Abstract
Molecular signatures are being increasingly integrated into predictive biology applications. However, there are limited studies comparing the overall predictivity of transcriptomic versus epigenomic signatures in relation to perinatal outcomes. This study set out to evaluate mRNA and microRNA (miRNA) expression and cytosine-guanine dinucleotide (CpG) methylation signatures in human placental tissues and relate these to perinatal outcomes known to influence maternal/fetal health; namely, birth weight, placenta weight, placental damage, and placental inflammation. The following hypotheses were tested: (1) different molecular signatures will demonstrate varying levels of predictivity towards perinatal outcomes, and (2) these signatures will show disruptions from an example exposure (ie, cadmium) known to elicit perinatal toxicity. Multi-omic placental profiles from 390 infants in the Extremely Low Gestational Age Newborns cohort were used to develop molecular signatures that predict each perinatal outcome. Epigenomic signatures (ie, miRNA and CpG methylation) consistently demonstrated the highest levels of predictivity, with model performance metrics including R2 (predicted vs observed) values of 0.36–0.57 for continuous outcomes and balanced accuracy values of 0.49–0.77 for categorical outcomes. Top-ranking predictors included miRNAs involved in injury and inflammation. To demonstrate the utility of these predictive signatures in screening of potentially harmful exogenous insults, top-ranking miRNA predictors were analyzed in a separate pregnancy cohort and related to cadmium. Key predictive miRNAs demonstrated altered expression in association with cadmium exposure, including miR-210, known to impact placental cell growth, blood vessel development, and fetal weight. These findings inform future predictive biology applications, where additional benefit will be gained by including epigenetic markers.
Keywords: computational toxicology, predictive biology, multi-omics, epigenomics, placenta, machine learning
Predictive biology efforts are rapidly expanding to address the need to accelerate patient diagnoses, drug development, and chemical safety assessments, spanning biomedical, pharmaceutical, and toxicological applications. Key to advancing predictive biology is the implementation of molecular-based approaches, which can be used to rapidly screen for disease state, drug efficacy, and chemical safety. As an example, transcriptomic signatures represent important molecular endpoints that are now being integrated into chemical toxicity testing as they provide important insight into disease mechanisms and toxicological responses through genome-wide approaches (Rager and Fry, 2013). Furthermore, epigenomic signatures represent underutilized endpoints that could be used to further inform potential toxicity outcomes (Chappell and Rager, 2017; Clark and Rager, 2020). To effectively integrate such molecular signatures into predictive biology applications, it is first necessary to identify which signatures are predictive of outcomes of interest to human health. Given the worldwide prevalence of low birth weight (14.6%) (Blencowe et al., 2019) and preterm birth (11%; Walani, 2020), among other adverse birth outcomes, molecular signatures predictive of perinatal outcomes should be prioritized.
Predictive-omics signatures were first developed in the medical field to diagnose and prognose diseases (Bao et al., 2019; Friedman et al., 2009; Lima et al., 2010). These signatures have also been implemented in precision medicine, informing therapeutic strategies for specific disease subtypes (Cohen et al., 2011). In the field of toxicology, recent efforts have been aimed at establishing transcriptomic signatures predictive of adverse outcomes. One specific example that has started to gain regulatory acceptance is the toxicogenomics-DNA damage-inducing biomarker gene set, which includes 65 genes that demonstrate high accuracy in predicting genotoxic versus nongenotoxic chemicals (FDA, 2017; Li et al., 2015). Continued development of signatures predictive of adverse outcomes will improve and expedite risk assessment processes (Pettit et al., 2010). Notably, signatures predictive of perinatal outcomes have yet to be tested.
Transcriptomic signatures are becoming integrated into predictive biology applications, whereas epigenetic signatures largely remain under development. Epigenetic signatures are potentially heritable molecular mediators that can modify the levels at which genes are expressed without altering the underlying DNA sequence (Clark and Rager, 2020). These changes occur in response to intrinsic and environmental stimuli and include the methylation of cytosine-guanine (CpG) dinucleotides and degradation of mRNA transcripts by microRNAs (miRNAs; Clark and Rager, 2020). Epigenetic signatures have been evaluated for over a decade as potential disease biomarkers and therapeutic targets (Deng et al., 2010; Wang et al., 2008), although validated signatures in cohort studies remain limited (Szejniuk et al., 2019). In the context of chemical safety evaluations, epigenetic signatures have only recently been considered in the context of chemical regulation and risk assessment (LaRocca et al., 2017; Rager et al., 2017). Still, to our knowledge there have been no studies comparing the predictivity of transcriptomic versus epigenomic signatures in relation to perinatal outcomes.
The placenta represents an ideal target for the testing of molecular signatures that may predict perinatal outcomes. The placenta serves as the master regulator of the fetal environment through transport of nutrients, metabolism, hormone secretion, and selective protection to ensure proper growth and development of the fetus (Burton et al., 2016). In linking mother and fetus, the placenta can be exposed to environmental factors present in maternal circulation, resulting in harmful molecular changes (Vlahos et al., 2019). Toxicant-induced alterations can lead to placental functional dysregulation and ultimately impact maternal/fetal health (Bianco-Miotto et al., 2017; Rager et al., 2020). Recent studies have identified placental transcriptomic and epigenomic signatures associated with select perinatal outcomes, such as changes in birth weight (Payton et al., 2020) and placental phenotypes (eg, inflammation; Konwar et al., 2018). These phenotypes are risk factors for adverse birth outcomes that may impact later-in-life health. For instance, low birth weight is associated with an increased risk of morbidity and later-life cardiovascular disease, among other diseases (Blencowe et al., 2019). In addition, acute placental inflammation is a risk factor for neonatal mortality and morbidity, bronchopulmonary dysplasia, and later-life neurocognitive and developmental outcomes (Goldstein et al., 2020). Once these signatures are tested for predictivity and further evaluated in additional cohorts, such signatures could be leveraged for the diagnosis of medical health outcomes, pharmaceutical development, and screening of chemicals with unknown toxicity.
This study aimed to identify placental transcriptomic and epigenomic signatures associated with perinatal outcomes and compare their levels of predictivity. Placental molecular signatures were evaluated in infants from the ongoing Extremely Low Gestational Age Newborns (ELGAN) cohort, and signatures were related to various perinatal health outcomes. Specifically, placental mRNA, miRNA, and CpG methylation profiles were evaluated in relation to birth weight, placenta weight, placental damage, and placental inflammation. These outcomes were selected for 2 reasons: first, these phenotypes are recognized to significantly influence maternal and fetal health outcomes (Burton et al., 2016; Rager et al., 2020); second, these phenotypes overlap with those that are commonly collected during the testing of pharmaceuticals and industrial chemicals for potential prenatal exposure-induced toxicity (Fry et al., 2019; Tabacova et al., 2003). Genomic and epigenomic signatures were ranked according to their ability to predict outcomes using a machine learning approach. Findings were then applied in an additional pregnancy cohort, where predictive signatures were evaluated in association to cadmium, a known perinatal toxicant. This study is significant in that it evaluated transcriptomic and epigenomic signatures specific to perinatal outcomes in humans, demonstrating methods and molecular information that can be used to inform medical applications, pharmaceutical development, and chemical screening strategies.
MATERIALS AND METHODS
Study overview
This study was designed to evaluate the predictivity of genomic (mRNA) and epigenomic (miRNA and CpG methylation) signatures towards perinatal outcomes in humans, based on placental molecular signatures. The overall steps are summarized in Figure 1, and further detailed below. As an overview, genome-wide molecular signatures were measured within placenta samples collected through the ongoing ELGAN cohort, alongside tissue pathology and demographic data. Data surrounding this cohort are expansive and have served as the basis of previous evaluations, including multi-omic analyses (Addo et al., 2019; Payton et al., 2020; Santos et al., 2020). This study leveraged this dataset to address new questions while employing new data processing and statistical methods, which are detailed below. The outcomes selected for evaluation focused on phenotypes that are recognized to significantly influence maternal and fetal health outcomes (Burton et al., 2016; Fry et al., 2019) and overlap with those that are commonly collected during the in vivo safety testing of pharmaceuticals and industrial chemicals (Fry et al., 2019; Tabacova et al., 2003). These outcomes included infant birth weight, placenta weight, placental damage (ie, infarction), and placental inflammation (ie, neutrophil influx). Molecular signatures were then used to train and test models to predict each phenotype, while carefully considering various predictor variable selection strategies. The overall performance of each molecular category was then compared. Select top-ranking signatures were further evaluated in a separate cohort, in which these signatures were related to exposure to the known perinatal toxicant, cadmium.
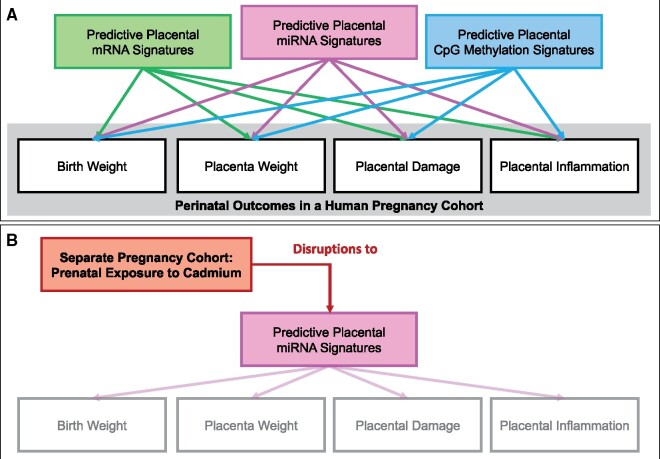
Figure 1.
Study overview separated into (A) identifying predictive -omic signatures, and (B) testing of an example signature to screen for harmful exposures. This study set-out to compare the predictivity between placental mRNA expression, microRNA (miRNA) expression, and cytosine-guanine (CpG) methylation signatures towards the predicting of perinatal outcomes in humans. A, First, models were trained and tested using mRNAs, miRNAs, or CpG methylation sites identified as associated with each outcome. We also trained and tested similar models using the top 100-ranking mRNAs, miRNAs, or CpG sites, based on significance, to evaluate whether the comparative predictivities would change based on the number of predictor variables. Results of these models were compared, and molecules identified to contribute to overall predictivity above noise. B, Second, we tested whether these predictive signatures could be used to screen for potential perinatal toxicants, using the identified predictive miRNA signature in relation to prenatal exposure to the known toxicant, cadmium, in a separate pregnancy cohort.
