Abstract.
Purpose: We propose a deep learning method for the automatic diagnosis of COVID-19 at patient presentation on chest radiography (CXR) images and investigates the role of standard and soft tissue CXR in this task.
Approach: The dataset consisted of the first CXR exams of 9860 patients acquired within 2 days after their initial reverse transcription polymerase chain reaction tests for the SARS-CoV-2 virus, 1523 (15.5%) of whom tested positive and 8337 (84.5%) of whom tested negative for COVID-19. A sequential transfer learning strategy was employed to fine-tune a convolutional neural network in phases on increasingly specific and complex tasks. The COVID-19 positive/negative classification was performed on standard images, soft tissue images, and both combined via feature fusion. A U-Net variant was used to segment and crop the lung region from each image prior to performing classification. Classification performances were evaluated and compared on a held-out test set of 1972 patients using the area under the receiver operating characteristic curve (AUC) and the DeLong test.
Results: Using full standard, cropped standard, cropped, soft tissue, and both types of cropped CXR yielded AUC values of 0.74 [0.70, 0.77], 0.76 [0.73, 0.79], 0.73 [0.70, 0.76], and 0.78 [0.74, 0.81], respectively. Using soft tissue images significantly underperformed standard images, and using both types of CXR failed to significantly outperform using standard images alone.
Conclusions: The proposed method was able to automatically diagnose COVID-19 at patient presentation with promising performance, and the inclusion of soft tissue images did not result in a significant performance improvement.
Keywords: COVID-19, chest radiography, computer-aided diagnosis, deep learning, early diagnosis
1. Introduction
The prolonged COVID-19 pandemic has profoundly impacted global public health and the economy. Early diagnosis of the COVID-19 disease is crucial not only for optimal implementation of treatment in individual patient care but also for disease containment and medical resource allocation from a public health perspective. Laboratory confirmation of SARS-CoV-2 is performed with a virus-specific reverse transcription polymerase chain reaction (RT-PCR) test. While there have been efforts to increase the RT-PCR testing capacity, shortages of test kits and long processing time remain a problem in resource-limited settings during surges, and the test has variable and moderate sensitivity (a systematic review reports 71% to 98% based on repeated testing).1,2 Chest radiography (CXR) is recommended for triaging at patient presentation and disease monitoring due to its fast speed, relatively low cost, wide availability, and portability.3,4 Characteristics, such as bilateral lower lobe consolidations, ground glass densities, peripheral air space opacities, and diffuse air space disease on CXR, have been related to COVID-19.5,6 Unfortunately, the nonspecificity of these features along with the shortage of radiological expertise due to the stress on healthcare resources during the pandemic make precise interpretation of such images challenging. Under such circumstances, deep learning can potentially assist in this task. In this study, we investigate the role of standard and soft tissue CXR images in the task of automated COVID-19 early diagnosis at patient presentation using deep learning.
There have been numerous studies on artificial intelligence (AI) applications for COVID-19 using CXR.7–14 However, due to difficulties in collecting sizeable datasets, many of these studies utilized publicly available datasets that consist of images extracted from publications,15,16 which by nature are not a representative selection of the patient population and may lead to biased results that cannot be recommended for clinical use.17 We therefore curated a large CXR database consecutively collected from our institution for this research. Moreover, large public CXR datasets established prior to the pandemic were usually utilized to enrich the training set.18–20 These images, all COVID-19 negative, differ from the COVID-19 positive cases in newly acquired datasets in image acquisition protocol, scanners, and patient population. Consequently, pooling them together would have introduced confounding variables into the classification task and potentially yielded overoptimistic results, because the models might learn to use these irrelevant factors to distinguish COVID-19 positive and negative, rather than identify disease presentations. To leverage prepandemic CXR datasets without adding confounding variables, in this study we designed a three-phase strategy to sequentially fine-tune the model during training, instead of pooling the datasets. Furthermore, while most current imaging AI research was developed on all COVID-19 cases available, including images acquired when the disease has progressed, our study tackled the challenge of COVID-19 early diagnosis at initial patient presentation, which is important for implementing isolation and treatment promptly. Finally, while most prior studies only considered standard CXR images, our work also investigated the role of soft tissue images in automated COVID-19 diagnosis using deep learning as they exhibit diagnostic utility in radiologists’ clinical assessment.
