse. Knowledge obtained from this study provides insight into the physiological mechanisms that underlie the tsetse-Spiroplasma symbiosis and may have translational implications with respect to controlling tsetse populations and the ability of the fly to transmit African trypanosomes.
Results
Overall gene expression profile
To understand the potential effects of Spiroplasma infection on tsetse’s reproductive physiology, we compared the RNA-seq data obtained from the male reproductive organs (testis, accessory gland and ejaculatory duct) of Spiroplasma infected (GffSpi+) and uninfected (GffSpi-) individuals obtained from wild populations in Uganda. We similarly obtained and compared RNA-seq data from GffSpi+ and GffSpi- female reproductive organs (ovaries and oocytes) isolated from the same populations. The percentage of reads from the different biological replicates that mapped to the Gff reference genome ranged from 49% to 81% (S1 Table in S1 Appendix).
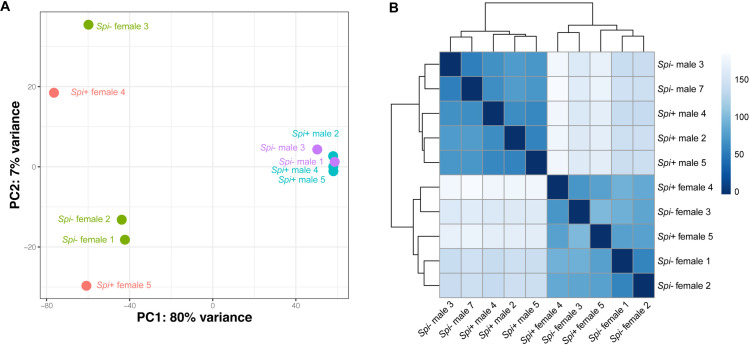
We used principal component (PC) and hierarchical cluster (HC) analyses to compare the overall gene expression profiles across all biological replicates according to sex and Spiroplasma infection status (Fig 1A and 1B). We found that PC1 and PC2 accounted for 80% and 7% of variance across all biological replicates, respectively. The variance in PC1 could be explained by differences in sex, with females clustering on the left side along the PC1 axis, and males clustering on the right side along the PC1 axis (Fig 1A). HC analyses also demonstrated a similar clustering result among different biological replicates (Fig 1B). Female and male clusters were distant from each other, and within the male samples, GffSpi+ were separated from the GffSpi- samples. However, within the female samples, one GffSpi+ (female 4) and one GffSpi- (female 3), were distant to the other three female samples (one GffSpi+ and two GffSpi-). This difference we noted within the female dataset could be due to the presence of greater variation in gene expression within the female reproductive tissues when compared to males, which is similar to the results shown in our PCA.
Fig 1.
Principal component analysis (A) and Hierarchical clustering (B) of expression data. The Principal Component Analysis (PCA) is based on differentially expressed genes from the female and male samples, while the Hierarchical Clustering (HC) reflects all genes within the dataset. Spiroplasma-infection status and sex are color-coded in the PCA.
The impact of Spiroplasma on sex-biased gene expression in males and females
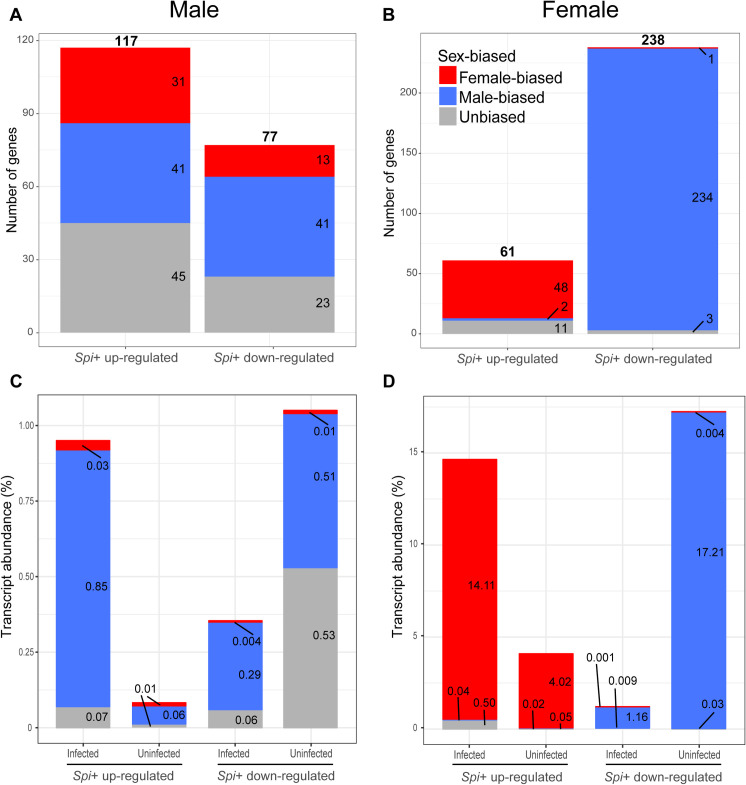
As our biological replicates clustered by sex, we first quantified the proportion of differentially expressed (DE) genes between females and males (sex-biased genes) using sex as a factor in the model we created in DESeq2 (see Materials and Methods for details). Of the 15,247 genes annotated in the Gff genome, we detected 10,540 genes expressed in our transcriptomes, with an adjusted p-value created by FDR correction (S1 Data) [24]. We observed that 21.7% of these genes (2,288/10,540) were preferentially expressed in females (female-biased genes; log2male/female<0 and adjusted p<0.05), while 20.5% (2,164/10,540) were preferentially expressed in males (male-biased genes; log2male/female>0 and adjusted p<0.05). Using pairwise comparisons for Spiroplasma infection status within each sex from the DESeq2 model, we found that a total of 194 and 299 genes were DE upon Spiroplasma infection in males and females, respectively. Within the male dataset, we determined that 117 (60.3%) and 77 (39.7%) of the DE genes were up- and down-regulated upon infection (adjusted p<0.05), respectively (Fig 2A, S2 Table in S1 Appendix and S2 Data). A similar analysis of the 299 DE genes in females showed that 61 (21.4%) and 238 (79.6%) were up- and down-regulated in the presence of Spiroplasma, respectively (Fig 2B and S3 Table in S1 Appendix).
Fig 2. Enrichment of genes with sex-biased expression that are differentially expressed between GffSpi+ and GffSpi- flies.
(A) The number of genes with sex-biased expression that are up- and down-regulated in GffSpi+ males. (B) The number of genes with sex-biased expression profiles that are up- and down-regulated in GffSpi+ females. (C) Transcript abundance (%) of up- and down-regulated genes relative to total transcript abundance in GffSpi+ and GffSpi- males. (D) Transcript abundance of up- and down-regulated genes relative to total transcript abundance in GffSpi+ and GffSpi- females. Female-biased genes are represented as red blue and male-biased genes as blue. Genes with no sex-bias are indicated as gray.
Spiroplasma infections can manipulate the reproductive physiology of their host insects [25]. In Drosophila, infections with the S. poulsonii strain MSRO affects gene regulation and dosage compensation machinery in males, resulting in male-killing during embryogenesis [22,26]. Because genes displaying sex-biased expression profiles in reproductive tissues can confer sexual phenotypes [27,28], we first identified the sex-biased genes expressed in male and female gonads, and next evaluated their DE status based on Spiroplasma infection. We determined that 41 of the 117 genes (35%) up-regulated, and 41 of the 77 genes (53%) down-regulated in GffSpi+ males are male-biased (Fig 2A and S2 Table in S1 Appendix). We also noted that the DE male-biased genes (n = 82) identified within the male DE gene set (n = 194) are enriched when compared to genome-wide male-biased genes (n = 2164) (Fig 2A and S2 Table S1 Appendix; p<0.001 for up-regulation and p<10−9 for down-regulation in Fisher’s exact test). We next examined the transcript abundance of the 41 up-regulated male-biased genes and determined their contribution to be 0.85% of the total transcriptome in the infected group, up from the 0.06% in the uninfected group (Fig 2C). We similarly analyzed the 41 down-regulated male-biased genes and determined their contribution to be 0.29% of the total transcriptome in the infected group, down from the 0.51% in the uninfected group (Fig 2C).
We next performed the same analysis with the female DE gene dataset. We determined that 48 of the 61 genes (78%) up-regulated in the GffSpi+ females were female-biased and enriched within the up-regulated genes when compared to genome-wide female-biased genes (n = 2288) (S3 Table in S1 Appendix; p<10−20 in Fisher’s exact test). Conversely, only one of the 238 genes down-regulated in GffSpi+ females was female-biased (Fig 2B and S3 Table in S1 Appendix), while 234 (98%) were male-biased (Fig 2B and S3 Table in S1 Appendix). The male-biased genes were thus overrepresented within the down-regulated genes (S3 Table in S1 Appendix; p<10−149 in Fisher’s exact test). We also examined the transcript abundance of the 48 up-regulated female-biased genes and determined their contribution to be 14.11% of the total transcriptome in the infected group, up from the 4.02% transcript abundance in the uninfected group (Fig 2D). The transcript abundance of the 234 down-regulated male-biased genes in the female dataset represented 1.16% of the total transcriptome in the infected group, down from 17.21% transcript abundance in the uninfected group (Fig 2D).
