Abstract
Hepatocellular carcinoma (HCC) is the most common liver cancer and one of the leading causes of cancer-related deaths in the world. Multiple immunotherapeutic approaches have been investigated to date, and immunotherapy has become the new standard of care therapy in HCC. However, the current role of immunotherapy in HCC remains non-curative. Given this context, a high priority for oncology is understanding the biomarkers that predict clinical response to immunotherapy, have the potential to improve patient selection to maximize the clinical benefit, and spare unnecessary toxicity. In this review, we summarize the key predictive and prognostic biomarkers investigated in immunotherapy clinical trials in HCC and the emerging biomarkers to serve as a roadmap for future clinical trials. Biomarkers from tumoral tissues including PDL-1 expression, tissue infiltrating lymphocytes, tumor mutational burden (TMB) and specific immune signatures, and from peripheral blood including neutrophil-to-lymphocytes ratio, platelet-to-lymphocytes ratio, circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), and specific cytokines, along with gut microbiota are among the studied biomarkers to date in the HCC era. More integrative approaches, including mathematical biomarkers to predict immunotherapy outcomes, are yet to be studied in HCC.
Keywords: hepatocellular carcinoma, immunotherapy, biomarker
Introduction
Hepatocellular carcinoma (HCC) is the most common liver cancer and the fourth leading cause of cancer-related deaths worldwide.1 Curative options (liver transplantation, resection, and ablation) remain limited to a small subset of patients because most patients are diagnosed at advanced stages and/or have underlying chronic liver disease, and there is a shortage of available organs for liver transplantation.2 Systemic treatment options for advanced HCC remained limited to targeted therapy and multitargeted tyrosine kinase inhibitors, in the form of sorafenib, lenvatinib, regorafenib, cabozantinib, and ramucirumab, until recently.3–7 In recent years, immune checkpoint inhibitors (ICIs) including nivolumab and pembrolizumab have shown survival benefit and have been approved as first or second line therapies.2 Most recently, the randomized Phase 3 IMbrave150 trial with sorafenib versus atezolizumab, anti-PD-L1 antibody, plus bevacizumab, anti-VEGF antibody, showed positive results, and atezolizumab plus bevacizumab became the new preferred standard first line treatment for advanced HCC.8 Results of the IMbrave150 trial presented at the Digital Liver Cancer Summit of the European Association for the Study of the Liver on February 2021 showed that atezolizumab plus bevacizumab treatment was superior in prolonging overall survival (OS) (19.2 months versus 13.4 months for sorafenib, HR=0.66, 95% CI=0.52–0.85, P=0.0009) and progression-free survival (PFS) (6.9 months versus 4.3 months for sorafenib, HR=0.65, 95% CI=0.53–0.81 P=0.0001) versus sorafenib.9
Although the clinical development of ICIs has been accelerating in HCC, treatment is considered non-curative. In addition, around 25% of patients develop grade 3–4 immune-related adverse events.10,11 Therefore, understanding which biomarkers predict clinical response to immunotherapy can potentially improve the patient selection and treatment outcomes. Furthermore, given the recent encouraging results of the Imbrave150 trial8 with ICI plus an anti-angiogenesis agent, there is an additional unmet need for biomarkers that could help to determine which patients should receive combination therapy, and which could be spared unnecessary toxicity. The biomarkers from longitudinal tumor and blood samples may also allow us to understand both intrinsic and acquired resistance mechanisms.
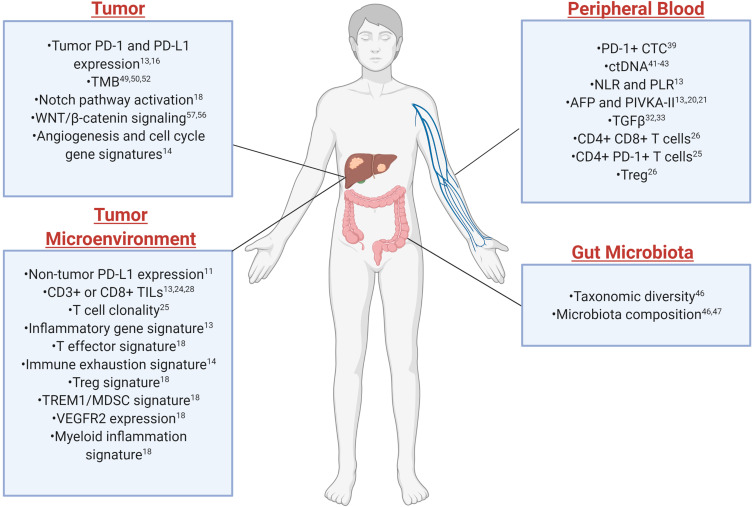
Biomarker studies in HCC are still in infancy given the later use of ICI and have primarily been led by results from other types of cancers. There is no established predictive biomarker for HCC, unlike some other cancer types where PD-L1 expression is used to select patients for pembrolizumab.12 Studies to identify possible predictive biomarkers for immunotherapy in HCC have recently begun. In this review, we summarize the key predictive and prognostic biomarkers studied in immunotherapy clinical trials in HCC (Table 1) and the emerging biomarkers from both tumoral tissue (tumor or tumor microenvironment) and extratumoral biospecimens such as peripheral blood and feces (Figure 1), to serve as a roadmap for designing future clinical and biomarker trials in HCC.
Table 1.
