Abstract
We conducted a multicenter evaluation of commercial and in-house PCR methods for the detection of enteroviruses. Three coded panels of test and control RNA samples, artificial clinical specimens, and representative enterovirus serotypes were used to assess amplification methods, RNA extraction methods, and reactivities with different enterovirus serotypes. Despite several differences between PCR methods, there was good agreement, although some variation in sensitivity was observed. Most PCR methods were able to detect enterovirus RNA derived from 0.01 50% tissue culture infective dose (TCID50) and were able to detect at least 1 TCID50 of enterovirus in cerebrospinal fluid, stool, or throat swab specimens. Most were also able to detect a wide range of enterovirus serotypes, although serotypic identification was not possible. Some laboratories experienced false-positive results due to PCR contamination, which appeared to result mainly from cross-contamination of specimens during RNA extraction. Provided that this problem is overcome, these PCR methods will prove to be a sensitive and rapid alternative to cell culture for the diagnosis of enterovirus infection.
Human enteroviruses include the polioviruses (PVs), group A and B coxsackieviruses (CVA and CVBs, respectively), echoviruses (ECVs), and enterovirus (ENV) types 68 to 71 (ENV 68 to 71). They cause a wide range of clinical syndromes including inapparent infection, aseptic meningitis, encephalitis, paralytic poliomyelitis, and myocarditis. Although there is currently no specific treatment for enterovirus infections, laboratory diagnosis is required to distinguish between enterovirus-induced disease and other potentially treatable conditions. Diagnosis or exclusion of PV infection is also important for monitoring the progress of the World Health Organization Poliomyelitis Eradication Initiative. Development of antiviral agents for the treatment of enterovirus infections may provide added impetus for laboratory diagnosis in the near future.
Laboratory diagnosis of disease caused by enterovirus is based on culture of an enterovirus from tissue samples from the target organ or associated body fluids such as cerebrospinal fluid (CSF). Detection of an enterovirus in stool or throat swab specimens provides circumstantial evidence of the etiology, and detection of PV in stool is the “gold standard” when investigating patients with suspected paralytic poliomyelitis (40). The enterovirus serotype can be identified by neutralization with pooled intersecting or monovalent antisera. This is usually of less clinical immediacy but is necessary for investigation of suspected PV infections and is useful for the study of enterovirus pathogenesis and epidemiology. However, serotyping methods are poorly standardized and are inconsistently used (38). Serological diagnosis is complicated by the large number of serotypes and is not frequently used.
In recent years reverse transcription-PCR assays have been described for the detection of enterovirus RNA in clinical material (8, 15, 19, 26, 27, 35, 45). One such assay, the Enterovirus Amplicor test, is commercially available from Roche Diagnostic Systems (35). These assays detect a wide range of enterovirus serotypes and are generally more sensitive than cell culture for enterovirus detection in clinical material. They do not, however, identify the enterovirus serotype present. There is at present no standardization of enterovirus PCR assays, and few comparative data on sensitivity and specificity among different laboratories were available until recently (23). A multicenter quality assessment of the enterovirus PCR assays currently used in research or diagnostic laboratories was therefore conducted within the framework of the European Union Concerted Action on Virus Meningitis and Encephalitis.
(This work was presented in part at the First Annual Meeting of the European Society for Clinical Virology, Bologna, Italy, September 1997.)
MATERIALS AND METHODS
Virus stocks.
Infected cell culture supernatants of the following viruses were used for this study. CVB type 3 (CVB 3) Nancy strain was provided by the Department of Virology, Guy’s, King’s College and St Thomas’ Hospitals’ School of Medicine, and was originally obtained from R. Kandolf, Tübingen, Germany. PV type 2 (PV 2) Sabin, CVA type 7 (CVA 7) Parker, CVA 21 Coe, CVA 24 Joseph, CVB 2 Pretorius, ECV type 16 (ECV 16) Harrington, ECV 20 JV-1, ECV 22 Harris, ECV 29 JV-10, and ENV 71 Br-Cr were provided by the Research Laboratory for Infectious Diseases, National Institute of Public Health and the Environmental, Bilthoven, The Netherlands. ECV 22 is now known to be genetically distinct from the other enteroviruses (17, 36) and is now classified in a different picornavirus genus together with ECV 23. Human coronavirus 229E and influenza virus type B were obtained from the American Type Culture Collection.
Quality assessment panels.
