Abstract
Purpose
Infection and transmission of carbapenem-resistant Aeromonas is a serious threat to public health. Rapid and accurate detection carbapenem-resistant of these organisms is essential for reasonable treatment and infection control. This study aimed to find a simple and effective method to detect carbapenem-resistant phenotype in Aeromonas.
Methods
A total of 131 clinical preserved Aeromonas strains were used in this study. The carbapenemase genes were detected by PCR. Modified carbapenem inactivation method (mCIM) in conjunction with EDTA-modified carbapenem inactivation method (eCIM) and simplified carbapenem inactivation method (sCIM) were performed to detect carbapenemases. We also designed a simple method, carbapenem inactivation method using supernatant (CIM-s), to detect the carbapenemase activity in the medium.
Results
Of the 131 Aeromonas strains, 79 contained carbapenemase genes, including 68 blaCphA, 6 blaKPC-2, 2 blaNDM-1 and 3 blaKPC-2+CphA. However, routine antibiotic susceptibility testing could not completely identify carbapenemase-producing Aeromonas. In phenotypic assays, the sensitivity and specificity of mCIM were 100%. The combined mCIM and eCIM could distinguish serine carbapenemase and metallo-β-carbapenemases except co-producing organisms. The sensitivity and specificity of sCIM were 92.4% and 100%, respectively, which could not detect CphA totally. CIM-s results indicate that these carbapenemases could secrete into the medium to perform their hydrolytic activities and had a sensitivity and specificity of 97.5% and 100%, respectively.
Conclusion
The combination of mCIM and eCIM can effectively detect and distinguish different types of carbapenemase in Aeromonas, and could be used as an important supplement approach to the antibiotic susceptibility testing.
Keywords: Aeromonas, modified carbapenem inactivation method, carbapenemase, multidrug resistant, phenotypic detection
Introduction
Bacterial resistance has become an urgent global concern. The increase in multidrug-resistant and even pan drug-resistant bacteria has brought serious challenges to clinical diagnosis and treatment.1 Gram-negative bacteria are important opportunistic pathogens of community-acquired infection and hospital-acquired infection. With the emergence of carbapenem-resistant Gram-negative bacteria, like non-fermenting Gram-negative bacteria and Enterobacterales, the clinical treatment options are very limited, which in turn prolong the length of stays in hospital, increase the cost of care and enhance the risk of mortality.2–4 Currently, it is widely accepted that the significant mechanism of carbapenem resistance in these organisms is producing carbapenemases. Carbapenemases are mainly categorized into Ambler class A, B, and D depending on the Ambler classification scheme. Class A and D carbapenemases are serine carbapenemases with serine hydrolysis mechanism. In addition, class B carbapenemases are metallo-β-lactamases (MBLs) requiring zinc ions catalysis to generate hydrolysis activity.4,5 Carbapenemase genes are frequently located on plasmids and mobile genetic elements, facilitating transfer between different strains, which make nosocomial infection control facing severe challenges.6,7
The genus Aeromonas, widely distributed in the natural environment, is a Gram-negative, oxidase-positive, facultative-anaerobic bacillus. There are 36 species in the genus Aeromonas,8 of which A. hydrophila, A. caviae and A. veronii bv. sobria are the major pathogens of infection.9 In addition to diarrhea, Aeromonas can cause a variety of extra-intestinal infections, such as skin and soft tissue infection, wound infection, intra-abdominal infection, pneumonia and bacteremia, indicating that Aeromonas isolates are significant human opportunistic pathogens.10–14 Therefore, infection and transmission of carbapenem-resistant Aeromonas pose a potential threat to public health. The main mechanism of Aeromonas resistance to carbapenems is to carry CphA MBL that is located in the chromosome and only has activity against carbapenems.15,16 Besides CphA, Aeromonas strains carrying other carbapenemases are also emerging. In 2007, Aeromonas caviae containing IMP-19 MBL was isolated in France.17 In 2008, the first multi-resistant Aeromonas hydrophila producing VIM-4 MBL was identified in Hungary.18 Additionally, Aeromonas-producing KPC-2 carbapenemase also successively emerged in Brazil, the United States and China.19–21 In recent years, Aeromonas harboring blaOXA-181, blaGES-24 and even blaGES/OXA-48/NDM-1 multiple carbapenemase-encoding genes have been detected.22–24
Molecular methods to detect carbapenemase-encoding genes are regarded as the gold standard for identification of carbapenemase-producing organisms. However, molecular methods require professional technicians, special detection equipment, expensive costs, time-consuming and multitudinous potential target genes, which limit their routine application.3,6,25 Various phenotypic assays based on carbapenemase characteristics have been developed, including modified Hodge test, Carba NP test, modified carbapenem inactivation method (mCIM), matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS), and other derivative experiments, which can be carried out in microbiological laboratory.4–6 For blaCphA-positive Aeromonas, the antimicrobial susceptibility testing by disk diffusion or MIC failed to fully display carbapenem-resistant phenotype. On the contrary, some phenotypic assays yield consistent results with genotypes.15,16 Here, it should be noted that the mCIM in conjunction with EDTA-modified carbapenem inactivation method (eCIM), recommended by CLSI, reveals perfect sensitivity and specificity in carbapenemase-producing Enterobacterales.5,26,27 However, whether mCIM and eCIM are suitable for the detection of carbapenemase in Aeromonas remains to be established.
In the present study, we systematically analyzed the antimicrobial resistance patterns of Aeromonas and compared the consistency of different methods for the antimicrobial susceptibility of carbapenems. Then, we detected the production of carbapenemase in Aeromonas by mCIM in conjunction with eCIM, simplified carbapenem inactivation method (sCIM),28 and compared them with polymerase chain reaction (PCR) to investigate the potential resistance mechanisms to carbapenems. To the best of our knowledge, this is the first confirmation that mCIM and eCIM are accurate and effective for the detection of multiple carbapenemases in Aeromonas. Moreover, we designed a simple method, carbapenem inactivation method using supernatant (CIM-s), to detect the activity of carbapenemase secreted in the medium.