Proposed Biomarkers in Recent Clinical Trials Using Immune Checkpoint Inhibitors for HCC
| Trial | Treatment | Line of Treatment | Number of Patients | Assay | Biomarker | Outcome | Ref. |
|---|---|---|---|---|---|---|---|
| KEYNOTE-224 (NCT02702414) | Pembrolizumab | Second line | N=52 | IHC | Baseline PD-L1 expression* | PD-L1 expression on tumor cells, lymphocytes and macrophages was associated with response. PD-L1 expression on tumor cells alone was not associated with response. | [11] |
| CheckMate 040 (NCT01658878) | Nivolumab and Ipilimumab | Second line | N=145 | IHC | Baseline tumor PD-L1 expression* | No association between baseline tumor PD-L1 expression and treatment response. | [15] |
| CheckMate 459 (NCT02576509) | Nivolumab vs Sorafenib | First line | N=743 | IHC | Baseline tumor PD-L1 expression* | Patients with PD-L1 ≥1% was found to be associated with longer median OS in those treated with Nivolumab vs Sorafenib. | [16] |
| CheckMate 040 | Nivolumab | First or second line | N=44 (Escalation phase), N=174 (Expansion phase) | IHC | Baseline tumor PD-L1 expression* | No association between baseline tumor PD-L1 expression and treatment response. | [10] |
| CheckMate 040 | Nivolumab | First or second line | N=195 (PD-L1), N=178 (PD-1) | IHC | Baseline tumor PD-L1 and PD-1 expressions* | Both were associated with improved OS. PD-1, but not PD-L1, was associated with ORR. |
[13] |
| N=189 (CD3), N=192 (CD8) |
IHC | Baseline CD3+ or CD8+ TILs* | CD3+ or CD8+ TILs exhibited a trend towards improved OS. | ||||
| N=189 (CD4), N=190 (FOXP3) | IHC | Baseline CD4+ or FOXP3+ TILs* | Not associated with OS. | ||||
| N=135 | IHC | Baseline CD68+ or CD163+ macrophages* | Not associated with OS. | ||||
| N=37 | RNA seq | Baseline inflammatory signature of tumor tissue* | Inflammatory signature consisting of CD274 (PD-L1), CD8A, LAG3, STAT1 was associated with both improved objective response rate and overall survival. | ||||
| N=242 (NLR), N=243 (PLR) | Flow cytometry | Baseline Neutrophil to lymphocyte ratio (NLR) or platelet to lymphocyte ratio (PLR)* | Lower NLR and PLR were found to be associated with PR and CR on nivolumab. | ||||
| N=149 | Serum AFP detection | Baseline Serum AFP | Low AFP (<400 µg/L) showed a numerical (although not statistically significant) association with response. | ||||
| GO30140 (NCT02715531) Imbrave150 (NCT03434379) |
Atezolizumab and Bevacizumab | First line | N=58 (GO30140) N=150 (Imbrave150) |
Serum AFP detection | Serum AFP | ≥75% decrease or ≤10% increase in AFP levels at 6 weeks after the treatment was significantly associated with improved OS and PFS. | [20] |
| GO300140 (NCT02715531) | Atezolizumab and Bevacizumab | First line | N=73 | WES | TMB# | Not associated with response or PFS. | [18] |
| N=90 | RNA seq | PD-L1 gene expression* | Associated with response and longer PFS. | ||||
| N=90 | RNA seq | T-effector signature (GZM, PRF1, CXCL9)* | Associated with response and longer PFS. | ||||
| N=90 | RNA seq | Notch pathway (HES1) activation genes# | Associated with lack of response and shorter PFS. | ||||
| GO300140 (NCT02715531) | Atezolizumab and Bevacizumab vs Atezolizumab alone | First line | N=91 | RNS seq | VEGFR2 expression^ | Associated with longer PFS in patients treated with atezolizumab + bevacizumab than in those treated with atezolizumab alone. | [18] |
| N=91 | RNA seq | Treg signature (CCR8, BATF, CTSC, TNFRSF4, FOXP3, TNFRSF18, IKZF2, and IL2RA)* | Associated with longer PFS in patients treated with atezolizumab + bevacizumab than in those treated with atezolizumab alone. | ||||
| N=91 | RNA seq | Myeloid inflammation signature (CXCL1, CXCL2, CXCL3, CXCL8, IL6, PTGS1)* | Associated with longer PFS in patients treated with atezolizumab + bevacizumab than in those treated with atezolizumab alone. | ||||
| N=91 | RNA seq | TREM1/MDSC signatures* | Associated with longer PFS in patients treated with atezolizumab + bevacizumab than in those treated with atezolizumab alone. | ||||
| NCT01853618 | Tremelimumab in combination with radiofrequency ablation or chemoablation | Adjuvant | N=9 | IHC | Post-treatment CD3+ and CD8+ TILs* | Responders had higher post-tremelimumab CD3+ and CD8+ TILs than non-responders. | [24] |
| NCT02658019 | Pembrolizumab | Second line | N=24 | ELISA | Baseline plasma IL‐1β, IL‐6, IL‐8, IL‐12, IL‐18, IFN‐γ, TGF‐β, IL‐10, CXCL9, CCL4, CCL5, PD‐L1, and PD‐L2* | High baseline plasma TGF‐β levels (≥200 pg/mL) significantly correlated with poor treatment outcomes. | [33] |
| NCT02407990, NCT04068519 | Tislelizumab | Second line | N=41 | NGS | Angiogenesis^ (TEK, KDR, HGF, and EGR1), Immune exhaustion* (CD274, CTLA4, TIGIT, and CD96), cell-cycle (E2F7, FOXA1, and FANCD2) gene signatures | Non-responders had elevated angiogenesis, immune exhaustion, and cell-cycle gene signature than responders. | [14] |
| NCT01853618 | Tremelimumab in combination with radiofrequency ablation or chemoablation | Adjuvant | N=34 | Flow Cytometry | Baseline CD4+PD1+ cells* | The frequency of CD4+ PD1+ cells in PBMC were higher in responder than in non-responder. |
[25] |
| N=19 (pre-treatment tissue), N=22 (post-treatment tissue) | IHC | Tumor CD3 and PD-1 expression* | Responders had higher CD3 and PD-1 expression in post-treatment tumor tissues. No statistically significant difference in pre-treatment tumor tissues. | ||||
| N=19 (pre-treatment tissue), N=22 (post-treatment tissue) | IHC | Tumor CD8 and CD68 expression* | No statistically significant difference between responder and non-responders in pre- and post-treatment tumor tissues. | ||||
| N=26 | TCR seq | T-cell repertoire* | Responders had high TIL clonality compared to non-responders. |
Notes: *Immune-related biomarkers. ^Angiogenesis-related biomarkers. #Genomic biomarkers.
