Abstract
Bacterial wound infections are a threat to public health. Although antibiotics currently provide front-line treatments for bacterial infections, the development of drug resistance coupled with the defenses provided through biofilm formation render these infections difficult, if not impossible, to cure. Antimicrobials from natural resources provide unique antimicrobial mechanisms and are generally recognized as safe and sustainable. Herein, an all-natural antimicrobial platform is reported. It is active against bacterial biofilms and accelerates healing of wound biofilm infections in vivo. This antimicrobial platform uses gelatin stabilized by photocrosslinking using riboflavin (vitamin B2) as a photocatalyst, and carvacrol (the primary constituent of oregano oil) as the active antimicrobial. The engineered nanoemulsions demonstrate broad-spectrum antimicrobial activity towards drug-resistant bacterial biofilms and significantly expedite wound healing in an in vivo murine wound biofilm model. The antimicrobial activity, wound healing promotion, and biosafety of these nanoemulsions provide a readily translatable and sustainable strategy for managing wound infections.
Graphical Abstract
All-natural cross-linked nanoemulsions provide effective treatment of wound biofilm infections
Antibiotics are the current treatment of choice for bacterial infections.1 Unfortunately, bacteria are rapidly acquiring resistance against these agents through different mechanisms they have developed over billions of years to remove and/or deactivate toxins or evade their activities.2–4 The ability of pathogenic bacteria, including Pseudomonas aeruginosa and Staphylococcus aureus, to form biofilms results in particularly problematic infections in wounds and on implanted devices.5,6 The extracellular polymeric substance (EPS) matrix of biofilms has evolved as a barrier to some antibiotics and host immune responses. The slow growth and presence of persister cells in biofilms further foster resistance against traditional antibiotics.7 Biofilms also impede the wound healing process, resulting in chronic wounds associated with increased morbidity, mortality, and decreased quality of life.8, 9 The lack of effective antibiofilm agents has led to an annual multibillion-dollar (US) burden to healthcare systems worldwide.10
Plant-derived essential oils provide a potential resource to combat bacterial biofilm-associated infections.11 Essential oils are produced by plants as protection against infections by bacteria.12 These oils have been extensively used in traditional medicine as antioxidant, anti-inflammatory and antibacterial agents, and their efficacy has been scientifically validated.13,14 The additional benefits of using essential oils, including aroma,15 safety,16 and sustainability, have contributed to their increasing use in therapeutics. The low aqueous solubility of essential oils, however, limits their use in combating planktonic bacterial infections.17 Moreover, these hydrophobic oils are not able to efficiently penetrate into the highly charged EPS of biofilms,18 making them of limited use against biofilm-associated infections.
Synthetic nanoparticles and polymers provide a strategy for enhancing the activity of essential oil-based antimicrobials.19,20 We set out to generate an all-natural platform that delivers active antimicrobials derived from essential oils for treatment of wound biofilms, with the potential for increased safety and community acceptance. In this platform, riboflavin (vitamin B2) is used to cross-link21, 22 gelatin that stabilizes nanodroplets of carvacrol, a key antimicrobial component of oregano oil. Our approach combines nature-derived ingredients to generate well-defined nanostructured-materials with size and stability required for potent antimicrobial and antibiofilm. applications.23,24 These nanoemulsions eradicated both Gram-positive and -negative bacterial biofilms in vitro. Significantly, the nanoemulsion system was highly active in vivo, reducing bacterial loads in the wound site and enhancing the rate of wound healing in a murine wound biofilm model. Taken together, the described nanoemulsions used only bio-derived GRAS (generally regarded as safe) components to provide a safe, sustainable and effective treatment for wound biofilms.
The choice of oil for fabricating the nanoemulsions plays a critical role in therapeutic effects.25 Oregano oil is among the most potent antimicrobial essential oils.19,26 Carvacrol is the primary active component of oregano oil,19 and was chosen to provide the GRAS benefits of oregano oil without the batch-to-batch variability observed with unpurified oils. Moreover, commercial gelatin was chosen as the scaffold for the oil-in-water nanoemulsion engineered to overcome the poor water solubility of carvacrol. Gelatin is naturally-derived, inherently biocompatible, biodegradable, non-cytotoxic, and has low antigenicity.27 Structurally, gelatin is hydrophilic with hydrophobic domains, allowing it to encapsulate and stabilize hydrophobic oils, such as carvacrol, in aqueous condition.28 The gelatin matrix was stabilized through a cross-linking technique used for therapeutic corneal collagen cross-linking.21 This strategy employs riboflavin (vitamin B2) as a photo-initiator for cross-linking gelatin fibers. Nanoemulsions were fabricated by emulsifying a suspension of riboflavin in carvacrol into an aqueous suspension of gelatin (Fig. 1a), generating a transiently stable emulsion. This emulsion was then irradiated (365 nm) to activate the cross-linking process, resulting in stable nanoemulsions.
Fig. 1.
