Abstract
The composition of the skin microbiome varies widely among individuals sampled at the same body site. A key question is which molecular factors determine strain-level variability within sub-ecosystems of the skin. We used a genomics-guided approach to identify an antibacterial biosynthetic gene cluster in Cutibacterium acnes (formerly Propionibacterium acnes) that is widely distributed across individuals and skin sites. Experimental characterization of this cluster identified of a new thiopeptide antibiotic, cutimycin. Analysis of individual human skin hair follicles revealed that cutimycin plays a role in the chemical ecology of follicular microbiota and in colonization resistance against Staphylococcus species.
One Sentence Summary:
Cutimycin, a thiopeptide antibiotic produced by a widespread skin commensal, reduces Staphylococcus colonization of human follicles.
Niche competition among resident microbiota is postulated to influence skin microbial community composition via colonization resistance. In conjunction with host environmental factors (e.g., desiccation, low pH, high salt and high lipid concentrations (1)) and the host immune response (2), this results in a distinctive skin microbiota with variations among sites that are characterized as being predominantly sebaceous, moist or dry. Species of Staphylococcus, Corynebacterium and Cutibacterium (formerly the cutaneous Propionibacterium (3)) are among the most prevalent and abundant members of the human skin microbiota.
Recent studies have begun to uncover mechanisms of competition among bacterial species that shape microbiota composition across human skin (4). Small molecules are one means by which bacteria interact with each other and their environment. The genes required to produce these small molecules co-localize in biosynthetic gene clusters (BGCs) (5). BGCs are abundant in the human microbiome (6), but relatively few have a proven function (6–8). For example, the nasal and skin colonizer Staphylococcus lugdunensis produces a nonribosomal peptide, lugdunin, that inhibits growth of and colonization by S. aureus (7). Similarly, some strains of coagulase-negative Staphylococcus produce antimicrobial peptides that kill S. aureus. These strains are reduced in atopic dermatitis when S. aureus predominates and their expansion decreases skin colonization by S. aureus (8). Other mechanisms of bacterial competition on skin and in the nostrils include protease activity (9), disruption of Staphylococcus quorum sensing (10, 11), competition for iron (12), bacterial release of antimicrobial free fatty acids from host triacylglycerols (13), and niche competition mediated by the host (14). Nonetheless, the examples of lugdunin (7) and Staphylococcus-derived lanthipeptides (8) highlight the key roles of BGCs and their products for members of the human microbiota, and the need to identify and characterize secreted antibacterial compounds from other members of human skin and nasal microbiota (15, 16).
One widely distributed family of BGCs in the human microbiome is predicted to encode a sub-class of ribosomally synthesized, post-translationally modified peptides (RiPPs) known as thiopeptides (6). Many thiopeptides, such as berninamycin from Streptomyces bernensis and the semi-synthetic LFF571, which was used in a Phase 2 clinical study, have potent anti-staphylococcal activity via inhibition of protein synthesis (17, 18). We noted that among the most widely distributed thiopeptide BGCs in the human microbiome (6), one was present in 8 of 219 (3.7%) sequenced isolates of Cutibacterium species (Table S1), all Cutibacterium acnes. We and others predict that the product of this BGC (ppa0859–0866 in C. acnes strain KPA171202; Fig. 1A) is a thiopeptide (19, 20) structurally related to the antibiotic berninamycin (21, 22).
Fig. 1.
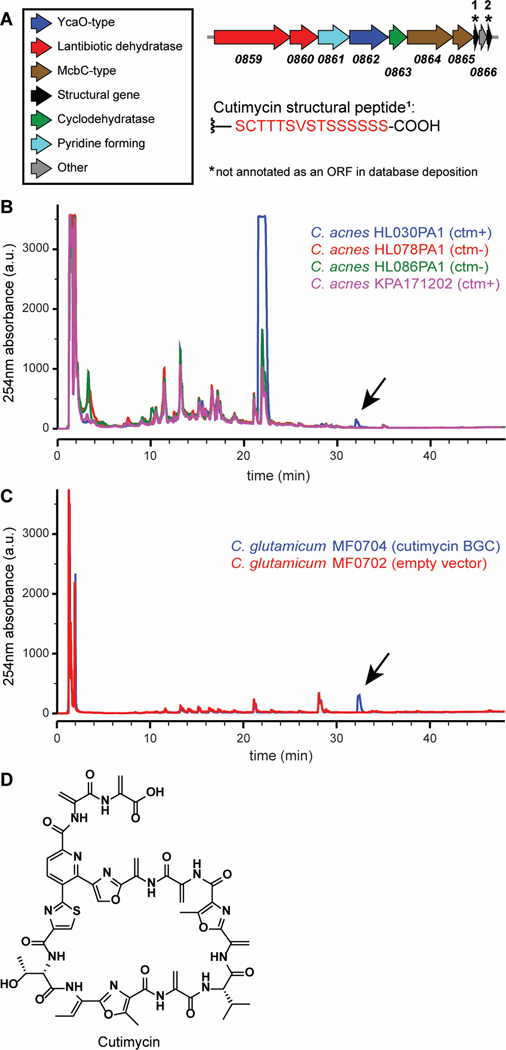
Detection of cutimycin (ctm), the product of BGC ppa0859–0866, in native and heterologous hosts. A) Arrow representation of the ppa0859–0866 BGC from C. acnes KPA171202. B) HPLC profiles for crude ethyl acetate extracts of select ppa0859–0866 BGC+ and - C. acnes strains. The thiopeptide product of ppa0859–0866, dubbed cutimycin, elutes at 73.5% MeCN as visible in the blue trace from C. acnes HL030PA1. Arrow indicates the cutimycin peak. C) Comparison of HPLC profiles of crude cell extracts of C. glutamicum hosting the cutimycin BGC on a plasmid (blue) versus hosting the empty vector control (red). Arrow indicates the cutimycin peak. D) Structure of the Cutibacterium-produced thiopeptide cutimycin.