Materials and Methods
Bacterial Isolates and Antibiotic Susceptibility Testing
A total of 131 clinical preserved Aeromonas strains from The Second Affiliated Hospital of Chongqing Medical University were used in this study. These isolates were mainly from the liver abscess drainage (36/131), followed by wound secretions (19/131), blood (18/131), pus (14/131), stool (13/131), bile (11/131), urine (8/131), sputum (7/131), and ascitic fluid (5/131). All isolates were stored in skim milk supplemented with 10% glycerine at −80°C. And these strains were subcultured twice at 35°C for 18–24 h using blood agar plates before testing. The organisms were identified to the species level by Vitek-2 Compact system (bioMérieux, Hazelwood, MO, USA) and were confirmed by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) according to the manufacturer’s instructions. The antibiotic susceptibility testing was performed by using Vitek-2 AST-GN67 and AST-XN04 cards (bioMérieux, Hazelwood, MO, USA). As the CLSI guideline M45 recommended, the susceptibilities of imipenem and meropenem were confirmed by the disk diffusion method on Mueller–Hinton agar (MHA) and broth microdilution method using cation-adjusted Mueller–Hinton broth (CAMHB).29 The results were interpreted according to the CLSI guideline M45.29 This study does not include any patient information.
Molecular Identification of Carbapenemase-Encoding Genes by PCR
For analysis of carbapenemase-encoding genes, bacterial genomic DNA was extracted using a Spin Column Bacterial Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China). Then, PCR was performed using gene-specific primers and PCR kit (Takara Bio Inc. Otsu, Japan) in BIO-RAD MyCycler PCR system (Bio-Rad Laboratories, Inc., California, USA). The PCR thermal cycling procedures were as follows: 30 seconds at 94°C for enzyme activation, 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute, finishing with a 10-minute step at 72°C for final extension. The sequences and product lengths of primers are summarized in Table 1. The PCR amplified products were analyzed by electrophoresis on 1.5% agarose gels and visualized in a BIO-RAD Gel Doc XR+ gel imaging system (Bio-Rad Laboratories, Inc., California, USA). All positive products of the carbapenemase-encoding genes were verified by commercial direct sequencing (Sangon Biotech, Shanghai, China).
Table 1.
Primer Sequences for Carbapenemase-Encoding Genes
| Gene | Primer | Nucleotide Sequence (5ʹ–3ʹ) | Size (bp) |
|---|---|---|---|
| CphA | F | GCTTAGAGCTCCTAAGGAGCAAGATGAAAGGTTGG | 720 |
| R | GCATAGGTACCTTATGACTGGGGTGCGGCCTTG | ||
| KPC-2 | F | CATTCAAGGGCTTTCTTGCTGC | 538 |
| R | ACGACGGCATAGTCATTTGC | ||
| NDM-1 | F | CAGCACACTTCCTATCTC | 292 |
| R | CCGCAACCATCCCCTCTT | ||
| VIM | F | GATGGTGTTTGGTCGCATA | 390 |
| R | CGAATGCGCAGCACCAG | ||
| SME | F | AACGGCTTCATTTTTGTTTAG | 820 |
| R | GCTTCCGCAATAGTTTTATCA | ||
| IMP | F | CATGGTTTGGTTGTTCTTGT | 488 |
| R | ATAATTTAGCGGACTTTGGC | ||
| GES | F | GCTTCATTCACGCACTATT | 323 |
| R | CGATGCTAGAAACCGCTC | ||
| IMI | F | TGCGGTCGATTGGAGATAAA | 399 |
| R | CGATTCTTGAAGCTTCTGCG | ||
| GIM | F | CGAATGGGTTGGTAGTTCTGGATAATAATC | 198 |
| R | ATGTGTATGTAGGAATTGACTTTGAATTTAGC | ||
| SIM | F | TACAAGGGATTCGGCATCG | 571 |
| R | TAATGGCCTGTTCCCATGTG | ||
| OXA-48-like | F | TTGGTGGCATCGATTATCGG | 438 |
| R | GAGCACTTCTTTTGTGATGGC |
mCIM in Conjunction with eCIM for Carbapenemase Production in Aeromonas
The mCIM and eCIM for Aeromonas were performed according to the method recommended by CLSI for Enterobacterales.30 Two 1-μL loopfuls of bacteria from an overnight blood agar plate were emulsified in 2 mL trypticase soy broth (TSB) or 2 mL TSB added 20 μL of the 0.5 M EDTA, respectively. Subsequently, a 10-μg meropenem disk (Oxoid Ltd, Basingstoke, UK) was immersed in each suspension and incubated at 35°C in ambient air for 4 h. A MHA plate was inoculated with a 0.5 McFarland suspension of E. coli ATCC 25922 as for the routine disk diffusion procedure. The meropenem disks were separately removed from the TSB or TSB-EDTA suspension and placed on the same MHA plate previously inoculated with the meropenem-susceptible E. coli ATCC 25922 indicator strain. All plates were incubated at 35°C in ambient air for 18–24 h. Following incubation, the zones of inhibition were measured as for the routine disk diffusion method. The results were interpreted according to the CLSI guideline M100 for mCIM and eCIM in Enterobacterales.30 The mCIM and eCIM using 10-μL loopful of Aeromonas were also carried out. The test was performed in triplicate, and the results were interpreted by three independent technicians.
sCIM for Carbapenemase Production in Aeromonas
According to the method described by Jing et al,28 we performed sCIM for carbapenemase production in Aeromonas. A MHA plate was inoculated with a 0.5 McFarland suspension of E. coli ATCC 25922 as for the routine disk diffusion procedure. One to three colonies of test organisms from an overnight blood agar plate were smeared onto one side of 10-μg imipenem disk (Oxoid Ltd, Basingstoke, UK). Then, the side of imipenem disk containing test bacteria was placed on the MHA plate previously inoculated with the imipenem-susceptible E. coli ATCC 25922 indicator strain. An imipenem disk without any bacteria was placed on the same MHA plate as the control. After the plates were incubated at 35°C in ambient air for 16–18 h, the zones of inhibition were measured as for the routine disk diffusion method. A zone diameter of 6–20 mm or presence of pinpoint colonies within a ≤22 mm zone diameter was considered to be carbapenemase positive. A zone diameter of ≥26 mm was considered to be carbapenemase negative. A zone diameter of 23–25 mm was considered to be carbapenemase indeterminate.28 All strains were tested in triplicates, and the results were interpreted by three independent technicians.
