Abstract
Purpose
The Coronavirus disease 2019 (COVID-19) has led to an unparalleled influx of patients. Prognostic scores could help optimizing healthcare delivery, but most of them have not been comprehensively validated. We aim to externally validate existing prognostic scores for COVID-19.
Methods
We used “COVID-19 Evidence Alerts” (McMaster University) to retrieve high-quality prognostic scores predicting death or intensive care unit (ICU) transfer from routinely collected data. We studied their accuracy in a retrospective multicenter cohort of adult patients hospitalized for COVID-19 from January 2020 to April 2021 in the Greater Paris University Hospitals. Areas under the receiver operating characteristic curves (AUC) were computed for the prediction of the original outcome, 30-day in-hospital mortality and the composite of 30-day in-hospital mortality or ICU transfer.
Results
We included 14,343 consecutive patients, 2583 (18%) died and 5067 (35%) died or were transferred to the ICU. We examined 274 studies and found 32 scores meeting the inclusion criteria: 19 had a significantly lower AUC in our cohort than in previously published validation studies for the original outcome; 25 performed better to predict in-hospital mortality than the composite of in-hospital mortality or ICU transfer; 7 had an AUC > 0.75 to predict in-hospital mortality; 2 had an AUC > 0.70 to predict the composite outcome.
Conclusion
Seven prognostic scores were fairly accurate to predict death in hospitalized COVID-19 patients. The 4C Mortality Score and the ABCS stand out because they performed as well in our cohort and their initial validation cohort, during the first epidemic wave and subsequent waves, and in younger and older patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00134-021-06524-w.
Keywords: COVID-19, SARS-CoV-2, Prognosis, Intensive care units, Mortality, Cohort studies
Take home message
| In this retrospective cohort study of 14,343 patients, seven out of 32 previously published prognostic scores were able to fairly predict 30-day in-hospital mortality using routinely collected clinical and biological data (area under the ROC curve > 0.75). The 4C Mortality Score and the ABCS stand out because they performed as well in our cohort and their initial validation cohort, during the first and subsequent epidemic waves, in younger and older patients, and showed satisfactory calibration. Their ability to guide clinical management decisions and appropriate resource allocation should now be evaluated in future studies. |
Introduction
Since the end of 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide [1]. At the end of May 2021, there were over 167 million confirmed cases and over 3.4 million deaths from the coronavirus disease 2019 (COVID-19) around the world [2]. Hospital facilities have, thus, faced an unparalleled influx of patients. The evolution of hospitalized patients varies widely, from those necessitating no or low level of oxygen to those evolving to acute respiratory or hemodynamic failure requiring admission to intensive care units (ICU) [3, 4]. Accurate outcome prediction with scores based on patient characteristics (age, sex, comorbidities, clinical state, laboratory and imaging results, etc.) help optimizing healthcare delivery in a limited medical resources context [5]. They can also be used to select patients with a homogeneous risk for a given outcome for inclusion in clinical studies.
Various scores have been developed since the beginning of the outbreak and older ones, routinely used in community acquired pneumonia and other conditions, have also been tested in the setting of COVID-19. A systematic review updated in July 2020 found 39 published prognostic scores estimating mortality risk in COVID-19 patients and 28 aimed to predict progression to severe or critical disease. All scores were rated at high or unclear risk of bias. Only a few had undergone external validation, with shortcomings including unrepresentative patient sets, small sizes of the derivation samples and insufficient numbers of outcome events [6]. Moreover, the worldwide applicability of these prediction scores remains an open question: healthcare systems and patient profiles differ between countries [7] and may impact these scores' performances.
The aim of this study was to evaluate the accuracy of published scores to predict in-hospital mortality or ICU admission in SARS-CoV-2-infected patients, using a large multicenter cohort from the Greater Paris University Hospitals (GPUH).
Methods
Study reporting
Our manuscript complies with the relevant reporting guidelines, namely the REporting of studies Conducted using Observational Routinely collected health Data (RECORD) statement [8] and the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement [9]. Completed checklists are available in Appendix 2.
Study design and setting
We conducted a retrospective cohort study using the GPUH’s Clinical Data Warehouse (CDW), an automatically filled database containing data collected during routine clinical care in the GPUH. GPUH is a public institution and count 39 hospitals (22,474 beds) spread across Paris and its region, accounting for 1.5 million hospitalizations each year (10% of all hospitalizations in France). The data of patients hospitalized for COVID-19 in GPUH was used to evaluate the accuracy of published prognostic scores for COVID-19. Final data extraction was performed on May 8th, 2021. The GPUH’s CDW Scientific and Ethics Committee (IRB00011591) granted access to the CDW for the purpose of this study and no linkage was made with other databases.
Inclusion and exclusion criteria
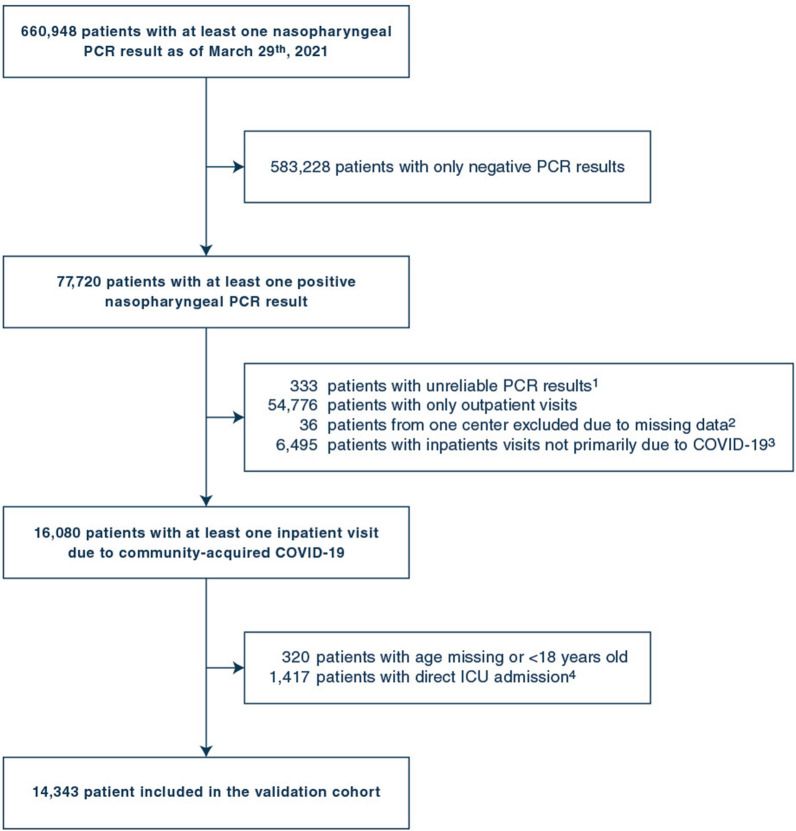
Patients’ selection process is summarized in Fig. 1. All patients with a result found in the database for reverse transcriptase-polymerase chain reaction (PCR) for SARS-CoV-2 in a respiratory sample were screened. Patients were included in the study if they met both following criteria:
A hospital stay with an International Classification of Diseases, 10th edition (ICD-10) code for COVID-19 (U07.1),
At least one positive respiratory PCR for SARS-CoV-2 from 10 days before to 3 days after hospital admission.
Fig. 1.
Flow chart of selected patients.
