Abstract
We investigated the requirements for enhancer-promoter communication by using the human β-globin locus control region (LCR) DNase I-hypersensitive site 2 (HS2) enhancer and the ɛ-globin gene in chromatinized minichromosomes in erythroid cells. Activation of globin genes during development is accompanied by localized alterations of chromatin structure, and CACCC binding factors and GATA-1, which interact with both globin promoters and the LCR, are believed to be critical for globin gene transcription activation. We found that an HS2 element mutated in its GATA motif failed to remodel the ɛ-globin promoter or activate transcription yet HS2 nuclease accessibility did not change. Accessibility and transcription were reduced at promoters with mutated GATA-1 or CACCC sites. Strikingly, these mutations also resulted in reduced accessibility at HS2. In the absence of a globin gene, HS2 is similarly resistant to nuclease digestion. In contrast to observations in Saccharomyces cerevisiae, HS2-dependent promoter remodeling was diminished when we mutated the TATA box, crippling transcription. This mutation also reduced HS2 accessibility. The results indicate that the ɛ-globin promoter and HS2 interact both structurally and functionally and that both upstream activators and the basal transcription apparatus contribute to the interaction. Further, at least in this instance, transcription activation and promoter remodeling by a distant enhancer are not separable.
A central question in developmental biology is how enhancers activate gene transcription in a tissue- and developmental-stage-specific fashion, a complex process which takes place in the chromatin environment of the nucleus. We have addressed this issue by studying components of the human β-globin gene locus, the locus control region (LCR) DNase I-hypersensitive site 2 (HS2) enhancer and the embryonic ɛ-globin gene. The locus consists of five genes expressed sequentially during development and a multicomponent, far-upstream regulatory element, the LCR (for a review, see reference 6). In naturally occurring thalassemias with large deletions encompassing the LCR and upstream sequences, expression of the downstream globin genes is abolished, and the chromatin structure of the locus becomes resistant to DNase I cleavage (20). Therefore, it has long been thought that the LCR mediates both decondensation of the chromatin of the globin locus and activation of transcription of the genes at different stages in development. However, recent experiments in which the mouse or human β-globin LCR was deleted in its natural chromosomal context demonstrate that while the LCR is required for high-level transcription, it may not be required for decondensation of the locus or correct developmental regulation of the globin genes (16, 51). Thus, the LCR, at a minimum, fulfills the role of a traditional enhancer.
The LCR contains four HS sites (HS1 to HS4) detected exclusively in the chromatin of erythroid cells (21, 55). Of these, only HS2 has enhancer activity in transient transfection assays, although in transgenic mice all four of the HS sites can activate the various β-globin genes to different extents (22, 56). During development in the mouse, HS2 and the other HS sites form before the globin genes are activated for transcription (33). Subsequently, when the genes are activated, their individual promoters become DNase I hypersensitive as well (31). A small number of transcription factor recognition sequences recur throughout the globin promoters and/or LCR cores including HS2. These include GATA-1 motifs, Maf recognition elements (recognized by NF-E2 and other factors), and CACCC class motifs (recognized by Krüppel-like proteins) (2, 8, 17, 24).
Two mechanistic explanations for the activation of globin promoters by the LCR have been proposed. In the dominant chromatin opening model, regulation of the individual genes is autonomous and dependent on the changing transcription factor milieu during development (39). In this view, the LCR is simply responsible for creating a decondensed, or otherwise favorable, chromatin environment in which the globin gene promoters can interact with stage-specific transcription factors. In the mutual interaction model, the LCR physically contacts the individual promoters and activates them in sequence, switching in response to stage-specific factors (14, 42). While there is evidence for autonomous regulation, several lines of investigation provide indirect support for a mutual interaction model for enhancer-promoter cross-talk. In situ hybridization data indicate that only a single globin gene on a chromosome is active at any one time (58). Studies of transgenic mice suggest that individual globin promoters may compete for interaction with the LCR (14). Moreover, it has been observed that the chick β/ɛ 3′ enhancer alone, without a promoter, is not sufficient to form an open chromatin structure (52). Finally, in vitro experiments show that the transcription factors which bind to HS2 and the other LCR HS sites and globin promoters can homo- and heterodimerize (13, 41, 61) and can interact with TAFII130 or CREB binding protein/p300, components of the transcriptional machinery (1, 9, 12, 64).
Enhancers work, at least in part, by altering repressive chromatin structure (19). In vitro approaches and transient transfection assays lack a physiological chromatin context, while random chromosomal integrants in transgenic mouse experiments may be confounded by position effects, particularly when mutant regulatory elements are studied (34). To circumvent these problems and to focus on the role of particular transcription factors in mediating enhancer-dependent promoter remodeling and transcription activation in chromatin, we have used Epstein-Barr virus-derived episomes that are stably maintained in human erythroid K562 cells (62). Such minichromosomes have been used extensively in Saccharomyces cerevisiae to obtain refined structural analyses of chromatin transitions accompanying transcription activation and in mammalian cells to dissect the immunoglobulin heavy-chain LCR enhancer (18, 38).
We have previously reported that transcription of the human ɛ-globin gene on minichromosomes is dependent on the HS2 enhancer, and the nucleosomal structure of the gene correlates well with that of the endogenous locus (28). In those studies we observed that the loss or alteration of a particular nucleosome in the ɛ-globin promoter depended on linking HS2 to ɛ-globin in the same minichromosome. The studies presented here demonstrate that the structure of HS2 depends on the ɛ-globin promoter and is mediated by GATA-1 and CACCC binding factors. Furthermore, the interaction of enhancer and globin promoter is dependent on the presence of an intact TATA box.
MATERIALS AND METHODS
Minichromosomes, cell culture conditions, and transfection.
The construction of minichromosomes carrying the ɛ-globin gene with or without HS2 (pɛA or pɛHS2A) has been described elsewhere (28). Briefly, the ɛ-globin gene was a 3.7-kb genomic EcoRI fragment (GenBank accession no. U01317, coordinates 17482 to 21233). The β-globin LCR HS2 was a 374-bp HindIII-to-XbaI fragment (GenBank accession no. U01317, coordinates 8486 to 8860). Clustered point mutations eliminating binding to GATA or CACCC motifs, as monitored by gel mobility shift assays, were introduced into minichromosomes by using QuikChange site-directed mutagenesis (Stratagene) and sequenced to verify the mutation. The GATA motif was mutated to TCGC, and the CACCC motif was mutated to CACAA (HS2 sites) or CACCG (promoter site). K562 cells were grown in RPMI 1640 medium with 10% fetal calf serum. Electroporation of K562 cells was done as described elsewhere (29). Single-cell clones were selected by limiting dilution in the presence of 200 μg of hygromycin B (Boehringer Mannheim) per ml, and multiple individual clones of each type were studied. Copy number and intactness of minichromosome structure were determined by Southern blot analysis of DNA from three nuclear isolations of each clone (27).