ELGAN study cohort recruitment and placenta tissue collection
Placenta samples were collected from the ongoing ELGAN cohort, consisting of mother and infant pairings from pregnancies that lasted <28 weeks of gestation (McElrath et al., 2008; Payton et al., 2020). Subjects were recruited and samples collected using methods that have previously been detailed (Beaumont et al., 2017). In brief, pregnant women scheduled to give birth at 1 of 14 participating ELGAN sites were recruited for participation between 2002 and 2004. Each of the participating institutions had study procedures that were approved by their respective Institutional Review Boards, with consent provided by each participant (Beaumont et al., 2017). Within the full study cohort, 1506 infants and 1249 mothers were enrolled. Demographic information and pregnancy-specific measures were collected by trained research nurses postdelivery through the application of structured questionnaires (McElrath et al., 2008). Infant anthropometric measures, including birth weight, were recorded (Streimish et al., 2012).
Study participants additionally consented to the collection and evaluation of placenta samples, using methods that have been previously detailed (Addo et al., 2019; Onderdonk et al., 2008). In brief, placentas were collected immediately postdelivery, placed into sterile basins, and transported to separate sampling rooms for biopsies. Placentas were further processed by pulling the amnion surrounding the embryonic sack apart from the fetally derived chorion. Tissue samples were trimmed from the chorion base after applying traction to the trophoblast tissue below. Collected tissues were placed into sterile 2 ml cryovials that were submerged in liquid nitrogen, frozen, and shipped to the University of North Carolina at Chapel Hill (UNC-Chapel Hill). The samples analyzed in the current evaluation included 390 placentas that were profiled for genomic and epigenomic signatures.
Perinatal phenotypic outcome measurements
Measurements that were collected during the ELGAN cohort evaluation included those that inform potential perinatal outcomes, namely, infant birth weight, placenta weight, placental damage, and placental inflammation. Infant birth weight (as a continuous variable) was measured using standard hospital protocols, where an infant is weighed immediately after delivery. Placenta weight (as a continuous variable) was measured using the placenta samples collected immediately postdelivery, which were placed inside sterile basins, and then evaluated by a pathologist within a separate examination room where placentas were trimmed and weighed (Hecht et al., 2008). Histological examinations of placentas were completed within 24 h of delivery according to the College of American Pathologists guidelines. Representative sections were collected from (1) all abnormal areas, (2) routine sections of the umbilical cord and membrane roll, and (3) 2 full thickness sections from the center and a paracentral zone of the placental disc. Placental damage (as a binary variable: 0 = absent, 1 = present) were noted as the absence or presence of lesions during examination, specifically defined as the presence of infarction. Placental inflammation (as a binary variable: 0 = none, 1 = moderate or severe) was measured through histopathological evaluation of the chorioamniotic section of the placenta based on neutrophil count (0, moderate = 1–19 neutrophils/20×; severe≥ 20 neutrophils/20×; Hecht et al., 2008).
Placental DNA and RNA extraction and molecular signature profiling
Placental tissues (approximately 0.02 g) were cut from the frozen biopsy samples that were transferred to UNC-Chapel Hill and rinsed with sterile 1 × PBS to remove residual blood. Washed samples were immediately snap frozen while in homogenization tubes and placed on dry ice to preserve DNA/RNA integrity. Samples were then homogenized in Buffer RLT with β-mercaptoethanol (Qiagen, Valencia, California). An AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) was used to extract DNA and RNA. Resulting DNA and RNA quantities were measured using the NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts) and tested for quality using the QIAxcel system (Qiagen) and the LabChip instrument (Perkin Elmer, Massachusetts), respectively. DNA quality DV200 values and RNA integrity numbers were within acceptable ranges for the implemented Infinium MethylationEPIC BeadChip (Weisenberger et al., 2021) and Lexogen sequencing technologies (Lexogen, 2021) as described in our previous analyses using the same cohort and corresponding samples (Eaves et al., 2020), with methods also detailed below.
Genome-wide mRNA expression profiles were measured using the QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen, Vienna, Austria). Libraries were prepared using Lexogen’s recommended protocol (Lexogen, 2021). Resulting libraries were pooled and sequenced (single-end 50 bp) with the Illumina HiSeq 2500 using one lane. Sequencing read counts per mRNA were aligned and organized using Salmon to the GENCODE database (v30) (Harrow et al., 2012; Patro et al., 2017), resulting in expression measures across 37,268 RNA transcripts, including protein coding and noncoding RNAs. Genome-wide miRNA expression profiles were measured using the HTG EdgeSeq miRNA Whole Transcriptome Assay (HTG Molecular Diagnostics, Tucson, Arizona). miRNA libraries were pooled and sequenced (single-end 50 bp) on one lane of the Illumina Hiseq 2500. Libraries were prepared by on the Sciclone G3 (Perkin Elmer) by automation to avoid potential batch to batch artifacts. Sequencing read counts per miRNA were aligned to miRbase (v20) and organized through Parser (HTG Molecular Diagnostics), yielding expression measures of 2083 human miRNA transcripts. Thus, the current analysis focused on miRNAs, as opposed to other small RNA types. To further ensure high-quality sequencing data, we carried out additional quality assessment/quality control steps, including (1) the filtering of lowly expressed mRNAs/miRNAs (described below); (2) the inclusion of surrogate variables within statistical models to account for additional sources of heterogeneity across samples (described below); (3) the confirmation that published mRNA and miRNA placental-specific clusters were captured in this dataset, as described in detail and published in our previous study using the same data (Eaves et al., 2020).
Extracted DNA samples were bisulfate-converted using the EZ DNA methylation kit (Zymo Research, Irvine, California) and associated methylation levels quantification using the Infinium MethylationEPIC BeadChip (Illumina, San Diego, California). To minimize batch effects, samples were randomly allocated to different plates and chips. This resulted in the profiling more than 850 000 total CpG sites across the human genome, 616 598 of which were annotated and associated with 26 642 unique genes. Resulting molecular signature data were used in downstream data processing and statistical analyses. Raw and processed signature data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository and are publicly available under GEO series GSE154829 and GSE167885 (NCBI, 2020).
Data processing and statistical analysis to identify mRNAs and miRNAs associated with perinatal outcomes
The mRNA and miRNA sequencing data were processed separately through similar pipelines in R Software (v3.6.2). Count data were first filtered to exclude universally lowly expressed transcripts, requiring that genes be expressed at signals above the overall median (equal to zero) in at least 25% of the samples, resulting in a final panel of 10 412 mRNAs and 1130 miRNAs, paralleling our previously published methods (Klaren et al., 2019; Payton et al., 2020; Rager et al., 2017). Potential sample outliers were evaluated using principal component analysis, with calculations and visualizations produced using the prcomp function in the basic statistical packages in R. Outliers were additionally assessed using hierarchical clustering, with distance metrics calculated and visualized using the hclust function. Sample outliers that were identified through both approaches were excluded. Resulting count data were normalized by median-of-ratio estimates based on sample-specific size factors enabled through the DESeq2 package (v1.24.0), yielding variance stabilized expression values (Love et al., 2014). Potential sources of sample heterogeneity and batch effects were addressed through surrogate variable analysis (SVA) using the SVA R package (version 3.36.0). Control probes were estimated using default parameters, and 3 significant surrogate variables were calculated and included in the final statistical models as covariates (Leek, 2014; Leek et al., 2012).
Covariates related to the evaluated perinatal outcomes were selected for inclusion in statistical models based on an approach that builds upon our previously published methods (Clark et al., 2019; Payton et al., 2020; Rager et al., 2017). First, nonparametric statistical tests were carried out to identify significant (p < .05), plausible associations between variables and each outcome. This list was then informed through directed acyclic graph approaches by extant literature on associations between (1) variables and each outcome and (2) variables and genomic/epigenomic alterations (Shrier and Platt, 2008). Using these lists, sensitivity analyses evaluating various combinations of potential confounders were performed to identify covariate combinations that balanced reducing bias with increasing precision (evaluated based on comparing fold change and standard error estimates), while reducing the potential for collinearity amongst variables. This resulted in the following list of covariates: for infant birth weight: gestational age (days), exposure to first- or secondhand smoking (0 = no exposure; 1 = any exposure), and multiple gestations (0 = singleton; 1 = nonsingleton); for placental weight: body mass index (BMI), gestational age (days), multiple gestations (0 = singleton; 1 = nonsingleton), and sex; for placental damage: BMI and maternal age; and for placental inflammation: BMI and maternal age. It is notable that we did not further stratify our models according to infant sex, as we recently carried out a similar analysis in the same cohort evaluating multi-omic signatures in relation to one of the outcomes (ie, birth weight) and found largely overlapping responses in mRNAs and miRNAs in male and female infants (Payton et al., 2020); though sex was still considered during covariate selection and included in the placental weight models, as detailed earlier.
Statistical models were then implemented to identify mRNA and miRNA signatures associated with each of the perinatal outcomes, separately. Outcomes were either continuous (for birth weight and placental weight, with values log-transformed) or binary (for placental damage and inflammation). Models were based on negative binomial generalized linear models enabled through the DESeq2 package in R (v3.12), using shrunken logarithmic fold changes in expression (Love et al., 2014). Z-statistics were calculated by dividing fold change values by standard errors and used to compare against standard normal distribution curves to generate Wald test p values, as previously described (Love et al., 2014). Resulting p values were adjusted for multiple testing through the Benjamini-Hochberg (BH) procedure (Benjamini and Hochberg, 1995). The mRNAs and miRNAs with BH-adjusted p < .10 were identified as significantly associated with a perinatal outcome.
Data processing and statistical analysis to identify CpG methylation sites associated with perinatal outcomes
DNA methylation data were processed and analyzed in R Software using methods similar to those previously implemented (Rager et al., 2015; Santos et al., 2019). In brief, CpG methylation data were preprocessed using the minfi package in R (Aryee et al., 2014; Teschendorff and Zheng, 2017). Quality control was carried out to identify sample outliers and those with inadequate signals across probes, excluding samples that failed. Functional normalization was performed using the normal-exponential out-of-band correction method, followed by the functional normalization method with the top 2 principal components of the control matrix (Fortin et al., 2014, 2017; Triche et al., 2013). A total of 856 832 CpG sites were available for downstream analyses following quality control. Lastly, the ComBat function was used from the sva package to adjust for potential batch effects across plates (Leek, 2014; Leek et al., 2012). Methylation levels were calculated and expressed as β values (β = intensity of the methylated allele (M))/(intensity of the unmethylated allele (U) + intensity of the methylated allele (M) + 100). β values were logit transformed to M values prior to analysis. To evaluate differential CpG methylation in relation to each phenotypic outcome, robust linear regression models were fit at each CpG site. This method protects against potential heteroscedasticity and incorporates test statistics modified using the Phipson’s robust empirical Bayes procedure, shrinking probe-wide sample variances towards a common value and controlling for test-statistic inflation (Phipson et al., 2016; Teschendorff and Zheng 2017). Perinatal outcomes were evaluated separately, adjusting for the same set of covariates included in miRNA and mRNA analyses. In controlling for cell-type heterogeneity, the top 10 component variables derived from SVA were selected, as described in the mRNA and miRNA analysis. Models were first fit at over 850 000 CpG sites. Results from CpGs not annotated or associated with genes were excluded prior to identifying statistically significant associations based on BH-adjusted p < .10.