1. Where validation by a biologist occured before or 20 days after recorded sample collection date and time. 2. Patients from Georges Pompidou European Hospital were excluded, as all biological and clinical data from this hospital were missing due to interoperability issues with the CDW. 3. Hospitalizations with no ICD-10 code for Covid-19, or with an ICD-10 code for Covid-19 and a first positive PCR sample obtained more than 10 days before or more than 3 days after admission. 4. Hospitalizations for Covid-19 with ICU transfer within 2 hours following hospital admission, and no visit in any other GPUH hospital in the preceding 24 hours
Patients were excluded from the study if they met at least one of the following criteria:
PCR result considered unreliable (i.e., time of validation by the biologist before the time of PCR sample collection, or more than 20 days after the time of sample collection),
Asymptomatic positive PCR result during a COVID-unrelated hospitalization or COVID considered as hospital-acquired (i.e., a first positive PCR sample collected more than 3 days after hospital admission),
Direct ICU admission (i.e., time between recorded hospital admission and recorded ICU admission less than 2 h and no visit in another GPUH hospital in the preceding 24 h),
Age < 18, not recorded or unknown,
Hospitalization in the Georges Pompidou European hospital, one of the 39 GPUH hospitals (all biological and clinical data from this hospital were missing, due to interoperability issues with the CDW).
To have a follow-up of 30 days or more for all hospitalized patients, only patients with a PCR performed before March 30th were considered.
Data collection
The reference date used for baseline characteristics was the date of hospital admission for COVID-19. The following data were collected:
Demographic data and data on hospital admission.
Medical history (based on ICD-10 codes for current or previous hospital visits; the list of codes used is based on a previously published work [10]).
Vital signs and biological values (the first value found in the database from 24 h before to 48 h after hospital admission was retrieved for each patient, as a delay can exist for logistical reasons between true and recorded admission date; values obtained in ICU were not considered).
Outcomes (in-hospital mortality, ICU admission and invasive mechanical ventilation within 30 days from admission).
Of note, invasive mechanical ventilation is always performed in ICU in France.
Selection of published scores
The selection of high-quality published scores was performed using “COVID-19 Evidence Alerts” (https://plus.mcmaster.ca/Covid-19/), a service provided by the McMaster University, in which evidence reports on COVID-19 published in all journals included in MEDLINE are critically appraised for scientific merit based on prespecified criteria (see https://hiru.mcmaster.ca/hiru/InclusionCriteria.html). All studies identified by the “Clinical Prediction Guide” filter were systematically screened by two independent investigators (L.A. and P.S.), and discrepancies were adjudicated by a third investigator (Y.L.). Studies were included if they met all the following criteria:
studies on prognostic scores predicting ICU transfer or in-hospital mortality for patients hospitalized for COVID-19, including scores primarily developed for other purposes prior to the pandemic,
meeting all the prespecified criteria for “higher quality” (i.e., generated in one or more sets of real patients; validated in another set of real patients; study providing information on how to apply the prediction guide); or studies excluded from this category only due to the lack of an independent validation cohort, but in which derivation and validation were performed in different samples from the same cohort (split validation),
computable with the data collected in the CDW.
The last search in “COVID-19 Evidence Alerts” was performed on April 3rd, 2021. The process for scores’ selection and reasons for exclusion are detailed in Appendix 3 and Figure S1, and information on scores included in the study in Table S1 and S2.
Statistical analysis
Aberrant values for biological tests and vital signs were treated as described in Table S3. Missing data were treated by multiple imputations (mice function of the mice package, 50 imputed datasets with 15 iterations, predictive means matching method for quantitative variables, after log or square-root transformation when needed to get a more normalized dataset), under the missing-at-random hypothesis. Outcome variables were included in the dataset used for imputation. Rubin’s rule was used to pool estimates obtained in each imputed dataset. Variables used for multiple imputations are detailed in Table S4.
For each score included in the analysis and each outcome, discrimination was assessed by drawing a receiver operating characteristics (ROC) curve and computing the corresponding area under the curve (AUC). DeLong’s method [11] was used to estimate the variance in each dataset, results were pooled with Rubin’s rule and used to compute pooled 95% confidence intervals.
First, we assessed the performance of each score to predict the available outcome closest to the one used in the original study, with the required adaptations to be computed with the available data. AUC in our cohort and in previously published studies were compared using a Z-test for independent samples. Second, we assessed the performance of each score to predict 30-day in-hospital mortality and the composite of 30-day in-hospital mortality or ICU transfer. Third, we used a Z-test for paired data following DeLong’s method [11] to compare the accuracy of scores with an AUC > 0.75 to predict 30-day hospital mortality. Sensitivity analyses were conducted on subgroups of age (≤ 65 or > 65 years old) or wave of admission (before or after June 15th, 2020, a graphically determined threshold), considering only complete cases (only patients with all data available to compute a given score), and considering the area under the precision-recall curve instead of under the ROC curve (pr.curve function of the PRROC package). Heterogeneity of AUC between subgroups was assessed using an interaction term between the score and the grouping variable in a logistic regression model predicting the outcome.
Post hoc analyses were performed to further characterize the best scores at predicting 30-day in-hospital mortality (AUC > 0.75). Calibration curves were drawn by plotting the observed mortality rate in each class as a function of the predicted probability of mortality, with patients grouped by deciles of predicted probability. For each score, a logistic regression model was built to predict 30-day in-hospital mortality with its predictors and fitted on our data. Variable importance was determined using the absolute value of the t-statistic for each predictor in this model (varImp function of the caret package). Calibration curves were drawn using probabilities predicted by the revised logistic regression models fitted on our data.
All tests are two-sided, and a p value < 0.05 was considered significant. Continuous variables are reported as mean (standard deviation) for normally distributed variables, and median [interquartile range] for non-normally distributed variables. Binary variables are reported as number of patients with a positive result (percentage of patients with a positive result). Analyses were performed using the R freeware version 4 (packages mice, pROC, psfmi, Amelia, PRROC, caret).
Results
Baseline characteristics and outcomes of patients included in the study
We included 14,343 patients in the validation cohort (Fig. 1). First hospital admission for COVID-19 was on January 29th, 2020 and last on April 6th, 2021. Patients’ baseline characteristics are summarized in Table 1 and outcomes are summarized in Table 2. Baseline characteristics appeared similar during the first wave and subsequent waves (Table S5). Initial care site appeared to be an important factor for vital signs or biological values to be missing (Table S6). Multiple imputations were therefore stratified by center. In-hospital mortality at day 30 was 18% overall, significantly lower during the first wave than in the subsequent waves, and significantly higher in patients older than 65 years old (Figure S2, p < 0.001 for Log-Rank test).
Table 1.