CIM-s for the Detection of Carbapenemase Activity in the Medium
A 10-μL loopful of bacteria from an overnight blood agar plate were emulsified in 10 mL TSB and incubated at 35°C in ambient air for 18–24 h. The suspension was centrifuged at 8000 rpm for 10 min to collect the supernatant. A 10-μg meropenem disk was immersed in each 2 mL centrifuged supernatant or 2 mL supernatant added 20 μL of the 0.5 M EDTA, respectively, instead of incubating the disk in the organism suspension as in the mCIM and eCIM. After incubation at 35°C in ambient air for 1 h, 2 h and 4 h, respectively, the meropenem disks were separately removed from the supernatant or supernatant-EDTA and placed on the MHA plate previously inoculated with a 0.5 McFarland suspension of E. coli ATCC 25922. Then, the plates were incubated at 35°C in ambient air for 18–24 h. The zone diameters were measured as for the routine disk diffusion method. A meropenem disk, immersed in 2 mL TSB, was placed on the MHA plate as the control. And, 100 μL of supernatant was inoculated onto the blood agar plate to check that the culture supernatants were bacteria free. The results were interpreted according to the CLSI M100 for mCIM and eCIM in Enterobacterales.30 The test was performed in triplicate, and three independent technicians interpreted the results.
Statistical Analysis
Statistical analyses were performed with the SPSS software version 22.0 (SPSS, Inc., Chicago, IL, USA). Descriptive statistics were presented as number and percentage. McNemar’s test was used to compare the difference of resistance rates of imipenem and meropenem detected by different methods. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy value were calculated using genes’ results as the gold standard. The indeterminate results were considered false negative in this study.
Results
Antibiotic Susceptibility of Carbapenem in Aeromonas
All 131 isolates were identified as A. hydrophila, A. caviae and A. sobria by Vitek-2 Compact system, and further confirmed as A. hydrophila, A. caviae and A. veronii by MALDI-TOF MS. However, the species results of the two methods were not completely consistent. The isolates demonstrated high resistance to imipenem (54.96%), meropenem (46.56%) and doripenem (43.51%), but were less so for ertapenem (17.56%). We also determined the susceptibilities of two main carbapenems, imipenem and meropenem, by the disk diffusion method and broth microdilution method. The resistance rates to imipenem and meropenem were 34.35% and 8.40% by the disk diffusion, and were 30.53% and 9.92% by the broth microdilution, respectively. The intermediate rates were dramatically increased. The antibiotic susceptibility results of imipenem and meropenem among the three methods were significant different (Tables 2 and 3, χ2=30.719, p<0.001 for imipenem and χ2=72.971, p<0.001 for meropenem).
Table 2.
Distribution of Imipenem Results Detected by Different Methods (n)
| PCR (n) | Vitek-2 Compact (mg/L) | Broth Microdilution (mg/L) | Disk Diffusion (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ≥4 | 2 | ≤1 | ≥4 | 2 | ≤1 | ≤19 | 20–22 | ≥23 | |
| Positive (79) | 72 | 3 | 4 | 40 | 24 | 15 | 45 | 26 | 8 |
| CphA (68) | 61 | 3 | 4 | 29 | 24 | 15 | 34 | 26 | 8 |
| KPC-2 (6) | 6 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 |
| NDM-1 (2) | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 |
| KPC-2+CphA (3) | 3 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 |
| Negative (52) | 0 | 0 | 52 | 0 | 0 | 52 | 0 | 0 | 52 |
Table 3.
Distribution of Meropenem Results Detected by Different Methods (n)
| PCR (n) | Vitek-2 Compact (mg/L) | Broth Microdilution (mg/L) | Disk Diffusion (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ≥4 | 2 | ≤1 | ≥4 | 2 | ≤1 | ≤19 | 20–22 | ≥23 | |
| Positive (79) | 68 | 2 | 9 | 13 | 7 | 59 | 11 | 7 | 61 |
| CphA (68) | 57 | 2 | 9 | 2 | 7 | 59 | 0 | 7 | 61 |
| KPC-2 (6) | 6 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 |
| NDM-1 (2) | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 |
| KPC-2+CphA (3) | 3 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 |
| Negative (52) | 0 | 0 | 52 | 0 | 0 | 52 | 0 | 0 | 52 |
Aeromonas Harboring Diverse Carbapenemase-Encoding Genes
We detected the main carbapenemase-encoding genes, including class A carbapenemases (blaKPC-2/blaIMI/blaGES), class B carbapenemases (blaNDM-1/blaIMP/blaVIM/blaCphA) and class D carbapenemases (blaOXA-48-like), by PCR. Of the 131 isolates, 79 isolates (60.3%) contained carbapenemase genes, and 52 isolates (39.7%) were carbapenemase negative without any of the tested carbapenemase genes. Of the 79 carbapenemase-positive isolates, 68 harbored the carbapenemase genes blaCphA, 6 contained blaKPC-2, 2 harbored blaNDM-1 and 3 were positive for both blaKPC-2 and blaCphA simultaneously. The PCR-positive products of the carbapenemase-encoding genes were verified by sequencing.