Predictive modeling of perinatal outcome-associated molecular signatures
Models were built using placental mRNA, miRNA, and CpG methylation signatures to predict perinatal outcomes through random forest modeling. Predictor variables were selected using a biologically driven approach, where molecular signatures were first filtered using a significance threshold of BH-adjusted p < .10 or top 100 ranking genes based on BH-adjusted p in association with each outcome. For comparison, predictor variables were also selected using an empirically driven approach, where molecular signatures were first filtered to reduce autocorrelation using the findCorrelation function in the caret R package (RDocumentation, 2021a). Here, molecules were selected to remove instances of molecular pairings having R > 0.80. The biological approach was found to out-perform the empirical approach based on model performance parameters, and because we were interested in ultimately interpreting the potential biological consequences of these predictor variables, the presented analyses focus on results produced using the statistical biological filters associated with each outcome.
Predictive models were built using random forest modeling, a machine learning method that builds many decision-tree models based on input predictor variables that work collectively to classify the output dependent variable (Breiman, 2001). We selected to use this machine learning method, as we have demonstrated significant utility in this specific method towards the training and testing of computational models aimed at predicting toxicological outcomes while incorporating -omics data (Klaren et al., 2019; Ring et al., 2021). Additionally, this method represents a relatively established method within the machine learning field, and our ultimate goal was to evaluate the relative predictivity of different -omic profiles, which could be directly addressed through this established approach.
In this study, predictor variables included mRNAs, miRNAs, or CpG methylation sites in combination with their respective covariates, and dependent variables included each of the perinatal outcomes, evaluated separately (Figure 1). Models were also built using covariates alone, as points of comparison. For the continuous outcome variables, namely infant birth weight and placenta weight, underlying decision trees were built using regression-based methods. For the binary outcome variables, namely placental damage and placental inflammation, underlying decision trees were build using classification-based methods.
Random forest modeling was implemented using methods similar to our previous analyses (Klaren et al., 2019; Ring et al., 2021). Here, the randomForest package in R (v4.6-14) was implemented to include 10 001 trees within each random forest (RDocumentation, 2021b). Prior to the development and testing of predictive models, data were split into training and test sets. Here, training and test sets were obtained by dividing the data randomly into 3 equal-sized folds. Two folds were used as the combined training dataset, and 1-fold was used as the test dataset. Data distributions are provided for example training and test sets used in models, where similar frequency distributions were observed between these randomly selected subsets of data (Supplementary Figure 1). Model performance parameters were obtained, depending on the outcome variable type, as further detailed below.
For the continuous outcome variables, birth weight and placenta weight, the overall predictive performance of each resulting regression model was evaluated based on four resulting parameters calculated from the test set. First, the root mean squared error (RMSE) across the test set was calculated, representing the square root of the mean squared error (MSE) (the difference between observed and predicted values for the test set). Second, the percent variance explained by the model was calculated, representing how well the out-of-bag (OOB) predictions explained the target variance of the test set, calculated as: (1−MSE)/(variance of the test set) * 100%. Third, a linear regression model was fit to the predicted versus observed values for the test set, and the R2 value of this linear regression was calculated used. Fourth, the RMSE of this regression model was calculated, representing the SD of the residuals resulting from the predictive model (ie, prediction errors).
For the binary outcome variables, placental damage and placental inflammation, the prediction of the random forest model is based on a “majority vote” of the ensemble of decision trees in the random forest. The overall predictive performance of each resulting model was evaluated based on the resulting confusion matrix: in how many cases the model (1) correctly predicted no outcome (ie, no damage or inflammation) in the actual absence of an outcome (true negatives); (2) incorrectly predicted no outcome in the actual presence of an outcome (false negatives); (3) correctly predicted an outcome in the actual presence of an outcome (true positives); and (4) incorrectly predicted an outcome in the actual absence of an outcome (false positives). Using this confusion matrix, model performance parameters of sensitivity, specificity, and precision were derived and used to rank overall model performance. Balanced accuracy was also calculated as the average across sensitivity and specificity values. Collectively, the parameters used to evaluate models designed to predict both continuous and categorical outcomes are well-documented and recognized metrics to evaluate model performance (CRAN, 2021; RDocumentation, 2021b). Example test data, source codes, and model results for this predictive modeling analysis are publicly available (Ragerlab, 2021).
Evaluating top-ranking model predictors to gain insight into underlying biology of perinatal outcomes
To evaluate the extent to which each predictor variable contributed to the overall performance of the predictive model, parallel models were built based on training datasets that included additional predictor variables representing noise. These “noise” variables included randomly permuted values generated using the Sample function in R, allowing for bootstrapping (ie, random sampling with replacement; RDocumentation, 2021c). Values were randomly selected from 5 of the molecular variables (ie, 5 random mRNAs, or 5 random miRNAs, or 5 random CpG sites, depending on the model) and all covariate variables included in each model. All predictor variables were then ranked by importance. For the continuous outcomes, variable-importance rankings were calculated based on the percent increase of the MSE when a given variable is randomly permuted (Breiman, 2001; Strobl et al., 2007). For the categorical outcomes, variable-importance rankings were calculated as the mean decrease in accuracy in predicting out-of-bag samples when each predictor variable was permuted. These resulting predictor-variable importance rankings summarized the overall contribution of each variable to the model performance, both alone and upon interacting with other predictors. Results were used to identify which of the variables were more important than noise variables. The variables that outranked noise were then selected, and new random forest models were trained using only the selected variables, to obtain final variable contribution measures for biological interpretation. Using this approach, the overall top-ranking predictors were identified for each outcome category.
Feature contribution measures were obtained for the top-ranking predictors and visualized using the forestFloor package in R (Palczewska et al., 2014; Welling et al., 2016). Results were plotted to understand the change in the overall probability that the model predicts an outcome that is attributable to a given predictor variable, over the range of observed values. As examples, we focused on the top-ranking miRNAs contributing to the models that predicted birth weight and placenta weight, as these models demonstrated some of the highest performance metrics. We specifically focused on the models built using molecules meeting the BH-adjusted p value < .10 statistical cutoff; though the variable importance and contribution measures were largely consistent with the “top 100” models. Resulting plots of the top-ranking miRNAs highlighted how much each miRNA contributed towards the probability of a change in birth weight or placenta weight.
The biological implications of top-ranking molecular predictors were also evaluated at the systems level. Here, we continued analyses on the miRNAs as an example, as these demonstrated high levels of predictivity. We also aggregated all top-ranking miRNA predictors (meaning those contributing to models above noise) across outcomes and analyzed those miRNAs as collective key regulators of perinatal outcomes. These miRNAs were analyzed for known relationships in canonical pathways, biological functions, and disease signatures, within the Ingenuity Pathway Analysis knowledgebase (Ingenuity Systems, Redwood City, California). Statistical enrichment of each pathway, function, and/or disease signature was determined using a modified Fischer’s Exact test, where over-represented categories were defined as those containing more miRNAs than expected by random chance based on BH-corrected p < .10 (Benjamini and Hochberg, 1995), paralleling our previous investigations (Klaren et al., 2019; Payton et al., 2020; Rager et al., 2017).
Identifying molecular changes in a separate pregnancy cohort in association with environmental cadmium exposure
The utility of top-ranking molecular predictors in identifying a known perinatal toxicant, cadmium, was evaluated in a separate pregnancy cohort, which has been detailed in our previous studies and was approved by the UNC Institutional Review Board (No. 11–2054) (Brooks et al., 2016; Laine et al., 2015; Martin et al., 2015). Briefly, women (n = 36) receiving obstetric care at UNC hospitals consented to collection of placental samples at the time of delivery. This cohort included those with and without symptoms of preeclampsia, as defined by the American College of Obstetricians and Gynecologists (ACOG, 2021). The purpose of this cohort study was to evaluate factors that influence preeclamptic risk, and therefore, when this study was implemented, any woman who had prediabetes, diabetes, or gestational diabetes were excluded. Placental miRNA expression signatures were measured using the Affymetrix GeneChip Human Gene 2.0 ST array, representing a different platform than what was used for the aforementioned analyses. Because of this platform difference, a portion of the top-ranking miRNA predictor variables identified in the preceding analyses overlapped in this evaluation and were thus the focus of this analysis. Cadmium concentrations were also measured in these same placenta tissues using ICP-MS methods, as previously described (Laine et al., 2015). The relationships between top-ranking miRNA predictor variables and cadmium were examined using linear regression models, which evaluated whether cadmium exposure was associated with perturbations in miRNA expression. Specifically, all top-ranking miRNA predictors (n = 32) (meaning those contributing to models above noise) across all outcomes were regressed onto placental cadmium levels (ng/g) (ln-transformed) while controlling for maternal age and smoking, representing covariates identified using methods that parallel the above statistical analyses. Multiple testing correction was carried out using the BH procedure, and statistical significance was defined as BH-adjusted p < .10, paralleling the previously detailed methods.
RESULTS
Study Cohort Characteristics
Placentas from 378 women were included in this study from the ELGAN cohort (McElrath et al., 2008), and participant information is summarized (Table 1). In general, the majority of women identified as White (n = 229, 61%), aged 14–45 years old, had a normal BMI (n = 199, 53%), completed 12–16 years of education (n = 184, 49%) and did not have smoke exposure (N = 289, 77%). With regards to the perinatal outcomes that were assessed in this study, all infants were considered to have low birth weight (<2500 g, range of 420–1418 g), some infants were considered growth-restricted based on birth weight-for-gestational age estimates (n = 20, 5%), mothers had an average placenta weight of 260.2 g (range of 79–1180 g), a portion had indications of placental inflammation (ie, histological chorioamnionitis; n = 117, 31%), and a portion had placental infarction (n = 60, 16%). For each perinatal outcome, a slightly different subset of placentas was included in the final mRNA, miRNA, and CpG methylation analyses depending on the availability of covariate data and molecular data QA/QC filters (Supplementary Table 1).
Table 1.