Baseline characteristics of patients included in the study
| Variable | No death within 30 daysa (n = 11,760) | Death within 30 days (n = 2583) | All patients (n = 14,343) | |||
|---|---|---|---|---|---|---|
| Missing | Missing | Missing | ||||
| Demographic data | ||||||
| Female sex, n (%) | 5175 (44) | 1014 (39.3) | 6189 (43.1) | |||
| Age, years (SD) | 66 (17.6) | 79.2 (12) | 68.4 (17.5) | |||
| Diagnosis of COVID-19 | ||||||
| Admission during “first wave”, n (%) | 4863 (41.4) | 1279 (49.5) | 6142 (42.8) | |||
| Time between PCR and admission, days | − 0.1 [− 0.1, 0] | 0 [− 0.1, 0] | − 0.1 [− 0.1, 0] | |||
| Medical history, n (%) | ||||||
| Modified Charlson comorbidity index, pts | 0 [0, 2] | 2 [0, 4] | 1 [0, 2] | |||
| Congestive heart failure | 1228 (10.4) | 637 (24.7) | 1865 (13) | |||
| Myocardial infarction | 666 (5.7) | 297 (11.5) | 963 (6.7) | |||
| Peripheral vascular disease | 620 (5.3) | 264 (10.2) | 884 (6.2) | |||
| Cerebrovascular disease | 985 (8.4) | 376 (14.6) | 1361 (9.5) | |||
| Hemiplegia | 442 (3.8) | 157 (6.1) | 599 (4.2) | |||
| Dementia | 1364 (11.6) | 638 (24.7) | 2002 (14) | |||
| Arterial hypertension | 4723 (40.2) | 1403 (54.3) | 6126 (42.7) | |||
| Diabetes | 2699 (23) | 716 (27.7) | 3415 (23.8) | |||
| Diabetes with end-organ damage | 1480 (12.6) | 542 (21) | 2022 (14.1) | |||
| Chronic pulmonary disease | 1366 (11.6) | 397 (15.4) | 1763 (12.3) | |||
| Moderate or severe renal disease | 1536 (13.1) | 660 (25.6) | 2196 (15.3) | |||
| Moderate or severe liver disease | 127 (1.1) | 33 (1.3) | 160 (1.1) | |||
| Any tumor | 1064 (9) | 480 (18.6) | 1544 (10.8) | |||
| Metastatic solid tumor | 261 (2.2) | 150 (5.8) | 411 (2.9) | |||
| Connective tissue disease | 241 (2) | 64 (2.5) | 305 (2.1) | |||
| HIV infection | 218 (1.9) | 20 (0.8) | 238 (1.7) | |||
| Obesity (ICD-10 codes only) | 2289 (19.5) | 426 (16.5) | 2715 (18.9) | |||
| Vital signs on admission | ||||||
| Heart rate, beats per minute | 2729 (23.2) | 88.7 (SD 17.5) | 615 (23.8) | 87.5 (SD 18.5) | 3344 (23.3) | 88.5 (SD 17.7) |
| Respiratory rate, cycles per minute | 4623 (39.3) | 24.4 (SD 7.3) | 992 (38.4) | 27 (SD 8.1) | 5615 (39.1) | 24.9 (SD 7.5) |
| Altered consciousness, n (%) | 7008 (59.6) | 133 (2.8) | 1573 (60.9) | 112 (11.1) | 8581 (59.8) | 245 (4.3) |
| Diastolic blood pressure, mmHg | 4934 (42) | 75.3 (SD 14.5) | 1046 (40.5) | 72.4 (SD 17.1) | 5980 (41.7) | 74.8 (SD 15.1) |
| Mean blood pressure, mmHg | 5201 (44.2) | 94.4 (SD 15.2) | 1276 (49.4) | 91.3 (SD 17.6) | 6477 (45.2) | 93.9 (SD 15.6) |
| Systolic blood pressure, mmHg | 4932 (41.9) | 131.4 (SD 21.3) | 1044 (40.4) | 130.7 (SD 24.8) | 5976 (41.7) | 131.2 (SD 22) |
| Pulse saturometry, % | 3767 (32) | 96 [93, 98] | 784 (30.4) | 94 [90, 97] | 4551 (31.7) | 96 [93, 98] |
| Temperature, °C | 2759 (23.5) | 37.5 (SD 0.9) | 615 (23.8) | 37.5 (SD 1) | 3374 (23.5) | 37.5 (SD 1) |
| Body mass index (BMI), kg/m2 | 4227 (35.9) | 27.2 (SD 6.4) | 1208 (46.8) | 26.6 (SD 7.1) | 5435 (37.9) | 27.1 (SD 6.5) |
| Biological values on admission | ||||||
| Hemoglobin, g/dl | 1376 (11.7) | 13.1 (SD 1.9) | 383 (14.8) | 12.7 (SD 2.2) | 1759 (12.3) | 13 (SD 2) |
| Leukocytes, G/l | 1378 (11.7) | 7 (SD 3.7) | 384 (14.9) | 8 (SD 5.1) | 1762 (12.3) | 7.2 (SD 4) |
| Neutrophils, G/l | 1574 (13.4) | 5.3 (SD 3.1) | 416 (16.1) | 6.4 (SD 4.1) | 1990 (13.9) | 5.5 (SD 3.4) |
| Lymphocytes, G/l | 1597 (13.6) | 1 [0.7, 1.4] | 423 (16.4) | 0.8 [0.5, 1.1] | 2020 (14.1) | 0.9 [0.7, 1.3] |
| Platelets count, G/l | 1385 (11.8) | 223.5 (SD 93) | 384 (14.9) | 201.9 (SD 92.9) | 1769 (12.3) | 219.7 (SD 93.4) |
| Sodium, mmol/l | 467 (4) | 135.9 (SD 4.3) | 132 (5.1) | 136.6 (SD 6.2) | 599 (4.2) | 136 (SD 4.7) |
| Potassium, mmol/l | 652 (5.5) | 4.1 (SD 0.6) | 196 (7.6) | 4.2 (SD 0.7) | 848 (5.9) | 4.1 (SD 0.6) |
| Bicarbonates, mmol/l | 5361 (45.6) | 24.4 (SD 3.7) | 1196 (46.3) | 23 (SD 4.4) | 6557 (45.7) | 24.2 (SD 3.9) |
| Proteins, g/l | 796 (6.8) | 71.8 (SD 7.1) | 186 (7.2) | 69.8 (SD 8.1) | 982 (6.8) | 71.5 (SD 7.3) |
| Urea, mmol/l | 663 (5.6) | 6 [4.3, 8.8] | 168 (6.5) | 10 [6.6, 15.3] | 831 (5.8) | 6.5 [4.6, 9.9] |
| Serum creatinine, µmol/l | 436 (3.7) | 80 [64, 103] | 124 (4.8) | 103 [77, 152] | 560 (3.9) | 82.4 [66, 110] |
| Alanine aminotransferase, IU/l | 1995 (17) | 30 [20, 47.5] | 482 (18.7) | 28 [18.6, 45] | 2477 (17.3) | 29.5 [20, 47] |
| Asparate aminotransferase, IU/l | 2366 (20.1) | 41 [29, 60] | 560 (21.7) | 51 [34, 78] | 2926 (20.4) | 42 [29.2, 63] |
| Total bilirubin, µmol/l | 1959 (16.7) | 8 [6, 11.5] | 468 (18.1) | 9 [6, 13] | 2427 (16.9) | 8 [6, 12] |
| Lactate dehydrogenase, IU/l | 5688 (48.4) | 352 [267, 477] | 1273 (49.3) | 430 [322, 581] | 6961 (48.5) | 362 [275, 499] |
| Creatinine phosphokinase, IU/l | 5470 (46.5) | 123 [64, 276] | 1200 (46.5) | 186 [85 480] | 6670 (46.5) | 132 [67, 300] |
| Troponine, ng/l | 6149 (52.3) | 15 [9, 24] | 1283 (49.7) | 34 [18, 76.1] | 7432 (51.8) | 15 [10, 31] |
| Activated partial thromboplastin time | 2555 (21.7) | 1.2 (SD 0.3) | 605 (23.4) | 1.3 (SD 0.4) | 3160 (22) | 1.2 (SD 0.3) |
| Prothrombin time, % | 2238 (19) | 87 [76, 98] | 535 (20.7) | 82 [69, 93] | 2773 (19.3) | 87 [75, 97] |
| Fibrinogen, g/l | 4248 (36.1) | 5.8 (SD 1.6) | 952 (36.9) | 5.8 (SD 1.6) | 5200 (36.3) | 5.8 (SD 1.6) |
| d-dimers, µg/l | 4918 (41.8) | 900 [557, 1560] | 1287 (49.8) | 1375 [828, 2560] | 6205 (43.3) | 964 [585, 1690] |
| C-reactive protein, mg/l | 1104 (9.4) | 65 [26, 121] | 261 (10.1) | 96 [49.1, 163.9] | 1365 (9.5) | 70 [30, 129] |
| Procalcitonin, µg/l | 5973 (50.8) | 0.1 [0.1, 0.3] | 1263 (48.9) | 0.3 [0.2, 1] | 7236 (50.4) | 0.2 [0.1, 0.4] |
| Albumin, g/l | 7792 (66.3) | 32.7 (SD 5.4) | 1659 (64.2) | 30.9 (SD 5.4) | 9451 (65.9) | 32.4 (SD 5.5) |
aEither patients discharged alive before day 30 (n = 8459), or patients still in hospital and alive at day 30 (n = 3301). SD: standard deviation. Continuous variables are reported as mean (SD) for normally distributed variables and median [interquartile range] for non-normally distributed variables
Table 2.
