Abstract
The present paper analyzes the histoplasmin electrophoretic profiles and the randomly amplified polymorphic DNA (RAPD) patterns of the fungus Histoplasma capsulatum isolated from Mexican patients with AIDS-associated histoplasmosis. Clinical isolates from Guatemala, Colombia, and Panama, as well as H. capsulatum isolates from different sources in nature, were also processed. All histoplasmin samples shared four antigenic fractions of 200, 49, 10.5, and 8.5 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). According to their percentage of relatedness, based on SDS-PAGE histoplasmin electrophoretic image analysis, H. capsulatum isolates were divided in two groups: group A contained all AIDS-associated isolates studied and two human reference strains from Mexican histoplasmosis patients without AIDS; group B included bat guano, infected bat, and cock excreta isolates from the State of Guerrero, Mexico, plus three human histoplasmosis strains from Guatemala, Panama, and Colombia. Polymorphic DNA patterns evaluated by RAPD-PCR showed three major bands of 4.4, 3.2, and 2.3 kb in most H. capsulatum isolates studied. Four groups were related by DNA polymorphisms: group I was formed by most of the AIDS-associated H. capsulatum isolates studied, one human histoplasmosis strain from Colombia, two human reference strains from Mexican patients without AIDS, and one human histoplasmosis strain from Guatemala. Group II consisted of only a single strain from Panama. Group III included three strains: one from a Mexican patient with AIDS and two isolated from nature in Guerrero (cock excreta and bat guano). The last, group IV, consisted of only one strain isolated from an infected bat, captured in Guerrero. A tight relationship between phenotypic and genotypic characterization was observed, and both analyses could be useful tools for typing H. capsulatum from different sources and geographic origins.
Histoplasmosis, a systemic mycosis, has been reported worldwide, although it is most frequently found in the American continents. Its causative agent, the dimorphic fungus Histoplasma capsulatum var. capsulatum, grows as an infectious mycelial phase in contaminated substrata at ambient temperatures, as a parasitic intracellular yeast phase in susceptible hosts, or in media that favor yeast growth at 37°C (14). In the infected host, the fungus finds several special microenvironments where it can survive, particularly within professional phagocytes (7, 8). Infection may or may not cause symptomatic illness, depending on the efficiency of the immune response (29). Failures in cellular immunity result in a disseminated form of the disease. Endogenous reactivation may occur when previously infected individuals become immunosuppressed, with AIDS, for example (17, 33). In Mexico, the number of H. capsulatum isolates associated with AIDS is rising, and information concerning their epidemiology is not yet available (5, 9, 21, 23). H. capsulatum isolates from the United States, Panama, and Puerto Rico have been grouped into six different classes according to their DNA polymorphisms (12). Several AIDS-associated H. capsulatum isolates were included in classes 5 and 6 as suggested by Keath et al. (12) and in class 1 by Spitzer et al. (26). These classifications have also been related to the virulence and geographic distribution of H. capsulatum strains (12, 26). Most non-AIDS-associated histoplasmosis clinical isolates have been included in class 2 (12, 26, 27, 31).
This study was aimed at determining the phenotypic and genotypic relatedness between H. capsulatum isolates from Mexican AIDS patients and isolates from other sources and geographic origins.
MATERIALS AND METHODS
Fungal isolates and cultures.
Sources and geographic origins of H. capsulatum isolates used in this study are provided in Table 1. Mycelial-phase precultures were grown in GYE broth medium (2% glucose and 1% yeast extract) at 28°C.
TABLE 1.