Cutibacterium species are particularly well adapted to life on human skin with their ability to thrive in the lipid-rich environment of the human skin hair follicle, with its associated sebaceous gland. Hypothesizing that the small molecule product of this predicted cutimycin BGC plays a role in skin microbial community composition, we set out to identify this compound and determine its structure.
Based on its computationally predicted similarity to berninamycin (23), we hypothesized that the Cutibacterium BGC encodes a thiopeptide that C. acnes uses to target Staphylococcus species (phylum Firmicutes) in their shared habitats on human skin, including the skin inside the nostrils. To test for production of a thiopeptide, we selected phylogenetically distinct C. acnes strains that differ with respect to the presence or absence of BGCs. By analyzing crude culture extracts from these strains by HPLC, we observed that the thiopeptide BGC+ isolate C. acnes HL030PA1 produces a compound with a retention time and UV absorption spectrum similar to that of the thiopeptide berninamycin (Fig. 1B, blue trace). To investigate whether production of this compound affects competition between C. acnes and Staphylococcus, we performed an in vitro competition assay with a producing and a nonproducing strain of C. acnes each versus S. aureus UAMS-01 under low pH, high lipid conditions, mimicking the natural follicle environment (Fig. S1). We observed a zone of inhibition for C. acnes HL030PA1, but not for C. acnes KPA17102, consistent with the detection of a berninamycin-like peak from HL030PA1 (Fig. 1B, blue trace) and not from KPL171202 (Fig. 1B, pink trace) when grown in vitro.
To determine whether the BGC from C. acnes HL030PA1 was sufficient to produce the observed compound, we expressed this cluster heterologously in a related Actinobacterium, Corynebacterium glutamicum. We analyzed organic extracts prepared from cell pellets of the wild-type and BGC+ strains of C. glutamicum, observing that the latter but not the former contained a molecule identical to the one produced by C. acnes HL030PA1 (Fig. 1C). Unlike in the cell pellet, we did not detect the molecule in an extract of the culture supernatant of the BGC+ C. glutamicum. These data establish that the Cutibacterium BGC is sufficient for the biosynthesis of the molecule detected, but not its export.
Because of higher production yields and the ability to grow under aerobic conditions, we scaled up cultivation of the cutimycin BGC+ C. glutamicum and purified the thiopeptide (Fig. S2). Determination of the accurate mass at 1131.3411 m/z allowed prediction of a formula of C51H51N14O15S+, with a predicted monoisotopic mass of 1131.3373 (Δtheoretical = 3.4 ppm) (Figs. S3A & S4). This is in accordance with the compound detected from C. acnes HL030PA1, with an accurate mass at 1131.3364 m/z (Δtheoretical = 0.8 ppm) (Fig. S3B). The planar structure of the thiopeptide was solved de novo based on 1D and 2D NMR experiments and HRMSe and is described in depth in the supplemental information (Figs. S5-S12; Table S2). The configuration of each stereogenic center was determined using Marfey’s analysis (Fig. S13). We defined atom position numbering and the amino acid numbering (Figs. 1D and S14) following the convention set in the previous publication of the structure of berninamycin for ease of comparison (24). We assigned the trivial name cutimycin to this Cutibacterium-derived thiopeptide.
