Abstract
Lonicerae japonicae flos (LJF) is widely used for the treatment of inflammation-related diseases in traditional Chinese medicine (TCM). To clarify the anti-inflammatory mechanism of LJF, 29 compounds with high content in LJF were selected for network pharmacology. Then, a comprehensive network pharmacology strategy was implemented, which involved compound-inflammation-target construction, protein-protein interaction (PPI) network analysis, and enrichment analysis. Finally, molecular docking and in vitro experiments were performed to verify the anti-inflammatory activity and targets of the key compound. As a result, 279 inflammation-associated proteins were identified, which are mainly involved in the AGE/RAGE signaling pathway in diabetic complications, the HIF-1 signaling pathway, the PI3K-AKT signaling pathway, and EGFR tyrosine kinase inhibitor resistance. A total of 12 compounds were linked to more than 35 targets, including apigenin, kaempferol, quercetin, luteolin, and ferulic acid. The results of molecular docking showed that AKT has the most binding activity, exhibiting certain binding activity with 10 compounds, including vanillic acid, protocatechuic acid, secologanic acid, quercetin, and luteolin; the results of qRT-PCR and WB confirmed that two key compounds, secologanic acid and luteolin, could significantly decrease the secretion of TNF-α and the AKT expression of RAW264.7 murine macrophages stimulated by LPS (lipopolysaccharide). These results demonstrate that the comprehensive strategy can serve as a universal method to illustrate the anti-inflammatory mechanisms of traditional Chinese medicine by identifying the pathways or targets.
1. Introduction
Inflammation is the body's protective response to injury or infection, but insufficient or excessive inflammation increases the incidence of many diseases [1]. In the process of the inflammatory response, cytokines play an important bidirectional regulatory role. In the early stage of inflammation, proinflammatory cytokines, such as TNF-α, IL-8, IL-1β, and IFN-γ, are predominant, which can eliminate the threat of infection or trauma through activating a variety of immune cells and promoting the inflammatory response. In the late stage of inflammation, anti-inflammatory cytokines such as IL-10 and IL-13 are dominant, which can weaken and control the inflammatory response [2]. However, in some special cases, the bidirectional regulation pathway of cytokines is damaged, and proinflammatory cytokines continue to play their roles, leading to a large number of immune cells being activated in some parts of the body and even forming inflammatory storms (also known as cytokine storms) in severe cases [3]. Inflammatory storms may be one of the leading causes of severe complications and death in severely affected patients for some acute respiratory infections, such as COVID-19, SARS-COV (SARS), MERS-COV (Middle East respiratory syndrome), and influenza [4–6].
Lonicerae japonicae flos (LJF), the dried flower buds of Lonicerae japonicae Thunb, also called Japanese honeysuckle or jīn yín huā, possesses the functions of clearing heat, removing toxins, and dispersing wind-heat, which has been widely used in traditional Chinese medicine to treat various diseases such as cough, fever, sore throat, and influenza infection [7]. LJF has significant anti-inflammatory effects, especially for the prevention and treatment of upper respiratory inflammation [8, 9]. LJF contains a variety of compounds, such as phenolic acids, flavonoids, and iridoids [10]. Most of the studies that have investigated the anti-inflammatory substance material of LJF have mainly focused on the single compound or crude extracts [11, 12]; however, reports are scarce pertaining to the complex interactions between the Chinese herb and cellular proteins or the influence of their interactions on the functions and behaviors of the body [13, 14].
Network pharmacology is an emerging subject involving the construction of multilayer networks of disease phenotypes, genes, and drugs. Network pharmacology aids in the prediction of new drug targets, deciphering the mode of action, and exploring new drugs [15]. Considering the complexity of the components and functions of TCM, network pharmacology is considered to be an effective approach for identifying key targets and signaling pathways. Ephedra has been widely used to treat asthma in Asia; a strategy of network pharmacology combined with molecular docking and in vitro experiments was performed to predict the anti-inflammatory targets of ephedra in treating asthma; the results showed that SELE, IL-2, and CXCL10 are critical targets for ephedra against inflammation due to asthma [16]. In addition, Cui et al. [17] predicted the therapeutic targets of tanshinone I and cryptotanshinone against inflammation and investigated the pharmacological molecular mechanism in vitro using a network pharmacology-based strategy. In this study, a comprehensive network pharmacology strategy was carried out, which included the prediction of potential targets, pathway enrichment analysis, and molecular docking. Finally, a murine macrophage cell inflammatory model was built to confirm the predicted result. The entire design of this study is shown in Figure 1.
Figure 1.
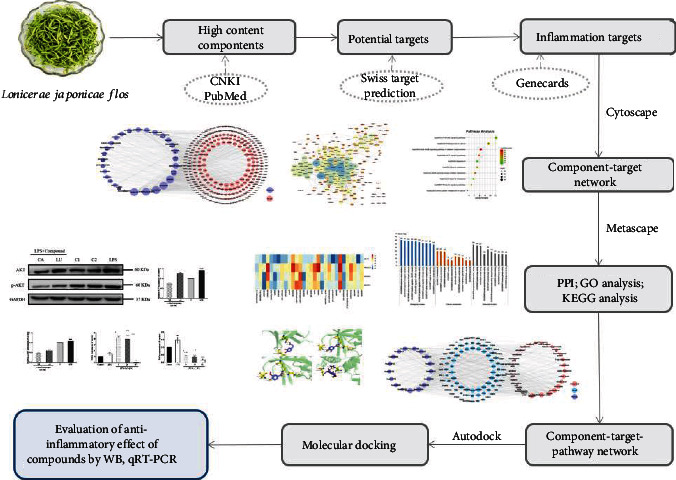
The integrated process of the network pharmacology-based method to identify the anti-inflammatory mechanism of Lonicerae japonicae flos.
2. Methods
2.1. Data Preparation and Construction
The anti-inflammatory chemical components of LJF were gathered from NCBI and PubMed, and a total of 29 components with high content in which were screened for network pharmacology and molecular docking analysis [18–39]. All compound structures were downloaded from the TCMSP (https://tcmspw.com/tcmsp.php) and NCBI PubChem databases (https://pubchem.ncbi.nlm.nih.gov/) and saved in mol2 format. ChemDraw software (v16.0) was used to draw 3D diagrams of the components that were not in the databases.
The online prediction platform SwissTargetPrediction (https://www.swisstargetprediction.ch/) can be used to identify potential targets of natural products and synthetic compounds. The mol2 format files of 29 compounds were imported into the SwissTargetPrediction and TCMSP database, respectively. Then, the prediction targets of the compounds were obtained by integrating the genes collected from the two platforms. The targets were converted into the UniProtKB format using Retrieve/ID mapping (http://www.uniprot.org/uploadlists/) for subsequent enrichment analysis. The GeneCards database (https://www.genecards.org/) was used for collecting inflammation-related targets by imputing in search item “inflammation.” The inflammation targets related to the 29 compounds of LJF were obtained by integrating the compound targets with the inflammation targets and taking the duplicate targets. The compound-target network was constructed using Cytoscape (v3.7.2) software.
2.2. Protein-Protein Interaction (PPI) Network
PPI is the process through which two or more proteins form a protein complex through noncovalent bonds [16]. The STRING (v11.0) database (https://string-db.org/) was used to build compound-inflammation target PPI network. “Homo sapiens” was chosen, and a scoring value of >0.9 was selected as the high confidence basis for protein interactions [40]. After eliminating the duplicates, the resultant data were imported into Cytoscape (v3.7.2) for establishing the protein-protein interaction (PPI) network; then, Cytohubba, a plugin of Cytoscape, was used to screen hub genes within the network.
2.3. GO and KEGG Pathway Enrichment Analysis
GO is a system widely used for the classification of gene functions and describing the functions of gene products [41]. KEGG pathway enrichment analysis links genomic information to higher-order functional information, suggesting the target that is mainly related to signal pathways. The Metascape database (https://metascape.org/gp/index.html) was used for performing GO and KEGG enrichment analysis. A P value of <0.01 is considered statistically significant, and a smaller P value indicates a more significant correlation.
2.4. Molecular Docking Analysis
Autodock 4.2.0 (http://autodock.scripps.edu/) was used to perform a docking simulation for verifying the credibility of predicted hub genes. All compound structures were downloaded from the ZINC database (http://zinc.docking.org/), and the 3D structures of the targets were downloaded from the RCSB PDB database (http://www.rcsb.org/). The simulation's scoring can be used for evaluating the degree of binding between the compound and the target; the smaller the score, the stronger the binding activity.
2.5. Cell Culture
The murine macrophage cell line RAW264.7 was purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. The cells were cultured in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 atmosphere.
2.6. Drugs and Reagents
Luteolin and secologanic acid were purchased from Shanghai Yuanye Biotechnology Co., Ltd. (Shanghai, China), dimethylsulfoxide (DMSO) and lipopolysaccharide (LPS) were purchased from Sigma (MO, USA), Dulbecco's modified Eagle's medium (DMEM) and trypsin were purchased from Gibco (USA), the BCA assay kit was purchased from SparkJade (Shandong, China), and antibodies were purchased from Wanleibio (China).
2.7. MTT Assay
The RAW264.7 cells in the treated groups were pretreated with secologanic acid (5, 10, 20, 40, and 80 μM) or luteolin (5, 10, 20, 40, and 80 μM) for 1 h, followed by treatment with LPS (1 μg/mL, Sigma) for 24 h. The cells in the control group or LPS group were incubated with only medium or LPS (1 μg/mL) for 24 h. After removing the culture solution, the cells were washed with PBS 3 times and incubated with 5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) at 37°C for 4 h. Then, 150 μL of DMSO was added to stop the reaction, and the reaction liquid was detected at 570 nm. Each experiment was repeated 3 times.