2. Methods
2.1. Database
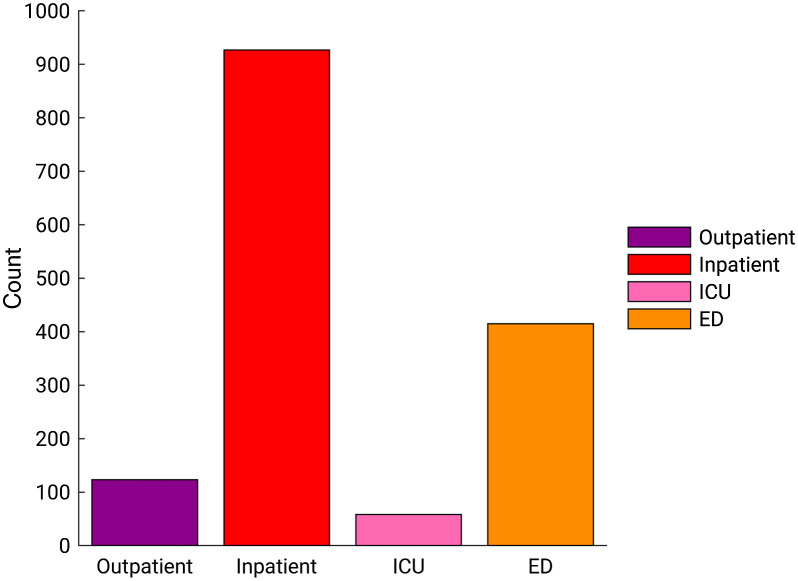
A database was retrospectively collected under a HIPAA-compliant, IRB-approved protocol during the COVID-19 outbreak, consisting of CXR exams acquired between January 30, 2020 and February 3, 2021, and their corresponding clinical reports. From adult patients who underwent the RT-PCR test for the SARS-CoV-2 virus at our institution, we consecutively collected their CXR exams after and up to a year prior to their initial RT-PCR tests. The first CXR exam after each patient’s initial RT-PCR test was selected for this study, as it is the earliest available image of the patient with confirmed COVID-19 status. Dual-energy subtraction (DES) exams and portable exams with a ClearRead bone suppression series (Riverain Technologies) were included, and exams whose standard image or soft tissue image was missing were excluded. Ultimately, the dataset for this study consisted of 9860 adult patients who had CXR exams acquired within 2 days after their initial RT-PCR tests, 1523 (15.5%) of whom tested positive and 8337 (84.5%) of whom tested negative for COVID-19. Figure 1 shows the distribution of the patient visit status, i.e., settings in which the CXR exams were acquired among COVID-19 positive patients in the dataset. Note that the COVID-19 positive patients who were hospitalized or in the intensive care unit (ICU) at the time of the CXR could have been receiving treatment for diseases other than COVID-19, and COVID-19 might not have been the primary reason for their hospital stay. Thus, we did not assume COVID-19 severity based on patient visit status.
Fig. 1.
Distribution of the patient visit status in which CXR exams were acquired among COVID-19 positive patients in the dataset. ICU, intensive care unit; ED, emergency department.
2.2. Classifier Training
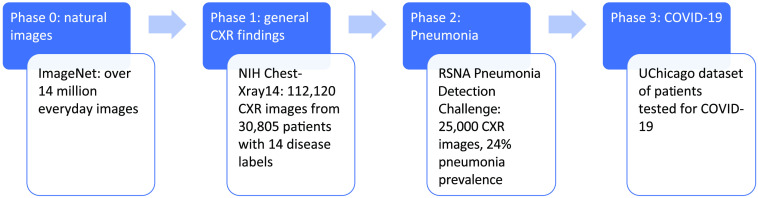
As shown in Fig. 2, a sequential transfer learning strategy was employed to train the model in three phases on gradually more specific and complex tasks, mimicking the human learning process.21 Instead of presenting the model with a random mixture of CXR examples and directly training it to diagnose COVID-19, the process was designed to fine-tune the model in a cascade approach in three phases: (1) first, the model was pretrained on natural images in ImageNet and fine-tuned on the National Institutes of Health (NIH) ChestX-ray14 dataset to diagnose a broad spectrum of 14 pathologies.20,22 (2) Then, the model was refined on the Radiological Society of North America (RSNA) Pneumonia Detection Challenge dataset, which has a high pneumonia prevalence, to detect opacities caused by pneumonia.18 (3) Finally, the model was fine-tuned further on the training set of the COVID-19 dataset. The final model was then evaluated on the held-out independent test set to distinguish between CXRs of COVID-19 positive and COVID-19 negative patients. The DenseNet-121 architecture was chosen for the task because of its success in diagnosing various diseases on CXR in previous publications.23,24
Fig. 2.
Sequential transfer learning strategy for the diagnosis of COVID-19, and information on the dataset for each phase of training.