RNase protection assay.
RNA was prepared from 5 × 106 to 6 × 106 cells of K562 clones carrying various minichromosomes by using PUREscript (Gentra Systems). Transcripts from the episomal copy of the ɛ-globin gene are marked by a mutation in the 5′ untranslated region such that RNase digestion produces a smaller protected fragment than do endogenous transcripts (28). RNase digestion and gel analysis were performed generally as described by the manufacturer of the reagents (Ambion). Digestion was carried out at a 1/30 dilution of RNase A/T1 for 60 min at 37°C. The ɛ-globin and γ-actin (loading control) probes used have been described elsewhere (7, 28). Transcription of the ɛ-globin gene was normalized to the γ-actin message level and corrected for copy number of the minichromosome. The transcription levels of wild-type ɛHS2 clones (in which the ɛ-globin promoter was linked to HS2; n = 12) were determined at least three times for separate RNA preparations, and the mean transcription was set at 100%. The mean and standard error of the mean (SEM) for multiple clones of each type of mutation determined in triplicate are presented in the figures. The significance of the difference between the grand mean for each mutant and for wild-type ɛHS2 was computed by the Dunnett multiple-comparison test using the software package InStat (Graphpad).
Preparation of nuclei and nuclease digestion.
Nuclei of K562 clones were prepared as described elsewhere (28). Aliquots of 1 ml of purified nuclei (from 2 × 107 to 4 × 107 cells) were digested with 0, 3, 6, 12, or 25 μg of DNase I (GibcoBRL) per ml for 10 min at room temperature in the presence of 3 mM CaCl2. Alternatively, digestion of nuclei was performed with 100 U of various restriction enzymes as indicated in the text and figures (New England Biolabs) for 30 min at 37°C. DNA purification and Southern analysis were done as described elsewhere (28). Southern blots were hybridized with probes labeled with [32P]dCTP by random priming to a specific activity of 109 cpm/μg of DNA. The probes used are indicated in the figures. For restriction enzyme accessibility experiments, the intensity of bands was quantitated with a PhosphorImager (Molecular Dynamics). The percent restriction enzyme cleavage (% cut/uncut + cut) for wild-type ɛHS2 clones (AvaII, n = 8; MscI, n = 4) was determined for three separate nuclear isolations of each clone, and the grand mean was set to 100% digestion. The mean percentage of wild-type cutting for a representative individual clone for each type of mutation is presented (see Fig. 2, 3, 5, and 6). The significance of the difference between the mean cutting for the mutant clones and for ɛHS2 was computed by the Dunnett multiple-comparison test, and the results are noted in the text and figure legends. When multiple clones carrying a particular mutation were studied, the mean percent cutting for these clones was compared to the wild-type mean by the Mann-Whitney test (see Fig. 5, 7, and 8). Statistical analyses were performed with the software package InStat (Graphpad).
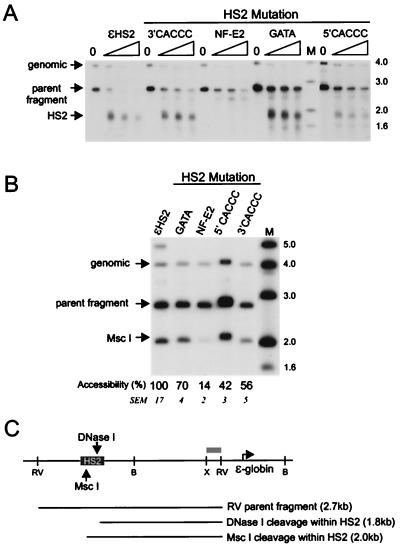
FIG. 2.
Effects of HS2 mutations on DNase I hypersensitivity and restriction enzyme accessibility at HS2. (A) Nuclei isolated from K562 clones containing the indicated minichromosomes were digested with DNase I, and the HS2 site was mapped by indirect end labeling after digestion with EcoRV. The amounts of DNase I used were (from left to right) 0, 12, 19, and 25 μg/ml. The genomic band indicated results from hybridization of the probe to an endogenous ɛ-globin 5′ flanking fragment of 4 kb which does not contain HS sites but which serves as a control for the extent of DNase I digestion. (B) Nuclei were digested with 100 U of MscI for 30 min, at which point maximal cutting was observed. DNA was then purified and cleaved to completion with EcoRV. The data were quantitated on a PhosphorImager. The percent MscI cleavage for wild-type ɛHS2 minichromosomes was determined as described in Materials and Methods and set at 100. A representative wild-type clone is shown on the gel. The mean percent cutting and SEM for the mutant clones compared to the wild-type level are shown below each lane. The percent cleavage for all mutants except the HS2GATA mutant differed significantly from the wild-type level (P < 0.05). (C) The positions of DNase I and MscI cleavage within HS2 are indicated. Lanes M, markers with sizes given in kilobases at the right. The probe fragment used for both experiments was an XbaI-EcoRV fragment from the 5′ flank of the ɛ-globin gene (gray bar). H, HindIII; X, XbaI; RV, EcoRV; B, BamHI.
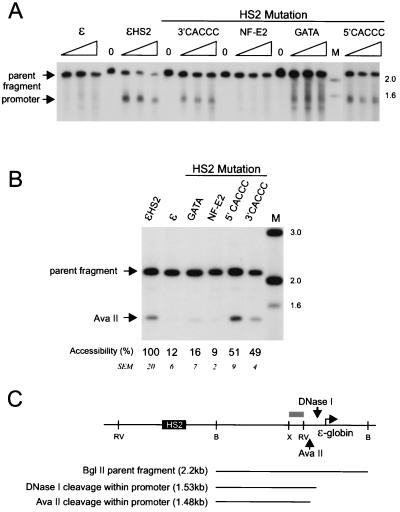
FIG. 3.
Effects of HS2 mutations on DNase I hypersensitivity and restriction enzyme accessibility at the ɛ-globin promoter. (A) Samples were digested with DNase I and processed as detailed in the legend to Fig. 2A except that the ɛ-globin promoter HS site was mapped after digestion with BglII. (B) Samples were processed as detailed in the legend to Fig. 2B except that primary digestion was with AvaII. The mean percent cutting and SEM for the mutant clones compared to wild-type levels are shown below each lane. The percent cleavage for the NF-E2 and HS2GATA mutants differed significantly from the wild-type level (P < 0.05) but was not significantly different for the HS2 3′ CACCC and 5′ CACCC mutants. (C) Positions of DNase I and AvaII cleavage within the ɛ-globin promoter are shown. Lanes M, markers with sizes given in kilobases at the right. The probe fragment used was the same as detailed in the legend to Fig. 2 (gray bar). X, XbaI; RV, EcoRV; B, BamHI.
