Abstract
Background
Mutations of the isocitrate dehydrogenase (IDH) gene occur in over 80% of low-grade gliomas and secondary glioblastomas. Despite considerable efforts, endogenous in vitro IDH-mutated glioma models remain scarce. Availability of these models is key for the development of new therapeutic interventions.
Methods
Cell cultures were established from fresh tumor material and expanded in serum-free culture media. D-2-Hydroxyglutarate levels were determined by mass spectrometry. Genomic and transcriptomic profiling were carried out on the Illumina Novaseq platform, methylation profiling was performed with the Infinium MethylationEpic BeadChip array. Mitochondrial respiration was measured with the Seahorse XF24 Analyzer. Drug screens were performed with an NIH FDA-approved anti-cancer drug set and two IDH-mutant specific inhibitors.
Results
A set of twelve patient-derived IDHmt cell cultures was established. We confirmed high concordance in driver mutations, copy numbers and methylation profiles between the tumors and derived cultures. Homozygous deletion of CDKN2A/B was observed in all cultures. IDH-mutant cultures had lower mitochondrial reserve capacity. IDH-mutant specific inhibitors did not affect cell viability or global gene expression. Screening of 107 FDA-approved anti-cancer drugs identified nine compounds with potent activity against IDHmt gliomas, including three compounds with favorable pharmacokinetic characteristics for CNS penetration: teniposide, omacetaxine mepesuccinate, and marizomib.
Conclusions
Our twelve IDH-mutant cell cultures show high similarity to the parental tissues and offer a unique tool to study the biology and drug sensitivities of high-grade IDHmt gliomas in vitro. Our drug screening studies reveal lack of sensitivity to IDHmt inhibitors, but sensitivity to a set of nine available anti-cancer agents.
Keywords: drug repurposing, glioma, IDH1, patient-derived cell culture, preclinical models
Key Points.
IDHmt glioma cultures closely resemble their parental tumors.
Microscopic monitoring of early passages and colony isolation increases IDH1mt culture success.
Drug screening identified nine repurposed candidate drugs for IDHmt glioma.
Importance of the Study.
IDH-mutations are highly prevalent in low-grade and secondary high-grade gliomas. Despite this high frequency, however, very few in vitro models have been reported for IDH-mutated gliomas. In this manuscript, we describe and characterize twelve primary cultures from IDH-mutant astrocytomas in detail. We show that these cultures retain most of the genetic, epigenetic, and metabolic features of their respective parental tumors. Because of these similarities, these independent model systems will not only help understand the molecular defects driven by the mutation, but are also vital to identify means to target these tumors. Screening of 107 FDA-approved anti-cancer agents on these cultures identified a set of highly effective agents that may offer candidates for either systemic or assisted delivery treatment of this tumor subtype.
Isocitrate dehydrogenase (IDH) mutations are present in approximately 80% of low-grade gliomas (grade II and III) and secondary glioblastomas.1IDH mutations result in a neomorphic gain-of-function of the mutant enzyme, which causes it to convert alpha-ketoglutarate (α-KG) to D-2-hydroxyglutarate (D-2-HG). D-2-HG is a competitive inhibitor of several α-KG dependent enzymes including the histone lysine demethylase JMJD2 and the methylcytosine dioxygenase TET2, which results in the global hypermethylated (G-CIMP, glioma CpG island methylator phenotype) state of IDH-mutant gliomas.2,3
The discovery of this somatic mutation in malignant glioma instigated the development of agents targeting IDH-mutant tumors.4,5 Unfortunately, preclinical research has been hampered by lack of IDHmt glioma model systems, as tumor samples from IDH-mutated glioma patients are notoriously difficult to culture.6,7 Worldwide, only a few endogenous IDH-mutant cell lines have been described thus far, some derived from fresh patient material, others from patient-derived xenografts that were serially transplanted in mice.8–14
Over the last ten years, our lab has attempted to establish cell cultures from over 275 low-grade and secondary high-grade gliomas resected in our clinic. Our continuous effort resulted in 12 cultures from IDH-mutant 1p19q noncodeleted astrocytomas, which is the largest set of patient-derived IDH-mutant (IDHmt) glioma cultures described to date. We show that these cell cultures retain the morphological, genetic, epigenetic, metabolic, and transcriptomic features of the primary tumor. Current developments in the neuro-oncology field, directed toward improving drug delivery to CNS tumors, have reignited interest in anti-cancer agents that are already available. We utilized our IDHmt cell cultures to screen 107 FDA-approved anti-cancer agents to identify potential candidates for either systemic or enhanced delivery applications.
Methods
Tumor Processing and Cell Culture
Fresh glioma tissue samples were obtained directly from the operating room of the Erasmus Medical Center or the Elizabeth Tweesteden Hospital, both in the Netherlands. The use of patient tissue for this study was approved by the local ethics committees of these hospitals and all patients signed informed consent forms according to the guidelines of the Institutional Review Boards of the respective hospitals.
Samples were processed essentially according to a further optimized protocol based on Balvers et al (see also Supplementary Methods).6 After 5–8 days, cultures were transferred to a new flask coated with 1:100 Cultrex PathClear RGF-BME (R&D Systems) for adherent expansion. Cell cultures were considered successful if they could be passaged at least five times while retaining their IDH1 mutation.
Growth rates and doubling times were assessed by seeding 2 × 105 cells in precoated T75 flasks (±10 µM AGI-5198, Agios), which were counted and split at ~90% confluency for at least three passages.
In Vivo Study
The animal experiment was performed in accordance with the local Animal Ethical Committee, Erasmus MC Rotterdam. Intrastriatal injections of 2.5 × 105 GS.0661 (N = 2) and GS.0580 (N = 2) cells were performed on 7-week-old NOD-SCID female mice (strain C.B-17/IcrHantmhsd-Prkdcs (Envigo, Italy) as described previously.15 The maximum age up to which NOD-SCID mice are permitted to be kept in experiment under the Dutch ethical guidelines for animal experimentation is 240 days.
Image Analysis of Low Passage Cell Populations
Cells were plated in 96 wells CellCarrier Ultra plates (Perkin Elmer). After 6 days of culturing, live cells were stained with Hoechst and cell tracker green (1:1000, Thermo Fisher) and imaged using a spinning disk Opera Phenix confocal high throughput microscope system (Perkin Elmer) equipped with a water immersion 40x objective. Images were analyzed using Harmony Analysis Software (PerkinElmer).
Sequencing
Genomic DNA was extracted from cell pellets (passages ranging from 0–12), cryosections of snap-frozen tumor material, or leukocytes using the DNeasy Blood & Tissue Kit (Qiagen).
All RNA was extracted from pellets of cultured cells (passage numbers ranging from 8–14) and cryosections of snap-frozen tumor material with the RNeasy Plus Mini Kit (Qiagen).
Presence of IDH1 mutations was confirmed by Sanger sequencing using 5’-GTG GCA CGG TCT TCA GAG A-3’ and 5’- TTC ATA CCT TGC TTA ATG GGT GT- 3’ primers.
Five hundred ng genomic DNA was fragmented to ~300 bp with the Kapa Hyperplus Library prep kit. The exome was captured by the SeqCap wash kit and SeqCap pure capture bead kit (both from Roche). The Illumina Novaseq platform was used to sequence 150 bases (paired-end sequencing) to obtain 6GB per sample.
