Abstract
Objective
Port-wine birthmark (PWB) is a common occurrence in the newborn, and general pediatricians, dermatologists, and ophthalmologists are often called on to make an assessment of risk for Sturge-Weber syndrome (SWS) due to workforce shortages in pediatric neurologists and MRI’s low sensitivity for SWS brain involvement in infants. We therefore aimed to develop a quantitative EEG (qEEG) approach to safely screen young infants with PWB for SWS risk and optimal timing of diagnostic MRI.
Methods
Forty-eight infants (prior to first birthday) underwent EEG recording. Signal processing methods compared voltage between left and right sides using a previously defined pipeline and diagnostic threshold. In this test sample, we compared sensitivity/specificity of the qEEG metric against MRI performed after the first birthday. We also used likelihood ratio testing to determine whether qEEG adds incremental information beyond topographical extent of PWB, another risk marker of brain involvement.
Results
qEEG helped predict SWS risk in the first year of life (p=0.031), with a sensitivity of 50% and a specificity of 81%. It added about 40% incremental information beyond PWB extent alone (p=0.042).
Conclusion
qEEG adds information to risk prediction in infants with facial PWB.
Keywords: ischemia, port-wine birthmark, Sturge-Weber syndrome, quantitative EEG, predictive model, prognostic biomarker, diagnostic biomarker
INTRODUCTION
Port-wine birthmarks (PWB)1 are common and occur in approximately 3 of 1,000 live births (Jacobs and Walton, 1976). While most PWBs are associated only with cosmetic consequences, PWBs occurring on the upper face signal an increased risk for Sturge-Weber syndrome (SWS), a vascular malformation syndrome that involves skin, eyes, and brain. SWS occurs in approximately 1 in 20,000 individuals (Dymerska et al., 2017). Leptomeningeal angiomata (capillary-venous malformations) cause chronic ischemia that can lead to intellectual disability/other cognitive alterations, visual field cuts, hemiparesis, stroke-like episodes, headaches and epilepsy. Whereas PWB are present at birth, the potential neurological manifestations of SWS develop over time, with signs typically evident by 1 year of age (Sujansky and Conradi, 1995).
Definitive diagnosis of SWS with intracranial involvement is achieved via gadolinium-enhanced MRI after 1 year of age; it is relatively costly (Zallmann et al., 2018a), has a low sensitivity for leptomeningeal angiomata in infants (<1 year)(Zallmann et al., 2018b), and repeat imaging exposes infants to the risks associated with gadolinium (Kanda et al., 2013) and sedation (Food and Drug Administration, 2016), making it suboptimal as a screening tool. Nevertheless, promising advances in presymptomatic treatment with low-dose aspirin or with anti-seizure drugs (Ville et al., 2002, Day et al., 2019), interventions nevertheless with potential side-effects (Lopez et al., 2013), motivate novel methods for early risk assessment. Within the Kennedy Krieger Sturge-Weber Center (KKI-SWC), assessment typically includes history, physical examination, and clinical EEG interpretation. However, each of these clinical measures is subjective and based on disease-focused clinical experience. Anecdotal evidence from two decades of second-opinion referrals suggests that subtle visual field cuts and hemipareses may be assessed differently by even by pediatric neurologists in the community. Even the development of epilepsy, which clinical experience tells us is a strong indication of SWS in an infant with facial PWB, is imperfect as we have seen alternative causes.
Based on the clinical use of EEG in SWS (Radermecker, 1951, Kossoff et al., 2014), we developed (Hatfield et al., 2007) and initially validated a quantitative-EEG-based prognostic/diagnostic biomarker (qEEG) to predict which infants with facial PWB were at highest risk of developing brain (Ewen et al., 2009). The accuracy was 100%, but the test sample was small. The goal of the current study was to assess performance using the same algorithm within a larger test sample (Bossuyt et al., 2015, Ewen, 2016, Ewen and Beniczky, 2018, Sahin et al., 2018, Ewen et al., 2019). When implemented in the community, the qEEG result could be integrated with risk information based on the extent and pattern of the PWB (Shirley et al., 2013, Dutkiewicz et al., 2015, Comi et al., 2016). We therefore also validated qEEG to a higher and more ecological standard, by assessing its ability to prognosticate beyond information based on extent of PWB. These results both validate the qEEG test independently and begin to develop a risk-calculator based on qEEG+PWB extent.
METHODS
Study Design
This study reflects the prospective validation (Cohen et al., 2016) of a prognostic/diagnostic, EEG-based (Ewen and Beniczky, 2018) biomarker (FDA-NIH Biomarker Working Group, 2017). In the first instance, it is a larger replication of Ewen et al. (Ewen et al., 2009), using the same methodology and diagnostic threshold. Parents gave informed consent and were part of a Johns Hopkins Institutional Review Board-approved larger prospective, longitudinal natural history study of SWS (“Multidisciplinary Protocol to Address the Pathophysiology of Sturge-Weber Syndrome”) which recruited infants seen in the KKI-SWSC with facial PWBs between September 2004 through November 2018. The MRI reference standards were performed through March 2019.