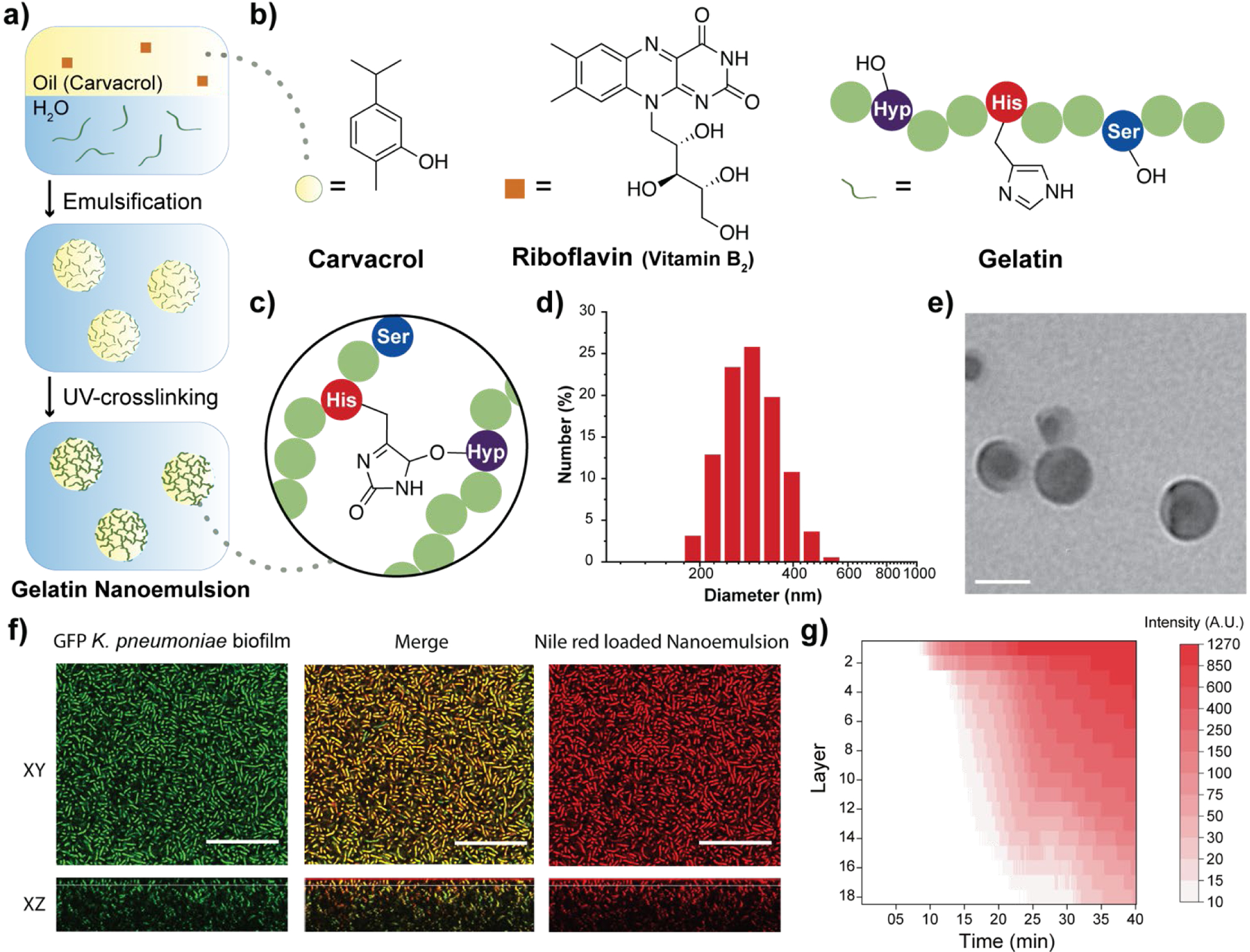
Fabrication and characterization of gelatin nanoemulsions. a) Riboflavin (UV-cross-linking initiator) was dissolved in carvacrol. The oil mixture was then emulsified in an aqueous gelatin solution and cross-linked using long wavelength UV-A light (365 nm) to fabricate gelatin nanoemulsions. b) Chemical structures of carvacrol, riboflavin, and the functional groups of gelatin participating in the cross-linking reaction. c) Proposed cross-linked structure of gelatin nanoemulsions. d) Dynamic light scattering histogram of nanoemulsions in phosphate buffered saline (150 mM). e) Transmission electron microscopy images of nanoemulsions. Scale bar is 100 nm. f) Confocal laser scanning microscopy images of green fluorescent protein-expressing Klebsiella pneumoniae (IDRL-1999) biofilm after 40 minutes treatment with Nile red-loaded nanoemulsions (XY top view, XZ side view). Scale bars are 50 μm and the thickness of this biofilm is ~37 μm. g) Spatial and time distribution of red fluorescence within the biofilm after the addition of Nile red-loaded nanoemulsions, indicating complete penetration after 35 min.