Cutimycin has potent activity in vitro against Staphylococcus but not against commensal Actinobacteria from skin. Based on cutimycin’s structural similarity to berninamycin and LFF571 (Fig. S15), and results of the in vitro competition assay (Fig. S1), we hypothesized it would display anti-staphylococcal activity but would lack activity against common Actinobacteria skin commensals. To test this, we determined the MICs for cutimycin and berninamycin against a selection of species commonly found on skin sites, including the nostrils (Table S3). Cutimycin exhibited potent inhibition of the USA300 community associated methicillin-resistant S. aureus strain NRS384 (MIC 0.2 μM), as well as strains of Staphylococcus epidermidis. In contrast, a panel of other C. acnes strains, with and without the cutimycin BGC, plus two common skin and two common nasal Corynebacterium species (phylum Actinobacteria) displayed increased resistance to cutimycin with MICs ≥ 3.2 μM. These data led us to hypothesize that cutimycin favors the growth of resident skin Actinobacteria, including C. acnes, over that of common skin staphylococcal species.
Cocultivation with susceptible Staphylococcus strains increased transcription of the cutimycin BGC. The production of secondary metabolites can be costly and BGCs often contain regulatory mechanisms for inducible rather than constitutive expression. Although we did not identify an obvious regulatory element in the cutimycin BGC, we hypothesized that cutimycin-susceptible species would induce transcription of cutimycin, whereas resistant species would not. To test this, we assayed transcription of the cutimycin BGC during in vitro cocultivation of C. acnes with S. aureus, S. epidermidis (both susceptible) or Corynebacterium striatum (resistant) compared to C. acnes monocultivation. For this qRT-PCR-based assay, we used C. acnes KPA171202 because the encoded cutimycin BGC (25) is transcribed during exponential growth in vitro (19) but production of cutimycin was not detectable in vitro (Fig. 1B) simplifying assay interpretation. Compared to when C. acnes was grown alone, transcripts of ppa0860 from the cutimycin BGC in C. acnes KPA171202 increased twofold in the presence of S. aureus or S. epidermidis but decreased in presence of C. striatum (0.16 fold) (Fig. 2). These results indicate that cutimycin transcription is selectively increased by the presence of Staphylococcus targets. To determine whether cutimycin is produced in vivo in C. acnes’ natural habitat, we used mass spectrometry to analyze pooled content from hair follicles of intact skin of individual adult volunteers. Due to the small quantity of each sample (25–80 follicles), it was not possible to observe cutimycin using untargeted mass spectrometry (26). Using targeted mass spectrometry, we detected cutimycin in 28 % of the samples (Fig. S16, Table S4). Based on an external standard and a typical hair follicle volume of 0.2 mm3 (27), the concentration of cutimycin was estimated to be to be 0.97 +/− 0.12 μM in the samples where the molecule was detected. However, the local concentrations likely exceed this value as cutimycin-producing strains are unlikely to be evenly distributed throughout the follicle. Based on these data, we hypothesized that cutimycin plays a role in modulating amounts of Staphylococcus in the context of human skin colonization.
Fig. 2.
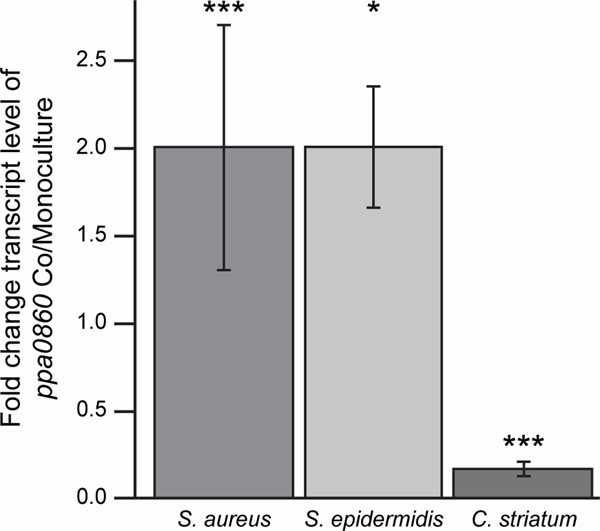
The C. acnes ppa0860 transcript increases during cocultivation with Staphylococcus. The ratio of qRT-PCR results for ppa0860 from co- vs. monoculture with S. aureus (Sau, n=8, p=0.0001), S. epidermidis (Sep, n=3, p=0.047) and C. striatum (Cst, n=3, p=0.00005). Paired t-test. Error bars are SD.