In both sexes, unbiased (non-sex biased) genes were underrepresented in the DE datasets when compared to the genome-wide unbiased genes (Fig 2, S2 and S3 Tables in S1 Appendix; p<10−4 for males and p<10−9 for females in Fisher’s exact test). Collectively, these results suggest that Spiroplasma infections strongly influence the expression of sex-biased genes in tsetse’s reproductive tissues. Given that sexual dimorphisms are largely shaped by sex-biased gene expressions [27,28], the significant number of sex-biased genes and transcript abundance affected by Spiroplasma in females relative to males suggests that the endosymbiont may affect female reproductive physiology to a greater extent.
Spiroplasma infection effects on genes that encode spermatophore constituents
When tsetse copulate, the ejaculate, which is composed of sperm and secretory products (i.e., seminal fluid) derived from the male testes and accessory glands, are transferred to the female uterus where they are encapsulated into a spermatophore. The spermatophore functions as a protective container for the ejaculate and ensures that sperm and seminal fluid are delivered to the female spermathecal ducts, which in turn modifies female behavior, including potential inhibition of further matings by competing males [3–5]. Within 24 h of the commencement of copulation, sperm are transferred to the female’s spermatheca and the spermatophore is discharged from the uterus [3]. We had previously identified the proteins detected in the G. morsitans morsitans (Gmm) spermatophore by collecting the structures from the female spermatheca shortly after completion of copulation [2]. We first identified the homologs of these Gmm protein encoding genes in Gff and then evaluated their expression status in the presence of Spiroplasma. We found that of the 287 genes whose products were detected in the Gmm spermatophore, seven were DE in GffSpi+ males and 12 were DE in GffSpi+ females relative to GffSpi—datasets (S1 Data). Of the seven genes encoding spermatophore proteins contributed by male gonads, six were up-regulated and one was down-regulated in the presence of Spiroplasma. Of the 12 genes encoding spermatophore proteins contributed by female reproductive organs, four were up-regulated and eight were down-regulated in the presence of Spiroplasma. Among the DE genes in females were two that encode transcription factors and three that encode serine protease inhibitors, while the up-regulated genes in males primarily encoded cuticle related proteins.
Immunity genes up-regulated in GffSpi+ males
Long term persistence of endosymbionts requires evasion of, and/or resistance against, host immune responses. Although the absence of metabolically costly immune responses would benefit host fitness, induced immunity can also be beneficial to the symbiosis as it confers resistance to other pathogens that could compete for limited nutritional resources. In Drosophila, Spiroplasma infections do not activate host immune responses, and the bacterium is not susceptible to either the cellular or humoral arms of the fly’s immune system [29]. To understand tsetse-Spiroplasma immune dynamics, we investigated whether DE genes in GffSpi+ males and females encoded immunity related functions. We found that in the male gonads, the presence of Spiroplasma significantly induces Toll pathway constituents, including toll-like receptor 7 (GFUI009072), the antimicrobial peptide defensin (GFUI031425), and easter (GFUI050711), which encodes a serine-protease required to process the extracellular Toll ligand Spätzle. We also noted that in addition to the Toll signaling pathway, several abundantly expressed Mucin encoding genes (GFUI039642, GFUI016405, GFUI054349, GFUI017943) were induced in GffSpi+ males. Mucins are produced by epithelial tissues where they function in different roles from lubrication to cell signaling to forming chemical barriers as well as binding and entrapping pathogens [30]. Taken together, this analysis indicates that Spiroplasma infection induces the upregulation of a highly restricted repertoire of immune related genes in the reproductive tract of Gff males. The bacterium had no impact on immune gene expression in Gff females.
Gene ontology (GO) analysis of DE gene products
To understand the major putative function(s) for the 493 DE gene products associated with Spiroplasma infection in Gff reproductive physiology, we performed GO (gene ontology) term enrichment analysis [31]. We found that within the up-regulated genes in GffSpi+ males, the significantly enriched GO terms are associated with cuticle development, chitin process, cell adhesion, defense response and receptor activity (S1A Fig and S3 Data). The down-regulated genes in GffSpi+ males were enriched for lipid catabolic process and peptidyl-dipeptidase activity (S1B Fig and S3 Data). The up-regulated genes in GffSpi+ females were enriched for lipid metabolic process and defense response (S1C Fig and S3 Data) while the down-regulated genes in GffSpi+ females were enriched for peptidase activity, proteolysis, and chitin metabolic process (S1D Fig and S3 Data).
Spiroplasma infection effects on female gene expression
The up-regulated female-biased genes in GffSpi+ females comprised over 14% of the entire transcriptome, while these same genes comprised only 4% of the total transcriptome in GffSpi- females. Among the up-regulated gene dataset, we noted the presence of several highly abundant tsetse milk proteins, including Mgp1 (GFUI006902), Mgp10 (GFUI050429), Mgp4 (GFUO050451). In addition, we detected high level expression of a gene annotated as Apolipoprotein (GFUI006901) located adjacent to the mgp1 (GFUI006902) locus.
We also noted that 234 male-biased genes were dramatically down-regulated in Spiroplasma infected females. These genes contributed a total of 1% of the transcriptome in the infected state, down from 17% in the uninfected state. Among the abundant and highly reduced genes were ones that encode multiple proteins annotated with digestive functions, such as those that had signatures of midgut trypsins (GFUI029963, GFUI029966, GFUI14886, GFUI024126, GFUI049688, GFUI006483, GFUI026212), serine proteases (GFUI026465, GFUI009869, GFUO026202), trypsin-like serine proteases (GFUI028730, GFUI028738), zinc-carboxypeptidases (GFUI030548, GFUI007477), chymotrypsin-like proteins (GFYI032994, GFUI032998, GFUI010855), and a carboxypeptidase (GFUI022995). The physiological consequences of this reduced proteolytic activity on GffSpi+ females remain to be determined.
Impact of Spiroplasma infection on Gff female metabolism and reproductive fitness
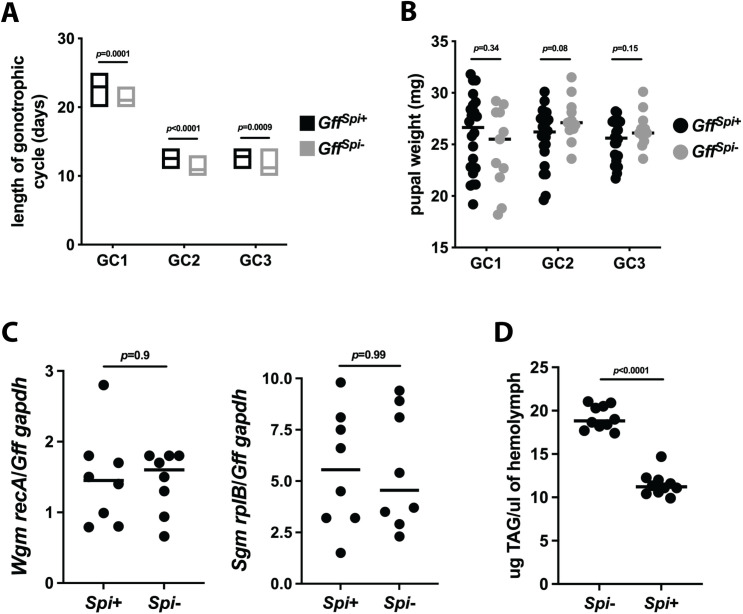
We next investigated whether Spiroplasma induced differences in gene expression regulate critical metabolic processes relevant for fecundity. To do so we measured several reproductive fitness parameters in GffSpi+ and GffSpi- pregnant females and males. We began by measuring the impact of Spiroplasma infection on the length of tsetse’s gonotrophic cycle (GC), which includes oogenesis, embryogenesis and larvigenesis. We observed that the1st, 2nd, and 3rd GCs of GffSpi+ females (23.0 ± 0.24, 12.6 ± 0.19, and 13.0± 0.2 days, respectively) were significantly longer than that of their age-matched counterparts that did not harbor Spiroplasma (21.0 ± 0.36, 11.0 ± 0.29, and 11.2± 0.33 days, respectively) (GC1, p = 0.0001; GC2, p<0.0001; GC3, p = 0.0009; log rank test) (Fig 3A and S4 Data). We next weighed pupae-stage offspring from GffSpi+ and GffSpi- females and observed no significant difference between groups across all three GCs (GC1, GffSpi+ = 26.0 ± 1.4 mg, GffSpi- = 24.6 ± 1.1 mg; GC2, GffSpi+ = 25.7 ± 1.0 mg, GffSpi- = 27.4 ± 1.8 mg; GC3, GffSpi+ = 25.4 ± 1.7 mg, GffSpi- = 26.4 ± 1.6 mg) (GC1, p = 0.34; GC2, p = 0.08; GC3, p = 0.15; multiple t tests) (Fig 3B). These findings indicate that an infection with Spiroplasma does not impact pupal weight but does increase the length of tsetse’s GC. As such, infection with this bacterium may result in the production of fewer offspring over the course of the female’s lifespan, thus exerting a detrimental impact on the fly’s overall reproductive fitness.