H. capsulatum isolates
| Isolatea | Sources and Geographic origin |
|---|---|
| EH-46 | Histoplasmosis patient without AIDS; Guerrero, Mexico |
| EH-53 | Histoplasmosis patient without AIDS; Hidalgo, Mexico |
| EH-313 | Bat guano, Guerrero, Mexico |
| EH-314 | Cock excreta; house yard in Guerrero, Mexico |
| EH-315 | Infected adult female bat; Guerrero, Mexico |
| EH-316 | Histoplasmosis patient with AIDS; Guerrero, Mexico |
| EH-317 | Histoplasmosis patient with AIDS; Morelos, Mexico |
| EH-318 | Histoplasmosis patient with AIDS; Morelos, Mexico |
| EH-319 | Histoplasmosis patient with AIDS; Mexico City, Mexico |
| EH-323 | Histoplasmosis patient with AIDS; Hidalgo, Mexico |
| EH-325 | Histoplasmosis patient with AIDS; Chiapas, Mexico |
| H.1.02.W | Human histoplasmosis; Guatemala City, Guatemala |
| GM | Human histoplasmosis; Medellín, Colombia |
| G-186B | Human histoplasmosis; Panama |
Strains EH-46 through EH-315 were obtained from Laboratorio Immunología de Hongos (LIH), Facultad de Medicina, Universidad Nacional Autónoma de México. Strain EH-53 is the LIH reference strain. Strain EH-313 was isolated from Juxtlahuaca cave. Strain EH-315 was obtained from an infected bat (Mormoops megalophylla), Coapala cave. Strains EH-316 through EH-325 were obtained from patients at the Instituto Nacional de la Nutrición “Salvador Zubirán.” Strain H.1.02.W was provided by H. Logemann, San Carlos University. Strain GM was provided by J. G. McEwen, Corporación para Investigaciones Biológicas. Strain G-186B was provided by G. S. Kobayashi, Washington University, St. Louis, Mo.
Fungal identification.
The identity of H. capsulatum isolates was determined morphologically. Mycelium-to-yeast conversion was obtained at 37°C in brain heart infusion (BHI) broth (Bioxón, Mexico City, Mexico) supplemented with 0.1% l-cysteine and 1% glucose. Identity was confirmed by the exoantigen test of Standard and Kaufman (11, 28), which was performed in double immunodiffusion (19). The positive reference antigen was the histoplasmin from our laboratory (strain EH-53). A positive human histoplasmosis serum sample and a negative serum sample from a healthy human volunteer were used as reference sera. The sera were previously standardized.
Antigen production.
For the Standard and Kaufman test (11, 28) a small volume of each exoantigen from a 15-day-old H. capsulatum strain in BHI broth culture was concentrated by ultrafiltration through membrane filters with a 10,000-molecular-weight cutoff (Millipore Corporation, Bedford, Mass.). For polyacrylamide electrophoresis profiles of the crude antigen histoplasmin, homogeneous inocula of each H. capsulatum strain in mid-log-phase culture were harvested from GYE precultures and grown in Smith’s synthetic asparagine broth at 28°C (24). All H. capsulatum strains were cultured at the same time with the same batch of Smith medium. Histoplasmin from each 3-month-old Smith medium culture was filtered on paper to remove the mycelium, dialyzed, and concentrated in the Amicon Cell System (Amicon, Lexington, Mass.) using a PM-10 membrane with a 10,000-molecular-weight cutoff. Each histoplasmin sample was stored at −80°C in the presence of 2 mM phenylmethylsulfonyl fluoride (Gibco Laboratories, Grand Island, N.Y.). Concentrations of proteins (16) and carbohydrates (6) were accurately determined prior to their use in the different assays.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of histoplasmins (500 μg/ml) diluted 1:2 in the sample buffer (10 mM Tris-HCl; 1 mM EDTA, pH 8.0; 2.5% SDS; 5% β-mercaptoethanol; 0.01% bromophenol blue) was performed following the method of Laemmli (15) in a Mini-Protean II electrophoresis cell apparatus (Bio-Rad Laboratories, Richmond, Calif.), using 12.5% homogeneous gels, at 100 V for 1 h. Protein size standards were obtained from 205- to 6.5-kDa molecular mass markers (Sigma Chemical Co., St. Louis, Mo.), and bands were silver stained as described by Heukeshoven and Dernick (10).
Isolation of H. capsulatum whole-cell DNA.