The cutimycin BGC is widely distributed on human skin but present in only a subset of strains at each body site. Humans harbor an abundance of Cutibacterium on their skin, with C. acnes being the overwhelmingly predominant species. In the absence of an animal model for C. acnes colonization of skin hair follicles, we began by examining isolate and metagenomic sequence data to explore a potential role for the cutimycin BGC in modulating the composition of the human skin community. The cutimycin BGC is present in the genomes of only about 4% (8/219 in Table S1) of sequenced C. acnes isolates, most of which belong to a common clade including strain KPA171202 (supplemental text). However, in a longitudinal high-resolution skin metagenomic dataset from 12 healthy volunteers (28), the cutimycin BGC could be detected in 11/12 individuals. When it was present in an individual, it could be identified across sampled sites stably over time (orange bars in Figs. 3 and S17). In spite of its broad distribution, in the majority of samples < 30% of the total C. acnes contained the BGC. C. acnes is also a common member of the human nostril microbiome, which is assayed from the skin of the nasal vestibule (29, 30). Because this site was absent in our metagenomic dataset, we assayed for the cutimycin BGC in nostril metagenomic data from the Human Microbiome Project, finding it in 8/75 samples (10.7%). From the 12 intensively sampled volunteers, we also examined the distribution of four other C. acnes BGCs predicted to code for interesting bioactive compounds (Table S5). Similar to the cutimycin BGC, we observed a wide distribution with strain-level variation for these other Cutibacterium BGCs on adult human skin (Figs. 3 and S17). There were, however, no significant correlations between the absence/presence of the cutimycin BGC, or any other predicted C. acnes BGC, and the abundance of specific skin commensals in these 12 volunteers. At first these results seemed incongruous with cutimycin’s anti-staphylococcal activity. However, we reasoned that the spatial scale of a skin swab is much larger than the scale at which C. acnes interacts with neighboring bacterial species. Therefore, we hypothesized that a much finer spatial resolution was required to test whether cutimycin has an effect on microbial community composition. Because C. acnes is a known resident of human skin follicles, we next assayed the distribution of the cutimycin BGC and its potential impact on community composition at the level of individual human skin hair follicles.
Fig. 3.
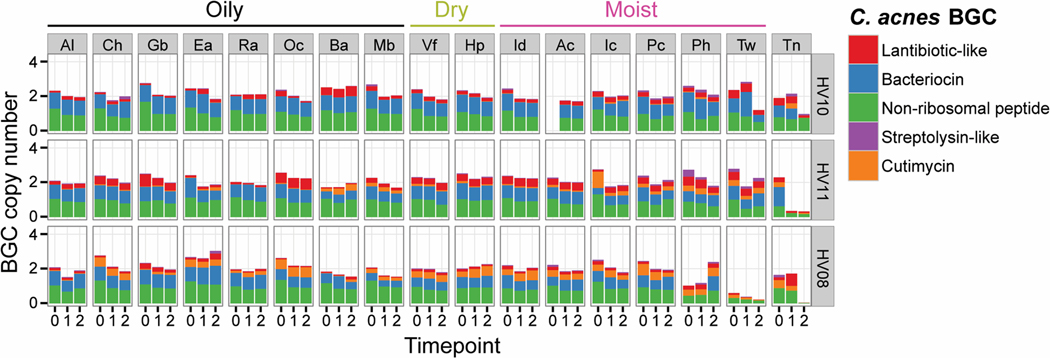
Example of spatial and temporal distribution of C. acnes BGCs in the skin metagenomes of three healthy individuals with low, medium or high relative abundance of the cutimycin BGC across 17 skin sites. Rows group samples from a single volunteer (coded Healthy Volunteer (HV) 01–12), columns represent samples across a specific body site (coded Ac through Vf). Cells contain bar graphs for each of the three time points, depicting the copy number of the C. acnes BGCs, which are standardized by comparing against 13 C. acnes housekeeping genes. The BGC types, bacteriocin, lantibiotic, non-ribosomal peptide, streptolysin-like peptide and cutimycin are color coded and their abundances are stacked on top of each other (a colored bar with a height of 1 means all of the C. acnes in the sample harbor that BGC, whereas a height of 0 means the BGC is absent in all of the C. acnes). Al = alar crease, Ch = cheek, Gb = glabella, Ea = external auditory canal, Ra = retroauricular crease, Oc = occiput, Ba = back, Mb = manubrium, Vf = volar forearm, Hp = hypothenar palm, Id = interdigital web space, Ac = antecubital fossa, Ic = inguinal crease, Pc = popliteal fossa, Ph = plantar heel, Tw = toe web space, Tn = toenail (see also diagram in Fig. S17A).
Skin follicles containing the cutimycin BGC have a higher ratio of C. acnes to S. epidermidis (Fig. 4). To address the distribution question across individual skin hair follicles, we sampled the contents of 6–10 healthy follicles from the outer surface of the nose of 16 human volunteers (Figs. 4A, 4B and Table S6). For each follicle, we quantified colony forming units (CFUs) to assay community composition and performed PCR of Cutibacterium isolates to determine the presence or absence of the cutimycin BGC. From 156 individual follicles, we identified three bacterial species by 16S rRNA gene sequencing of representative colonies: C. acnes, Cutibacterium granulosum, and S. epidermidis. These three species are readily cultivable and were easily distinguished by colony morphology (Fig. 4C), permitting quantification of each in follicular content (Fig. S18). Since follicles differed in the total number of CFUs (possibly due to size, sampling or true variation), we measured the ratio of C. acnes/S. epidermidis in follicles with and without the cutimycin BGC (Figs. 4D, 4E). Strikingly, follicles that contained C. acnes strains that were cutimycin BGC-positive had, on average, a greater than an order of magnitude higher C. acnes/S. epidermidis ratio than those that were cutimycin BGC negative (Wilcoxon signed-rank test, p=0.006) (Figs. 4D, 4E). This finding suggests a role for cutimycin in modulating the composition of the skin microbiome, and it highlights that the characteristic spatial scale of this interaction is tiny – that of an individual follicle.