Three panels of coded samples were prepared in the coordinating laboratory (Guy’s, King’s College & St Thomas’ Hospitals’ School of Medicine, London, United Kingdom). Panel A (nine samples) consisted of total RNA derived from serial dilutions of a CVB 3-infected Vero cell culture supernatant in sterile water. RNA was prepared with RNAzol B (Biogenesis, Poole, Dorset, United Kingdom) as described previously (26). Panel B (15 samples) consisted of pooled clinical specimens (CSF, stool filtrate, throat swab transport medium) or cell culture medium that previously tested negative for enteroviruses by cell culture and PCR and that were spiked with dilutions of CVB 3-infected cell culture supernatant. Panel C (24 samples) consisted of dilutions of representative enterovirus serotypes and other viruses, as described above. Each panel included virus-negative controls which were prepared in a separate room.
Enterovirus PCR.
The panels described above were distributed to 14 participating laboratories. Panel A was distributed as ethanol precipitates. Participants were asked to collect RNA precipitates by centrifugation, wash the pellets in 70% ethanol, and then dissolve them in 20 μl of sterile water and to use 1/10 volume of this solution for PCR. Panels B and C were distributed on dry ice in 100-μl volumes. Participants were asked to prepare the RNA by their own methods and to use 1/10 volume of this RNA extract for PCR. Sensitivity limits for each assay were based on the amount of RNA present in 1/10 volume of the reconstituted sample. However, participants using the Enterovirus Amplicor test were asked to resuspend the pellets in 100 μl of sample buffer and to use 30 μl of RNA extract for PCR in accordance with the manufacturer’s protocol. Details of the laboratory methods used are given in the Results section.
Nucleotide sequence analysis.
One participant performed nucleotide sequence analysis of the PCR products from panel C samples in an effort to identify the enterovirus serotypes detected. Nested PCR products corresponding to nucleotides 166 to 463 of the enterovirus genome (19) were sequenced directly with Dyedeoxy terminators and an ABI 377 automated sequencer (Applied Biosystems). The sequences were compared with enterovirus sequences deposited in the GenBank and EMBL databases with the GCG package (Genetics Computer Group, Madison, Wis.).
Analysis of results.
Participants sent their results to the European Union Concerted Action on Virus Meningitis and Encephalitis office (Manchester Royal Infirmary, Manchester, United Kingdom), which served as a neutral party, where the results from each participant were assigned a code and forwarded to the coordinating laboratory. After the results, had been entered into a database, the laboratory code was broken to allow analysis of the results and evaluation of enterovirus PCR methods. The code breaker for the three panels of samples was also sent to the participants at this stage.
Nucleotide sequence accession numbers.
The nucleotide sequences of PCR products derived from panel C samples have been deposited in GenBank under accession nos. AF068878 to AF068885 and AF076999.
RESULTS
Of the 14 laboratories that received the three quality assessment panels, 11 laboratories produced 13 datum sets for panel A and 11 laboratories produced 12 datum sets for panels B and C. Three data sets were generated by the Enterovirus Amplicor assay (35). The remainder were generated by in-house methods. All in-house methods used guanidine thiocyanate-based RNA extraction methods, including methods that used RNAzol B and RNeasy (Qiagen GmbH, Hilden, Germany) and in-house protocols (6, 9). Most in-house methods used separate reverse transcription steps and PCRs. PCRs used single or nested primer pairs, most of which have been published previously (8, 10, 15, 19, 27, 33, 45), with total cycle numbers ranging from 35 to 80. Most used gel electrophoresis with ethidium bromide staining for the detection of PCR products. The results of the PCR analyses are given in Fig. 1 to 3.
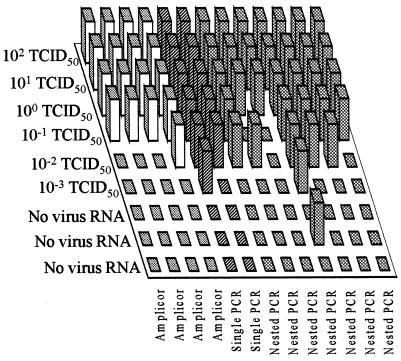
FIG. 1.
Performance of participating laboratories in detecting enterovirus RNA dilutions included in panel A samples. The datum sets obtained by the Roche Amplicor test are shown as white columns, while the datum sets obtained by in-house single PCR or nested PCR assays are shown as diagonally striped and cross-hatched columns, respectively. A positive result is indicated by a raised column.