Driver mutations were derived from Brennan et al.16 CopyNumbers were calculated using the CNVKit package.17
RNA was isolated from cell cultures (± 5 µM AGI-5198) using the RNeasy kit (Qiagen). We performed paired-end sequencing of 2x100 with the Illumina Novaseq platform to obtain 8–10 GB per sample. For details on sequencing, data processing, and analysis, see Supplementary Methods.
DNA methylation levels were assessed using the Infinium methylationEPIC beadchip arrays (Illumina) according to standard protocols and classified as described18 or using the TCGAbiolinks Bioconductor package. Further analysis was done using the Bioconductor Minfi package.19 The sequencing and gene expression data is available at Zenodo.org (doi: 10.5281/zenodo.4498024).
Mitochondrial Oxidative Respiration Assay
We measured cellular oxidative respiration with the Seahorse XF24 Analyzer as described previously.20 Cell cultures were seeded in ten-plo at a density of 40 000 cells per well and grown overnight in standard culture medium. One hour before the start of the experiment we replaced the culture medium with XF Assay Medium (Agilent Technologies) pH7.4, supplemented with 10 mM glucose, 2 mM glutamin, and 1 mM sodium pyruvate, and incubated at 37°C without CO2. After baseline measurements, cellular response after sequential injections of 1 μM oligomycin, 0.5 μM FCCP, and 1 μM antimycin were measured. Basal respiration, mitochondrial ATP production, proton leakage, and maximal respiration rates were calculated with Seahorse Wave software.
D/L-2-hydroxyglutarate Measurements
Intracellular D-2-HG and L-2-HG was measured in cell pellets after five days of culture as described.21
Drug screening
Drug screening was done using IDH inhibitors AGI-5198 (Agios) or BAY-1436032 (Bayer) or the FDA-approved Oncology Drug Set II library (National Cancer Institute) containing 107 compounds. The compounds Gemcitabine hydrochloride (Sigma-Aldrich), Paclitaxel (Sigma-Aldrich), Teniposide (Santa Cruz Biotechnology, Inc), Daunorubicin hydrochloride (SelleckChem), Romidepsin (MedChemExpress), Dactinomycin (BioViotica), Regorafenib (MedChemExpress), Omacetaxine mepesuccinate (Sigma-Aldrich), and Marizomib (Sigma-Aldrich), were selected for the validation studies. Initial NIH compound screens were carried out in a 96-well format on 7 IDH mutant cultures and validation studies were performed in 384-well plates on all 12 IDH mutant cultures using the STARlet automated pipetting system (Hamilton). Serial dilutions of the selected compounds were added after 24 hours and viability was assessed by CellTiter GLO 2.0 after five days. For details see Supplementary Methods.
Gene Expression Analysis
Differential gene expression analysis and normalization were performed with DESeq2 and further visualizations of expression profiles used the DESeq2 VST transformed read counts.22 Only genes with ≥4 read counts across the tested samples were included.
Principal component analysis was performed on the top 500 genes with the highest variance. We performed cluster analysis based on the 1000 most variably expressed genes using the pheatmap package in R, and scaled by row.
Pathway analysis was done using the DAVID Bioinformatics Resources 6.8 (Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57) using a false discovery rate (FDR) of 0.05 as cut-off and using Gene Set Enrichment Analysis (GSEA) using an FDR < 0.25.23 Statistical significance of pathway enrichment scores was ascertained by permutation testing with matched random gene sets.
Statistical analysis
The unpaired Student’s T-test was used for the comparison of two groups (statistical significance was defined as P < .05). To determine the significance of contingency tables we used the Fisher exact test (statistical significance was defined as P < .05). The IC50 values were calculated by applying a nonlinear regression (curve-fit) analysis and selecting the dose-response inhibition equation in Graphpad Prism.
Results
Establishment of Successful IDHmt Cell Cultures
Glioma resection material of over 275 low-grade or secondary high-grade gliomas was transported to our lab to establish patient-derived cell cultures. We sequenced all successful cultures (N = 80) for IDH mutations and identified twelve cell cultures derived that retained their IDH-R132H mutation, from eleven patients. Nine cultures were derived from secondary glioblastomas, one from a grade III anaplastic astrocytoma, and two from grade II astrocytomas. All cultures were derived from 1p/19q noncodeleted gliomas. Patient and cell line characteristics are listed in Supplementary Table S1. Supplementary Table S2 summarizes on which levels each IDHmt cell culture has been characterized.
The doubling time of IDHmt cultures had a mean of 4.9 days (N = 12, range 1.8–16.4) and the IDH-wildtype (IDHwt) glioblastoma cultures had a mean of 2.6 days (N = 13, range 1.6–3.6, P = .07) (Supplementary Figure S1A). We passaged GS.0580 up to passage 50, without any notable changes in growth rate or morphology and verified the continued presence of the IDH1 mutation. We determined intracellular levels of the enantiomers D-2-HG and L-2-HG in all our IDHmt, and seven IDHwt cell cultures. All IDHmt cell cultures revealed dramatically increased levels of D-2-HG over L-2-HG (mean value 859; range 21–2700) compared to those of IDHwt cell cultures (mean value 1.2; range 0.3–5.3, P < .05), indicating the presence of mutant IDH enzyme activity (Supplementary Figure S1B).
In Vivo Expansion
We performed a pilot experiment to test whether the IDH-mutant cultures can be propagated in vivo. For this, NOD-SCID mice were orthotopically injected with GS.0661 (N = 2) and GS.0580 (N = 2). The mice did not develop tumors within the timeframe up to which they were permitted to be kept in experiment (190 days post-tumor cell injection). In all four cases, H&E staining and immunohistochemistry with an antibody against IDH1-R132H did not show tumor formation.
IDHmt Cultures Resemble Parental Tumors on Genomic Level
We performed whole-exome sequencing on seven IDHmt cultures and their parental tumors to determine whether they retained the classical features of IDHmt astrocytic gliomas. Apart from the IDH1 mutation, TP53 was most frequently mutated in the cell cultures (N = 7), followed by ATRX (Figure 1A). Both driver mutations were retained in cell cultures if they were present in parental tumors.