The phase 1 test set was specified by the original database curators of NIH ChestX-ray14, and the rest of the dataset was randomly divided at the patient level into for training and 20% for validation. The DenseNet-121 model was initialized with weights optimized for ImageNet (phase 0), and the final classification layer was replaced with a 14-node fully connected layer with sigmoid activation. Images were downsampled by a factor of four to , gray-scale normalized by sample, and randomly augmented by horizontal flipping, shifting by up to 10% of the image size, and rotation of up to 8 deg. The model was trained with a batch size of 64, weighted cross-entropy loss function, and Adam optimizer with an initial learning rate of 0.001. Step decay on learning rate and early stopping were employed. The misclassification penalty for cases in a class was assigned to be inversely proportional to its class prevalence to address the problem of class imbalance.
The phase 2 test set containing 1000 images was specified by the database curators of the RSNA Pneumonia Detection Challenge dataset, and the rest was split randomly at the patient level into 80% for training and 20% for validation, holding the class prevalence constant in the subsets. The DenseNet-121 model was initialized with the weights from phase 1, and the final classification layer was replaced with a one-node fully connected layer with sigmoid activation to differentiate whether CXR images contain evidence of pneumonia. Methods for image preprocessing and model training followed that in phase 1, except the initial learning rate was reduced to 0.0001.
In phase 3 (Fig. 3), a U-Net-based model was used to segment the lung region on all images in our dataset in this phase, to reduce the influence on the classification model from irrelevant regions in the images.25 The U-Net model architecture was augmented with inception blocks and residual blocks.26 We used the weights that had been pretrained on a prepandemic public CXR dataset for lung segmentation and fine-tuned on an external CXR dataset that included COVID-19 patients.12,25,27 Open and close operations were performed in the postprocessing steps to fill holes and reduce noise in the predicted masks. Then, the smallest rectangular region that was able to enclose the predicted lung mask was cropped from each image. The masks were predicted using standard CXR images, and their corresponding soft tissue CXR images were cropped using the same masks. The cropped images were resized to .
Fig. 3.
Illustration of phase 3 in the sequential training process, fine-tuning the model on the pandemic-era CXR dataset to distinguish between COVID-19 positive and negative patients. The model architectures shown are for illustration purposes and are not the precise or complete architectures of the modified U-Net and the DenseNet models.
The COVID-19 dataset collected from our institution was split at the patient level into 6310 (64%) for training, 1578 (16%) for validation, and 1972 (20%) for testing, keeping the COVID-19 disease prevalence constant across all three subsets. Image preprocessing besides the cropping step and model training followed the methods in phase 2, and the model was initialized with the weights from phase 2. The fine-tuning and evaluation in this phase were performed on the full standard CXR images, the cropped standard CXR images, and the cropped soft tissue images in the dataset. The combined use of cropped standard and soft tissue images was then investigated in a feature fusion manner, which showed superior results in a prior study using multiparametric images.28 Specifically, the standard and soft tissue images were input to two DenseNet-121 models trained with shared weights, whose activation maps prior to the final fully connected layer were concatenated, forming the ensemble of features extracted from the two types of input for classification. Training of the parallel models was split across four GPUs memory allocation.
2.3. Evaluation
In each phase, the classification performance for each task label was evaluated using receiver operating characteristic (ROC) curve analysis with the area under the ROC curve (AUC) as the figure of merit. The 95% confidence intervals (CIs) of the nonparametric Wilcoxon–Mann–Whitney AUCs were calculated by bootstrapping (2000 bootstrap samples).29 Proper binormal model was used to plot the ROC curves.30 All reported classification performance metrics pertain to the held-out independent test set in each phase ( for phase 1, for phase 2, patients for phase 3). For the in-house COVID-19 dataset in phase 3, comparisons of performance in terms of AUC were performed using the DeLong test and equivalence test.31,32 Bonferroni–Holm corrections were used to account for multiple comparisons.33 A corrected -value was considered to indicate a statistically significant difference in performance, and an equivalence margin of was chosen prima facie. Additional evaluation metrics including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and score were calculated and reported at four sensitivity levels for the different methods in phase 3. Bland–Altman analysis was used as an adjunct method to compare the estimated COVID-19 probabilities for individual patients for the different classification approaches. Gradient-weighted class activation mapping (Grad-CAM) was generated to provide a visual explanation of the model’s classification.34 The test set evaluation was also performed on the DES exam subset and the portable exam subset separately.
3. Results
The ROC curves for the classification tasks in the first two phases are shown in Fig. 4. Phase 1 and phase 2 yielded AUC values similar to recent publications on the same tasks using these datasets.23,24,35
Fig. 4.