Abbreviations: AFP, alpha feto protein; IHC, immunohistochemistry; NGS, next generation sequencing; NLR, neutrophil-to-lymphocyte ratio; OS, overall survival; PLR, platelet-to-lymphocyte ratio; RNA seq, RNA sequencing; TCR, T cell receptor; TGFβ, transforming growth factor β; TILs, tumor infiltrating lymphocytes; TMB, tumor mutational burden; WES, whole exome sequencing.
Figure 1.
Spectrum of biomarkers from different biological sources for response to immune checkpoint inhibitors in HCC. Created with BioRender.com.
Current Landscape of Biomarker Analysis in Clinical Trials Studying Immunotherapy of HCC
Nivolumab was the first approved immune checkpoint inhibitor in HCC based on the CheckMate 040 trial (NCT01658878), where the safety and clinical benefit were assessed across multiple HCC etiologies leading to conditional approval of nivolumab in the second-line setting.10 This study included four groups of patients: 1) those who did not receive or were intolerant of sorafenib without viral hepatitis; 2) those who progressed on sorafenib without viral hepatitis; 3) those who were HCV infected; and 4) patients who were HBV infected. Objective response rates (ORRs) were similar across different etiologies, including both sorafenib-naïve and sorafenib-treated patients, and were reported to be between 15 and 20%. Notably, response rates occurred regardless of PD-L1 expression on tumor cells.10 In a recent study, tumor samples from dose-escalation and dose-expansion phases of the CheckMate 040 trial were analyzed to explore biomarkers with a goal of identifying patients who may benefit the most from immunotherapy.13 This study showed that in the overall population (sorafenib-naïve and sorafenib-treated), high PD-L1 expression on tumor cells was associated with improved survival. However, PD-L1 expression was not predictive of ORR, suggesting that PD-L1 may not serve as an ideal biomarker.13 Furthermore, high PD-1 (a T-cell exhaustion marker) expression in tumor was associated with ORR. Higher densities of CD3+ or CD8+ tumor infiltrating lymphocytes (TILs), but not CD4+ or FOXP3+ TILs, exhibited a trend towards improved OS (overall survival). Unlike T-cells, no association was found between survival and either CD68+ (all macrophages) or CD163+ (alternatively activated) macrophages in tumor tissues. Additionally, gene expression profiling (GEP) of tumor tissues revealed that an inflammatory signature consisting of CD274 (PD-L1), CD8α, LAG3, and STAT1 was associated with both improved ORR and OS, suggesting that underlying inflammation within the tumor microenvironment may favor improved clinical outcome. Moreover, lower expression of systemic inflammatory markers including the neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) was found to be associated with ORR with nivolumab treatment.
Another study, where tislelizumab, an anti-PD-1 monoclonal antibody, was given to patients with advanced HCC previously treated with sorafenib (NCT02407990 and NCT04068519), showed that tislelizumab demonstrated anti-tumor activity with an ORR of 13% and PFS of 3.3 months.14 Notably, sorafenib exposure appeared to change the PD-L1 expression and gene signatures within the tumor microenvironment: sorafenib-exposed samples were associated with more immune-suppressive signatures. In contrast, sorafenib-naïve samples were associated with high PD-L1 expression and an immune-cell activation signature along with higher expression of co-inhibitory molecules. PD-L1 expression on tumor cells was associated with improved response and PFS from tislelizumab. Non-responders showed elevated expression of genes related to angiogenesis (TEK, KDR, HGF, and EGR1), immune exhaustion (CD274, CTLA-4, TIGIT, and CD96), and cell cycle (E2F7, FOXA1, and FANCD2), compared to responders.14
In the KEYNOTE-224 trial, where the PD-1 inhibitor pembrolizumab was given to patients with HCC in the second-line setting, PD-L1 expression on both tumor cells and non-tumor cells (lymphocytes and macrophages) was found to be associated with improved responses to treatment, whereas PD-L1 expression on tumor cells alone was not associated with clinical response.11 Similarly, in the CheckMate 040 trial, where nivolumab and ipilimumab were given to patients with advanced HCC who were previously treated with sorafenib, no association was found between treatment response and baseline PD-L1 expression.15 In the ChekMate 459 study (NCT02576509), where the PD-1 inhibitor, nivolumab versus sorafenib were given to HCC patients, nivolumab showed consistent survival benefit over sorafenib regardless of baseline PD-L1 expression (PD-L1 ≥1% HR=0.80 [95% CI=0.54–1.17]; PD-L1 <1% HR=0.84 [95% CI=0.70–1.01).16 However, expression of PD-L1 ≥1% was found to be associated with longer median OS in those treated with nivolumab versus sorafenib (16.1 versus 8.6 months).16 In a multicohort study, where the PD-L1 inhibitor, durvalumab was given to HCC patients in combination with the anti-VEGFR2 antibody, ramucirumab, the PD-L1 high subgroup was found to be associated with a better response to treatment in terms of both OS and PFS.17
In the Phase 1b GO300140 trial (NCT02715531), archival pre-treatment tumor tissues or fresh biopsies were obtained from patients with unresectable HCC receiving atezolizumab plus bevacizumab or atezolizumab monotherapy to investigate molecular features associated with clinical response or improved progression-free survival (PFS). Gene expression analysis demonstrated that high PD-L1 and T-effector gene (GZM, PRF1, and CXCL9) signatures were associated with improved responses and longer PFS, whereas gene expression related to Notch pathway activation (ie, high expression of HES1) was associated with a lack of response and shorter PFS.18 In contrast, whole exome sequencing showed that tumor mutation burden (TMB) was not associated with a response to treatment or PFS. Importantly, high expression of VEGFR2, genes related to regulatory T-cells (Tregs; CCR8, BATF, CTSC, TNFRSF4, FOXP3, TNFRSF18, IKZF2, and IL2RA), myeloid inflammation (CXCL1, CXCL2, CXCL3, CXCL8, IL6, and PTGS1), and TREM1/MDSC (Myeloid-derived suppressor cells) signatures were associated with longer PFS in patients treated with atezolizumab plus bevacizumab than in those treated with atezolizumab monotherapy.