Subject Characteristics
| n (%) Average [Range] | |
|---|---|
| Maternal age (years) | 30 [14–45] |
| Maternal race | |
| White | 229 (61.2) |
| Nonwhite | 145 (38.8) |
| Maternal BMI (kg/m2) | |
| Underweight (<18.5) | 26 (7.0) |
| Normal (18.5 to <25.0) | 199 (53.3) |
| Overweight (25.0 to <30.0) | 67 (18.0) |
| Overweight or obese (>30.0) | 81 (21.7) |
| Smoke exposurea | |
| No | 289 (76.5) |
| Yes | 89 (23.5) |
| Multiple gestation pregnancy | |
| No | 276 (73.0) |
| Yes | 102 (27.0) |
| Highest level of educational attainment | |
| <12 years | 47 (12.5) |
| Between 12 and 16 years | 184 (49.1) |
| >16 years | 144 (38.4) |
| Newborn sex | |
| Male | 198 (52.4) |
| Female | 180 (47.6) |
| Intrauterine growth-restricted | |
| Yes | 20 (5.3) |
| No | 358 (94.7) |
| Newborn birth weight (g) | 834.2 [420–1418] |
| Gestational age (days) | 182.6 [161–195] |
| Chorioamnionitis | |
| No | 239 (67.1) |
| Yes | 117 (32.9) |
| Placental weight (g) | 260.2 [79–1180] |
| Placental infarction | |
| No | 297 (83.2) |
| Yes | 60 (16.8) |
Maternal demographic data, pregnancy characteristics, and perinatal outcome data for the current cohort. The overall count of included subjects was n = 378.
First- or secondhand smoke exposure.
Molecular Signatures Predicting Perinatal Outcomes
We identified mRNA, miRNA, and CpG methylation signatures from human placentas associated with a suite of four perinatal outcomes. These signatures included molecules identified as significantly (BH-adjusted p < .10) associated with each of the outcomes: specifically, 254 mRNAs, 268 miRNAs, and 414 CpG methylation sites associated with birth weight (Supplementary Tables 2–4); 12 mRNAs and 70 miRNAs associated with placental weight (note that 0 CpG methylation sites met the statistical cutoff for association to placental weight; Supplementary Tables 5–7); 1 mRNA, 125 miRNAs, and 84 CpG methylation sites associated with placental damage (Supplementary Tables 8–10); and 20 mRNAs, 195 miRNAs, and 253 CpG methylation sites associated with placental inflammation (Supplementary Tables 11–13). These molecular lists were used separately to train and test predictive models, leveraging the expression/methylation levels across placenta samples in combination with applicable covariates towards predicting each of the perinatal outcomes.
Model performance was evaluated through the use of several metrics dependent upon the type of dependent variable (ie, continuous vs categorical). To evaluate whether model performance may differ depending on the number of included predictor variables, models were built and tested using those that met the aforementioned statistical association filter (based on BH-adjusted p value < .10), and also those that ranked among the top 100 most significant molecules, regardless of statistical filtering (Supplementary Tables 2–13). Performance results from these models are summarized in Tables 2 and 3. Overall, for the models built to predict the continuous outcomes, birth weight or placenta weight, the resulting performance metrics were as follows: the RMSE across the test set ranged between 124 and 161 (average = 140); R2 of the regression fit between the predicted versus observed values across the test set ranged between 0.08 and 0.57 (average = 0.39); RMSE of the regression fit between the predicted versus observed values ranged between 38 and 73 (average = 57); and the percent variance explained by the model was estimated between 7% and 46% (average = 34%). For the models built to predict the categorical outcomes, placental damage and inflammation, the resulting performance metrics were as follows: the sensitivity ranged between 0.75 and 1.00 (average = 0.93); the specificity ranged between 0 and 0.56 (average = 0.21); the balanced accuracy ranged between 0.49 and 0.77 (average = 0.57); and the precision ranged between 0.67 and 0.90 (average = 0.80).
Table 2.
Predictive Model Results Describing the Performance of Random Forest Models at Predicting the Continuous Outcome Variables, (A) Birth Weight and (B) Placenta Weight
| Molecular Signature Filter | Model Predictors a | Number of Molecular Predictors | Model Rank | RMSE Across Test Set | R2 of Regression Fit Between Predicted Versus Observed Values | RMSE of Regression Fit Between Predicted Versus Observed Values | % Variance Explained by Model |
|---|---|---|---|---|---|---|---|
| (A) Birth weight | |||||||
| Molecules meeting BH-adjusted p < .10 | Covariates aloneb | 0 | — | 161 | 0.27 | 57 | 23% |
| mRNAs | 254 | 3 | 141 | 0.45 | 71 | 41% | |
| miRNAs | 268 | 2 | 137 | 0.46 | 69 | 44% | |
| CpG methylation sites | 414 | 1 | 146 | 0.57 | 57 | 45% | |
| Top 100 most significant molecules | Covariates aloneb | 0 | — | 161 | 0.27 | 57 | 23% |
| mRNAs | 100 | 3 | 140 | 0.45 | 73 | 42% | |
| miRNAs | 100 | 1 | 134 | 0.48 | 70 | 46% | |
| CpG Methylation Sites | 100 | 2 | 148 | 0.52 | 65 | 44% | |
| (B) Placenta weight | |||||||
| Molecules meeting BH-adjusted p < .10 | Covariates alonec | 0 | – | 124 | 0.41 | 46 | 35% |
| mRNAs | 11 | 2 | 135 | 0.32 | 54 | 29% | |
| miRNAs | 70 | 1 | 128 | 0.36 | 48 | 31% | |
| CpG methylation sites | — | — | — | — | — | — | |
| Top 100 Most Significant Molecules | Covariates alonec | 0 | — | 124 | 0.41 | 46 | 35% |
| mRNAs | 100 | 3 | 155 | 0.08 | 63 | 7% | |
| miRNAs | 100 | 1 | 126 | 0.40 | 43 | 33% | |
| CpG methylation sites | 100 | 2 | 141 | 0.36 | 38 | 26% | |
Molecular signatures were filtered using a significance threshold of BH-adjusted p < .10 or top 100 ranking genes based on BH-adjusted p in association with each outcome. Models were ranked based on their performance across the prediction accuracy measures below, with 1 indicating the highest-ranking model and 3 indicating the lowest ranking model across molecular categories.
Abbreviation: RMSE, root mean squared error.
Note that all models included their corresponding covariates.
Covariates for birth weight included gestational age, exposure to first- or secondhand smoking, sex, and multiple gestations.
Covariates for placenta weight included BMI, multiple gestations, gestational age, and sex.
Table 3.
Predictive Model Results Describing the Performance of Random Forest Models at Predicting the Categorical Outcome Variables, (A) Placental Damage and (B) Placental Inflammation
| Molecular SignatureFilter | ModelPredictors a | Number ofMolecular Predictors | ModelRank | Sensitivity | Specificity | BalancedAccuracy | Precision |
|---|---|---|---|---|---|---|---|
| (A) Placental damage | |||||||
| Molecules meeting BH-adjusted p < .10 | Covariates aloneb | 0 | — | 0.96 | 0.14 | 0.55 | 0.90 |
| mRNAs | 1 | 3 | 0.97 | 0.04 | 0.51 | 0.81 | |
| miRNAs | 125 | 2 | 0.98 | 0.00 | 0.49 | 0.88 | |
| CpG methylation sites | 84 | 1 | 0.99 | 0.05 | 0.52 | 0.83 | |
| Top 100 most significant molecules | Covariates aloneb | 0 | — | 0.96 | 0.14 | 0.51 | 0.90 |
| mRNAs | 100 | 2 | 1.00 | 0.09 | 0.63 | 0.82 | |
| miRNAs | 100 | 3 | 0.98 | 0.00 | 0.63 | 0.88 | |
| CpG methylation sites | 100 | 1 | 1.00 | 0.10 | 0.77 | 0.84 | |
| (B) Placental inflammation | |||||||
| Molecules meeting BH-adjusted p < .10 | Covariates aloneb | 0 | — | 0.75 | 0.28 | 0.55 | 0.67 |
| mRNAs | 20 | 2 | 0.89 | 0.38 | 0.54 | 0.74 | |
| miRNAs | 195 | 2 | 0.93 | 0.33 | 0.49 | 0.74 | |
| CpG Methylation Sites | 253 | 1 | 0.98 | 0.56 | 0.55 | 0.83 | |
| Top 100 most significant molecules | Covariates aloneb | 0 | — | 0.75 | 0.28 | 0.51 | 0.67 |
| mRNAs | 100 | 3 | 0.94 | 0.28 | 0.61 | 0.72 | |
| miRNAs | 100 | 2 | 0.91 | 0.31 | 0.61 | 0.73 | |
| CpG methylation sites | 100 | 1 | 0.95 | 0.42 | 0.68 | 0.79 | |
Molecular signatures were filtered using a significance threshold of BH-adjusted p < .10 or top 100 ranking genes based on BH-adjusted p in association with each outcome. Models were ranked based on their performance across the prediction accuracy measures of sensitivity, specificity, and precision (as balanced accuracy is based upon sensitivity and specificity), with 1 indicating the highest-ranking model and 3 indicating the lowest ranking model across molecular categories.
Note that all models included their corresponding covariates.
Covariates for damage and inflammation included BMI and maternal age.
In general, whether molecular signatures were selected based on a statistical cutoff, or whether they were selected based on a general “top 100” most significant molecular ranking, did not impact the overall model performance and associated rankings. The exception to this trend was when molecules failed to meet the statistical cutoff, and thus the model could not include any predictor variables. For example, no CpG methylation sites were identified to pass the statistical cutoff in relation to placenta weight. However, when using the top 100 most significant CpG sites regardless of meeting the statistical cutoff, these molecules still demonstrated predictive power in range of the other observations (Table 2), demonstrating biological relevance and predictivity in the absence of traditional statistical filters.
Comparing the Predictivity of Each Molecular Signature
The overall performance of each model built to predict perinatal outcomes was compared across molecular categories. Specifically, each metric of performance was combined into an average ranking and used to compare each model’s performance, which was combined into an overall performance rank per outcome, as detailed in Tables 2 and 3. The results clearly indicated that the epigenetic signatures based on miRNA expression or CpG methylation out-performed mRNA expression signatures in relation to the perinatal outcomes. In general, the miRNA expression signatures performed the best when predicting birth weight and placenta weight; and the CpG methylation signatures performed the best when predicting placental damage and inflammation. Additionally, in the majority of cases, model performance improved when adding molecular signatures to the predictor variables, as opposed to only using subject covariate data.