Fig. 4.
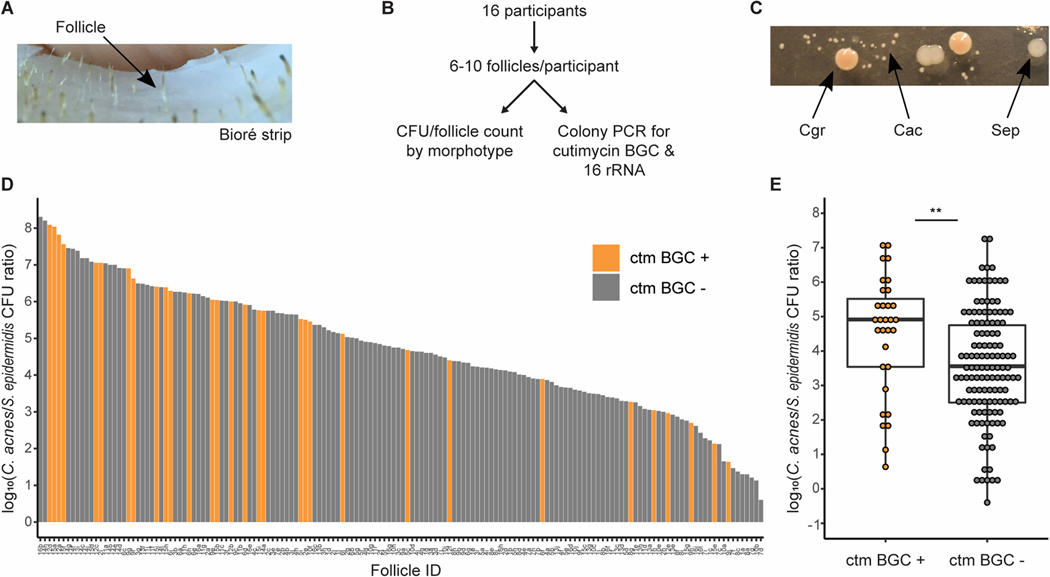
The presence of the cutimycin BGC has a significant impact on the C. acnes/S. epidermidis ratio in human skin hair follicles. (A) Picture of Bioré pore strip with harvested follicular content. (B) Schematic of the experimental design. (C) Picture of culture isolates from follicular content showing the distinct colony morphologies of C. acnes (Cac), S. epidermidis (Sep) and C. granulosum (Cgr). D) Ranked histogram of the ratio of C. acnes/S. epidermidis from each sampled follicle by the presence and absence of the cutimycin BGC. The Follicle ID denotes a participant by a number from 1–16 and pores by a letter from a-j (e.g. 10b). E) Box plots of the impact of the cutimycin (ctm) BGC presence (+) or absence (–) on the log10 ratio of C. acnes/S. epidermidis CFUs from individual human skin follicular plugs (n=156, each follicle is represented by a dot) collected from 16 participants in total. For statistical analysis, data were pooled based on the assumption that follicles within an individual are independent. There was a statistically significant difference in Cac/Sep (Wilcoxon signed-rank test, p=0.006) between ctm+ and ctm- samples.
We also observed some cutimycin BGC-negative follicles with a high C. acnes/S. epidermidis ratio. One possible explanation is that alternative C. acnes-produced anti-Staphylococcus activities are present within these follicles, possibly encoded by one of the other predicted C. acnes BGCs (Figs. 3 and S17). However, other bacterial molecules/mechanisms could be mediating the competitive interactions, e.g., nutrient competition, toxic primary metabolites (31) or antimicrobial free fatty acids released from host triacylglycerols (32), as well as possible host-mediated effects.
In addition to inferences regarding the impact of cutimycin on community composition, these data on the presence or absence of the cutimycin BGC in individual human skin follicles indicate that follicles might be colonized by at least 2 different C. acnes strains and that C. acnes strain-level colonization is punctate, sometimes varying from follicle to follicle in an individual (Fig. 4D; Table S6). Thus, future exploration of the effect of C. acnes small molecules on human skin microbiota requires investigating at the fine-scale resolution of the individual skin follicle (Figs. 4A, 4C; Table S6).
