ABSTRACT
Biological soil crusts (biocrusts) are communities of microbes that inhabit the surface of arid soils and provide essential services to dryland ecosystems. While resistant to extreme environmental conditions, biocrusts are susceptible to anthropogenic disturbances that can deprive ecosystems of these valuable services for decades. Until recently, culture-based efforts to produce inoculum for cyanobacterial biocrust restoration in the southwestern United States focused on producing and inoculating the most abundant primary producers and biocrust pioneers, Microcoleus vaginatus and members of the family Coleofasciculaceae (also called Microcoleus steenstrupii complex). The discovery that a unique microbial community characterized by diazotrophs, known as the cyanosphere, is intimately associated with M. vaginatus suggests a symbiotic division of labor in which nutrients are traded between phototrophs and heterotrophs. To probe the potential use of such cyanosphere members in the restoration of biocrusts, we performed coinoculations of soil substrates with cyanosphere constituents. This resulted in cyanobacterial growth that was more rapid than that seen for inoculations with the cyanobacterium alone. Additionally, we found that the mere addition of beneficial heterotrophs enhanced the formation of a cohesive biocrust without the need for additional phototrophic biomass within native soils that contain trace amounts of biocrust cyanobacteria. Our findings support the hitherto-unknown role of beneficial heterotrophic bacteria in the establishment and growth of biocrusts and allow us to make recommendations concerning biocrust restoration efforts based on the presence of remnant biocrust communities in disturbed areas. Future biocrust restoration efforts should consider cyanobacteria and their beneficial heterotrophic community as inoculants.
IMPORTANCE The advancement of biocrust restoration methods for cyanobacterial biocrusts has been largely achieved through trial and error. Successes and failures could not always be traced back to particular factors. The investigation and application of foundational microbial interactions existing within biocrust communities constitute a crucial step toward informed and repeatable biocrust restoration methods.
KEYWORDS: biological soil crust, cyanobacteria, cyanosphere, soil microbiology, soil restoration, biocrust
INTRODUCTION
Biological soil crusts (biocrusts) are globally occurring phototroph-driven microbial communities (1) composed primarily of cyanobacteria (2), algae (3), lichens (4), and bryophytes (5) that support a diverse community of heterotrophic bacteria (6), archaea (7), and fungi (8) contained within the top layer of soils throughout dryland ecosystems. These communities are estimated to cover 12% of Earth’s terrestrial surface (9) and provide essential ecosystem services wherever present (10–12). Incipient biocrust communities are formed when pioneer cyanobacteria, typically filamentous bundle-forming Microcoleus vaginatus and members of the family Coleofasciculaceae (formerly referred to as the Microcoleus steenstrupii complex) (13, 14), which includes newly characterized genera such as Funiculus, Parifilum, Arizonema, Crustifilum, and Crassifilum, provide initial soil stabilization against erosion (2, 12). Depending on environmental conditions, other key biocrust organisms, such as heterocystous cyanobacteria, lichens, and bryophytes (15–17), can colonize and further contribute to biocrust functionality through increased soil fertilization (18). While naturally resistant to the high temperatures and drought common to dryland ecosystems (19, 20), biocrusts are particularly vulnerable to disruptive forces associated with human activities, especially during periods of desiccation (21, 22). Compressional forces from activities such as ranching and foot/vehicle traffic can cause fragmentation of biocrust communities that leads to the degradation and loss of associated ecosystem services (21, 22). Given that the growth of biocrusts is constrained to short periods following sparse precipitation events (20, 23, 24), the natural recovery of degraded communities can range from years to centuries (25, 26).
Given biocrusts’ important role in dryland ecosystem processes, developing methods that enable the rapid restoration of degraded biocrust communities and their associated services has become a focus of restoration efforts in dryland ecosystems. Because biocrust communities are driven by their phototrophic members, current advances in biocrust restoration have centered on the isolation and/or scale-up of these components for eventual transplantation into degraded areas (27–31). However, the survival of cultivated biocrust inoculum in the field is notoriously unreliable (32, 33) and, in general, lower than that reported for inoculation strategies based on slurries of remnant biocrust communities (25, 34, 35). This suggests that nonphototrophic microbial components of biocrusts may also play an important role in survival of biocrust inoculum.
Recent studies have provided evidence that the pioneer biocrust cyanobacterium Microcoleus vaginatus is capable of spatially arranging neighboring soil bacteria into a cyanosphere microbiome (analogous to the rhizosphere), outsourcing necessary functional traits, such as nitrogen fixation, to other bacteria in exchange for photosynthates (36–39). Resource trading within this pioneer consortium facilitates the initial establishment of biocrust communities on nutrient-poor bare soils, and future restoration efforts should consider this relationship. In this regard, the growth-promoting effects of cyanosphere heterotrophs on M. vaginatus under nitrogen limitation have been demonstrated through coculturing experiments in a laboratory setting (37), but whether these benefits are transferable to soil substrates, harsh climatic conditions, or other pioneer cyanobacteria, such as those from the family Coleofasciculaceae, remains to be tested. Here, we tested the potential use of this pioneer consortium, containing pedigreed heterotrophic and phototrophic components, in the restoration of degraded biocrust communities and the production of biocrust inoculum by inoculating soil substrates under both optimized and representative environmental conditions and monitoring biocrust development. We hypothesized that coinoculation of pioneer cyanobacteria with heterotrophic cyanosphere partners will reduce the time necessary to form a resource trading relationship, thus accelerating the development of a cohesive biocrust.
RESULTS
Effect of beneficial heterotrophs on M. vaginatus growth on sterile soil substrate.
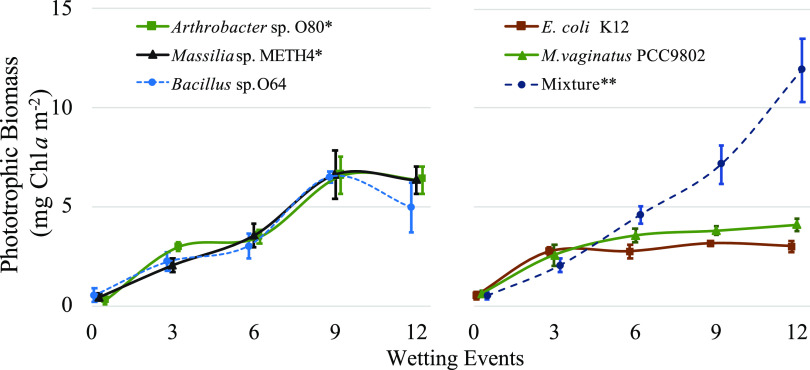
We cocultured M. vaginatus PCC9802, an axenic strain originally isolated from biocrust from the U.S. Southwest, on sterilized soil substrate with each beneficial heterotrophic isolate (Arthrobacter sp. strain O80, Massilia sp. strain METH4, and/or Bacillus sp. strain O64) to determine their effect on phototrophic growth under laboratory conditions (Fig. 1). After 12 growth/desiccation cycles, yields of cocultures with Arthrobacter sp. strain O80 and Massilia sp. strain METH4 were significantly higher (55% higher yield for both) than those of noncocultured controls (analysis of variance [ANOVA], P < 0.05 for both), while yields of cocultures with Bacillus sp. strain O64 and Escherichia coli K-12 were similar to those of noncocultured controls (ANOVA, P = 0.90 for O64 and 0.75 for K-12). Yields of individual cyanosphere isolates showed no difference in growth promotion (ANOVA, P > 0.21 for all). Yields of cocultures with a mixture of cyanosphere heterotrophs were significantly higher than those obtained by either inoculating with M. vaginatus alone (190% higher yield) (ANOVA, P < 0.001) or coculturing with individual isolates (∼100% higher yield) (ANOVA, P < 0.001 for all). Full data sets of chlorophyll a (Chl a) determinations can be found in Table S1 in the supplemental material. Since incubations were carried out in an open container, it was important to monitor for adventitious colonization (40). Microscopic examination of experimental and control plates showed no cyanobacterial contaminants across the duration of the experiment.