Outcomes of patients included in the study
| Outcome | All patients (n = 14,343) |
|---|---|
| In-hospital mortalitya, n (%) | 2583 (18) |
| Time between hospital admission and death, days | 8.1 [4.2, 13.7] |
| ICU admissiona, n (%) | 3289 (22.9) |
| Time between hospital and ICU admission, days | 1 [0.2, 2.8] |
| Invasive mechanical ventilationb, n (%) | 1634 (11.4) |
| In-hospital mortality or ICU admission, n (%) | 5067 (35.3) |
aOnly deaths or ICU admissions within 30 days following hospital admission were considered linked to COVID-19
bAll patients requiring invasive mechanical ventilation were admitted in ICU in GPUH’s hospitals. Time delays are reported as median [interquartile range]
Selected scores and their performance to predict the original outcome
Thirty-two scores [12–37] were included in the study: 23 were specifically derived in COVID-19 patients and 9 were pre-existing scores developed for other purposes and tested in COVID-19 patients (Table 3, Table S1 and S2, Appendix 3). Among 27 scores with available 95% CI to estimate AUC variance in previous reports, 19 (70%) had an AUC significantly lower in our cohort (Table 3). The 4C Mortality Score was the only one with an AUC significantly higher in our cohort compared to the previously published value (p < 0.001).
Table 3.
Summary of scores included in the study and comparison to previously published data
| Score name | Data from previously published studies | Current study | p value | |||
|---|---|---|---|---|---|---|
| Sample size for validation | Outcome | AUROC [95% CI] | Outcome used for comparison | AUROC [95% CI] | ||
| 4C Mortality Score [12] | 22,361 | Death (in-hospital) | 0.767 [0.760–0.773] | Death (in-hospital) | 0.785 [0.775–0.795] | 0.003 |
| ABC-GOALSc [13]a | 240 | ICU admission | 0.770 [0.710–0.830] | ICU admission | 0.628 [0.616–0.640] | < 0.001 |
| ABCS [14] | 188 | Death (30 days) | 0.838 [0.777–0.899] | Death (in-hospital, 30 days) | 0.790 [0.780–0.801] | 0.128 |
| A-DROP [12]a | 15,572 | Death (in-hospital) | 0.736 [0.728–0.744] | Death (in-hospital) | 0.730 [0.718–0.741] | 0.415 |
| ANDC [15] | 125 | Death | 0.975 [0.947–1.000] | Death (in-hospital, 30 days) | 0.751 [0.741–0.761] | < 0.001 |
| Bennouar et al. [16] | 247 | Death (28 days) | 0.900 [0.870–0.940] | Death (in-hospital, 28 days) | 0.724 [0.713–0.736] | < 0.001 |
| CHA(2)DS(2)-VASc [17] | 864 | Death | 0.690 [0.650–0.730] | Death (in-hospital) | 0.687 [0.677–0.697] | 0.887 |
| COPS [18]a | 1865 | Death (28 days) | 0.896 [0.872–0.911] | Death (in-hospital, 28 days) | 0.745 [0.734–0.756] | < 0.001 |
| CORONATION-TR [19]a | 37,377 | Death (30 days) | 0.896 [0.890–0.902] | Death (in-hospital, 30 days) | 0.769 [0.757–0.780] | < 0.001 |
| COVID-19 SEIMC [20]a | 2126 | Death (in-hospital, 30 days) | 0.831 [0.806–0.856] | Death (in-hospital, 30 days) | 0.752 [0.743–0.762] | < 0.001 |
| COVID-AID [21]a | 265 | Death (7 days) | 0.851 [0.781–0.921] | Death (in-hospital, 7 days) | 0.775 [0.762–0.788] | 0.036 |
| COVID-GRAM [22]a | 710 | Composite: death, ICU admission, invasive mechanical ventilation | 0.880 [0.840–0.930] | Composite: death (in-hospital), ICU admission, invasive mechanical ventilation | 0.700 [0.690–0.711] | < 0.001 |
| COVID-NoLab [23] | 537 | Death (in-hospital) | 0.803 [Unknown] | Death (in-hospital) | 0.693 [0.683–0.704] | NA |
| COVID-SimpleLab [23] | 295 | Death (in-hospital) | 0.833 [Unknown] | Death (in-hospital) | 0.707 [0.696–0.718] | NA |
| CURB-65 [12] | 15,560 | Death (in-hospital) | 0.720 [0.713–0.728] | Death (in-hospital) | 0.724 [0.711–0.736] | 0.595 |
| Hachim et al. [24] | 289 | ICU admission | Unknown [Unknown] | ICU admission | 0.514 [0.503–0.526] | NA |
| Hu et al. [25] | 64 | Death | 0.881 [Unknown] | Death (in-hospital) | 0.724 [0.713–0.735] | NA |
| KPI Score [26] | 309 | Composite: death (in-hospital), ICU, invasive mechanical ventilation, NIV, oxygen, steroids, IVIg, ECMO, CRRT, dyspnea, X-ray consolidation | 0.888 [0.854–0.922] | Composite: death (in-hospital), ICU admission, invasive mechanical ventilation | 0.597 [0.588–0.606] | < 0.001 |
| LOW-HARM Score [27]a | 400 | Death (in-hospital) | 0.960 [0.940–0.980] | Death (in-hospital) | 0.603 [0.588–0.618] | < 0.001 |
| Mei et al. (Full) [28]a | 276 | Death (60 days) | 0.970 [0.960–0.980] | Death (in-hospital, 60 days) | 0.730 [0.719–0.741] | < 0.001 |
| Mei et al. (Simple) [28] | 276 | Death (60 days) | 0.880 [0.800–0.960] | Death (in-hospital, 60 days) | 0.717 [0.706–0.729] | < 0.001 |
| NEWS2 [29]a | 66 | Composite: death or ICU admission | 0.822 [0.690–0.953] | Composite: death (in-hospital), ICU admission | 0.639 [0.626–0.651] | 0.006 |
| PLANS [30] | 1031 | Death (in-hospital) | 0.870 [0.850–0.890] | Death (in-hospital) | 0.739 [0.729–0.750] | < 0.001 |
| PREDI-CO [31] | 526 | Composite: invasive mechanical ventilation, NIV, oxygen saturation < 93% with FiO2 = 1 | 0.850 [0.810–0.880] | ICU admission, invasive mechanical ventilation | 0.646 [0.635–0.657] | < 0.001 |
| PRESEP [32] | 557 | Death (60 days) | 0.607 [0.555–0.652] | Death (in-hospital, 60 days) | 0.586 [0.571–0.600] | 0.447 |
| qSOFA [12] | 19,361 | Death (in-hospital) | 0.622 [0.615–0.630] | Death (in-hospital) | 0.583 [0.566–0.601] | < 0.001 |
| RISE UP [33] | 642 | Death (30 days) | 0.770 [0.680–0.760] | Death (in-hospital, 30 days) | 0.770 [0.759–0.782] | 1.000 |
| SIMI [34] | 275 | Composite: death, NIV, invasive mechanical ventilation | 0.800 [Unknown] | Composite: death (in-hospital), ICU admission, invasive mechanical ventilation | 0.664 [0.655–0.674] | NA |
| SIRS [35] | 175 | Death (in-hospital) | 0.700 [0.610–0.800] | Death (in-hospital) | 0.538 [0.526–0.551] | < 0.001 |
| STSS [36] | 100 | Death (30 days) | 0.962 [0.903–0.990] | Death (in-hospital, 30 days) | 0.697 [0.683–0.712] | < 0.001 |
| Wang et al. (clinical) [37] | 44 | Death | 0.830 [0.680–0.930] | Death (in-hospital) | 0.729 [0.720–0.738] | 0.188 |
| Wang et al. (laboratory) [37] | 44 | Death | 0.880 [0.750–0.960] | Death (in-hospital) | 0.628 [0.616–0.640] | < 0.001 |
aAlterations were used to compute these scores. Previously published values used are those from the validation cohorts of the initial studies (external if available, otherwise internal). Z-test was used to compare previously published values and values in our cohort
AUROC area under the receiver operating characteristic curve, CI confidence interval, IVIg intravenous immunoglobulins, NIV non-invasive ventilation, CRRT continuous renal replacement therapy, ECMO extracorporeal membrane oxygenation
Performance to predict 30-day in-hospital mortality and the composite of 30-day in-hospital mortality or ICU admission
Results are summarized in Table S7, and Figure S3 shows the ROC curves of the three most accurate scores for each outcome. None of the included scores had a very high accuracy to predict 30-day in-hospital mortality alone, or the composite of 30-day in-hospital mortality or ICU admission (all AUC < 0.8). AUC was higher to predict 30-day in-hospital mortality alone than 30-day in-hospital mortality or ICU admission for 25/32 scores (78%).