AFP (alpha-fetoprotein) is the most widely used and accepted serum tumor biomarker in HCC.19 Zhu et al20 investigated the role of AFP as a potential biomarker for atezolizumab plus bevacizumab efficacy on patients from the phase Ib study GO30140 and Imbrave150. The optimal time for AFP measurement and optimal AFP cutoffs were determined based on the GO30140 study and further validated on data from Imbrave150. This analysis demonstrated that a 6-week interval was found to be optimal for AFP measurement. A 75% or greater decrease in AFP was found to be predictive of tumor response, and 10% or less increase correlated with disease control. The data from Imbrave150 demonstrated that in patients with baseline AFP levels greater than 20 ng/mL who received atezolizumab plus bevacizumab, ≥75% decrease or ≤10% increase in AFP levels at 6 weeks were significantly associated with OS and PFS improvements.20 Furthermore, another study in which lenvatinib and an anti-PD-1 antibody were given to patients with unresectable HCC as front-line therapy showed that a more than 50% decrease in AFP and protein induced by vitamin K absence or antagonist-II (PIVKA-II) levels measured at 2–3 weeks after the treatment was associated with a higher rate of radiological response.21
Notably, both transarterial chemoembolization (TACE) and locoregional ablative therapies have been shown to induce a peripheral immune response that can be potentially augmented by immune-modulating agents.22,23 The study by Duffy et al24 highlighted the benefit from tremelimumab (CTLA-4 inhibitor) in combination with locoregional treatments in patients with advanced HCC (NCT01853618). In this pilot trial, 32 patients who underwent either subtotal radiofrequency ablation (RFA), cryoablation (CA), or TACE received tremelimumab. Out of 19 evaluable patients who had measurable target lesions outside of the areas treated with ablation or TACE, five (26.3%) achieved confirmed partial response. Median OS and time-to-progression (TTP) were 12.3 and 7.4 months, respectively. Interestingly, the percentage of activated CD4+ and CD8+ T-cells in the peripheral blood mononuclear cells (PBMCs) increased after the treatment. Additionally, evaluable tumor lesions showed increased CD3+CD8+ tumor-infiltrating lymphocytes following treatment. Importantly, responders, defined as stable disease or partial response of at least 4 months, had higher post-tremelimumab CD3+ and CD8+ T-cell infiltration in tumor tissue, compared with non-responders, defined as disease progression or partial response of under 4 months.24 This study was followed by another cohort in which comprehensive immune monitoring on PBMCs and tumors was published from patients in the same trial.25 Tremelimumab therapy appeared to increase CD8+ HLA-DR+, CD8+ PD-1+, CD8+ ICOS+, CD4+ HLA-DR+, CD4+ PD-1+, and CD4+ ICOS+ T-cells in the PBMCs of both responder and non-responder patients. Importantly, responder patients had a higher frequency of CD4+ PD-1+ cells in PBMCs prior to therapy than patients not responding, suggesting that the CD4+ PD1+ cell count may be used as a predictive biomarker of response to ICIs. Although there were no differences in this study between responders and non-responders regarding the expression of PD-1, CD3, CD8, and CD68 in pre-treatment tumor tissues, responders showed significantly increased CD3 and PD-1 expression after treatment compared to non-responders. Furthermore, T-cell receptor sequencing indicated that tremelimumab appeared to decrease peripheral clonality, showing a broadening of the T-cell repertoire.25 Another study with a total of 14 stage 4 HCC patients who received PD-1/PD-L1 inhibitor treatment showed that responders had a lower percentage of Tregs, a lower ratio of effector memory T-cells to Tregs, and a higher proportion of CD4+ CD8+ double-positive T-cells in peripheral blood after the treatment than in non-responders.26 Moreover, the expression levels of TIM3 and OX40 were lower in responders than non-responders.26
Taken together, immunotherapy approaches with combined systemic and local therapies in advanced unresectable HCC are rapidly evolving, along with exploratory studies of potential predictive and prognostic biomarkers. However, the retrospective nature of the analysis, along with the small number of patients, are limiting the ability to validate potential important biomarkers in this setting. Therefore, future studies should focus on prospective incorporation of biomarkers strategies to obtain pre-treatment sampling from tissue and/or blood, which is becoming critical to the success of identifying and validating predictive and prognostic immune biomarkers in HCC. This strategy will facilitate the development of immune biomarker strategies to select patient candidates for trials entry and eventually identify predictive biomarkers of therapeutic response.
Notably, recent immunotherapy studies in operable disease in different tumor types suggested that the therapeutic efficacy of neoadjuvant immune checkpoint inhibition was greater than that of the same treatment in the adjuvant setting.27 In HCC, Kaseb et al28 showed an illustrative case report of a series of patients from a randomized Phase II trial of perioperative immunotherapy for HCC (NCT03222076) and reported pathologic complete response in three of nine initial cases on the trial. Subsequently, 20 patients underwent surgical resection after receiving neoadjuvant nivolumab alone (Arm A) or nivolumab plus ipilimumab (Arm B). Six out of 20 patients who underwent surgery (30%, three and three in Arms A and B) achieved major pathologic responses, and no recurrence was observed in these patients. This study demonstrates that neoadjuvant immunotherapy-based treatment may contribute to a paradigm shift in the pre-operative setting of resectable HCC. The cases with pathologic complete response presented in this study showed an increase in CD8+ T-cell infiltration in tumor tissues, specifically with two T-cell clusters (CD3+CD8+CD45RO+Eomes+CD57+CD38low and CD3+CD8+CD45RO+Eomes+). Interestingly, Tregs (CD3+CD4+CD45RO+FOXP3+ICOS+ cells), which are known to be associated with poor prognosis and decreased survival in HCC by suppressing anti-tumor immune response,29 also showed an increase in post-treatment tumor tissue of the patient, compared to a pre-treatment tumor tissue.28 Responders on the study were found to have an increased ratio of CD8+ T-cells to T-regulatory cells. This is particularly important in HCC, since the intratumoral balance between cytotoxic effector T-cells and inhibitory regulatory cells has been strongly associated with HCC tumor recurrence after resection and also overall survival in HCC.30 Therefore, future studies in the operable setting in HCC represent a unique opportunity to identify predictive and prognostic immune biomarkers to neoadjuvant or adjuvant immunotherapy in resectable HCC. CD8+ T-cells to Tregs ratio is a specially attractive biomarker in this setting.30 This study is ongoing, and final results may provide further insights into the use of TILs as a biomarker of response to neoadjuvant ICIs in resectable HCC.