2.8. Quantitative Real-Time PCR
The RAW264.7 cells in the drug groups were pretreated with caffeic acid (5, 20, and 80 μM) or luteolin (5, 20, and 80 μM) for 1 h, followed by treatment with LPS (1 μg/mL) for 24 h. The cells in the control group or LPS group were incubated with only medium or LPS (1 μg/mL) for 24 h. The total RNA was collected using TRIzol (Invitrogen, CA, USA) reagent; the mRNA was reverse-transcribed to cDNA with a PrimeScript RT Reagent Kit (Tiangen, China); cDNA was mixed with Universal SYBR Green Fast qPCR Mix kits (ABclonal, China) and each primer pair to produce a 20 μL reaction mixture. The levels of β-actin were considered to be an endogenous control, and the expansion conditions were as follows: heating to 95°C for 3 min, 45 cycles of 95°C for 5 s, and 60°C for 32 s, and the 2−ΔΔCT method was used to calculate the relative fold changes. The primers are listed as follows: TNF-α, forward: 5′-CTCTTCTGCCTGCTGCACTTTG-3′ and reverse: 5′-ATGGGCTACAGGCTTGTCACTC-3′; β-actin, forward: 5′-CATTGCTGACAGGATGCAGAAGG-3′ and reverse: 5′-TGCTGGAAGGTGGACAGTGAGG-3′.
2.9. Western Blot
Western blotting was performed as reported previously [42]. The RAW264.7 cells in the drug groups were pretreated with secologanic acid or luteolin (20 μM) for 24 h and then treated with LPS (1 μg/mL) for 0.5 h. The cells in the control group or LPS group were separately incubated with medium or LPS (1 μg/mL) for 24 h. The cells were washed with ice-cold PBS and lysed in ice-cold RIPA lysis buffer containing PMSF. Homogenates were centrifuged at 12,000 rpm for 20 min at 4°C, filtered through a Millipore filter with a pore size of 0.45 μm, and stored at −80°C until use. Protein concentrations were quantified by BCA assay. The proteins (20 μg) were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked for 2 h at room temperature with 5% nonfat milk/TBST, probed with the appropriate primary antibodies (AKT, p-AKT, and GAPDH) (1 : 1000 dilution), and incubated overnight at 4°C. Then, the membranes were incubated with an HRP-conjugated secondary antibody (1 : 5000 dilution) for 1 h at room temperature, and bands were detected by enhanced chemiluminescence.
2.10. Statistical Analysis
Integrated band intensities in the western blot analysis were determined using the ImageJ software. All data are presented as X ± SD. Differences between the groups were analyzed by one-way analysis of variance, followed by the least significant difference test using IBM SPSS Statistics (v.25.0) software. A P value of <0.05 was considered statistically significant.
3. Results and Discussion
3.1. Data Preparation and Construction
More than 217 compounds were isolated and identified from LJF. Among them, 58 compounds have anti-inflammatory effects. A total of 29 compounds (Figure 2) with high content, including flavonoids (compounds 1-9), iridoids (compounds 10-18), volatile oils (compounds 19-21), and organic acids (compounds 22-29), were selected for network pharmacology and molecular docking research. Collectively, 1298 target proteins were retrieved from the SwissTargetPrediction database. After overlapping, 317 protein targets were converted to 317 UniProtKB identifiers by employing ID mapping (http://www.uniprot.org/uploadlists/) for the following enrichment analysis. A total of 279 repeated targets (Figure 3) were selected as LJF inflammation-related targets by comparing the 317 target proteins with the 9921 inflammation-related targets obtained from the GeneCards database. The compound-target (C-T) network was built in Cytoscape (3.7.2) software by converting the 29 compounds (purple ellipses) and the 279 targets (pink circles) to nodes, totaling 308 nodes and 1119 edges (Figure 4). The different sizes of the target nodes indicated the degree of compounds with the common target protein. The larger the degree, the more the number of corresponding targets or the more corresponding components of this target. Compounds 8 (apigenin), 1 (3-O-methylquercetin), 4 (kaempferol), 7 (quercetin), 5 (luteolin), 24 (ferulic acid), 19 (alpha-terpineol), 22 (caffeic acid), 11 (adinoside G), 29 (isochlorogenic acid C), 13 (loganin), and 28 (isochlorogenic acid A) were linked to more than 35 targets and can be considered the main active compounds of LJF (Table 1). Critically, the degree values of 25 compounds were higher than 19, and most of them had anti-inflammatory effects. The degree values of 22 targets, including CA2, CA1, CA9, CA4, and AKR1B1, were higher than 10, indicating that these targets perhaps play a key role in the anti-inflammatory effect for LJF (Table 2). The results of this study show that multiple targets can connect with the same compound, and a single target can interact with multiple compounds, indicating that LJF plays a therapeutic role through the synergy of multiple compounds and targets.
Figure 2.
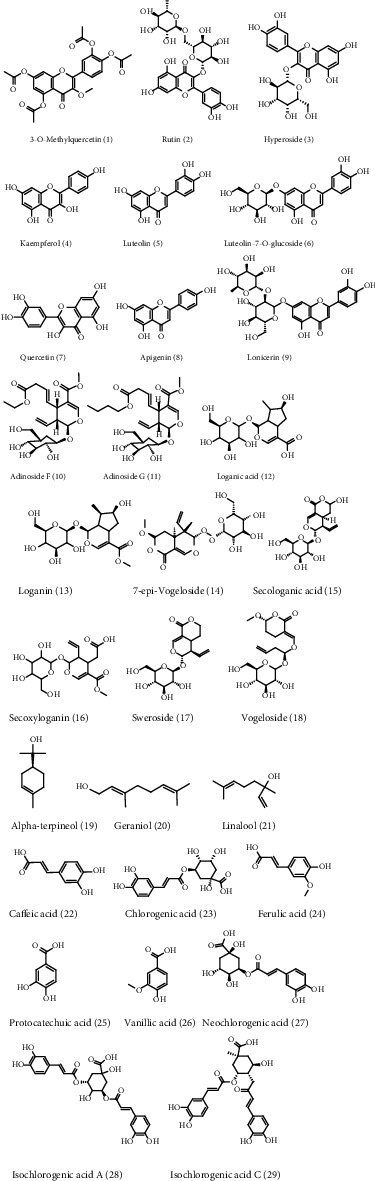
The structure of the 29 active compounds in Lonicerae japonicae flos.
Figure 3.
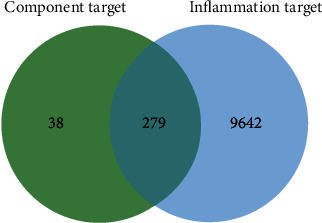
Venn diagram of 317 component targets and 9921 inflammation targets. The 279 inflammatory targets overlap in the middle.
Figure 4.
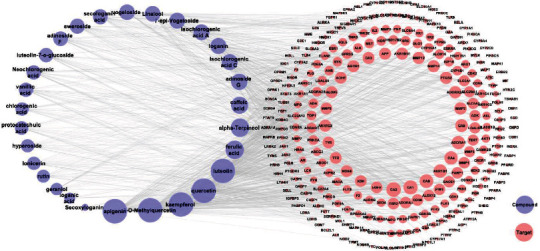
Component-target (C-T) interaction network of Lonicerae japonicae flos. The purple ellipses represent the component, the pink nodes represent the target, and the edges represent the relationship between components and the targets. The size of the nodes in the figure is associated with the degree in the network.
Table 1.
29 components of Lonicerae japonicae flos corresponding to the inflammation targets.
| No. | Component | Target | Counts |
|---|---|---|---|
| 8 | Apigenin | NOX4, AKR1B1, XDH, MAOA, FLT3, CYP19A1, ESR1, ACHE, ADORA1, PTGS2, ESR2, CDK6, ADORA2A, SYK, GSK3B, ABCC1, HSD17B1, TTR, CSNK2A1, CFTR, CYP1B1, ABCG2, AKR1B10, TNKS2, TNKS, ALOX5, PARP1, CA2, ABCB1, ALOX12, CA4, PTPRS, GLO1, APP, MMP9, MMP2, MMP12, CD38, TOP1, ARG1, ESRRA, PFKFB3, GRK6, ALOX15, TYR, HSD17B2, AHR, CA1, CA9, CBR1, AR, TERT, PIM1, EGFR, CDK1, LCK, AURKB, TBXAS1, IGF1R, KDR, PLK1, MET, ALK, AXL, BCHE, ADORA3, CDK2, HTR2C, GPR35, DAPK1, MPG, SLC22A12, F2, ST6GAL1, PLG, AVPR2, DRD4, MPO, PIK3R1, SRC, PTK2, MMP13, MMP3, CA3, CA6, PKN1, NEK2, CXCR1, CAMK2B, AKT1 | 90 |
| 1 | 3-O-Methylquercetin | NOX4, CYP1B1, APP, AKR1B1, XDH, MCL1, CA2, CA4, ABCG2, ABCC1, PLG, ABCB1, IGF1R, EGFR, ADORA1, ACHE, ALOX15, ALOX12, AVPR2, MAOA, FLT3, CYP19A1, F2, PIM1, ALOX5, AURKB, DRD4, GLO1, MPO, PIK3R1, ADORA2A, DAPK1, CA1, GSK3B, SRC, PTK2, HSD17B2, KDR, MMP13, MMP3, CA3, PLK1, CA6, CDK1, MMP9, MMP2, PKN1, CA9, CSNK2A1, MET, NEK2, CXCR1, CAMK2B, ALK, AKT1, PLA2G1B, BACE1, AXL, AKR1C2, AKR1C1, AKR1C3, AKR1C4, AKR1A1, GPR35, CDK6, CDK2, ARG1, SYK, MAPT, TOP2A, INSR, MYLK, PIK3CG, APEX1, TYR, HSD17B1, AHR, ESRRA, TERT, PTPRS, ESR2, MPG, SLC22A12, ADORA3, PARP1, TTR, MMP12, CD38, AKR1B10 | 89 |