Here, we have identified a molecular mechanism of niche competition between two of the most common members of the human skin microbiome: C. acnes and Staphylococcus species. With this work, cutimycin becomes one of the few BGCs from the human skin microbiome with a known molecular function. Our elucidation of cutimycin’s function will facilitate exploration into possible clinical applications of cutimycin, or cutimycin-producing C. acnes strains, to selectively inhibit Staphylococcus colonization, while leaving commensal Actinobacteria undisturbed. S. aureus nasal colonization is a risk for invasive infection (33) and, in the absence of an effective anti-staphylococcal vaccine (34), there is a need to identify strains of beneficial bacteria and their bioactive products that could be used to generate nasal and skin microbiota resistant to colonization by S. aureus. Such approaches might also have application in preventing or treating skin diseases that include a shift in either microbiome composition, such as in atopic dermatitis flares, or in host-microbe interactions, such as acne vulgaris (35, 36).
One limitation of this study is that natural microbial ecosystems are governed by a complex combination of multiple factors. Although C. acnes cutimycin production significantly contributes to bacterial community composition in the human follicle, our data indicate that additional bacterial or host mechanisms are at play. Another limitation pertains to the differences between in vitro testing conditions and the natural skin environment. The interpretation of MICs might differ for bacteria residing in an environment with high lipid content or growing as a biofilm. Also, the regulation of the cutimycin BGC might be influenced by both other colonizing bacteria as well as host factors. These limitations offer opportunities for future studies and would greatly benefit from the availability of an animal model to study C. acnes skin hair follicle colonization.
In conclusion, this successful elucidation of the cutimycin-mediated competition of C. acnes with Staphylococcus in vivo on humans demonstrates the power of combining systems-level approaches (e.g., in silico mining to characterize an antibiotic from the skin microbiome) with reductionist approaches (e.g., the in vitro cultivation of known bacterial colonizers of human skin follicles to explore of the impact of cutimycin at the fine-scale resolution of the individual skin follicle) to discover new mechanisms that underlie the ecology of the microbiome.
Supplementary Material
Competition assay between C. acnes and S. aureus under conditions mimicking the natural follicle environment.
HPLC chromatogram of cutimycin purification.
Low energy mass spectrum for cutimycin (calculated ([M+H]+ 1131.3374 for C51H51N14O15S+) isolated from A) the heterologous C. glutamicum producer ([M+H]+ 1131.3411) and B) from C. acnes HL030PA1 ([M+H]+ 1131.3364).
High energy mass spectrum for cutimycin labeled with predicted fragments by Unifi.
1H NMR Spectrum of cutimycin taken in DMSO-d6 at 600 MHz.
13C NMR Spectrum of cutimycin taken in DMSO-d6 at 151 MHz.
1H-1H COSY NMR in DMSO-d6.
1H-13C HSQC NMR in DMSO-d6.
1H-13C HMBC NMR in DMSO-d6.
Zoom in of 1H-13C HMBC NMR in DMSO-d6 identifying dehydroalanine residues through presence of diastereotopic terminal alkene protons (position 20, δH 5.74, 5.75 and position 24 δH 5.73, 6.35) with HMBC correlations to carbon atoms 19 and 23 respectively.
Zoom in of 1H-13C HMBC NMR in DMSO-d6 showing the weak four-bond HMBC correlation from position 13 to quaternary carbon 16 at δC 139. 13.
Marfey’s analysis of cutimycin. Individual amino acid Marfey’s analysis derivatives, cutimycin hydrolysis product derivatives, and coinjections.
The subunits of cutimycin with key HMBC (arrows) and COSY (bold bonds) correlations.
Structure of cutimycin
The structures of A) cutimycin, B) berninamycin A, and C) LFF-571.
Mass spectrometric detection of cutimycin from pooled human follicular content (n=60) for one of the tested samples.
Box plots of the impact of the cutimycin BGC presence/absence on C. acnes, C. granulosum and S. epidermidis CFUs in human skin follicles.
Linear depiction of cutimycin with the key mass spec fragments labeled (Table S9).
Metagenomic analysis of C. acnes BGCs at 17 skin sites from sites from 12 healthy participants.
The key fragments confirming the order of the amino acids in cutimycin.
Cutimycin BGC presence/absence in 219 Cutibacterium genomes
NMR Data of cutimycin taken in DMSO-d6 at 600 MHz and 151 MHz.
Minimal inhibitory concentrations for cutimycin and bernamycin
Cutimycin in human follicular samples has been detected in 28% of all samples across two separate experiments.
Locus tags for the C. acnes BGCs assayed for in skin metagenomic data.