Mycelial-phase H. capsulatum in GYE agar cultures was inoculated into 50 ml of GYE broth precultures and incubated at 28°C, with shaking at 120 rpm. Each preculture was added to 500 ml of fresh GYE broth and incubated at 28°C for 2 to 3 weeks, with shaking at 120 rpm. Procedures were followed as described elsewhere (25). Briefly, homogeneous log-phase growth mycelia were collected and ground with a mortar and pestle in liquid N2, suspended in 50 mM Tris (pH 8.0), with 62.5 mM EDTA and 2% SDS, and stirred for 1 h at room temperature. After centrifugation at 4,000 × g for 10 min at 4°C, 1/4 volume of 8 M potassium acetate was added to the collected supernatant. The mixture was incubated on ice for 30 min and then centrifuged at 16,300 × g for 10 min at 4°C. Nucleic acids were precipitated by adding 1/4 volume of 8 M ammonium acetate and an equal volume of isopropanol to the supernatant. Separated DNA was further treated with RNase A and proteinase K and subjected to phenol-chloroform extraction by a standard method (22).
RAPD-PCR assay.
Randomly amplified polymorphic DNA-PCR (RAPD-PCR) is a rapid, PCR-based amplification method and was performed as previously described for H. capsulatum strains by Kersulyte et al. (13) and Woods et al. (34). The assay was done in a 20-μl reaction mixture with 1 ng of H. capsulatum DNA, 10 mM Tris-HCl at pH 8.3, 2 mM MgCl2, 200 μM deoxynucleoside triphosphates, 15 pmol of random primer 1281 (5′-AACGCGCAAC) (Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, Mexico), and 1 U of Taq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.). The reaction mixture was overlaid with 1 drop of mineral oil. The PCR amplification was performed for 35 cycles in a thermal cycler (Perkin-Elmer Cetus), where each cycle consisted of 10 s at 94°C, 30 s at 36°C, and 1 min at 72°C. The PCR product was resolved by 1.5% agarose gel electrophoresis and stained with ethidium bromide. Molecular size standards were derived from phage λ cut with HindIII and φX174 cut with HaeIII (Stratagene Cloning Systems, La Jolla, Calif.).
Image analyses.
The SDS-PAGE histoplasmin band profiles, as well as the DNA amplified fragment patterns obtained by RAPD-PCR, were photographed to record their relative migration (Rf). The Rf values of the histoplasmin antigenic fractions and of the DNA fragments were compiled and converted into 1-0 data matrices.
The molecular weight (Mr) of each histoplasmin band and the size (in kilobases) of each DNA band were calculated by reference to the standards (22, 32).
Analyses using SPSS/PC+.
SPSS/PC+ software, version 2.3, was used. Matrices based on Rf values were analyzed by an unweighted group method with average linkage between groups to elaborate each dendrogram (18).
RESULTS
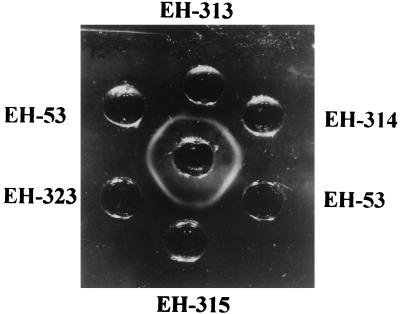
The fungal isolates (Table 1) were identified according to several criteria: development of the conventional colonial morphology of H. capsulatum with white to buff brown colonies, the presence of typical microscopic characteristics with a variable number of microconidia, and observation of abundant round to pyriform macroconidia with finger-like projections; conversion to the yeast phase in 1 to 3 weeks; and finally, production of precipitin lines by exoantigens reacting with human histoplasmosis immune serum. Lines of identity can be seen for selected H. capsulatum isolates in Fig. 1.
FIG. 1.
Exoantigens of H. capsulatum strains. Center well, positive human histoplasmosis immune serum; EH-53, human reference strain; EH-313, H. capsulatum isolated from bat guano; EH-314, H. capsulatum isolated from cock excreta; EH-315, H. capsulatum isolated from an infected bat; EH-323, H. capsulatum isolated from Mexican histoplasmosis patient with AIDS.
Histoplasmin electrophoretic profiles.