Fig 3. Impact of Spiroplasma infection on the reproductive and nutritional fitness of female tsetse flies.
(A) Gonotrophic cycle (GC) length of offspring from GffSpi+ and GffSpi- females. Age-matched, pregnant females from each group (n = 34 per group) were housed in individuals cages and monitored daily to observe frequency of pupal deposition. Statistical significance was determined via log-rank test. (B) The weight of pupae deposited by GffSpi+ and GffSpi- females. Each dot represents an individual pupa, bars represent median values of pupae form each GC. Statistical significance was determined via multiple t-tests with correction via the Holm-Šídák method. (C) Relative densities of Wigglesworthia and Sodalis in GffSpi+ and GffSpi- females. Relative Wigglesworthia recA and Sodalis rplB copy number was quantified using genomic DNA derived from GffSpi+ or GffSpi- female midguts (including the Wigglesworthia harboring bacteriome organ). Wigglesworthia recA and Sodalis rplB was normalized relative to Gff gapdh copy number in each sample. Each dot represents one biological replicate (three midguts per replicate), and bars indicate median values. Statistical significance was determined via students t-test. (D) Amount of triacylglyceride (TAG) circulating in the hemolymph of GffSpi+ and GffSpi- females. Three microliters of hemolymph was extracted from two-week-old pregnant females and TAG was quantified colorimetrically via comparison to triolein standard curve (S2 Fig). Each dot represents an individual pupa, bars represent median values of pupae form each GC. Statistical significance was determined via unpaired t-test.
We have previously demonstrated that experimental depletion of Wigglesworthia density (via treatment with antibiotics) significantly impairs tsetse’s reproduction [12,32–34]. With this in mind, we measured the relative densities of endosymbiotic Wigglesworthia and Sodalis in GffSpi+ and GffSpi- females to determine if these symbionts are impacted by the presence of Spiroplasma. We determined that GffSpi+ and GffSpi- females house similar densities of Wigglesworthia (Fig 3C, left graph) and Sodalis (Fig 3C, right graph). Because Spiroplasma infection did not impact the density of these tsetse symbionts, we next investigated whether the prolonged GC length presented by GffSpi+ females reflected a competition for resources between the bacterium and pregnant female flies. This theory is supported by the fact that in Drosophila, Spiroplasma proliferation is limited by the availability of circulating lipids [20]. The gut of 3rd instar tsetse larvae contains high levels of triacylglyceride (TAG) [35], which originate in tsetse’s fat body and are transferred through hemolymph to the milk gland where they are incorporated into maternal milk secretions [36,37]. To determine if Spiroplasma hijacks tsetse TAG, at the nutritional expense of developing intrauterine larvae, we compared TAG levels in the hemolymph of pregnant Spi+ and Spi- females. We found that pregnant Spi- females had significantly more TAG circulating in their hemolymph (19.2 ± 0.4 μg/μl) than did their age and pregnancy stage-matched Spi+ counterparts (11.5 ± 0.4 μg/μl) (p<0.0001; t test) (Fig 3D). These findings suggest that, like in Drosophila, Spiroplasma uses tsetse lipids as an energy source. This competition between tsetse and Spiroplasma for dietarily limited yet metabolically important lipids may result in the relatively long GC we observed in Spi+ mothers because less of this nutrient is available for incorporation into milk secretions.
Impact of Spiroplasma infection on Gff male reproductive fitness
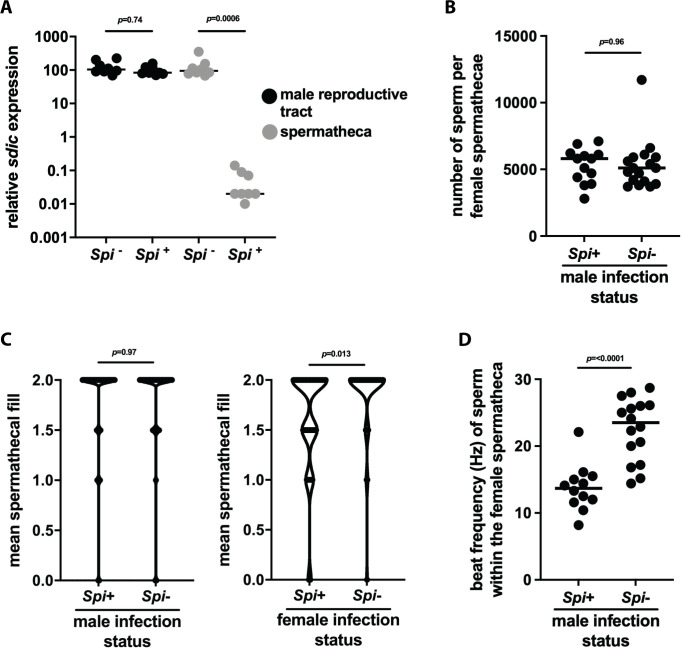
Our transcriptomic data informed us that infection with Spiroplasma impacts the expression of several genes that encode spermatophore-associated proteins. These proteins, many of which arise from the male accessory gland and are transferred in seminal fluid, play a prominent role in sperm fitness [2,38]. We thus compared the fitness of sperm derived from Spi+ compared to Spi- males to determine if infection with the bacterium impacts the fitness off Gff sperm. To do so we quantified the transcript abundance of sperm-specific dynein intermediate chain (sdic; VectorBase gene ID GFUI025244) in the spermathecae of 7 day old female flies two days after having mated with either Spi+ or Spi- males. The Drosophila homologue of this gene is expressed exclusively in sperm cells [39] and has been used to quantify sperm abundance and competitiveness (as a function of motility) [40–42]. We observed that sdic transcript abundance in the spermathecae of females that mated with Spi- males was significantly higher than in that of their counterparts that mated with Spi+ males (Fig 4). This conspicuous difference in the abundance of sdic transcripts expressed by sperm within the spermathecae of females that mated with Spi- versus Spi+ males was surprising considering the fact that expression of this gene in the reproductive tract of field captured Spi- and Spi+ males was not significantly different when measured by RNAseq (S1 Data). To validate this RNAseq data, we used RT-qPCR to measure sdic expression in sperm housed in the reproductive tract of 7 days old Spi- and Spi+ lab reared Gff males (these sperm were the same age as those used to measure sdic transcript abundance in the spermathecae of mated females). Similar to what we observed via RNAseq, sdic expression was not significantly different in sperm housed in the male reproductive tract (Fig 4).
Fig 4. The impact of Spiroplasma infection on male Gff reproductive fitness.
(A) Relative expression of sperm-specific dynein intermediate chain (sdic) in sperm located within the reproductive tract of GffSpi+ and GffSpi- males or within the female spermatheca following copulation. Sdic expression in each sample was normalized relative to tsetse’s constitutively expressed pgrp-la gene. Each dot represents one biological replicate, and bars indicate median values. Statistical significance was determined via one-way ANOVA followed by Tukey’s HSD post-hoc analysis. (B) Quantification of sperm within the spermatheca of female flies that mated with either Spi+ or Spi- males. Measurements were made using a Neubauer counting chamber. Each dot represents one spermatheca, and bars indicate median values. Statistical significance was determined via student’s t-test. (C) Spermathecal fill of female flies that mated with either Spi+ or Spi- males. Spermatheca fill data were analyzed using Kruskal-Wallis rank test using R companion and FSA software packages [75,76]. (D). Motility, as a measure of flagellar beat frequency in hertz (Hz), of sperm within the spermatheca of female flies that mated with either Spi+ or Spi- males. Video recordings of sperm were acquired at a rate of 50 frames per second, and beat frequency was analyzed using FIJI and the ImageJ plugin SpermQ. Each dot on the graph represents the mean beat frequency of two sperm tails from each spermatheca. Statistical significance was determined via student’s t-test.
The above data indicate that sdic expression decreases significantly in sperm that originate from Spi+ males following transfer to the female spermatheca, thus suggesting that the bacterium plays a role in either regulating the number of sperm transferred during mating and/or sperm motility (and thus competitiveness) after mating. To investigate further we quantified both the quantity and motility of sperm in the spermathecae of females 24 hrs after having mated with either Spi+ or Spi- males. Manual counting indicated no significant difference in the number of stored sperm when females had mated with either Spi+ (5375 ± 537 sperm per spermathecae) or Spi- (5358 ± 632 sperm per spermathecae) males (Fig 4B). We additionally quantified the abundance of stored sperm as a reflection of spermathecal fill, which measures the quantity of sperm and seminal fluid transferred from males to the spermatheca of their female mates. Male Spiroplasma infection status had no impact on spermathecal fill. However, when female mates were infected with Spiroplasma we observed a significant reduction in the size of their spermatheca compared to that of Spi- females (Kruskal-Wallis, X2 = 6.1763, df = 1, p = 0.0129) (Fig 4C). We also investigated whether the precipitous drop in sdic expression in sperm transferred from Spi+ compared to Spi- males was representative of a decrease in the motility of sperm derived from males in the latter group. We observed that sperm derived from Spi+ males exhibited a mean beat frequency of 13.4 (±1.7) hertz (Hz), while those derived from Spi- males exhibited a mean beat frequency of 22.1 (±1.4) Hz (Fig 4D).