Cutimycin BGC presence/absence; the CFUs of C. acnes, C. granulosum and S. epidermidis; and the C. acnes/S. epidermidis CFU ratio in the content of individual human skin follicles.
Bacterial strains and plasmids used in this study.
Primers used in this study.
Acknowledgments:
We are deeply grateful to the participants who provided sebaceous samples from their skin hair follicles.
Funding: R01 AI101018 (KPL and MAF), U41 AT008718 (RGL), NSERC Discovery RGPIN-2016-03962 (RGL), NIH grants DP1 DK113598 (MAF), R01 DK110174 (MAF); an HHMI-Simons Faculty Scholars Award (MAF), an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Foundation (MAF), JC is supported by Seed Funding from the Cleveland Clinic Foundation.
Footnotes
Competing Interests: Declaration of Interests: A.L.B. and K.L.K. are currently employees of Genentech Inc. Management/Advisory/Consulting Activities: M.A.F. is a co-founder and director of Federation Bio, a company developing microbiome-based therapeutics. A.A.A. is currently a consultant for Ometa labs LLC. Financial Interests: M.A.F. is a co-founder and director of Federation Bio, a company developing microbiome-based therapeutics.
Data and materials availability: All data are available in the main text or the supplementary materials. Bacterial strains that are not available from a stock center will be made available via an MTA upon written request to K.P.L.
References and Notes:
- 1.Chen YE, Fischbach MA, Belkaid Y, Skin microbiota-host interactions. Nature 553, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd AL, Belkaid Y, Segre JA, The human skin microbiome. Nat Rev Microbiol 16, 143–155 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Scholz CF, Kilian M, The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol 66, 4422–4432 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Cogen AL, Nizet V, Gallo RL, Skin microbiota: a source of disease or defence? Br J Dermatol 158, 442–455 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA, Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA, A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brotz-Oesterhelt H, Grond S, Peschel A, Krismer B, Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, Shafiq F, Kotol PF, Bouslimani A, Melnik AV, Latif H, Kim JN, Lockhart A, Artis K, David G, Taylor P, Streib J, Dorrestein PC, Grier A, Gill SR, Zengler K, Hata TR, Leung DY, Gallo RL, Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y, Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, Van Dyke MJ, Cech NB, Horswill AR, Coagulase-Negative Staphylococcal Strain Prevents Staphylococcus aureus Colonization and Skin Infection by Blocking Quorum Sensing. Cell Host Microbe 22, 746–756 e745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP, Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front Microbiol 7, 1230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbendieck RM, May DS, Chevrette MG, Temkin MI, Wendt-Pienkowski E, Cagnazzo J, Carlson CM, Gern JE, Currie CR, Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl Environ Microbiol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP, Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. MBio 7, e01725–01715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto M, Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 5, 183–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janek D, Zipperer A, Kulik A, Krismer B, Peschel A, High Frequency and Diversity of Antimicrobial Activities Produced by Nasal Staphylococcus Strains against Bacterial Competitors. PLoS Pathog 12, e1005812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Sullivan JN, Rea MC, O’Connor PM, Hill C, Ross RP, Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol Ecol 95, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Citron DM, Tyrrell KL, Merriam CV, Goldstein EJ, Comparative in vitro activities of LFF571 against Clostridium difficile and 630 other intestinal strains of aerobic and anaerobic bacteria. Antimicrob Agents Chemother 56, 2493–2503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullane K, Lee C, Bressler A, Buitrago M, Weiss K, Dabovic K, Praestgaard J, Leeds JA, Blais J, Pertel P, Multicenter, randomized clinical trial to compare the safety and efficacy of LFF571 and vancomycin for Clostridium difficile infections. Antimicrob Agents Chemother 59, 1435–1440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brzuszkiewicz E, Weiner J, Wollherr A, Thurmer A, Hupeden J, Lomholt HB, Kilian M, Gottschalk G, Daniel R, Mollenkopf HJ, Meyer TF, Bruggemann H, Comparative genomics and transcriptomics of Propionibacterium acnes. PLoS One 6, e21581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA, Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci U S A 106, 2549–2553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe H, Kushida K, Shiobara Y, Kodama M, The structures of sulfomycin I and berninamycin A. Tetrahedron Lett 29, 1401–1404 (1988). [Google Scholar]
- 22.Lau RC, Rinehart KL, Berninamycins B C, and D, minor metabolites from Streptomyces bernensis. J Antibiot (Tokyo) 47, 1466–1472 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Bergy ME, Coats JH, Reusser F, Antibiotic berninamycin and process for making same. US3689639 A. (1972). [Google Scholar]