FIG 1.
Biomass dynamics (mean biomass ± standard deviation [SD], n = 3) of M. vaginatus PCC9802 cocultured with individual heterotrophic cyanosphere bacteria (left) or with an equal-proportion cyanosphere heterotroph mixture (right) on sterile hot-desert soil substrates under laboratory conditions for 12 wetting events. Controls for growth trials were M. vaginatus PCC9802 grown alone and in coculture with a noncyanosphere heterotrophic isolate, Escherichia coli K-12. Asterisks indicate treatments yielding significantly higher biomass than M. vaginatus PCC9802 grown alone (*, P < 0.05; **, P < 0.001).
Beneficial heterotrophs’ effect on hot-desert biocrust inoculum.
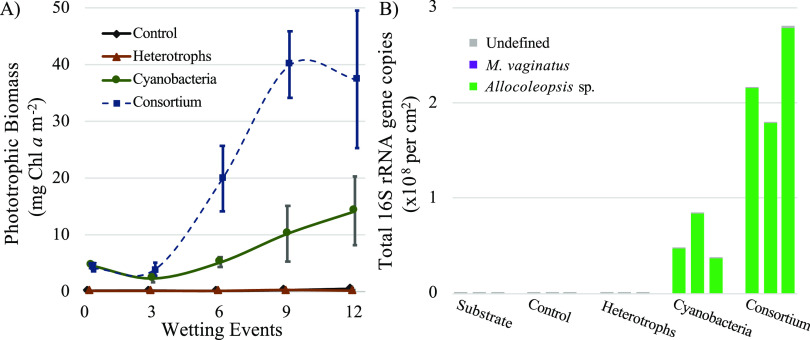
We tested the application of pioneer cyanobacterial isolates (M. vaginatus N8 and Allocoleopsis sp. strain N19) and beneficial cyanosphere heterotrophs, both separately (cyanobacteria and heterotrophs) and together as a phototroph/heterotroph consortium, on the development of biocrust on unsterilized native soils from a hot-desert location under field conditions. After 12 growth/desiccation cycles, both cyanobacteria and consortium treatments grew significantly from the initial inoculation levels (ANOVA, P < 0.05 for cyanobacteria and P < 0.001 for consortium) yielding averages of 8.0 and 35.8 mg Chl a m−2, respectively, with consortium yields significantly exceeding those of the cyanobacterial treatment (ANOVA, P < 0.001) (Fig. 2A). The heterotroph-alone treatment and the uninoculated control yielded averages of 0.07 and 0.09 mg Chl a m−2, respectively, and did not grow significantly (ANOVA, P = 0.26 for heterotrophs and 0.07 for the control) (Fig. 2A). Full data sets of Chl a determinations and visual aspects of treatments can be found in Table S2 and Fig. S2, respectively. The bacterial community composition already existing in the hot-desert soil substrate harbored Actinobacteria, Proteobacteria, Crenarchaeota, and Acidobacteria as major components, with minimal cyanobacterial abundance at the phylum level (<0.03% of total bacterial community) (Fig. S1A). Final bacterial community compositions showed an increase in cyanobacterial abundance where cyanobacterial biomass was added (cyanobacteria and consortium) (Fig. S1A). In particular, the inoculated cyanobacterial isolate Allocoleopsis sp. strain N19 accounted for up to 33% and 60% of the reads bacterial community reads in cyanobacterial and consortium treatments, respectively (Fig. S1A), and over 99% of cyanobacterial reads in both cyanobacteria and consortium treatments. (Fig. 2B). We detected the presence of two of the three inoculated heterotrophs (Bacillus sp. strain O64 and Arthrobacter sp. strain O80) (Table S3) in the appropriate addition treatments; however, M. vaginatus N8 was entirely absent from all treatments and controls at the final time point (Fig. 2B).
FIG 2.
(A) Biomass dynamics (mean biomass ± standard deviation [SD], n = 5) of unsterilized hot-desert soil substrates inoculated with beneficial heterotrophs, pioneer cyanobacteria, or both beneficial heterotrophs and cyanobacteria (consortium) under field conditions. (B) Cyanobacterial abundance and community structure of initial substrate compared to treatments and control incubated for 12 wetting events, as determined by high-throughput 16S rRNA gene analysis coupled to qPCR.
Beneficial heterotrophs effect on cold-desert biocrust inoculum.
We similarly tested the application of pioneer cyanobacterial isolates (M. vaginatus HS016 and Arizonema sp. strain HS024) and beneficial cyanosphere heterotrophs, both separately (cyanobacteria and heterotrophs) and together as a phototroph/heterotroph consortium, on the development of biocrust on unsterilized native soils from a cold-desert location under field conditions. After 12 growth/desiccation cycles, all treatments grew significantly from the initial inoculation levels (t test, P < 0.003 for all treatments), yielding 6.8, 6.9, and 8.1 mg Chl a m−2 for heterotroph, cyanobacterial, and consortium treatments, respectively. Only the consortium yielded significantly more phototrophic biomass than the uninoculated control (ANOVA, P < 0.05), though differences in final yields were not significant among inoculation treatments (ANOVA, P = 0.17) (Fig. 3A). The controls, however, also grew significantly (t test, P < 0.001) from the initial levels, yielding 3.9 mg Chl a m−2. Full data sets of Chl a determinations and visual aspects of treatments can be found in Table S2 and Fig. S3, respectively.
FIG 3.