mCIM in Conjunction with eCIM Can Accurately Detect Carbapenemase in Aeromonas
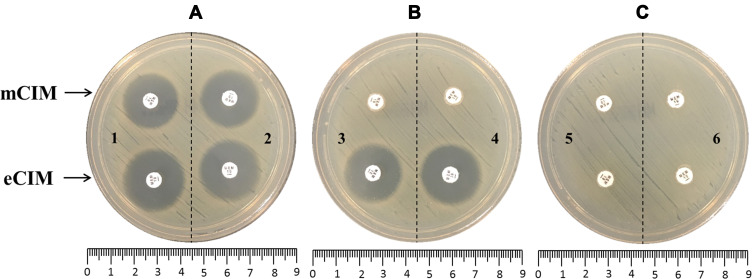
Of the 131 isolates, 79 isolates were carbapenemase positive and 52 isolates were carbapenemase negative by mCIM that were consistent with the PCR-based gene testing results. The inhibition zone diameters of 52 non-carbapenemase producers were 21–23 mm. In 79 PCR-positive isolates, 68 Aeromonas with blaCphA and 2 Aeromonas with blaNDM-1 exhibited mCIM and eCIM positive. Six Aeromonas contained blaKPC-2 and three Aeromonas harbored both blaKPC-2 and blaCphA displayed mCIM positive and eCIM negative (Figure 1A–C and Table 4). Both sensitivity and specificity of mCIM were 100% (Table 5). The test results using 1-μL and 10-μL loopfuls of bacteria were identical (data not shown).
Figure 1.
The representative photograph of mCIM and eCIM results in Aeromonas. (A) Negative results. 1, blank control, without bacteria; 2, Aeromonas without carbapenemase production. (B) mCIM positive and eCIM positive. 3, Aeromonas-producing CphA; 4, Aeromonas-producing NDM-1. (C) mCIM positive and eCIM negative. 5, Aeromonas-producing KPC-2; 6, Aeromonas-producing both KPC-2 and CphA.
Abbreviations: mCIM, modified carbapenem inactivation method; eCIM, EDTA-modified carbapenem inactivation method.
Table 4.
Comparison of mCIM, sCIM and CIM-s in the Detection of Carbapenemase
| PCR (n) | Zone (mm) | Positive (n, %) | Indeterminate (n, %) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| mCIM | sCIM | CIM-s* | mCIM | sCIM | CIM-s* | mCIM | sCIM | CIM-s* | |
| Positive (79) | 6–10 | 6–25 | 6–20 | 79(100) | 73(92.4) | 77(97.5) | 0(0) | 6(7.6) | 2(2.5) |
| CphA (68) | 6–10 | 6–25 | 6–20 | 68(100) | 62(91.2) | 66(97.1) | 0(0) | 6(8.2) | 2(2.9) |
| KPC-2 (6) | 6 | 6 | 6 | 6(100) | 6(100) | 6(100) | 0(0) | 0(0) | 0(0) |
| NDM-1 (2) | 6 | 6–10 | 6 | 2(100) | 2(100) | 2(100) | 0(0) | 0(0) | 0(0) |
| KPC-2+CphA (3) | 6 | 6 | 6 | 3(100) | 3(100) | 3(100) | 0(0) | 0(0) | 0(0) |
| Negative (52) | 21–23 | 26–30 | 21–23 | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
Notes: *The results were determined after incubation at 35°C in ambient air for 4h.
Abbreviations: mCIM, modified carbapenem inactivation method; sCIM, simplified carbapenem inactivation method; CIM-s, carbapenem inactivation method using supernatant.
Table 5.
Sensitivity and Specificity of Phenotypic Tests and IMP (95% CI)
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | |
|---|---|---|---|---|---|
| mCIM | 100 (95.4–100) | 100 (93.2–100) | 100 (95.4–100) | 100 (93.2–100) | 100 (97.2–100) |
| sCIM | 92.4 (84.2–97.2) | 100 (93.2–100) | 100 (95.1–100) | 89.7 (78.8–96.1) | 95.4 (90.3–98.3) |
| CIM-s* | 97.5 (91.2–99.7) | 100 (93.2–100) | 100 (95.3–100) | 96.3 (87.3–99.6) | 98.5 (94.6–99.8) |
| IMP** | 91.1 (82.6–96.4) | 100 (93.2–100) | 100 (95.0–100) | 88.1 (77.1–95.1) | 94.7 (89.3–97.8) |
Notes: *The results were determined after incubation at 35°C in ambient air for 4 h. **Resistance to imipenem tested by Vitek-2 compact system.
Abbreviations: mCIM, modified carbapenem inactivation method; sCIM, simplified carbapenem inactivation method; CIM-s, carbapenem inactivation method using supernatant; IMP, imipenem; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
The Sensitivity and Specificity of sCIM
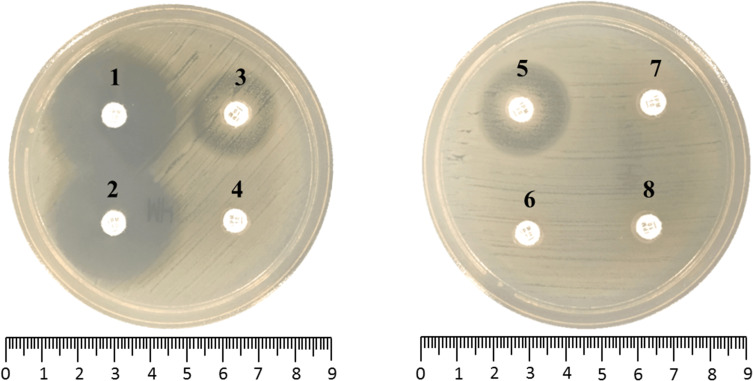
The zone diameters of 52 carbapenemase gene-negative isolates were all ≥26 mm. In 79 carbapenemase gene-positive isolates, 73 isolates of zone diameters were 6–20 mm or presence of pinpoint colonies within a ≤22 mm zone diameter that was considered to be carbapenemase positive by sCIM. Among the 73 isolates that were positive by sCIM, the zone diameters of Aeromonas expressing blaKPC-2, blaNDM-1 and blaKPC-2+CphA were all 6mm. However, the zone diameters of Aeromonas, harboring blaCphA alone, ranged from 6 to 20 mm. The other six gene-positive isolates displayed the zone diameters of 23–25 mm that were considered to be carbapenemase indeterminate (Figure 2 and Table 4). The carbapenemase indeterminate isolates were all blaCphA positive. The sensitivity and specificity were 92.4% and 100%, respectively (Table 5).