Notably, transforming growth factor-β (TGF-β) is known to be an immunosuppressive and fibrotic cytokine.31 The highly activated TGFβ signature was found to be associated with fibrosis and activated stromal signatures in HCC and might mediate immunotherapy resistance.32 Out of 11 cytokines and chemokines tested, only baseline TGF-β cytokine level in peripheral blood was significantly higher in non-responders than in responders in a study with 24 patients with unresectable HCC receiving pembrolizumab. This suggests that baseline TGF-β could be a predictive biomarker for response to pembrolizumab,33 and that the integration of anti-TGF-β agents into immunotherapy strategies could be an attractive approach to enhance immunotherapy activity.
Importantly, HCC has different underlying etiologies that can lead to carcinoma by distinctly regulating the hepatic microenvironment and immune responses which could eventually affect immunotherapy efficacy in patients.34 A recent study by Pfister et al35 showed that immunotherapy did not improve survival in patients with non-viral HCC. Interestingly, patients with NASH-induced HCC who received ICIs showed reduced survival compared to patients with other etiologies, possibly due to accumulation of exhausted, unconventionally activated CD8+PD1+ T-cells in NASH-effected liver.35 Therefore, patients stratification according to underlying etiologies should be taken into consideration for future immunotherapy-based clinical trials in HCC.
Emerging Biomarkers for Immunotherapy in HCC
Imaging
Tumor stiffness and fibrosis could be another factor in predicting immunotherapy response in HCC. A total of 25 patients with HCC were evaluated for stiffness by magnetic resonance elastography (MRE) and enhancement by magnetic resonance imaging (MRI) before and after immunotherapy.36 This study showed that treatment response was associated with the absence of portal venous phase capsular enhancement and an increase in HCC stiffness.36 Moreover, stiffness was significantly correlated with intratumoral T lymphocytes on tumor biopsy.37
Circulating Tumor Cells
Liquid biopsies are non-invasive and provide rapid real-time information for prediction of treatment response or disease monitoring. Liquid biopsy entails an analysis of circulating tumor components such as circulating tumor cells (CTCs), extracellular vesicles, or nucleic acids released from tumor cells.38 A recent study by Winogrand et al39 showed the presence of PD-L1+CTCs was associated with favorable immunotherapy outcome (n=10), but it was also a negative prognostic biomarker and survival predictor overall (n=87). Among ten patients with HCC receiving immune checkpoint inhibitors (nivolumab or pembrolizumab), all patients who responded had PD-L1+ CTCs, and all patients without PD-L1+ CTCs failed treatment. In the same study, PD-L1+ CTCs were evaluated in 87 patients with HCC at different stages and their presence in blood accurately discriminated patients with more advanced disease from those with early-stage HCC, hence it was proposed to be a prognostic biomarker. PD-L1+ CTCs were also found to be an independent survival predictor as patients with PD-L1+ CTCs had significantly inferior OS compared to those without PD-L1+ CTCs, even after controlling for other factors.
Circulating Tumor DNA (ctDNA)
ctDNA is released from apoptotic and necrotic tumor cells and can be used to determine information about tumor burden and cancer mutational profile,40 thus it could be a feasible biomarker for predicting treatment outcome. A study with a large cohort of 313 patients with eight tumor types including HCC who received various treatments, including immunotherapy, showed that either the absolute value of ctDNA content fraction at the time of clinical imaging or the dynamic changes in ctDNA were highly correlated with clinical outcome.41 Importantly, changes in ctDNA were found to be consistent with corresponding imaging-based evaluations of the same patients.41 A recent prospective phase II clinical trial was conducted to assess the feasibility of using ctDNA in five different groups of patients with advanced solid tumors treated with pembrolizumab.42 The study showed that both baseline and changes in ctDNA levels from baseline were correlated with OS and PFS. The latter was also found to be predictive of ICI response across cancer types.42 In a cohort of 48 unresectable HCC patients receiving atezolizumab plus bevacizumab, a personalized ctDNA assay for each patient was designed based on tumor mutational signatures, and a longitudinal detection of ctDNA was performed to monitor treatment outcome.43 This study showed that higher baseline ctDNA was associated with greater tumor burden, and dynamic changes in ctDNA levels post-treatment were associated with response. The patients whose ctDNA became undetectable after the treatment showed longer PFS.43 Importantly, across a variety of solid tumors, including colorectal cancer,44 ctDNA studies have gained major interest following resection of primary colorectal cancer supporting the correlation between the presence of ctDNA post-resection and the existence of microscopic residual disease and a subsequent recurrence. Thus, mutational signatures of the primary HCC tumor may emerge as a potential noninvasive biomarker for monitoring HCC tumor recurrence, and, therefore, warrant future testing and validation studies in resectable HCC.