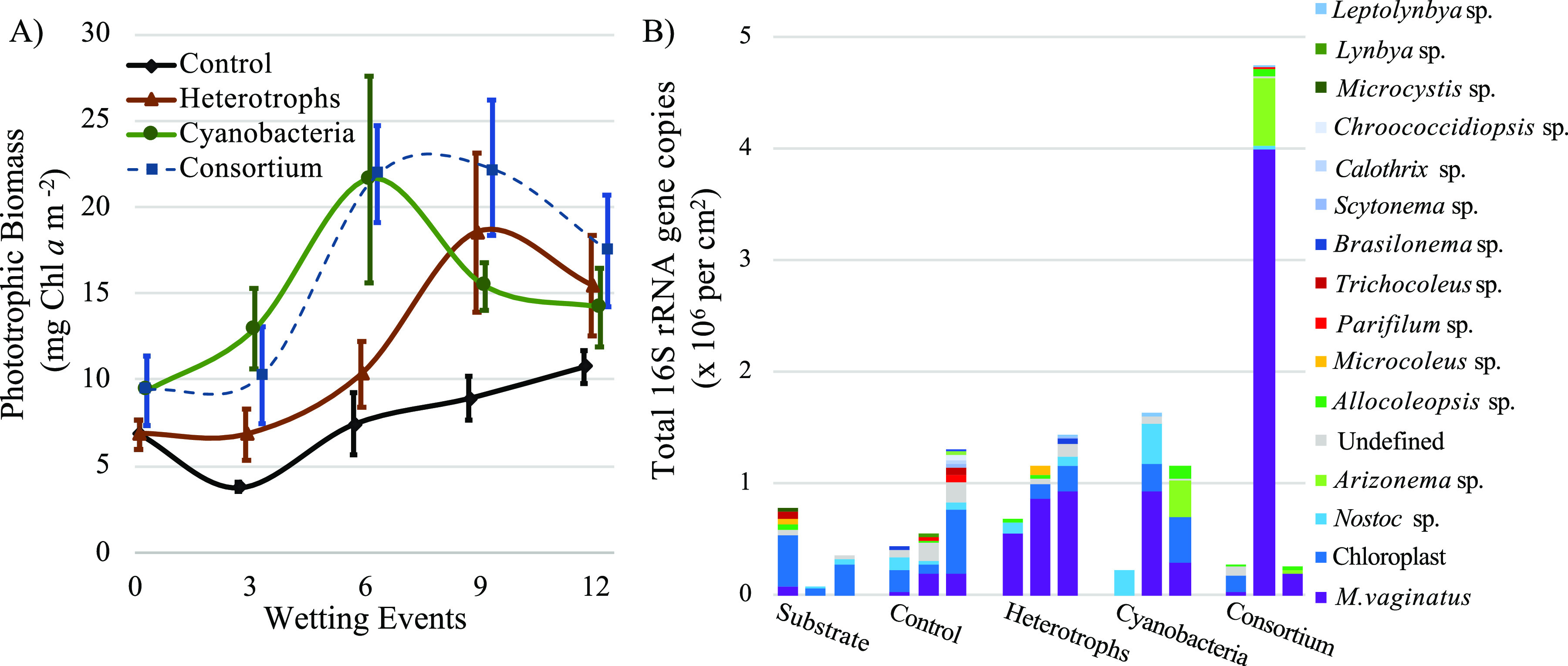
(A) Biomass dynamics (mean biomass ± standard deviation [SD], n = 5) of unsterilized cold-desert soil substrates inoculated with beneficial heterotrophs, pioneer cyanobacteria, or both beneficial heterotrophs and cyanobacteria (consortium) under field conditions. (B) Cyanobacterial abundance and community structure of initial substrate compared to treatments and controls incubated for 12 wetting events, as determined by high-throughput 16S rRNA gene analysis coupled to qPCR.
The composition of the initial bacterial community of the cold-desert soil used as the substrate showed the presence of Proteobacteria, Actinobacteria, and Cyanobacteria as major components at the phylum level (Fig. S1B). The cyanobacterial community of the cold-desert soil substrate was dominated by chloroplast sequences (>42% of cyanobacterial community), indicating the presence of eukaryotic algae, followed by trace amounts of typical cyanobacterial biocrust organisms such as Microcoleus vaginatus, Allocoleopsis, Parifilum, Nostoc, and Trichocoleus (Fig. 3B). The composition of the bacterial community at the final time point showed a modest increase in cyanobacterial abundance across all addition treatments (Fig. S1B). Within the cyanobacterial community, there were shifts toward an incipient biocrust community. In particular, increases of pioneer cyanobacteria such as M. vaginatus and genera of the Coleofasciculaceae, including Allocoleopsis and Arizonema, were observed compared to the uninoculated control, and in the case of the heterotroph treatment, this shift in abundance was significant (t test, P < 0.02) (Fig. 3B). Additionally, two of the three inoculated heterotrophic bacteria (Arthrobacter sp. strain O80 and Massilia sp. strain METH4) were detected in the heterotroph treatment, while all three inoculated heterotrophic bacteria were detected within the consortium treatment (Table S3). Interestingly, sequences undistinguishable from that of Arthrobacter sp. strain O80 were also detected in the initial soil substrate and were present in the final bacterial community of all treatments and controls.
Evaluation of effect of beneficial heterotrophs on soils with various levels of degradation.
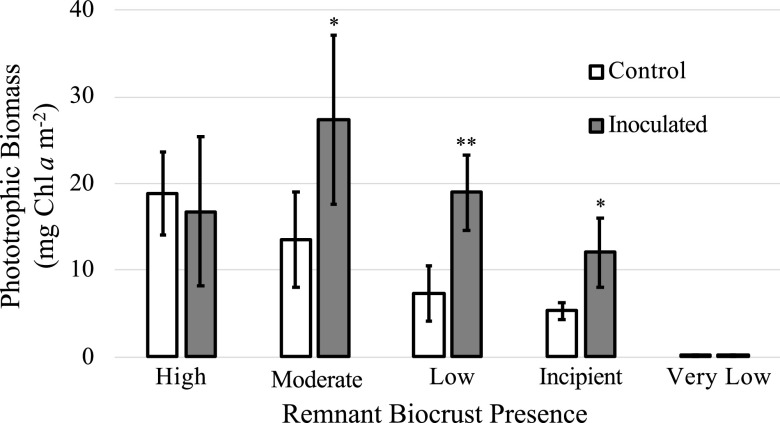
Beneficial heterotrophs were applied to unsterilized native soil substrates simulating a gradient of remnant biocrust presence to investigate whether heterotroph additions alone could accelerate biocrust regeneration. After 12 growth/desiccation cycles, we observed no significant differences in phototrophic biomass yield between the inoculation treatment and uninoculated controls in soils that had high (10 mg Chl a m−2) and very low (<0.25 mg Chl a m−2) levels of remnant biocrust (t test, P = 0.68 for high levels and P = 0.79 for very low levels) (Fig. 4). At moderate, low, and incipient remnant community levels, where initial substrates contained 5, 2.5, and 1.25 mg Chl a m−2, respectively, inoculation treatments yielded significantly more phototrophic biomass than the uninoculated controls (t test, P < 0.05 for moderate levels, P < 0.003 for low levels, and P < 0.04 for incipient levels) (Fig. 4).
FIG 4.
Phototrophic biomass yield of remnant hot-desert biocrust communities incubated with or without (control) beneficial heterotrophs after 12 wetting events (mean biomass ± SD, n = 4) as a function of initial phototrophic biomass. Asterisks indicate significance differences in yield (*, P < 0.05; **, P < 0.005).
DISCUSSION
Cyanosphere heterotrophs accelerate establishment of biocrust inoculum.
In hot-desert trials, coinoculation of heterotrophic isolates with locally sourced cyanobacteria resulted in increases of phototrophic biomass wherever sufficient initial amounts (>0.25 mg Chl a m−2) were present, with phototrophic yields of the consortium being 4-fold higher than that of cyanobacteria alone (Fig. 2A and Fig. S2). Cyanosphere heterotrophs effectively doubled the speed of biocrust development, as the consortium treatment required half the wetting events to reach biomass levels of the cyanobacterium-only treatment at the final time point (Fig. 2A). However, no significant phototrophic growth was detected with the addition of cyanosphere heterotrophs without cyanobacteria, likely because insufficient cyanobacterial components were natively present in the soil substrate. Interestingly, while these hot-desert soils were inoculated with both M. vaginatus N8 and Allocoleopsis sp. strain N19 isolated from these very soils, only Allocoleopsis sp. strain N19 was present at the final time point (Fig. 2B). This finding is consistent with the temperature-driven niche differentiation known for these cyanobacteria, as members of the Coleofasciculaceae (formerly M. steenstrupii complex) tend to be more thermotolerant than M. vaginatus (41), which does not tolerate mean temperatures above 15°C. The clear growth-promoting effects of heterotroph additions on Allocoleopsis sp. strain N19 also suggest that filamentous biocrust cyanobacteria other than M. vaginatus can benefit from interactions with heterotrophs from the M. vaginatus cyanosphere, a concept that requires further testing.