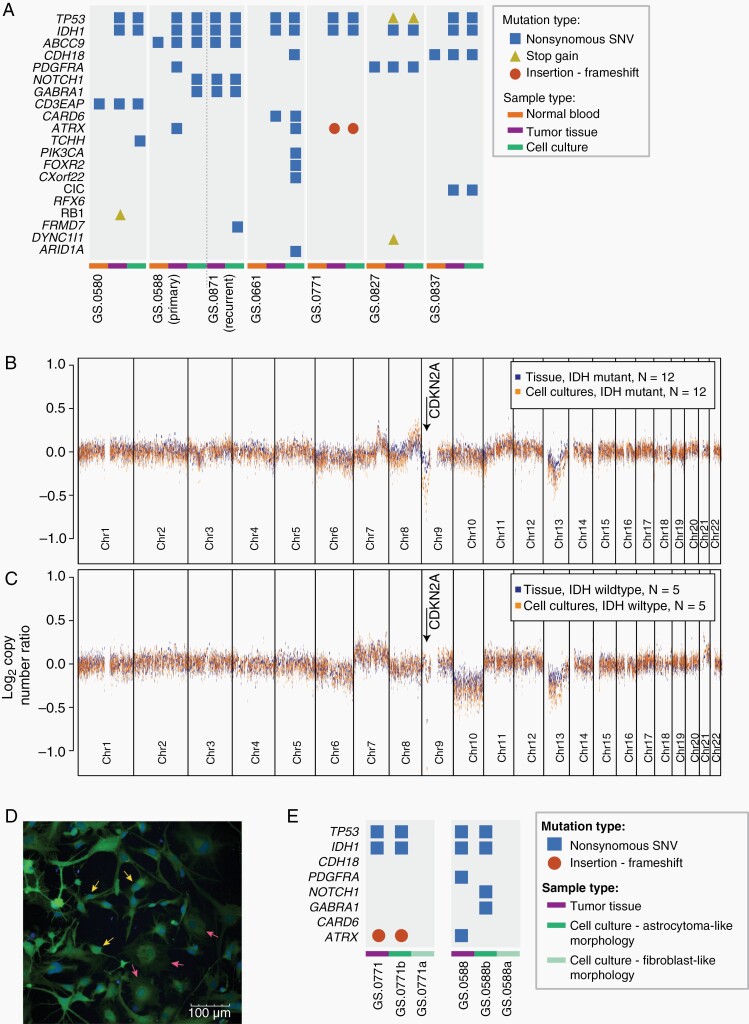
Figure 1.
IDHmt cell cultures represent the genotype of IDHmt gliomas. (A) Presence of glioma-associated mutations in non-neoplastic control tissue (orange bars), tumor tissue (purple bars), or cell cultures (green bars), determined by whole exome sequencing. Mutation types included are nonsynonymous SNVs (blue squares), stop gains (yellow triangles) and frameshift (red circles). (B, C) Median copy number log ratio plots of IDHmt tissues and cultures (N = 12), and IDHwt tissues and cultures (N = 5) based on global methylation data. The X-axes show chromosome 1–22, the Y-axes show the median ratio of the log2 copy number change. Chromosomal gains and losses are more pronounced in cultures than in tissues, probably due to the absence of noncancerous cells in cultures. (D) Fluorescent image of an early IDH mutant cell culture with two distinct phenotypes: astrocytoma-like cells (three individual cells pointed out by yellow arrows) and fibroblast-like cells (three individual cells pointed out by pink arrows). The scale bar represents 100 μm. (E) Mutations identified in IDHmt tumors GS.0771 and GS0588, and their respective derived cultures. Each tumor resulted in one culture with astrocytoma-like cells and one culture with a fibroblast-like morphology.
Interestingly, mutations in NOTCH1 and GABRA1 were not identified in one primary tumor sample, while they were found in the primary cell culture (GS.0588b). At tumor recurrence, however, both mutations were present in both tumor and cell culture, which indicates subclonal expansion of mutations already detectable in our primary cell culture. Furthermore, in copy number analysis we found a striking homozygous loss of the CDKN2A/B locus in all IDHmt cell cultures, whereas this loss was detected in only eight out of twelve IDHmt tumors (Figure 1B, Supplementary Figure S2A-L). When compared to the glioblastomas present in the TCGA database, CDKN2A/B loss in combination with IDH1/2 mutations was significantly more common in our cell cultures (12/12 compared to 6/15, P = .001). We did not note such specific copy number changes when we compared IDHwt glioma cultures to their parental tumors.
Overall, copy number analysis shows similarity of copy number alterations between the tumor tissues and corresponding cell cultures, for both IDHmt and IDHwt cultures (Supplementary Figure S2).
2D Culture Morphology Reveals IDH Status
In early passages of the IDHmt cell cultures two types of cells were often visible: round-bodied cells with thin protrusions and bright edges (astrocytic phenotype) and dark, large flat cells (fibroblast-like phenotype). We hypothesized that IDHmt glioma cells would be of the astrocytic phenotype. We separated morphologically different colonies from low-passage (P ≤ 1) cell cultures. Colony isolation and expansion from a mixed population of cells, yielded two distinct cell cultures: the original culture (designated “a,” eg, GS.0771a) that evolved to almost exclusively contain fibroblast-like cells, and an astrocytoma-like culture (designated “b”). The two distinct cell types can be observed in a mixed culture in Figure 1D). Sequencing revealed that only GS.0771b maintained its IDH mutation and copy numbers of the parental tumor; whereas GS.0771a did not retain the IDH mutation and showed a diploid genome (Supplementary Figure S3 A-C). Mutations in TP53 and ATRX from the parental tumor were preserved in GS.0771b but not GS.0771a (Figure 1E). Similarly, clone separation of GS.0588, also showed retention of IDH1 and TP53 mutations in the astrocytic GS.0588b culture but not in the fibroblast-like GS0588a culture. All successful cultures with verified IDH mutations had astrocytoma-like morphologies, although some degree of heterogeneity between cultures was observed (Supplementary Figure S4A-L). They are readily distinguishable from the fibroblast-like cultures, that no longer harbor the IDH mutation, by brightfield microscopy (Supplementary Figure S4M-O). Quantification of morphologic parameters of the fibroblast-like and astrocytoma-like phenotype are summarized in Supplementary Table S3. The most distinguishing factor is cell area (6.1 × 103 μm2 for fibroblast-like vs. 1.2 × 103 μm2 for astrocytoma-like), which is the main feature that can be identified by eye. Furthermore, the nuclei of IDHmt glioma cultures are significantly smaller, and the GFP intensity is higher when live-staining the cells (Figure 1D). Thus, astrocytoma-like morphological features can be used as preselection for establishing IDHmt cell cultures.