Figure 2.
The representative photograph of sCIM results in Aeromonas. 1, blank control. 2, negative result, non-carbapenemase-producing Aeromonas. 3–4, positive results, CphA-producing Aeromonas. 5, indeterminate result, CphA-producing Aeromonas. 6–8, positive results, Aeromonas-producing KPC-2 (6), NDM-1 (7) or KPC-2+CphA (8).
Abbreviation: sCIM, simplified carbapenem inactivation method.
The Results of CIM-s in Aeromonas
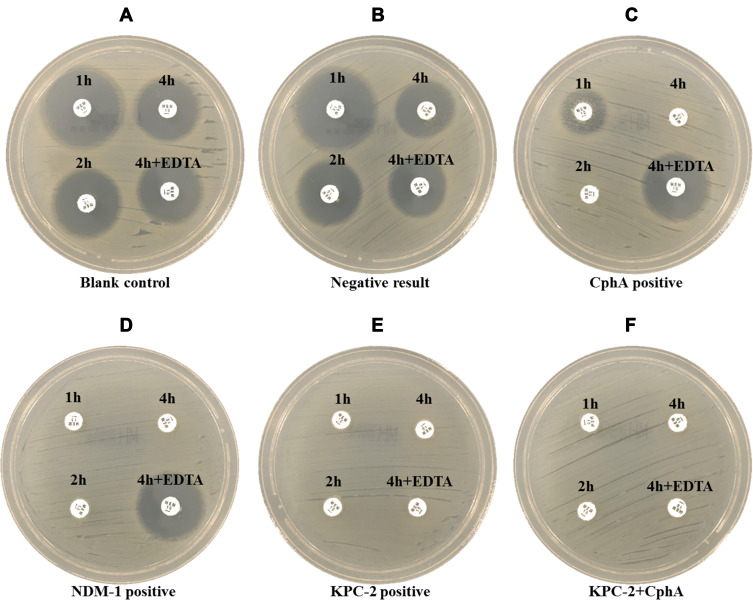
According to the results of CIM-s, we speculated that KPC-2, NDM-1 and CphA were secreted carbapenemases. The culture supernatants of isolates that were positive for carbapenemase genes were screened for secreted carbapenemase activity. With the prolongation of incubation time, the zone diameters were decreased. After incubation for 1 h, the zone diameters of 36 isolates were in the positive range. After incubation for 2 h, the positive results increased to 58 strains. At the end of 4 h incubation, 77 isolates displayed carbapenemase activity, yet there were still two isolates with blaCphA that showed indeterminate results. Adding 0.5M EDTA, the zone diameters of 66 isolates with blaCphA and 2 isolates with blaNDM-1 were expanded ≥5 mm. Six isolates with blaKPC-2 and 3 isolates with blaKPC-2+CphA had no significant change with the zone diameters expanding <4 mm. In this process, the zone diameters of 52 isolates without carbapenemase genes were larger than 21 mm, which was interpreted as carbapenemase negative (Figure 3A–F and Table 4). The CIM-s results after 4 h incubation were more consistent with carbapenemase gene testing results. The sensitivity reached 97.5%, and the specificity was 100% (Table 5).
Figure 3.
The representative photograph of CIM-s results in Aeromonas. The meropenem disks were immersed in 2mL supernatant or supernatant with 0.5M EDTA and incubated for 1h, 2h and 4h respectively. (A) Blank control. (B) Negative result. (C–F) Positive results, CphA (C) NDM-1 (D), KPC-2 (E) or KPC-2 + CphA (F).
Abbreviation: CIM-s, carbapenem inactivation method using supernatant.
Discussion
The taxonomy of the genus Aeromonas is quite complicated. It is difficult to accurately identify Aeromonas to species level by traditional biochemical tests and automated systems,31,32 which were confirmed by evaluating the accuracy of six commercial systems for identifying clinical Aeromonas isolates.33 MALDI-TOF MS is a rapid and effective method for identification of Aeromonas genus. However, the species level results are inconsistent with the housekeeping gene sequencing.34,35 In this study, we utilized Vitek-2 Compact system and MALDI-TOF MS to identify 131 clinical isolates as Aeromonas, although there were some differences at the species level. In subsequent studies, we did not further distinguish the species, referring to all isolates as Aeromonas.
The carbapenemase gene detection results demonstrated that blaCphA, accounting for 60%, was the major carbapenemase gene in Aeromonas. In other studies, the prevalence of blaCphA was even higher, reaching 69%–77%.15,16,36 As blaCphA is an MBL located in the bacterial chromosome, it reduces the risk of horizontal transmission of resistance genes compared to plasmid-mediated genes.37 Except blaCphA, we also detected nine strains of Aeromonas carrying blaKPC-2-encoding gene (three of which contained both blaKPC-2 and blaCphA) and two strains carrying blaNDM-1-encoding gene. BlaKPC-2 and blaNDM-1 are the two major carbapenemases genes in Enterobacteriaceae. These two genes are usually located on plasmids and easily spread between different bacteria. The plasmid-mediated horizontal transmission of carbapenem resistance genes is the main reason for the global prevalence of carbapenem-resistant Enterobacteriaceae (CRE).3,6 We speculate that these two carbapenem-resistant genes of blaKPC-2 and blaNDM-1 in Aeromonas may originate from Enterobacteriaceae. However, so far, the specific source and transmission mechanism remains to be determined.