The results were less clear in the cold-desert trials. Treatments had very small beneficial effects over the controls after 12 wetting events (Fig. 3A and Fig. S3), and none surpassed biomass levels of approximately 20 mg Chl a mg−1, suggesting that a nutrient limitation to growth in all treatments was at play. However, the nitrogen content of this cold-desert substrate (27) was more than 10-fold higher than those previously reported for the hot-desert substrate (see above). As such, nitrogen limitation is an unlikely culprit. While phototrophic biomass grew only modestly, benefits due to characteristics of the cyanobacterial community composition did occur (Fig. 3B). Pioneer cyanobacteria, such as M. vaginatus and members of the Coleofasciculaceae such as Allocoleopsis and Arizonema, which were absent or in very low abundance in the original substrate, increased in abundance to dominate the cyanobacterial community across all treatments. This might have been expected where M. vaginatus and Arizonema sp. were interventionally added, but interestingly, it also occurred in the heterotroph addition treatment where no additional phototrophic biomass was added. Here, these crust-forming cyanobacteria increased to represent >70% of the cyanobacterial community, a significant increase compared to the uninoculated controls. This suggests that even in the absence of significant biocrust yields, beneficial heterotrophs could still shift the community in a way that poises it for the eventual formation of an incipient biocrust.
Cyanosphere heterotrophs perform better as a consortium.
In all cases, the presence of cyanosphere heterotrophs, either individually or as a mixture, led to improved performance of M. vaginatus on soil substrates over M. vaginatus grown axenically or cocultured with a noncyanosphere heterotroph. The beneficial effects were similar in all three isolates, but when they were inoculated as a mixture, the beneficial effects were even greater (Fig. 1). This consortium synergy could be due to different strains occupying particular environmental niches associated with diurnal fluctuations in temperature and oxygen concentration (42). Indeed, some of the isolates used here seemed better suited to the particular temperature ranges and soil types used in outdoor experimental trials (Table S3). This suggests that the effective range of environmental conditions for the mutualistic relationship may be increased by adding a diverse consortium of beneficial heterotrophs.
Heterotroph additions as an alternative for restoring moderately degraded soils.
The remnant biocrust communities within soils of degraded sites constitute a significant source of native phototrophic biomass and were previously reported to significantly influence overall biocrust community composition (43). Our findings indicate that when sufficient phototrophic biomass is present (>0.25 mg Chl a m−2), merely adding beneficial heterotrophs can accelerate the consolidation of fragmented communities into a whole biocrust (Fig. 4). These beneficial bacteria tend to be copiotrophs that are otherwise rare in dryland soils outside their associations with carbon-rich environments such as plant rhizospheres (44, 45) and M. vaginatus cyanospheres (36). Our results indicate that by increasing the presence of these beneficial heterotrophs within disturbed remnant communities through additions, existing cyanobacteria might require less time to recruit and sustain copiotrophic populations necessary for the formation of a resource trading cyanosphere. Under certain circumstances, heterotroph additions could be considered an effective alternative to the production and inoculation of phototrophic components, which are time intensive to grow (27, 46), difficult to scale up (28, 43), and vulnerable to biocrust pathogens (46). Unlike filamentous cyanobacteria, many heterotrophic soil bacteria are simple to isolate, grow rapidly in liquid culture, and are easily scalable, properties that are invaluable to large-scale restoration efforts.
New recommendations for biocrust restoration strategies.
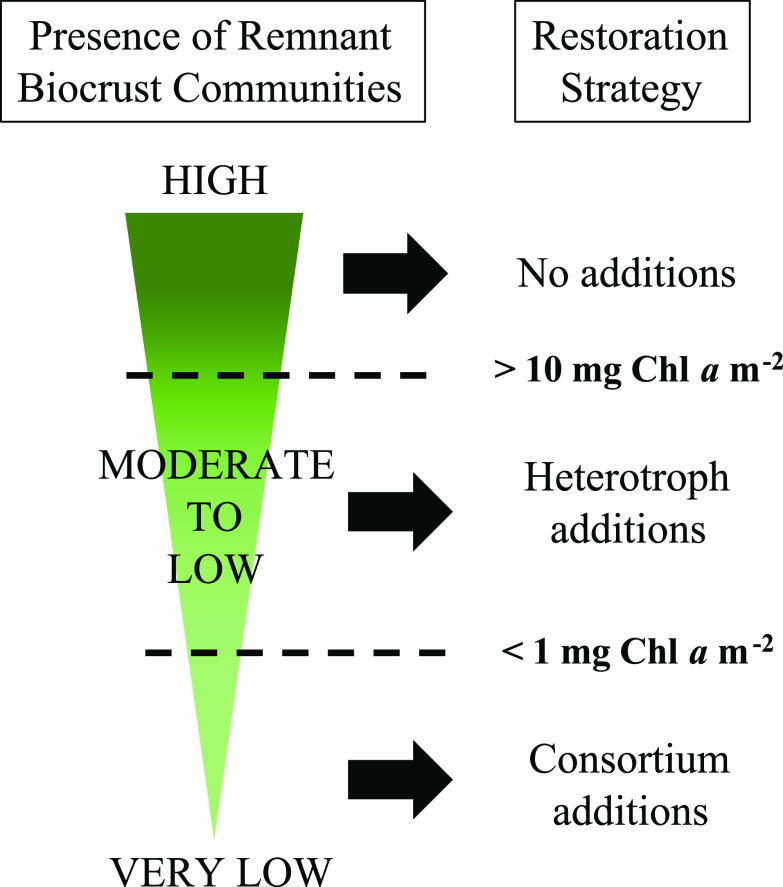
Here, we recommend that future biocrust restoration efforts use a tiered system for determining the best restoration strategy based on the presence of existing phototrophic biomass (Fig. 5). Minimally disturbed areas with high levels of phototrophic biomass need no additional intervention, requiring only precipitation, time, and protection from further disturbance. Disturbed areas with moderate to low levels of phototrophic biomass may need only application of beneficial heterotrophic bacteria to facilitate the rapid consolidation of fragmented biocrust communities into full-fledged biocrusts. Severely disturbed or uncolonized areas with very low levels of phototrophic biomass will still require significant intervention, including but not limited to increasing the abundances of appropriate cyanobacterial pioneers as well as the addition of beneficial heterotrophic bacteria. Therefore, we recommend that a simple, initial assessment of both the initial presence and composition of the phototrophic community in soils of degraded areas be conducted in order to determine the level of intervention required on a case-by-case basis.
FIG 5.
Recommendation for restoration of cyanobacterial biocrusts based on presence of phototrophic biomass at degraded sites.
Conclusion.
We drew on knowledge of fundamental microbial interactions within biocrusts to demonstrate the underexplored potential of heterotrophic bacteria in the establishment of cyanobacterial biocrust inoculum and evaluate their role in enhancing the consolidation of disturbed biocrust communities, leading to new recommendations for biocrust restoration strategies for drylands. This study provides evidence that shifts the paradigm of restoration approaches from an exclusive focus on phototrophic components to one that includes selected heterotrophic components, often considered secondary contributors to crust functionality.