Gut Microbiota
Microbiota, commensal microorganisms, are known to influence immune responses and could be used as an emerging biomarker in immunotherapy. Accumulated evidence has shown that the gut microbiota may support immunotherapy efficacy or lack thereof in various cancer types.45 A study with eight HCC patients receiving anti-PD-1 treatment showed that fecal samples from responders had higher taxonomic diversity and more gene counts in 20 species including Akkermansia and Ruminococcaceae than those from non-responders, and the dissimilarity was evident as early as the 6th week after treatment initiation. This suggests that gut microbiota composition may provide an early prediction of treatment outcome.46 Another study with 74 advanced stage gastrointestinal cancer patients receiving anti-PD-1/PD-L1 treatment showed that fecal samples from responders had a higher abundance of Prevotellaceae and a lesser abundance of Bacteroidacea.47 Moreover, one particular subgroup of responders harbored a significantly higher abundance of Prevotella, Ruminococcaceae, and Lachnospiraceae. In this study, no significant difference was seen regarding taxonomic diversity between responders and non-responders.47
Tumor Mutational Burden and Specific Mutational Alterations
High TMB, non-synonymous single nucleotide variants (nsSNVs) in a tumor, is hypothesized to produce more neoantigens which foster high frequencies and greater diversity of tumor-specific T-cell responses, thus making immunotherapy more effective.48 Higher TMB predicts favorable outcome to PD-1/PD-L1 blockage across diverse tumors including HCC.49,50 Multiple studies have shown that TMB is generally low (median <10 mut/Mb) in HCC.51 For example, one small case series in HCC (n=17) found a median TMB of 4 mut/Mb. There was no significant difference between immunotherapy responders and non-responders regarding TMB.52
With the implementation of next generation sequencing, several driver mutations in genes including TERT, CTNNB1, TP53, AXIN1, ARID1A, and ARID1B were identified in HCC.53 Based on these mutations and epigenetic alterations, further molecular subclassification was developed.54 Although these mutations were “non-druggable”, they might potentially affect response to immunotherapy.55 Importantly, activating mutations in WNT/β-catenin signaling were associated with a lower disease control rate (DCR), shorter median PFS, and shorter median OS for patients treated with ICIs.56 No other pathway alterations were found to be associated with resistance or responsiveness to immunotherapy in HCC.56 This suggests that the presence of activating WNT/β-actin mutation confers innate resistance to ICIs which was consistent with the study in melanoma where lack of T-cell infiltration was correlated with altered β-catenin signaling.57,58
Mathematical Biomarkers and Combined Multiparametric Biomarkers
The complex interactions of multiple biological systems dictate the response to ICI. Mathematical modeling is one approach to bridge the scientific knowledge gaps that exist for these interactions. Butner et al59,60 have recently shown in a series of papers that ICI response can be modeled on “super parameters” that describe the “Anti-tumor immune state”, the “tumor cell kill rate” of ICI, and the tumor proliferation rate. The investigators demonstrated that some of these super parameters could be estimated by taking measurements of the tumor volume from standard computed tomography (CT) scans over time, and that these could be used to predict outcomes and long-term benefits of ICI in many solid tumors. Application of physical oncology to HCC may reveal how traditional biomarkers could be used in a mechanistic mathematical model to help identify patients who are likely to benefit from combination ICIs and other novel therapeutic approaches.
Multiparametric approaches to biomarker studies that combine different aspects of tumors have been recently started since no single robust biomarker has been identified for most cancer so far. Recently, in a study with metastatic non-small cell lung cancer patients, the multiparametric analysis gave a better diagnostic performance than single parameters like radiomic signature, PD-L1, or IFN-γ expression regarding clinical response, sensitivity, and specificity.61 In another interesting study, an astronomical algorithm was applied to multiplex immunofluorescence labeling of pathology specimens to establish spatial relationships between tumor cells and multiple immune elements and immunoarchitectural characterization of the tumor.62 With only six markers, they identified key features in pretreatment melanoma specimens that predicted response to immunotherapy, suggesting that this approach might give more accuracy regarding treatment response prediction than individual IHC markers like PD-L1 expression.
Conclusion
We have described the data for ICIs and the associated biomarker work in recent landmark studies for HCC. Although multiple potential candidates that predict response and guide therapy decisions have emerged, there is currently no standard biomarker that predict ICI outcome in patients with HCC. Capturing the complexities of ICI response and resistance through non-invasive means will be the subject of intense investigations in the coming years. Notably, HCC is often diagnosed without the need for a biopsy based on characteristic imaging pattern per American Association for Study of Liver Diseases (AASLD) guidelines.63 However, obtaining tissue biopsies in patients receiving ICI might give invaluable information about possible predictive biomarkers and the mechanism of resistance to immunotherapy. Tumor PD-L1 expression is the most widely studied biomarker in HCC, but there are conflicting results on its predictive potential as in other cancer types. Integrative multiparametric approaches that combine histopathology, imaging, and immune signatures appear to be the most comprehensive way to assess treatment outcomes and seem to be promising in the future. Personalized therapy through a biomarker-driven approach is expected to result in improved outcomes for patients and transition this approach from non-curative to potentially curative.
Abbreviations
AFP, alpha feto protein; CT, computed tomography; CTC, circulating tumor cell; ctDNA, circulating tumor DNA; DCR, disease control rate; GEP, gene expression profiling; HCC, hepatocellular carcinoma; ICIs, Immune checkpoint inhibitors; IHC, immunohistochemistry; MDSC, myeloid-derived suppressor cell; MRE, magnetic resonance elastography; MRI, magnetic resonance imaging; NGS, next generation sequencing; nsSNVs, non-synonymous single nucleotide variants; ORR, objective response rate; OS, overall survival; PFS, progression-free survival; PIVKA-II, protein induced by vitamin K absence or antagonist-II; TACE, transarterial chemoembolization; TGFβ, transforming growth factor β; TILs, tumor infiltrating lymphocytes; TMB, tumor mutational burden; Treg, regulatory T-cell; TTP, time to progression.