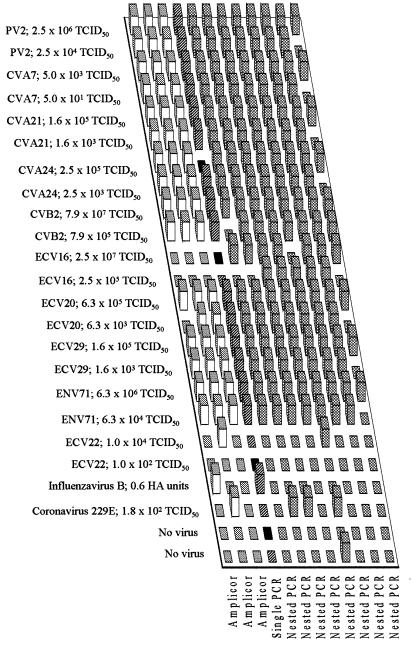
FIG. 3.
Performance of participating laboratories in detecting enterovirus serotypes included in panel C samples. See legend to Fig. 1 for interpretation of the columns. Samples identified in black were not tested.
Panel A.
The PCR detection limits of enterovirus PCR assays ranged from 0.001 to 1 50% tissue culture infective dose (TCID50) (Fig. 1). Of a total of 78 enterovirus RNA-positive samples tested in all laboratories, 60 (77%) were conclusively identified as positive. A single false-positive result occurred for 1 of 12 datum sets, i.e., for 1 of 39 negative control samples tested (2.6%).
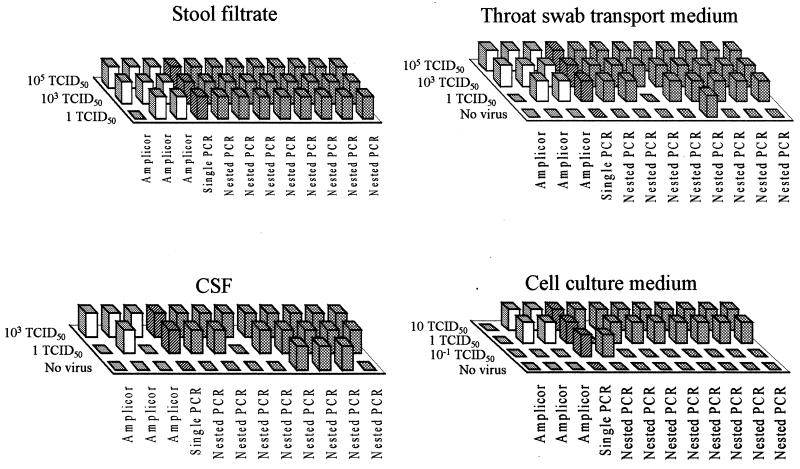
Panel B.
One negative control sample in panel B was subsequently found to be contaminated with enterovirus. The results obtained with this sample were therefore discarded. One TCID50 of CVB 3 was detected in stool filtrate, throat swab, CSF, or cell culture fluid samples in 11, 10, 9, and 10 of 12 datum sets, respectively (Fig. 2). Of a cumulative total of 121 enterovirus RNA-positive samples tested in all laboratories, 103 (85%) were conclusively identified as positive. False-positive results were obtained for 3 of 12 datum sets and 4 of 36 (11%) negative control samples.
FIG. 2.
Performance of participating laboratories in detecting enterovirus in different specimen types included in panel B samples. See legend to Fig. 1 for interpretation of the columns.
Panel C.
Eight of the 10 enterovirus serotypes used in panel C were detected at one or both dilutions tested in all datum sets (Fig. 3). The results for one datum set indicated reduced sensitivity, failing to detect the higher dilution of 9 of 10 serotypes. Two of 10 datum sets tested positive for ECV 22 at the lower dilution only. ECV 16 was detected in only 7 of 11 datum sets at the lower dilution and in only 5 of 12 datum sets at the higher dilution. If the ECV 22-containing samples are regarded as negative controls, 194 of 214 enterovirus-positive samples (91%) were correctly identified. False-positive results occurred for 6 of 12 datum sets and 10 of 70 (14%) negative control samples. In an attempt to identify the serotypes of the viruses in panel C samples, PCR product sequences were compared with known enterovirus sequences. Satisfactory nucleotide sequence data were obtained for 13 of 18 enterovirus-positive samples (excluding samples containing ECV 22), and a correct identification of serotype, based on maximum sequence similarity, was achieved for only 6 of these 13 samples (Table 1).
TABLE 1.