MATERIALS AND METHODS
Biocrust and soil substrate sourcing.
Hot-desert biocrusts and soil substrates were collected in May 2019 from the Chihuahuan Desert (New Mexico, Jornada Basin Long-Term Ecological Research Network [LTER] site; 32.56378N, 106.75522W). Hot-desert biocrust communities were sampled using petri plates (15-cm diameter, 1-cm depth) that were utilized to cut intact biocrust from the field (47). Cold-desert soil substrates were collected in May 2014 from the Great Basin Desert (Hill Air Force Base, Utah Test and Training Range; 41.10419N, 113.00820W). Additional cold-desert location details are described in a previous work (28). For both locations, soil substrate was taken at a depth of 5 to 20 cm. Samples were air dried and maintained inactive at a low relative humidity (RH; 15%) in darkness until experimentation in spring/fall 2020.
Cyanobacterial isolation and growth conditions for cyanobacterial and heterotrophic strains.
To isolate site-specific pioneer cyanobacterial strains, hot-desert biocrusts were wetted with reverse osmosis (RO) water to allow cyanobacterial bundles to migrate toward the soil surface. These bundles were pulled from the biocrust and incubated on 50% BG11o plus 1% gellan gum (48) for 14 days. Outgrowth of cyanobacterial biomass from bundles was transferred to 24-well plates with 1 ml BG11 in culture room conditions (23°C, ∼20 μE m−2 s−1, 14-h light cycle) and slowly scaled up to 20-ml, 150-ml, and 750-ml cell culture flasks over several weeks as growth permitted. After determination of initial purity and taxonomy, cyanobacterial strains were assigned based on microscopy. Genomic DNA was extracted using a PowerSoil Pro kit (Qiagen, Hilden, Germany) and submitted for Sanger sequencing to the ASU Genomics core facilities, using the cyanobacterium-specific primers CYA106F/CYA805R (49) as previously described (28). Forward and reverse sequences were aligned in Geneious version 8.0 (50), and the 600- to 700-bp consensus sequences were phylogenetically assigned taxonomy using our own curated cyanobacterial database/tree version 2 (https://github.com/FGPLab/cydrasil/releases/tag/v2.0) via RAxML (51) and displayed using ITOL (52). Cyanobacterial strains chosen for experimentation were those with 16S rRNA sequences most closely related to the most abundant cyanobacterial sequences from field communities as previously described (28). Details of the isolation and selection of cyanobacterial strains for the cold-desert location are described in a previous work (28). All cyanobacterial strains were maintained in BG11 medium under culture room conditions (23°C, ∼20 μE m−2 s−1, 14-h light cycle). Isolation and selection of heterotrophic strains used in experimentation (Bacillus sp. strain O64, Massilia sp. strain METH4, and Arthrobacter sp. strain O80) are described in a previous work (37). Heterotrophic isolates were maintained on Burk’s medium plus 1% gellan gum (53) under culture room conditions (23°C).
Assessment of beneficial effects of cyanosphere heterotrophs on axenic Microcoleus vaginatus in sterile conditions.
To quantitatively assess the growth-promoting effects of cyanosphere heterotrophs on the growth of M. vaginatus, we inoculated heterotroph strains, both separately and as a mixture on sterilized soil substrate, with an axenic strain of M. vaginatus (PCC9802) originally isolated from biocrusts in the U.S. Southwest and available through the Pasteur Culture Collection of Cyanobacteria (Paris, France). A total of 115 g of dry sterilized hot-desert soil substrate (autoclaved 5 times for 20 min at 121°C) was distributed in 21 petri plates (10-cm diameter, 2.5 cm deep).
All strains were harvested and washed three times prior to inoculation by pelleting cells in a centrifuge at 8,000 rpm for 8 min, removing supernatant, and resuspending cells in sterile RO water in order to remove remnant nutrients from previous medium as previously described (37). The washed cells were then mixed to create coculture mixtures that were pipetted evenly onto the soil surface at 0.54 ± 0.18 mg Chl a m−2 for M. vaginatus PCC9802 and at ∼108 cells of heterotroph isolates per plate. Treatments (n = 3) consisted of cocultures of M. vaginatus PCC9802 with individual beneficial heterotrophic strains: Bacillus sp. strain O64, Arthrobacter sp. strain O80, Massilia sp. strain METH4, or an equal concentration mixture of heterotrophs (O64, O80, and METH4), as well as a nonbiocrust strain, E. coli K-12. The controls consisted of M. vaginatus PCC9802 grown alone and uninoculated soil substrate. All plates were incubated by simulating natural wet/dry cycles, with 12 consecutive wet/dry cycles as previously described (43), providing additions of 25 ml sterile RO water followed by 72 h of desiccation by ambient evaporation. All plates were incubated at 25°C, under 100 to 120 μE m−2 s−1 of white light and with a 14-h illumination/10-h dark cycle. For biomass measurements, a cork borer was used to randomly obtain three soil cores (0.9-cm diameter, 0.5 cm deep) from each replicate plate 24 h after initial wetting and at the 3rd, 6th, 9th, and 12th wetting events. Sampled cores were air dried and stored at 4°C in the dark until extraction.
Assessment of beneficial effects of cyanosphere heterotrophs on biocrust pioneer species from two desert locations.
To determine if the growth-promoting effects of beneficial heterotrophs were retained under field-like conditions and whether this can be applied to climatic and edaphically different locales, we tested the effect of heterotroph additions on the growth of locally sourced cyanobacterial pioneer species from hot- and cold-desert locations using unsterilized native soil substrates under field-like conditions. A total of 225 g of unsterilized dry native substrate from each location was distributed into 20 petri plates (15-cm diameter, 1 cm deep). Cyanobacterial strains from hot desert (M. vaginatus N8 and Allocoleopsis sp. strain N19) and cold desert (M. vaginatus HS016 and Arizonema sp. strain HS024) as well as heterotrophic strains used for inoculation were harvested and washed three times prior to inoculation by pelleting cells in a centrifuge at 8,000 rpm for 8 min, removing supernatant, and resuspending cells in sterile RO water, to remove remnant nutrients from previous medium as previously described (37). Due to the clumping tendency of filamentous cyanobacterial strains, each strain used was homogenized by forcing the culture through a sterile 60-ml syringe until observed clumps were broken apart (28, 54).
For each location, three treatment mixtures were created and pipetted onto their respective soil surfaces: a cyanobacterial mixture (cyanobacteria), a heterotrophic mixture (heterotrophs), and a cyanobacterium-heterotroph mixture (consortium). A total of 5 ml of this mixture was pipetted evenly onto the respective soil substrates for the following treatments (n = 5): cyanobacteria only, heterotrophs only, and a cyanobacterium-heterotroph mixture (consortium). Cyanobacterial mixtures containing equal proportions of cyanobacterial pioneer strains were inoculated onto soil substrates at 4.22 ± 0.64 and 2.25 ± 1.06 mg Chl a m−2 for hot- and cold-desert locations, respectively. Heterotroph bacteria used for heterotroph and consortium treatments were inoculated at ∼109 cells of each heterotroph isolate per plate. The controls for each location consisted of uninoculated soil substrates receiving 5 ml of sterile RO water.