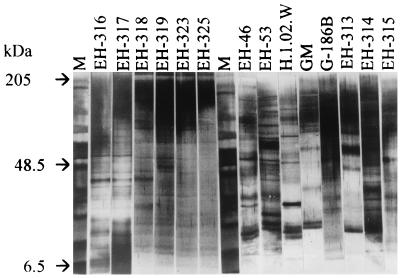
The phenotypic relationships of the studied H. capsulatum isolates were estimated by using the SDS-PAGE protein electrophoretic profiles of histoplasmin samples. Results showed four common bands of 200, 49, 10.5, and 8.5 kDa in all H. capsulatum isolates studied (Fig. 2). Homogeneous electrophoretic profiles, including eleven common bands of 200, 77, 54, 49, 43, 28, 16.5, 14, 11.5, 10.5, and 8.5 kDa, were observed in repeated assays with histoplasmin samples by using H. capsulatum isolated in Mexico from histoplasmosis patients with AIDS. The relatedness of 14 histoplasmin electrotypes was analyzed by the SPSS/PC+ program to elaborate a dendrogram (Fig. 3) defining two groups (A and B) of H. capsulatum strains, which showed 75% relatedness. Group A contained eight strains that exhibited 80% relatedness and had two subgroups. Subgroup A1 contained six Mexican AIDS-associated isolates (EH-316 to 319, EH-323, and EH-325), which were 86% related. Subgroup A2 contained the two Mexican non-AIDS-associated isolates (EH-46 and EH-53), which were 87% related. Group B included six strains with 80% relatedness among them and had three subgroups. Subgroup B1 contained strains isolated from bat guano (EH-313) and an infected bat (EH-315) in Guerrero, which were 89% related. Subgroup B2 had the clinical isolates from Panama (G-186B) and Guatemala (H.1.02.W), as well as the strain isolated from cock excreta (EH-314) in Guerrero, all of which were 87% related. Finally, subgroup B3 contained the Colombian strain (GM), which was 82% related to the former subgroup.
FIG. 2.
SDS-PAGE of histoplasmin from H. capsulatum isolates from different sources and geographic origins. Lanes 1 and 8, molecular weight markers. The gel was silver stained as described elsewhere (10). Image analysis was performed as described in Materials and Methods.
FIG. 3.
Dendrogram showing relatedness among H. capsulatum isolates from different sources and geographic origins (Table 1) according to their histoplasmin electrophoretic profiles. The SPSS/PC+ computer program was used to elaborate the dendrogram.
RAPD patterns of H. capsulatum isolates.
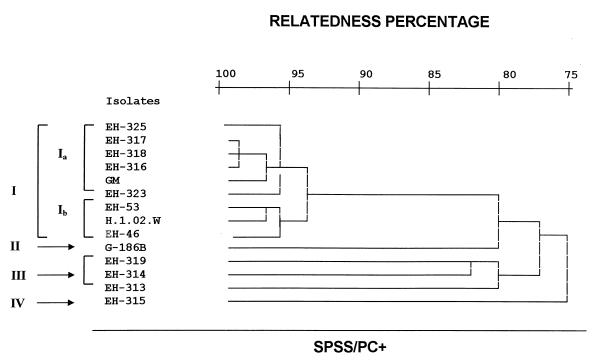
The genotypic relation of the H. capsulatum isolates was tested by using polymorphic DNA band patterns obtained by the RAPD-PCR method. Results showed three major bands of 4.4, 3.2, and 2.3 kb in most H. capsulatum isolates studied (Fig. 4). The same polymorphic DNA patterns were observed in repeated assays of H. capsulatum DNA isolated from Mexican patients with AIDS-associated histoplasmosis. Relatedness among isolates through the polymorphic DNA patterns was analyzed by using the SPSS/PC+ program to elaborate a dendrogram (Fig. 5) and revealed 75% relatedness among four groups (I to IV) of H. capsulatum strains. Group I was formed by nine strains with 94% relatedness and showed two subgroups. Subgroup Ia contained five of the Mexican AIDS-associated isolates (EH-316 to -318, EH-323, and EH-325) and the Colombian strain (GM), which were 96% related. Subgroup Ib contained the two Mexican non-AIDS isolates (EH-46 and EH-53) and the Guatemalan strain (H.1.02.W), which were also 96% related. Group II contained only one strain from Panama (G-186B), which was 80% related to group I strains. Group III included three strains, which were 80% related. Two group III strains were isolated from nature in the state of Guerrero, one from bat guano (EH-313) and another from cock excreta (EH-314); the third strain was isolated from a Mexican AIDS patient (EH-319). Finally, group IV contained only the strain isolated from an infected bat (EH-315) captured in a cave in the state of Guerrero, which was 75% related to all former strains.