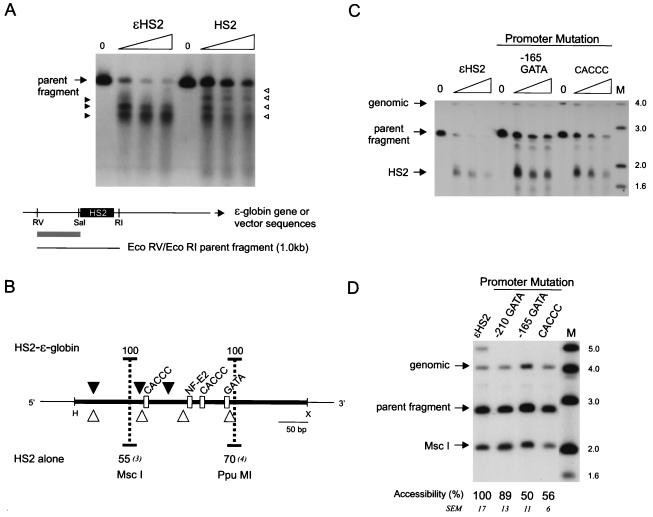
FIG. 5.
Nuclease sensitivity of HS2 in the absence of a linked globin gene and effects of promoter mutations on HS2. (A) Nuclei of K562 clones containing HS2 alone on minichromosomes were digested with DNase I as detailed in the legend to Fig. 2A, and the cleavage sites were mapped after double digestion with EcoRI and EcoRV. (B) Summary of DNase I cleavage sites in wild-type ɛHS2 and in HS2 alone. The percent accessibility and SEM at the HS2 MscI and PpuMI sites for six clones of each type are shown. The probe fragment used for both experiments was an EcoRV-to-SalI fragment flanking HS2 (gray bar). RV, EcoRV; RI, EcoRI; S, SalI. (C) Samples were processed as detailed in the legend to Fig. 2A, and the HS2 site was mapped after digestion with EcoRV. (D) Samples were processed as detailed in the legend to Fig. 2B. For the positions of DNase I and MscI cleavage within the EcoRV parent band, see Fig. 2C. Lanes M, markers with sizes given in kilobases at the right. The probe fragment used for both experiments was as detailed in the legend to Fig. 2.
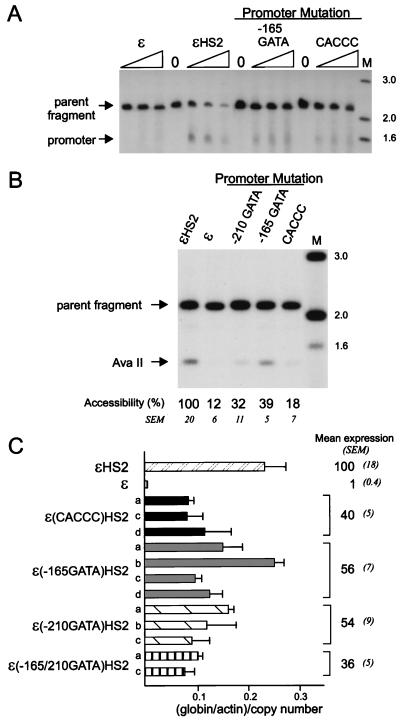
FIG. 6.
Effects of promoter mutations on nuclease sensitivity and transcription of the ɛ-globin promoter. (A) Samples were digested with DNase I and processed as detailed in the legend to Fig. 3A, mapping the ɛ-globin promoter HS site after digestion with BglII. (B) Samples were digested with AvaII and processed as detailed in the legend to Fig. 3B. The mean percent cutting and SEM for the mutant clones compared to wild-type levels are shown below each lane and differed significantly from the wild-type level (P < 0.05) for all mutants. For the positions of DNase I and AvaII cleavage within the ɛ-globin promoter, see Fig. 3C. Lanes M in panel A and B, markers with sizes given in kilobases at the right. The probe fragment used in panels A and B was as detailed in the legend to Fig. 2A. (C) Mean levels of expression of ɛ-globin RNA and SEM for several individual clones with each type of promoter mutation. The relative amount of ɛ-globin RNA was determined as detailed in Materials and Methods and in the legend to Fig. 4C. The differences from the wild-type grand mean were all statistically significant (P < 0.05).
FIG. 7.
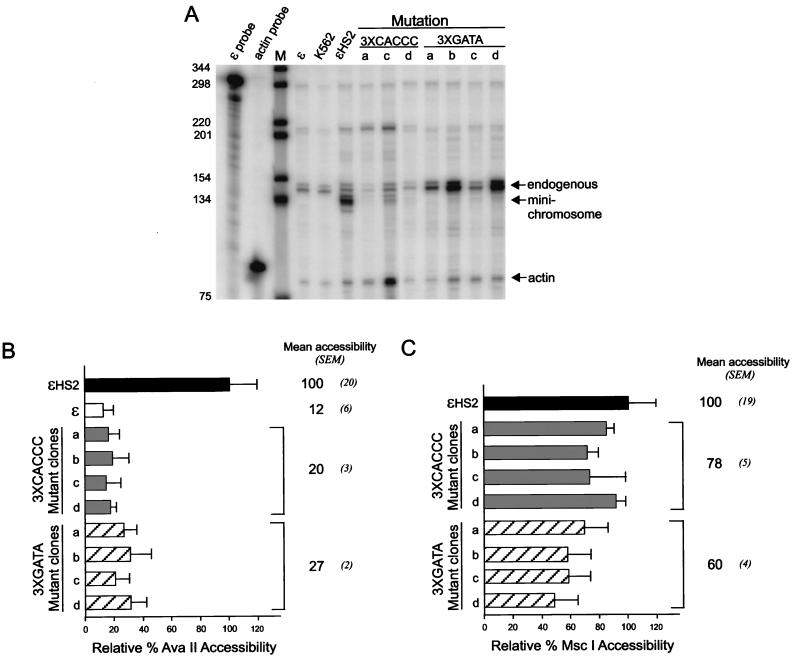
Transcription and restriction enzyme accessibility with multiple HS2 and promoter mutations. (A) RNase protection was used to measure the abundance of ɛ-globin transcripts, and the results for clones a to d with multiple CACCC site or GATA site mutations are shown. Lane M is as detailed in the legend to Fig. 4A. (B) Samples were processed as detailed in the legend to Fig. 3B by digestion with AvaII to determine promoter accessibility. The results for clones a to d are compared to the values for ɛ and ɛHS2 wild-type minichromosomes (see Materials and Methods). (C) Samples were processed as described above except that digestion was with MscI to determine HS2 accessibility.
FIG. 8.