Top-Ranking Predictor Variables Provide Insight into Underlying Biology of Perinatal Outcomes
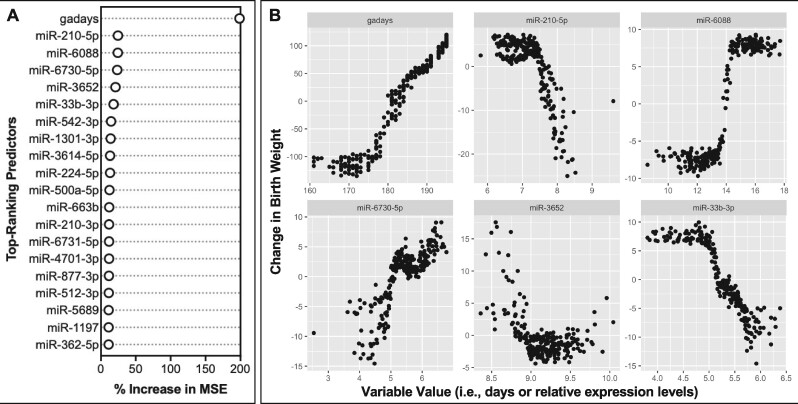
Top-ranking predictor variables were identified and further interpreted to gain biological insight into the molecular mediators of perinatal outcomes. These top-ranking variables were defined as those that contribute to each model’s overall performance above random noise, obtained through random permutations across predictor variables (see Materials and Methods). Examples discussed here focus on 257 top-ranking miRNAs (Supplementary Table 14), as these molecular signatures demonstrated some of the highest levels of predictivity across models, and we were also able to test these signatures in relation to an exogenous exposure in a separate cohort. For illustrative purposes, the relative importance of each top-ranking predictor variable in the miRNA-based models are shown for birth weight (Figure 2A), as based on the percent increase of the mean squared error when a given variable is randomly permuted (Breiman, 2001; Strobl et al., 2007). This ranking demonstrated that, not surprisingly, the covariate of gestational age contributed the most significantly to the overall model. Then, select miRNAs were found to contribute to the model’s overall performance, including miR-210-5p, miR-6088, miR-6730-5p, and miR-3652. These top-ranking predictors were then viewed according to individual changes in variable expression contributing to the individual changes in birth weight predictions (Figure 2B). These findings indicated that as gestational age increased, the predicted change in birth weight increased, as one would expect. Trends between individual miRNAs varied, with some miRNAs showing increased expression alongside predicted increases in birth weight (miR-6088 and miR-6730-5p), some miRNAs showing decreased expression alongside predicted increases in birth weight (miR-210-5p and miR-33b-3p), and a miRNA showing a U-shaped relationship with predicted changes in birth weight (miR-3652).
Figure 2.
Contributions of the top-ranking predictor variables in microRNA (miRNA)-based predictions of infant birth weight. A, The relative importance of the 20 predictor variables with the highest importance is shown in terms of the percent increase of the mean squared error when a given variable is randomly permuted. All of these variables contributed towards model performance at levels above random noise. B, The relative contribution of each variable towards model predictions is shown, focusing on the 6 predictor variables with the highest importance. Values for each predictor variable are plotted against their relative impact on birth weight predictions in the resulting model. Results focus on the miRNA models developed using molecules identified as significantly associated with birth weight; although similar results were obtained from the miRNA models developed using molecules identified as the “top 100” most significantly associated with birth weight. Abbreviation: gadays, gestational age in days.
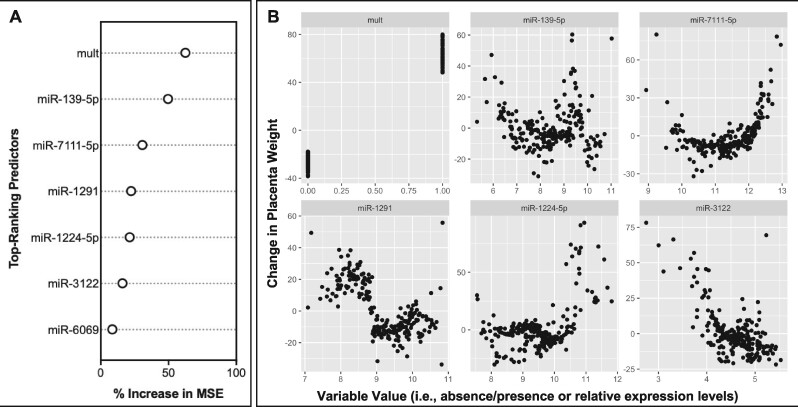
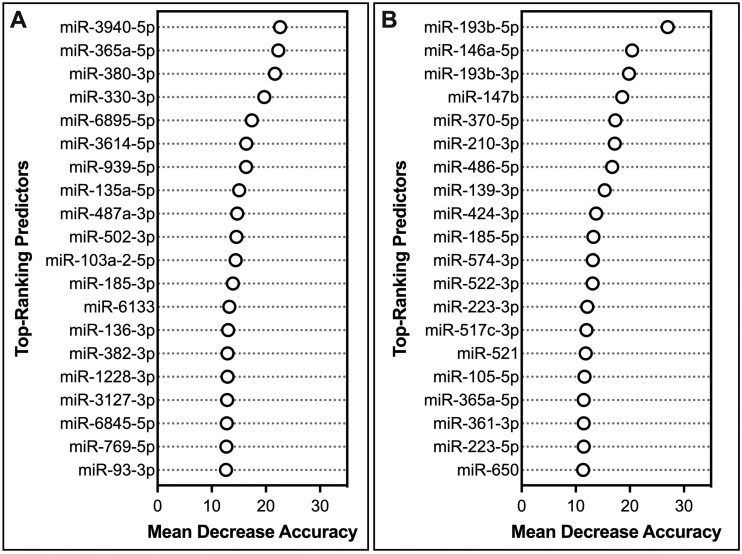
A similar example was produced for placenta weight, in which the relative importance of each top-ranking predictor variable in the miRNA-based model performance was evaluated (Figure 3A). Here, the covariate of multiparity was identified to most significantly contribute to model performance, followed by select miRNAs, including miR-139-5p, miR-7111-5p, miR-1291, miR-1224-5p, and miR-3122. These top-ranking predictors were then viewed according to individual changes in variable expression contributing to the individual changes in placenta weight predictions (Figure 3B). Trends between individual miRNAs varied, with miRNAs showing increased expression alongside predicted increases in placenta weight (eg, miR-1224-5p), miRNAs showing decreased expression alongside predicted increases in placenta weight (eg, miR-3122), and several miRNAs demonstrating more complex relationships (eg, U-shaped and sigmoidal curves) in association with predicted changes in placenta weight (eg, miR-139-5p, miR-7111-5p, and miR-1291). Examples of top-ranking miRNA predictors were also included for placental damage (Figure 4A) and placental inflammation (Figure 4B), which notably included miR-210-3p.
Figure 3.
Contributions of the top-ranking predictor variables in microRNA (miRNA)-based predictions of placenta weight. A, The relative importance of the 7 predictor variables that rank above random noise is shown in terms of the percent increase of the mean squared error when a given variable is randomly permuted. B, The relative contribution of each variable towards model predictions is shown, focusing on the 6 predictor variables with the highest importance. Values for each predictor variable are plotted against their relative impact on placenta weight predictions in the resulting model. Results focus on the miRNA models developed using molecules identified as significantly associated with placenta weight; although similar results were obtained from the miRNA models developed using molecules identified as the “top 100” most significantly associated with placenta weight. Abbreviation: mult, multiparity.
Figure 4.
Contributions of the top-ranking miRNA predictor variables in microRNA (miRNA)-based predictions of (A) placental damage and (B) placental inflammation. The relative importance of the 20 miRNA predictor variables with the highest importance is shown in terms of the mean decrease in model accuracy when a given variable is randomly permuted. All of these variables contributed towards model performance at levels above random noise. Results focus on the miRNA models developed using molecules identified as significantly associated with the phenotypic outcomes; although similar results were obtained from the miRNA models developed using molecules identified as the “top 100” most significantly associated with each outcome.
An overall functional enrichment analysis was carried out at the systems-level on all top-ranking miRNAs that demonstrated predictivity above noise for at least 1 perinatal outcome (Supplementary Table 14). These miRNAs showed an enrichment for certain pathways and associated biological function and disease signatures, including organismal injury and abnormalities, reproductive system disease, inflammatory disease, and inflammatory response (Table 4 and Supplementary Table 15). These findings provide further biological interpretation of the functional involvement of these miRNAs on important cellular processes and disease-relevant outcomes.
Table 4.
Biological Functions and Disease Signatures That Were the Most Significantly Associated With Top-Ranking miRNAs Identified to Predict Perinatal Outcomes
| Category | BH-Adjusted p Value |
|---|---|
| Organismal injury and abnormalities | 6.0E-45 |
| Reproductive system disease | 6.0E-45 |
| Neurological disease | 1.4E-26 |
| Psychological disorders | 1.4E-26 |
| Inflammatory disease | 1.3E-21 |
| Inflammatory response | 1.3E-21 |
| Renal and urological disease | 1.3E-21 |
| Cancer | 4.9E-18 |
| Gastrointestinal disease | 1.2E-14 |
| Respiratory disease | 1.2E-14 |
| Hematological disease | 8.9E-14 |
| Immunological disease | 8.9E-14 |
| Connective tissue disorders | 3.8E-13 |
| Hereditary disorder | 8.4E-08 |
| Infectious diseases | 1.4E-07 |
| Organismal development | 1.4E-07 |
| Endocrine system disorders | 2.0E-07 |
| Metabolic disease | 2.0E-07 |
| Skeletal and muscular systemdevelopment and function | 6.5E-07 |
| Tissue morphology | 6.5E-07 |
Abbreviations: BH, Benjamini-Hochberg. For full listings of miRNAs with known roles in each category, see Supplementary Table 15.
Predictive miRNA Signatures Also Show Alterations in Association with the Known Prenatal Toxicant, Cadmium
One of the utilities of identifying molecular signatures predictive of perinatal outcomes is to use these same signatures as a screen for exogenous insults that induce similar molecular changes, and thus, leverage these signatures as a tool for the screening of potential perinatal toxicants that may impact maternal and fetal health. This strategy was evaluated here using one of the top-molecular predictors (ie, placental miRNAs) in a separate pregnancy cohort, and relating these signatures to cadmium concentrations measured in the same tissues. Cadmium was selected as the focus of this evaluation for the following lines of reasoning: (1) cadmium is known to induce changes in birth weight, placenta weight, placental damage, and placental inflammation (Erboga and Kanter, 2016; Hu et al., 2018; Huang et al., 2019; Johnston et al., 2014; Punshon et al., 2019); (2) prenatal exposure to cadmium is recognized to influence maternal/fetal health outcomes (Erboga and Kanter, 2016; Geng and Wang, 2019; Hu et al., 2018; Huang et al., 2019; Johnston et al., 2014; Punshon et al., 2019); and (3) we have previously published data surrounding its concentrations within the placenta (Brooks et al., 2016; Laine et al., 2015). The specific demographics of this cohort have been described in our previous studies (Brooks et al., 2016; Martin et al., 2015), though it is of note that these pregnancies were on average longer in duration (mean = 35.5 weeks [range = 22–41]), in comparison to the cohort used in developing the predictive signatures. Molecular signatures were also measured in placenta tissues from this cohort using different platforms that were microarray-based. As a result, 32 of the top-ranking miRNAs identified as predictors of various perinatal outcomes (ranking above noise) overlapped with miRNAs measured in this additional cohort (Supplementary Table 14).