Global Methylation Profiles are Retained in IDHmt Glioma Cultures
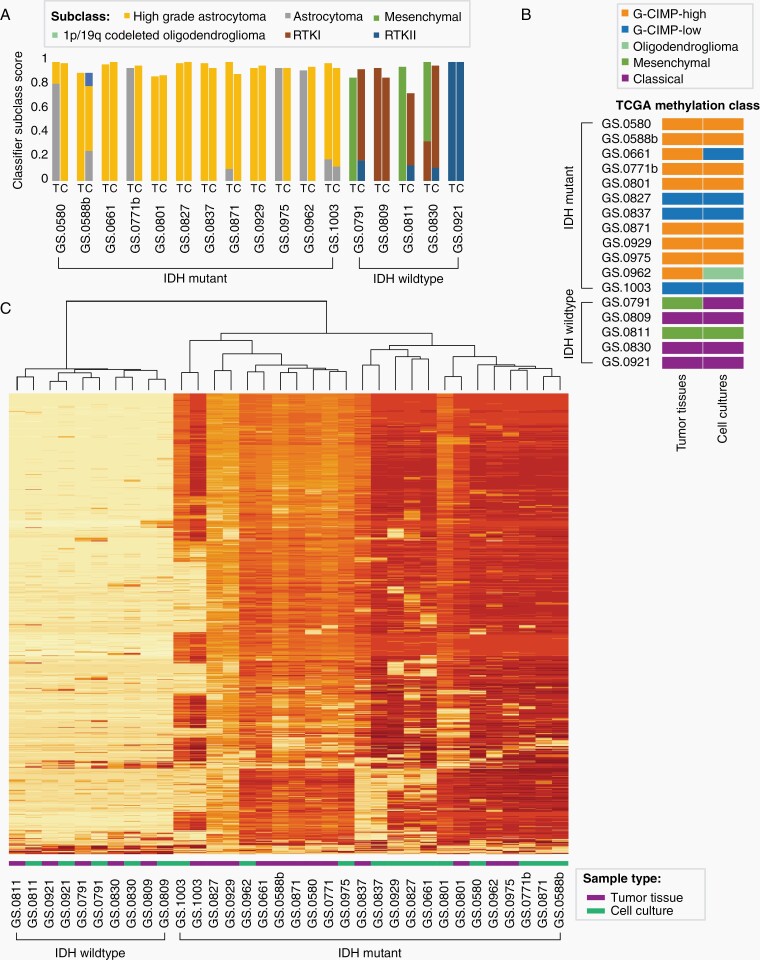
To determine whether the epigenetic profiles of our cultures were retained, we assessed the DNA methylation class of twelve IDHmt and five IDHwt cell cultures and their parental tumors. In a publicly available classifier that can predict all CNS tumor types, all cell cultures were classified as gliomas and received the same overall class as their parental tumors with a classifier score above the cutoff of 0.9, except for glioma cell culture GS.0811 which received a score of 0.74.18 All IDHmt tumors were classified as IDHmt with either subclass A_IDH (astrocytoma, IDH mutant, N = 4) or A_IDH_HG (astrocytoma, IDHmt high grade, N = 8) (Figure 2A). Interestingly, all IDHmt cultures were assigned to methylation subclass A_IDH_HG, which points towards tumor progression in culture or to the selection of more aggressive subclones.
Figure 2.
Global methylation profiles of astrocytoma tumor tissues and derived cell cultures. One sample was analysed for each tumor and derived culture (N = 12 IDHmt, N = 5 IDHwt). (A) Bar graph of subclass scores from the global methylation-based CNS tumor classifier defined by Capper at al.18 The origin of the DNA sample, tumor tissue (T) or cell culture (C), is indicated on the X-axis. The Y-axis represents the methylation class family member classifier score (positive match score ≥ 0.5, maximum score of 1). (B) Representation of TCGA classification of tumor tissue and derived cell culture samples, based on seven probes associated with G-CIMP high or G-CIMP low methylation class. (C) Unsupervised clustering heatmap of DNA methylation profiles of IDHwt tissue samples and cell cultures, and IDHmt tissue samples and cell cultures, based on the 1000 most variable methylated probes. The two main clusters separate IDHwt from IDHmt.
TCGA-based classification of G-CIMP methylation profiles of our samples classified nine tumors as G-CIMP-high, while the remaining three gliomas were classified as G-CIMP-low (Figure 2B).24 One of the G-CIMP-high tumors was G-CIMP-low in its corresponding cell culture, another switched to the oligodendroglioma subclass. Of the five IDHwt cultures all but one retained the G-CIMP classification.
Unsupervised clustering of the 1000 most variably methylated CpG sites separated IDHwt tumors and cell cultures from IDHmt tumors and cell cultures (Figure 2C), with IDHwt samples revealing lower overall methylation intensity. All IDHwt cultures cluster closest to their respective parental tumor sample. IDHmt tumor tissue samples tend to cluster together rather than with their matched cell culture; only four out of twelve IDHmt sample sets group together. Indeed, the primary (GS.0588b) and recurrent (GS.0871) tumor tissue samples derived from the same patient, cluster together, as do the derived cell cultures. This indicates longitudinal preservation of methylation profiles, both in vitro and in situ.
Gene Expression Shows Correlation Between Tumor and Cell Cultures
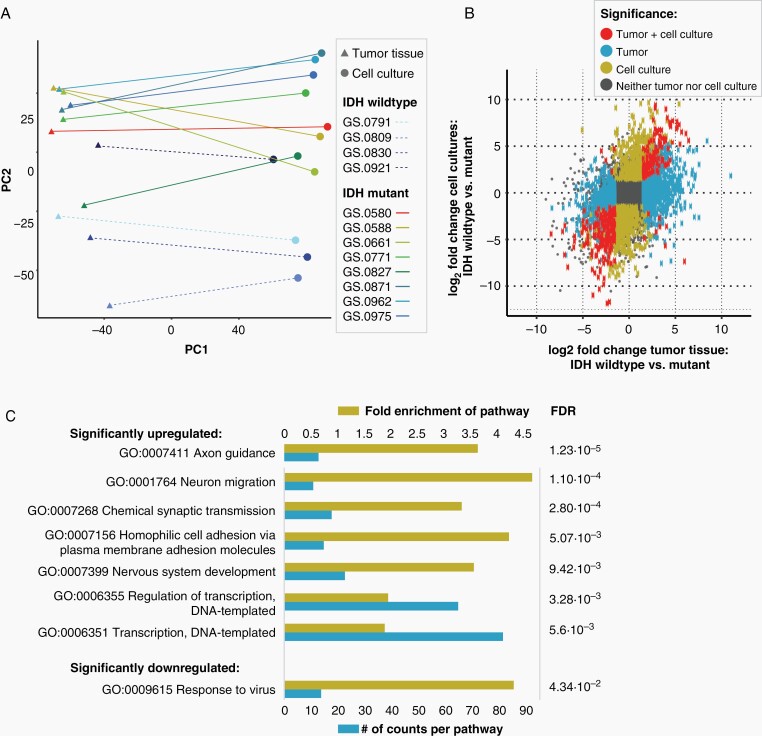
We performed RNA sequencing to evaluate the changes in expression profiles between tumor tissue and cell cultures. A PCA plot shows a very clear separation between tumor tissues and cell cultures in the first dimension (Figure 3A). The second dimension separates IDHwt samples from IDHmt samples.
Figure 3.
Correlation of transcriptomics of IDHmt cell cultures and parental tumor tissue. One sample was analysed from each tumor and derived culture (N = 8 IDHmt, N = 4 IDHwt) (A) Clustering of glioma cell cultures and tumor tissues based on RNA expression through principal component analysis. Components 1 and 2 are shown. (B) Scatterplot showing the correlation between the log2 fold change of IDHwt versus mutant in tumor tissues (X-axis) and IDHwt versus mutant in cell cultures (Y-axis). The grey square represents the cut-off for the identification of differentially expressed genes, set at a log2 fold change larger than –1.5 or smaller than 1.5. (C) Gene ontology pathway analysis of significantly upregulated or downregulated pathways in IDHmt versus IDHwt cell cultures (adjusted P-value < .05). The size of the blue bars represents the number of DEGs in that pathway (bottom X-axis), and yellow bars represents the fold enrichment (FE) of the pathway (top X-axis). The false discovery rate values are shown on the right side of the graph.
We correlated the log2 fold changes of IDHmt versus IDHwt tumor tissue with the log2 fold changes of IDHmt versus IDHwt cultures (adjusted P-value < .05, log2 [fold change] ≥ 0.58 for upregulated genes or ≤–0.58 for downregulated genes) (Figure 3B). There is an overall correlation, with a total of 161 genes significantly differentially expressed in both IDHmt tumor tissue and IDHmt cell cultures compared to their wild-type counterparts. Gene ontology enrichment analysis identified upregulated differentially expressed genes involved in transcription, synaptic transmission, and neural migration, and downregulated genes associated with response to virus infection (Figure 3C).