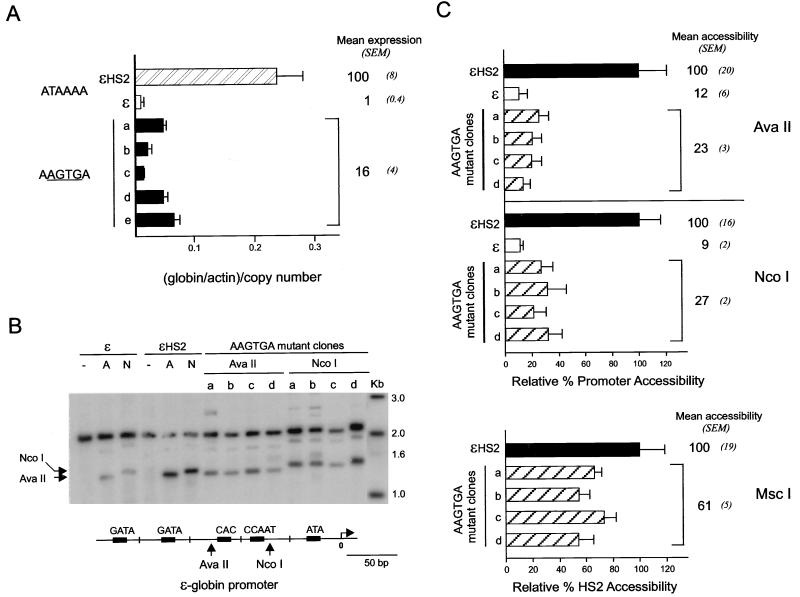
Effects of TATA box mutations on transcription and promoter restriction enzyme accessibility. (A) RNase protection was used to measure the abundance of ɛ-globin transcripts. The relative amount of ɛ-globin RNA was determined as detailed in the legend to Fig. 4B. Mean levels of expression of ɛ-globin RNA and SEM for several individual clones with the TATA box mutation are shown. (B) Nuclei were digested as detailed in the legend to Fig. 3B but with AvaII or NcoI to determine promoter accessibility. The positions of AvaII and NcoI cleavage within the ɛ-globin promoter are shown in the diagram at the bottom. Lane Kb, markers with sizes given in kilobases at the right. (C) Accessibility at promoter AvaII and NcoI sites and accessibility at the HS2 MscI site for clones a to d are compared to the values for ɛ and ɛHS2 wild-type minichromosomes (see Materials and Methods).
RESULTS
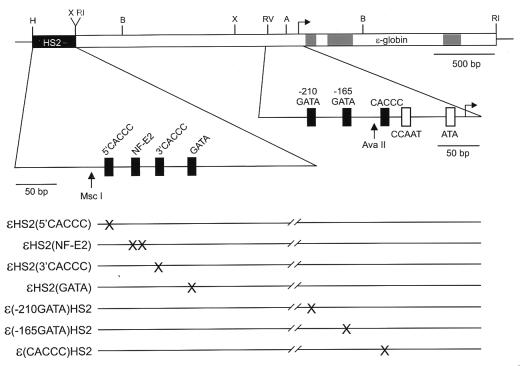
In transient transfection assays using a chloramphenical acetyltransferase (CAT) reporter gene linked to a 294-bp ɛ-globin promoter and HS2, the two HS2 tandem NF-E2 sites and an ɛ-globin promoter GATA-1 site are required for high-level transcription (26). To investigate the role of erythroid transcription factors on enhancer-dependent transcription of the gene promoter in its natural sequence context in chromatin, we introduced minichromosomes carrying the Epstein-Barr virus origin of replication and the ɛ-globin gene, with or without HS2, into human erythroid K562 cells which actively express the endogenous ɛ-globin gene (28). HS2 and the proximal ɛ-globin promoter region are shown in Fig. 1 in an expanded format to indicate the positions of the NF-E2, GATA, and CACCC motifs in these sequences; the sites mutated in the various minichromosomes are also shown.
FIG. 1.
Mutations introduced into ɛHS2 minichromosomes. The regions of the minichromosomes containing the ɛ-globin gene (gray rectangles represent exons) and HS2 are shown with HS2 and the proximal ɛ-globin promoter region in an expanded format to indicate the positions of the NF-E2, GATA, and CACCC motifs in these sequences. The sites that are mutated in the various minichromosomes are shown (×) below. H, HindIII; X, XbaI; RV, EcoRV; B, BamHI; RI, EcoRI.
Mutation of GATA and CACCC motifs in the HS2 enhancer primarily affects promoter structure and transcription of the linked globin gene.
We first investigated the role of the HS2 CACCC and GATA sites in determining chromatin structure at HS2 as judged by DNase I hypersensitivity. Individual K562 clones containing minichromosomes with HS2 mutations as illustrated in Fig. 1 were isolated, and nuclei from the clones were digested with DNase I or with MscI to examine HS2 chromatin structure. Figure 2A shows that DNase I cleavage at HS2 results in a band at 1.8 kb for wild-type ɛHS2 minichromosomes. CACCC and GATA mutations appeared to reduce DNase I sensitivity, as the parent band is more resistant to cleavage. In contrast, mutating the NF-E2 site abolished DNase I hypersensitivity (28). To make a quantitative comparison of the effects of these mutations on HS2 chromatin structure, nuclei from the cell clones were digested with MscI, which has a recognition site in HS2 (Fig. 2B). The results of MscI digestion are consistent with Fig. 2A. Of note, cutting at MscI remained at 70% of the wild-type level for the HS2 GATA mutant. The extent of MscI digestion for all HS2 mutants except GATA was statistically different from the wild-type level. Thus, in contrast to NF-E2, the binding of GATA-1 is not required for the formation of HS2.
We next investigated the ability of these mutant enhancers to alter the chromatin structure of the ɛ-globin promoter. Nuclei of cells carrying minichromosomes were treated with DNase I and then cleaved with BglII to map the ɛ-globin promoter HS site (Fig. 3A). Nontranscribed wild-type ɛ and actively transcribed ɛHS2 minichromosomes are included as controls. Mutation of single CACCC sites in HS2 decreased somewhat DNase I sensitivity at the promoter compared to wild-type HS2. However, the promoter linked to the HS2 GATA mutant was only weakly DNase I sensitive. Restriction enzyme accessibility at the AvaII site in the ɛ-globin promoter was used to quantitate remodeling by HS2 mutants (Fig. 3B), as this site is accessible only when the promoter is remodeled by HS2 (28). Accessibility was reduced by half when single CACCC sites in HS2 were mutated. In contrast, AvaII accessibility at the promoter was reduced to 10 to 20% of the wild-type level for the NF-E2 and GATA HS2 mutants, which correlates well with their decreased sensitivity to DNase I. Although HS2 formation is not significantly impaired when the HS2 GATA site is mutated, promoter remodeling is markedly diminished.