Seven scores had an AUC > 0.75 to predict 30-day in-hospital mortality (Table 4). The 4C Mortality score and the ABCS had the highest AUC to predict 30-day in-hospital mortality (4C Mortality score: 0.793, 95% CI 0.783–0.803; ABCS: 0.790, 95% CI 0.780–0.801). Their AUC did not differ significantly from each other (p = 0.61) but were significantly higher than that of the following scores (p < 0.01 for all comparisons). The CORONATION-TR score had the highest AUC to predict 30-day in-hospital mortality or ICU admission (AUC 0.724, 95% CI 0.714–0.733). Table S8 provides the sensitivities and specificities for these scores to predict in-hospital mortality using cut-off values from previous reports, and Figure S4 shows the Kaplan–Meier curves for in-hospital mortality of the three scores that performed best to predict in-hospital mortality.
Table 4.
Detailed characteristics of scores with an AUROC > 0.75 to predict 30-day in-hospital mortality in the analysis using multiple imputed data
| Score name | Information needed to compute the score | AUROC [95% CI] | Accuracy to predict in-hospital mortality | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient’s characteristics | Medical history | Initial presentation | Biology | In-hospital mortality | In-hospital mortality or ICU admission | Performed as well or better than in the first published validation cohort | Performed equally well in patients < 65 years old | Performed equally well in all epidemic waves | |
| 4C Mortality Score | Age, sex | Chronic cardiac disease, chronic respiratory disease (excluding asthma), chronic renal disease, mild to severe liver disease, dementia, chronic neurological conditions, connective tissue disease, diabetes mellitus, HIV infection, malignancy | Respiratory rate, oxygen saturation, consciousness | Urea, CRP | 0.793† [0.783–0.803] | 0.659 [0.649–0.670] | Yes | Yes | Yes |
| ABCS | Age, sex | COPD | – | CRP, white blood cells, lymphocytes, d-dimer, AST, Troponin I, procalcitonin | 0.790† [0.780–0.801] | 0.682 [0.672–0.692] | Yes | Yes | Yes |
| COVID-GRAMa | Age | COPD, hypertension, diabetes, coronary artery disease, chronic kidney disease, cancer, cerebrovascular disease, hepatitis B, immunodeficiency | Abnormalities on chest radiography, hemoptysis, dyspnea, consciousness | Neutrophils, lymphocytes, LDH, bilirubin | 0.771 [0.760–0.783] | 0.688 [0.677–0.699] | No | Yes | Yes |
| RISE UP | Age | – | Heart rate, mean blood pressure, respiratory rate, oxygen saturation, temperature, Glasgow coma scale | Albumin, urea, LDH, bilirubin | 0.770 [0.759–0.782] | 0.660 [0.650–0.671] | Yes | No | Yes |
| CORONATION-TRa | Age | Heart failure, diabetes, coronary artery disease, peripheral artery disease, collagen tissue disorders, malignancy, lymphoma, heart failure, COPD, cerebrovascular disease, hypertension, diabetes mellitus, valvular heart disease, chronic liver disease | Pneumonia on chest tomography | Neutrophils, lymphocytes, platelets, d-dimer, LDH, CRP, hemoglobin, creatinine, albumin | 0.769 [0.757–0.780] | 0.724 [0.714–0.733] | No | Yes | Yes |
| ANDC | Age | – | – | Neutrophils, lymphocytes, d-dimer, CRP | 0.759 [0.748–0.769] | 0.642 [0.632–0.652] | No | Yes | Yes |
| COVID-19 SEIMCa | Age, sex | – | Dyspnea, oxygen saturation | Neutrophils, lymphocytes, eGFR | 0.752 [0.743–0.762] | 0.587 [0.578–0.597] | No | No | Yes |
Scores are ordered by performance to predict in-hospital mortality
AUROC area under the receiver operating characteristic curve, CI confidence interval, CRP C-reactive protein, LDH lactate dehydrogenase, AST aspartate transaminase, COPD chronic obstructive pulmonary disease, eGFR estimated glomerular filtration rate
aAlterations were used to compute these scores
†p < 0.01 for AUC comparison between these scores and the other scores
Sensitivity and post hoc analyses
Among the seven scores with an AUC > 0.75 to predict 30-day in-hospital mortality: accuracy was not significantly altered by wave of admission for any of them (Table S9); accuracy was significantly lower in the subgroup of patients > 65 years old for two of them (RISE-UP and COVID-19 SEIMC; Table S10); AUC was < 0.75 in the analysis using complete cases for one of them (CORONATION-TR; Table S7); the 4C Mortality Score ranked first to predict in-hospital mortality in analyses using multiple imputed data and analyses using complete cases (Table S7).
Main results were unchanged when using the area under the precision-recall curve instead of under the ROC curve to measure discriminative ability: the 4C Mortality score and the ABCS ranked first and second to predict 30-day in-hospital mortality, and the CORONATION-TR score ranked first to predict 30-day in-hospital mortality or ICU transfer (Table S11).
As shown by calibration curves (Figure S5), the risk of 30-day in-hospital mortality was overestimated by 6/7 scores (all but the CORONATION-TR), and most notably so for the COVID-GRAM and ANDC scores. Overestimation was overall less important during the first epidemic wave than subsequent waves (Figure S5) and was corrected after logistic coefficients revision (Figure S6).
In variable importance analysis, age was the most important factor to predict 30-day in-hospital mortality in 5 scores (4C Mortality, ANDC, CORONATION-TR, COVID-GRAM, RISE UP), troponin positivity in 1 score (ABCS) and low estimated glomerular filtration rate in 1 score (COVID-19 SEIMC) (Figure S7).
Discussion
Key results
Most scores (19/27 with available data for comparison) had a significantly lower accuracy in our study compared to previously published studies, and most scores (25/32) had a lower accuracy to predict the composite outcome of 30-day in-hospital mortality or ICU admission, compared to 30-day in-hospital mortality alone. Seven scores had a high accuracy (AUC > 0.75) for the prediction of 30-day in-hospital mortality: the 4C Mortality and ABCS scores had significantly higher AUC values compared to the other scores; the CORONATION-TR score was the most accurate to predict in-hospital mortality or ICU admission; the RISE-UP and COVID-19 SEIMC scores were less accurate in the subgroup of patients > 65 years old. The discriminative performance of these scores was not altered by wave of admission despite changes in clinical care such as larger use of corticosteroids and lower use of invasive ventilation during the subsequent waves. On the opposite, calibration was poorer during the second and subsequent waves than in the first wave.