Dynamic light scattering (DLS), transmission electron microscopy (TEM) and attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) were used to characterize the nanoemulsions. DLS analysis showed that the hydrodynamic diameter of nanoemulsions is ~310 nm with a narrow size distribution (polydispersity index = 0.017) (Fig. 1d). These nanoemulsions are stable at room temperature for at least 30 days and are degraded in the presence of collagenase (Fig. S1, Supplementary Information). TEM photographs revealed a spherical morphology of the nanoemulsions (Fig. 1e and Fig. S2, Supplementary Information). The size observed in TEM (~100 nm) is smaller than observed by DLS, presumably due to partial collapse upon removal of carvacrol under vacuum during TEM. Finally, the chemical nature of the cross-linking was demonstrated by the emergence of a band at 1033 cm−1 arising from aliphatic-aromatic ether formation, an additional aromatic ether signature at 1242 cm−1, and appearance of sp3 C-H stretches at 2957 cm−1 (Fig. 1c and Fig. S3, Supplementary Information).
Carvacrol and other hydrophobic materials fail to penetrate biofilms.18 We hypothesized that the cationic charge of gelatin at the low pH29 found in biofilms30 would facilitate transport of nanoemulsions into biofilms. Confocal laser scanning microscopy (CLSM) was used to demonstrate effective and complete penetration of gelatin nanoemulsions into biofilms. Transport was tracked by loading the vehicle with the hydrophobic dye Nile red. Nanoemulsions were incubated with 4-day old biofilms of green fluorescent protein (GFP)-expressing K. pneumoniae IDRL-11999. As shown in Fig. 1f, gelatin nanoemulsions penetrated the biofilm matrix completely, as indicated by visualization of Nile red throughout the thickness of the biofilm, with colocalization of red and green signals. Time-dependent z-stack scanning demonstrated that complete penetration of nanoemulsions occurred within 40 minutes (Fig. 1g)
We next probed the mechanism of action of gelatin nanoemulsions. We hypothesized that gelatin nanoemulsions kill bacteria the same manner as carvacrol, namely through disrupting the cell membrane. We used propidium iodide (PI) staining to monitor the membrane permeability of bacteria.31 Planktonic bacteria (P. aeruginosa, ATCC-27853) were treated with gelatin nanoemulsions or antibiotic Ceftazidime (from 0.125X to 4X of their respective minimum inhibitory concentrations (MICs)) in the presence of PI. Gelatin nanoemulsion treatment immediately generated fluorescence inside bacteria, indicating that PI penetrated through the compromised cell membrane and bound to DNA (Fig. S4, Supplementary Information). In contrast, bacteria treated with Ceftazidime did not generate fluorescence, as Ceftazidime does not directly act on the membrane. Significantly, studies have shown that therapeutics causing membrane disruption are unlikely to induce resistance,32, as demonstrated in a recent effective treatment of burn infection models. 33
After validating the biofilm penetration profile and the mechanism of action, we used Alamar blue assay to assess antimicrobial activity of nanoemulsions against biofilms of four clinical bacterial isolates (P. aeruginosa CD-1006, methicillin-resistant S. aureus [MRSA] CD-489, Escherichia coli CD-2, and Enterobacter cloacae complex CD-1412). Treatment of these biofilms with nanoemulsions for 3 hours eliminated bacteria within the biofilms at 5% v/v (1.95 mM of carvacrol) (Fig. 2), with individual components of the nanoemulsions having minimal effect. Notably, treatment of bacteria with only gelatin (nutrient34) or riboflavin (nutrient/potential quorum sensing signal35) can enhance biofilm viability. We further quantified the minimum biofilm inhibitory concentrations (MBICs) and minimum biofilm eradication concentrations (MBECs) towards several clinical isolates (Table S1, Supplementary Information), including P. aeruginosa (CD-1006, IDRL-11442, ATCC 27583) and MRSA (IDRL-6169) to represent bacterial species that are common constituents of wound biofilm infections.36 All biofilms were suppressed or eradicated by nanoemulsions at concentrations ranging from 4% v/v (1.56 mM) to 8% v/v (3.12 mM). This activity was mirrored in more challenging dual-species biofilm models (MRSA/P. aeruginosa and MRSA/E. coli) (Table S1, Supplementary Information).37
Fig. 2.