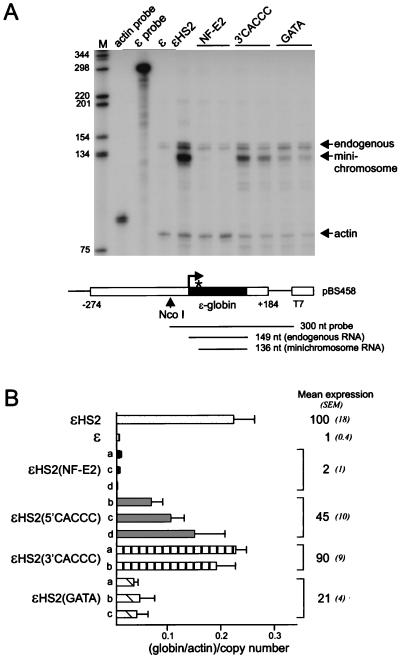
To determine whether the HS2 mutations affected transcription, ɛ-globin RNA levels were measured by RNase protection as illustrated in Fig. 4A. The minichromosomal ɛ-globin gene is not transcribed in the absence of an enhancer, while inclusion of HS2 results in transcription of the linked gene. Transcription was not activated by the HS2 NF-E2 mutant enhancer (28), shown here for comparison, while the GATA and CACCC mutations diminished transcription to various extents. Figure 4B shows ɛ-globin RNA expression for several clones with each type of HS2 mutation compared to wild-type ɛ (n = 3) and ɛHS2 (n = 12) clones. Mutation of the HS2 5′ CACCC site reduced transcription to 45% of the wild-type level, while mutation of the 3′ CACCC site resulted in transcript levels not statisically different from the wild-type level. Transcription was severely reduced in the HS2 GATA mutant.
FIG. 4.
Transcription of the ɛ-globin gene linked to HS2 mutants. (A) RNase protection was used to measure the abundance of ɛ-globin transcripts. The episomal copy of the ɛ-globin gene has been marked by a mutation in the 5′ untranslated region (∗) and produces a shorter protected fragment than endogenous transcripts (shown at the bottom). The bands produced by the endogenous and minichromosomal copies of the ɛ-globin gene are indicated by arrows. An RNA probe for γ-actin was included as a loading control. Lane M, markers in base pairs as indicated at the left. (B) Mean levels of expression of ɛ-globin RNA and SEM for several individual clones with each type of HS2 mutation are shown at the right. The amount of ɛ-globin RNA was determined for three separate RNA preparations for each clone. The results are compared with the mean levels of expression of ɛ-globin RNA for wild-type ɛ clones (n = 3) and wild-type ɛHS2 clones (n = 12). The differences from the wild-type grand mean were statistically significant (P < 0.05) except for the HS2 3′ CACCC mutant.
The data in Fig. 3 and 4 indicate that the extent of altered chromatin structure at the ɛ-globin promoter correlates well with the level of transcription of the gene. They further show that although HS2 exhibits strong DNase I hypersensitivity when the HS2 GATA-1 site is mutated, this mutant HS2 does not efficiently remodel the promoter and supports greatly reduced transcription. This observation suggests that GATA-1 bound to HS2 is critical both for remodeling and activating the distant promoter. Mutation of this site does not affect the activity of HS2 in transient transfection assays which lack a natural chromatin context (26), suggesting GATA-1 bound to HS2 may be involved in enhancer-dependent disruption of a promoter nucleosome (28).
A wild-type ɛ-globin promoter is required for proper HS2 formation.
In contrast to the ɛ-globin promoter, which is structurally altered by the action of the HS2 enhancer, the enhancer could potentially form its distinctive nuclease-sensitive structure in chromatin in an autonomous fashion (48). Alternatively, an enhancer may not be sufficient to form a disrupted chromatin structure in the absence of a promoter (52). As a preliminary test of whether a promoter could affect HS2 structure, we created K562 clones with minichromosomes that contained HS2 without a linked globin gene and examined the DNase I sensitivity of HS2 (Fig. 5A). Although HS2 was sensitive to DNase I in the absence of a globin gene, the site was considerably weakened, as judged by the greatly increased resistance to cleavage of the parent band, and the fine structure of DNase I cutting was altered. Figure 5B summarizes the location of DNase I cleavage sites for ɛHS2 and for HS2 alone. Because of the reduced DNase I sensitivity of HS2 in the absence of a globin gene, cleavage at sites close to the HS2 GATA-1 and NF-E2 binding motifs could be visualized at the lowest enzyme concentration (Fig. 5A, uppermost open arrowheads). In ɛHS2, this region has already been digested by DNase I at the same concentration, and the longest band visualized terminated between the NF-E2 and 5′ CACCC sites. To further examine the structure of HS2 without a linked globin gene, we analyzed the accessibility of the MscI and PpuMI sites in HS2. The data (not shown) are summarized in Fig. 5B and indicate that MscI accessibility was reduced to a mean of 55% relative to ɛHS2, while the mean relative PpuMI accessibility was 70%. Thus, HS2 structure is clearly different in the absence of a linked globin gene.
To further probe the interrelationship between the promoter and HS2, we examined whether mutation of promoter GATA or CACCC motifs affected the chromatin structure of HS2. Nuclei from the K562 clones mutated as depicted in Fig. 1 were digested with DNase I or with MscI to examine HS2 chromatin structure. Mutation of the GATA site at −210 did not affect HS2 structure (Fig. 5D). However, DNase I sensitivity at HS2 was reduced when either the promoter −165 GATA site or the CACCC site was mutated (Fig. 5C). A quantitative assessment of these changes by MscI digestion of nuclei revealed that accessibility at HS2 for these promoter mutants was reduced to about half and differed significantly from the wild-type level (P < 0.05) (Fig. 5D). Thus, the structural effect on HS2 of each of these mutations is equivalent to the effect of removing the globin gene entirely.
Multiple GATA or CACCC mutations are required to prevent transcription.
Transient transfection assays indicated that the GATA-1 site at −165 but not the site at −210 in the ɛ-globin promoter is required for enhanced transcription of a CAT reporter gene (26). We concluded that this site is important for promoter-enhancer communication. To determine the role of promoter transcription factor motifs when the promoter resides in its natural sequence context in a chromatin environment, nuclei from individual K562 clones with minichromosomes mutated as illustrated in Fig. 1 were digested with DNase I or with AvaII to examine promoter chromatin structure. The −165 GATA and CACCC mutant promoters were less sensitive to DNase I than the wild-type promoter (Fig. 6A) and had decreased accessibility to AvaII (Fig. 6B, 39 and 18%, respectively). Likewise, mutation of the −210 GATA site reduced the AvaII accessibility of the promoter (32%). There was a moderate but statistically significant (P < 0.05) reduction in transcription from these mutant promoters, as determined by RNase protection experiments and shown in Fig. 6C. However, even when both the −165 and −210 promoter GATA-1 sites were mutated, transcription remained at 36% of wild-type levels. In contradistinction to transient transfection assays where mutation of the single −165 GATA site prevented enhanced transcription from the ɛ-globin promoter, in chromatin no single promoter mutation, not even the double GATA site mutation, is sufficient to totally disrupt transcription. The results suggest that in chromatin, multiple interactions contribute to enhancer-promoter communication.