As an opportunistic pathogen, Aeromonas can cause a variety of infections. Effective treatment of Aeromonas infection has become an urgent issue to be faced. Quinolones and the third- or fourth-generation cephalosporins could be used as the favorite antibiotics to empirically treat Aeromonas infection.38,39 Despite the fact that carbapenems can be effective drugs in the treatment of Gram-negative bacterial infection, they should be used prudently for Aeromonas. CphA, high prevalent in Aeromonas, has substrate specificity for carbapenem antibiotics, which can hydrolyze carbapenems, resulting in treatment failure. Moreover, for the blaCphA-positive strains, the routine antibiotic susceptibility testing could not completely detect the carbapenem resistance phenotype, which had a high false-negative rate.15,16,40 Our results are consistent with previous studies. In 79 carbapenemase producers, the result of imipenem testing by VITEK2 was the best indicator with only 6 blaCphA-positive strains not detected as resistant. The resistance rates of imipenem and meropenem, tested by disk diffusion or broth microdilution, were significantly reduced, and only by increasing the inoculum size could the resistance results be shown.15,40 The possible reason is that with standard inoculum, it cannot express enough CphA to inactivate carbapenem, or there might be gene modification to influence the expression and activity of CphA. The specific mechanism needs further study. Therefore, routine disk diffusion or broth microdilution is not very suitable for conventional application. It is very important to find a simple and effective method to identify carbapenem resistance phenotype in Aeromonas.
Various phenotypic tests have been used in carbapenemase-producing Gram-negative bacilli with the advantages of user-friendly, affordable, accurate, and feasible for implementation in clinical microbiological laboratories.4,27 The application of phenotypic test in Aeromonas may be an ideal method to identify carbapenemase producer. Some researchers have found that MHT and Carba NP could detect carbapenemase activity in 97% of blaCphA-positive isolates.15,16 However, MHT and Carba NP test cannot distinguish specific carbapenemase types. Moreover, the above studies only tested the efficacy for CphA, and it was not known whether other types of carbapenemases in Aeromonas could be detected. mCIM, based on the growth test in the presence of carbapenem antibiotics,4 is a phenotypic screening test recommended by CLSI to detect CRE with high specificity and sensitivity. mCIM in combination with eCIM can distinguish serine carbapenemase and MBL except when co-produced serine carbapenemase and MBL organism that will emerge false negative of eCIM. Our results were consistent with the above description. In view of the high prevalence of blaCphA in Aeromonas, it cannot be ruled out whether CphA MBL is co-produced, when the phenotypic test is interpreted as serine carbapenemase production. In our study, the sensitivity and specificity of mCIM for Aeromonas were both 100%, which were better than MHT and Carba NP test. The volume of bacteria used in Enterobacterales and Pseudomonas aeruginosa by mCIM is different. Therefore, we used 1-μl and 10-μl loopfuls of bacteria to carry out mCIM and eCIM in Aeromonas at the same time. The two results were completely consistent, so the 1-μl was considered sufficient.
sCIM is another phenotypic test for the detection of carbapenemase based on mCIM with improvement of experimental technique.28 In the sCIM, the organism is directly smeared onto an imipenem disk. The carbapenemases-producing isolate released carbapenemases to hydrolyze imipenem immediately, causing a decrease in the zone diameter. sCIM seems to be an effective method to identify the carbapenemase-producing isolates in the Gram-negative bacilli.28 We applied this method to Aeromonas. Whereas for blaCphA-positive isolates, there were six results interpreted as carbapenemase indeterminate result by sCIM. We speculate that the expression levels of CphA carbapenemase were different in Aeromonas, and the hydrolytic activity of CphA was weaker than KPC-2 and NDM-1. These strains could not effectively inactivate imipenem to reduce the inhibition zones. For those isolates of blaCphA positive with sCIM indeterminate results, the results of mCIM were all positive. The cultural environment of the two methods was distinct. In mCIM, the organisms were grown in liquid TSB medium, which might be more conducive to release of CphA carbapenemase to effectively inactivate the carbapenem antibiotics. In sCIM, it was not beneficial to standardize as the bacteria to be tested were directly smeared onto an imipenem disk. The smeared bacteria quantity will be different by distinct operators, and the concentration of bacteria on antibiotic disk could affect the size of zone.28 Compared with sCIM, which cannot completely detect the Aeromonas harboring blaCphA, mCIM is a more suitable method to identify and detect the carbapenemase in Aeromonas.
To further verify the hydrolytic activity of carbapenemase, especially CphA, secreted into the medium, we made further improvement on the basis of mCIM test, called CIM-s. We used the supernatant of bacteria cultured overnight in TSB broth, instead of bacterial suspension, to inactivate meropenem disk, and observed the change of inhibition zone in E. coli ATCC 25922 plates. We found that the supernatant containing KPC-2 and NDM-1 showed positive results after inactivation for 1 h. However, the positive rate of CphA supernatant was only 36.8% in 1 h. As the inactivation time extended to 4 h, the positive rate reached 97.1%. Only two strains showed indeterminate results. The carbapenem hydrolytic activity of KPC-2 and NDM-1 appeared significantly higher than that of CphA. By observing the inactivation of meropenem in the supernatant of blaCphA-positive strains in CIM-s, it was confirmed that the expression and secretion ability of CphA was different in Aeromonas. There were strains with weak expression of CphA. After adding EDTA, the activity of NDM-1 and CphA could be inhibited, while KPC-2 was not affected. Therefore, the carbapenemase type could be preliminarily distinguished by CIM-s in combination with eCIM-s. Like eCIM, isolates containing both serine carbapenemase and MBL, such as KPC-2 and CphA, could not be detected accurately, only showing CIM-s positive.
By comparing mCIM, sCIM and CIM-s, the operation of sCIM is the most convenient as it does not require standard strains, TSB medium and extra incubation process. However, the sensitivity of sCIM is low, and sCIM cannot distinguish carbapenemase type. CIM-s needs a longer time and complex operation, which is not an ideal method to detect the carbapenemase, but it can be confirmed that the carbapenemase is secreted into the medium to exhibit hydrolytic activity. mCIM can detect a variety of carbapenemases in Aeromonas, including the weak expression of CphA, and the results are consistent with the gene detection. CphA is an inducible carbapenemase.41,42 After the addition of carbapenem antibiotics, the expression and secretion of CphA were promoted, and then effectively inactivate meropenem disk. For patients infected with Aeromonas, there is a high amount of bacteria in the tissue, combined with the complex microenvironment, which may be conducive to the release of CphA, resulting in the continuous presence of CphA in the surrounding medium. The use of carbapenem drugs may further induce and promote the expression of CphA, leading to treatment failure.15,42 When it is considered to treat Aeromonas infection with carbapenems, it is recommended to perform mCIM combined with eCIM to detect whether it is a carbapenemase-producing strain.