The IDH Mutation Suppresses Bioenergetic Metabolism in Cultured Glioma
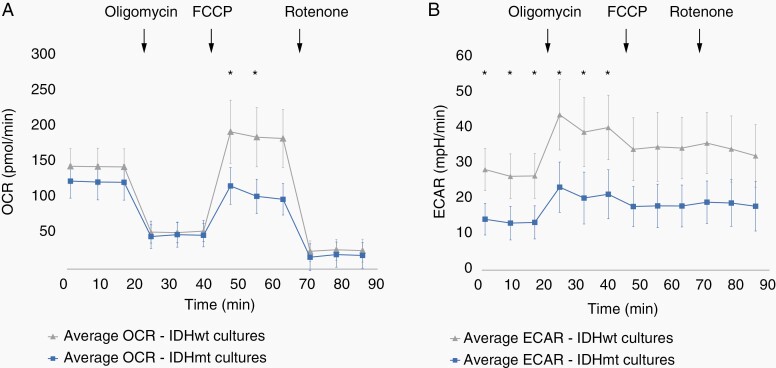
Oncometabolite D-2-HG has a myriad of effects on epigenetics and metabolism.25 Mitochondrial respiration was assessed by measuring extracellular oxygen consumption rate (OCR) and glycolysis through the acidification rate (ECAR), in six IDHmt and seven IDHwt cultures. Although basal OCR did not differ between IDHwt and IDHmt glioma cultures, mitochondrial reserve respiratory capacity was significantly reduced in IDHmt cultures (Figure 4A). Moreover, mutant cells also displayed reduced ECAR, in both basal conditions and after inhibition of mitochondrial ATP synthase (adjusted P-value < .05) (Figure 4B). Canonical pathway analysis (KEGG) of RNA expression data, comparing IDHmt cell cultures with IDHwt cultures, revealed that IDHmt cultures have downregulation of glycolysis and of other processes crucially involved in bioenergetics, such as pyruvate metabolism, the citric acid cycle, and nicotinate and nicotinamide metabolism, which all contribute to respiration substrates production (Supplementary Table S4). Conversely, no differences were observed in the expression of genes of the electron transport chain complexes. Overall, evidence at transcription level is consistent with biochemical analyses showing reduced bioenergetic function in mutants and we conclude that IDHmt cell cultures have an intact electron transport chain but lower metabolite supply.
Figure 4.
Live cell metabolic analysis of IDHmt glioma cultures. (A) Mean OCR of IDHmt cultures (blue, N = 6) and IDHwt cultures (grey, N = 7). (B) Mean ECAR of IDHmt cultures (blue, N = 6) and IDHwt cultures (grey, N = 7). Error bars represent the pooled standard deviations. Asterisks indicate significantly different mean OCR or ECAR values at the indicated timepoint (Independent Student’s T-test, P < .5). Error bars represent the SD. Ten technical replicates were tested for each culture.
IDH Mutant-Specific Inhibitors Affect Enzyme Activity But Not Growth Rate of IDHmt Cells
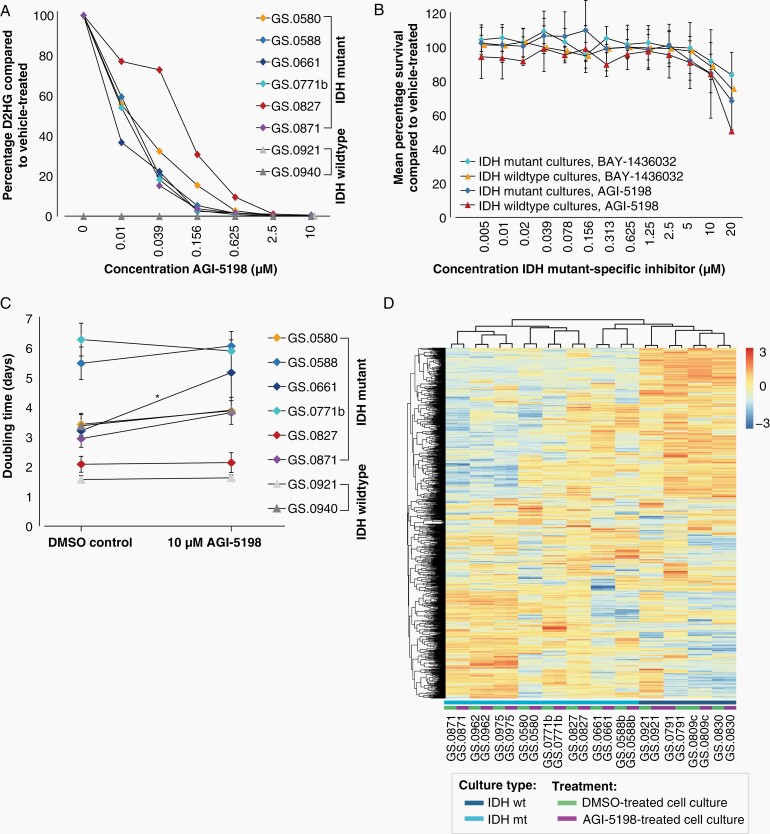
Addition of IDH mutant-specific inhibitor AGI-5198 to the culture medium for seven days resulted in a dose-dependent decrease of D-2-HG levels in all IDHmt cell cultures (Figure 5A). A concentration of 2.5 µM AGI-5198 was sufficient to reduce the D-2-HG levels by over 99%. The D-2-HG levels of the two IDHwt cultures were below the detection limit. Despite this strong D-2-HG reduction, we did not observe any effect on cell viability. Similar results were obtained using an alternative inhibitor (Bay-1436032) (Figure 5B). For both inhibitors, doses over 10 µM resulted in a decrease in cell viability, but this effect was also observed in IDHwt cultures, indicating more general toxicity. We also examined the effect on doubling times over three consecutive passages (Figure 5C). Only IDHmt cell culture GS.0661 showed a significantly extended doubling time (P = .02) in the presence of the 10 µM inhibitor, from 3.2 days for controls to 5.2 days in the presence of AGI-5198. Moreover, we did not observe any overt morphological differences between treated and untreated IDHwt or IDHmt glioma cultures microscopically. These data suggest that the IDHmt cultures are not dependent on high D2-HG levels for viability or growth, at least not for the time spans investigated.
Figure 5.
In vitro effect of IDH mutant-specific inhibitors on IDHmt glioma cultures. (A) Percentage of D2-HG in IDHmt (N = 6) and IDHwt (N = 2) cell cultures treated with increasing concentrations of IDH-mutant specific inhibitor AGI-5198 compared to DMSO-controls. Each datapoint represents a single measurement. (B) Mean average survival of IDHmt (N = 6) and IDHwt (N = 2) cultures treated with increasing concentrations of BAY-1438032 or AGI-5198. Data are presented as mean percentage survival ± SD. Each culture was tested in three technical replicates. (C) Mean doubling times of IDHmt (N = 6) and IDHwt (N = 2) cell cultures, cultured with and without 10 µM IDH mutant-specific inhibitor AGI-5198. Error bars represent standard deviations over three consecutive passages. (*P < .05, paired T-test). Three technical replicates were used for each culture and each time point. (D) Heatmap showing unsupervised cluster analysis of RNA expression of IDHmt (N = 8) and IDHwt (N = 4) glioma cultures, cultured with and without IDH mutant-specific inhibitor AGI-5198 for seven days. Unsupervised clustering was performed on the 1000 most variably expressed genes. The heatmap was scaled by row.