- 24.Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT, The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc Natl Acad Sci U S A 110, 8483–8488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A, Hujer S, Durre P, Gottschalk G, The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305, 671–673 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Melnik AV, da Silva RR, Hyde ER, Aksenov AA, Vargas F, Bouslimani A, Protsyuk I, Jarmusch AK, Tripathi A, Alexandrov T, Knight R, Dorrestein PC, Coupling Targeted and Untargeted Mass Spectrometry for Metabolome-Microbiome-Wide Association Studies of Human Fecal Samples. Anal Chem 89, 7549–7559 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Otberg N, Richter H, Schaefer H, Blume-Peytavi U, Sterry W, Lademann J, Variations of hair follicle size and distribution in different body sites. J Invest Dermatol 122, 14–19 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA, Temporal Stability of the Human Skin Microbiome. Cell 165, 854–866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. Human Microbiome Project, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP, New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, Gallo RL, Huang CM, Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One 8, e55380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marples RR, Downing DT, Kligman AM, Control of free fatty acids in human surface lipids by Corynebacterium acnes. J Invest Dermatol 56, 127–131 (1971). [DOI] [PubMed] [Google Scholar]
- 33.von Eiff C, Becker K, Machka K, Stammer H, Peters G, Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344, 11–16 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Giersing BK, Dastgheyb SS, Modjarrad K, Moorthy V, Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine 34, 2962–2966 (2016). [DOI] [PubMed] [Google Scholar]
- 35.O’Neill AM, Gallo RL, Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 6, 177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, Li H, Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol 133, 2152–2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO, Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Eggeling L, Bott M, Handbook of Corynebacterium glutamicum. (Taylor & Francis, 2005), pp. 616 [Google Scholar]
- 39.Zhu A, Sunagawa S, Mende DR, Bork P, Inter-individual differences in the gene content of human gut bacterial species. Genome Biol 16, 82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenblum S, Carr R, Borenstein E, Extensive strain-level copy-number variation across human gut microbiome species. Cell 160, 583–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes O, Guyonvarch A, Bonamy C, Salti V, David F, Leblon G, ‘Integron’-bearing vectors: a method suitable for stable chromosomal integration in highly restrictive corynebacteria. Gene 107, 61–68 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Competition assay between C. acnes and S. aureus under conditions mimicking the natural follicle environment.
HPLC chromatogram of cutimycin purification.
Low energy mass spectrum for cutimycin (calculated ([M+H]+ 1131.3374 for C51H51N14O15S+) isolated from A) the heterologous C. glutamicum producer ([M+H]+ 1131.3411) and B) from C. acnes HL030PA1 ([M+H]+ 1131.3364).
High energy mass spectrum for cutimycin labeled with predicted fragments by Unifi.
1H NMR Spectrum of cutimycin taken in DMSO-d6 at 600 MHz.
13C NMR Spectrum of cutimycin taken in DMSO-d6 at 151 MHz.
1H-1H COSY NMR in DMSO-d6.
1H-13C HSQC NMR in DMSO-d6.
1H-13C HMBC NMR in DMSO-d6.
Zoom in of 1H-13C HMBC NMR in DMSO-d6 identifying dehydroalanine residues through presence of diastereotopic terminal alkene protons (position 20, δH 5.74, 5.75 and position 24 δH 5.73, 6.35) with HMBC correlations to carbon atoms 19 and 23 respectively.
Zoom in of 1H-13C HMBC NMR in DMSO-d6 showing the weak four-bond HMBC correlation from position 13 to quaternary carbon 16 at δC 139. 13.
Marfey’s analysis of cutimycin. Individual amino acid Marfey’s analysis derivatives, cutimycin hydrolysis product derivatives, and coinjections.
The subunits of cutimycin with key HMBC (arrows) and COSY (bold bonds) correlations.
Structure of cutimycin
The structures of A) cutimycin, B) berninamycin A, and C) LFF-571.
Mass spectrometric detection of cutimycin from pooled human follicular content (n=60) for one of the tested samples.
Box plots of the impact of the cutimycin BGC presence/absence on C. acnes, C. granulosum and S. epidermidis CFUs in human skin follicles.
Linear depiction of cutimycin with the key mass spec fragments labeled (Table S9).
Metagenomic analysis of C. acnes BGCs at 17 skin sites from sites from 12 healthy participants.
The key fragments confirming the order of the amino acids in cutimycin.
Cutimycin BGC presence/absence in 219 Cutibacterium genomes
NMR Data of cutimycin taken in DMSO-d6 at 600 MHz and 151 MHz.
Minimal inhibitory concentrations for cutimycin and bernamycin
Cutimycin in human follicular samples has been detected in 28% of all samples across two separate experiments.
Locus tags for the C. acnes BGCs assayed for in skin metagenomic data.
Cutimycin BGC presence/absence; the CFUs of C. acnes, C. granulosum and S. epidermidis; and the C. acnes/S. epidermidis CFU ratio in the content of individual human skin follicles.
Bacterial strains and plasmids used in this study.
Primers used in this study.