In an attempt to completely disrupt putative protein-protein interactions required for activated transcription, we simultaneously mutated multiple factor binding sites in HS2 and in the ɛ-globin promoter. In these constructions, either the three GATA sites (3×GATA clones) or the three CACCC sites (3×CACCC clones) were mutated (Fig. 1). The results in Fig. 7A indicate that for both types of multiple mutants, transcription of the ɛ-globin gene was very low (average of 3% of wild-type transcription for 3×CACCC clones) or undetectable (3×GATA clones). Restriction enzyme accessibility was used to probe promoter and HS2 chromatin structure in these mutants (Fig. 7B and C). The average AvaII accessibility at the promoter was reduced to 27% relative to the wild type for the 3×GATA mutant clones and 20% for the 3×CACCC clones, similar to the alteration in structure observed for some of the single mutations (Fig. 2D and 4B). As judged by MscI accessibility, the structure of HS2 was affected in the 3×GATA mutants (60% of wild-type accessibility) and somewhat less so in the 3×CACCC mutants (78%). MscI accessibility at HS2 for the 3×CACCC mutants was not statistically significantly different from that of the wild type or of single CACCC mutants in this data set. However, unlike the single mutations, the multiple ones eliminate transcription, suggesting that at least one role of the multiple contacts in HS2 and the ɛ-globin promoter by GATA-1 and CACCC factors is to provide sufficient stability to enhancer-promoter communication that transcription initiation will occur.
An intact TATA box is required for promoter remodeling and transcription activation.
In several studies in yeast and mammalian systems, promoter remodeling and transcription per se can be separated, suggesting that remodeling is a prerequisite for formation of the TATA-bound transcription complex (18, 35, 44, 45, 59). If promoter remodeling and transcription activation are sequential events, then promoter remodeling should still occur if the TATA box were mutated to cripple transcription. Alternatively, these processes might occur in concert, and remodeling and transcription would be similarly affected by TATA box mutation. We created HS2 ɛ-globin minichromosomes with four clustered point mutations eliminating the ɛ-globin TATA site. These mutants displayed very low levels of transcription (Fig. 8A). We studied the accessibility of the promoter chromatin for these mutant clones at both the AvaII and NcoI sites (Fig. 8B and C). Accessibility was severely reduced to 23% of the wild-type level at the AvaII site and 27% at the NcoI site. Thus, remodeling of the promoter requires components of the complex formed over the TATA box, suggesting that remodeling and transcription activation by a distant enhancer are mechanistically linked. Interestingly, the TATA box mutation also affected the structure of HS2, which was reduced to 61% accessibility at the MscI site (Fig. 8C). This further illustrates, along with the data in Fig. 5, that the promoter and enhancer help to establish each other as nuclease-sensitive structures.
DISCUSSION
In this work, we tested the importance of particular transcription factor binding sites in enhancer-dependent promoter remodeling and the mechanistic linkage between promoter remodeling and active transcription. We addressed these questions in a chromatin environment by using virus-based minichromosomes. Genes carried on such independent episomes should not be subject to position effects which may influence the activity of transgenes integrated randomly into the genomes of cell culture lines or transgenic mice. We placed HS2 of the β-globin LCR and a large genomic fragment of the human ɛ-globin gene into minichromosomes, preserving the natural sequence environment of the promoter and separating HS2 from the promoter by about 2 kb. Our data support a model in which the structures of HS2 and the ɛ-globin promoter are functionally interdependent and in which communication between the two regulatory elements is mediated at least in part by reiterated GATA-1 and CACCC motifs. We further find that enhancer-dependent promoter remodeling and transcription initiation are not separable in this system.
Two general mechanisms can be envisioned to account for the observed communication between HS2 and the ɛ-globin promoter (see reference 50 for a review). Either the two regulatory elements physically contact one another or mutual interaction is effected by the propagation of signals between the two elements without physical contact (for example, by tracking along the intervening DNA). However, our observation that communication is bidirectional in that mutation of either regulatory element affects the structure of the other is most easily understood in terms of a physical contact model, particularly since (i) we observe no changes in the chromatin structure of the 2-kb region between HS2 and the ɛ-globin promoter (28) and (ii) individual globin promoters have been observed to compete for interaction with the LCR (see the introduction).
HS2 structure depends on the ɛ-globin promoter.
We found that mutations introduced into the ɛ-globin promoter at the CACCC or −165 GATA site alter the chromatin structure of the distant HS2 enhancer (restriction enzyme accessibility of 56 and 50% of the wild-type level, respectively). Mutation of the ɛ-globin TATA element in the context of an otherwise intact promoter similarly results in an altered HS2 structure (61% accessibility). These observations illustrate the interdependence of formation of the promoter and enhancer chromatin structures and provide a new line of evidence in support of the mutual interaction model. When HS2 was studied without a linked globin gene, a reduction in restriction enzyme accessibility was also observed (55 and 70% accessibility at two different restriction sites), as well as an alteration in the pattern of DNase I subbands. In contrast, the chicken β/ɛ-globin enhancer/LCR is not hypersensitive in the absence of a linked globin gene in transgenic mouse chromosomes, unless it integrates near an active mouse promoter (52). Since there are two genes on the minichromosomes which are required for its maintenance, the remaining HS2 nuclease sensitivity may result from interaction with one or the other of these functional nonerythroid promoters. If such an interaction is responsible for HS2 nuclease sensitivity, it is clearly different from the interaction of HS2 with the native ɛ-globin promoter. Alternatively, the continued occupancy of the NF-E2 sites may account for the remaining nuclease sensitivity.
Role of transcription factors at HS2.
Mutation of the NF-E2 sites in HS2 severely reduces expression of a linked β-globin gene in transgenic mice (11, 37, 53, 54). Likewise, we found that mutation of the NF-E2 sites abolishes transcription of a linked ɛ-globin gene and that nuclease sensitivity at HS2 is no longer detectable (this work and reference 28). Taken together with the observation that ɛ-globin transcription requires linked HS2 sequences, these data suggest that transcription fails to occur because the enhancer complex at HS2 fails to form properly in the absence of NF-E2 site occupancy. Indeed, transcription factor binding at the NF-E2 sites may be the primary or an early step in the formation of the HS2 enhancer. The related transcription factor AP1 can disrupt a nucleosome in vitro in the absence of remodeling activities (46), and in vitro studies indicate that NF-E2 can disrupt a nucleosome, although it is not clear whether this is an intrinsic activity of NF-E2, or whether chromatin remodeling complexes are involved (3).