FIG. 4.
RAPD-PCR patterns obtained for H. capsulatum isolates from different sources and geographic origins, generated by primer 1281. Lanes 1, 8, and 17 size markers; lane 18, negative control. The gel was ethidium bromide stained. Image analysis was performed as described in Materials and Methods.
FIG. 5.
Dendrogram showing relatedness among H. capsulatum isolates from different sources and geographic origins (Table 1) according to their polymorphic DNA patterns. The SPSS/PC+ computer program was used to elaborate the dendrogram.
DISCUSSION
The diversity of DNA polymorphisms detected by restriction fragment length polymorphisms and Southern blotting with mitochondrial, ribosomal, or yps-3 nuclear gene probes have allowed medical mycologists to propose classifications of environmental and clinical H. capsulatum isolates that correlate with the geographic distribution of the strains (12, 26, 27, 31). The present paper establishes relatedness among strains by using SDS-PAGE protein electrophoretic profiles of histoplasmin antigenic fractions and DNA polymorphisms based on RAPD patterns. Image analyses of the findings of both methods showed that the strains studied had a 75 to 99% relatedness. The analyses also revealed relationships among the H. capsulatum strains correlating with their sources of isolation and geographic origins (Table 2). Based on these analyses, it is possible that strains EH-316-318, EH-323 and EH-325, isolated from Mexican patients with AIDS-associated histoplasmosis, share similarities with H. capsulatum isolates from Mexican histoplasmosis patients without AIDS, such as the EH-46 and the EH-53 strains (Table 2). The relationship of EH-53 strain to the above-mentioned H. capsulatum isolates is especially important due to its isolation from a miner infected in an abandoned silver mine in the state of Hidalgo, which supports a direct correlation between source of infection and development of the disease. Furthermore, a particularly important aspect that could explain their close relatedness is the geographic proximity of the residences of the patients listed in Table 1 (sources of strains EH-316, EH-317, EH-318, EH-323, EH-46, and EH-53). Of course, it is sometimes difficult to know the precise geographic location where a human being contracted an infection. The different behavior shown by the EH-319 clinical isolate, detected through the more sensitive typing method (RAPD-PCR), could be accounted for by its probably erroneous allocation to Mexico City. However, most of the H. capsulatum isolates from Mexican histoplasmosis patients have been associated with known areas of endemic histoplasmosis infection in Mexico (20, 30).
TABLE 2.
Phenotypic and genotypic comparison among H. capsulatum strains
| Source | Isolate | Electrophoresis groupa | RAPD-PCR groupa |
|---|---|---|---|
| Patients | |||
| Mexican | |||
| With AIDS | EH-316 | A1 | Ia |
| EH-317 | A1 | Ia | |
| EH-318 | A1 | Ia | |
| EH-319 | A1 | III | |
| EH-323 | A1 | Ia | |
| EH-325 | A1 | Ia | |
| Without AIDS | EH-46 | A2 | Ib |
| EH-53 | A2 | Ib | |
| Non-Mexican, without AIDS | H.1.02.W | B2 | Ib |
| GM | B3 | Ia | |
| G-186B | B2 | II | |
| Other | |||
| Bat guano | EH-313 | B1 | III |
| Cock excreta | EH-314 | B2 | III |
| Infected bat | EH-315 | B1 | IV |
Based on relatedness obtained from the dendrograms elaborated with SDS-PAGE and RAPD-PCR data.