Outdoor growth trials for both hot- and cold-desert locations were performed in an open area in Phoenix, AZ, in May-June 2020 and November-December 2020, respectively. All plates were incubated simulating natural wet/dry cycles, with 12 consecutive wet/dry cycles, providing additions of 50 ml RO water followed by 72 h of desiccation by ambient evaporation. Growth conditions for the outdoor trials were a maximal light intensity of ∼400 to 600 μE m−2 s−1 and temperatures ranging from 21 to 39°C and 6 to 26°C in May-June 2020 and November-December 2020. Extreme summer temperatures were avoided, as previously suggested (40, 46). During midday hours of clear days, the experiment was shaded with a thin white cotton sheet to moderate soil temperature by diffusing light intensity to overcast levels. Chl a determinations were performed by using a cork borer to randomly collect three (1.7-cm diameter, 0.5 cm deep) cores from each replicate plate 24 h after initial wetting, and at the 3rd, 6th, 9th, and 12th wetting events. Sampled cores were air dried and stored at 4°C in darkness until extraction.
Assessment of beneficial effects of cyanosphere heterotrophs on various levels of phototrophic biomass in native soil substrates.
Heterotroph additions were made to soil substrates with various levels of diluted biocrust communities in order to determine if the effect of additions of cyanosphere heterotrophs alone was dependent on the existing cyanobacterial biomass. Whole biocrust communities from a hot desert were lightly homogenized with a mortar and pestle and thoroughly mixed with unsterilized soil substrate to decrease the phototrophic biomass in the mixture to 10 mg Chl a m−2. This mixture was then further diluted with unsterilized soil substrate to obtain soil mixtures with 5, 2.5, and 1.25 mg Chl a m−2. The controls consisted of both sterilized and nonsterilized soil substrate representing bare soil substrates. A total of 225 g of each mixed soil was aliquoted into eight petri plates (15-cm diameter, 1 cm deep), half with 5 ml of the heterotrophic mixture (n = 4) described above and half with 5 ml of sterile RO water (n = 4). Outdoor trials took place in an open area in Phoenix, AZ, and growth conditions were a maximal light intensity of ∼400 to 600 μE m−2 s−1 and temperatures ranging from 6 to 26°C in November-December 2020. All plates were incubated simulating natural wet/dry cycles, with 12 consecutive wet/dry cycles, providing additions of 50 ml RO water followed by 72 h of desiccation by ambient evaporation. On clear days, a thin white cotton sheet was used to moderate soil temperature by diffusing light intensity to overcast levels. Chl a sampling was performed by using a cork borer to randomly collect three (1.7-cm diameter, 0.5 cm deep) cores from each replicate plate initially and 24 h after final wetting. Cores from each plate were pooled, air dried, and stored at 4°C in darkness until extraction.
Microbial community composition.
Pooled cores from each plate were lightly homogenized with a mortar and pestle, and a weighted aliquot was used for DNA extraction. Soil DNA was extracted with a PowerSoil Pro extraction kit (Qiagen) using the standard protocol. Before sequencing, DNA extracts from the five replicate plates were pooled into three samples. Bacterial/archaeal community analysis was performed via commercial next-generation sequencing in a MiSeq Illumina platform. Amplicon sequencing of the V4 region of the 16S rRNA gene was performed with the barcoded primer set 515F/806R (55) following the Earth Microbiome Project (EMP) protocol (56) for library preparation. PCR amplifications were done in triplicate, then pooled, and quantified using a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen). Two hundred forty nanograms of DNA from each replicate was pooled and cleaned using a QIAquick PCR purification kit (Qiagen). The DNA in the pooled amplification product was quantified using an Illumina library quantification kit ABI Prism (Kapa Biosystems) and diluted with NaOH to a final concentration of 4 nM, then denatured, and diluted to a final concentration of 4 pM, and 30% of PhiX was added to the solution. The library was then loaded in the sequencer using the chemistry version 2 (2 × 250 paired-end reads) and following the manufacturer’s specifications (56). Sequencing was performed in the Microbiome Analysis Laboratory at Arizona State University (Tempe, AZ, USA), yielding raw FASTQ sequence files.
Bioinformatic analysis.
The raw FASTQ file was demultiplexed within the MiSeq Illumina workflow under default parameters. Paired sequences were demultiplexed and analyzed via QIIME 2.10 (57), using the DADA2 plugin (58) where sequences were trimmed to include 250 bases from the V4 region, bound by 515F/806R primers (59), to create a feature table with representative sequences (features) and their frequency of occurrence. To remove highly variable positions, sequences were aligned with the MAFFT program (60). FastTree (61) was used to generate a tree. Taxonomy was assigned with the naive Bayes classifier trained on the Greengenes 13.8 release. While all phyla were analyzed, additional steps were taken to identify cyanobacteria due to the poor taxonomic resolution obtained with Greengenes (62). Cyanobacterial sequences were filtered out from the feature table, and cyanobacterial sequences that attained at least 0.05% of the total number of cyanobacterial features were then phylogenetically assigned to the level of greatest taxonomic resolution, generally genus level, using our own curated cyanobacterial database/tree version 2 via RAxML (51) and displayed using ITOL (52). For analyses on comparisons of Chl a content and total bacterial abundance, statistical tests were performed using R (version 3.4.3) (63). P values from statistical tests were corrected for multiple comparisons.
Chl a determinations.
Chl a was used as a proxy for photosynthetic biomass. Pooled samples were extracted per plate and time point by grinding the soil-cyanobacterium mixture in 90% acetone with a mortar and pestle for 3 min, as previously described (28, 40, 54). Extracts were then transferred to a 2-ml microcentrifuge tube, the volume was adjusted to 2 ml with 90% acetone, and the mixture was vortexed for 30 s and then stored in the dark for 24 h at 4°C. Absorbance spectra were recorded on a UV-visible spectrophotometer (Shimadzu UV-1601). Interference from scytonemin and carotenoids was corrected using a trichromatic equation (64), where appropriate. Chl a yield was calculated by subtracting initial values from values at the time point of interest.
16S rRNA gene copy number determinations.
Absolute abundance of bacteria, used as an additional proxy for biomass, was determined by qPCR (quantitative real-time PCR) using aliquoted DNA extracted from homogenized cores for each treatment. After fluorometric determination of DNA concentration in the extract (Qubit, Life Technologies, NY, USA), we used qPCR with a universal (bacterial/archaeal) 16S rRNA gene primer set (338F [5′-ACTCCTACGGGAGGCAGCAG-3′] and 518R [5′-GTATTACCG CGGCTGCTGG-3′]) to determine the number of 16S rRNA gene copies present in each extract. The PCR was performed in triplicate using the Sso Fast mix (Bio-Rad, Hercules, CA, USA) under conditions published previously (15). The final 16S rRNA gene copy number per unit volume of biocrust was determined from the qPCR data (copies/extract) and the total soil volume used for extraction. The number of 16S rRNA genes obtained by qPCR was later used to arrive at total population sizes for each phylum or cyanobacterial taxon, by multiplying the total number of genes by the relative abundance of the taxa, as determined by Illumina sequencing and bioinformatic analyses as previously described (43, 65).
Data availability.
Raw sequence data have been submitted to NCBI and are publicly available under BioProject number PRJNA740061 for experimental trials and under BioProject number PRJNA705202 for remnant hot-desert biocrust communities.