To address the effect of D2-HG on RNA expression, eight IDHmt and four IDHwt cultures were treated for seven days with the IDH mutant-specific inhibitor AGI-5198. Treatment with AGI-5198 does not result in a distinct expression signature and clustering analysis showed that treated and untreated cell culture counterparts remain nearest neighbors in all cases (Figure 5D). In IDHmt glioma cultures, upon AGI-5198-treatment seven genes were significantly downregulated, and 17 upregulated (FDR < 0.00001, log2 [fold change] ≥ 1 or ≤ –1).
Drug Screening of 107 Approved Anti-Cancer Agents on IDHmt Cell Cultures
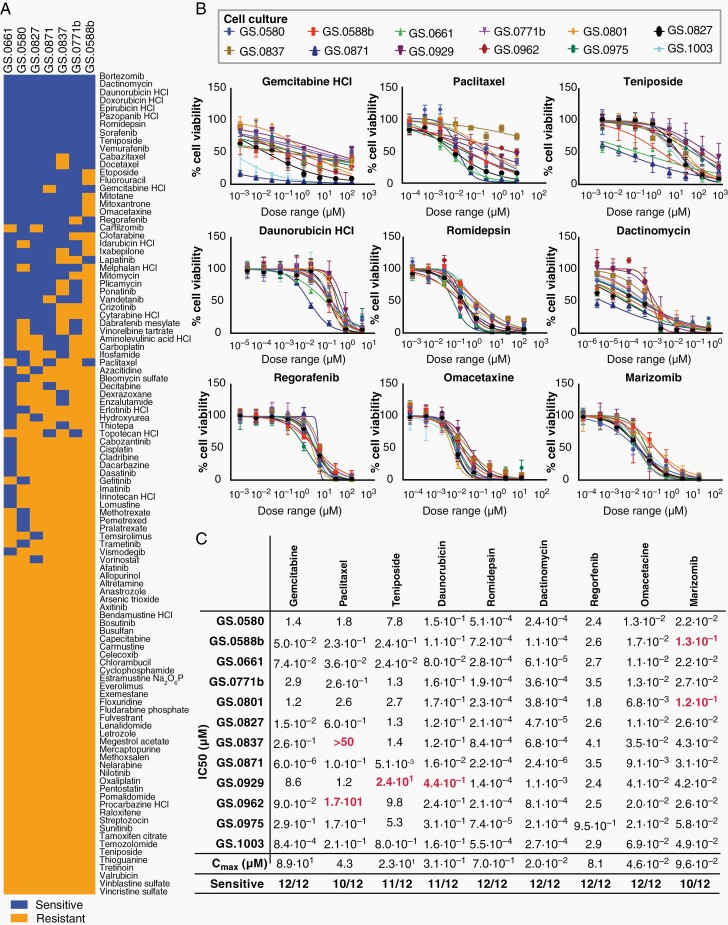
The seven first available IDHmt glioma cultures were screened for sensitivity to 107 FDA-approved oncology drugs. As an initial cut-off, we defined cultures as sensitive if IC50 values of the compounds were below reported Cmax plasma values in patients.26 Nineteen compounds were identified that were effective in at least six out of seven cultures (Figure 6A). These compounds were grouped into nine drug subclasses and ranked within each class according to the CNS multi-parameter optimization (MPO) score, which predicts blood-brain barrier (BBB) crossing based physiochemical properties (Supplementary Table S5).27 However, compounds with low MPO scores were still considered as possible candidates if enhanced or local delivery systems are in development for these drugs. Based on these data, we selected one compound from each subclass for further validation: gemcitabine, teniposide, daunorubicin, romidepsin, dactinomycin, regorafenib, and omacetaxine. We replaced the proteasome inhibitor bortezomib with marizomib, which has a similar mechanism of action and is currently under investigation in clinical GBM trials.28,29 For the taxanes we opted to include paclitaxel, as this compound is currently under investigation using focused ultrasound delivery (Supplementary Table S5).30
Figure 6.
Drug screen with FDA-approved anti-cancer compounds on IDHmt glioma cultures. (A) Heatmap of response of IDHmt glioma cultures (N = 7) to an FDA-approved anti-cancer drug set. Sensitive cell cultures (blue) were defined as having an IC50 < Cmax plasma, while resistant cultures (yellow) had IC50 > Cmax. All drugs were tested with a 10-fold dilution series of six concentrations, in technical duplicates. (B) Dose-response curves of the anti-cancer compounds gemcitabine, paclitaxel, teniposide, daunorubicin, romidepsin, dactinomycin, regorafenib, omacetaxine and marizomib, on all IDHmt cell cultures (N = 12). The X-axes represent increasing concentrations of the respective compounds. The Y-axes indicate the percentage cell viability compared to DMSO-treated controls. All drugs were tested in 3-fold dilution series with 8 concentrations. Error bars represent standard deviation of technical quadruplicates. At least two biological replicates of the screens were performed for each cell culture. (C) IC50 values were calculated based on the dose-response curves shown in Figure 6B. All values represent the drug concentration in µM. Red numbers indicate IC50 > Cmax and are labelled as resistant.
Using smaller-stepped dilution series, IC50 values of these drugs were determined on all twelve IDHmt cultures. This validation set revealed high overall sensitivity to each of the compounds (Figure 6B), although a degree of intertumoral variability was observed. Notably, low-grade glioma culture GS.0962 showed most resistance to therapy. Mean IC50 values remained well-below reported plasma Cmax values for most of the compounds, in particular for romidepsin and dactinomycin (Figure 6C). These results present a set of available anti-cancer compounds that are effective against IDHmt glioma cells and which are either expected to cross the BBB, such as omacetaxin mepesuccinate and marizomib, or which are being developed for local or enhanced delivery, such as paclitaxel.
Discussion
In this study, we present a panel of twelve cell cultures derived from 1p19q noncodeleted astrocytic tumors, and an in-depth characterization of these cultures. Previously, we reported that EGF/FGF supplemented serum-free culture conditions do not support growth of IDHmt tumors.6 However, with adapted tumor dissociation and culture protocols, combined with careful monitoring of morphological features of the cells and in some cases isolation of astrocytic subpopulations, we demonstrate that such cultures can be established and maintained for a prolonged period of time. We identified morphological features that are an indicator of the stem-like nature of IDHmt glioma cells in adherent cell culture with brightfield microscopy, namely elongated cells with significantly smaller cell bodies, thin protrusions, and bright edges. The relatively low overall success rate may be attributed to slow growth rate of most IDHmt gliomas, while culture success is based on expanding the fastest-growing cells, which in some cases can result in stromal cells overcrowding the tumor cells in culture. Furthermore, culture conditions are very different from the local micro-environment within a tumor. This transition can be facilitated by the addition of growth factors to the culture medium, use of extracellular matrix coatings as we do in our protocol, or facilitating ex-situ adaptation through serial xenografts.14,31
Homozygous loss of CDKN2A/B is a relatively common occurrence in gliomas and a predictor of poorer patient survival in IDHmt astrocytomas.32 Interestingly, without exception, we observed loss of CDKN2A/B in all IDHmt glioma cultures, whereas this was detected in only eight of the twelve parental tumors. Loss of CDKN2A/B thus appears to be a prerequisite for successful culture establishment of IDHmt gliomas in our culture conditions. CDKN2A/B loss points towards a more aggressive phenotype and our cultures should therefore be considered to represent of more advanced stages of astrocytomas, even when derived from low-grade tumors.