| 4 | Kaempferol | NOX4, AKR1B1, XDH, TYR, FLT3, CA2, ALOX5, HSD17B2, ABCC1, HSD17B1, AHR, ESRRA, ABCB1, CYP1B1, ABCG2, ADORA1, CA4, ACHE, MAOA, GLO1, SYK, GSK3B, MMP9, MMP2, ALOX15, ALOX12, PTPRS, ADORA2A, ARG1, GPR35, ESR2, DAPK1, MPG, SLC22A12, CDK6, CDK2, TTR, AKR1B10, TNKS2, TNKS, CYP19A1, CSNK2A1, EGFR, AVPR2, IGF1R, F2, PIM1, AURKB, DRD4, MPO, PIK3R1, CA1, SRC, PTK2, KDR, MMP13, MMP3, CA3, PLK1, CA6, CDK1, PKN1, CA9, MET, NEK2, CXCR1, CAMK2B, ALK, AKT1, PLA2G1B, BACE1, AXL, AKR1C2, AKR1C1, AKR1C3, AKR1C4, AKR1A1, APP, PARP1, MMP12, CD38, TOP1, ESR1, PTGS2, CFTR, PFKFB3, GRK6, TERT, BCHE | 89 |
| 7 | Quercetin | NOX4, AVPR2, AKR1B1, XDH, MAOA, IGF1R, FLT3, CYP19A1, EGFR, F2, CA2, PIM1, ALOX5, AURKB, DRD4, ADORA1, GLO1, MPO, PIK3R1, ADORA2A, DAPK1, CA1, GSK3B, SRC, PTK2, HSD17B2, KDR, MMP13, MMP3, CA3, ALOX15, ABCC1, PLK1, CA6, CDK1, MMP9, MMP2, PKN1, CA9, CSNK2A1, ALOX12, MET, CA4, NEK2, CXCR1, CAMK2B, ALK, AKT1, ABCB1, PLA2G1B, BACE1, CYP1B1, AXL, ABCG2, AKR1C2, AKR1C1, AKR1C3, AKR1C4, AKR1A1, GPR35, SYK, MAPT, TOP2A, INSR, ACHE, MYLK, PIK3CG, APEX1, ARG1, PTPRS, ESR2, MPG, SLC22A12, CDK6, CDK2, TYR, HSD17B1, AHR, ESRRA, APP, PARP1, TTR, MMP12, CD38, AKR1B10, TNKS2, TNKS, TOP1, TERT | 89 |
| 5 | Luteolin | NOX4, AKR1B1, XDH, MAOA, FLT3, CA2, ALOX5, ADORA1, GLO1, APP, SYK, GSK3B, PARP1, TTR, MMP9, MMP2, CA4, MMP12, CD38, CYP1B1, ABCG2, AKR1B10, TNKS2, TNKS, TOP1, ARG1, PTPRS, ABCC1, HSD17B1, ACHE, CDK6, ABCB1, HSD17B2, ALOX15, ALOX12, ESR2, CYP19A1, ADORA2A, CSNK2A1, ESR1, PTGS2, CFTR, GRK6, CDK2, TERT, CA1, CA9, CDK1, TYR, AHR, ESRRA, GPR35, DAPK1, AVPR2, IGF1R, EGFR, F2, PIM1, AURKB, DRD4, MPO, PIK3R1, SRC, PTK2, KDR, MMP13, MMP3, CA3, PLK1, CA6, PKN1, MET, NEK2, CXCR1, CAMK2B, ALK, AKT1, PLA2G1B, BACE1, AXL, AKR1C2, AKR1C1, AKR1C3, AKR1C4, AKR1A1, PFKFB3, PLG, AR | 88 |
| 24 | Ferulic acid | CA2, CA1, CA6, CA9, MAOB, ALOX5, MMP9, MMP1, MMP2, PTPN1, CA3, AKR1B1, APP, NFE2L2, STAT3, HSD11B1, ESR2, CA4, TLR4, MET, CYP1A1, CYP1A2, CYP1B1, PTGS1, EGFR, TTR, PTGS2, TUBB1, RELA, ADORA1, ADORA2A, ADORA2B, TLR9, AKR1B10, ALOX15, PRKCE, F3, NOS2, FYN, LCK, SLC16A1, ABCB1, TOP2A, FBP1, BACE1, GLO1, CPA1, KDM4C, AHR, AMPD3, PARP1 | 51 |
| 19 | Alpha-terpineol | AR, CYP19A1, CA2, CA1, CA4, CHRM2, SLC6A4, TRPM8, NR1H3, PTPN1, NR1I3, SREBF2, NPC1L1, BCHE, ACHE, SQLE, ESR1, SLC6A2, DRD2, ESR2, CYP17A1, CYP2C19, NR3C2, PTPRF, PTPN2, PLA2G1B, ACP1, AKR1B10, SIGMAR1, TRPV3, NR3C1, ATP12A, PTPN6, SHBG, FABP4, PPARA, FABP3, FABP5, PPARD, FABP1, RORA, HMOX1, HMGCR, PGR, CD81, G6PD, SCD, ADRA2C, HSD11B1, SLC6A3 | 50 |
| 22 | Caffeic acid | CA2, ALOX5, CA1, CA6, MMP9, MMP1, MMP2, PTPN1, CA9, CA3, AKR1B1, ESR2, CA4, AKR1B10, HCAR2, MIF, TLR4, ERBB2, ESR1, SLC6A2, TTR, MAPK1, AKR1C3, AKR1C4, AKR1C2, SYK, APP, EGFR, FYN, LCK, PTGS1, PIK3CB, CYP1A2, CYP2C9, CYP3A4, CYP2C19, PIK3CA, ELANE, F3, HSD11B1, MAOB, NFE2L2, STAT3 | 43 |
| 11 | Adinoside G | ADORA2A, ADORA1, ADORA3, SLC29A1, MMP3, MMP9, ADAM17, ADORA2B, ADK, ST6GAL1, SLC5A2, MAPK14, SLC5A1, TOP1, IMPDH1, LGALS3, LGALS7, PARP1, TNKS2, TNKS, HSPA5, MMP1, LGALS9, MMP2, CA2, CA1, CA9, OGA, NRAS, PTGS2, MMP13, MMP7, MMP12, MMP8, GPR55, GBA, HK2, HK1 | 38 |
| 29 | Isochlorogenic acid C | AKR1B1, APP, AKR1B10, MMP12, MMP2, MMP13, PRKCD, CA4, PRKCA, CA6, CA2, CA1, ABCB1, SLC37A4, CA9, FYN, TTR, MMP1, PDE5A, ELANE, SELE, SELP, PDE4D, PDE9A, PDE1B, HCAR2, PTGDR2, CASP3, MGLL, PIM1, FOLH1, CASP6, CASP7, CASP8, CASP1, EDNRA | 36 |
| 13 | Loganin | TYR, ADORA1, CA2, CA1, CA9, HSP90AA1, ADORA3, ADORA2A, EPHX2, CA4, SLC5A2, ADA, HRAS, LGALS3, LGALS9, CA6, ADK, FUCA1, AKR1B1, TYMP, AKR1C3, SLC29A1, SLC5A1, IGFBP3, ADORA2B, ATIC, ALOX12, PNP, PABPC1, SLC5A4, MMP13, AMPD3, MMP1, MMP7, MMP12, MMP8 | 36 |
| 28 | Isochlorogenic acid A | AKR1B1, MMP2, MMP12, APP, MMP13, AKR1B10, ELANE, SLC37A4, CA4, PRKCD, CA2, CA1, CA9, ABCB1, PDE9A, PDE1B, POLB, PDE5A, CASP3, ABL1, EPHA2, SRC, KDR, MAP3K9, FGFR1, AURKA, BTK, PRKCA, BACE1, MME, ECE1, HCAR2, PTGDR2, EDNRA, PIM1 | 35 |
| 14 | 7-epi-Vogeloside | LGALS3, LGALS9, ADORA1, ADORA2A, ADK, OGA, ADORA3, CA1, CA9, CA2, SLC5A2, HK2, HK1, EGFR, SLC29A1, SLC5A4, SLC5A1, AKR1B1, GBA, ADORA2B, PTGS2, GAPDH, GAA, PYGM, EDNRA, TYR, ADA, PNP, LGALS7, MAPK10, HSPA5, HSPA8, PTPN11, SLC28A2 | 34 |
| 21 | Linalool | CA2, CA1, CA4, TRPV3, TRPM8, NR3C2, NR3C1, PGR, SLC6A3, SIGMAR1, HSD17B2, SQLE, HMOX1, IDO1, DRD2, ADRA2C, PTGS2, OPRM1, OPRD1, OPRK1, SCN5A, SCN9A, PTAFR, PARP1, ADRA1A, JAK1, JAK2, AR, MAPK8, LRRK2, TYMS, HRH3, HRH4, LTA4H | 34 |
| 18 | Vogeloside | LGALS3, LGALS9, ADORA1, ADORA2A, ADK, OGA, ADORA3, CA1, CA9, CA2, SLC5A2, HK2, HK1, EGFR, SLC29A1, SLC5A4, SLC5A1, AKR1B1, GBA, ADORA2B, PTGS2, GAPDH, GAA, PYGM, EDNRA, TYR, ADA, PNP, LGALS7, MAPK10, HSPA5, HSPA8, PTPN11, SLC28A2 | 34 |
| 15 | Secologanic acid | ADORA1, ADORA2A, FUCA1, TYR, ADK, CA2, CA1, CA9, CA6, CA4, CA3, AKR1C3, LGALS3, LGALS9, ADORA3, FOLH1, AKR1B1, PNP, HK2, HK1, ADA, HRAS, SLC5A2, HSP90AA1, ATIC, ALOX12, TYMP, SLC5A1, SLC5A4 | 29 |
| 17 | Sweroside | ADORA1, ADORA2A, FUCA1, TYR, ADK, CA2, CA1, CA9, CA6, CA4, CA3, AKR1C3, LGALS3, LGALS9, ADORA3, FOLH1, AKR1B1, PNP, HK2, HK1, ADA, HRAS, SLC5A2, HSP90AA1, ATIC, ALOX12, TYMP, SLC5A1, SLC5A4 | 29 |
| 10 | Adinoside F | IMPDH1, MMP3, MMP9, MMP1, ADAM17, ADORA1, ADORA2A, ADORA3, ADORA2B, MMP13, MMP7, MMP12, MMP8, SLC5A2, SLC29A1, ST6GAL1, MMP2, SLC5A1, IL2, ADA, CA2, CA1, CA9, TOP1, HSPA8, DNMT1, ADK, HRAS | 28 |
| 6 | Luteolin-7-o-glucoside | TNF, IL2, AKR1B1, ADORA1, XDH, CA2, NOX4, ADRA2C, ALDH2, NMUR2, ADRA2A, ACHE, RPS6KA3, CA4, CD38, PRKCA, MMP1, MMP7, MMP8, CA1, CA9, ALOX5, PTGS2, SLC29A1, HSP90AA1, PLG | 26 |
| 27 | Neochlorogenic acid | AKR1B1, AKR1B10, MMP13, MMP2, APP, MMP12, ELANE, SLC37A4, PRKCD, PRKCA, BACE1, PDE4D, PDE9A, PDE1B, CA6, ABCB1, CA2, CA1, CA9, NEU2, CASP3, CASP6, CASP7, CASP8, CASP1 | 25 |
| 26 | Vanillic acid | CA2, CA1, CA9, CA3, CA6, CA4, TPMT, TTR, FUT7, KDM6B, FTO, KDM4C, FYN, LCK, FBP1, AKR1C3, MMP9, MMP1, MMP2, MMP8, SQLE, POLA1, POLB, SERPINE1, TUBB1 | 25 |
| 23 | Chlorogenic acid | AKR1B1, AKR1B10, MMP12, MMP13, MMP2, APP, ELANE, SLC37A4, PRKCD, PRKCA, CA2, CA1, CA9, BACE1, PDE4D, PDE9A, PDE1B, CA6, ABCB1, NEU2 | 20 |
| 25 | Protocatechuic acid | CA2, CA1, CA6, CA9, CA4, CA3, FUT7, SQLE, LDHA, LDHB, TTR, ESR2, COMT, BCL2L1, IGF1R, ALK, SERPINE1, AKR1C3, GPR35, ALB | 20 |
| 3 | Hyperoside | NOX4, ADRA2C, AKR1B1, CA2, CA4, ACHE, RPS6KA3, NMUR2, ADRA2A, PTGS2, CD38, PDE5A, TNF, IL2, ADORA1, XDH, ALOX5, SLC29A1, TERT | 19 |
| 9 | Lonicerin | IL2, XDH, TNF, ADORA1, AKR1B1, NMUR2, ADRA2A, ADRA2C, ACHE, NOX4, CA2, RPS6KA3, PTGS2, CD38, PRKCA, CA4, ALDH2, PDE5A, CA1 | 19 |
| 2 | Rutin | NMUR2, ADRA2A, ADRA2C, ACHE, AKR1B1, CA4, NOX4, CA2, RPS6KA3, XDH, CD38, PTGS2, PDE5A, TNF, IL2, ADORA1, ALOX5, TERT | 18 |
| 20 | Geraniol | SQLE, PTGS1, PTGS2, PGR, HMGCR, KCNH2, UGT2B7, EPHX2, JAK1, JAK2, CYP11B1, CYP11B2, PIM1, PIM3 | 14 |
| 12 | Loganic acid | NEU2, ADORA1, SELP, SELL, CA2, CA1, CA9 | 7 |
| 16 | Secoxyloganin | SELP, ADORA1, LGALS3, NOD2 | 4 |
Table 2.