Taken together, these results indicate that the Spiroplasma infection status of male Gff does not impact the number of sperm they transfer during mating, but female infection status of the mating pairs does impact the number of sperm stored in their spermatheca. Additionally, Spiroplasma exerts a significant impact on the competitiveness of sperm stored in the spermatophore, as cells that originate from infected males are significantly less motile than are those that originate from their uninfected counterparts.
Vertical transmission of Spiroplasma
Spiroplasma is maternally transmitted in several insect systems, including the tsetse fly, as evidenced by the presence of the endosymbiont in the intrauterine larva [14]. However, not all tsetse populations, nor individuals within distinct populations, house the bacterium [14,15]. This heterogeneity in infection prevalence suggests that vertical transmission of the bacterium may be imperfect and thus occur at a frequency of less than 100%. We performed three distinct experiments as a means of quantitating the fidelity of Spiroplasma vertical transmission (see Materials and Methods for experimental details).
For experiment 1, we observed that 86% of adult offspring (from three GCs combined) derived from matings between Spi+ mothers and Spi+ fathers were Spi+ as were 97% of adult offspring from matings between Spi+ mothers and Spi- fathers. Also in experiment 1 we observed no Spi+ adult offspring when mothers lacked the bacterium, regardless of their mate’s infection status (Table 1 and S5 Data). For experiment 2, 67% of pupal offspring (from three GCs combined) derived from matings between Spi+ mothers and Spi+ fathers were Spi+ as were 44% of pupal offspring from matings between Spi+ mothers and Spi- fathers (Table 1 and S5 Data). In experiment 2, four matings between Spi- mothers and Spi+ fathers collectively resulted in the deposition of nine pupae, four of which carried Spiroplasma infections. Interestingly, in three out of four mating pairs, the second offspring (GC2) was positive for the Spiroplasma infection while the first offspring (GC1) was negative (Table 1 and S5 Data), indicating imperfect paternal transmission at best. The varying results we obtained between these experiments could reflect the different developmental stages of the offspring we tested (experiment 1, newly eclosed adults; experiment 2, newly deposited pupae). Spiroplasma density decreases during development as tsetse age from larvae through pupation and eclosion to adulthood [14]. Hence, it remains to be seen whether the Spiroplasma present in the pupae analyzed in experiment 2 would persist through the lengthy pupal stage (~ 30 days) and into adulthood.
Table 1. Vertical transmission of Spiroplasma across three gonotrophic cycles (GCs).
| Spiroplasma infection status | # of Spiroplasma infected offspring/total offspring sample/GC | ||||||
|---|---|---|---|---|---|---|---|
| experiment #a | mother | father | number of mating pairsb | GC1 | GC2 | GC3 | total % transmissionc |
| Exp 1 | + | + | 7 | 6/7 | 6/7 | 6/7 | 86% (18/21) |
| - | 13 | 13/13 | 13/13 | 12/13 | 97% (38/39) | ||
| - | + | 9 | 0/9 | 0/9 | 0/9 | 0% (0/27) | |
| - | 5 | 0/5 | 0/5 | 0/5 | 0% (0/15) | ||
| Exp 2 | + | + | 5 | 4/5 | 2/3 | 0/1 | 67% (6/9) |
| - | 7 | 6/7 | 3/5 | 2/2 | 79% (11/14) | ||
| - | + | 4 | 1/4 | 3/3 | 0/2 | 44% (4/9) | |
| - | 1 | 0/1 | 0/1 | 0/0 | 0% (0/2) | ||
aIn experiment 1 Spiroplasma infection prevelance was determined using adult offspring, while in experiment 2 Spiroplasma infection prevelance was determined using pupal offspring.
bNumber of mating pairs at the beginning of each experiment. Not all mothers survived for three GCs.
cPercentage of Spiroplasma infected offspring derived from each parental mating pair over the course of all three GCs combined.
Finally, in experiment 3, we determined Spiroplasma infection status of mothers only. Of the Spi+ mothers observed, 73% (11/15) and 100% (9/9) of their pupal offspring from GCs 1 and 2, respectively, were infected with Spiroplasma (no Spi+ mothers in this experiment survived long enough to produce GC 3 offspring). Nine out of the 22 total progeny from Spi- mothers were Spi+, although transmission did not occur across sequential GCs (S5 Table in S1 Appendix and S5 Data). While this experiment suggests possible paternal Spiroplasma transmission, we cannot confirm the route of infection because we did not test the Spiroplasma status of these fathers.
Taken together, these transmission data indicate that Spiroplasma is vertically transmitted via maternal lineages with high frequency, although paternal transmission may provide a less frequent route.
Discussion
Reproductive tissue-associated heritable endosymbionts affect their arthropod host’s physiology to facilitate their transmission. In tsetse, both natural populations and laboratory Gff lines are found to house heterogenous infections with parasitic Spiroplasma, but little is known about the physiological impact(s) of this microbe on tsetse reproduction. Our transcriptomic analyses revealed that Spiroplasma infection significantly alters gene expression in reproductive tissues of both male and female Gff. In particular, amongst the genes impacted by Spiroplasma infection in females, significantly more female-biased genes are up-regulated, and male-biased genes are down-regulated when compared to their uninfected counterparts. Using a laboratory line of Gff that carries a heterogenous infection with Spiroplasma, we discovered that infection with the bacterium results in a reduction in fecundity as evidenced by a significantly longer gonotrophic cycle in infected females compared to their uninfected counterparts. Loss of fecundity likely results from the decreased levels of hemolymph lipids in Spiroplasma infected females, which suggests that the bacterium competes with its host during pregnancy for nutrients that are critical for larval development. Additionally, we observed a dramatic reduction in the abundance of a sperm-specific sdic transcripts following transfer of spermatozoa from Spi+ males to their mate’s spermatheca. Upon further investigation we determined that these sperm cells exhibit compromised motility and thus likely decrease competitiveness. Finally, our Spiroplasma transmission studies suggest maternal transmission of the bacterium occurs with high fidelity from infected mothers to each of their offspring. However, we also observed evidence of paternal transmission of the bacterium, which could explain the heterogenous infections we observed in this laboratory line. Collectively, our findings significantly enhanced fundamental knowledge on the tsetse-Spiroplasma symbiosis with implications for other arthropods. Additionally, the information obtained in this study may be applicable to the development of novel tsetse control strategies for population reduction.
Sex-biased gene expression strongly influences sexual phenotypes in most animals [27,28], and a large proportion of sex-biased genes are expressed in reproductive tissues [43,44]. With this in mind, the differences in sex-biased gene expression we observed in between GffSpi- and GffSpi+ females suggest that housing the bacterium could induce beneficial reproductive phenotypes. Our transcriptomic data from GffSpi+ females showed that female-biased genes are up-regulated while male-biased genes are down-regulated relative to their uninfected (GffSpi-) counterparts. Notably, we observed that Spiroplasma infection induced a significant upregulation of genes that encode tsetse milk gland proteins (MGPs). These tsetse specific molecules are abundantly expressed in the accessory gland of pregnant females and are a prominent component of tsetse milk [1,45]. MGPs are thought to be mostly lipid carriers, and experimental evidence suggests that they act in this capacity as lipid emulsification agents and possible phosphate carrier molecules [37]. Thus, when faced with depleted levels of circulating TAG, which comprises a substantial fraction of the lipids found in tsetse milk [35], Spiroplasma infected females may adaptively increase MGP production in an effort to scavenge and transport other lipids to developing intrauterine larvae. This outcome may reflect one Spiroplasma induced change in sex-biased gene expression that confers a fitness benefit to GffSpi+ females.
The depletion of circulating TAG by hemolymph borne Spiroplasma likely accounts for the increase we observed in GC length in pregnant mothers that harbor the bacterium. Despite the longer larval development period we observed for progeny of Spi+ compared to Spi- mothers, the weight of pupal offspring deposited was similar between the two groups. Because female tsetse produce unusually few offspring (6–8) over the course of their lifespan (compared to other insects), an increase in GC length would likely result in a significant reduction in population size over time. In fact, infection with a trypanosome strain that induces a metabolically costly immune response also lengthens tsetse’s GC by a duration similar to that (approximately 2 days) induced by infection with Spiroplasma [46]. Mathematical modelling indicates that this increase in GC length would theoretically reduce tsetse fecundity by approximately 30% over the course of a female’s reproductive lifespan [46]. Thus, a moderate trypanosome infection prevalence of 26%, which is similar to the Spiroplasma infection prevalence in field captured Gff (5–34%, depending on population geographic location) [14,15], could significantly decrease fly population size [46]. The models also predict that infection prevalence above these frequencies would result in a population crash.