Results of attempts to identify enterovirus serotypes by nucleotide sequence analysis of PCR products
| Enterovirus serotypes in panel C
|
Published enterovirus sequence of greatest similarity
|
||
|---|---|---|---|
| Serotype | TCID50 | Serotype (accession no.) | % Similarity |
| PV 2 | 2.5 × 106 | PV 2 (X00595) | 98 |
| PV 2 | 2.5 × 104 | PV 2 (X00595) | 98 |
| CVA 7 | 5.0 × 103 | CVA 16 (U05876) | 92 |
| ECV 7 (L76401) | 92 | ||
| CVA 7 | 5.0 × 101 | CVA 16 (U05876) | 92 |
| ECV 7 (L76401) | 92 | ||
| CVA 21 | 1.6 × 105 | CVA 21 (D00538) | 100 |
| CVA 21 | 1.6 × 103 | CVA 21 (D00538) | 97 |
| CVA 24 | 2.5 × 105 | No sequence obtained | |
| CVA 24 | 2.5 × 103 | PV 1 (L76404) | 94 |
| CVB 2 | 7.9 × 107 | CVB 2 (Y09512) | 94 |
| CVB 2 | 7.9 × 105 | No sequence obtained | |
| ECV 16 | 2.5 × 107 | No sequence obtained | |
| ECV 16 | 2.5 × 105 | No sequence obtained | |
| ECV 20 | 6.3 × 105 | CVB 3 (M33854) | 96 |
| ECV 20 | 6.3 × 103 | CVB 3 (M33854) | 96 |
| ECV 29 | 1.6 × 105 | CVB 3 (M33854) | 96 |
| ECV 29 | 1.6 × 103 | CVB 3 (M33854) | 96 |
| ENV 71 | 6.3 × 106 | ENV 71 (U22521) | 99 |
| ENV 71 | 6.3 × 104 | No sequence obtained | |
DISCUSSION
There was considerable variation between the different PCR protocols used by the participants. However, variation in enterovirus RNA detection sensitivity was not apparently related to any of these variables. It is more likely that variation is related to the degree of optimization of laboratory methods, the experience of laboratory personnel, or random sampling effects with samples containing high dilutions of virus or viral RNA close to assay detection limits. The experience of laboratory personnel is likely to be a major influence on test performance, and this is illustrated by the variation in results obtained by participants using the Roche Enterovirus Amplicor test. The low level of variation in sensitivity for tests with panel B samples indicates that all RNA extraction methods used are satisfactory for the treatment of stool, CSF, and throat swab specimens for enterovirus PCR. These are the most common types of specimens submitted for diagnostic evaluation for patients with suspected enterovirus infection. Most PCR methods were at least as sensitive as cell culture for the detection of enterovirus in these specimens.
In evaluating panel C samples, tests by one laboratory had a reduced sensitivity of detection of most serotypes, possibly due to suboptimal primer recognition of these viral RNA sequences. However, most datum sets indicated adequate performance with this panel. The failure to detect ECV 22 in most cases was expected, since this virus is genetically dissimilar to other enteroviruses (17, 36). ECV 22-containing samples were therefore considered enterovirus-negative controls, and positive PCR results were regarded as false positive for the purposes of analysis of the results. The high rate of false-negative results for ECV 16 is more surprising. Only nested PCR protocols were able to detect this serotype, and all nested PCR protocols detected at least one of the dilutions tested. The ECV 16 strain used may be less readily detected by some enterovirus primers. Alternatively, partial deterioration of the original stock may have occurred.
False-positive results occurred mainly with panels B and C, suggesting that most false positivity resulted from cross-contamination during RNA extraction, although the difference in the false-positivity rate between the results for panel A and those for panels B and C was not statistically significant (P = 0.07; Fisher’s exact test). The occurrence of false-positive results for 4 of 30 negative control samples (13%) from panels B and C with the Enterovirus Amplicor test, which includes enzymatic elimination of PCR product carryover, also supports this conclusion. Nested PCR methods, which were used by several participants, may be particularly sensitive to PCR contamination. Other PCR quality assessment schemes have also recorded false-positive results (7, 12, 14, 18, 23, 31, 37, 43, 44). This problem is unlikely to be eliminated until sample processing can be contained and automated, as suggested by Damen et al. (12). The high proportion of enterovirus-positive samples and the high levels of virus in some samples (10−3 to 107 TCID50s) included in these panels makes this quality assessment exercise a stringent measure of the level of cross-contamination. However, the range of viral titers in these samples is representative of that observed in clinical samples. The use of highly sensitive assays may thus prove problematic when testing samples with potentially high virus titers, such as throat swab and stool specimens.