Top 20 targets of degree value in component-target interaction network.
| Target | Degree | Target | Degree |
|---|---|---|---|
| CA2 | 27 | MMP12 | 12 |
| CA1 | 25 | MMP13 | 12 |
| CA9 | 22 | CA3 | 11 |
| CA4 | 20 | APP | 11 |
| AKR1B1 | 20 | TTR | 10 |
| ADORA1 | 19 | TYR | 10 |
| CA6 | 15 | AKR1C3 | 10 |
| MMP2 | 14 | MMP9 | 10 |
| PTGS2 | 13 | ALOX5 | 10 |
| ADORA2A | 13 | ACHE | 10 |
| AKR1B10 | 12 | ABCB1 | 10 |
3.2. Protein-Protein Interaction (PPI) Network
The compound-inflammation target PPI network was constructed by inputting all the 279 LJF inflammation-related targets into the database of STRING and then inputting the PPI network into Cytoscape 3.7.2 software to be visualized. As shown in Figure 5, the PPI network contained 224 functional nodes and edges. The bigger the nodes, the higher the degree of interactivity, indicating a stronger interaction among the proteins. Then, Cytohubba (a plugin of Cytoscape) was used for screening hub genes in the PPI network. As shown in Table 3, the degree values of 20 targets were higher than 23, which indicates that they had more interaction with other targets and might play key roles in treating inflammation. The top 10 targets, including PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha), MAPK1 (mitogen-activated protein kinase 1), PIK3R1 (phosphoinositide-3-kinase regulatory subunit 1), SRC (SRC proto-oncogene), APP (amyloid beta precursor protein), HRAS (HRas proto-oncogene), STAT3 (signal transducer and activator of transcription 3), HSP90AA1 (heat shock protein 90 alpha family class A member 1), NRAS (NRAS proto-oncogene), and FYN (FYN proto-oncogene), were selected according to their degree values (Figure 6). Among them, target PIK3CA interacted the most with other proteins.
Figure 5.
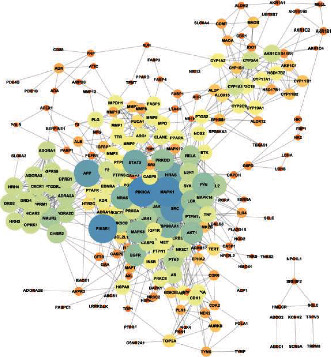
Protein-protein interaction (PPI) network analysis of 224 potential targets. The nodes indicate proteins, and edges represent protein-protein associations. The closer and the larger the nodes are, the higher the degree of freedom they have.
Table 3.
Top 20 targets of the protein-protein interaction network.
| No. | Targets | Degree | Betweenness centrality | Closeness centrality |
|---|---|---|---|---|
| 1 | PIK3CA | 45 | 0.056863 | 0.420118 |
| 2 | MAPK1 | 44 | 0.146963 | 0.441909 |
| 3 | PIK3R1 | 43 | 0.045671 | 0.418468 |
| 4 | SRC | 41 | 0.048756 | 0.416016 |
| 5 | APP | 38 | 0.124074 | 0.394444 |
| 6 | HRAS | 35 | 0.020650 | 0.375661 |
| 7 | STAT3 | 34 | 0.097739 | 0.408829 |
| 8 | HSP90AA1 | 31 | 0.087902 | 0.407266 |
| 9 | NRAS | 30 | 0.017837 | 0.365352 |
| 10 | FYN | 29 | 0.009133 | 0.381720 |
| 11 | AKT1 | 27 | 0.048552 | 0.397388 |
| 12 | EGFR | 27 | 0.044610 | 0.404175 |
| 13 | JAK2 | 27 | 0.011099 | 0.370435 |
| 14 | MAPK8 | 27 | 0.031883 | 0.401887 |
| 15 | LCK | 26 | 0.005706 | 0.376991 |
| 16 | PRKCD | 26 | 0.040957 | 0.387273 |
| 17 | PTPN11 | 24 | 0.005269 | 0.371080 |
| 18 | NMUR2 | 24 | 0.017057 | 0.345779 |
| 19 | JAK1 | 23 | 0.004552 | 0.372378 |
| 20 | RELA | 23 | 0.036432 | 0.401130 |
Figure 6.
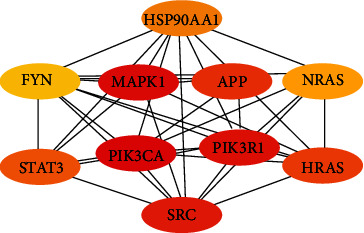
Top 10 genes' interaction network of Lonicerae japonicae flos. The nodes indicate proteins, and edges represent protein-protein associations. The depth of the color shade indicates the high degree of the node.
3.3. GO (Gene Ontology) Analysis
GO is a widely used system for the classification of gene functions and describing the functions of gene products [41]. GO analysis of the anti-inflammatory targets of LJF was executed for three factors: biological process (BP), molecular function (MF), and cell composition (CC). The P values of 1844 biological processes, 41 cell components, and 236 molecular functions were less than 0.01. As shown in Figure 7, the top 10 enrichment results of BP were as follows: response to toxic substance, response to inorganic substance, cellular response to nitrogen compound, positive regulation of MAPK cascade, regulation of MAPK cascade, positive regulation of transferase activity, positive regulation of kinase activity, cellular response to organonitrogen compound, response to antibiotic, and response to oxidative stress. The top 10 enrichment results of MF were phosphotransferase activity (alcohol group as acceptor), protein kinase activity, kinase activity, protein serine/threonine kinase activity, oxidoreductase activity, oxidoreductase activity acting on paired donors with the incorporation or reduction of molecular oxygen, cofactor binding, heme binding, monooxygenase activity, and tetrapyrrole binding.
Figure 7.
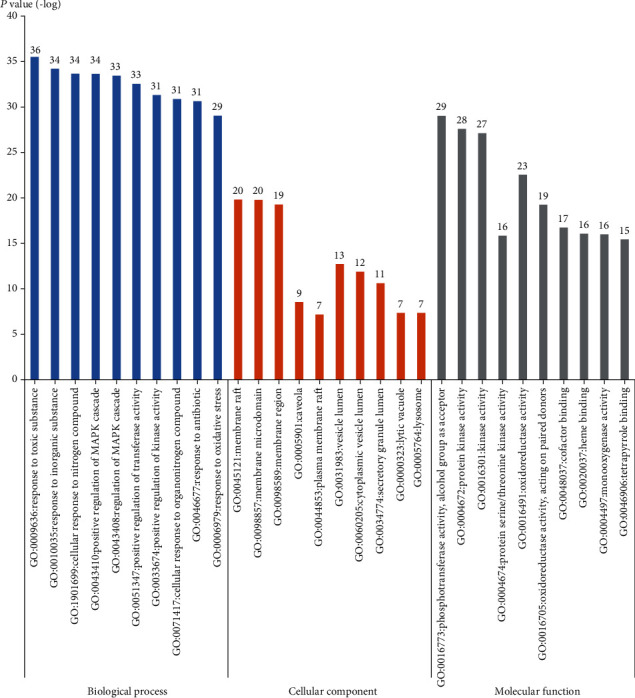
Gene ontology enrichment with top 10 P value for each item. The blue columns, the orange columns, and the gray columns are biological process, cellular component, and molecular function, respectively. The y-axis stands for the P values of fold change.