Fitted proper binormal ROC curves for classification tasks in the first two phases of training. The legend gives the AUC with 95% CI for each classification task.
In phase 3, when full standard CXR images were used as input, the model achieved an AUC of 0.74 (95% CI: 0.70, 0.77), which was significantly lower than an AUC of 0.76 (95% CI: 0.73, 0.79) obtained when cropped standard CXR images were used (Table 1). Figure 5 shows examples of Grad-CAM heatmaps demonstrating that when full images were used as input, areas outside the patient body [e.g., the text label on the image in Fig. 5(a)] and areas outside the lungs [e.g., abdominal region and chest walls in Fig. 5(a) and shoulder and neck region in both Figs. 5(a) and 5(b)] contributed to the classification model’s prediction, whereas influence from these irrelevant regions was eliminated or reduced using cropped images. Due to the superior classification performance and the reduced influence from areas outside the lungs, cropped images were used in our subsequent analysis.
Table 1.
AUCs for using full and cropped standard CXR, and the -value and 95% CI of the difference in AUCs. Asterisks denote statistical significance.
| Full standard CXR | Cropped standard CXR | |
|---|---|---|
| AUC [95% CI] | 0.74 [0.70, 0.77] | 0.76 [0.73, 0.79] |
| -value 95% CI for | 0.04* [0.001, 0.049] | |
Fig. 5.
Example standard CXR and their Grad-CAM heatmap overlays of (a) a COVID-19 positive case and (b) a COVID-19 negative case. The model prediction scores () are noted. Both examples show influence on model predictions from irrelevant areas outside the lungs when the full images were used, which was reduced when the cropped images derived from automatic lung segmentation were used.
For the three classification schemes, using copped images for standard CXR, soft tissue CXR, and the combination of both, yielded AUC values on the held-out test set of 0.76 (95% CI: 0.73, 0.79), 0.73 (95% CI: 0.70, 0.76), and 0.78 (95% CI: 0.74, 0.81), respectively (Fig. 6 and Table 2). Using soft tissue CXR yielded a significantly lower AUC value than using standard CXR and using the combination of both types of CXR. Using the combination of both types of CXR appeared to achieve a higher AUC value than when using standard CXR alone, but this improvement failed to reach statistical significance and the performance was statistically equivalent to using standard CXR alone with an equivalence margin of .
Fig. 6.
Fitted proper binormal ROC curves for the COVID-19 classification task for the held-out test set in the third phase when using cropped standard CXR and/or cropped soft tissue CXR images.
Table 2.
Comparisons of classification performances using the DeLong test when standard CXR images, soft tissue images, or both were used. The -value (before multiple comparison corrections) and CIs of the difference in AUCs are presented for each comparison. The significance levels () and the widths of the CIs are adjusted based on Bonferroni–Holm corrections.
| Comparison | -value for | CI of | |
|---|---|---|---|
| Standard versus soft tissue | 0.01a | 0.017 | 98.3% CI: [−0.061, −0.001] |
| Standard versus both | 0.18 | 0.050 | 95% CI: [−0.008, 0.041] |
| Soft tissue versus both | 0.02a | 0.025 | 97.5% CI: [0.001, 0.058] |
Statistical significance after correcting for multiple comparisons.
The desired operating range is in the high sensitivity regime for diagnosing COVID-19 on CXR exams, not only due to the harm of having false-negative diagnosis for COVID-19 but also because CXR exams are useful for identifying COVID-19 positive patients who had a false-negative RT-PCR test given the test's moderate sensitivity. Table 3 presents the additional evaluation metrics on the held-out test set, including sensitivity, specificity, PPV, NPV, and score at two sensitivity levels, 0.90 and 0.95, for the three methods in phase 3.
Table 3.
Additional evaluation metrics for the COVID-19 classification task for the held-out test set in the third phase when using cropped standard CXR and/or cropped soft tissue CXR images. The metrics are calculated at two sensitivity levels. The 95% CIs are shown in brackets.