One participant used direct nucleotide sequence analysis of nested PCR products obtained from panel C samples. Serotypes for which no sequence data are available could not be identified in this way, and correct identification could be achieved only when published sequences were available for the same virus strain as that included in the quality control trial. Thus, for CVA 24 an incorrect serotype was assigned because the PCR product sequence of CVA 24 Joseph determined in this study showed greater similarity to published PV 1 sequences (96% similarity) than to the published sequence of the CVA 24 variant, strain EH24/70 (accession no. D90457; 89% similarity [data not shown]). Thus, sequence analysis of this conserved genomic region (nucleotides 166 to 463) cannot be used to identify enterovirus serotypes. Others have found that nucleotide sequence analysis of the 5′ nontranslated region permits classification of enteroviruses into two broad groups of PV-like and CVB-like enteroviruses, while analysis of sequences encoding viral capsid protein 2 (VP2) permits classification of enteroviruses into four major phylogenetic clusters (4, 29). Since the serotype is determined by antigenic determinants located within VP1, VP2, and VP3 (24), serotypic identification would probably require nucleotide sequence analysis of this genomic region. A molecular typing system would complement PCR-based diagnostic methods and may prove to be more reliable than current serotyping methods but would require considerable research and development (25). However, several PV-specific PCR assays which may prove to be useful in distinguishing between PV and other enteroviruses (1, 11, 13, 22), for differentiation of PV serotypes (21), or for intratypic differentiation of vaccine and wild-type PV strains (5, 41) in patients with suspected PV infection have recently been described.
Use of a commercially available assay such as the Enterovirus Amplicor test would contribute to standardization. Our results indicate that this test has performance characteristics comparable to those of most in-house methods, and the time required to perform this test is less than that required to perform in-house methods. The Enterovirus Amplicor test has been validated for the detection of enteroviruses in CSF, where its superior sensitivity over cell culture methods has repeatedly been demonstrated (3, 16, 20, 23, 30, 32, 39, 42). It has also been successfully used to detect enteroviruses in serum and throat swabs and, with somewhat reduced sensitivity compared to that of virus culture, in urine (2, 3, 28, 34). By evaluating the Enterovirus Amplicor test alongside in-house PCR methods in a multicenter study, we now provide further evidence of the utility of this assay.
The major problem identified in this study is the risk of false positive results, which was more pronounced in some laboratories. Provided that adequate measures are taken to monitor for and exclude false positivity, enterovirus PCR is likely to prove to be a reliable means of enterovirus detection in clinical specimens. However, an ongoing mechanism for quality assessment will be required to ensure that test sensitivity, specificity, and standardization are maintained.
ACKNOWLEDGMENTS
The assistance of the following individuals in this study is gratefully acknowledged: I. Casas C.N.M., Instituto de Salud Carlos III, Madrid, Spain; H. Nordei, and L. Sundqvist, Swedish Institute for Infectious Disease Control, Stockholm, Sweden; L. Cantero-Aguilar, Service de Bacteriologie, Virologie et Hygiene, Hôpital Saint Vincent de Paul, Paris, France; and U. Kämmerer and R. Kandolf, Department of Molecular Pathology, University of Tübingen, Tübingen, Germany.
Appendix
Other members of the European Union Concerted Action on Virus Meningitis and Encephalitis are M. Ciardi, Instituto di Malattie Infettive, University of Rome, Rome, Italy; P. Cinque, Division di Malattie Infettive, Ospedale San Raffaele, Milan, Italy; G. Gerna, Viral Diagnostic Service, IRCCS Policlinico San Matteo, Pavia, Italy; J. M. Echevarría, C.N.M., Instituto de Salud Carlos III, Madrid, Spain; B. Faber Vestergaard, Department of Virology, Statens Seruminstitut, Copenhagen, Denmark; M. Forsgren, Clinical Virology F68, Huddinge University Hospital, Huddinge, Sweden; A. Linde and M. Grandien, Swedish Institute for Infectious Disease Control, Stockholm, Sweden; T. Hovi, Enterovirus Laboratory, National Public Health Institute, Helsinki, Finland; M. Koskiniemi, Department of Virology, University of Helsinki, Helsinki, Finland; P. Lebon, Service de Bacteriologie, Virologie et Hygiene, Hôpital Saint Vincent de Paul, Paris, France; P. Monteyne and C. Sindic, Laboratoire de Neurochimie, Université Catholique de Louvain, Brussels, Belgium; E. Puchhammer-Stockl, Institute of Virology, University of Vienna, Vienna, Austria; C. Taylor, Department of Virology, Public Health Laboratory, Newcastle General Hospital, Newcastle-upon-Tyne, United Kingdom; V. ter Meulen, Institut für Virologie und Immunobiologie, Universität Würzburg, Würzburg, Germany; and T. Weber, Department of Neurology, Marienkrankenhaus Hamburg, Hamburg, Germany.