Global methylation profiling is rapidly gaining momentum in the classification of central nervous system tumors.18 Using this classifier, all IDHmt astrocytoma cell cultures were assigned to the class of high-grade astrocytomas (A_IDH_HG), which, is in agreement with homozygous deletion of the CDKN2A/B locus in all IDHmt glioma cultures. This indicates a higher tumor grade compared to the parental tumors, which in some cases classified as low grade and did not show loss of CDKN2A/B. In a methylation-based classifier defined by the TCGA, such large differences were not identified and most cultures remained G-CIMP-high.
The functions of IDH mutations in metabolism are an important research focus, from a mechanistic point of view as well as a way to identify metabolic vulnerabilities that can be targeted for therapy.22,33 We measured mitochondrial respiration and glycolysis rate in our set of IDHmt cultures and correlated these parameters with gene expression data. This revealed that IDHmt glioma cultures have compromised bioenergetics and reduced mitochondrial reserve capacity, however, the electron transport chain remains unperturbed, suggesting that IDHmt cultures may have a lower metabolite supply. This is consistent with other reports that show reduced glycolysis in IDHmt patient and PDX tumors. Alternative carbon sources (such as glutamate, acetate, and lactate) may supplement the bioenergetic fuel in IDHmt gliomas.10,34,35 The availability of cell cultures with endogenous IDH mutation allows for further studies into these processes.
IDH mutations are early events in gliomagenesis and usually remain clonal as the tumor progresses.36–38 Occasional loss of mutant IDH in recurrent gliomas has been reported, suggesting that the presence of the mutation is not essential for tumor survival or tumor progression.39 Indeed, inhibition of the mutant enzyme and D2HG production did not decrease short-term viability, and only one cell culture showed a small decrease in growth rate when exposed for several passages. This contrasts with previous research that reports decreased growth rates in vitro and in xenografted mouse models.40,41 The source of this discrepancy may lie in the advanced grade of our cell cultures. If so, this may have important clinical implications as it suggests that patients with newly diagnosed low-grade glioma (rather than secondary glioblastoma) may be better candidates for the use of IDH mutant-specific inhibitors. Alternatively, longer exposure to the inhibitor may be required, or the tumor microenvironment may play a role in the anti-glioma effect of the inhibitor.
IDH mutant glioma cells revealed downregulated gene expression associated with response to virus infection. This is an interesting finding, as results from early clinical trials testing oncolytic viruses in GBM have indeed found IDH mutant glioma patients to be overrepresented among the responding patients.42 It has been suggested that this may relate to, (among other intrinsic features) the presence of the IDHmt neoantigen, which has high uniformity and penetrance. However, our data suggests that IDHmt glioma cells may also possess increased viral sensitivities.43
Although our lab is experienced in intracranial injection of glioma cultures to create orthotopic xenografts,44,45 the tested IDHmt cell cultures failed to grow in vivo. Previous data have demonstrated that the rate at which orthotopically injected IDH mutant cultures form tumors is variable, but can take more than six months.8,12,14 Future experiments should determine whether the other IDH mutant cultures form tumors in vivo.
Drug screening of 107 FDA-approved anti-cancer compounds identified 19 compounds that showed effective killing at IC50 values well-below Cmax plasma values. Although plasma Cmax values do not accurately reflect drug concentrations in the tumor due to many factors, including protein-binding, BBB penetration, and the activity of drug efflux pumps, we used this value to discriminate between potentially effective compounds and those which are unlikely to reach effective concentrations in the brain. Interestingly, a number of the identified compounds have also been identified in previous in vitro compound screens on smaller sets of glial tumors.46–50 Several of the compounds identified in our screen have also been tested in clinical trials for glioma in the past (see Supplementary Table S5), but none of the compounds were tested specifically in IDH-mutant patients. The current developments in local or enhanced CNS delivery methods open up new treatment possibilities for highly potent compounds that do not have favorable characteristics to pass the BBB, such as romidepsin and dactinomycin.51,52 Of particular interest are the compounds teniposide, omacetaxine, and marizomib, which are reported to cross the BBB and may offer candidates for systemic delivery.53–55
In conclusion, we established a set of twelve patient-derived IDHmt glioma cell cultures from an astrocytoma background and different WHO grades. Based on the genetic, epigenetic, transcriptomic, and metabolic similarities with the parental tumor, our IDHmt glioma cultures offer a versatile model system to test new therapeutic strategies and to advance research on this important subset of gliomas.
Supplementary Material
Acknowledgments
We thank all patients for contributing to this research and the neurosurgeons involved for providing the tissue, Maurice de Wit, Alicia van der Ploeg, Judith van der Burg and Stanley Van for their contribution to data acquisition, and the NIH/NCI Developmental Therapeutics Program (http://dtp.cancer.gov) for providing the Oncology Drug Set.
Funding
This work was supported by the Stichting STOPHersentumoren (to M.L.M.L.); Strijd van Salland (to P.F.); European Union’s Horizon 2020 Research and Innovation programme under the Marie Skłodowska-Curie (No 766069 to S.L. and M.L.M.L.), Erasmus Foundation-Brain Tumor Survival Marathon (to M.L.M.L.).
Authorship Statement. Conceptualization: C.V., M.L.M.L. and P.J.F.; Methodology: C.V.; M.L.M.L., P.J.F.; Software: Y.H.; R.T.C.Y.; Formal analysis: Y.H., C.V., P.J.F., R.T.C.Y., C.P.G.; Investigation: C.V., I.N., T.V.K., S.B., E.A.S.; Resources: P.G.M., E.A.S.; Writing—Original Draft: C.V., I.N., M.L.M.L.; Writing—Review and editing: M.L.M., P.J.F., P.G.M., C.P.G., C.M.F.D., S.L.W.K., S.L.; Visualization: C.V.; Supervision: M.L.M.L., P.J.F., S.L.; Funding acquisition: M.L.M.L., P.J.F and S.L.
Conflict of interest statement. The authors declare no competing interest.