Our results indicate that infection with Spiroplasma also exerts a significant impact on reproductive processes in laboratory reared Gff males. We observed that sperm from both Spi+ and Spi- males expressed the same abundance of sperm-specific sdic transcripts prior to mating, but that following insemination and transfer to the female spermatheca, sperm from Spi+ males expressed conspicuously fewer transcripts of this gene than did sperm that had originated from Spi- males. The Drosophila homolog of this gene encodes a cytoplasmic dynein intermediate chain that is necessary for the proper function of the cytoplasmic dynein motor protein complex [39], which is involved in sperm motility [47]. Accordingly, one prominent phenotype we observed was that following insemination and transfer to the female spermatheca, the motility of sperm that had originated from Spi+ males was impaired when compared to that of sperm that had originated from Spi- males. This finding implies that sdic exhibits the same function in Gff as it does in Drosophila. Motility is of paramount importance to sperm competitiveness [48], and sperm with impaired motility have less fertilization success than heathy sperm [49]. Field data indicate that wild populations of Gff females can exhibit polyandry and store sperm from more than one mate [7]. Our data suggest that sperm transferred by Spi+ males may be at a competitive disadvantage in comparison to sperm derived from Spi- males because they exhibit reduced motility. It remains to be determined if, as a means of promoting polyandry, females seek additional mates if they first copulate with a Spi+ male that transfers motility compromised sperm.
Spiroplasma mediated mechanisms that influence the regulation of sdic expression are currently unknown. Spiroplasma could mark sperm in the reproductive tract of Gff males [50], possibly via post-transcriptional modification of sdic, such that sperm motility is compromised following insemination and transfer to the female spermatheca. Alternatively, female contributions to the spermatophore could also influence sdic expression after sperm are packaged into the structure. In Spiroplasma infected females, we have observed 12 DE genes that encode products identified as constituents spermatophore. Further experiments are necessary to determine if any of these Spiroplasma modified female products, including two transcription factors and three serine protease inhibitors, play a role in the sperm modifications we observed here.
Infections with heritable endosymbionts can come at a significant metabolic cost to their host [51]. This outcome appears to also be the case with the Spiroplasma and tsetse symbiosis, and results in a significant reduction in the reproductive fitness of both female and male flies. However, from an evolutionary perspective, these microbes, including Spiroplasma, usually also provide an overall fitness advantage in order for infections to be maintained. In the tsetse model system, the fly, the commensal symbiont Sodalis, as well as pathogenic trypanosomes, typically compete for host nutrients as they are auxotrophic for metabolically critical B vitamins [12,52–54]. Interestingly, these nutrients are present in low quantities in tsetse’s vertebrate blood specific diet, and they are instead supplemented via the mutualistic endosymbiont, Wigglesworthia [11–13]. Despite the negative impacts of Spiroplasma on tsetse’s fecundity, this bacterium also benefits its host by creating an environment within the fly that is hostile to parasitic trypanosomes [15]. The parasite resistance phenotype conferred by Spiroplasma infections could arise from induced host immune responses, or could reflect competition for critical nutrients (i.e., found in the bloodmeal and/or produced by tsetse) that both organisms require to sustain their metabolic needs [55–57]. This effect of inhibiting metabolically costly trypanosome infections could thus offset the cost of housing similarly detrimental Spiroplasma infections. Additionally, we demonstrate that while Spiroplasma can be both matrilineally and patrilineally transmitted, vertical transmission of the bacterium within our Gff colony does not occur at a frequency of 100%. This data corroborates observations from the field, which reveal that individual flies between and within distinct populations also present heterogeneous Spiroplasma infections [14,15]. In our study we observed a high frequency of maternal transmission into pupal and adult offspring as well as low frequency paternal transmission into pupal offspring. If Spiroplasma detected in the pupal stage survive and colonize adults, this outcome could contribute to the heterogenous infection prevalence we observe in the Gff lab line. The absence of steadfast vertical transmission of Spiroplasma by Gff could facilitate fly survival by reducing the population level detrimental impact (i.e., prolonged GC length and reduced sperm motility) associated with housing the bacterium. This may represent another advantageous factor that facilitates Spiroplasma’s ability to sustain infections within the fly across multiple generations.
Gff is the prominent vector of pathogenic African trypanosomes in east and central Africa, and reducing the size of fly populations is currently the most effective method for controlling parasite transmission. One means of accomplishing this is through the use of sterile insect technique (SIT), which involves the sequential release of a large number of sterilized males into a target environment [58,59]. Because of their large number, these infertile males outcompete wild males for resident female mates, and the population size drops significantly, or the population is eliminated [60,61]. Knowledge gained from this study, in conjunction with that from previous studies, will have a significant impact on tsetse rearing efficiency for the SIT programs. Specifically, decreased fecundity can have a negative impact on the success of colony maintenance. However, enhanced resistance to trypanosomes is advantageous for SIT applications as the released males, which also feed exclusively on vertebrate blood, have the potential to serve as vectors of disease. Thus, Spiroplasma has the potential to reduce the impact of this shortcoming by inducing a trypanosome refractory phenotype in released males such that they would be less likely vector disease, thus increasing the overall safety and efficacy of the control program.
Materials and methods
Field sampling
Glossina fuscipes fuscipes (Gff) were collected from the Albert Nile river drainage in Northwest Uganda. Sampling sites included three villages from the Amuru district: Gorodona (GOR; 3°15’57.6"N, 32°12’28.8"E), Okidi (OKS; 3°15’36.0"N, 32°13’26.4"E), and Toloyang (TOL; 3°15’25.2"N, 32°13’08.4"E). Flies were sampled using biconical traps and wing fray data were recorded (stage 2 for the majority of samples, stage 3 for a small subset of samples, corresponding approximately to 3 and 4 week-old adults, respectively). All females analyzed had been mated based on the visual presence of intrauterine larva or a developing oocyte in their ovaries. The Spiroplasma infection prevalence in flies from the Goronda and Okidi regions was about 46% [15]. Reproductive tissues (may have been contaminated with milk gland tissue, which is composed of an extensive tubular network that is difficult to separate from female reproductive tissues) from all flies were dissected in the field in sterile 1x phosphate buffered saline (PBS) and flash frozen in liquid nitrogen for later RNA extraction.
Data generation (Illumina HiSeq), transcriptome assembly and quality assessment
Total RNA from Gff female (ovaries, spermathecae, uterus and oocysts) and male (testes, accessory glands and ejaculatory duct) reproductive tracts was extracted according to TRIzol reagent manufacturer’s instructions (Thermo Fisher Scientific, USA). Total RNA was treated with Ambion TURBO DNA-free DNase (Thermo Fisher Scientific, USA) and RNA quality was evaluated using an Agilent 2100 Bioanalyzer. An aliquot from each RNA was reverse transcribed into cDNA and screened for Spiroplasma using PCR amplification assay with symbiont-specific primers as described [15]. Prior to pooling RNA for sequencing (see below), the same cDNAs were used to confirm the sex of the field collected tissues by performing PCR using primers that amplify sex specific genes (see S4 Table in S1 Appendix for gene names and amplification primers). RNA from three female or male reproductive tracks were pooled into distinct biological replicates for RNA-seq analysis. We analyzed two biological replicates from GffSpi+ females and three biological replicates from GffSpi+ males, and three biological replicates from GffSpi- females and two biological replicates from GffSpi- males. Ribosomal reduction libraries were prepared with the Ribo-Zero Gold rRNA removal kit (human/mouse/rat) MRZG12324 (Illumina, USA) and sequenced at the Yale University Center of Genome Analysis (YCGA) on an Illumina HiSeq 2500 machine (75bp paired-end reads). In order to obtain information on Spiroplasma transcripts as well as the tsetse host, we chose Ribosomal Reduction libraries over mRNA (poly A) libraries.
RNA-seq data analysis
RNA-seq sequencing produced an average of 96 million reads across all biological samples. Reads were mapped to the Gff reference genome (accession # JACGUE000000000 from NCBI BioProject PRJNA596165) using HISAT2 v2.1.0 with default parameters [62,63]. The annotations for the assembly were ported over from the annotation of the previous assembly (https://vectorbase.org/vectorbase/app/) using the UCSC liftOver software suite of programs [64]. The function ‘htseq-count’ in HTSeq v0.11.2 [65] was then used to count the number of reads aligned to the annotated genes in the reference genome with the option “-s reverse” because we generated strand-specific forward reads in RNA-seq libraries. We evaluated the number of DE genes between groups of female and male reproductive organs from GffSpi+ and GffSpi- using the HTSeq output as input into DESeq2 v1.22.1 [66]. We first developed a DESeq2 model to predict differential gene expression as a function of sex and Spiroplasma-infection status, and the interaction between sex and Spiroplasma-infection status. We then extracted the log2 fold changes (log2FCs) for each gene with false discovery rate (FDR) adjusted p-values [24]. Finally, we defined genes as DE if the log2 fold change (log2FC) was significantly different from 0, and we only considered genes with adjusted p-values as expressed for downstream analysis.
We next conducted a PC and HC analysis to analyze normalized expression count data from the DESeq2 program. We used a regularized log transformation of the normalized data created by “rlog” function DESeq2 [66]. For the HC by sample-to-sample distance, we used “pheatmap” function in R. We measured the expression abundance as the sum of TPM that were calculated using the TPM Calculator [67].
Because the function of genes in Gff are not well annotated, we performed ‘tblastx’ in CLC Genomics Workbench (CLC Bio, Cambridge, MA) with Gff transcript to search for predicted protein sequences bearing the closest homology to those from G. m. morsitans, Musca domestica, and Drosophila melanogaster. Using gene-associated GO terms [31], we performed enrichment analysis of gene ontology (GO) terms with the topGO R package [68].