Limitations and strengths
We conducted a large, multicentre, independent study to validate systematically selected prognostic scores for COVID-19, using routine clinical care data. Selection criteria were chosen to identify the most promising scores, although many of them had not yet been externally validated or had been validated in small cohorts only. Outcomes used in our study (in-hospital mortality, ICU admission and invasive mechanical ventilation) are of high clinical importance, objective and reliably collected in the CDW.
The main limitations of our study are consequences of its retrospective design, with a risk for selection and information bias. Selection bias was controlled using objective and reproducible inclusion and exclusion criteria, based on both administrative (ICD-10 codes for COVID-19) and microbiological information (PCR for SARS-CoV-2). This information is exhaustively recorded in the database, as ICD-10 codes for all hospital stays are independently assessed by a trained physician or technician before transmission to the national health insurance service for billing. Information bias for comorbidities and medical history was controlled by collecting ICD-10 codes for both index and previous visits, using a systematic procedure that was independently validated in a medico-administrative database whose structure is similar to ours [10]. Missing physiological values, such as oxygen saturation, respiratory rate, are explained by several templates available to record them in electronic health records. Only a limited number of these templates are used to gather and aggregate these data in the CDW. Missing biological values, such as d-dimers, CRP or ferritin, are explained by unstandardized practices across GPUH hospitals. As a result, the rate of missing values varied across centers for physiological and biological values (see Table S6), and was high for several important variables such as the Glasgow coma scale. To control these biases, we used multiple imputations under the missing-at-random hypothesis [38], taking centers into account, and we performed a confirmatory sensitivity analysis using complete data.
Several scores, based on machine- or deep-learning algorithms, or using data rarely collected for initial evaluation of patients in clinical practice (such as myoglobin or interleukins) could not be computed in our cohort (see Appendix 3). Although for many of them discriminative performance seemed high in previous studies, their use in clinical practice is more difficult, as they would require changing protocols for patients’ initial evaluation to add costly biological tests, and, for machine- or deep-learning based algorithms, to set an automatic system for computation. Further prospective pragmatic studies are needed on these matters.
Interpretation and generalizability
Our cohort includes patients from Paris and its suburbs, with various ethnicities and socioeconomic backgrounds [39]. Patients are treated in various hospitals, each of them having different resources and practices. Our validation study is strengthened by the number and diversity of included patients and settings, and by the independence from all cohorts used for the derivation and first validation of investigated prognostic scores. Patients were consecutively recruited, and the number of outcome events was very large, overcoming two major shortcomings of previous validation studies. For example, several included scores were previously validated in less than 100 patients (Table 3). The waste of time and money on inappropriately designing or validating COVID-19 prognostic scores has been stressed in a living systematic review [6].
Using a cut-off value of 0.75 for AUC to predict in-hospital death, seven scores were identified as having a high accuracy. They differ in characteristics that may influence their choice for a given use in a given clinical context. For example, some scores use costly biological tests and are not appropriate for countries with limited resources; some use many variables and may be hard to compute at the bedside; some are less accurate in older patients; some are more accurate to predict ICU admission and therefore more suitable to predict the demand on healthcare systems. For the seven fairly accurate scores identified, we provide detailed characteristics that can help clinicians choose the best suited to their needs (Table 4). The 4C Mortality and ABCS scores appear to be the most promising ones, as they use a limited number of variables that are available in routine clinical care, had a fair accuracy in our external validation study, performed equally well during the first epidemic wave and subsequent waves, and in younger and older patients.
The risk of 30-day in-hospital mortality was overestimated by 6/7 scores (all but the CORONATION-TR), and more so during the second and subsequent waves. This can be explained by overall better outcomes during these later waves, as seen in our study and in other ones [40]. Many published scores were derived and validated on first wave data. Revising the scores using local and current data is necessary if accurate estimations of the mortality risk are needed. Likewise, the thresholds indicating a high risk of poor outcome should be locally defined.
In variable importance analysis, age was the most influential factor in 5/7 scores, even in those including many clinical and biological variables (for example, the CORONATION-TR score), underlining the importance of age in driving severity among hospitalized COVID-19 patients. Elevated baseline troponin was the most important factor in the ABCS, which discriminated and calibrated well in our cohort. Troponin has been previously shown to be independently associated with mortality in both non-ICU [41] and ICU [42] patients, stressing its potential relevance for risk stratification at bedside.
The place these scores could have to guide therapeutic strategies is yet to be determined. Their most promising use may be as a tool to guide hospital admission, in the context of a pandemic with a high demand and a low offer for hospital beds, especially in low-income countries [43, 44]. Further studies should be conducted on this important issue.
Scores specifically derived for COVID-19 outperformed generic scores for infectious pneumonia or for sepsis. This highlights the specificity of COVID-19 in comparison to other forms of pneumonia, with a key role for the inflammatory and pro-thrombotic status to drive severity [45–47]. However, given their simplicity of use and their good performance to predict in-hospital mortality in our cohort, scores such as the CURB-65 or A-DROP scores could still be considered for risk stratification in COVID-19 patients. On the opposite, scores used in sepsis such as qSOFA or SIRS seemed to offer no clear benefit for risk stratification. Low specificity can be explained by a limited number of factors used for initial evaluation, as many patients present with abnormal vital signs or white blood cells counts, and those factors alone are insufficient to identify patients at high risk for critical illness. Low sensitivity can be explained as patients truly at risk for critical illness (particularly the elderly or patients with many comorbidities) may initially appear clinically stable before suddenly and dramatically worsening.
Accuracy was lower in our cohort to predict ICU admission compared to in-hospital mortality, even for scores specifically aimed at predicting this endpoint. This could partly be explained by the complexity of ICU admission criteria, which may differ across countries, according to local guidelines and demography, and may vary with time given the pressure on ICU beds [48]. In France for example, during the first wave of the pandemic, some patients with invasive mechanical ventilation urgently initiated in the emergency room or in general wards could not be transferred to the hospital-related ICU due to shortage of beds, and were transferred to other hospitals, either in the Paris region or in other regions [49].
In conclusion, several scores using routinely collected clinical and biological data have a fair accuracy to predict in-hospital death. The 4C Mortality Score and the ABCS stand out because they performed as well in our cohort and their initial validation cohort, during the first epidemic wave and subsequent waves, and in younger and older patients. Their use to guide appropriate clinical care and resource utilization should be evaluated in future studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Data used in preparation of this article were obtained from the AP-HP COVID CDW Initiative database. A complete listing of the members can be found at https://eds.aphp.fr/covid-19. We thank the four anonymous reviewers for their insightful comments and constructive suggestions.