| Sensitivity | Input | Specificity | PPV | NPV | F1 score |
|---|---|---|---|---|---|
| 0.95 | Standard | 0.15 [0.10, 0.26] | 0.17 [0.16, 0.19] | 0.95 [0.92, 0.97] | 0.29 [0.28, 0.32] |
| Soft tissue | 0.11 [0.07, 0.20] | 0.16 [0.16, 0.18] | 0.92 [0.89, 0.96] | 0.28 [0.27, 0.30] | |
| Both | 0.18 [0.14, 0.29] | 0.17 [0.17, 0.20] | 0.95 [0.94, 0.97] | 0.30 [0.29, 0.32] | |
| 0.90 | Standard | 0.30 [0.22, 0.39] | 0.19 [0.17, 0.21] | 0.94 [0.92, 0.95] | 0.31 [0.29, 0.34] |
| Soft tissue | 0.23 [0.16, 0.33] | 0.18 [0.16, 0.20] | 0.93 [0.90, 0.95] | 0.30 [0.28, 0.32] | |
| Both | 0.34 [0.25, 0.43] | 0.20 [0.18, 0.22] | 0.95 [0.93, 0.96] | 0.33 [0.30, 0.36] |
Figure 7 shows the standard and soft tissue CXR images in four example cases and their Grad-CAM heatmaps from the penultimate layer of their respective models. These examples were selected to illustrate the differences in model prediction and/or heatmaps that arose when the two types of CXR images were used. In both the positive case [Fig. 7(a)] and the negative case [Fig. 7(b)], using standard CXR images resulted in more accurate predictions, possibly due to undesirable alterations to the anatomy presentation when the soft tissue images were generated by postprocessing algorithms and/or the fact that the datasets used for pretraining do not contain soft tissue images. On the other hand, the examples in Figs. 7(c) and 7(d) both show activations in the shoulder region when standard CXR images were used, whereas in the soft tissue images the bones were removed and hence did not contribute to the model prediction. In both cases, the soft tissue model yielded accurate predictions with reasonable activation areas shown in the heatmaps. The influence from the shoulder bones led to a false-positive prediction in the negative case in Fig. 7(d) when standard CXR was used but did not greatly affect the prediction score in the positive case in Fig. 7(c).
Fig. 7.
Standard and soft tissue CXR of four example cases (post-cropping) and their Grad-CAM heatmap overlays. The model prediction scores () are noted. In all four cases, model predictions and/or heatmaps show differences when the two types of CXR images are used. For cases shown in (a) and (b), standard CXR resulted in more accurate predictions, whereas for cases shown in (c) and (d), soft tissue CXR resulted in more accurate predictions.
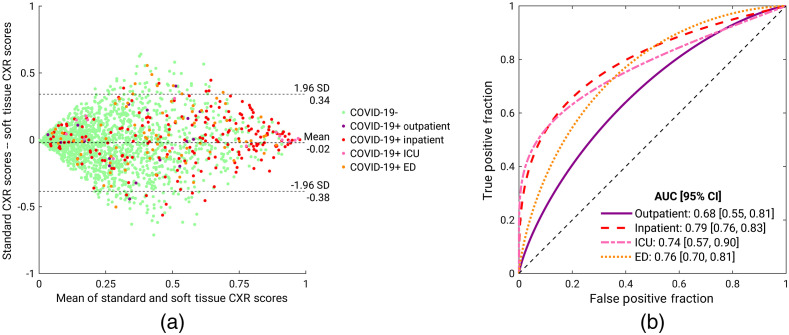
The Bland–Altman plot in Fig. 8(a) shows a notable discrepancy between the model predictions based on standard and soft tissue CXR images. It also shows that predictions, especially for COVID-19 positive cases, spanned a wide range. The patient visit status of COVID-19 positive patients is indicated by different colors. While COVID-19 early diagnosis on CXR scans is challenging in all categories, outpatient cases appear to be more challenging than other categories in our data. This observation is confirmed by Fig. 8(b), the ROC curves for COVID-19 classification using both standard and soft tissue CXR combined by feature fusion, presented by patient visit status. The outpatient category yielded the lowest AUC value, and the inpatient category yielded the highest AUC value.
Fig. 8.
(a) Bland–Altman plot for the model predictions based on standard and soft tissue CXR images. The patient visit status of COVID-19 positive patients is indicated by different colors. (b) ROC curves for COVID-19 classification using both standard and soft tissue CXR combined by feature fusion, presented by patient visit status. ICU, intensive care unit; ED, emergency department.
The separate test set evaluation on the portable exam subset and the DES exam subset is presented in Table 4. AUC values were higher for the DES subset than the portable subset when using standard images or the fusion of standard and soft tissue images, potentially attributed to the higher image quality in DES exams than portable or differences in the patient groups that received these two types of exams. However, AUC values were slightly lower for the DES subset than the portable subset when using just soft tissue images, potentially because the model was trained mostly on synthetic soft tissue images as the majority of the dataset was portable exams and thus performed better on this type of images. To study the utility and the contribution of soft tissue images in these two types of CXR exams in COVID-19-related image interpretation, separate analyses and controlled experiments are needed in future work.
Table 4.