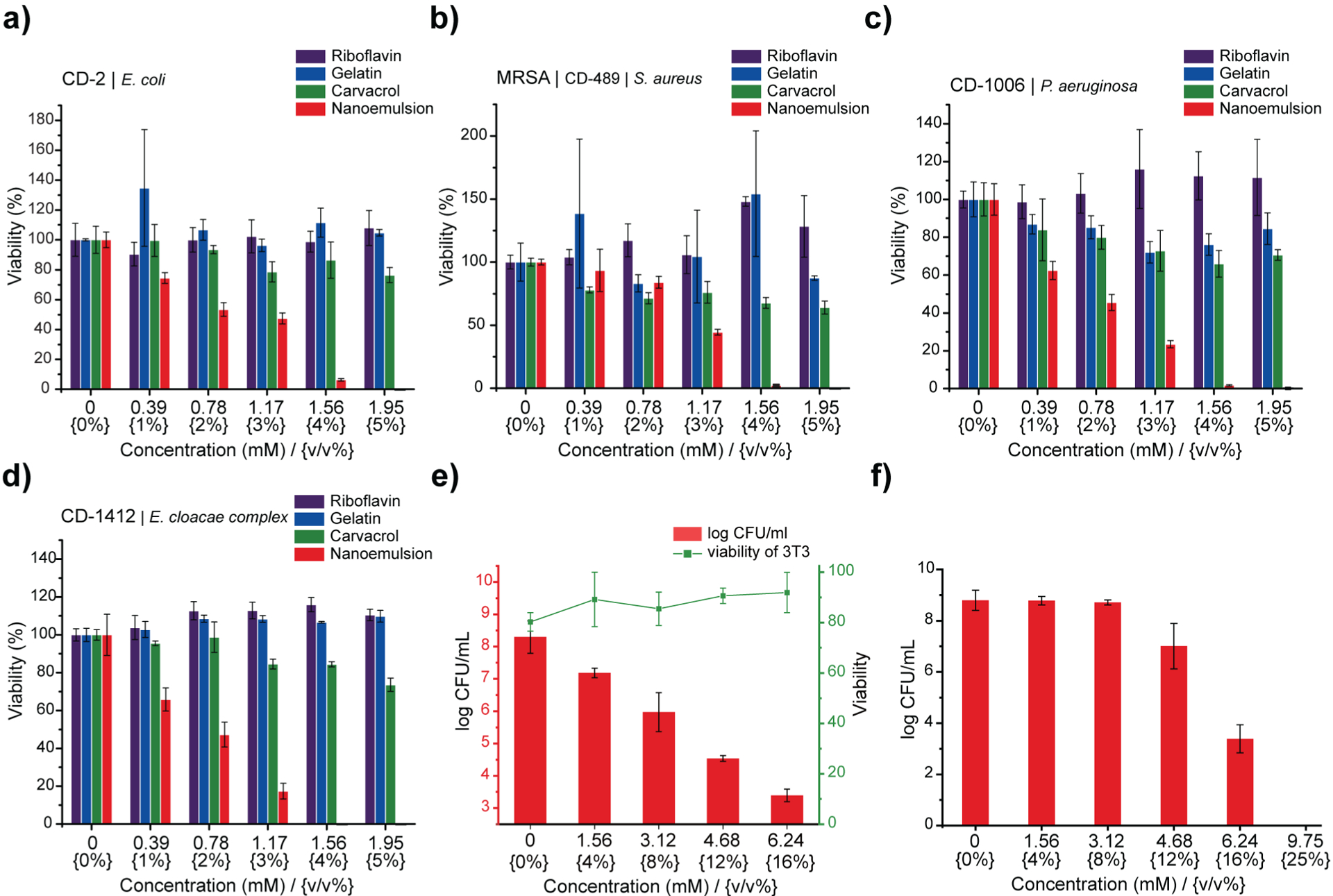
Gelatin nanoemulsions kill the bacteria within biofilms. Viability of a) Escherichia coli CD-2, b) methicillin-resistant Staphylococcus aureus CD-489, c) Pseudomonas aeruginosa CD-1006, and d) Enterobacter cloacae complex CD-1412 biofilms after 3 hours of treatment with riboflavin, gelatin, carvacrol, or nanoemulsions. e) Viability of 3T3 fibroblast cells and P. aeruginosa biofilms in the co-culture model after 3 h treatment with nanoemulsions. Scatters and lines represent 3T3 fibroblast cell viability. Bars represent log10 of colony forming units of bacteria in biofilms. f) Gelatin nanoemulsions kill the biofilm bacteria in simulated wound conditions. Log colony forming units (CFU) of methicillin-resistant Staphylococcus aureus IDRL-6169 biofilms after 3 hours of treatment with gelatin nanoemulsions in simulated wound fluid. Data are presented as mean ± standard deviation and represent three independent experiments.
We next used an in vitro bacterial biofilm-mammalian cell coculture model to evaluate antimicrobial activity and biocompatibility of gelatin nanoemulsions. P. aeruginosa (ATCC-27853) was seeded on the monolayer of NIH 3T3-fibroblast cells for 6 hours to mimic biofilm infections on mammalian cells. Subsequently, the cocultures were treated with gelatin nanoemulsion for three hours. The viabilities of mammalian cells and bacteria were determined using LDH assay and colony counting, respectively. As shown in fig. 2e, gelatin nanoemulsions effectively reduce bacterial colonies up to 5 log10 colony forming units (CFUs), while the toxicity toward 3T3 fibroblast cells remained negligible. Moreover, the activity of the nanoemulsions was determined using a simulated wound fluid (SWF) model, mimicking the interference/deactivation of antimicrobial activity by wound fluids.38 In this experiment, 4-day old MRSA (IDRL-6169) biofilms were grown using tryptic soy broth (TSB)/SWF (1:1) solution. Gelatin nanoemulsions were serially diluted with SWF solution and added to the biofilms; three hours after, antimicrobial efficacy was determined using quantitative colony counting. Nanoemulsions were active against the biofilms in SWF, with 12% v/v (4.68 mM) and 16% v/v (6.24 mM) of nanoemulsions resulting in ~2 and ~5 log10 CFU reduction, respectively (Fig. 2f).