Our study also had several limitations. All the isolates were from The Second Affiliated Hospital of Chongqing Medical University. The number of isolates to be verified was relatively insufficient. We did not identify Aeromonas to species level by housekeeping gene sequencing to compare interspecific differences. The carbapenemase gene profiles in this study were limited, only containing KPC-2, NDM-1 and CphA. A multicenter and large sample size verification is required to further confirm our results.
Conclusion
The infections caused by Aeromonas are complex and diverse. The use of carbapenem antibiotics should be cautious as the routine antibiotic susceptibility testing cannot detect carbapenem resistance phenotype completely for carbapenemase-producing Aeromonas. Our research expands the application of mCIM and eCIM. mCIM in combination with eCIM can effectively detect multiple carbapenemases in Aeromonas, which is suitable for carrying out in the microbiological laboratory. CIM-s reveals that these carbapenemases are secreted into the medium to perform their hydrolytic activities. In consideration of the high prevalence of carbapenemase, especially CphA, it is suggested to routinely apply mCIM and eCIM in Aeromonas as a supplement to the results of antibiotic susceptibility testing.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81802062 and 31600135), the Chongqing Municipal Education Commission Grant (KJQN201800402) and the Chongqing Science and Technology Commission Grant (cstc2017JcyjAX0065).
Ethical Approval
The clinical samples were part of the routine hospital laboratory procedure, not specifically isolated for this study, so an institutional review board was not required.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gajdács M. The concept of an ideal antibiotic: implications for drug design. Molecules. 2019;24(5):892. doi: 10.3390/molecules24050892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajdács M, Bátori Z, Ábrók M, Lázár A, Burián K. Characterization of resistance in gram-negative urinary isolates using existing and novel indicators of clinical relevance: a 10-year data analysis. Life. 2020;10(2):16. doi: 10.3390/life10020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonomo RA, Burd EM, Conly J, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2018;66(8):1290–1297. doi: 10.1093/cid/cix893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamma PD, Simner PJ. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol. 2018;56(11):e01140–e01218. doi: 10.1128/JCM.01140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sfeir MM, Hayden JA, Fauntleroy KA, et al. EDTA-modified carbapenem inactivation method: a phenotypic method for detecting metallo-β-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2019;57(5):e01757–e01818. doi: 10.1128/JCM.01757-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: detection and antimicrobial therapy. Front Microbiol. 2019;10:1823. doi: 10.3389/fmicb.2019.01823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juhász J, Ligeti B, Gajdács M, et al. Colonization dynamics of multidrug-resistant Klebsiella pneumonia are dictated by microbiota-cluster group behavior over individual antibiotic susceptibility: a metataxonomic analysis. Antibiotics. 2021;10(3):268. doi: 10.3390/antibiotics10030268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khor WC, Puah SM, Koh TH, Tan JAMA, Puthucheary SD, Chua KH. Comparison of clinical isolates of Aeromonas from Singapore and Malaysia with regard to molecular identification, virulence, and antimicrobial profiles. Microb Drug Resist. 2018;24(4):469–478. doi: 10.1089/mdr.2017.0083 [DOI] [PubMed] [Google Scholar]
- 9.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23(1):35–73. doi: 10.1128/CMR.00039-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadi N, Mahmoodi Z, Emami A, Malekzadegan Y, Valadbeygi T. Isolation and molecular identification of Aeromonas wound infection in Iranian burn patients. Infect Disord Drug Targets. 2019;19(3):269–273. doi: 10.2174/1871526518666180903165823 [DOI] [PubMed] [Google Scholar]
- 11.Olszewski AE, Karandikar MV, Surana NK. Aeromonas as a cause of purulent folliculitis: a case report and review of the literature. J Pediatric Infect Dis Soc. 2017;6(1):e1–e3. doi: 10.1093/jpids/piw073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouquet H, Guimas M, Teulier S, Testaert H, Bergot E, Reviron-Rabec L. Pneumopathie à Aeromonas liée aux noyades [Near-drowning associated Aeromonas pneumonia]. Rev Mal Respir. 2018;35(9):959–962. doi: 10.1016/j.rmr.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 13.Nolla-Salas J, Codina-Calero J, Vallés-Angulo S, et al. Clinical significance and outcome of Aeromonas spp. infections among 204 adult patients. Eur J Clin Microbiol Infect Dis. 2017;36(8):1393–1403. doi: 10.1007/s10096-017-2945-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajdács M. Resistance trends and epidemiology of Aeromonas and Plesiomonas infections (RETEPAPI): a 10-year retrospective survey. Infect Dis. 2019;51(9):710–713. doi: 10.1080/23744235.2019.1640389 [DOI] [PubMed] [Google Scholar]
- 15.Wu CJ, Chen PL, Wu JJ, et al. Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic Aeromonas isolates. J Med Microbiol. 2012;61(5):712–719. doi: 10.1099/jmm.0.038323-0 [DOI] [PubMed] [Google Scholar]
- 16.Sinclair HA, Heney C, Sidjabat HE, et al. Genotypic and phenotypic identification of Aeromonas species and CphA-mediated carbapenem resistance in Queensland, Australia. Diagn Microbiol Infect Dis. 2016;85(1):98–101. doi: 10.1016/j.diagmicrobio.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 17.Neuwirth C, Siebor E, Robin F, Bonnet R. First occurrence of an IMP metallo-beta-lactamase in Aeromonas caviae: IMP-19 in an isolate from France. Antimicrob Agents Chemother. 2007;51(12):4486–4488. doi: 10.1128/AAC.01462-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libisch B, Giske CG, Kovács B, Tóth TG, Füzi M. Identification of the first VIM metallo-beta-lactamase-producing multiresistant Aeromonas hydrophila strain. J Clin Microbiol. 2008;46(5):1878–1880. doi: 10.1128/JCM.00047-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montezzi LF, Campana EH, Corrêa LL, et al. Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int J Antimicrob Agents. 2015;45(2):174–177. doi: 10.1016/j.ijantimicag.2014.10.016 [DOI] [PubMed] [Google Scholar]