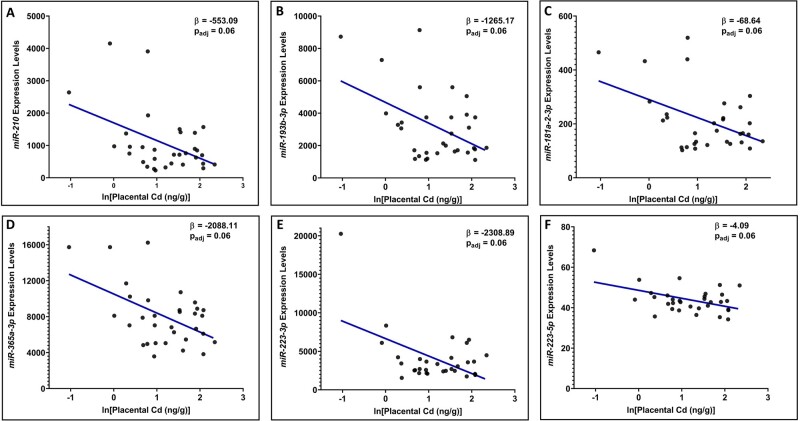
We then used these 32 miRNAs to test whether expression levels were associated with placental cadmium levels. In total, the expression levels of n = 6 (19%) miRNAs predictive of perinatal phenotypes were also significantly (BH-adjusted p < .10) associated with placental cadmium levels. These included miR-181a-2-3p, miR-193b-3p, miR-210, miR-223-3p, miR-223-5p, and miR-365a-3p (Figure 5 and Supplementary Table 16). It is of importance to note that miR-210 was queried within this cohort through array-based technologies, though this miRNA maps to mature miRNA sequences of miR-210-3p and miR-210-5p (miRBase, 2021); and both miR-210-3p and miR-210-5p were identified as top-ranking contributors towards perinatal outcomes (ie, birth weight, placenta weight, and placental inflammation) within the previously detailed cohort using more advanced sequencing technologies.
Figure 5.
Scatterplots showing the relationship between placental microRNAs (miRNAs) associated with environmental cadmium exposure in a separate pregnancy cohort. miRNAs displayed include (A) miR-210, (B) miR-193b-3p, (C) miR-181a-2-3p, (D) miR-365a-3p, E) miR-223-3p, and (F) miR-223-5p. These miRNAs notably overlap with those identified as top-ranking predictors of perinatal outcomes in humans. Beta values represent the change in miRNA expression for each doubling of placental cadmium (ng/g).
DISCUSSION
The field of predictive biology is placing increased emphasis on the use of molecular data to predict health outcomes, representing an initiative that is of high importance throughout clinical, pharmaceutical, and industrial chemical research and applications. Transcriptomic and epigenomic signatures, however, have yet to be compared and evaluated for utility towards informing perinatal outcomes. This study set out to test the hypotheses that placental transcriptomic (mRNA) and epigenomic (miRNA and CpG methylation) signatures can be used to predict perinatal outcomes in humans, and different molecular (-omic) categories demonstrate varying levels of predictivity. Here, we showed that molecular signatures can be used to predict apical outcomes using human placental tissue, with improved performance in comparison to the use of human covariate data alone. We found that in all evaluated outcomes, molecular signatures based on epigenomic signatures showed the highest predictivity, consistently outranking mRNA expression signatures. Last, using placental miRNA signatures from an additional pregnancy cohort, we found that the expression levels of top-ranking miRNA predictors were also altered in association with the known prenatal toxicant, cadmium. Together, these findings suggest epigenomic signatures outperform transcriptomic signatures in predicting perinatal outcomes in humans and highlight the general utility of multi-omic signatures in the evaluation of potential toxicants that may impact maternal and fetal health.
Signatures based on placental miRNA expression were found to predict birth weight and placenta weight with the highest performance metrics; and signatures based on CpG methylation were found to predict placental damage and inflammation with the highest performance metrics, based on overall findings across multiple statistical prefilters that were tested. miRNAs are molecules of high interest, as they play a significant role in regulating the expression of protein-coding genes and have been shown to impact a variety of diseases, including those relevant to pregnancy and fetal developmental outcomes (Gross et al., 2017; Liu et al., 2018; Payton et al., 2020; Rager et al., 2014; Williams et al., 2012; Zhu et al., 2009). These molecules show promise as biomarkers of exposure, biomarkers of disease, and therapeutic targets, due to their high degree of stability as evidenced by studies finding robust detection of miRNAs in various tissues and biofluids, even in degraded samples (Condrat et al., 2020; Liu et al., 2009; Peiro-Chova et al., 2013). Whereas mRNA profiles are subject to endogenous RNase activity, miRNAs are less susceptible to these catalytic processes and are thus more stable over time, demonstrating half-lives that are estimated to be 10 times longer than mRNAs (Aryani and Denecke, 2015; Gantier et al., 2011). In terms of CpG methylation, previous studies have demonstrated utility in using CpG methylation levels to predict health outcomes, including a recent study from our group showing that placental CpG sites predict social cognition scoring later-in-life (Santos et al., 2020). Our general finding that epigenomic signatures consistently out-predict mRNA expressions is novel, as there is an overall lack of studies comparing predictive capabilities across multiple -omic platforms. This is important, as this research field is in the midst of developing molecular signatures for elucidating molecular key events and eventual health outcomes, that largely focus on genome-based measures (eg, DNA damage; Li et al., 2015). Therefore, the inclusion of epigenetic signatures alongside mRNA signatures will likely improve the utility of molecular signatures in predictive biology applications.
Machine learning approaches have been criticized for their seemingly limited ability to provide information describing the underlying relationships between the variables being analyzed and are thus often referred to as “black boxes” (Palczewska et al., 2014). In an effort to address this potential limitation and gain insight into the biological underpinnings of the modeled relationships, we evaluated how much each predictor variable contributed to the overall model performance, and thus identified the “importance” of each predictor. We further interpreted our findings through a feature contribution method, which elucidated the relative contribution of each variable towards predicting changes in a perinatal outcomes (Palczewska et al., 2014). As an example, we used these methods to evaluate predictor variables contributing to miRNA-based predictions of birth weight and identified key miRNAs that significantly contributed to birth weight predictions, including those involved in pregnancy pathologies. For example, miR-210-5p showed increased expression in association with predicted decreases in birth weight. This miRNA has previous implications in placental-mediated influences on pregnancy outcomes (Awamleh and Han, 2020; Frazier et al., 2020) and birth weight (Bian et al., 2021). Reduced expression of miR-210 in the mouse placenta has specifically been shown to exacerbate hypoxia-induced reductions in fetal weight (Bian et al., 2021). This finding was noted to parallel reductions in the placental spongiotrophoblast layer and impaired development of labyrinth fetal blood vessels (Bian et al., 2021). At the cellular-level, miR-210 is largely recognized for its role in protecting tissues in response to conditions of hypoxia and inflammation, and its involvement in angiogenesis (Bavelloni et al., 2017). This specific miRNA has notably been identified at increased levels in maternal blood samples from patients with preeclamptic outcomes across several studies (Frazier et al., 2020), demonstrating its potential detection in samples that may be easier to collect in the clinical setting. Therefore, our finding that miR-210 expression can inform the prediction of multiple perinatal outcomes has potential utility towards toxicological and clinical applications.
An important application of the identified molecular signatures is to screen for potential exposures that cause similar changes in molecular signatures, and thereby, use these signatures as tools for the screening of potential perinatal toxicants that may impact maternal/fetal health outcomes. This strategy was tested here using an additional pregnancy cohort consisting of women exposed to varying levels of cadmium in the environment (Brooks et al., 2016; Laine et al., 2015). The evaluation of prenatal cadmium exposure is of high relevance to the current analysis, as cadmium is a recognized perinatal toxicant known to induce changes in birth weight, placenta weight, placental damage, and placental inflammation (Erboga and Kanter, 2016; Hu et al., 2018; Huang et al., 2019; Johnston et al., 2014; Punshon et al., 2019), and it is recognized to influence maternal/fetal health outcomes (Erboga and Kanter, 2016; Geng and Wang, 2019; Hu et al., 2018; Huang et al., 2019; Johnston et al., 2014; Punshon et al., 2019). Here, we find overlapping miRNAs that demonstrate predictive capabilities towards perinatal outcomes are also altered by cadmium exposure in the placenta. These include critical miRNAs involved in inflammation, injury, and hypoxia-relevant signaling, including miR-210. The general finding that top-ranking miRNAs predictive of perinatal outcomes are also modified by cadmium further demonstrate the biological relevance and application of these signatures towards capturing necessary biology to predict important health outcomes in humans.
This study advances knowledge surrounding placental mRNA, miRNA, and CpG methylation signature predictivity and implications towards perinatal outcomes; though there remain additional steps that could further enhance this research area. To begin with, placental multi-omics signatures were derived from a cohort of pregnancies ending in births prior to 28 weeks of gestation, which likely impacted baseline transcription and methylation signatures. It will be informative to carry out similar analyses in other cohorts to test the reproducibility of these signatures across additional full-term birth cohorts, as well as additional tissues (eg, maternal blood and cord blood). This study also focused on the analysis of mRNAs, miRNAs, and CpG methylation sites; though future studies could incorporate additional types of understudied molecules (eg, noncoding RNAs). We also carried out 33 separate models in this study, representing a robust analysis; though future studies will be carried out to evaluate the potential influence of sex on these signatures. An additional aspect to expand upon in the future is the inclusion of additional cohorts and in vitro models to further evaluate the utility of these predictive signatures. For example, we did not have access to a separate pregnancy cohort with similar perinatal outcomes and placenta multi-omic signatures. As these data become available, we will carry out additional tests to refine these molecular signatures across populations, as well as additional chemical exposure domains.
In conclusion, this study implemented a novel approach to compare the overall predictivity of placental transcriptomic and epigenomic signatures towards in relation to perinatal outcomes in humans. We identified important signatures that contributed to the prediction of perinatal outcomes, and also demonstrated that epigenetic signatures based on miRNA expression or CpG methylation out-performed signatures based on mRNA expression. Top-ranking predictors were highlighted for their involvement in important cellular processes, including organismal injury and inflammatory processes. Overlapping signatures showed alterations in association with environmental cadmium exposure in an additional pregnancy cohort, highlighting the functionality of such signatures in screening for potential exogenous insults that may be contributing to perinatal toxicity which may, in turn, impact maternal/fetal health. Together, these findings support the utility of placental molecular signatures in informing biological mechanisms and predicting health outcomes, providing groundwork for future predictive biology applications to better elucidate what causes toxicity and potentially identify therapeutics to alleviate these adverse health outcomes.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DATA AVAILABILITY STATEMENT
Molecular signature data are publicly available (NCBI, 2020). Example test data, source codes, and model results in this manuscript are available at https://github.com/Ragerlab.