COVID-19 classification performance by CXR exam type: portable or dual-energy exam. The 95% CIs are shown in brackets.
| Portable (80%) | Dual-energy (20%) | All | ||
|---|---|---|---|---|
| COVID-19+ prevalence | 16% | 12% | 15% | |
| AUC | Standard | 0.74 [0.70, 0.78] | 0.86 [0.80, 0.91] | 0.76 [0.73, 0.79] |
| Soft tissue | 0.73 [0.62, 0.80] | 0.71 [0.70, 0.77] | 0.73 [0.70, 0.76] | |
| Both | 0.77 [0.73, 0.80] | 0.83 [0.77, 0.89] | 0.78 [0.74, 0.81] | |
4. Discussion
We curated a large CXR database during the COVID-19 pandemic and designed a sequential transfer learning strategy to pretrain and fine-tune a model on increasingly specific and complex tasks with the final goal of distinguishing COVID-19 positive and negative patients using their initial CXR exam within 2 days of their initial RT-PCR test for COVID-19. We identified the necessity to reduce the influence from irrelevant regions of the images on model predictions and incorporated automatic lung segmentation and cropping in the pipeline. We also investigated the role of soft tissue images in CXR exams in addition to the standard CXR images. We achieved promising performance, , using both types of CXR images combined via feature fusion for diagnosing COVID-19 on CXR at patient presentation. In our study, the performance using feature fusion was equivalent to that obtained when analyzing standard cropped CXRs alone, but given the improved performance observed in prior non-COVID studies, we will continue investigating this approach in future studies. Further investigation on the potential benefit of soft tissue images in the task of automated COVID-19 diagnosis is needed.
Two alternative fusion methods for the combined use of the two types of CXR images achieved similar results as the feature fusion method with shared weights reported in this paper. Averaging the prediction scores given by the standard CXR model and the soft tissue CXR model for each case yielded an AUC of 0.76 (95% CI: 0.73, 0.79). Feature fusion without weight sharing between the two parallel models achieved an AUC of 0.77 (95% CI: 0.73, 0.80) and was more computationally expensive with twice as many parameters in the models as when weight sharing was employed.
It is worth noting that all patients in our dataset had medical indications for receiving a CXR exam, such as being symptomatic for possible COVID-19, receiving medical care related to pneumonia of unknown or known origin, or undergoing diagnosis or treatment for other diseases. As such, our dataset does not represent the entire population that undergoes RT-PCR testing for COVID-19. This increases the difficulty of our task, since many patients in our dataset presented with non-COVID lung abnormalities, which made the distinction between COVID and non-COVID patients more challenging than if more of the non-COVID patients had been healthy and presented with no abnormal lung findings.
We also note that the prepandemic public databases used for pretraining in this study only contain standard CXR images, which might partly contribute to the inferior performance of soft tissue images in the COVID-19 classification stage. When the first and second pretraining phases are removed, standard images, soft tissue images, and the fusion of both yielded similar AUC values in COVID-19 classification (0.73 [0.69, 0.67], 0.72 [0.69, 0.75], and 0.72 [0.68, 0.75]). The results show that pretraining stages led to larger improvements in AUC for standard images than soft tissue images and significantly improved the fusion model’s performance ().
There are several other limitations of our study. First, the database was collected from a single institution. We will contribute to multi-institutional databases and perform independent evaluations when such datasets become available in the future to assess the robustness of our approach. Such high-quality publicly available databases will also allow the research community to compare performances and establish reference standards, which are currently lacking. Second, training was performed on a combination of both dual-energy subtraction (DES) CXR exams and portable CXR exams in our database. The soft tissue image in a DES exam is obtained from two physically acquired images, whereas the synthetic soft tissue image in a portable exam is generated from the standard image using postprocessing algorithms. Therefore, in future work, we will evaluate the contribution of each type of soft tissue image when using deep-learning-based methods in tasks, such as COVID-19 detection, classification, and prognosis. Third, a single CXR exam at patient presentation was used to diagnose evidence of COVID-19 for each patient. In future work, we will investigate the role of temporal analysis, utilizing previous CXR exams of patients suspected of having COVID-19, instead of only using images at a single time point. In addition, while the earliest available CXR images with confirmed COVID-19 status of each patient were used, there were varying degrees of disease presentation on the images. For example, some patients did not have a CXR acquired at earlier stages of their disease, and some were asymptomatic. The dataset, however, supported the research goal of investigating the identification of COVID-19 using patients’ earliest possible CXR. Finally, while the Grad-CAM technique, which we used to visualize and explain model predictions, is one of the commonly used explainability techniques for convolutional neural networks, the created heated maps are not intended for precise localization tasks, especially in the medical imaging domain. Future studies will investigate methods to more precisely localize COVID-19 presentations on CXR images.