3.4. KEGG Pathway Enrichment Analysis
The inflammatory process can trigger different diseases depending on the specific inflamed tissue or organ involved. However, all disorders have common features or a conjoint cellular process, such as the activation of a stress signaling pathway and the concomitant production of inflammatory cytokines [43]. A total of 279 LJF inflammation-related targets were input to the Metascape database, and 173 pathways that had statistical significance were input for pathway enrichment analysis. The 10 pathways with the lowest P values are documented in Table 4 and include the following: the AGE-RAGE signaling pathway in diabetic complications, the HIF-1 signaling pathway, the EGFR tyrosine kinase inhibitor resistance, the proteoglycans in cancer, the insulin resistance, the prolactin signaling pathway, the PI3K-AKT signaling pathway, the endocrine resistance, the hepatitis B, and central carbon metabolism in cancer.
Table 4.
Top 10 pathways ranked according to P value.
| Pathway | P value | Count |
|---|---|---|
| hsa04933: AGE-RAGE signaling pathway in diabetic complications | 7.94E − 24 | 23 |
| hsa04066: HIF-1 signaling pathway | 3.56E − 22 | 22 |
| hsa01521: EGFR tyrosine kinase inhibitor resistance | 1.09E − 21 | 20 |
| hsa05205: proteoglycans in cancer | 5.5E − 21 | 27 |
| hsa04931: insulin resistance | 3.31E − 20 | 21 |
| hsa04917: prolactin signaling pathway | 8.97E − 20 | 18 |
| hsa04151: PI3K-Akt signaling pathway | 5.81E − 19 | 31 |
| hsa01522: endocrine resistance | 1.8E − 18 | 19 |
| hsa05161: hepatitis B | 2.05E − 17 | 21 |
| hsa05230: central carbon metabolism in cancer | 2.22E − 17 | 16 |
The top three pathways containing the most LJF inflammation-related targets were as follows: the PI3K-AKT signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, and the HIF-1 signaling pathway (Figure 8), which indicates that the three pathways play a crucial role in the anti-inflammation properties of LJF. The cascade pathway of pharmacological mechanism of LJF acting on inflammation is illustrated in Figure 9.
Figure 8.
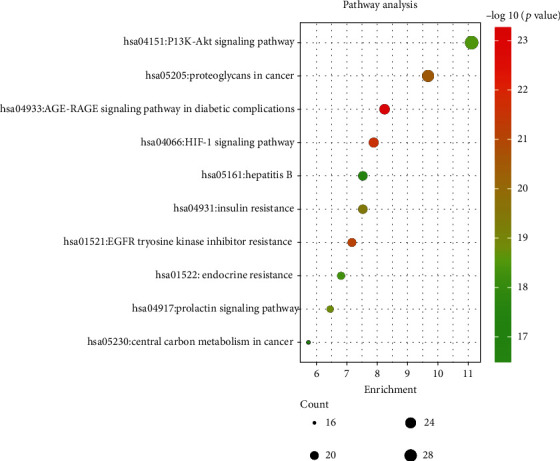
KEGG pathway enrichment with the top 10 P value. The y-axis stands for enriched pathways of the targets. The color of the bubble is associated with the P value, and the size is related to the enrichment number of targets.
Figure 9.
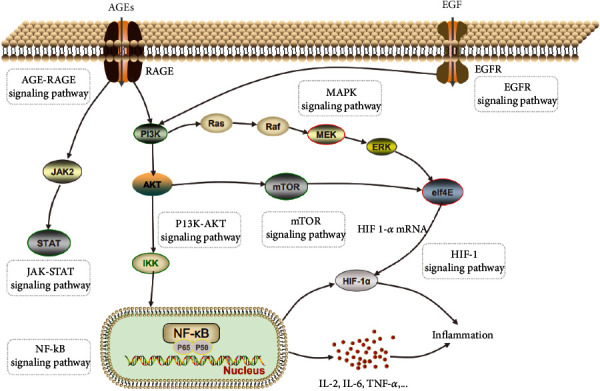
Pharmacological mechanism cascade pathway of Lonicerae japonicae flos impact on inflammation.
AKT is a key downstream signaling protein of PI3K. After PI3K is activated, it binds to AKT in the plasma membrane. Then, the activated AKT can promote the expression and secretion of proinflammatory cytokines by activating the NF-κB pathway, resulting in an imbalance of cytokine secretion and a series of inflammatory reactions [44–46]. Several studies have shown that the inhibition of the PI3K signal can inhibit the secretion of proinflammatory factors in macrophages and dendritic cells and increase the secretion of anti-inflammatory factor IL-10 when a Toll-like receptor- (TLR-) mediated inflammatory response occurs [47–52]. The important role of PI3K/AKT/NF-κB signaling pathways in the anti-inflammatory effects of LJF was confirmed in a previous report [53].
In the AGE-RAGE signaling pathway in diabetic complications, highly abundant AGEs in the diabetic milieu of the kidneys upregulate RAGE expression, and ligand-evoked RAGE stimulation leads to the activation of intracellular signaling pathways, including JAK/STAT, MAPK/ERK, PI3K/AKT/mTOR, and NF-κB, of which the common end is the activation of nuclear transcription factors involved in the inflammatory and fibrotic processes [54].
Crosstalk between HIF-1α and NF-κB regulates essential inflammatory functions in myeloid cells. HIF-1α increases macrophage aggregation, invasion, and motility and drives the expression of proinflammatory cytokines, such as TNR-α and IL-6 [55].
The 17 pathways with P values of less than 3 × 10−15 were selected, and the compound-target-pathway (C-T-P) network was constructed using Cytoscape software (Figure 10). In a network, nodes with high degree values indicate high interconnectedness [56]. The targets AKT1, PIK3CA, PIK3CB, and PIK3R1 were related to all 17 pathways; thus, they were recognized as key targets. Target MAPK connected to 16 pathways simultaneously and had more interactions with other targets in the PPI network. The top five components with higher degree values, including compounds 8 (apigenin), 7 (quercetin), 4 (kaempferol), 1 (3-O-methylquercetin), and 5 (luteolin), were recognized as the key components, with degree values of 90, 89, 89, 89, and 88, respectively, indicating that these components are correlated with at least 88 of the 90 targets within the 17 pathways. In addition, another seven components, including compounds 24 (ferulic acid), 19 (α-terpineol), 22 (caffeic acid), 11 (adinoside G), 18 (vogeloside), 14 (7-epi-vogeloside), and 13 (loganin), had degree values higher than 30, which suggests that they also play important roles in the anti-inflammation effects of LJF.
Figure 10.
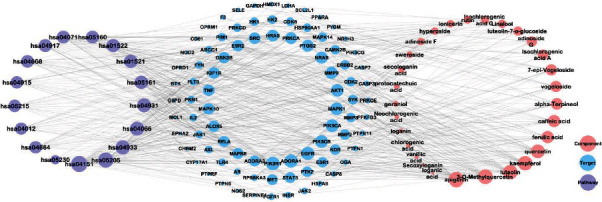
Component-target-pathway (C-T-P) interaction network. The purple, pink, and blue nodes are the pathway, the component, and the target, respectively. The edges represent the relationship between pathway, component, and target. The size of the nodes in the figure is associated with the degree in the network.
3.5. Molecular Docking Analysis
The dock results of 29 components and hub genes (AKT, PIK3R1, PIK3CA, and MAPK1) are shown in Figure 11, and the docking ligand-protein binding energy is summarized in Table 5. It is generally considered that the value of the Autodock Vina score indicates the binding activity between a compound and a protein [16]. Molecular docking was used for the verification of the interactions between ingredients and target genes [57]. Among the four targets, AKT has the most binding activity, which exhibited binding activity with 10 compounds, and their Vina scores were all less than -5. These 10 compounds are as follows: vanillic acid, protocatechuic acid, caffeic acid, secologanic acid, quercetin, apigenin, adinoside F, luteolin, ferulic acid, and alpha-terpineol. In addition, there were four components, including secologanic acid, adinoside F, apigenin, and kaempferol, which exhibited binding activity (binding energy ≤ −5) with the targets PIK3R1 and MAPK1. In addition, luteolin and target MAPK1 also had binding activity. The top four compound-target complexes were screened by sorting the docking scores in descending order, as shown in Figure 12. Compounds 26 (vanillic acid), 25 (protocatechuic acid), and 22 (caffeic acid) exhibited binding activity with the target AKT1, and compound 10 (adinoside F) showed binding activity to the target MAPK1. Previous studies showed that protocatechuic acid may inhibit the LPS-stimulated inflammatory mediator production in keratinocytes by reducing the Toll-like receptor-4-dependent activation of AKT, mTOR, and NF-κB pathways and the activation of JNK and p38-MAPK [58]. Caffeic acid inhibited the inflammatory response by downregulating the phosphorylation of several important transcriptional factors, such as NF-κB and STAT-3 [59]. In addition, vanillic acid inhibited the LPS-induced production of tumor necrosis factor TNF-α and interleukin IL-6 by suppressing the activation of NF-κB and caspase-1 [60].
Figure 11.
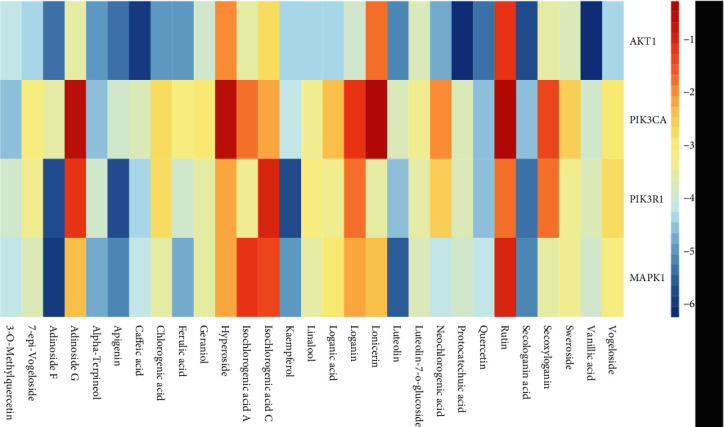
Heat maps show docking scores of hub genes combining to 29 components of Lonicerae japonicae flos. Color represents binding energy score.
Table 5.