Encouraged by the in vitro efficacy of the nanoemulsions, their in vivo activity was evaluated using a murine wound biofilm model (Fig. 3a) designed to assess efficacy against established biofilms found in chronic wound infections.8,9 In this model, MRSA IDRL-6169 biofilms were established in a wound created using a 5-mm skin puncture, and allowed to mature for 4 days. Mice were then separated into three groups: 100% v/v nanoemulsions (39 mM carvacrol), vancomycin (110 mg/kg), and saline solution only. Treatments were administered every other day (topical application of nanoemulsions and PBS, and intraperitoneal injection of vancomycin) until the day of sacrifice. Photographs were taken daily, and wound sizes and weights of the mice were monitored every day. The degree of purulence was also evaluated daily using a purulence reaction scoring system described in Fig. S5, Supplementary Information.
Fig. 3.
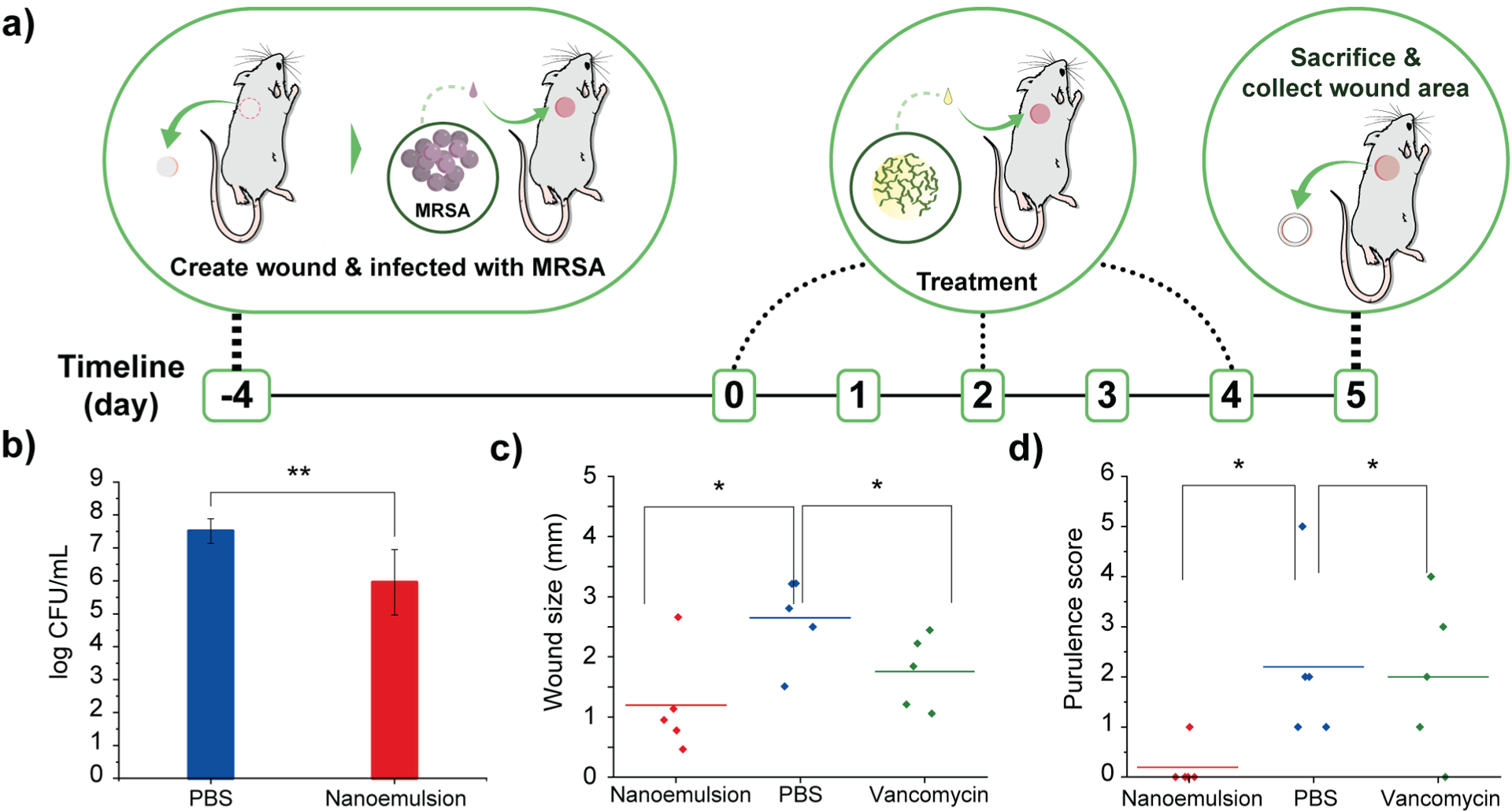
Gelatin nanoemulsions reduce bacterial load and expedite wound healing in a murine model in vivo. a) Schematic overview of the murine biofilm-associated wound infection model. b) Colony counts from the infected wounds treated with PBS and nanoemulsions. c) Wound size at the day of sacrifice. d) Purulence score at the day of sacrifice (*, ** = P values < 0.05 or 0.01, respectively
Treatment with nanoemulsions significantly reduced bacterial load in the wound as compared with PBS controls, with ~1.5 log10 unit reduction after administration of two treatments (Fig. 3b), a reduced degree of killing relative to in vitro studies owing to the much greater complexity of the in vivo environment. Significantly, vancomycin only had ~0.5 log10 bacterial reduction (Figure S6, Supplementary Information). The antimicrobial effect of the nanoemulsions was mirrored by their enhanced wound healing. The group treated with nanoemulsions showed a greater reduction in wound size after treatment as compared to those treated with vancomycin or PBS (Fig. 3c), with the two control group wound beds still containing pus (Fig. S7–9, Supplementary Information). The wounds were likewise better healed: mice in the nanoemulsion group had a purulence score 0, with normal-appearing healed wound beds. In contrast, vancomycin and PBS groups had purulence scores ~2, indicating the presence of pus (Fig. 3d and Figure S10, Supplementary Information).