REFERENCES
- 1.Abraham R, Chonmaitree T, McCombs J, Prabhakar B, Lo Verde P T, Ogra P L. Rapid detection of poliovirus by reverse transcription and polymerase chain reaction amplification: application for differentiation between poliovirus and nonpoliovirus enteroviruses. J Clin Microbiol. 1993;31:395–399. doi: 10.1128/jcm.31.2.395-399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abzug M J, Loeffelholz M, Rotbart H A. Diagnosis of neonatal enterovirus infection by polymerase chain reaction. J Pediatr. 1995;126:447–450. doi: 10.1016/s0022-3476(95)70466-3. [DOI] [PubMed] [Google Scholar]
- 3.Andréoletti L, Blassel-Damman N, Dewilde A, Vallée L, Cremer R, Hober D, Wattré P. Comparison of use of cerebrospinal fluid, serum, and throat swab specimens in diagnosis of enteroviral acute neurological infection by a rapid RNA detection PCR assay. J Clin Microbiol. 1998;36:589–591. doi: 10.1128/jcm.36.2.589-591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arola A, Santti J, Ruuskanen O, Halonen P, Hyypiä T. Identification of enteroviruses in clinical specimens by competitive PCR followed by genetic typing using sequence analysis. J Clin Microbiol. 1996;34:313–318. doi: 10.1128/jcm.34.2.313-318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of polioviruses analysed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bootman J S, Kitchin P A. Reference preparations in the standardisation of HIV-1 PCR—an international collaborative study. J Virol Methods. 1994;49:1–8. doi: 10.1016/0166-0934(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 8.Casas I, Klapper P E, Cleator G M, Echevarría J E, Tenorio A, Echevarría J M. Two different PCR assays to detect enteroviral RNA in CSF samples from patients with acute aseptic meningitis. J Med Virol. 1995;47:378–385. doi: 10.1002/jmv.1890470414. [DOI] [PubMed] [Google Scholar]
- 9.Casas I, Powell L, Klapper P E, Cleator G M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J Virol Methods. 1995;53:25–36. doi: 10.1016/0166-0934(94)00173-e. [DOI] [PubMed] [Google Scholar]
- 10.Casas I, Tenorio A, Echevarría J M, Klapper P E, Cleator G M. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J Virol Methods. 1997;66:39–50. doi: 10.1016/s0166-0934(97)00035-9. [DOI] [PubMed] [Google Scholar]
- 11.Chezzi C. Rapid diagnosis of poliovirus infection by PCR amplification. J Clin Microbiol. 1996;34:1722–1725. doi: 10.1128/jcm.34.7.1722-1725.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damen M, Cuypers H T M, Zaaijer H L, Reesink H W, Schaasberg W P, Gerlich W H, Niesters H G M, Lelie P N. International collaborative study on the second EUROHEP HCV-RNA reference panel. J Virol Methods. 1996;58:175–185. doi: 10.1016/0166-0934(96)02011-3. [DOI] [PubMed] [Google Scholar]
- 13.Egger D, Pasamontes L, Ostermayer M, Bienz K. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J Clin Microbiol. 1995;33:1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French Study Group for the Standardization of Hepatitis C Virus PCR. Improvement of hepatitis C virus RNA polymerase chain reaction through a multicenter quality control study. J Virol Methods. 1994;49:79–88. doi: 10.1016/0166-0934(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Glimåker M, Johansson B, Olcén P, Ehrnst A, Forsgren M. Detection of enteroviral RNA by polymerase chain reaction in cerebrospinal fluid from patients with aseptic meningitis. Scand J Infect Dis. 1992;25:547–557. doi: 10.3109/00365549309008542. [DOI] [PubMed] [Google Scholar]