Disclosure
Dr Michael A Curran reports personal fees and/grants from ImmunoGenesis, Inc., Alligator Bioscience, Inc., ImmunOS, Inc.; ImmunoMet, Inc., Oncoresponse, Inc., Pieris, Inc., Nurix, Inc., Aptevo, Inc., Servier, Inc., Kineta, Inc., Salarius, Inc., Xencor, Inc., Agenus, Inc., Mereo, Inc., Amunix, Inc., outside the submitted work; In addition, Dr Michael A Curran has a patent Methods and Composition for Localized Secretion of Anti-CTLA-4 Antibodies licensed to multiple licensees, a patent Dual specificity antibodies which bind both PD-L1 and PD-L2 and prevent their binding to PD-1 with royalties paid to ImmunoGenesis, Inc. Dr Eugene J Koay reports grant funding from NIH, GE Healthcare, Philips Healthcare, Elekta, SU2C. The authors declare that they have no other conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Demir T, Lee SS, Kaseb AO. Systemic therapy of liver cancer. Adv Cancer Res. 2021;149:257–94. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857 [DOI] [PubMed] [Google Scholar]
- 4.Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi: 10.1016/s0140-6736(18)30207-1 [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Qin S, Merle P, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389(10064):56–66. doi: 10.1016/s0140-6736(16)32453-9 [DOI] [PubMed] [Google Scholar]
- 6.Abou-Alfa GK, Meyer T, Cheng A-L, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379(1):54–63. doi: 10.1056/NEJMoa1717002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu AX, Park JO, Ryoo B-Y, et al. Ramucirumab versus placebo as second-line treatment in patients with advanced hepatocellular carcinoma following first-line therapy with sorafenib (REACH): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2015;16(7):859–870. doi: 10.1016/S1470-2045(15)00050-9 [DOI] [PubMed] [Google Scholar]
- 8.Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382(20):1894–1905. doi: 10.1056/NEJMoa1915745 [DOI] [PubMed] [Google Scholar]
- 9.Finn RS. Digital liver cancer summit, European association for the study of the liver. 2021. [Google Scholar]
- 10.El-Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi: 10.1016/s0140-6736(17)31046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu AX, Finn RS, Edeline J, et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label Phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi: 10.1016/s1470-2045(18)30351-6 [DOI] [PubMed] [Google Scholar]
- 12.Gridelli C, Ardizzoni A, Barberis M, et al. Predictive biomarkers of immunotherapy for non-small cell lung cancer: results from an experts panel meeting of the Italian association of thoracic oncology. Transl Lung Cancer Res. 2017;6(3):373–386. doi: 10.21037/tlcr.2017.05.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangro B, Melero I, Wadhawan S, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73(6):1460–1469. doi: 10.1016/j.jhep.2020.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou -M-M, Rau K-M, Kang Y-K, et al. 77 Association between programmed death-ligand 1 (PD-L1) expression and gene signatures of response or resistance to tislelizumab monotherapy in hepatocellular carcinoma (HCC). J ImmunoTher Cancer. 2020;8(Suppl 3):A47–A48. doi: 10.1136/jitc-2020-SITC2020.0077 [DOI] [Google Scholar]
- 15.Yau T, Kang Y-K, Kim T-Y, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the checkmate 040 randomized clinical trial. JAMA Oncol. 2020;6(11):e204564–e204564. doi: 10.1001/jamaoncol.2020.4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sangro B, Park J, Finn R, et al. LBA-3 CheckMate 459: long-term (minimum follow-up 33.6 months) survival outcomes with nivolumab versus sorafenib as first-line treatment in patients with advanced hepatocellular carcinoma.Ann Oncol. 2020;31:S241–S242. doi: 10.1016/j.annonc.2020.04.078 [DOI] [Google Scholar]
- 17.Bang YJ, Golan T, Dahan L, et al. Ramucirumab and durvalumab for previously treated, advanced non-small-cell lung cancer, gastric/gastro-oesophageal junction adenocarcinoma, or hepatocellular carcinoma: an open-label, phase Ia/b study (JVDJ). Eur J Cancer. 2020;137:272–284. doi: 10.1016/j.ejca.2020.06.007 [DOI] [PubMed] [Google Scholar]
- 18.Zhu AX, Guan Y, Abbas AR, et al. Abstract CT044: genomic correlates of clinical benefits from atezolizumab combined with bevacizumab vs. atezolizumab alone in patients with advanced hepatocellular carcinoma (HCC). Cancer Res. 2020;80:CT044–CT044. doi: 10.1158/1538-7445.Am2020-ct044 [DOI] [Google Scholar]
- 19.Galle PR, Foerster F, Kudo M, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39(12):2214–2229. doi: 10.1111/liv.14223 [DOI] [PubMed] [Google Scholar]
- 20.Zhu AX. International liver cancer association (ILCA). 2020. [Google Scholar]
- 21.Sun H-C, Zhu X-D, Huang C, et al. Early tumor marker decrease to predict the efficacy of combination therapy with lenvatinib plus anti-PD-1 antibodies in unresectable hepatocellular carcinoma (uHCC). J Clin Oncol. 2021;39(3_suppl):abstr304. doi: 10.1200/JCO.2021.39.3_suppl.304 [DOI] [Google Scholar]
- 22.Hansler J, Wissniowski TT, Schuppan D, et al. Activation and dramatically increased cytolytic activity of tumor specific T lymphocytes after radio-frequency ablation in patients with hepatocellular carcinoma and colorectal liver metastases. World J Gastroenterol. 2006;12(23):3716–3721. doi: 10.3748/wjg.v12.i23.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayaru L, Pereira SP, Alisa A, et al. Unmasking of α-fetoprotein-specific CD4+ T cell responses in hepatocellular carcinoma patients undergoing embolization. J Immunol. 2007;178(3):1914–1922. doi: 10.4049/jimmunol.178.3.1914 [DOI] [PubMed] [Google Scholar]
- 24.Duffy AG, Ulahannan SV, Makorova-Rusher O, et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J Hepatol. 2017;66(3):545–551. doi: 10.1016/j.jhep.2016.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agdashian D, ElGindi M, Xie C, et al. The effect of anti-CTLA4 treatment on peripheral and intra-tumoral T cells in patients with hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68(4):599–608. doi: 10.1007/s00262-019-02299-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ting VZ, Chew V, Wong CC, Yau TC. Abstract 3171: biomarkers of responses to immunotherapy in advanced hepatocellular carcinoma. Cancer Res. 2020;80:3171. doi: 10.1158/1538-7445.Am2020-3171 [DOI] [Google Scholar]
- 27.Liu J, Blake SJ, Yong MCR, et al. Improved efficacy of neoadjuvant compared to adjuvant immunotherapy to eradicate metastatic disease. Cancer Discov. 2016;6(12):1382–1399. doi: 10.1158/2159-8290.Cd-16-0577 [DOI] [PubMed] [Google Scholar]