- 20.Hughes HY, Conlan SP, Lau AF, et al. Detection and whole-genome sequencing of carbapenemase-producing Aeromonas hydrophila isolates from routine perirectal surveillance culture. J Clin Microbiol. 2016;54(4):1167–1170. doi: 10.1128/JCM.03229-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Wang X, Yu X, et al. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ Pollut. 2018;235:931–937. doi: 10.1016/j.envpol.2017.12.084 [DOI] [PubMed] [Google Scholar]
- 22.Anandan S, Gopi R, Devanga Ragupathi NK, et al. First report of blaOXA-181-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: threat for rapid dissemination. J Glob Antimicrob Resist. 2017;10:310–314. doi: 10.1016/j.jgar.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 23.Uechi K, Tada T, Sawachi Y, et al. A carbapenem-resistant clinical isolate of Aeromonas hydrophila in Japan harbouring an acquired gene encoding GES-24 β-lactamase. J Med Microbiol. 2018;67(11):1535–1537. doi: 10.1099/jmm.0.000842 [DOI] [PubMed] [Google Scholar]
- 24.Banerjee T, Pal S, Das A. Emergence of Aeromonas spp. harboring multiple carbapenemase-encoding genes from hospital sewage. J Lab Physicians. 2017;9(1):64–65. doi: 10.4103/0974-2727.187924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrabák J, Chudáčková E, Papagiannitsis CC. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect. 2014;20(9):839–853. doi: 10.1111/1469-0691.12678 [DOI] [PubMed] [Google Scholar]
- 26.Pierce VM, Simner PJ, Lonsway DR, et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol. 2017;55(8):2321–2333. doi: 10.1128/JCM.00193-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gajdács M, Ábrók M, Lázár A, et al. Detection of VIM, NDM and OXA-48 producing carbapenem resistant Enterobacterales among clinical isolates in Southern Hungary. Acta Microbiol Immunol Hung. 2020;67(4):209–215. doi: 10.1556/030.2020.01181 [DOI] [PubMed] [Google Scholar]
- 28.Jing X, Zhou H, Min X, et al. The Simplified Carbapenem Inactivation Method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative bacilli. Front Microbiol. 2018;9:2391. doi: 10.3389/fmicb.2018.02391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. 3rd ed. M45. PA: CLSI; 2016 [DOI] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: 30th Edition CLSI Supplement M100. PA: CLSI; 2020. [Google Scholar]
- 31.Puah SM, Khor WC, Kee BP, Tan JA, Puthucheary SD, Chua KH. Development of a species-specific PCR-RFLP targeting rpoD gene fragment for discrimination of Aeromonas species. J Med Microbiol. 2018;67(9):1271–1278. doi: 10.1099/jmm.0.000796 [DOI] [PubMed] [Google Scholar]
- 32.Gonçalves Pessoa RB, de Oliveira WF, Marques DSC, Dos Santos Correia MT, de Carvalho EVMM, Coelho LCBB. The genus Aeromonas: a general approach. Microb Pathog. 2019;130:81–94. doi: 10.1016/j.micpath.2019.02.036 [DOI] [PubMed] [Google Scholar]
- 33.Lamy B, Laurent F, Verdier I, et al. Accuracy of 6 commercial systems for identifying clinical Aeromonas isolates. Diagn Microbiol Infect Dis. 2010;67(1):9–14. doi: 10.1016/j.diagmicrobio.2009.12.012 [DOI] [PubMed] [Google Scholar]
- 34.Lamy B, Kodjo A, Laurent F; ColBVH Study Group. Identification of Aeromonas isolates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Diagn Microbiol Infect Dis. 2011;71(1):1–5. doi: 10.1016/j.diagmicrobio.2011.04.014 [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Sancho M, Cerdá I, Fernández-Bravo A, et al. Limited performance of MALDI-TOF for identification of fish Aeromonas isolates at species level. J Fish Dis. 2018;41(10):1485–1493. doi: 10.1111/jfd.12837 [DOI] [PubMed] [Google Scholar]
- 36.Wu CJ, Chen PL, Hsueh PR, et al. Clinical implications of species identification in monomicrobial Aeromonas bacteremia. PLoS One. 2015;10(2):e0117821. doi: 10.1371/journal.pone.0117821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams RJ, Mathys DA, Mollenkopf DF, Whittle A, Daniels JB, Wittum TE. Carbapenemase-producing Aeromonas veronii disseminated in the environment of an equine specialty hospital. Vector Borne Zoonotic Dis. 2017;17(6):439–442. doi: 10.1089/vbz.2016.2083 [DOI] [PubMed] [Google Scholar]
- 38.Rhee JY, Jung DS, Peck KR. Clinical and therapeutic implications of Aeromonas bacteremia: 14 years nation-wide experiences in Korea. Infect Chemother. 2016;48(4):274–284. doi: 10.3947/ic.2016.48.4.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker JL, Shaw JG. Aeromonas spp. clinical microbiology and disease. J Infect. 2011;62(2):109–118. doi: 10.1016/j.jinf.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 40.Rossolini GM, Zanchi A, Chiesurin A, Amicosante G, Satta G, Guglielmetti P. Distribution of cphA or related carbapenemase-encoding genes and production of carbapenemase activity in members of the genus Aeromonas. Antimicrob Agents Chemother. 1995;39(2):346–349. doi: 10.1128/AAC.39.2.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segatore B, Massidda O, Satta G, Setacci D, Amicosante G. High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob Agents Chemother. 1993;37(6):1324–1328. doi: 10.1128/AAC.37.6.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sánchez-Céspedes J, Figueras MJ, Aspiroz C, et al. Development of imipenem resistance in an Aeromonas veronii biovar sobria clinical isolate recovered from a patient with cholangitis. J Med Microbiol. 2009;58(4):451–455. doi: 10.1099/jmm.0.47804-0 [DOI] [PubMed] [Google Scholar]