FUNDING
This study was supported by grants from the National Institutes of Health (NIH) the Office of the NIH Director [UG3OD023348, UH3OD023348] (RCF, TO), the National Institute of Child Health and Human Development [R01HD092374 (RCF, TO); R03HD101413 (HS)], the National Institute of Nursing Research [K23NR017898 (HS); R01NR019245 (HS)], and the National Institute of Environmental Health Sciences [P42ES031007] (RCF). Funding was also provided through the Institute for Environmental Health Solutions at the University of North Carolina Gillings School of Global Public Health.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
The authors certify that all research involving human subjects was done under full compliance with all government policies and the Helsinki Declaration.
REFERENCES
- ACOG. (2021). American college of obstetricians and gynecologists (ACOG) current guidelines for identifying preeclampsia. Available at: https://www.labce.com/spg1839185_american_college_of_obstetricians_and_gynecologist.aspx#:∼:text=For%20diagnosing%20PE%2C%20ACOG%20recommends,pre-ssure%20has%20previously%20been%20normal. Accessed January 1, 2021.
- Addo K. A., Bulka C., Dhingra R., Santos H. P. Jr, Smeester L., O'Shea T. M., Fry R. C. (2019). Acetaminophen use during pregnancy and DNA methylation in the placenta of the Extremely Low Gestational Age Newborn (ELGAN) cohort. Environ. Epigenet. 5, dvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryani A., Denecke B. (2015). In vitro application of ribonucleases: Comparison of the effects on mrna and mirna stability. BMC Res. Notes 8, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee M. J., Jaffe A. E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A. P., Hansen K. D., Irizarry R. A. (2014). Minfi: A flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awamleh Z., Han V. K. M. (2020). Identification of mir-210-5p in human placentae from pregnancies complicated by preeclampsia and intrauterine growth restriction, and its potential role in the pregnancy complications. Pregnancy Hypertens. 19, 159–168. [DOI] [PubMed] [Google Scholar]
- Bao X., Anastasov N., Wang Y., Rosemann M. (2019). A novel epigenetic signature for overall survival prediction in patients with breast cancer. J. Transl. Med. 17, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavelloni A., Ramazzotti G., Poli A., Piazzi M., Focaccia E., Blalock W., Faenza I. (2017). Mirna-210: A current overview. Anticancer Res. 37, 6511–6521. [DOI] [PubMed] [Google Scholar]
- Beaumont R. N., Horikoshi M., McCarthy M. I., Freathy R. M. (2017). How can genetic studies help us to understand links between birth weight and type 2 diabetes? Curr. Diab. Rep. 17, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bian X., Liu J., Yang Q., Liu Y., Jia W., Zhang X., Li Y. X., Shao X., Wang Y. L. (2021). MicroRNA-210 regulates placental adaptation to maternal hypoxic stress during pregnancy†. Biol. Reprod. 104, 418–429. [DOI] [PubMed] [Google Scholar]
- Bianco-Miotto T., Craig J. M., Gasser Y. P., van Dijk S. J., Ozanne S. E. (2017). Epigenetics and dohad: From basics to birth and beyond. J. Dev. Orig. Health Dis. 8, 513–519. [DOI] [PubMed] [Google Scholar]
- Blencowe H., Krasevec J., de Onis M., Black R. E., An X., Stevens G. A., Borghi E., Hayashi C., Estevez D., Cegolon L., et al. (2019). National, regional, and worldwide estimates of low birthweight in 2015, with trends from 2000: A systematic analysis. Lancet Glob. Health 7, e849–e860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32. [Google Scholar]
- Brooks S. A., Martin E., Smeester L., Grace M. R., Boggess K., Fry R. C. (2016). MiRNAs as common regulators of the transforming growth factor (tgf)-beta pathway in the preeclamptic placenta and cadmium-treated trophoblasts: Links between the environment, the epigenome and preeclampsia. Food Chem. Toxicol. 98, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton G. J., Fowden A. L., Thornburg K. L. (2016). Placental origins of chronic disease. Physiol. Rev. 96, 1509–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell G. A., Rager J. E. (2017). Epigenetics in chemical-induced genotoxic carcinogenesis. Curr. Opin. Toxicol. 6, 10–17. [Google Scholar]
- Clark J., Martin E., Bulka C. M., Smeester L., Santos H. P., O’Shea T. M., Fry R. C. (2019). Associations between placental CpG methylation of metastable epialleles and childhood body mass index across ages one, two and ten in the Extremely Low Gestational Age Newborns (ELGAN) cohort. Epigenetics 14, 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J., Rager J. E.. 2020. Epigenetics: An overview of CpG methylation, chromatin remodeling, and regulatory/non-coding RNAs. In Environmental Epigenetics in Toxicology and Public Health (Fry R. C., Ed.), pp. 3–32. Elsevier. [Google Scholar]
- Cohen A. L., Soldi R., Zhang H., Gustafson A. M., Wilcox R., Welm B. E., Chang J. T., Johnson E., Spira A., Jeffrey S. S., et al. (2011). A pharmacogenomic method for individualized prediction of drug sensitivity. Mol. Syst. Biol. 7, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat C. E., Thompson D. C., Barbu M. G., Bugnar O. L., Boboc A., Cretoiu D., Suciu N., Cretoiu S. M., Voinea S. C. (2020). Mirnas as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 9, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAN. (2021). Package ‘randomforest’. Available at: https://cran.r-project.org/web/packages/randomForest/randomForest.pdf. Accessed January 1, 2021
- Deng D., Liu Z., Du Y. (2010). Epigenetic alterations as cancer diagnostic, prognostic, and predictive biomarkers. Adv. Genet. 71, 125–176. [DOI] [PubMed] [Google Scholar]
- Eaves L. A., Phookphan P., Rager J. E., Bangma J., Santos H. P. Jr, Smeester L., O’Shea T. M., Fry R. C. (2020). A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics 12, 1543–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erboga M., Kanter M. (2016). Effect of cadmium on trophoblast cell proliferation and apoptosis in different gestation periods of rat placenta. Biol. Trace Elem. Res. 169, 285–293. [DOI] [PubMed] [Google Scholar]
- FDA. (2017). Biomarker letter of support. Center for Drug Evaluation and Research, Food and Drug Administration. Available at: https://www.fda.gov/media/112682/download. Accessed July 1, 2021.
- Fortin J. P., Labbe A., Lemire M., Zanke B. W., Hudson T. J., Fertig E. J., Greenwood C. M., Hansen K. D. (2014). Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J. P., Triche T. J. Jr,, Hansen K. D. (2017). Preprocessing, normalization and integration of the illumina humanmethylationepic array with minfi. Bioinformatics 33, 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier S., McBride M. W., Mulvana H., Graham D. (2020). From animal models to patients: The role of placental microRNAs, mir-210, mir-126, and mir-148a/152 in preeclampsia. Clin. Sci. (Lond) 134, 1001–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. R., Weinberg J. B., Barry W. T., Goodman B. K., Volkheimer A. D., Bond K. M., Chen Y., Jiang N., Moore J. O., Gockerman J. P., et al. (2009). A genomic approach to improve prognosis and predict therapeutic response in chronic lymphocytic leukemia. Clin Cancer Res 15, 6947–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry R. C., Bangma J., Szilagyi J., Rager J. E. (2019). Developing novel in vitro methods for the risk assessment of developmental and placental toxicants in the environment. Toxicol. Appl. Pharmacol. 378, 114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier M. P., McCoy C. E., Rusinova I., Saulep D., Wang D., Xu D., Irving A. T., Behlke M. A., Hertzog P. J., Mackay F., et al. (2011). Analysis of microRNA turnover in mammalian cells following dicer1 ablation. Nucleic Acids Res. 39, 5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H. X., Wang L. (2019). Cadmium: Toxic effects on placental and embryonic development. Environ. Toxicol. Pharmacol. 67, 102–107. [DOI] [PubMed] [Google Scholar]
- Goldstein J. A., Gallagher K., Beck C., Kumar R., Gernand A. D. (2020). Maternal-fetal inflammation in the placenta and the developmental origins of health and disease. Front. Immunol. 11, 531543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross N., Kropp J., Khatib H. (2017). MicroRNA signaling in embryo development. Biology (Basel) 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., Kokocinski F., Aken B. L., Barrell D., Zadissa A., Searle S., et al. (2012). Gencode: The reference human genome annotation for the encode project. Genome Res. 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J. L., Allred E. N., Kliman H. J., Zambrano E., Doss B. J., Husain A., Pflueger S. M., Chang C. H., Livasy C. A., Roberts D., Elgan Study Investigators., et al. (2008). Histological characteristics of singleton placentas delivered before the 28th week of gestation. Pathology 40, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang H., Hu Y. F., Xu X. F., Chen Y. H., Xia M. Z., Zhang C., Xu D. X. (2018). Cadmium induces inflammatory cytokines through activating akt signaling in mouse placenta and human trophoblast cells. Placenta 65, 7–14. [DOI] [PubMed] [Google Scholar]
- Huang S., Kuang J., Zhou F., Jia Q., Lu Q., Feng C., Yang W., Fan G. (2019). The association between prenatal cadmium exposure and birth weight: A systematic review and meta-analysis of available evidence. Environ. Pollut. 251, 699–707. [DOI] [PubMed] [Google Scholar]
- Johnston J. E., Valentiner E., Maxson P., Miranda M. L., Fry R. C. (2014). Maternal cadmium levels during pregnancy associated with lower birth weight in infants in a North Carolina cohort. PLoS One 9, e109661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaren W. D., Ring C., Harris M. A., Thompson C. M., Borghoff S., Sipes N. S., Hsieh J. H., Auerbach S. S., Rager J. E. (2019). Identifying attributes that influence in vitro-to-in vivo concordance by comparing in vitro tox21 bioactivity versus in vivo drugmatrix transcriptomic responses across 130 chemicals. Toxicol. Sci. 167, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konwar C., Price E. M., Wang L. Q., Wilson S. L., Terry J., Robinson W. P. (2018). DNA methylation profiling of acute chorioamnionitis-associated placentas and fetal membranes: Insights into epigenetic variation in spontaneous preterm births. Epigenet. Chromatin 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine J. E., Ray P., Bodnar W., Cable P. H., Boggess K., Offenbacher S., Fry R. C. (2015). Placental cadmium levels are associated with increased preeclampsia risk. PLoS One 10, e0139341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocca J., Johnson K. J., LeBaron M. J., Rasoulpour R. J. (2017). The interface of epigenetics and toxicology in product safety assessment. Curr. Opin. Toxicol. 6, 87–92. [Google Scholar]
- Leek J. T. (2014). Svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D. (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexogen. (2021). Quantseq 3’ mrna-seq library prep kit user guide. Available at: https://www.lexogen.com/wp-content/uploads/2015/11/015UG009V0211_QuantSeq-Illumina.pdf. Accessed January 1, 2021.