Laboratory maintenance of Gff and determination of Spiroplasma infection status
Gff pupae were obtained from Joint FAO/IAEA IPCL insectary in Seibersdorf, Austria and reared at Yale University insectary at 26°C with 70–80% relative humidity and a 12 hr light:dark photo phase. Flies received defibrinated bovine blood every 48 hr through an artificial membrane feeding system [69].
Spiroplasma infection status of all lab-reared female, male and pupal Gff was determined by extracting genomic DNA (gDNA) from individual whole organisms or fly legs using a DNeasy Blood and Tissue kit according to the manufacturer’s (Qiagen) protocol. To confirm that intact gDNA was successfully extracted, all samples were subjected to PCR analysis using primers that specifically amplify Gff tubulin or microsatellite region GpCAG (as controls for genomic DNA quality) and Spiroplasma 16S rRNA. All PCR primers used in this study are listed in S4 Table in S1 Appendix. The Spiroplasma 16S rRNA locus was amplified by PCR (as described in [14]) using the following parameters: initial denaturation at 94°C for 5 min; 34 cycles of 94°C for 45 s, 59°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. Spiroplasma PCR reactions were carried out in a volume of 25 μl containing 1.5 μl gDNA. PCR products were analyzed on 2% agarose gels, and samples were considered infected with Spiroplasma if the expected PCR product of 455 bp was detected.
To confirm the sensitivity of this leg snip PCR assay, and thus eliminate the possibly of false negative outcomes, we randomly selected 10 Spiroplasma negative samples and two positive controls (as determined by endpoint PCR described above) and subjected them RT-qPCR using the Spiroplasma ropB. All RT-qPCR results were normalized to tsetse’s constitutively expressed pgrp-la gene (S2 Fig). RT-qPCR confirmed our endpoint PCR data (RT-qPCR output shown in S6 Data).
Impact of Spiroplasma infection on Gff metabolism and reproductive fitness
Hemolymph triacylglyceride (TAG) assay: Hemolymph (3 μl/fly) was collected (as described in [70]) from two-week-old pregnant female Gff, centrifuged (4°C, 3000xg for 5 minutes) to remove bacterial cells, diluted 1:10 in PBS containing 1.2 μl/ml of 0.2% phenylthiourea (to prevent hemolymph coagulation) and immediately flash frozen in liquid nitrogen. Hemolymph TAG levels were quantified colorimetrically by heating samples to 70°C for 5 min followed by a 10 min centrifugation 16,000xg. Five μl of supernatant was added to 100 μl of Infinity Triglycerides Reagent (Thermo Scientific) and samples were incubated at 37C for 10 min. Absorbance was measured at 540nm using a BioTek Synergy HT plate reader [71]. All Gff sample spectra data were compared to that generated from a triolein standard curve (0–50 μg, 10 μg increments; S3 Fig).
Impact on fecundity and endosymbiont density
The effect of Spiroplasma infection on Gff fecundity was measured by quantifying the length of three gonotrophic cycles (GC) and by weighing pupal offspring. To measure GC length, Gff females were mated as five days old adults and thereafter maintained in individual cages. All females were monitored daily to determine when they deposited larvae, and all deposited larvae were weighed.
To confirm the sensitivity of this leg snip PCR assay, and thus eliminate the possibly of false negative outcomes, we randomly selected 10 Spiroplasma negative samples and two positive controls (as determined by endpoint PCR described above) and subjected them RT-qPCR using the Spiroplasma ropB. All RT-qPCR results were normalized to tsetse’s constitutively expressed pgrp-la gene (S2 Fig). RT-qPCR confirmed our endpoint PCR data (RT-qPCR output shown in S6 Data).
Sperm-specific gene expression
Individual five-day old male and female flies (each fed twice) were placed together into tubular cages (height, 12.7 cm; diameter 6 cm) and allowed to mate for two days. Individual mating pairs were subsequently separated, and RNA (using Trizol reagent) was isolated from male and female reproductive tracts. RNA was then DNase treated and reverse transcribed into cDNA (using a Bio-Rad iScript cDNA synthesis kit) by priming the reaction with random hexamers. All males were then screened by PCR as described above to determine their Spiroplasma infection status.
Gff sperm-specific dynein intermediate chain (sdic; GFUI025244) transcript abundance (measured via RT-qPCR, primers used are listed in S4 Table in S1 Appendix) was used as a proxy measurement of sperm density and sperm fitness in male reproductive tracts and the spermathecae of pregnant females [39,47]. All RT-qPCR results were normalized to tsetse’s constitutively expressed pgrp-la gene, and relative expression of sdic was compared between male reproductive tracts and sperm (in the female spermathecae) that originated from Spi+ and Spi- males. All RT-qPCR assays were carried out in duplicate, and replicates quantities are indicated as data points on corresponding figures. Negative controls were included in all amplification reactions.
Sperm quantification and spermathecal fill assays
Sperm abundance in the spermathecae of females that had mated with either Spi+ and Spi- males was quantified by both direct cell counting and by measuring spermathecal fill. For direct counting, spermathecae were excised from mated females 24 hrs post-copulation with either Spi+ or Spi- males and placed into 10 μl of HEPES-buffered saline solution [145 mM NaCl, 4 mM KCl, 1 mM MgCl2, 1.3 mM CaCl2, 5 mM D-glucose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethane- sulfonic acid (HEPES), pH 7.4] in the well of a concave glass microscope slide. Spermathecae were gently poked with a fine needle and sperm were allowed to exude from the organ for 5 minutes. Samples were subsequently diluted 1:100 in HEPES-buffered saline and counted using a Neubauer counting chamber. To measure spermathecal fill, spermathecae from mated females were microscopically dissected 24h post-copulation in physiological saline solution (0.9% NaCl) and assessed subjectively at 100x magnification. Spermathecal fill of each individual organ was scored to the nearest quarter as empty (0), partially full (0.25, 0.50, or 0.75), or full (1.0) (S4 Fig), and the amount of sperm transferred was then computed as the mean spermathecal filling values of the spermathecae pairs [72,73].
Sperm motility assays
Spermathecae were excised from females 24 hrs post-copulation with either Spi+ and Spi- males and placed into 10 μl of HEPES-buffered saline solution [145 mM NaCl, 4 mM KCl, 1 mM MgCl2, 1.3 mM CaCl2, 5 mM D-glucose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethane- sulfonic acid (HEPES), pH 7.4] in the well of a concave glass microscope slide. Spermathecae were gently poked with a fine needle and sperm were allowed to exude from the organ for 5 minutes. Sperm beating was recorded using an inverted microscope (10x phase contrast, Zeiss Primovert) that housed a charge-coupled camera (Zeiss Axiocam ERc 5s). Two sperm tails per sample were analyzed. Recordings were acquired at a rate of 30 frames per second, and beat frequency was analyzed using FIJI and the ImageJ plugin SpermQ [74].
Vertical transmission efficiency
Three separate experiments, performed by different researchers in different insectaries, were implemented to monitor the dynamics of Spiroplasma transmission.
Experiment 1: Individual five-day old male and female flies (each fed twice) were placed together into small tubular cages (height, 12.7 cm; diameter 6.0 cm) and allowed to mate for two days. Males were subsequently removed from the cages (and frozen for future analysis) and pregnant females were fed every other day throughout the course of three GCs. Pupae were collected from each female and housed separately until adult emergence. Genomic DNA from legs of parent flies and all offspring was purified and subjected to PCR analysis to determine Spiroplasma infection status, as indicated above in subsection ‘Laboratory maintenance of Gff and determination of Spiroplasma infection status’.
Experiment 2: Forty 7–9 day old male Gff were released into large mating cages (45x45x45 cm), and 15 minutes later an equal number of 3–5 day old females were introduced. Mating was observed under standard rearing conditions. As soon as copulation began, mating pairs were collected and placed into individual cages (4 cm diameter x 6 cm height) where they were kept together for 24 h. Males were subsequently removed (and conserved in absolute ethanol at -20°C for further analysis), and mated females were pooled together in a larger cage and maintained under normal rearing conditions for 10 days. Finally, pregnant females were again separated into individual cages and maintained under normal conditions over the course of three GCs (~40 days). Individual pupa were collected from each female following each GC and conserved in ethanol at -20°C for further analysis 24h and 72 h post deposition.
Experiment 3: This experiment was performed the same in the same manner as was experiment 2, with the exception that male flies were disposed of after mating.
Statistical analyses
All statistical analyses were carried out using GraphPad Prism (v.9), Microsoft Excel or RStudio (v.1.2.5033 and v.1.3.1073). All statistical tests used, and statistical significance between treatments, and treatments and controls, are indicated on the figures or in their corresponding legends. All sample sizes are provided in corresponding figure legends or are indicated graphically as points on dot plots. Biological replication implies distinct groups of flies were collected on different days.
Supporting information
(DOCX)
The bar diagrams show the significantly enriched GO terms among (A) the up-regulated and (B) the down-regulated genes in GffSpi+ males, and among (C) the up-regulated and (D) the down-regulated genes in GffSpi+ females. The number of genes associated with the corresponding GO terms to the number of genes belonging to that GO term within the entire set of genes in the genome is shown for each bar. The colors associated with the different bars denote the two different GO categories; BP: Biological Process, MF: Molecular Function.