Members of AP-HP/Universities/INSERM COVID-19 Research Collaboration AP-HP COVID CDR Initiative: Pierre-Yves Ancel (APHP Paris University Center), Alain Bauchet (APHP Saclay University), Nathanael Beeker (APHP Paris University Center), Vincent Benoit (WIND Department APHP Greater Paris University Hospital), Romain Bey (WIND Department APHP Greater Paris University Hospital), Aurélie Bourmaud (APHP Paris University North), Stéphane Bréant (WIND Department APHP Greater Paris University Hospital), Anita Burgun (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Fabrice Carrat (APHP Sorbonne University), Charlotte Caucheteux (Université Paris-Saclay, Inria, CEA), Julien Champ (INRIA Sophia-Antipolis – ZENITH team, LIRMM, Montpellier, France), Sylvie Cormont (WIND Department APHP Greater Paris University Hospital), Julien Dubiel (WIND Department APHP Greater Paris University Hospital), Catherine Duclos (APHP Paris Seine Saint Denis Universitary Hospital), Loic Esteve (SED/SIERRA, Inria Centre de Paris), Marie Frank (APHP Saclay University), Nicolas Garcelon (Imagine Institute), Alexandre Gramfort (Université Paris-Saclay, Inria, CEA), Nicolas Griffon ("WIND Department APHP Greater Paris University Hospital UMRS1142 INSERM"), Olivier Grisel (Université Paris-Saclay, Inria, CEA), Martin Guilbaud (WIND Department APHP Greater Paris University Hospital), Claire Hassen-Khodja (Direction of the Clinical Research and Innovation, AP-HP), François Hemery (APHP Henri Mondor University Hospital), Martin Hilka (WIND Department APHP Greater Paris University Hospital), Anne Sophie Jannot (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Jerome Lambert (APHP Paris University North), Richard Layese (APHP Henri Mondor University Hospital), Léo Lebouter (WIND Department APHP Greater Paris University Hospital), Damien Leprovost (Clevy.io), Ivan Lerner (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Kankoe Levi Sallah (APHP Paris University North), Aurélien Maire (WIND Department APHP Greater Paris University Hospital), Marie-France Mamzer (President of the AP-HP IRB), Patricia Martel (APHP Saclay University), Arthur Mensch (ENS, PSL University), Thomas Moreau (Université Paris-Saclay, Inria, CEA), Antoine Neuraz (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Nina Orlova (WIND Department APHP Greater Paris University Hospital), Nicolas Paris (WIND Department APHP Greater Paris University Hospital), Bastien Rance (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Hélène Ravera (WIND Department APHP Greater Paris University Hospital), Antoine Rozes (APHP Sorbonne University), Pierre Rufat (APHP Sorbonne University), Elisa Salamanca (WIND Department APHP Greater Paris University Hospital), Arnaud Sandrin (WIND Department APHP Greater Paris University Hospital), Patricia Serre (WIND Department APHP Greater Paris University Hospital), Xavier Tannier (Sorbonne University), Jean-Marc Treluyer (APHP Paris University Center), Damien Van Gysel (APHP Paris University North), Gael Varoquaux (Université Paris-Saclay, Inria, CEA, Montréal Neurological Institute, McGill University), Jill-Jênn Vie (SequeL, Inria Lille), Maxime Wack (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital), Perceval Wajsburt (Sorbonne University), Demian Wassermann (Université Paris-Saclay, Inria, CEA), Eric Zapletal (Department of Biomedical Informatics, HEGP, APHP Greater Paris University Hospital.
Author contributions
YL: conceptualization, methodology, software, formal analysis, investigation, writing—original draft, writing—review and editing, visualization; LA, PS: validation, methodology, writing—review and editing; GL, MB: software, writing—review and editing; JL, AB: software, methodology, writing—review and editing; QR: methodology, writing—original draft, writing—review and editing; OS: conceptualization, methodology, formal analysis, writing—original draft, writing—review and editing, supervision, project administration.
Funding
None.
Availability of data and material
Raw data cannot be transmitted to non-GPUH staff without specific authorization from the GPUH CDW Scientific and Ethics Committee.
Code availability
R scripts are available at request to the corresponding author.
Declarations
Conflicts of interest
None for any of the authors.
Ethical approval
The GPUH’s CDW Scientific and Ethics Committee (IRB00011591) granted access to the CDW for the purpose of this study (authorization n°200063).
Footnotes
The members of AP-HP/Universities/INSERM COVID-19 Research Collaboration AP-HP COVID CDR Initiative are listed in Acknowledgements.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Loris Azoyan and Piotr Szychowiak contributed equally to this work.
Contributor Information
Olivier Steichen, Email: olivier.steichen@aphp.fr.
on behalf of the AP-HP/Universities/INSERM COVID-19 Research Collaboration AP-HP COVID CDR Initiative:
Pierre-Yves Ancel, Alain Bauchet, Nathanael Beeker, Vincent Benoit, Romain Bey, Aurélie Bourmaud, Stéphane Bréant, Anita Burgun, Fabrice Carrat, Charlotte Caucheteux, Julien Champ, Sylvie Cormont, Julien Dubiel, Catherine Duclos, Loic Esteve, Marie Frank, Nicolas Garcelon, Alexandre Gramfort, Nicolas Griffon, Olivier Grisel, Martin Guilbaud, Claire Hassen-Khodja, François Hemery, Martin Hilka, Anne Sophie Jannot, Jerome Lambert, Richard Layese, Léo Lebouter, Damien Leprovost, Ivan Lerner, Kankoe Levi Sallah, Aurélien Maire, Marie-France Mamzer, Patricia Martel, Arthur Mensch, Thomas Moreau, Antoine Neuraz, Nina Orlova, Nicolas Paris, Bastien Rance, Hélène Ravera, Antoine Rozes, Pierre Rufat, Elisa Salamanca, Arnaud Sandrin, Patricia Serre, Xavier Tannier, Jean-Marc Treluyer, Damien Van Gysel, Gael Varoquaux, Jill-Jênn Vie, Maxime Wack, Perceval Wajsburt, Demian Wassermann, and Eric Zapletal
References
- 1.WHO (2021) Novel coronavirus—China. In: WHO. http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/. Accessed 21 Feb 2021
- 2.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 4.COVID-ICU Group on behalf of the REVA Network and the COVID-ICU Investigators Clinical characteristics and day-90 outcomes of 4244 critically ill adults with COVID-19: a prospective cohort study. Intensive Care Med. 2020;47:60–73. doi: 10.1007/s00134-020-06294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinberg E, Balakrishna A, Habboushe J, et al. Calculated decisions: COVID-19 calculators during extreme resource-limited situations. Emerg Med Pract. 2020;22:CD1–CD5. [PubMed] [Google Scholar]
- 6.Wynants L, Calster BV, Bonten MMJ, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balmford B, Annan JD, Hargreaves JC, et al. Cross-country comparisons of Covid-19: policy, politics and the price of life. Environ Resour Econ. 2020;76:525–551. doi: 10.1007/s10640-020-00466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benchimol EI, Smeeth L, Guttmann A, et al. The REporting of studies Conducted using Observational Routinely-collected health Data (RECORD) Statement. PLOS Med. 2015;12:e1001885. doi: 10.1371/journal.pmed.1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [DOI] [PubMed] [Google Scholar]
- 10.Bannay A, Chaignot C, Blotière P-O, et al. The best use of the Charlson Comorbidity Index with electronic health care database to predict mortality. Med Care. 2016;54:188–194. doi: 10.1097/MLR.0000000000000471. [DOI] [PubMed] [Google Scholar]
- 11.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 12.Knight SR, Ho A, Pius R, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC who clinical characterisation protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mejía-Vilet JM, Córdova-Sánchez BM, Fernández-Camargo DA, et al. A risk score to predict admission to the intensive care unit in patients with Covid-19: the ABC-GOALS score. Salud Publica Mex. 2020;63:1–11. doi: 10.21149/11684. [DOI] [PubMed] [Google Scholar]