- 28.Kaseb AO, Vence L, Blando J, et al. Immunologic correlates of pathologic complete response to preoperative immunotherapy in hepatocellular carcinoma. Cancer Immun Res. 2019;7(9):1390–1395. doi: 10.1158/2326-6066.Cir-18-0605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu JF, Ding Y-H, Ying X-H, et al. Regulatory T cells, especially ICOS(+) FOXP3(+) regulatory T cells, are increased in the hepatocellular carcinoma microenvironment and predict reduced survival. Sci Rep. 2016;6(1):35056. doi: 10.1038/srep35056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Q, Qiu S-J, Fan J, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25(18):2586–2593. doi: 10.1200/jco.2006.09.4565 [DOI] [PubMed] [Google Scholar]
- 31.Schon H-T, Weiskirchen R. Immunomodulatory effects of transforming growth factor-β in the liver. Hepatobiliary Surg Nutr. 2014;3:386–406. doi: 10.3978/j.issn.2304-3881.2014.11.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Su X. Abstract 4286: TGF-β signature is a therapeutic biomarker for combination immunotherapy for hepatocellular carcinoma. Cancer Res. 2020;80:4286. doi: 10.1158/1538-7445.Am2020-4286 [DOI] [Google Scholar]
- 33.Feun LG, Li -Y-Y, Wu C, et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer. 2019;125(20):3603–3614. doi: 10.1002/cncr.32339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roderburg C, Wree A, Demir M, Schmelzle M, Tacke F. The role of the innate immune system in the development and treatment of hepatocellular carcinoma. Hepat Oncol. 2020;7(1):HEP17. doi: 10.2217/hep-2019-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfister D, Núñez NG, Pinyol R, et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature. 2021;592(7854):450–456. doi: 10.1038/s41586-021-03362-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qayyum A, Avritscher R, Aslam R, et al. Immune checkpoint blockade (ICB) response evaluation with MRI/MR elastography (MRE) in surgical and nonsurgical patients with HCC. J Clin Oncol. 2020;38(4_suppl):480. doi: 10.1200/JCO.2020.38.4_suppl.48031815584 [DOI] [Google Scholar]
- 37.Qayyum A, Hwang K-P, Stafford J, et al. Immunotherapy response evaluation with magnetic resonance elastography (MRE) in advanced HCC. J ImmunoTher Cancer. 2019;7(1):329. doi: 10.1186/s40425-019-0766-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut. 2020;69(11):2025–2034. doi: 10.1136/gutjnl-2019-320282 [DOI] [PubMed] [Google Scholar]
- 39.Winograd P, Hou S, Court CM, et al. Hepatocellular carcinoma-circulating tumor cells expressing PD-L1 are prognostic and potentially associated with response to checkpoint inhibitors. Hepatol Commun. 2020;4(10):1527–1540. doi: 10.1002/hep4.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 41.Li J, Jiang W, Wei J, et al. Patient specific circulating tumor DNA fingerprints to monitor treatment response across multiple tumors. J Transl Med. 2020;18(1):293. doi: 10.1186/s12967-020-02449-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1(9):873–881. doi: 10.1038/s43018-020-0096-5 [DOI] [PubMed] [Google Scholar]
- 43.Hsu C-H, Lu S, Abbas A, et al. Longitudinal and personalized detection of circulating tumor DNA (ctDNA) for monitoring efficacy of atezolizumab plus bevacizumab in patients with unresectable hepatocellular carcinoma (HCC). J Clin Oncol. 2020;38(15_suppl):3531. doi: 10.1200/JCO.2020.38.15_suppl.3531 [DOI] [Google Scholar]
- 44.Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–1131. doi: 10.1001/jamaoncol.2019.0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. doi: 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Y, Wang T, Tu X, et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J ImmunoTher Cancer. 2019;7(1):193. doi: 10.1186/s40425-019-0650-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng Z, Cheng S, Kou Y, et al. The gut microbiome is associated with clinical response to anti–PD-1/PD-L1 immunotherapy in gastrointestinal cancer. Cancer Immun Res. 2020;8(10):1251–1261. doi: 10.1158/2326-6066.Cir-19-1014 [DOI] [PubMed] [Google Scholar]
- 48.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.Mct-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46(12):1267–1273. doi: 10.1038/ng.3126 [DOI] [PubMed] [Google Scholar]
- 52.Ang C, Klempner SJ, Ali SM, et al. Prevalence of established and emerging biomarkers of immune checkpoint inhibitor response in advanced hepatocellular carcinoma. Oncotarget. 2019;10(40):4018–4025. doi: 10.18632/oncotarget.26998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez DA, Wang XW. Impact of next-generation sequencing on outcomes in hepatocellular carcinoma: how precise are we really? J Hepatocell Carcinoma. 2020;7:33–37. doi: 10.2147/JHC.S217948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rebouissou S, Nault J-C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72(2):215–229. doi: 10.1016/j.jhep.2019.08.017 [DOI] [PubMed] [Google Scholar]
- 55.Rizzo A, Brandi G. Biochemical predictors of response to immune checkpoint inhibitors in unresectable hepatocellular carcinoma. Cancer Treat Res Commun. 2021;27:100328. doi: 10.1016/j.ctarc.2021.100328 [DOI] [PubMed] [Google Scholar]
- 56.Harding JJ, Nandakumar S, Armenia J, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019;25(7):2116–2126. doi: 10.1158/1078-0432.Ccr-18-2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pinyol R, Sia D, Llovet JM. Immune exclusion-Wnt/CTNNB1 class predicts resistance to immunotherapies in HCC. Clin Cancer Res. 2019;25(7):2021–2023. doi: 10.1158/1078-0432.Ccr-18-3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 59.Butner JD, Wang Z, Elganainy D, et al. A mathematical model for the quantification of a patient’s sensitivity to checkpoint inhibitors and long-term tumour burden. Nat Biomed Eng. 2021;5(4):297–308. doi: 10.1038/s41551-020-00662-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butner JD, Elganainy D, Wang CX, et al. Mathematical prediction of clinical outcomes in advanced cancer patients treated with checkpoint inhibitor immunotherapy. Sci Adv. 2020;6(18):eaay6298. doi: 10.1126/sciadv.aay6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Del Re M, Cucchiara F, Rofi E, et al. A multiparametric approach to improve the prediction of response to immunotherapy in patients with metastatic NSCLC. Cancer Immunol Immunother. 2021;70(6):1667–1678. doi: 10.1007/s00262-020-02810-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berry S, Giraldo NA, Green BF, et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science. 2021;372(6547):eaba2609. doi: 10.1126/science.aba2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933 [DOI] [PubMed] [Google Scholar]