It could be suggested that the phenotypic expression of the different electrophoretic profiles of the analyzed histoplasmin samples may be influenced by habitat interactions either with the infected host or with the environment. Such an influence has been observed in cases of Candida albicans infection, where isolates from patients with AIDS-associated candidiasis express a different serotype from those isolated from candidiasis patients without AIDS (2). Furthermore, it has also been observed that some factors, such as the pH of the environment in which the fungus grows, can exert a marked effect on the expression of the antigens responsible for the serotype specificity in Candida (1).
Analyses with biallelic markers by Carter et al. (4) of population structure in clinical H. capsulatum isolates from Indianapolis, Ind., revealed diversities in class 2 isolates. No associations between isolates from immunocompromised patients or from other clinical manifestations of histoplasmosis were found (4). Although molecular or phenotypic studies with H. capsulatum isolates found outside the United States are scarce, multiallelic markers were reported recently as useful tools for distinguishing some Colombian isolates from U.S. isolates (3). Considering these findings and the present results, it is important to continue these studies in order to elaborate a representative classification of H. capsulatum populations distributed in the American continents.
ACKNOWLEDGMENTS
We gratefully acknowledge Alice Piaget and Ingrid Mascher for their appropriate criticisms and editorial assistance.
This research was supported by Dirección General de Asuntos del Personal Académico (DGAPA), grant DGAPA-UNAM-IN203294.
REFERENCES
- 1.Baturen B, Bikandi J, San Millán R, Morangues M D, Regulez P, Quindós G, Pontón J. Variability in expression of antigens responsible for serotype specificity in Candida albicans. Microbiology. 1995;141:1535–1543. doi: 10.1099/13500872-141-7-1535. [DOI] [PubMed] [Google Scholar]
- 2.Brawner D L, Cutler J E. Oral Candida albicans isolates from nonhospitalized normal carriers, immunocompetent hospitalized patients, and immunocompromised patients with or without acquired immunodeficiency syndrome. J Clin Microbiol. 1989;27:1335–1341. doi: 10.1128/jcm.27.6.1335-1341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter D A, Burt A, Taylor J W, Koening G L, Dechairo B M, White T J. A set of electrophoretic molecular markers for strain typing and population genetic studies of Histoplasma capsulatum. Electrophoresis. 1997;18:1047–1053. doi: 10.1002/elps.1150180703. [DOI] [PubMed] [Google Scholar]
- 4.Carter D A, Burt A, Taylor J W, Koenig G L, White T J. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casanova-Cardiel L J, Ruíz-Ordaz I. Histoplasma capsulatum en sangre periférica de pacientes con SIDA. Informe de cuatro casos con elevación de deshidrogenasa láctica. Rev Investig Clin. 1993;45:67–70. [PubMed] [Google Scholar]
- 6.Dubois M, Giles K, Hamilton J K, Reber P A, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 7.Eissenberg L G, Goldman W E. Histoplasma variation and adaptive strategies for parasitism: new perspectives on histoplasmosis. Clin Microbiol Rev. 1991;4:411–421. doi: 10.1128/cmr.4.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eissenberg L G, Goldman W E. The interplay between Histoplasma capsulatum and its host cells. Bailliere’s Clin Infect Dis. 1994;1:265–283. [Google Scholar]
- 9.Gómez-Campos G, Villarreal-Urenda C, Robles-Romo M, Cano-Domínguez C. Hallazgos histopatológicos en 102 autopsias de pacientes con síndrome de inmunodeficiencia adquirida. Rev Med Inst Mex Seguro Soc. 1992;30:171–176. [Google Scholar]
- 10.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman L, Standard P G. Specific and rapid identification of medically important fungi by exoantigen detection. Annu Rev Microbiol. 1987;41:209–225. doi: 10.1146/annurev.mi.41.100187.001233. [DOI] [PubMed] [Google Scholar]