- 16.Gorgievski-Hrisoho M, Schumacher J-D, Vilimonovik N, Germann D, Matter L. Detection by PCR of enteroviruses in cerebrospinal fluid during a summer outbreak of aseptic meningitis in Switzerland. J Clin Microbiol. 1998;36:2408–2412. doi: 10.1128/jcm.36.9.2408-2412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyypiä T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci USA. 1992;89:8847–8851. doi: 10.1073/pnas.89.18.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson J B, Drew J, Lin H J, Otto P, Bremer J W, Hollinger F B, Wolinsky S M the ACTG PCR Working Group; the ACTG PCR Virology Laboratories. Establishment of a quality assurance program for human immunodeficiency virus type 1 DNA polymerase chain reaction assays by the AIDS Clinical Trials Group. J Clin Microbiol. 1993;31:3123–3128. doi: 10.1128/jcm.31.12.3123-3128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kämmerer U, Kunkel B, Korn K. Nested polymerase chain reaction for specific detection and rapid identification of human picornaviruses. J Clin Microbiol. 1994;32:285–291. doi: 10.1128/jcm.32.2.285-291.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler H H, Santner B, Rabenau H, Berger A, Vince A, Lewinski C, Weber B, Pierer K, Stuenzer D, Marth E, Doerr H W. Rapid diagnosis of enterovirus infection by a new one-step reverse transcription-PCR assay. J Clin Microbiol. 1997;35:976–977. doi: 10.1128/jcm.35.4.976-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, Da Silva E, Peñaranda S, Pallansch M, Kew O. Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1998;36:352–357. doi: 10.1128/jcm.36.2.352-357.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, Mulders M N, Holloway B P, Pallansch M A, Kew O M. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1996;34:2990–2996. doi: 10.1128/jcm.34.12.2990-2996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina B, Pozzetto B, Andreoletti L, Beguier E, Bourlet T, Dussaix E, Grangeot-Keros L, Gratacap-Cavillier B, Henquell C, Legrand-Quillien M C, Novillo A, Palmer P, Petitjean J, Sandres K, Dubreuil P, Fleury H, Freymuth F, Leparc-Goffart I, Hober D, Izopet J, Kopecka H, Lazizi Y, Lafeuille H, Lebon P, Roseto A, Marchadier E, Masquelier B, Picard B, Puel J, Siegneurin J M, Wattre P, Aymard M. Multicenter evaluation of a commercially available PCR assay for diagnosing enterovirus infection in a panel of cerebrospinal fluid specimens. J Clin Microbiol. 1996;34:3002–3006. doi: 10.1128/jcm.34.12.3002-3006.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minor P D, Ferguson M, Evans D M A, Almond J W, Icenogle J P. Antigenic structure of polioviruses of serotypes 1, 2, and 3. J Gen Virol. 1986;67:1283–1291. doi: 10.1099/0022-1317-67-7-1283. [DOI] [PubMed] [Google Scholar]
- 25.Muir P, Kämmerer U, Korn K, Mulders M N, Pöyry T, Weissbrich B, Kandolf R, Cleator G M, van Loon A M for the European Union Concerted Action on Virus Meningitis and Encephalitis. Molecular typing of enteroviruses: current status and future requirements. Clin Microbiol Rev. 1997;11:202–227. doi: 10.1128/cmr.11.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muir P, Nicholson F, Jhetam M, Neogi S, Banatvala J E. Rapid diagnosis of enterovirus infection by magnetic bead extraction and polymerase chain reaction detection of enterovirus RNA in clinical specimens. J Clin Microbiol. 1993;31:31–38. doi: 10.1128/jcm.31.1.31-38.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholson F, Meetoo G, Aiyar S, Banatvala J E, Muir P. Detection of enterovirus RNA in clinical samples by nested polymerase chain reaction for rapid diagnosis of enterovirus infection. J Virol Methods. 1994;48:155–166. doi: 10.1016/0166-0934(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen L P, Modlin J F, Rotbart H A. Detection of enteroviruses by polymerase chain reaction in urine samples of patients with aseptic meningitis. Pediatr Infect Dis J. 1996;15:625–627. doi: 10.1097/00006454-199607000-00013. [DOI] [PubMed] [Google Scholar]
- 29.Pöyry T, Kinnunen L, Hyypiä T, Brown B, Horsnell C, Hovi T, Stanway G. Genetic and phylogenetic clustering of enteroviruses. J Gen Virol. 1996;77:1699–1717. doi: 10.1099/0022-1317-77-8-1699. [DOI] [PubMed] [Google Scholar]