Consistent with analyses of transgenic mice (10), we observed that mutation of the 5′ HS2 CACCC motif reduces transcription (45% of the wild-type level). The 3′ CACCC mutation was less deleterious in transgenic mice and not significantly different from wild type in our studies (90% of wild-type transcription). The CACCC mutations affected remodeling of the linked ɛ-globin gene (about 50% of wild-type accessibility). Nuclease sensitivity at HS2 is also reduced in these mutants (42 and 56% for the 5′ and 3′ mutations, respectively). Because only one of these mutations affects transcription, and because it is not known what CACCC box binding protein(s) is relevant here, it is unclear what role the CACCC binding activities play in HS2 enhancer activity. However, it is provocative that Sp1, one candidate CACCC binding activity, can bind nucleosomes in vitro, and that another, EKLF, can recruit chromatin remodeling activities to the β-globin promoter in vitro (4, 36). Conceivably, the HS2 CACCC boxes are more important for communication with other LCR HSs or other globin promoters.
In contrast to the NF-E2 and CACCC mutations, the HS2 GATA mutation reduces neither DNase I sensitivity nor restriction enzyme accessibility (70% of wild-type accessibility; not statistically different from the wild-type level) at HS2 markedly. However, nuclease sensitivity at the promoter and transcription of the ɛ-globin gene are much reduced (16% accessibility and 21% transcription compared to wild-type levels). The transcription results are similar to those in transgenic mice (15). We previously observed that for minichromosomes with wild-type HS2 linked to ɛ-globin, DNase I digestion at the promoter and at HS2 releases doubly cleaved fragments for a large fraction of minichromosomes, indicating that both sites on the same molecule are simultaneously hypersensitive (28). Despite HS2 hypersensitivity in the HS2 GATA mutant, essentially no doubly cleaved fragments were observed, indicating that GATA-1 is required for cis HS2 effects on the promoter (unpublished data). This activity is not apparent in transient transfection assays where a chromatin context is absent (26). We propose that assembly of the regulatory structure at HS2 and its activation of the ɛ-globin promoter may be a stepwise process in which NF-E2 nucleates formation of HS2, GATA-1 principally mediates communication with the promoter, and CACCC sites contribute to stabilizing the functional interaction.
Promoter-enhancer communication is stabilized by multiple factor interactions.
In transient transfection experiments using a CAT reporter gene, mutation of the ɛ-globin promoter CACCC site substantially reduces transcription (27, 63), and the GATA motif at −165 is essential for HS2-enhanced but not basal transcription (26). In contrast, in the present work, mutation of the CACCC or −165 GATA motif in the ɛ-globin promoter reduces transcription by only about 50% in the presence of HS2. The experiments presented here analyze the function of factors interacting at these sites in the context of chromatin with 2 kb of upstream ɛ-globin sequence. The presence of a more natural sequence in a chromatin environment likely explains the differences in the results of the experiments (see also reference 49). Although the HS2 GATA mutation severely reduces transcription (20% of wild-type transcription), neither any single nor any double (unpublished data) GATA or CACCC mutation in HS2 or in the ɛ-promoter completely disrupts transcription of the gene. Only mutations which eliminated all the CACCC or all the GATA motifs reduced transcription to negligible levels. Loss of a single factor binding site may not be sufficient to disrupt multicomponent regulatory complexes (57). The stability of such complexes may be robust enough to tolerate the loss of even a substantial number of protein-protein contacts. We speculate that only when a critical number of structural determinants has been eliminated from both promoter and enhancer do chromatin remodeling activities and/or the basal transcription complex fail to interact sufficiently stably for transcription to occur. Further, these results are consistent with the prediction that communication of enhancer and promoter in chromatin may be mediated by homotypic and heterotypic interactions involving CACCC factors and GATA-1 observed in vitro (13, 41, 61).
Role of the TATA box.
We failed to observe remodeling of the ɛ-globin promoter linked to HS2 when the TATA box was mutated. This result contrasts with studies in yeast in which promoter remodeling is observed when transcription initiation is prevented by deletion or mutation of the TATA box (5, 18, 35). For example, when the TATA sequence is deleted from the yeast PHO5 gene promoter, PHO4 activator-dependent remodeling still occurs (18). Interestingly, the residues in the activation domains of both PHO4 and GAL4 which are required for transcription activation are the same ones required for promoter remodeling, raising the possibility that these may not be separate events in vivo and may be mediated by a single entity recruited by the activator (5, 40). Further dissection of promoter remodeling in yeast revealed that recruitment of RNA polymerase II holoenzyme (or an associated component) via the activation domain of PHO4 is sufficient to remodel chromatin (23). Thus, upstream activators (or enhancers) may alter promoter structure by recruiting chromatin modifying activities which are components of the RNA polymerase II machinery (30, 43).
Like that of the uninduced PHO5 promoter, the TATA sequence of the ɛ-globin gene is nucleosomal in the absence of HS2 (28). However, we observe that an intact TATA sequence is required for both ɛ-globin promoter remodeling and transcriptional activation, although nucleosomal TATA sequences are not normally accessible to TATA binding protein (TBP) (32, 60). We envision two possible explanations for this phenomenon. The nucleosome containing the TATA sequence may lack histone H1, permitting nucleosome sliding which could transiently expose the TATA site (47). Alternatively, TBP may be able to access the TATA site in the ɛ-globin promoter which lies near the edge of a nucleosome (28). Others have shown that TBP may gain access to its site at such a position within a nucleosome if histone tails are acetylated (25). These possibilities are testable in our system. Our data suggest that the TBP-containing TFIID complex or factors which interact with TFIID play an important role in recruitment of remodeling activities to the promoter. Thus, for a developmentally regulated gene which responds to a distant enhancer/LCR, disruption of the nucleosome at the promoter may occur in concert with transcription activation.
ACKNOWLEDGMENTS
We are grateful to David Clark, David Jackson, Gary Felsenfeld, and Marc Reitman for valuable discussions and comments on the manuscript.