(TIF)
Pgrp-la expression in each sample was normalized relative to geometrical mean of tsetse’s constitutively expressed gapdh and β-tubulin genes. Each dot represents one biological replicate, and bars indicate median values. Statistical significance was determined via students t-test.
(TIF)
0–50 μg aliquots of triolein were mixed with 100 μl of Infinity Triglycerides Reagent (Thermo Scientific) and samples were incubated at 37C for 10 min. Absorbance was measured at 540nm using a BioTek Synergy HT plate reader.
(TIF)
Image generated by Dr. Güler Demirbas-Uzel.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Ten Spiroplasma negative samples and two Spiroplasma positive samples (as determined by endpoint PCR described in the Materials and Methods) were randomly selected for this analysis (describe in the Materials and Methods, subsection ‘Laboratory maintenance of Gff and determination of Spiroplasma infection status’).
(XLSX)
Acknowledgments
We thank Dr. Güler Demirbas-Uzel (Insect Pest Control Laboratory, Joint FAO/IAEA Programme of Nuclear Techniques in Food & Agriculture, Vienna, Austria) for technical assistance with spermathecal fill assays. We are thankful for support from the United Nations, International Atomic Energy Association sponsored Coordinated Research Project entitled ‘Improvement of Colony Management in Insect Mass-Rearing for SIT Applications’. We thank Dr. Peter Takac, at the Slovakian Academy of Sciences, for supplying tsetse pupae.
Data Availability
Most of the data are contained within the manuscript and/or supporting information files. RNA-seq data has been submitted to NCBI, GEO accession number GSE183197 and BioProject accession number PRJNA759598.
Funding Statement
Funding was generously provided by the NIH/NIAID (RO1AI068932), the Ambrose Monell Foundation (monellfoundation.org), and the Li Foundation (lifoundation.org) to SA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Benoit JB, Attardo GM, Baumann AA, Michalkova V, Aksoy S. Adenotrophic viviparity in tsetse flies: potential for population control and as an insect model for lactation. Annu Rev Entomol 2015, 60:351–371. doi: 10.1146/annurev-ento-010814-020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scolari F, Benoit JB, Michalkova V, Aksoy E, Takac P, Abd-Alla AM, et al. The Spermatophore in Glossina morsitans morsitans: Insights into Male Contributions to Reproduction. Sci Rep 2016, 6:20334. doi: 10.1038/srep20334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollock JN. Sperm transfer by spermatophores in Glossina austeni Newstead. Nature 1970, 225:1063–1064. doi: 10.1038/2251063b0 [DOI] [PubMed] [Google Scholar]
- 4.Saunders DS, Dodd CWH. Mating, insemination, and ovulation in the tsetse fly, Glossina morsitans. J. Insect Physiol. 1972, 18:187–198. [Google Scholar]
- 5.Odhiambo T, Kokwaro E, Sequeira L. Histochemical and ultrastructural studies of the male accessory reproductive glands and spermatophore of the tsetse, Glossina morsitans morsitans Westwood. Insect Science and Its Application 1983, 4:227–236. [Google Scholar]
- 6.Attardo GM, Tam N, Parkinson D, Mack LK, Zahnle XJ, Arguellez J, et al. Interpreting Morphological Adaptations Associated with Viviparity in the Tsetse Fly Glossina morsitans (Westwood) by Three-Dimensional Analysis. Insects 2020, 11:651. doi: 10.3390/insects11100651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonomi A, Bassetti F, Gabrieli P, Beadell J, Falchetto M, Scolari F, et al. Polyandry is a common event in wild populations of the tsetse fly Glossina fuscipes fuscipes and may impact population reduction measures. PLoS Negl Trop Dis 2011, 5:e1190. doi: 10.1371/journal.pntd.0001190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vreysen MJ, Saleh K, Mramba F, Parker A, Feldmann U, Dyck VA, et al. Sterile insects to enhance agricultural development: the case of sustainable tsetse eradication on Unguja Island, Zanzibar, using an area-wide integrated pest management approach. PLoS Negl Trop Dis 2014, 8:e2857. doi: 10.1371/journal.pntd.0002857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabayo JP. Aiming to eliminate tsetse from Africa. Trends Parasitol 2002, 18:473–475. doi: 10.1016/s1471-4922(02)02371-1 [DOI] [PubMed] [Google Scholar]
- 10.Bing X, Attardo GM, Vigneron A, Aksoy E, Scolari F, Malacrida A, et al. Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proc Biol Sci 2017, 284:20170360. doi: 10.1098/rspb.2017.0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 2002, 32:402–407. doi: 10.1038/ng986 [DOI] [PubMed] [Google Scholar]
- 12.Snyder AK, Rio RV. "Wigglesworthia morsitans" folate (vitamin B9) biosynthesis contributes to tsetse host fitness. Appl Environ Microbiol 2015, 81:5375–5386. doi: 10.1128/AEM.00553-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rio RV, Symula RE, Wang J, Lohs C, Wu YN, Snyder AK, et al. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. mBio 2012, 3:e00240–11. doi: 10.1128/mBio.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doudoumis V, Blow F, Saridaki A, Augustinos A, Dyer NA, Goodhead I, et al. Challenging the Wigglesworthia, Sodalis, Wolbachia symbiosis dogma in tsetse flies: Spiroplasma is present in both laboratory and natural populations. Sci Rep 2017, 7:4699. doi: 10.1038/s41598-017-04740-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider DI, Saarman N, Onyango MG, Hyseni C, Opiro R, Echodu R, et al. Spatio-temporal distribution of Spiroplasma infections in the tsetse fly (Glossina fuscipes fuscipes) in northern Uganda. PLoS Negl Trop Dis 2019, 13:e0007340. doi: 10.1371/journal.pntd.0007340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukasik P, Guo H, van Asch M, Ferrari J, Godfray HC. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J Evol Biol 2013, 26:2654–2661. doi: 10.1111/jeb.12260 [DOI] [PubMed] [Google Scholar]
- 17.Hamilton PT, Peng F, Boulanger MJ, Perlman SJ. A ribosome-inactivating protein in a Drosophila defensive symbiont. Proc Natl Acad Sci U S A 2016, 113:350–355. doi: 10.1073/pnas.1518648113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballinger MJ, Perlman SJ. The defensive Spiroplasma. Curr Opin Insect Sci 2019, 32:36–41. doi: 10.1016/j.cois.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 19.Perlmutter JI, Bordenstein SR. Microorganisms in the reproductive tissues of arthropods. Nat Rev Microbiol 2020, 18:97–111. doi: 10.1038/s41579-019-0309-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herren JK, Paredes JC, Schupfer F, Arafah K, Bulet P, Lemaitre B. Insect endosymbiont proliferation is limited by lipid availability. Elife 2014, 3:e02964. doi: 10.7554/eLife.02964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst GD, Bandi C, Sacchi L, Cochrane AG, Bertrand D, Karaca I, et al. Adonia variegata (Coleoptera: Coccinellidae) bears maternally inherited flavobacteria that kill males only. Parasitology 1999, 118 (Pt 2):125–134. doi: 10.1017/s0031182098003655 [DOI] [PubMed] [Google Scholar]
- 22.Harumoto T, Lemaitre B. Male-killing toxin in a bacterial symbiont of Drosophila. Nature 2018, 557:252–255. doi: 10.1038/s41586-018-0086-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harumoto T, Fukatsu T, Lemaitre B. Common and unique strategies of male killing evolved in two distinct Drosophila symbionts. Proc Biol Sci 2018, 285:20172167. doi: 10.1098/rspb.2017.2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B 1995, 57:289–300. [Google Scholar]
- 25.Engelstadter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annual Review of Ecology Evolution and Systematics 2009, 40:127–149. [Google Scholar]
- 26.Harumoto T, Anbutsu H, Lemaitre B, Fukatsu T. Male-killing symbiont damages host’s dosage-compensated sex chromosome to induce embryonic apoptosis. Nat Commun 2016, 7:12781. doi: 10.1038/ncomms12781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet 2007, 8:689–698. doi: 10.1038/nrg2167 [DOI] [PubMed] [Google Scholar]
- 28.Mank JE. The transcriptional architecture of phenotypic dimorphism. Nat Ecol Evol 2017, 1:6. doi: 10.1038/s41559-016-0006 [DOI] [PubMed] [Google Scholar]
- 29.Herren JK, Lemaitre B. Spiroplasma and host immunity: activation of humoral immune responses increases endosymbiont load and susceptibility to certain Gram-negative bacterial pathogens in Drosophila melanogaster. Cell Microbiol 2011, 13:1385–1396. doi: 10.1111/j.1462-5822.2011.01627.x [DOI] [PubMed] [Google Scholar]
- 30.Dias RO, Cardoso C, Pimentel AC, Damasceno TF, Ferreira C, Terra WR. The roles of mucus-forming mucins, peritrophins and peritrophins with mucin domains in the insect midgut. Insect Mol Biol 2018, 27:46–60. doi: 10.1111/imb.12340 [DOI] [PubMed] [Google Scholar]
- 31.Attardo GM, Abd-Alla AMM, Acosta-Serrano A, Allen JE, Bateta R, Benoit JB, et al. Comparative genomic analysis of six Glossina genomes, vectors of African trypanosomes. Genome Biol 2019, 20:187. doi: 10.1186/s13059-019-1768-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nogge G. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia 1976, 32:995–996. doi: 10.1007/BF01933932 [DOI] [PubMed] [Google Scholar]