ACKNOWLEDGMENTS
This work was supported in part by the Jornada Basin Graduate Research Fellowship Program (NSF grant DEB 2025166), the Center for Bio-mediated and Bio-inspired Geotechnics (NSF grant ING 1449501), and NSF grant DEB 2129537 to F.G.-P.
C.N. and F.G.-P. conceived the research; C.N. designed and performed experiments and analyzed data; C.N. and F.G.-P. discussed results; C.N. and F.G.-P. wrote and edited the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Ferran Garcia-Pichel, Email: ferran@asu.edu.
Alfons J. M. Stams, Wageningen University
REFERENCES
- 1.Belnap J, Weber B, Büdel B. 2016. Biological soil crusts as an organizing principle in drylands, p 3–13. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 2.Garcia-Pichel F, López-Cortés A, Nübel U. 2001. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol 67:1902–1910. 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu C, Zhang D, Huang Z, Liu Y. 2003. The vertical microdistribution of cyanobacteria and green algae within desert crusts and the development of the algal crusts. Plant Soil 257:97–111. 10.1023/A:1026253307432. [DOI] [Google Scholar]
- 4.Bates ST, Nash TH, Sweat KG, Garcia-Pichel F. 2010. Fungal communities of lichen-dominated biological soil crusts: diversity, relative microbial biomass, and their relationship to disturbance and crust cover. J Arid Environ 74:1192–1199. 10.1016/j.jaridenv.2010.05.033. [DOI] [Google Scholar]
- 5.Lange OL, Belnap J, Reichenberger H, Meyer A. 1997. Photosynthesis of green algal soil crust lichens from arid lands in southern Utah, USA: role of water content on light and temperature responses of CO2 exchange. Flora 192:1–15. 10.1016/S0367-2530(17)30749-1. [DOI] [Google Scholar]
- 6.Nunes da Rocha U, Cadillo-Quiroz H, Karaoz U, Rajeev L, Klitgord N, Dunn S, Truong V, Buenrostro M, Bowen BP, Garcia-Pichel F, Mukhopadhyay A, Northen TR, Brodie EL. 2015. Isolation of a significant fraction of non-phototroph diversity from a desert biological soil crust. Front Microbiol 6:277. 10.3389/fmicb.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soule T, Anderson IJ, Johnson SL, Bates ST, Garcia-Pichel F. 2009. Archaeal populations in biological soil crusts from arid lands in North America. Soil Biol Biochem 41:2069–2074. 10.1016/j.soilbio.2009.07.023. [DOI] [Google Scholar]
- 8.Bates ST, Garcia-Pichel F, NashTH, III.. 2010. Fungal components of biological soil crusts: insights from culture-dependent and culture-independent studies. Bibliotheca Lichenol 105:197–210. [Google Scholar]
- 9.Rodriguez-Caballero E, Belnap J, Büdel B, Crutzen PJ, Andreae MO, Pöschl U, Weber B. 2018. Dryland photoautotrophic soil surface communities endangered by global change. Nat Geosci 11:185–189. 10.1038/s41561-018-0072-1. [DOI] [Google Scholar]
- 10.Elbert W, Weber B, Burrows S, Steinkamp J, Büdel B, Andreae MO, Pöschl U. 2012. Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nat Geosci 5:459–462. 10.1038/ngeo1486. [DOI] [Google Scholar]
- 11.Sancho LG, Belnap J, Colesie C, Raggio J, Weber B. 2016. Carbon budgets of biological soil crusts at micro-, meso-, and global scales, p 287–304. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 12.Belnap J, Gillette D. 1997. Disturbance of biological soil crusts: impacts on potential wind erodibility of sandy desert soils in southeastern Utah. Land Degrad Dev 8:355–362. . [DOI] [Google Scholar]
- 13.Garcia-Pichel F, Wojciechowski MF. 2009. The evolution of a capacity to build supra-cellular ropes enabled filamentous cyanobacteria to colonize highly erodible substrates. PLoS One 4:e7801. 10.1371/journal.pone.0007801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes MC, Giraldo‐Silva A, Roush D, Garcia‐Pichel F. 2021. Coleofasciculaceae, a monophyletic home for the Microcoleus steenstrupii complex and other desiccation‐tolerant filamentous cyanobacteria. J Phycol 10.1111/jpy.13199. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Couradeau E, Karaoz U, Lim HC, Nunes Da Rocha U, Northen T, Brodie E, Garcia-Pichel F. 2016. Bacteria increase arid-land soil surface temperature through the production of sunscreens. Nat Commun 7:10373–10377. 10.1038/ncomms10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeager C, Kornosky J, Housman DC, Grote EE, Belnap J, Kuske CR. 2004. Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado Plateau and Chihuahuan Desert. Appl Environ Microbiol 70:973–983. 10.1128/AEM.70.2.973-983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeager CM, Kornosky JL, Morgan RE, Cain EC, Garcia-Pichel F, Housman DC, Belnap J, Kuske CR. 2007. Three distinct clades of cultured heterocystous cyanobacteria constitute the dominant N2-fixing members of biological soil crusts of the Colorado Plateau, USA. FEMS Microbiol Ecol 60:85–97. 10.1111/j.1574-6941.2006.00265.x. [DOI] [PubMed] [Google Scholar]
- 18.Barger NN, Weber B, Garcia-Pichel F, Zaady E, Belnap J. 2016. Patterns and controls on nitrogen cycling of biological soil crusts, p 257–285. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 19.Garcia-Pichel F, Pringault O. 2001. Cyanobacteria track water in desert soils. Nature 413:380–381. 10.1038/35096640. [DOI] [PubMed] [Google Scholar]
- 20.Pringault O, Garcia-Pichel F. 2004. Hydrotaxis of cyanobacteria in desert crusts. Microb Ecol 47:366–373. 10.1007/s00248-002-0107-3. [DOI] [PubMed] [Google Scholar]
- 21.Belnap J, Eldridge D. 2001. Disturbance and recovery of biological soil crusts, p 363–383. In Belnap J, Lange O (ed), Biological soil crusts: structure, function and management. Springer, Berlin, Germany. [Google Scholar]
- 22.Zaady E, Eldridge DJ, Bowker MA. 2016. Effect of local-scale disturbance on biocrusts, p 429–450. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 23.Rajeev L, Nunes U, Klitgord N, Luning EG, Fortney J, Axen SD, Shih PM, Bouskill NJ, Bowen BP, Kerfeld CA, Garcia-Pichel F, Brodie EL, Northen TR, Mukhopadhyay A. 2013. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J 7:2178–2191. 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Pichel F, Belnap J. 1996. Microenvironments and microscale productivity of cyanobacterial desert crusts. J Phycol 32:774–782. 10.1111/j.0022-3646.1996.00774.x. [DOI] [Google Scholar]
- 25.Belnap J. 1993. Recovery rates of cryptobiotic crusts: inoculant use and assessment methods. Gt Basin Nat 53:89–95. [Google Scholar]
- 26.Weber B, Bowker M, Zhang Y, Belnap J. 2016. Natural recovery of biological soil crusts after disturbance, p 479–498. In Weber B, Büdel B, Belnap J (ed), Biological soil crusts: an organizing principle in drylands. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 27.Velasco Ayuso S, Giraldo-Silva A, Nelson C, Barger NN, Garcia-Pichel F. 2017. Microbial nursery production of high-quality biological soil crust biomass for restoration of degraded dryland soils. Appl Environ Microbiol 83:e02179-16. 10.1128/AEM.02179-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraldo-Silva A, Nelson C, Barger N, Garcia-Pichel F. 2019. Nursing biocrusts: isolation, cultivation and fitness test of indigenous cyanobacteria. Restor Ecol 27:793–803. 10.1111/rec.12920. [DOI] [Google Scholar]