- 12.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea and Febiger; 1992. pp. 464–513. [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 17.McKinsey D S, Spiegel R A, Hutwagner L, Stanford J, Driks M R, Brewer J, Gupta M R, Smith D L, O’Connor C, Dall L. Prospective study of histoplasmosis in patients infected with human immunodeficiency virus: incidence, risk factors, and pathophysiology. Clin Infect Dis. 1997;24:1195–1203. doi: 10.1086/513653. [DOI] [PubMed] [Google Scholar]
- 18.Norusis M J. SPSS/PC+ for the IBM PC/XT/AT. Chicago, Ill: SPSS Inc.; 1986. [Google Scholar]
- 19.Ouchterlony O, Nilsson L A. Immunodiffusion and immunoelectrophoresis. In: Weir D M, editor. Handbook of experimental immunology. Oxford, England: Blackwell Scientific Publications; 1978. pp. 19.1–19.44. [Google Scholar]
- 20.Pedroza-Serés M, Quiroz-Mercado H, Granados J, Taylor M L. The syndrome of presumed ocular histoplasmosis in Mexico: a preliminary study. J Med Vet Mycol. 1994;32:83–92. doi: 10.1080/02681219480000131. [DOI] [PubMed] [Google Scholar]
- 21.Robles-Romo M, Villarreal-Urenda C, Cano-Domínguez C, Gómez-Campos G, Ruíz-Rodríguez A. Infección por Histoplasma capsulatum en pacientes con SIDA. Aspectos epidemiológicos, clínicos y terapéuticos en 34 casos. Rev Med Inst Mex Seguro Soc. 1992;30:195–199. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 6.1–6.62. [Google Scholar]
- 23.Sánchez-Mejorada G, Ponce de León S, Ruíz-Palacios G M. Historia natural de la infección por VIH. In: Sepúlveda-Amor J, Brofman M, Ruíz-Palacios G, editors. SIDA, ciencia y sociedad en México. Mexico City, Mexico: Fondo de Cultura Económica; 1989. pp. 131–159. [Google Scholar]
- 24.Smith C E, Whiting E G, Bake E E, Rosenberger H G, Beard R R, Saito M T. The use of coccidioidin. Am Rev Tuberc. 1948;57:330–360. doi: 10.1164/art.1948.57.4.330. [DOI] [PubMed] [Google Scholar]
- 25.Spitzer E D. RFLP and its application to epidemiology of Histoplasma capsulatum. In: Maresca B, Kobayashi G S, editors. Molecular biology of pathogenic fungi: a laboratory manual. 2nd ed. New York, N.Y: Telos Press; 1994. pp. 375–379. [Google Scholar]
- 26.Spitzer E D, Keath E J, Travis S J, Painter A A, Kobayashi G S, Medoff G. Temperature-sensitive variants of Histoplasma capsulatum isolated from patients with acquired immunodeficiency syndrome. J Infect Dis. 1990;162:258–261. doi: 10.1093/infdis/162.1.258. [DOI] [PubMed] [Google Scholar]
- 27.Spitzer E D, Lasker B A, Travis S J, Kobayashi G S, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Standard P G, Kaufman L. Specific immunological test for the rapid identification of members of the genus Histoplasma. J Clin Microbiol. 1976;3:191–199. doi: 10.1128/jcm.3.2.191-199.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor M L, Díaz S, González P A, Sosa A C, Toriello C. Relationship between pathogenesis and immune regulation mechanisms in histoplasmosis: a hypothetical approach. Rev Infect Dis. 1984;6:775–782. doi: 10.1093/clinids/6.6.775. [DOI] [PubMed] [Google Scholar]
- 30.Taylor M L, Pérez-Mejía A, Yamamoto-Furusho J K, Granados J. Immunologic, genetic and social human risk factors associated to histoplasmosis: studies in the State of Guerrero, Mexico. Mycopathologia. 1997;138:137–141. doi: 10.1023/a:1006847630347. [DOI] [PubMed] [Google Scholar]
- 31.Vincent R D, Goewert R, Goldman W E, Kobayashi G S, Lambowitz A M, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 33.Wheat J. Endemic mycoses in AIDS: a clinical review. Clin Microbiol Rev. 1995;8:146–159. doi: 10.1128/cmr.8.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woods J P, Kersulyte D, Goldman W E, Berg D E. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J Clin Microbiol. 1993;31:463–464. doi: 10.1128/jcm.31.2.463-464.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]