- Li H. H., Hyduke D. R., Chen R., Heard P., Yauk C. L., Aubrecht J., Fornace A. J. Jr. (2015). Development of a toxicogenomics signature for genotoxicity using a dose-optimization and informatics strategy in human cells. Environ. Mol. Mutagen. 56, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S. C., Hernandez-Vargas H., Herceg Z. (2010). Epigenetic signatures in cancer: Implications for the control of cancer in the clinic. Curr. Opin. Mol. Ther. 12, 316–324. [PubMed] [Google Scholar]
- Liu A., Tetzlaff M. T., Vanbelle P., Elder D., Feldman M., Tobias J. W., Sepulveda A. R., Xu X. (2009). MicroRNA expression profiling outperforms mrna expression profiling in formalin-fixed paraffin-embedded tissues. Int. J. Clin. Exp. Pathol. 2, 519–527. [PMC free article] [PubMed] [Google Scholar]
- ΡΡLiu D. F., Li S. M., Zhu Q. X., Jiang W. (2018). The involvement of mir-155 in blood pressure regulation in pregnant hypertension rat via targeting FOXO3a. Eur. Rev. Med. Pharmacol. Sci. 22, 6591–6598. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for rna-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Ray P. D., Smeester L., Grace M. R., Boggess K., Fry R. C. (2015). Epigenetics and preeclampsia: Defining functional epimutations in the preeclamptic placenta related to the tgf-beta pathway. PLoS One 10, e0141294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElrath T. F., Hecht J. L., Dammann O., Boggess K., Onderdonk A., Markenson G., Harper M., Delpapa E., Allred E. N., Leviton A., et al. ; ELGAN Study Investigators. (2008). Pregnancy disorders that lead to delivery before the 28th week of gestation: An epidemiologic approach to classification. Am. J. Epidemiol. 168, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- miRBase. (2021). Mirbase: Stem-loop sequence hsa-mir-210. Available at: http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0000286. Accessed January 1, 2021.
- NCBI. (2020). Geo gene expression omnibus. Placental genomic and epigenomic signatures in infants borns at extremely low gestational age. Available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154829. Accessed July 22, 2020.
- Onderdonk A. B., Delaney M. L., DuBois A. M., Allred E. N., Leviton A, Extremely Low Gestational Age Newborns Study I. (2008). Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am. J. Obstet. Gynecol. 198, 110.e111–7. [DOI] [PubMed] [Google Scholar]
- Palczewska A., Palczewki J., Robinson R. M., Neagu D. (2014). Interpreting random forest classification models using a feature contribution method. In Integration of Resuable Systems (T. Bouabana-Tebibel and S. H. Rubin, Eds.), pp. 193–218. Springer International Publishing, Switzerland. [Google Scholar]
- Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton A., Clark J., Eaves L., Santos H. P. Jr, Smeester L., Bangma J. T., O'Shea T. M., Fry R. C., Rager J. E. (2020). Placental genomic and epigenomic signatures associated with infant birth weight highlight mechanisms involved in collagen and growth factor signaling. Reprod. Toxicol. 96, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro-Chova L., Pena-Chilet M., Lopez-Guerrero J. A., Garcia-Gimenez J. L., Alonso-Yuste E., Burgues O., Lluch A., Ferrer-Lozano J., Ribas G. (2013). High stability of microRNAs in tissue samples of compromised quality. Virchows Arch. 463, 765–774. [DOI] [PubMed] [Google Scholar]
- Pettit S., des Etages S. A., Mylecraine L., Snyder R., Fostel J., Dunn R. T. 2nd, Haymes K., Duval M., Stevens J., Afshari C., et al. (2010). Current and future applications of toxicogenomics: Results summary of a survey from the hesi genomics state of science subcommittee. Environ. Health Perspect. 118, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Lee S., Majewski I. J., Alexander W. S., Smyth G. K. (2016). Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10, 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punshon T., Li Z., Jackson B. P., Parks W. T., Romano M., Conway D., Baker E. R., Karagas M. R. (2019). Placental metal concentrations in relation to placental growth, efficiency and birth weight. Environ. Int. 126, 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Auerbach S. S., Chappell G. A., Martin E., Thompson C. M., Fry R. C. (2017). Benchmark dose modeling estimates of the concentrations of inorganic arsenic that induce changes to the neonatal transcriptome, proteome, and epigenome in a pregnancy cohort. Chem. Res. Toxicol. 30, 1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Bailey K. A., Smeester L., Miller S. K., Parker J. S., Laine J. E., Drobna Z., Currier J., Douillet C., Olshan A. F., et al. (2014). Prenatal arsenic exposure and the epigenome: Altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 55, 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Bangma J., Carberry C., Chao A., Grossman J., Lu K., Manuck T. A., Sobus J. R., Szilagyi J., Fry R. C. (2020). Review of the environmental prenatal exposome and its relationship to maternal and fetal health. Reprod. Toxicol. 98, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Fry R. C. (2013). Systems biology and environmental exposures. In Network Biology: Theories, Methods, and Applications (W. J. Zhang, Ed.), pp. 81–130. Nova Publishers, New York. [Google Scholar]
- Rager J. E., Tilley S. K., Tulenko S. E., Smeester L., Ray P. D., Yosim A., Currier J. M., Ishida M. C., Gonzalez-Horta Mdel C., Sanchez-Ramirez B., et al. (2015). Identification of novel gene targets and putative regulators of arsenic-associated DNA methylation in human urothelial cells and bladder cancer. Chem. Res. Toxicol. 28, 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragerlab. (2021). Julia e. Rager’s research group github webpage.Available at: https://github.com/Ragerlab. Accessed January 1, 2021.
- RDocumentation. (2021a). Findcorrelation: Determine highly correlated variables. Available at: https://www.rdocumentation.org/packages/caret/versions/6.0-88/topics/findCorrelation. Accessed May 15, 2021
- RDocumentation. (2021b). Randomforest: Classification and regression with random forest. https://www.rdocumentation.org/packages/randomForest/versions/4.6-14/topics/randomForest. Accessed January 1, 2021.
- RDocumentation. (2021c). Sample: Random samples and permutation. https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/sample. Accessed January 1, 2021.
- Ring C., Sipes N. S., Hsieh J. H., Carberry C., Koval L. E., Klaren W. D., Harris M. A., Auerbach S. S., Rager J. E. (2021). Predictive modeling of biological responses in the rat liver using in vitro tox21 bioactivity: Benefits from high-throughput toxicokinetics. Comp. Toxicol. 18, 100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos H. P. Jr, Bhattacharya A., Joseph R. M., Smeester L., Kuban K. C. K., Marsit C. J., O'Shea T. M., Fry R. C. (2020). Evidence for the placenta-brain axis: Multi-omic kernel aggregation predicts intellectual and social impairment in children born extremely preterm. Mol. Autism 11, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos H. P. Jr, Bhattacharya A., Martin E. M., Addo K., Psioda M., Smeester L., Joseph R. M., Hooper S. R., Frazier J. A., Kuban K. C., et al. (2019). Epigenome-wide DNA methylation in placentas from preterm infants: Association with maternal socioeconomic status. Epigenetics 14, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrier I., Platt R. W. (2008). Reducing bias through directed acyclic graphs. BMC Med. Res. Methodol. 8, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streimish I. G., Ehrenkranz R. A., Allred E. N., O’Shea T. M., Kuban K. C., Paneth N., Leviton A.; ELEGAN Study Investigators. (2012). Birth weight- and fetal weight-growth restriction: Impact on neurodevelopment. Early Hum. Dev. 88, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl C., Boulesteix A. L., Zeileis A., Hothorn T. (2007). Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinformatics 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szejniuk W. M., Robles A. I., McCulloch T., Falkmer U. G. I., Roe O. D. (2019). Epigenetic predictive biomarkers for response or outcome to platinum-based chemotherapy in non-small cell lung cancer, current state-of-art. Pharmacogenomics J. 19, 5–14. [DOI] [PubMed] [Google Scholar]
- Tabacova S., Kimmel C. A., Wall K., Hansen D. (2003). Atenolol developmental toxicity: Animal-to-human comparisons. Birth Defects Res. A Clin. Mol. Teratol. 67, 181–192. [DOI] [PubMed] [Google Scholar]
- Teschendorff A. E., Zheng S. C. (2017). Cell-type deconvolution in epigenome-wide association studies: A review and recommendations. Epigenomics 9, 757–768. [DOI] [PubMed] [Google Scholar]
- Triche T. J. Jr, Weisenberger D. J., Van Den Berg D., Laird P. W., Siegmund K. D. (2013). Low-level processing of illumina infinium DNA methylation beadarrays. Nucleic Acids Res. 41, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahos A., Mansell T., Saffery R., Novakovic B. (2019). Human placental methylome in the interplay of adverse placental health, environmental exposure, and pregnancy outcome. PLoS Genet. 15, e1008236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walani S. R. (2020). Global burden of preterm birth. Int. J. Gynaecol. Obstet. 150, 31–33. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liang Y., Lu Q. (2008). MicroRNA epigenetic alterations: Predicting biomarkers and therapeutic targets in human diseases. Clin. Genet. 74, 307–315. [DOI] [PubMed] [Google Scholar]
- Weisenberger D. J., Berg D. V., Pan F., Berman B. P., Laird P. W. (2021). Comprehensive DNA methylation analysis on the illumina® infinium® assay platform. Available at: https://support.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/appnote_dna_methylation_analysis_infinium.pdf. Accessed June 1, 2021.
- Welling S. H., Refsgaard H. F., Brockhoff P. B., Clemmensen L. H. (2016). Forest floor visualizations of random forest. Available at: https://arxiv.org/abs/1605.09196. Accessed April 1, 2021.
- Williams K. C., Renthal N. E., Condon J. C., Gerard R. D., Mendelson C. R. (2012). MicroRNA-200a serves a key role in the decline of progesterone receptor function leading to term and preterm labor. Proc. Natl. Acad. Sci. U.S.A. 109, 7529–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. M., Han T., Sargent I. L., Yin G. W., Yao Y. Q. (2009). Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 200, e661–e667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Molecular signature data are publicly available (NCBI, 2020). Example test data, source codes, and model results in this manuscript are available at https://github.com/Ragerlab.