Histological analysis of the wound beds similarly indicated enhanced healing with the nanoemulsions. Hematoxylin and eosin staining (H&E staining) revealed regeneration of keratin and epithelial layers, and collagen matrix for the nanoemulsion group. In contrast, for the vancomycin and PBS groups, inflammatory cells were still abundantly found in the area, indicating that the wounds were in the early stages of the wound healing cascade (Fig. 4 and Fig. S11, Supplementary Information). Notably, the healed skin had a normal appearing epidermis and dermis, suggesting that nanoemulsions do not alter morphology of the skin undergoing repair. The relative lack of inflammatory cells could also suggest an anti-inflammatory role for the nanoemulsions in wound healing.
Fig. 4.
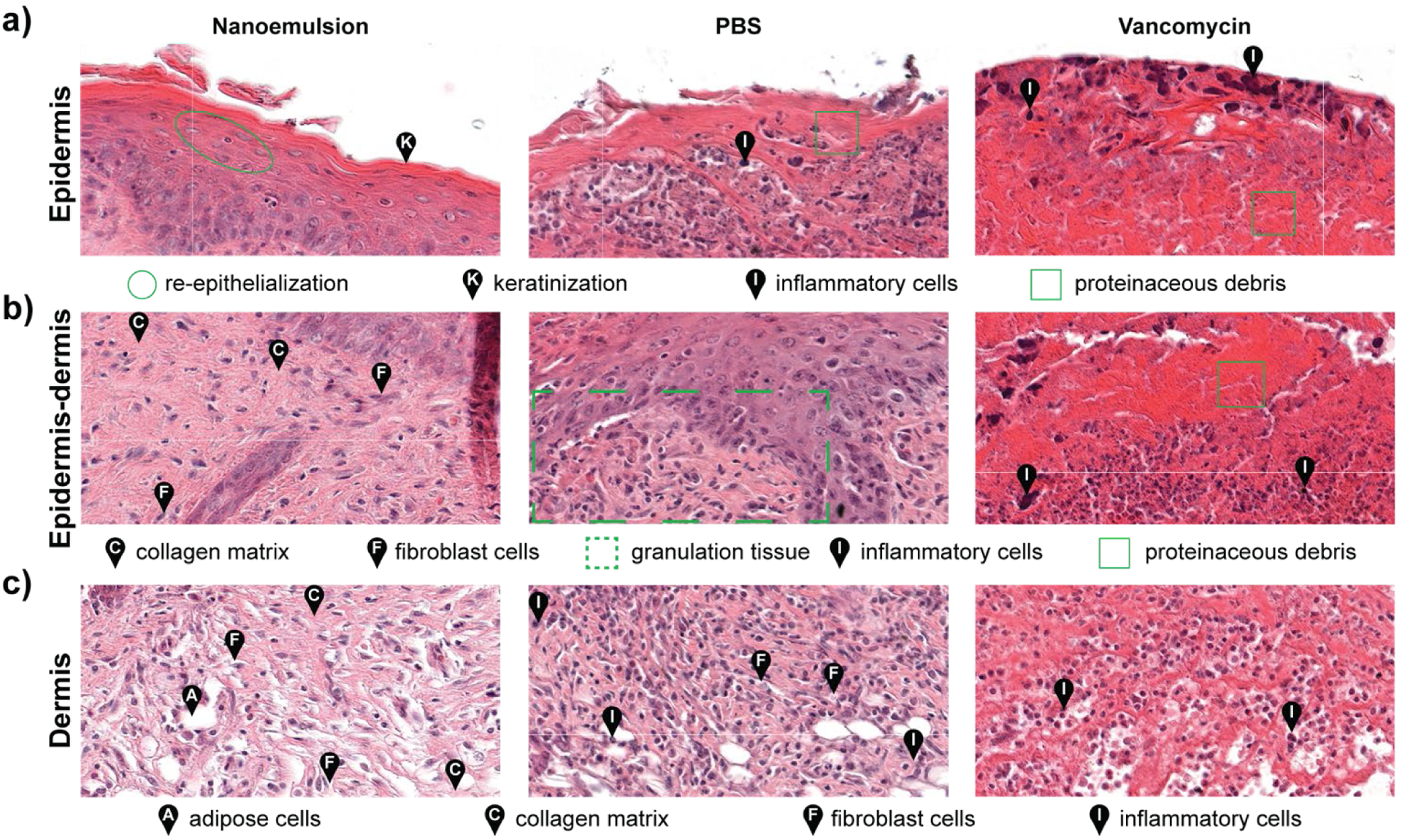
Histological analysis of the tissues surrounding infected wounds show enhanced healing with nanoemulsions. a) Epidermis samples showed regeneration of keratin and the epithelial layer with nanoemulsion treatment. In contrast, inflammatory cells and proteinaceous debris were observed with PBS and vancomycin treatments. b) Substantial formation of collagen matrix at the epidermis-dermis junction was observed after nanoemulsion treatment, whereas immature epidermis and granulation were observed with PBS. Necrosis and cell debris were also detected in the vancomycin-treated sample. c) The dermis was restored with nanoemulsion treatment, while inflammatory cells were still present in the PBS and vancomycin controls.