5. Conclusions
In summary, we curated a large CXR database during the COVID-19 pandemic and designed a sequential transfer learning strategy to fine-tune a convolutional neural network on increasingly specific and complex tasks to distinguish between COVID-19 positive and negative patients using the CXR exams acquired within 2 days of their initial RT-PCR test for COVID-19. We incorporated automatic lung segmentation and cropping in the analysis pipeline to reduce the contribution from irrelevant regions of the images to model predictions. The additional benefit provided by the inclusion of soft tissue images for improving the performance of diagnosing COVID-19 failed to show statistical significance in our classification framework. Future work will investigate the robustness of the method across various imaging sites and populations.
Acknowledgments
The authors acknowledge other lab members and collaborators within the Department of Radiology, the University of Chicago, including Chun-Wai Chan, MS; Jordan Fuhrman; Feng Li, MD; Hui Li, PhD; Tarik Rashada; Geneva Schlafly, BS; and the Human Imaging Research Office for their contributions. This work was partially supported by a C3.AI Digital Transformation Institute award, NIBIB COVID-19 Contract 75N92020D00021, and NIH S10 OD025081 Shared Instrument Grant.
Biographies
Qiyuan Hu is a PhD graduate in medical physics at the University of Chicago. She received her BA degrees in physics and mathematics from Carleton College in 2017. Her research interests include radiomics and deep learning methodologies for computer-aided diagnosis. She is a student member of SPIE and a former officer of the University of Chicago SPIE Student Chapter.
Karen Drukker is a research associate professor at the University of Chicago, where she has been involved in medical imaging research for 20+ years. She received her PhD in physics from the University of Amsterdam. Her research interests include machine learning applications in the detection, diagnosis, and prognosis of breast cancer and, more recently, of COVID-19 patients, focusing on rigorous training/testing protocols, generalizability, and performance evaluation of machine learning algorithms.
Maryellen L. Giger is the A.N. Pritzker Professor of Radiology for the Committee on Medical Physics, and the College at the University of Chicago. She has conducted research on computer-aided diagnosis, quantitative image analysis (radiomics), and deep learning in the areas of breast cancer, lung cancer, prostate cancer, and bone diseases. She is a fellow of SPIE and the 2018 SPIE president.
Disclosures
M. L. G. is a stockholder in R2 technology/Hologic and QView, receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi and Toshiba, and is a cofounder of and equity holder in Quantitative Insights (now Qlarity Imaging). K. D. received royalties from Hologic. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interests that would reasonably appear to be directly and significantly affected by research activities.
Contributor Information
Qiyuan Hu, Email: qhu@uchicago.edu.
Karen Drukker, Email: kdrukker@uchicago.edu.
Maryellen L. Giger, Email: m-giger@uchicago.edu.
References
- 1.Arevalo-Rodriguez I., et al. , “False-negative results of initial RT-PCR assays for COVID-19: a systematic review,” PLoS One 15(12), e0242958 (2020). 10.1371/journal.pone.0242958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson J., Whiting P. F., Brush J. E., “Interpreting a covid-19 test result,” BMJ 369, m1808 (2020). 10.1136/bmj.m1808 [DOI] [PubMed] [Google Scholar]
- 3.Rubin G. D., et al. , “The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society,” Chest 158, 106–116, (2020). 10.1016/j.chest.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American College of Radiology, “ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection,” 2020, https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection (accessed 17 June 2020).
- 5.Jacobi A., et al. , “Portable chest x-ray in coronavirus disease-19 (COVID-19): a pictorial review,” Clin. Imaging 64, 35–42 (2020). 10.1016/j.clinimag.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng M.-Y., et al. , “Imaging profile of the COVID-19 infection: radiologic findings and literature review,” Radiol. Cardiothorac. Imaging 2(1), e200034 (2020). 10.1148/ryct.2020200034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy K., et al. , “COVID-19 on the chest radiograph: a multi-reader evaluation of an AI system,” Radiology 296, E166–E172 (2020). 10.1148/radiol.2020201874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan A. I., Shah J. L., Bhat M. M., “CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images,” Comput. Methods Programs Biomed. 196, 105581 (2020). 10.1016/j.cmpb.2020.105581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozturk T., et al. , “Automated detection of COVID-19 cases using deep neural networks with x-ray images,” Comput. Biol. Med. 121, 103792 (2020). 10.1016/j.compbiomed.2020.103792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panwar H., et al. , “Application of deep learning for fast detection of COVID-19 in x-rays using nCOVnet,” Chaos Solitons Fractals 138, 109944 (2020). 10.1016/j.chaos.2020.109944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury M. E. H., et al. , “Can AI help in screening viral and COVID-19 pneumonia?” arXiv:2003.13145 (2020).