- 29.Antoninka A, Bowker MA, Reed SC, Doherty K. 2016. Production of greenhouse-grown biocrust mosses and associated cyanobacteria to rehabilitate dryland soil function. Restor Ecol 24:324–335. 10.1111/rec.12311. [DOI] [Google Scholar]
- 30.Bowker MA, Antoninka AJ. 2016. Rapid ex situ culture of N-fixing soil lichens and biocrusts is enhanced by complementarity. Plant Soil 408:415–428. 10.1007/s11104-016-2929-7. [DOI] [Google Scholar]
- 31.Antoninka A, Bowker MA, Chuckran P, Barger NN, Reed S, Belnap J. 2018. Maximizing establishment and survivorship of field-collected and greenhouse-cultivated biocrusts in a semi-cold desert. Plant Soil 429:213–225. 10.1007/s11104-017-3300-3. [DOI] [Google Scholar]
- 32.Chandler DG, Day N, Madsen MD, Belnap J. 2019. Amendments fail to hasten biocrust recovery or soil stability at a disturbed dryland sandy site. Restor Ecol 27:289–297. 10.1111/rec.12870. [DOI] [Google Scholar]
- 33.Faist AM, Antoninka AJ, Belnap J, Bowker MA, Duniway MC, Garcia-Pichel F, Nelson C, Reed SC, Giraldo-Silva A, Velasco-Ayuso S, Barger NN. 2020. Inoculation and habitat amelioration efforts in biological soil crust recovery vary by desert and soil texture. Restor Ecol 28:S96–S105. 10.1111/rec.13087. [DOI] [Google Scholar]
- 34.Maestre FT, Martín N, Díez B, López-Poma R, Santos F, Luque I, Cortina J. 2006. Watering, fertilization, and slurry inoculation promote recovery of biological crust function in degraded soils. Microb Ecol 52:365–377. 10.1007/s00248-006-9017-0. [DOI] [PubMed] [Google Scholar]
- 35.St Clair LL, Johansen JR, Webb BL. 1986. Rapid stabilization of fire-disturbed sites using a soil crust slurry: inoculation studies. Reclam Reveg Res 4:261–269. [Google Scholar]
- 36.Couradeau E, Giraldo-Silva A, De Martini F, Garcia-Pichel F. 2019. Spatial segregation of the biological soil crust microbiome around its foundational cyanobacterium, Microcoleus vaginatus, and the formation of a nitrogen-fixing cyanosphere. Microbiome 7:55–122. 10.1186/s40168-019-0661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson C, Giraldo-Silva A, Garcia-Pichel F. 2020. A symbiotic nutrient exchange within the cyanosphere microbiome of the biocrust cyanobacterium, Microcoleus vaginatus. ISME J 15:282–292. 10.1038/s41396-020-00781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baran R, Brodie EL, Mayberry-Lewis J, Hummel E, Nunes U, Rocha D, Chakraborty R, Bowen BP, Karaoz U, Cadillo-Quiroz H, Garcia-Pichel F, Northen TR. 2015. Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun 6:8289–8289. 10.1038/ncomms9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baran R, Lau R, Bowen BP, Diamond S, Jose N, Garcia-Pichel F, Northen TR. 2017. Extensive turnover of compatible solutes in cyanobacteria revealed by deuterium oxide (D2O) stable isotope probing. ACS Chem Biol 12:674–681. 10.1021/acschembio.6b00890. [DOI] [PubMed] [Google Scholar]
- 40.Sorochkina K, Velasco Ayuso S, Garcia-Pichel F. 2018. Establishing rates of lateral expansion of cyanobacterial biological soil crusts for optimal restoration. Plant Soil 429:199–211. 10.1007/s11104-018-3695-5. [DOI] [Google Scholar]
- 41.Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM. 2013. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science 340:1574–1577. 10.1126/science.1236404. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Pichel F, Belnap J. 2001. Small-scale environments and distribution of biological soil crusts, p 193–201. In Biological soil crusts: structure, function, and management. Springer, Berlin, Germany. [Google Scholar]
- 43.Nelson C, Giraldo-Silva A, Garcia-Pichel F. 2020. A fog-irrigated soil substrate system unifies and optimizes cyanobacterial biocrust inoculum production. Appl Environ Microbiol 86:e00624-20. 10.1128/AEM.00624-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dennis PG, Miller AJ, Hirsch PR. 2010. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327. 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- 45.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 46.Bethany J, Giraldo-Silva A, Nelson C, Barger NN, Garcia-Pichel F. 2019. Optimizing the production of nursery-based biological soil crusts for restoration of arid land soils. Appl Environ Microbiol 85:e00735-19. 10.1128/AEM.00735-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Büdel B, Darienko T, Deutschewitz K, Dojani S, Friedl T, Mohr KI, Salisch M, Reisser W, Weber B. 2009. Southern African biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Microb Ecol 57:229–247. 10.1007/s00248-008-9449-9. [DOI] [PubMed] [Google Scholar]
- 48.Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35:171–205. 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nübel U, Garcia-Pichel F, Muyzer G. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol 63:3327–3332. 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I, Bork P. 2016. Interactive Tree Of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson PW, Knight SG. 1952. Experiments in bacterial physiology, 3rd ed, p 53. Burgess, Minneapolis, MN. [Google Scholar]
- 54.Giraldo‐Silva A, Nelson C, Penfold C, Barger NN, Garcia‐Pichel F. 2020. Effect of preconditioning to the soil environment on the performance of 20 cyanobacterial strains used as inoculum for biocrust restoration. Restor Ecol 28:5187–5193. 10.1111/rec.13048. [DOI] [Google Scholar]
- 55.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert JA, Meyer F, Jansson J, Gordon J, Pace N, Tiedje J, Ley R, Fierer N, Field D, Kyrpides N, Glöckner F-O, Klenk H-P, Wommack KE, Glass E, Docherty K, Gallery R, Stevens R, Knight R. 2010. The Earth Microbiome Project: meeting report of the “1 EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6 2010. Stand Genomic Sci 3:249–253. 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl 1):4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Development Core Team. 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org [Google Scholar]
- 64.Garcia-Pichel F, Castenholz RW. 1991. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. J Phycol 27:395–409. 10.1111/j.0022-3646.1991.00395.x. [DOI] [Google Scholar]
- 65.Fernandes VMC, Machado de Lima NM, Roush D, Rudgers J, Collins SL, Garcia-Pichel F. 2018. Exposure to predicted precipitation patterns decreases population size and alters community structure of cyanobacteria in biological soil crusts from the Chihuahuan Desert. Environ Microbiol 20:259–269. 10.1111/1462-2920.13983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3, Fig. S1 to S3. Download AEM.01236-21-s0001.pdf, PDF file, 4.3 MB (4.3MB, pdf)
Data Availability Statement
Raw sequence data have been submitted to NCBI and are publicly available under BioProject number PRJNA740061 for experimental trials and under BioProject number PRJNA705202 for remnant hot-desert biocrust communities.