Binding energy (kcal/mol) of Lonicerae japonicae flos molecular docking.
| No. | Component | Target | |||
|---|---|---|---|---|---|
| AKT1 | PIK3CA | PIK3R1 | MAPK1 | ||
| C1 | 3-O-Methylquercetin | -4.27 | -4.73 | -4.17 | -4.39 |
| C15 | Secologanic acid | -5.98 | -4.69 | -5.52 | -5.24 |
| C11 | Adinoside F | -5.4 | -3.73 | -5.9 | -6.01 |
| C2 | Apigenin | -5.48 | -4.2 | -5.88 | -5.2 |
| C6 | Luteolin | -5.37 | -3.98 | -4.79 | -5.78 |
| C4 | Kaempferol | -4.55 | -4.23 | -5.8 | -5.16 |
| C8 | Quercetin | -5.54 | -4.69 | -4.66 | -4.26 |
| C22 | Caffeic acid | -6.11 | -3.91 | -4.52 | -4.25 |
| C19 | Alpha-terpineol | -5 | -4.69 | -4.2 | -4.85 |
| C29 | Vanillic acid | -6.52 | -4.04 | -3.89 | -4.07 |
| C28 | Protocatechuic acid | -6.39 | -3.91 | -3.93 | -4.15 |
| C24 | Ferulic acid | -5.19 | -3.39 | -4.08 | -4.8 |
| C10 | 7-epi-Vogeloside | -4.54 | -3.21 | -3.42 | -3.84 |
| C7 | Luteolin-7-o-glucoside | -3.86 | -3.58 | -3.74 | -3.82 |
| C21 | Linalool | -4.44 | -3.43 | -3.31 | -3.64 |
| C23 | Chlorogenic acid | -5.17 | -2.82 | -3 | -3.68 |
| C20 | Geraniol | -4.12 | -3.12 | -3.67 | -3.71 |
| C27 | Neochlorogenic acid | -4.94 | -2.06 | -2.78 | -4.34 |
| C13 | Loganic acid | -4.59 | -2.6 | -3.48 | -3.15 |
| C17 | Sweroside | -3.93 | -2.72 | -3.54 | -3.41 |
| C18 | Vogeloside | -4.5 | -3.05 | -2.82 | -3.22 |
| C16 | Secoxyloganin | -3.75 | -1.42 | -1.95 | -3.7 |
| C25 | Isochlorogenic acid A | -3.72 | -1.92 | -3.51 | -1.28 |
| C14 | Loganin | -4.12 | -1.27 | -1.97 | -2.32 |
| C5 | Lonicerin | -1.93 | -0.45 | -3.76 | -2.42 |
| C12 | Adinoside G | -3.69 | -0.76 | -1.39 | -2.45 |
| C26 | Isochlorogenic acid C | -2.91 | -2.34 | -1.13 | -1.51 |
| C3 | Hyperoside | -2.19 | -0.75 | -2.39 | -2.24 |
| C9 | Rutin | -1.24 | -0.42 | -1.82 | -1.19 |
Figure 12.
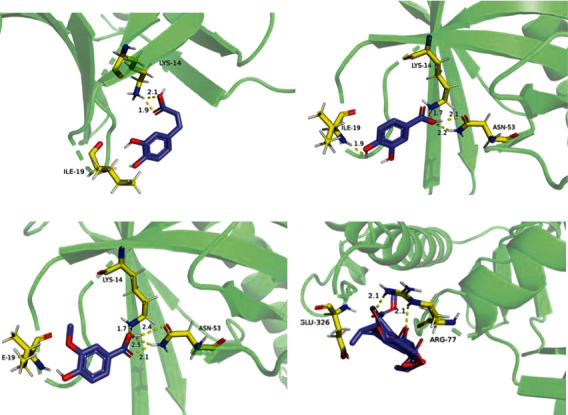
Component-target docking combination: (a) caffeic acid-AKT1 (score -6.11); (b) protocatechuic acid-AKT1 (score -6.39); (c) vanillic acid-AKT1 (score -6.52); (d) adinoside F-MAPK1 (score -6.01).
3.6. The Appropriate Concentrations of Luteolin and Secologanic Acid
According to the results of the compound-target-inflammation network (Figure 10) analysis, secologanic acid and luteolin are the main active compounds of LJF. MTT was performed to detect the viability of RAW264.7 cells, and the suitable concentrations of luteolin and secologanic acid in treating RAW264.7 cells were obtained. Figure 13 shows that treatment with various concentrations of luteolin (5, 10, 20, 40, and 80 μM) and secologanic acid (5, 10, 20, 40, and 80 μM) for 24 h had no cytotoxicity on the vitality of RAW264.7 cells. Therefore, concentrations of 5, 20, and 80 μM for luteolin and secologanic acid were selected as the treating concentrations in this study.
Figure 13.
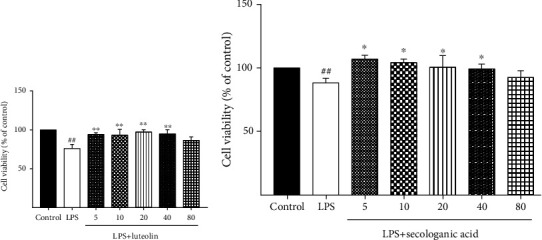
Effect of different concentrations of luteolin and secologanic acid on RAW264.7 macrophage viability. (a) RAW264.7 cells were incubated with luteolin (5, 10, 20, 40, and 80 μM) for 24 hours after being treated with 1 μg/mL LPS. (b) RAW264.7 cells were incubated with secologanic acid (5, 10, 20, 40, and 80 μM) for 24 hours after being treated with 1 μg/mL LPS. Data are expressed as the mean ± SD of three independent experiments. ##P < 0.01 compared with the control group; ∗P < 0.05 and ∗∗P < 0.01 compared with the model group.
3.7. Luteolin and Secologanic Acid Inhibit the Production of TNF-α
Tumor necrosis factor-alpha (TNF-α) is one of the major mediators of inflammation. Induced by a wide range of pathogenic stimuli, TNF-α induces other inflammatory mediators and proteases that orchestrate inflammatory responses. The proinflammatory effects of TNF-α are primarily due to its ability to activate NF-κB. Studies showed that downregulating the expression of TNF-α could inhibit inflammatory reactions, improve cardiac function, and inhibit oxidative stress [61]. To further confirm the anti-inflammatory activity of secologanic acid and luteolin, the levels of TNF-α in LPS-induced RAW264.7 cells were detected. As shown in Figure 14, LPS significantly increased the secretion of TNF-α compared to the control group. Luteolin, at 5, 20, and 80 μM, significantly inhibited the generation of TNF-α in a dose-dependent manner. Similarly, the expression of TNF-α in the 5, 20, and 80 μM secologanic acid-treated groups also significantly decreased compared to the LPS groups, but not in a dose-dependent manner.
Figure 14.
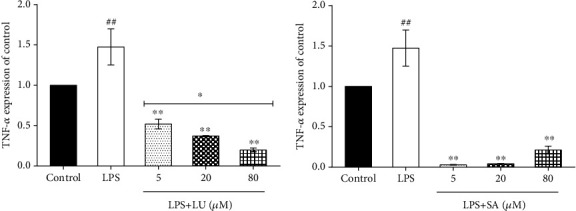
Effect of luteolin and secologanic acid on the mRNA levels of TNF-α. TNF-α levels of RAW264.7 were analyzed by qRT-PCR after incubation with LPS by 24 h. LU: luteolin; SA: secologanic acid. ##P < 0.01 versus the control group. ∗∗P < 0.01 versus the LPS group. ∗Above the horizontal line P < 0.05 versus other dose groups. The data were represented as the mean ± SD of three independent experiments.
3.8. Luteolin and Secologanic Acid Downregulated the Expression of AKT
AKT is a subset of the AGC protein Ser/Thr kinase family and plays an important role in cell growth, metabolic regulation, cancer, and other diseases [62]. In the process of inducing inflammation, AKT can be activated through phosphorylation; mediate the activation of downstream NF-KB, mTOR, and other signaling pathways; promote the secretion of proinflammatory cytokine TNF-a; and aggravate inflammation [44]. Studies found that the flavonoid extracts of LJF can reduce the expression level of p-AKT, participate in the PI3K/AKT signaling pathway, and inhibit the secretion of proinflammatory factors [53]. The PPI network and KEGG results all show that LJF's anti-inflammatory functions are involved with AKT, which was further confirmed by molecular docking technology. To further verify the role of AKT, the protein levels of AKT and p-AKT in RAW264.7 macrophagocytes treated with secologanic acid (20 μM) or luteolin (20 μM), separately, were determined in vitro. As shown in Figure 15, luteolin and secologanic acid significantly inhibited the relative expression of AKT and p-AKT compared to the LPS treatment (P < 0.01). Among them, secologanic acid showed better inhibition effects on the expression and activation of AKT than luteolin, which is consistent with the result of AKT binding (the Vina score of secologanic acid and luteolin with AKT was -5.98 and -5.37, respectively) in molecular docking. This study reveals that network pharmacology and molecular docking are powerful approaches for searching active compounds and hub targets for TCM.
Figure 15.
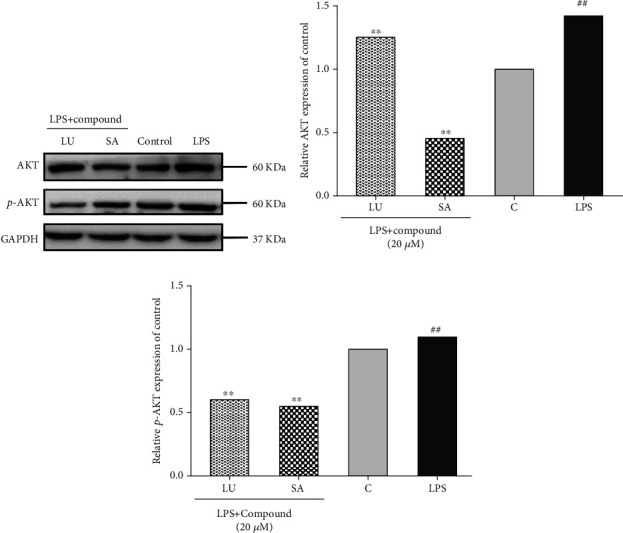
Effect of luteolin and secologanic acid on the expression of AKT and p-AKT in LPS-induced RAW264.7. (a) AKT and p-AKT levels of RAW264.7 were analyzed by western blotting after incubation with LPS for 0.5 h. (b) Relative AKT expression of control. (c) Relative p-AKT expression of control. LU: luteolin; SA: secologanic acid; C: control. ##P < 0.01 versus untreated macrophage control. ∗∗P < 0.01 versus the LPS group.