Conclusions
In summary, integration of nanostructured biomaterials with essential oil payloads provides effective treatment of challenging wound biofilms. Emulsification of oregano oil and gelatin followed by vitamin B2 cross-linking provided stable nanoemulsions comprised solely of naturally-occurring components. These described nanoemulsions effectively penetrate into biofilms, and kill embedded Gram-positive and -negative bacteria effectively in vitro. This antibacterial effect is observed in an in vivo murine model, and translates into enhancement in wound healing both in terms of wound size and degree of purulence. Crucially, this platform is highly modular, providing a general platform for the delivery of a wide variety of oils and other payloads. Overall, integration of the inherent bactericidal activity of essential oils with materials properties provided by biomaterials presents a new path of treatment for wound biofilms, with potential for treating other life-threatening bacterial infections.
Supplementary Material
Acknowledgements
This research was supported by the NIH (AI134770). Clinical samples obtained from the Cooley Dickinson Hospital Microbiology Laboratory (Northampton, MA) were kindly provided by Dr. Margaret Riley. The microscopy data was gathered in the Light Microscopy Facility and Nikon Center of Excellence at the Institute for Applied Life Sciences, UMass Amherst with support from the Massachusetts Life Sciences Center.
Footnotes
Conflicts of interest
The authors declare no competing final interest.
Electronic Supplementary Information (ESI) available: Detailed experimental methods, MBICs and MBBCs of gelatin nanoemulsions against bacterial biofilms, information of bacterial strains, stability and biodegradation of gelatin nanoemulsions determined by DLS, IR spectra of nanoemulsions, photographs of infected wounds, purulence scoring system, purulence scores of wounds, images of histological samples. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Col NF and O’Connor RW, Rev. Infect. Dis, 1987, 9, S232–S243. [DOI] [PubMed] [Google Scholar]
- 2.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO and V Piddock LJ, Nat. Rev. Microbiol, 2015, 13, 42–51. [DOI] [PubMed] [Google Scholar]
- 3.Aminov R, Front. Microbiol, 2010, 1, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Wang Z, Feng X, Bai E, Xiong Y, Zhu X, Shen B, Duan Y and Huang Y, Bioconjug. Chem, 2020, 31, 1425–1437. [DOI] [PubMed] [Google Scholar]
- 5.James GA, Swogger E, Wolcott R, deLancey Pulcini E, Secor P, Sestrich J, Costerton JW and Stewart PS, Wound Repair Regen, 2008, 16, 37–44. [DOI] [PubMed] [Google Scholar]
- 6.Arciola CR, Campoccia D, Speziale P, Montanaro L and Costerton JW, Biomaterials, 2012, 33, 5967–5982. [DOI] [PubMed] [Google Scholar]
- 7.Lewis K, ed. Romeo T, Springer Berlin Heidelberg, Berlin, Heidelberg, 2008, pp. 107–131.