REFERENCES
- 1.Amrolia P J, Ramamurthy L, Saluja D, Tanese N, Jane S M, Cunningham J M. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the α- and β-globin gene loci in an erythroid cell line. Proc Natl Acad Sci USA. 1997;94:10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vitro. Mol Cell Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 5.Axelrod J D, Reagan M S, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993;7:857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- 6.Baron M H. Developmental regulation of the vertebrate globin multigene family. Gene Expr. 1996;6:129–137. [PMC free article] [PubMed] [Google Scholar]
- 7.Baron M H, Maniatis T. Regulated expression of human α- and β-globin genes in transient heterokaryons. Mol Cell Biol. 1991;11:1239–1247. doi: 10.1128/mcb.11.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieker J J, Southwood C M. The erythroid Krüppel-like factor transactivation domain is a critical component for cell-specific inducibility of a β-globin promoter. Mol Cell Biol. 1995;15:852–860. doi: 10.1128/mcb.15.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caterina J J, Ciavatta J, Donze D, Behringer R R, Townes T M. Multiple elements in human β-globin locus control region 5′ HS 2 are involved in enhancer activity and position-independent transgene expression. Nucleic Acids Res. 1994;22:1006–1011. doi: 10.1093/nar/22.6.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caterina J J, Ryan T M, Pawlik K M, Palmiter R D, Brinster R L, Behringer R R, Townes T M. Human β-globin locus control region: analysis of the 5′ DNase I hypersensitive site HS 2 in transgenic mice. Proc Natl Acad Sci USA. 1991;88:1626–1630. doi: 10.1073/pnas.88.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng X, Reginato M J, Andrews N C, Lazar M A. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crossley M, Merika M, Orkin S H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- 15.Ellis J, Talbot D, Dillon N, Grosveld F. Synthetic human β-globin 5′HS2 constructs function as locus control regions only in multicopy transgene concatamers. EMBO J. 1993;12:127–134. doi: 10.1002/j.1460-2075.1993.tb05638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 17.Evans T, Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 18.Fascher K D, Schmitz J, Horz W. Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J Mol Biol. 1993;231:658–667. doi: 10.1006/jmbi.1993.1317. [DOI] [PubMed] [Google Scholar]
- 19.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 20.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 21.Forrester W C, Thompson C, Elder J T, Groudine M. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser P, Pruzina S, Antoniou M, Grosveld F. Each hypersensitive site of the human beta-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- 23.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 24.Gillemans N, Tewari R, Lindeboom F, Rottier R, de Wit T, Wijgerde M, Grosveld F, Philipsen S. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the β-globin locus control region in vivo. Genes Dev. 1998;12:2863–2873. doi: 10.1101/gad.12.18.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godde J S, Nakatani Y, Wolffe A P. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Q, Dean A. Enhancer-dependent transcription of the ɛ-globin promoter requires promoter-bound GATA-1 and enhancer-bound AP-1/NF-E2. Mol Cell Biol. 1993;13:911–917. doi: 10.1128/mcb.13.2.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Q, Dean A. Enhancer dependent transcription of the human ɛ-globin gene on a stably maintained minichromosome. In: Stamatoyannopoulos G, editor. Molecular biology of hemoglobin switching. Andover, United Kingdom: Intercept; 1995. pp. 279–288. [Google Scholar]
- 28.Gong Q H, McDowell J C, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the ɛ-globin gene in vivo by 5′ hypersensitive site 2 of the β-globin locus control region. Mol Cell Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Q H, Stern J, Dean A. Transcriptional role of a conserved GATA-1 site in the human ɛ-globin gene promoter. Mol Cell Biol. 1991;11:2558–2566. doi: 10.1128/mcb.11.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 31.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Human fetal to adult hemoglobin switching: changes in chromatin structure of the β-globin gene locus. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez G, Griffiths S D, Ford A M, Greaves M F, Enver T. Activation of the β-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci USA. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kioussis D, Festenstein R. Locus control regions: overcoming heterochromatin-induced gene inactivation in mammals. Curr Opin Genet Dev. 1997;7:614–619. doi: 10.1016/s0959-437x(97)80008-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee M S, Garrard W T. Uncoupling gene activity from chromatin structure: promoter mutations can inactivate transcription of the yeast HSP82 gene without eliminating nucleosome-free regions. Proc Natl Acad Sci USA. 1992;89:9166–9170. doi: 10.1073/pnas.89.19.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Adams C C, Workman J L. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994;269:7756–7763. [PubMed] [Google Scholar]
- 37.Liu D, Chang J C, Moi P, Liu W, Kan Y W, Curtin P T. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci USA. 1992;89:3899–3903. doi: 10.1073/pnas.89.9.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madisen L, Krumm A, Hebbes T R, Groudine M. The immunoglobulin heavy-chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D I, Fiering S, Groudine M. Regulation of β-globin gene expression: straightening out the locus. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 40.McAndrew P C, Svaren J, Martin S R, Horz W, Goding C R. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol Cell Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 43.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 44.Morgan J E, Whitlock J P., Jr Transcription-dependent and transcription-independent nucleosome disruption induced by dioxin. Proc Natl Acad Sci USA. 1992;89:11622–11626. doi: 10.1073/pnas.89.23.11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mymryk J S, Archer T K. Dissection of progesterone receptor-mediated chromatin remodeling and transcriptional activation in vivo. Genes Dev. 1995;9:1366–1376. doi: 10.1101/gad.9.11.1366. [DOI] [PubMed] [Google Scholar]
- 46.Ng K W, Ridgway P, Cohen D R, Tremethick D J. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 1997;16:2072–2085. doi: 10.1093/emboj/16.8.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 48.Pomerantz O, Goodwin A J, Joyce T, Lowrey C H. Conserved elements containing NF-E2 and tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human beta-globin locus control region. Nucleic Acids Res. 1998;26:5684–5691. doi: 10.1093/nar/26.24.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pondel M D, Murphy S, Pearson L, Craddock C, Proudfoot N J. Sp1 functions in a chromatin-dependent manner to augment human α-globin promoter activity. Proc Natl Acad Sci USA. 1995;92:7237–7241. doi: 10.1073/pnas.92.16.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 51.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reitman M, Lee E, Westfall H, Felsenfeld G. An enhancer/locus control region is not sufficient to open chromatin. Mol Cell Biol. 1993;13:3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talbot D, Grosveld F. The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J. 1991;10:1391–1398. doi: 10.1002/j.1460-2075.1991.tb07659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talbot D, Philipsen S, Fraser P, Grosveld F. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 1990;9:2169–2177. doi: 10.1002/j.1460-2075.1990.tb07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuan D, Solomon W, Li Q, London I M. The “beta-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuan D Y, Solomon W B, London I M, Lee D P. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “β-like” globin genes. Proc Natl Acad Sci USA. 1989;86:2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weintraub H. Summary: genetic tinkering—local problems, local solutions. Cold Spring Harbor Symp Quant Biol. 1993;58:819–836. doi: 10.1101/sqb.1993.058.01.089. [DOI] [PubMed] [Google Scholar]
- 58.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 59.Wong J, Shi Y B, Wolffe A P. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 61.Yang H Y, Evans T. Homotypic interactions of chicken GATA-1 can mediate transcriptional activation. Mol Cell Biol. 1995;15:1353–1363. doi: 10.1128/mcb.15.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 63.Yu C Y, Motamed K, Chen J, Bailey A D, Shen C K. The CACC box upstream of human embryonic ɛ globin gene binds Sp1 and is a functional promoter element in vitro and in vivo. J Biol Chem. 1991;266:8907–8915. [PubMed] [Google Scholar]
- 64.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]