- 33.Hill P, Saunders DS, Campbell JA. Letter: The production of "symbiont-free" Glossina morsitans and an associated loss of female fertility. Trans R Soc Trop Med Hyg 1973, 67:727–728. doi: 10.1016/0035-9203(73)90051-5 [DOI] [PubMed] [Google Scholar]
- 34.Michalkova V, Benoit JB, Weiss BL, Attardo GM, Aksoy S. Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl Environ Microbiol 2014, 80:5844–5853. doi: 10.1128/AEM.01150-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cmelik SH, Hurrell DP, Lunat M. Lipid content and composition of the tse-tse fly, Glossina morsitans Westwood. Comp Biochem Physiol 1969, 31:65–78. doi: 10.1016/0010-406x(69)92169-0 [DOI] [PubMed] [Google Scholar]
- 36.Attardo GM, Benoit JB, Michalkova V, Yang G, Roller L, Bohova J, et al. Analysis of lipolysis underlying lactation in the tsetse fly, Glossina morsitans. Insect Biochem Mol Biol 2012, 42:360–370. doi: 10.1016/j.ibmb.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benoit JB, Yang G, Krause TB, Patrick KR, Aksoy S, Attardo GM. Lipophorin acts as a shuttle of lipids to the milk gland during tsetse fly pregnancy. J Insect Physiol 2011, 57:1553–1561. doi: 10.1016/j.jinsphys.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sepil I, Hopkins BR, Dean R, Bath E, Friedman S, Swanson B, et al. Male reproductive aging arises via multifaceted mating-dependent sperm and seminal proteome declines, but is postponable in Drosophila. Proc Natl Acad Sci U S A 2020, 117:17094–17103. doi: 10.1073/pnas.2009053117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature 1998, 396:572–575. doi: 10.1038/25126 [DOI] [PubMed] [Google Scholar]
- 40.Yeh SD, Do T, Chan C, Cordova A, Carranza F, Yamamoto EA, et al. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci U S A 2012, 109:2043–2048. doi: 10.1073/pnas.1121327109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss BL, Maltz MA, Vigneron A, Wu Y, Walter KS, O’Neill MB, et al. Colonization of the tsetse fly midgut with commensal Kosakonia cowanii Zambiae inhibits trypanosome infection establishment. PLoS Pathog 2019, 15:e1007470. doi: 10.1371/journal.ppat.1007470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayaswal V, Jimenez J, Magie R, Nguyen K, Clifton B, Yeh S, et al. A species-specific multigene family mediates differential sperm displacement in Drosophila melanogaster. Evolution 2018, 72:399–403. doi: 10.1111/evo.13417 [DOI] [PubMed] [Google Scholar]
- 43.Ingleby FC, Flis I, Morrow EH. Sex-biased gene expression and sexual conflict throughout development. Cold Spring Harb Perspect Biol 2014, 7:a017632. doi: 10.1101/cshperspect.a017632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 2003, 299:697–700. doi: 10.1126/science.1079190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang G, Attardo GM, Lohs C, Aksoy S. Molecular characterization of two novel milk proteins in the tsetse fly (Glossina morsitans morsitans). Insect Mol Biol 2010, 19:253–262. doi: 10.1111/j.1365-2583.2009.00987.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu C, Rio RV, Medlock J, Haines LR, Nayduch D, Savage AF, et al. Infections with immunogenic trypanosomes reduce tsetse reproductive fitness: potential impact of different parasite strains on vector population structure. PLoS Negl Trop Dis 2008, 2:e192. doi: 10.1371/journal.pntd.0000192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeh SD, Do T, Abbassi M, Ranz JM. Functional relevance of the newly evolved sperm dynein intermediate chain multigene family in Drosophila melanogaster males. Commun Integr Biol 2012, 5:462–465. doi: 10.4161/cib.21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Civetta A, Ranz JM. Genetic factors influencing sperm competition. Front Genet 2019, 10:820. doi: 10.3389/fgene.2019.00820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh SR, Singh BN, Hoenigsberg HF. Female remating, sperm competition and sexual selection in Drosophila. Genet Mol Res 2002, 1:178–215. [PubMed] [Google Scholar]
- 50.Shropshire JD, Leigh B, Bordenstein SR. Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years? Elife 2020, 9:e61989. doi: 10.7554/eLife.61989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathe-Hubert H, Kaech H, Ganesanandamoorthy P, Vorburger C. Evolutionary costs and benefits of infection with diverse strains of Spiroplasma in pea aphids. Evolution 2019, 73:1466–1481. doi: 10.1111/evo.13740 [DOI] [PubMed] [Google Scholar]
- 52.Rio RVM, Jozwick AKS, Savage AF, Sabet A, Vigneron A, Wu Y, et al. Mutualist-provisioned resources impact vector competency. mBio 2019, 10:e00018–19. doi: 10.1128/mBio.00018-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snyder AK, Deberry JW, Runyen-Janecky L, Rio RV. Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc Biol Sci 2010, 277:2389–2397. doi: 10.1098/rspb.2010.0364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snyder AK, Rio RV. Interwoven biology of the tsetse holobiont. J Bacteriol 2013, 195:4322–4330. doi: 10.1128/JB.00487-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr Biol 2019, 29:R485–R495. doi: 10.1016/j.cub.2019.03.032 [DOI] [PubMed] [Google Scholar]
- 56.Paredes JC, Herren JK, Schupfer F, Lemaitre B. The role of lipid competition for endosymbiont-mediated protection against parasitoid wasps in Drosophila. mBio 2016, 7:e01006–16. doi: 10.1128/mBio.01006-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol 2017, 25:375–390. doi: 10.1016/j.tim.2017.02.014 [DOI] [PubMed] [Google Scholar]
- 58.Percoma L, Sow A, Pagabeleguem S, Dicko AH, Serdebeogo O, Ouedraogo M, et al. Impact of an integrated control campaign on tsetse populations in Burkina Faso. Parasit Vectors 2018, 11:270. doi: 10.1186/s13071-017-2609-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vreysen MJ, Seck MT, Sall B, Bouyer J. Tsetse flies: their biology and control using area-wide integrated pest management approaches. J Invertebr Pathol 2013, 112 Suppl:S15–25. doi: 10.1016/j.jip.2012.07.026 [DOI] [PubMed] [Google Scholar]
- 60.Lees RS, Gilles JR, Hendrichs J, Vreysen MJ, Bourtzis K. Back to the future: the sterile insect technique against mosquito disease vectors. Curr Opin Insect Sci 2015, 10:156–162. doi: 10.1016/j.cois.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 61.McGraw EA, O’Neill S. Beyond insecticides: new thinking on an ancient problem. Nat Rev Microbiol 2013, 11:181–193. doi: 10.1038/nrmicro2968 [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015, 12:357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019, 37:907–915. doi: 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, et al. The UCC genome browser database: update 2006. Nucleic Acids Res 2006, 34:D590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vera Alvarez R, Pongor LS, Marino-Ramirez L, Landsman D. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 2019, 35:1960–1962. doi: 10.1093/bioinformatics/bty896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexa A, Rahnenfuhrer, J. topGO: Enrichrichment analysis for gene ontology. R package version 2.42.0. Edited by; 2020.
- 69.Moloo SK. An artificial feeding technique for Glossina. Parasitology 1971, 63:507–512. doi: 10.1017/s0031182000080021 [DOI] [PubMed] [Google Scholar]
- 70.Benoit JB, Vigneron A, Broderick A, Wu Y, Sun JS, Carlson JR, et al. Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. Elife 2017, 6:e19535. doi: 10.7554/eLife.19535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agbu P, Cassidy JJ, Braverman J, Jacobson A, Carthew RW. MicroRNA miR-7 Regulates Secretion of Insulin-Like Peptides. Endocrinology 2020, 161:bqz040. doi: 10.1210/endocr/bqz040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mutika GN, Kabore I, Parker AG, Vreysen MJ. Storage of male Glossina palpalis gambiensis pupae at low temperature: effect on emergence, mating and survival. Parasit Vectors 2014, 7:465. doi: 10.1186/s13071-014-0465-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mutika GN, Marin C, Parker AG, Boucias DG, Vreysen MJ, Abd-Alla AM. Impact of salivary gland hypertrophy virus infection on the mating success of male Glossina pallidipes: consequences for the sterile insect technique. PLoS One 2012, 7:e42188. doi: 10.1371/journal.pone.0042188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen JN, Rassmann S, Jikeli JF, Wachten D. SpermQ-A simple analysis software to comprehensively study flagellar beating and sperm steering. Cells 2018, 8:10. doi: 10.3390/cells8010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mangiafico S. Rcompanion: functions to support extension education program evaluation. Edited by; 2020. vol R package version 2.3.26.]
- 76.Ogle DH, Wheeler, P., Dinno, A. FSA: fisheries stock analysis. R package version 0.8.32. Edited by; 2021.