- 8.Percival SL, McCarty SM and Lipsky B, Adv. Wound Care, 2014, 4, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, Akakuru OU, Ma X, Zheng J, Zheng J and Wu A, Bioconjug. Chem, 2020, 31, 1708–1723. [DOI] [PubMed] [Google Scholar]
- 10.Järbrink K, Ni G, Sönnergren H, Schmidtchen A, Pang C, Bajpai R and Car J, Syst. Rev, 2017, 6, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simões M, Bennett RN and Rosa EAS, Nat. Prod. Rep, 2009, 26, 746–757. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari BK, Valdramidis VP, O’ Donnell CP, Muthukumarappan K, Bourke P and Cullen PJ, J. Agric. Food Chem, 2009, 57, 5987–6000. [DOI] [PubMed] [Google Scholar]
- 13.Baratta MT, Dorman HJD, Deans SG, Biondi DM and Ruberto G, J. Essent. Oil Res, 1998, 10, 618–627. [Google Scholar]
- 14.Farzaei MH, Bahramsoltani R, Abbasabadi Z and Rahimi R, J. Pharm. Pharmacol, 2015, 67, 1467–1480. [DOI] [PubMed] [Google Scholar]
- 15.Etman M, Amin M, Nada AH, Shams-Eldin M and Salama O, Drug Discov. Ther, 2011, 5, 150–156. [DOI] [PubMed] [Google Scholar]
- 16.Freire Rocha Caldas G, Araújo AV, Albuquerque GS, Silva-Neto JDC, Costa-Silva JH, De Menezes IRA, Leite ACL, Da Costa JGM, Wanderley AG, Caldas GFR, Araújo AV, Albuquerque GS, Silva-Neto JDC, Costa-Silva JH, de Menezes IRA, Leite ACL, da Costa JGM and Almir Gonçalves Wanderley, Evidence-Based Complement. Altern. Med, 2013, 2013, No. 856168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samperio C, Boyer R, Eigel WN, Holland KW, McKinney JS, O’Keefe SF, Smith R and Marcy JE, J. Agric. Food Chem, 2010, 58, 12950–12956. [DOI] [PubMed] [Google Scholar]
- 18.Iannitelli A, Grande R, Di Stefano A, Di Giulio M, Sozio P, Bessa LJ, Laserra S, Paolini C, Protasi F and Cellini L, Int. J. Mol. Sci, 2011, 12, 5039–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landis RF, Li C-H, Gupta A, Lee Y-W, Yazdani M, Ngernyuang N, Altinbasak I, Mansoor S, Khichi MAS, Sanyal A and Rotello VM, J. Am. Chem. Soc, 2018, 140, 6176–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarda A, Rubilar JF, Miltz J and Galotto MJ, Int. J. Food Microbiol, 2011, 146, 144–150. [DOI] [PubMed] [Google Scholar]
- 21.Wollensak G, Spoerl E and Seiler T, Am. J. Ophthalmol, 2003, 135, 620–627. [DOI] [PubMed] [Google Scholar]
- 22.Ribes J, Beztsinna N, Bailly R, Castano S, Rascol E, Taib-Maamar N, Badarau E and Bestel I, Bioconjug. Chem, , DOI: 10.1021/acs.bioconjchem.1c00028. [DOI] [PubMed] [Google Scholar]
- 23.V Makabenta JM, Nabawy A, Li C-H, Schmidt-Malan S, Patel R and Rotello VM, Nat. Rev. Microbiol, 2021, 19, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Shi L, Su L, Van der Mei HC, Jutte PC, Ren Y and Busscher HJ, Chem. Soc. Rev, 2019, 48, 428–446. [DOI] [PubMed] [Google Scholar]
- 25.Terjung N, Löffler M, Gibis M, Hinrichs J and Weiss J, Food Funct, 2012, 3, 290–301. [DOI] [PubMed] [Google Scholar]
- 26.Nostro A, Blanco AR, Cannatelli MA, Enea V, Flamini G, Morelli I, Roccaro AS and Alonzo V, FEMS Microbiol. Lett, 2004, 230, 191–195. [DOI] [PubMed] [Google Scholar]
- 27.Elzoghby AO, Control J. Release, 2013, 172, 1075–1091. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson E, Food Hydrocoll, 2009, 23, 1473–1482. [Google Scholar]
- 29.Gaihre B, Khil MS, Lee DR and Kim HY, Int. J. Pharm, 2009, 365, 180–189. [DOI] [PubMed] [Google Scholar]
- 30.Vroom JM, De Grauw KJ, Gerritsen HC, Bradshaw DJ, Marsh PD, Watson GK, Birmingham JJ and Allison C, Appl. Environ. Microbiol, 1999, 65, 3502–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arndt-Jovin DJ and Jovin TM, in Fluorescence Microscopy of Living Cells in Culture Part B. Quantitative Fluorescence Microscopy—Imaging and Spectroscopy, eds. Taylor DL and Y.-L. B. T.-M. in Wang CB, Academic Press, 1989, vol. 30, pp. 417–448. [Google Scholar]
- 32.Landis RF, Li C-H, Gupta A, Lee Y-W, Yazdani M, Ngernyuang N, Altinbasak I, Mansoor S, Khichi MAS, Sanyal A and Rotello VM, J. Am. Chem. Soc, 2018, 140, 6176–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, Wang S, Wang T, Hu S, Bhayana B, Ishii M, Kong Y, Cai Y, Dai T, Cui W and Wu MX, Sci. Transl. Med, 2021, 13, eaba3571. [DOI] [PubMed] [Google Scholar]
- 34.Koser SA, Chinn BD and Saunders F, J. Bacteriol, 1938, 36, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajamani S, Bauer WD, Robinson JB, Farrow JM, Pesci EC, Teplitski M, Gao M, Sayre RT and Phillips DA, Mol. Plant-Microbe Interact, 2008, 21, 1184–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra R, Grande R, Butrico L, Rossi A, Settimio UF, Caroleo B, Amato B, Gallelli L and de Franciscis S, Expert Rev. Anti. Infect. Ther, 2015, 13, 605–613. [DOI] [PubMed] [Google Scholar]
- 37.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR and Rumbaugh KP, Infect. Immun, 2014, 82, 4718–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig WA and Welling PG, Clin. Pharmacokinet, 1977, 2, 252–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