- 14.Jiang M, Li C, Zheng L, et al. A biomarker-based age, biomarkers, clinical history, sex (ABCS)-mortality risk score for patients with coronavirus disease 2019. Ann Transl Med. 2021 doi: 10.21037/atm-20-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng Z, Chen Q, Li S, et al. ANDC: an early warning score to predict mortality risk for patients with coronavirus disease 2019. J Transl Med. 2020;18:328. doi: 10.1186/s12967-020-02505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennouar S, Bachir Cherif A, Kessira A, et al. Development and validation of a laboratory risk score for the early prediction of COVID-19 severity and in-hospital mortality. Intensive Crit Care Nurs. 2021 doi: 10.1016/j.iccn.2021.103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruocco G, McCullough PA, Tecson KM, et al. Mortality risk assessment using CHA(2)DS(2)-VASc scores in patients hospitalized with coronavirus disease 2019 infection. Am J Cardiol. 2020;137:111–117. doi: 10.1016/j.amjcard.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho S-Y, Park S-S, Song M-K, et al. Prognosis score system to predict survival for COVID-19 cases: a Korean Nationwide Cohort Study. J Med Internet Res. 2021;23:e26257. doi: 10.2196/26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanboğa IH, Canpolat U, Çetin EHÖ, et al. Development and validation of clinical prediction model to estimate the probability of death in hospitalized patients with COVID-19: Insights from a nationwide database. J Med Virol. 2021;93:3015–3022. doi: 10.1002/jmv.26844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berenguer J, Borobia AM, Ryan P, et al. Development and validation of a prediction model for 30-day mortality in hospitalised patients with COVID-19: the COVID-19 SEIMC score. Thorax. 2021 doi: 10.1136/thoraxjnl-2020-216001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajifathalian K, Sharaiha RZ, Kumar S, et al. Development and external validation of a prediction risk model for short-term mortality among hospitalized U.S. COVID-19 patients: a proposal for the COVID-AID risk tool. PLoS ONE. 2020;15:e0239536. doi: 10.1371/journal.pone.0239536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebell MH, Cai X, Lennon R, et al. Development and validation of the COVID-NoLab and COVID-SimpleLab risk scores for prognosis in 6 US health systems. J Am Board Fam Med. 2021;34:S127–S135. doi: 10.3122/jabfm.2021.S1.200464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hachim MY, Hachim IY, Naeem KB, et al. d-Dimer, troponin, and urea level at presentation with COVID-19 can predict ICU admission: a single centered study. Front Med. 2020 doi: 10.3389/fmed.2020.585003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, Yao N, Qiu Y. Comparing rapid scoring systems in mortality prediction of critical ill patients with novel coronavirus disease. Acad Emerg Med. 2020 doi: 10.1111/acem.13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamal MH, Doi SA, AlYouha S, et al. A Biomarker based severity progression indicator for COVID-19: the Kuwait Prognosis Indicator Score. Biomarkers. 2020 doi: 10.1080/1354750X.2020.1841296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soto-Mota A, Marfil-Garza BA, Rodríguez EM, et al. The low-harm score for predicting mortality in patients diagnosed with COVID-19: a multicentric validation study. J Am Coll Emerg Physicians Open. 2020;1:1436–1443. doi: 10.1002/emp2.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mei Y, Weinberg SE, Zhao L, et al. Risk stratification of hospitalized COVID-19 patients through comparative studies of laboratory results with influenza. EClinicalMedicine. 2020;26:100475. doi: 10.1016/j.eclinm.2020.100475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myrstad M, Ihle-Hansen H, Tveita AA, et al. National Early Warning Score 2 (NEWS2) on admission predicts severe disease and in-hospital mortality from Covid-19 - a prospective cohort study. Scand J Trauma Resusc Emerg Med. 2020;28:66. doi: 10.1186/s13049-020-00764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Chen Y, Chen S, et al. Derivation and validation of a prognostic model for predicting in-hospital mortality in patients admitted with COVID-19 in Wuhan, China: the PLANS (platelet lymphocyte age neutrophil sex) model. BMC Infect Dis. 2020;20:959. doi: 10.1186/s12879-020-05688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartoletti M, Giannella M, Scudeller L, et al. Development and validation of a prediction model for severe respiratory failure in hospitalized patients with SARS-CoV-2 infection: a multicentre cohort study (PREDI-CO study) Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2020;26:1545–1553. doi: 10.1016/j.cmi.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saberian P, Tavakoli N, Hasani-Sharamin P, et al. Accuracy of the pre-hospital triage tools (qSOFA, NEWS, and PRESEP) in predicting probable COVID-19 patients’ outcomes transferred by Emergency Medical Services. Casp J Intern Med. 2020;11:536–543. doi: 10.22088/cjim.11.0.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dam PM, Zelis N, Stassen P, et al. Validating the RISE UP score for predicting prognosis in patients with COVID-19 in the emergency department: a retrospective study. BMJ Open. 2021;11:e045141. doi: 10.1136/bmjopen-2020-045141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ageno W, Cogliati C, Perego M, et al. Clinical risk scores for the early prediction of severe outcomes in patients hospitalized for COVID-19. Intern Emerg Med. 2021 doi: 10.1007/s11739-020-02617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holten AR, Nore KG, Tveiten CEVWK, et al. Predicting severe COVID-19 in the Emergency Department. Resusc Plus. 2020;4:100042. doi: 10.1016/j.resplu.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demir MC, Ilhan B. Performance of the Pandemic Medical Early Warning Score (PMEWS), Simple Triage Scoring System (STSS) and Confusion, Uremia, Respiratory rate, Blood pressure and age ≥ 65 (CURB-65) score among patients with COVID-19 pneumonia in an emergency department triage setting: a retrospective study. Sao Paulo Med J. 2021;139:170–177. doi: 10.1590/1516-3180.2020.0649.R1.10122020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Zuo P, Liu Y, et al. Clinical and laboratory predictors of in-hospital mortality in patients with coronavirus disease-2019: a cohort study in Wuhan, China. Clin Infect Dis. 2020;71:2079–2088. doi: 10.1093/cid/ciaa538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterne JAC, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jannot A-S, Coutouris H, Burgun A et al (2020) COVID-19, a social disease in Paris: a socio-economic wide association study on hospitalized patients highlights low-income neighbourhood as a key determinant of severe COVID-19 incidence during the first wave of the epidemic. medRxiv 2020.10.30.20222901. 10.1101/2020.10.30.20222901
- 40.Kurtz P, Bastos LSL, Dantas LF, et al. Evolving changes in mortality of 13,301 critically ill adult patients with COVID-19 over 8 months. Intensive Care Med. 2021;47:538–548. doi: 10.1007/s00134-021-06388-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du R-H, Liang L-R, Yang C-Q, et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020 doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azoulay E, Fartoukh M, Darmon M, et al. Increased mortality in patients with severe SARS-CoV-2 infection admitted within seven days of disease onset. Intensive Care Med. 2020;46:1714–1722. doi: 10.1007/s00134-020-06202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma X, Vervoort D. Critical care capacity during the COVID-19 pandemic: Global availability of intensive care beds. J Crit Care. 2020;58:96–97. doi: 10.1016/j.jcrc.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen-Crowe B, Sutherland M, McKenney M, Elkbuli A. A closer look into global hospital beds capacity and resource shortages during the COVID-19 pandemic. J Surg Res. 2021;260:56–63. doi: 10.1016/j.jss.2020.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McElvaney OJ, McEvoy NL, McElvaney OF, et al. Characterization of the inflammatory response to severe COVID-19 illness. Am J Respir Crit Care Med. 2020;202:812–821. doi: 10.1164/rccm.202005-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goligher EC, Ranieri VM, Slutsky AS. Is severe COVID-19 pneumonia a typical or atypical form of ARDS? And does it matter? Intensive Care Med. 2020 doi: 10.1007/s00134-020-06320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helms J, Tacquard C, Severac F, et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med. 2020;46:1089–1098. doi: 10.1007/s00134-020-06062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sprung CL, Joynt GM, Christian MD, et al. Adult ICU triage during the coronavirus disease 2019 pandemic: who will live and who will die? Recommendations to improve survival. Crit Care Med. 2020 doi: 10.1097/CCM.0000000000004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Painvin B, Messet H, Rodriguez M, et al. Inter-hospital transport of critically ill patients to manage the intensive care unit surge during the COVID-19 pandemic in France. Ann Intensive Care. 2021;11:54. doi: 10.1186/s13613-021-00841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data cannot be transmitted to non-GPUH staff without specific authorization from the GPUH CDW Scientific and Ethics Committee.
R scripts are available at request to the corresponding author.