- 12.Wang L., Lin Z. Q., Wong A., “COVID-Net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images,” Sci. Rep. 10(1), 19549 (2020). 10.1038/s41598-020-76550-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minaee S., et al. , “Deep-COVID: predicting COVID-19 from chest x-ray images using deep transfer learning,” arXiv:2004.09363 (2020). [DOI] [PMC free article] [PubMed]
- 14.Wehbe R. M., et al. , “DeepCOVID-XR: an artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large US clinical dataset,” Radiology 299, 203511 (2020). 10.1148/radiol.2020203511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen J. P., Morrison P., Dao L., “COVID-19 image data collection,” arXiv:2003.11597 (2020).
- 16.Zhao J., et al. , “COVID-CT-dataset: a CT scan dataset about COVID-19,” arXiv:2003.13865 (2020).
- 17.Wynants L., et al. , “Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal,” BMJ 369, m1328 (2020). 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.“RSNA Pneumonia Detection Challenge,” 2018, https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data (accessed 18 June 2020).
- 19.Irvin J., et al. , “CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison,” in Proc. AAAI Conf. Artif. Intell., Vol. 33, pp. 590–597 (2019). [Google Scholar]
- 20.Wang X., et al. , “Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. IEEE Conf. Comput. Vision Pattern Recognit., pp. 2097–2106 (2017). 10.1109/CVPR.2017.369 [DOI] [Google Scholar]
- 21.Hu Q., Drukker K., Giger M. L., “Role of standard and soft tissue chest radiography images in COVID-19 diagnosis using deep learning,” Proc. SPIE 11597, 1159704 (2021). 10.1117/12.2581977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J., et al. , “ImageNet: a large-scale hierarchical image database,” in IEEE Conf. Comput. Vision Pattern Recognit., pp. 248–255 (2009). 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 23.Rajpurkar P., et al. , “CheXNet: radiologist-level pneumonia detection on chest x-rays with deep learning,” arXiv1711.05225 (2017).
- 24.Rajpurkar P., et al. , “Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists,” PLoS Med. 15(11), e1002686 (2018). 10.1371/journal.pmed.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motamed S., Rogalla P., Khalvati F., “RANDGAN: randomized generative adversarial network for detection of COVID-19 in chest x-ray,” Sci. Rep. 11, 8602 (2021). 10.1038/s41598-021-87994-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark T., et al. , “Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks,” J. Med. Imaging 4(4), 041307 (2017). 10.1117/1.JMI.4.4.041307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaeger S., et al. , “Two public chest x-ray datasets for computer-aided screening of pulmonary diseases,” Quant. Imaging Med. Surg. 4(6), 475–477 (2014). 10.3978/j.issn.2223-4292.2014.11.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Q., Whitney H. M., Giger M. L., “A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI,” Sci. Rep. 10(1), 10536 (2020). 10.1038/s41598-020-67441-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efron B., “Better bootstrap confidence intervals,” J. Am. Stat. Assoc. 82(397), 171–185 (1987). 10.1080/01621459.1987.10478410 [DOI] [Google Scholar]
- 30.Metz C. E., Pan X., “Proper’ binormal ROC curves: theory and maximum-likelihood estimation,” J. Math. Psychol. 43(1), 1–33 (1999). 10.1006/jmps.1998.1218 [DOI] [PubMed] [Google Scholar]
- 31.DeLong E. R., DeLong D. M., Clarke-Pearson D. L., “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics 44(3), 837–845 (1988). 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 32.Ahn S., Park S. H., Lee K. H., “How to demonstrate similarity by using noninferiority and equivalence statistical testing in radiology research,” Radiology 267(2), 328–338 (2013). 10.1148/radiol.12120725 [DOI] [PubMed] [Google Scholar]
- 33.Holm S., “A simple sequentially rejective multiple test procedure,” Scand. J. Stat. 6(2), 65–70 (1979). [Google Scholar]
- 34.Selvaraju R. R., et al. , “Grad-CAM: visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vision, pp. 618–626 (2017). 10.1109/ICCV.2017.74 [DOI] [Google Scholar]
- 35.Pan I., Cadrin-Chênevert A., Cheng P. M., “Tackling the radiological society of North America pneumonia detection challenge,” Am. J. Roentgenol. 213(3), 568–574 (2019). 10.2214/AJR.19.21512 [DOI] [PubMed] [Google Scholar]