- 30.Pozo F, Casas I, Tenorio A, Trallero G, Echevarria J M. Evaluation of a commercially available reverse transcription-PCR assay for diagnosis of enteroviral infection in archival and prospectively collected cerebrospinal fluid specimens. J Clin Microbiol. 1998;36:1741–1745. doi: 10.1128/jcm.36.6.1741-1745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quint W G, Heijtink R A, Schirm J, Gerlich W H, Niesters H G. Reliability of methods for hepatitis B virus DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson D W, Kociuba K R. Evaluation of the Roche Amplicor polymerase chain reaction assay for detection of enteroviruses in cerebrospinal fluid and its potential impact on patient management. Clin Microbiol Infect. 1997;3:672–676. doi: 10.1111/j.1469-0691.1997.tb00477.x. [DOI] [PubMed] [Google Scholar]
- 33.Rotbart H A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotbart H A, Ahmed A, Hickey S, Dagan R, McCracken G H, Jr, Whitley R J, Modlin J F, Cascino M, O’Donnel J F, Menegus M A, Blum D. Diagnosis of enterovirus infection by polymerase chain reaction of multiple specimen types. Pediatr Infect Dis J. 1997;16:409–411. doi: 10.1097/00006454-199704000-00014. [DOI] [PubMed] [Google Scholar]
- 35.Rotbart H A, Sawyer M H, Fast S, Lewinski C, Murphy N, Keyser E F, Spadoro J, Kao S-Y, Loeffelholz M. Diagnosis of enteroviral meningitis by using PCR with a colorimetric microwell detection assay. J Clin Microbiol. 1994;32:2590–2592. doi: 10.1128/jcm.32.10.2590-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanway G, Kalkkinen N, Roivainen M, Ghazi F, Khan M, Smyth M, Meurman O, Hyypiä T. Molecular and biological characteristics of echovirus 22, a representative of a new picornavirus group. J Virol. 1994;68:8232–8238. doi: 10.1128/jvi.68.12.8232-8238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandamme A M, Fransen K, Debaisieux L, Marissens D, Sprecher S, Vaira D, Vandenbroucke A T, Verhofstede C the Belgian AIDS Reference Laboratories. Standardisation of primers and an algorithm for HIV-1 diagnostic PCR evaluated in patients harbouring strains of diverse geographical origin. J Virol Methods. 1995;51:305–316. doi: 10.1016/0166-0934(94)00126-2. [DOI] [PubMed] [Google Scholar]
- 38.Van Loon A M, Cleator G M, Ras A. External quality assessment of enterovirus detection and typing. Bull W H O. 1999;77:217–223. [PMC free article] [PubMed] [Google Scholar]
- 39.Van Vliet K E, Glimåker M, Lebon P, Klapper P E, Taylor C E, Ciardi M, van der Avoort H G A M, Diepersloot R J A, Kurtz J, Peeters M F, Cleator G M, van Loon A M for the European Union Concerted Action on Viral Meningitis and Encephalitis. Multicenter evaluation of the Amplicor enterovirus PCR test with cerebrospinal fluid from patients with aseptic meningitis. J Clin Microbiol. 1998;36:2652–2657. doi: 10.1128/jcm.36.9.2652-2657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Manual for the virological investigation of poliomyelitis. Geneva, Switzerland: World Health Organization; 1990. [Google Scholar]
- 41.Yang C F, De L, Holloway B P, Pallansch M A, Kew O M. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 1991;20:159–179. doi: 10.1016/0168-1702(91)90107-7. [DOI] [PubMed] [Google Scholar]
- 42.Yerly S, Gervaix A, Simonet V, Caflisch M, Perrin L, Wunderli W. Rapid and sensitive detection of enteroviruses in specimens from patients with aseptic meningitis. J Clin Microbiol. 1996;34:199–201. doi: 10.1128/jcm.34.1.199-201.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaaijer H L, Cuypers H T M, Reesink H W, Winkel I N, Gerken G, Lelie P N. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet. 1993;341:722–724. doi: 10.1016/0140-6736(93)90488-3. [DOI] [PubMed] [Google Scholar]
- 44.Zeuzem S, Ruster B, Roth W K. Clinical evaluation of a new polymerase chain reaction assay (Amplicor HCV) for detection of hepatitis C virus. Z Gastroenterol. 1994;32:342–347. [PubMed] [Google Scholar]
- 45.Zoll G J, Melchers W J G, Kopecka H, Jambroes G, van der Poel H J A, Galama J M D. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J Clin Microbiol. 1992;30:160–165. doi: 10.1128/jcm.30.1.160-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]