References
- 1.Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Eng J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Yu J, Tu L, Huang N, Li H, Luo Y. Isocitrate dehydrogenase mutations in glioma: from basic discovery to therapeutics development. Front Oncol. 2019;9:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golub D, Iyengar N, Dogra S, et al. Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. 2019;9:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellinghoff IK, Cloughesy TF, Wen PY, et al. A phase I, open label, perioperative study of AG-120 and AG-881 in recurrent IDH1 mutant, low-grade glioma: Results from cohort 1. J Clin Oncol. 2019;37(15_suppl):2003–2003. [Google Scholar]
- 6.Balvers RK, Kleijn A, Kloezeman JJ, et al. Serum-free culture success of glial tumors is related to specific molecular profiles and expression of extracellular matrix-associated gene modules. Neuro Oncol. 2013;15(12):1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 8.Luchman HA, Stechishin OD, Dang NH, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14(2):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laks DR, Crisman TJ, Shih MY, et al. Large-scale assessment of the gliomasphere model system. Neuro Oncol. 2016;18(10):1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett M, Sperry J, Braas D, et al. Metabolic characterization of isocitrate dehydrogenase (IDH) mutant and IDH wildtype gliomaspheres uncovers cell type-specific vulnerabilities. Cancer Metab. 2018;6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly JJ, Blough MD, Stechishin OD, et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10). Neuro Oncol. 2010;12(7):745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones LE, Hilz S, Grimmer MR, et al. Patient-derived cells from recurrent tumors that model the evolution of IDH-mutant glioma. Neurooncol Adv. 2020;2(1):vdaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Núñez FJ, Mendez FM, Kadiyala P, et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Translat Med. 2019;11(479):eaaq1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakimoto H, Tanaka S, Curry WT, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20(11):2898–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamfers ML, Fulci G, Gianni D, et al. Cyclophosphamide increases transgene expression mediated by an oncolytic adenovirus in glioma-bearing mice monitored by bioluminescence imaging. Mol Ther. 2006;14(6):779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milanese C, Payán-Gómez C, Galvani M, et al. Peripheral mitochondrial function correlates with clinical severity in idiopathic Parkinson’s disease. Mov Disord. 2019;34(8):1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin Chem. 2004;50(8):1391–1395. [DOI] [PubMed] [Google Scholar]
- 22.Tateishi K, Wakimoto H, Iafrate AJ, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28(6):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceccarelli M, Barthel FP, Malta TM, et al. ; TCGA Research Network . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker SJ, Metallo CM. Metabolic consequences of oncogenic IDH mutations. Pharmacol Ther. 2015;152:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liston DR, Davis M. Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin Cancer Res. 2017;23(14):3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wager TT, Hou X, Verhoest PR, Villalobos A. Moving beyond rules: the development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem Neurosci. 2010;1(6):435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NCT03345095 Cg. A phase III trial of with marizomib in patients with newly diagnosed glioblastoma (MIRAGE). 2017; https://clinicaltrials.gov/ct2/show/NCT03345095?term=marizomib&cond=glioblastoma&draw=2&rank=1. Accessed on 04-11-2020.
- 29.Roth P, Reijneveld JC, Gorlia T, et al. EORTC 1709/CCTG CE.8: a phase III trial of marizomib in combination with standard temozolomide-based radiochemotherapy versus standard temozolomide-based radiochemotherapy alone in patients with newly diagnosed glioblastoma. J Clin Oncol. 2019;37(15_suppl):TPS2072-TPS2072. [Google Scholar]
- 30.NCT04528680. Ultrasound-based blood-brain barrier opening and albumin-bound paclitaxel for recurrent glioblastoma (SC9/ABX).2020; https://clinicaltrials.gov/ct2/show/NCT04528680. Accessed 22-10-2020.
- 31.Balvers RK, Kleijn A, Kloezeman JJ, et al. Serum-free culture success of glial tumors is related to specific molecular profiles and expression of extracellular matrix-associated gene modules. Neuro Oncol. 2013;15(12):1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appay R, Dehais C, Maurage CA, et al. ; POLA Network . CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019;21(12):1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Chou AP, Chen W, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dekker LJM, Wu S, Jurriëns C, et al. Metabolic changes related to the IDH1 mutation in gliomas preserve TCA-cycle activity: an investigation at the protein level. FASEB J. 2020;34(3):3646–3657. [DOI] [PubMed] [Google Scholar]
- 35.Fack F, Tardito S, Hochart G, et al. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med. 2017;9(12):1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazor T, Chesnelong C, Pankov A, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc Natl Acad Sci U S A. 2017;114(40):10743–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pusch S, Krausert S, Fischer V, et al. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017;133(4):629–644. [DOI] [PubMed] [Google Scholar]
- 41.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiocca EA, Nassiri F, Wang J, Peruzzi P, Zadeh G. Viral and other therapies for recurrent glioblastoma: is a 24-month durable response unusual? Neuro Oncol. 2019;21(1):14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Accomando WP, Rao AR, Hogan DJ, et al. Molecular and immunologic signatures are related to clinical benefit from treatment with vocimagene amiretrorepvec (Toca 511) and 5-fluorocytosine (Toca FC) in patients with glioma. Clin Cancer Res. 2020;26(23):6176–6186. [DOI] [PubMed] [Google Scholar]
- 44.Balvers RK, Belcaid Z, van den Hengel SK, et al. Locally-delivered T-cell-derived cellular vehicles efficiently track and deliver adenovirus delta24-RGD to infiltrating glioma. Viruses. 2014;6(8):3080–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belcaid Z, Berrevoets C, Choi J, et al. Low-dose oncolytic adenovirus therapy overcomes tumor-induced immune suppression and sensitizes intracranial gliomas to anti-PD-1 therapy. Neurooncol Adv. 2020;2(1):vdaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donson AM, Amani V, Warner EA, et al. Identification of FDA-approved oncology drugs with selective potency in high-risk childhood ependymoma. Mol Cancer Ther. 2018;17(9):1984–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. [DOI] [PubMed] [Google Scholar]
- 48.Dao Trong P, Jungwirth G, Yu T, et al. Large-scale drug screening in patient-derived IDHmut glioma stem cells identifies several efficient drugs among FDA-approved antineoplastic agents. Cells. 2020;9(6):1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang P, Mukthavaram R, Mukthavavam R, et al. Novel anti-glioblastoma agents and therapeutic combinations identified from a collection of FDA approved drugs. J Transl Med. 2014;12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor JT, Ellison S, Pandele A, et al. Actinomycin D downregulates Sox2 and improves survival in preclinical models of recurrent glioblastoma. Neuro Oncol. 2020;22(9):1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albrecht KW, de Witt Hamer PC, Leenstra S, et al. High concentration of daunorubicin and daunorubicinol in human malignant astrocytomas after systemic administration of liposomal daunorubicin. J Neurooncol. 2001;53(3):267–271. [DOI] [PubMed] [Google Scholar]
- 52.Zhang DY, Dmello C, Chen L, et al. Ultrasound-mediated delivery of paclitaxel for glioma: a comparative study of distribution, toxicity, and efficacy of albumin-bound versus cremophor formulations. Clin Cancer Res. 2020;26(2):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeiner PS, Kinzig M, Divé L, et al. Regorafenib CSF penetration, efficacy, and MRI patterns in recurrent malignant glioma patients. J Clin Med. 2019;8(12):2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di K, Lloyd GK, Abraham V, et al. Marizomib activity as a single agent in malignant gliomas: ability to cross the blood-brain barrier. Neuro Oncol. 2016;18(6):840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feun LG, Savaraj N, Landy H, Levin H, Lampidis T. Phase II study of homoharringtonine in patients with recurrent primary malignant central nervous system tumors. J Neurooncol. 1990;9(2):159–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.