4. Conclusions
In this study, the key anti-inflammation compounds and targets of LJF were screened using a network pharmacology strategy and then verified by molecular docking and in vitro experiments. The main results are as follows. First, 279 targets were selected as LJF inflammation-related targets by comparing compound-related targets with inflammation-related targets. Second, the network analysis of compounds and targets shows that 12 compounds are linked to more than 35 targets, including apigenin, 3-O-methylquercetin, kaempferol, quercetin, luteolin, ferulic acid, alpha-terpineol, caffeic acid, adinoside G, isochlorogenic acid C, loganin, and isochlorogenic acid A. Third, hub genes, such as PIK3CA, MAPK1, PIK3R1, AKT1, SRC, APP, HRAS, and STAT3, were screened from the protein-protein interaction network. Fourth, the bioactivity of 279 inflammation targets spreads widely, involving response to toxic substance, response to inorganic substance, cellular response to nitrogen compound, positive regulation of MAPK cascade, positive regulation of kinase activity, cellular response to organonitrogen compound, response to antibiotic, and response to oxidative stress. Through building and analyzing the C-T-P network, the top four pathways, including the PI3K-AKT signaling pathway, the AGE-RAGE signaling pathway in diabetic complications, the EGFR tyrosine kinase inhibitor resistance, and the HIF-1 signaling pathway, are assumed to play a crucial role in the anti-inflammation effects of LJF. Fifth, the results of molecular docking show that 10 compounds, including vanillic acid, protocatechuic acid, caffeic acid, secologanic acid, quercetin, apigenin, adinoside F, luteolin, ferulic acid, and alpha-terpineol, exhibited binding activity with target AKT1. Finally, the anti-inflammatory effects and anti-inflammatory mechanisms of two key components, including secologanic acid and luteolin, were tested in vitro for further confirming the network pharmacological screening results. The results of qRT-PCR show that both secologanic acid and luteolin can inhibit TNF-α generation at the mRNA level. The western blotting results show that the expression levels of AKT and p-AKT were significantly decreased after secologanic acid and luteolin treatment. Secologanic acid showed better inhibition effects than luteolin on the activation of AKT, which is consistent with the results of AKT binding in molecular docking. Notably, secologanic acid is the most abundant iridoid glycoside in LJF [7]. In this study, the anti-inflammatory activity and mechanism of secologanic acid were verified by molecular docking and in vitro experiments for the first time.
In conclusion, the results of our study suggest that network pharmacology is a powerful tool for discovering the active compounds and mechanisms of action in TCM.
Acknowledgments
This work was supported by the Shandong Provincial Science and Technology of Traditional Chinese Medicine Project (2020Z02), Shandong Provincial Major Science and Technology Innovation Project (2019JZZY011020), National Key Research and Development Program of China (2017YFC1701503), and National Natural Science Foundation of China (81872963 and 81903750).
Contributor Information
Fang Zhang, Email: zfang_819@163.com.
Yongqing Zhang, Email: zyq622003@126.com.
Data Availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Xiaoying Guo and Xiao Yu contributed equally to this work.
References
- 1.Tracey K. J. The inflammatory reflex. Nature . 2002;420(6917):853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 2.Xia S., Zhang X., Zheng S., et al. An update on inflamm-aging: mechanisms, prevention, and treatment. Journal of Immunology Research . 2016;2016:12. doi: 10.1155/2016/8426874.8426874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B., Li X., Xue J., et al. Mechanism of Inflammatory Storm Induced by Novel Coronavirus Pneumonia and Intervention Measures of Traditional Chinese Medicine. Chinese Journal of Experimental Traditional Medical Formulae . 2020;26(13):32–38. [Google Scholar]
- 4.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Seminars in Immunopathology . 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R., Wang X., Ni L., et al. COVID-19: melatonin as a potential adjuvant treatment. Life Sciences . 2020;250, article 117583 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huo C., Jin Y., Zou S., et al. Lethal influenza A virus preferentially activates TLR3 and triggers a severe inflammatory response. Virus Research . 2018;257:102–112. doi: 10.1016/j.virusres.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F., Shi P., Liu H., et al. A simple, rapid, and practical method for distinguishing Lonicerae japonicae flos from Lonicerae flos. Molecules . 2019;24(19) doi: 10.3390/molecules24193455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Chi Q., Xiong S., Zeng C., Wan L. Effects of Lonicera japonica extract on lung inflammation in LPS-induced ARDS rats. Journal of Guangdong Pharmaceutical University . 2017;33(3):379–382. [Google Scholar]
- 9.Park Y. C., Jin M., Kim S. H., Kim M. H., Namgung U., Yeo Y. Effects of inhalable microparticle of flower of _Lonicera japonica_ in a mouse model of COPD. Journal of Ethnopharmacology . 2014;151(1):123–130. doi: 10.1016/j.jep.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Shang X., Pan H., Li M., Miao X., Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Journal of Ethnopharmacology . 2011;138(1):1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu K. H., Rhee H. I., Kim J. H., et al. Anti-inflammatory and analgesic activities of SKLJI, a highly purified and injectable herbal extract ofLonicera japonica. Bioscience, Biotechnology, and Biochemistry . 2010;74(10):2022–2028. doi: 10.1271/bbb.100279. [DOI] [PubMed] [Google Scholar]
- 12.Tzeng T. F., Tzeng Y. C., Cheng Y. J., Liou S. S., Liu I. M. The ethanol extract from Lonicera japonica Thunb. regresses nonalcoholic steatohepatitis in a methionine- and choline-deficient diet-fed animal model. Nutrients . 2015;7(10):8670–8684. doi: 10.3390/nu7105423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y., Yin L., Zhang Y., et al. Study on anti-inflammatory efficacy and correlative ingredients with pharmacodynamics detected in acute inflammation rat model serum from Caulis _Lonicerae japonicae_. Phytomedicine . 2016;23(6):597–610. doi: 10.1016/j.phymed.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Lu H., Zhang L., Huang H. Study on the isolation of active constituents in Lonicera japonica and the mechanism of their anti-upper respiratory tract infection action in children. African Health Sciences . 2015;15(4):1295–1301. doi: 10.4314/ahs.v15i4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M., Chen J. L., Xu L. W., Ji G. Navigating traditional Chinese medicine network pharmacology and computational tools. Evidence-based Complementary and Alternative Medicine . 2013;2013:23. doi: 10.1155/2013/731969.731969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X. F., Cheng W. B., Jiang Y., et al. A network pharmacology-based strategy for predicting anti-inflammatory targets of ephedra in treating asthma. International Immunopharmacology . 2020;83, article 106423 doi: 10.1016/j.intimp.2020.106423. [DOI] [PubMed] [Google Scholar]
- 17.Cui S., Chen S., Wu Q., Chen T., Li S. A network pharmacology approach to investigate the anti-inflammatory mechanism of effective ingredients from Salvia miltiorrhiza. International Immunopharmacology . 2020;81:p. 106040. doi: 10.1016/j.intimp.2019.106040. [DOI] [PubMed] [Google Scholar]
- 18.Park S. Y., Jin M. L., Yi E. H., Kim Y., Park G. Neochlorogenic acid inhibits against LPS-activated inflammatory responses through up-regulation of Nrf2/HO-1 and involving AMPK pathway. Environmental Toxicology and Pharmacology . 2018;62:1–10. doi: 10.1016/j.etap.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Nie J., Chang Y., Li Y., et al. Caffeic acid phenethyl ester (propolis extract) ameliorates insulin resistance by inhibiting JNK and NF-κB inflammatory pathways in diabetic mice and HepG2 cell models. Journal of Agricultural and Food Chemistry . 2017;65(41):9041–9053. doi: 10.1021/acs.jafc.7b02880. [DOI] [PubMed] [Google Scholar]
- 20.Bai F., Fang L., Hu H., Yang Y., Feng X., Sun D. Vanillic acid mitigates the ovalbumin (OVA)-induced asthma in rat model through prevention of airway inflammation. Bioscience, Biotechnology, and Biochemistry . 2019;83(3):531–537. doi: 10.1080/09168451.2018.1543015. [DOI] [PubMed] [Google Scholar]
- 21.de Oliveira M. G., Marques R. B., de Santana M. F., et al. α-Terpineol reduces mechanical hypernociception and inflammatory response. Basic & Clinical Pharmacology & Toxicology . 2012;111(2):120–125. doi: 10.1111/j.1742-7843.2012.00875.x. [DOI] [PubMed] [Google Scholar]
- 22.Devi K. P., Malar D. S., Nabavi S. F., et al. Kaempferol and inflammation: from chemistry to medicine. Pharmacological Research . 2015;99:1–10. doi: 10.1016/j.phrs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Jiang K., Zhang T., Yin N., et al. Geraniol alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis. Oncotarget . 2017;8(41):71038–71053. doi: 10.18632/oncotarget.20298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khajevand-Khazaei M. R., Mohseni-Moghaddam P., Hosseini M., Gholami L., Baluchnejadmojarad T., Roghani M. Rutin, a quercetin glycoside, alleviates acute endotoxemic kidney injury in C57BL/6 mice via suppression of inflammation and up-regulation of antioxidants and SIRT1. European Journal of Pharmacology . 2018;833:307–313. doi: 10.1016/j.ejphar.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Kim M. G., Kim S. M., Min J. H., et al. Anti-inflammatory effects of linalool on ovalbumin-induced pulmonary inflammation. International Immunopharmacology . 2019;74, article 105706 doi: 10.1016/j.intimp.2019.105706. [DOI] [PubMed] [Google Scholar]
- 26.Ko W. C., Chen M. C., Wang S. H., Lai Y. H., Chen J. H., Lin C. N. 3-O-Methylquercetin more selectively inhibits phosphodiesterase subtype 3. Planta Medica . 2003;69(4):310–315. doi: 10.1055/s-2003-38874. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Yao J., Han C., et al. Quercetin, inflammation and immunity. Nutrients . 2016;8(3):p. 167. doi: 10.3390/nu8030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Y., Dong S., Kiyama R., Cai P., Liu L., Shen H. Flos lonicerae extracts and chlorogenic acid protect human umbilical vein endothelial cells from the toxic damage of perfluorooctane sulphonate. Inflammation . 2013;36(3):767–779. doi: 10.1007/s10753-013-9603-5. [DOI] [PubMed] [Google Scholar]
- 29.Liu D., Yu X., Sun H., Zhang W., Liu G., Zhu L. Flos lonicerae flavonoids attenuate experimental ulcerative colitis in rats via suppression of NF-κB signaling pathway. Naunyn-Schmiedeberg's Archives of Pharmacology . 2020;393(12):2481–2494. doi: 10.1007/s00210-020-01814-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y. M., Shen J. D., Xu L. P., Li H. B., Li Y. C., Yi L. T. Ferulic acid inhibits neuro-inflammation in mice exposed to chronic unpredictable mild stress. International Immunopharmacology . 2017;45:128–134. doi: 10.1016/j.intimp.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Ormazabal P., Scazzocchio B., Varì R., et al. Effect of protocatechuic acid on insulin responsiveness and inflammation in visceral adipose tissue from obese individuals: possible role for PTP1B. International Journal of Obesity . 2018;42(12):2012–2021. doi: 10.1038/s41366-018-0075-4. [DOI] [PubMed] [Google Scholar]
- 32.Park C. M., Song Y. S. Luteolin and luteolin-7-O-glucoside protect against acute liver injury through regulation of inflammatory mediators and antioxidative enzymes in GalN/LPS-induced hepatitic ICR mice. Nutrition Research and Practice . 2019;13(6):473–479. doi: 10.4162/nrp.2019.13.6.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian W., Kang A., Peng L., et al. Gas chromatography-mass spectrometry based plasma metabolomics of H1N1-induced inflammation in mice and intervention with Flos Lonicerae Japonica-Fructus Forsythiae herb pair. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences . 2018;1092:122–130. doi: 10.1016/j.jchromb.2018.05.047. [DOI] [PubMed] [Google Scholar]
- 34.Rakariyatham K., Wu X., Tang Z., Han Y., Wang Q., Xiao H. Synergism between luteolin and sulforaphane in anti-inflammation. Food & Function . 2018;9(10):5115–5123. doi: 10.1039/C8FO01352G. [DOI] [PubMed] [Google Scholar]
- 35.Ren K., Jiang T., Zhou H. F., Liang Y., Zhao G. J. Apigenin retards atherogenesis by promoting ABCA1-mediated cholesterol efflux and suppressing inflammation. Cellular Physiology and Biochemistry . 2018;47(5):2170–2184. doi: 10.1159/000491528. [DOI] [PubMed] [Google Scholar]
- 36.Sozański T., Kucharska A. Z., Rapak A., et al. Iridoid-loganic acid _versus_ anthocyanins from the _Cornus mas_ fruits (cornelian cherry): common and different effects on diet-induced atherosclerosis, PPARs expression and inflammation. Atherosclerosis . 2016;254:151–160. doi: 10.1016/j.atherosclerosis.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q., Xiao L. Isochlorogenic acid A attenuates acute lung injury induced by LPS via Nf-κB/NLRP3 signaling pathway. American Journal of Translational Research . 2019;11(11):7018–7026. [PMC free article] [PubMed] [Google Scholar]
- 38.Yang R., Fang L., Li J., Zhao Z., Zhang H., Zhang Y. Separation of five iridoid glycosides from Lonicerae japonicae flos using high-speed counter-current chromatography and their anti-inflammatory and antibacterial activities. Molecules . 2019;24(1) doi: 10.3390/molecules24010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y. Q., Zhao Y. T., Zhao X. Y., et al. Hyperoside suppresses lipopolysaccharide-induced inflammation and apoptosis in human umbilical vein endothelial cells. Curr Med Sci . 2018;38(2):222–228. doi: 10.1007/s11596-018-1869-2. [DOI] [PubMed] [Google Scholar]
- 40.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research . 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martucci D., Masseroli M., Pinciroli F. Gene ontology application to genomic functional annotation, statistical analysis and knowledge mining. Studies in Health Technology and Informatics . 2004;102:108–131. [PubMed] [Google Scholar]
- 42.Zhou Y., Zhang C. Y., Duan J. X., et al. Vasoactive intestinal peptide suppresses the NLRP3 inflammasome activation in lipopolysaccharide-induced acute lung injury mice and macrophages. Biomedicine & Pharmacotherapy . 2020;121, article 109596 doi: 10.1016/j.biopha.2019.109596. [DOI] [PubMed] [Google Scholar]
- 43.Haegeman G. Inhibition of signal transduction pathways involved in inflammation. European Respiratory Journal . 2003;22(Supplement 44):16s–19s. doi: 10.1183/09031936.03.00000503a. [DOI] [PubMed] [Google Scholar]
- 44.Shi Z. M., Han Y. W., Han X. H., et al. Upstream regulators and downstream effectors of NF-κB in Alzheimer’s disease. Journal of the Neurological Sciences . 2016;366:127–134. doi: 10.1016/j.jns.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Koorella C., Nair J. R., Murray M. E., Carlson L. M., Watkins S. K., Lee K. P. Novel regulation of CD80/CD86-induced phosphatidylinositol 3-kinase signaling by NOTCH1 protein in interleukin-6 and indoleamine 2,3-dioxygenase production by dendritic cells∗♦. The Journal of Biological Chemistry . 2014;289(11):7747–7762. doi: 10.1074/jbc.M113.519686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhandapani K. M., Wade F. M., Wakade C., Mahesh V. B., Brann D. W. Neuroprotection by stem cell factor in rat cortical neurons involves AKT and NFkappaB. Journal of Neurochemistry . 2005;95(1):9–19. doi: 10.1111/j.1471-4159.2005.03319.x. [DOI] [PubMed] [Google Scholar]
- 47.Bai W., Liu H., Ji Q., et al. TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cellular Signalling . 2014;26(5):942–950. doi: 10.1016/j.cellsig.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Aksoy E., Taboubi S., Torres D., et al. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nature Immunology . 2012;13(11):1045–1054. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fallah M. P., Chelvarajan R. L., Garvy B. A., Bondada S. Role of phosphoinositide 3-kinase-Akt signaling pathway in the age-related cytokine dysregulation in splenic macrophages stimulated via TLR-2 or TLR-4 receptors. Mechanisms of Ageing and Development . 2011;132(6-7):274–286. doi: 10.1016/j.mad.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown J., Wang H., Hajishengallis G. N., Martin M. TLR-signaling networks: an integration of adaptor molecules, kinases, and cross-talk. Journal of Dental Research . 2011;90(4):417–427. doi: 10.1177/0022034510381264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan W., Morinaga H., Kim J. J., et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. The EMBO Journal . 2010;29(24):4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laird M. H., Rhee S. H., Perkins D. J., et al. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. Journal of Leukocyte Biology . 2009;85(6):966–977. doi: 10.1189/jlb.1208763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han M. H., Lee W. S., Nagappan A., et al. Flavonoids isolated from flowers of Lonicera japonica Thunb. Inhibit inflammatory responses in BV2 microglial cells by suppressing TNF-α and IL-β through PI3K/Akt/NF-kb signaling pathways. Phytotherapy Research . 2016;30(11):1824–1832. doi: 10.1002/ptr.5688. [DOI] [PubMed] [Google Scholar]
- 54.Sanajou D., Ghorbani Haghjo A., Argani H., Aslani S. AGE-RAGE axis blockade in diabetic nephropathy: current status and future directions. European Journal of Pharmacology . 2018;833:158–164. doi: 10.1016/j.ejphar.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Palazon A., Goldrath A. W., Nizet V., Johnson R. S. HIF transcription factors, inflammation, and immunity. Immunity . 2014;41(4):518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Zhou Z., Han M., et al. The anti-inflammatory components from the effective fraction of Syringae Folium (ESF) and its mechanism investigation based on network pharmacology. Bioorganic Chemistry . 2020;99, article 103764 doi: 10.1016/j.bioorg.2020.103764. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen N. T., Nguyen T. H., Pham T. N. H., et al. Autodock Vina adopts more accurate binding poses but Autodock4 forms better binding affinity. Journal of Chemical Information and Modeling . 2020;60(1):204–211. doi: 10.1021/acs.jcim.9b00778. [DOI] [PubMed] [Google Scholar]
- 58.Nam Y. J., Lee C. S. Protocatechuic acid inhibits toll-like receptor-4-dependent activation of NF- κB by suppressing activation of the Akt, mTOR, JNK and p38-MAPK. International Immunopharmacology . 2018;55:272–281. doi: 10.1016/j.intimp.2017.12.024. [DOI] [PubMed] [Google Scholar]
- 59.Lu M., Dai Y., Xu M., et al. The attenuation of 14-3-3ζ is involved in the caffeic acid-blocked lipopolysaccharide-stimulated inflammatory response in RAW264.7 macrophages. Inflammation . 2017;40(5):1753–1760. doi: 10.1007/s10753-017-0618-1. [DOI] [PubMed] [Google Scholar]
- 60.Huang X., Xi Y., Mao Z., et al. Vanillic acid attenuates cartilage degeneration by regulating the MAPK and PI3K/AKT/NF-κB pathways. European Journal of Pharmacology . 2019;859, article 172481 doi: 10.1016/j.ejphar.2019.172481. [DOI] [PubMed] [Google Scholar]
- 61.Jiang T., Zhang L., Ding M., Li M. Protective effect of vasicine against myocardial infarction in rats via modulation of oxidative stress, inflammation, and the PI3K/Akt pathway. Drug Design, Development and Therapy . 2019;13:3773–3784. doi: 10.2147/DDDT.S220396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cole P. A., Chu N., Salguero A. L., Bae H. AKTivation mechanisms. Current Opinion in Structural Biology . 2019;59:47–53. doi: 10.1016/j.sbi.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.