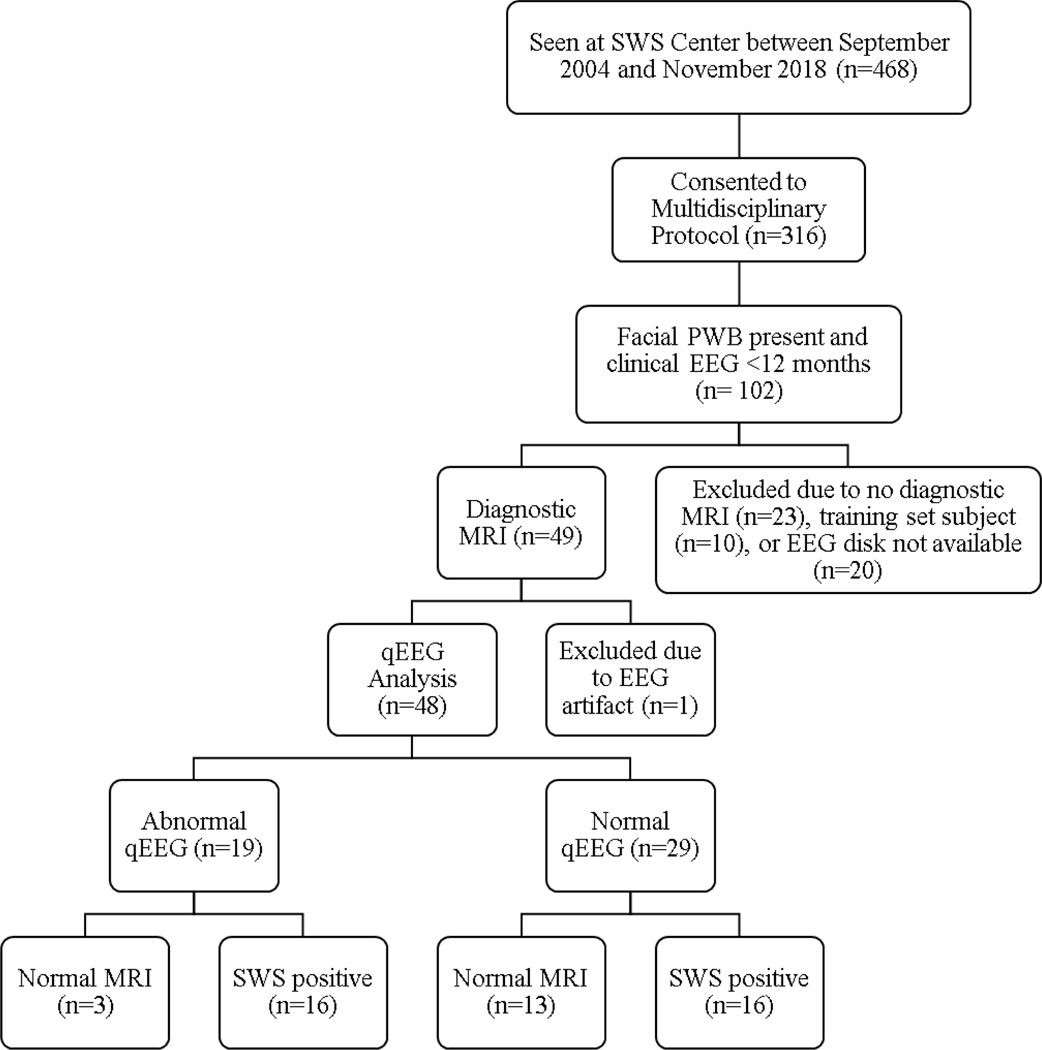
Recruitment was consecutive, and no sample size estimation was conducted prior to analysis. All participants were until first birthday), born with a PWB that involved skin from the frontonasal placode and were therefore considered to be at increased risk for development of SWS (Zallmann et al., 2018a). A total of 316 patients referred to the clinic and enrolled in this study were preemptively determined to have an increased-risk PWB, and further subjected to eligibility screening as outlined in the flow chart (Fig. 1). Infants were excluded if they did not have an EEG performed before their first birthday or a gadolinium-enhanced brain MRI. Patients with negative MRIs were included only if at least one MRI occurred after their first birthday. All nine of the subjects used in the test set of Ewen et al. (Ewen et al., 2009) were included; those in the training set were not included. MRI with contrast served as the reference standard: either negative after the first birthday (SWS-), or positive for leptomeningeal enhancement at any time (SWS+). A final cohort of 48 subjects met criteria and had analyzable qEEG. One was excluded because of persistent EKG artifact in the EEG. No patient had an initial negative MRI after the first birthday that was later followed by a positive MRI. The pediatric neuroradiologist (DML, for studies read at the KKI-SWSC) was not necessarily blinded to participant identity, symptoms or extent of PWB but was unaware of the qEEG result. A pediatric neurologist (AMC) reviewed all available imaging discs (47/48) and a radiology report for the remaining subject to confirm SWS brain involvement or its absence, as well as side of (maximal) involvement. She was blinded to qEEG results at the time of review. There were no indeterminant findings. PWB extent measurement was based upon documented physical examination by AMC and retrospectively placed into one of four descriptive categories that were ranked ordinally: 1=small unilateral, 2=central, 3=bilateral, and 4=complete hemifacial (unilateral or bilateral).
Figure 1.
STARD flowchart for participants (Bossyut et al., 2015). PWB, port-wine birthmark; qEEG, quantitative EEG; STARD, Standards for the Reporting of Diagnostic Accuracy Studies; SWS, Sturge-Weber syndrome.
Each included participant underwent at least one EEG recording at some point between birth and the first birthday (Fig. 2). Although some patients had more than 1 EEG performed in the first year of life, we used the patients’ first available EEG for this investigation. Thirty-four first EEGs were obtained within the institution, while 14 were obtained from outside. All EEGs recorded at Johns Hopkins Hospital or Kennedy Krieger Institute were at least 30- to 40-minute recordings performed in line with ACNS guidelines (Kuratani et al., 2016), using Nihon Kohden (Tokyo, Japan) or Bio-logic Ceegraph (Mundelein, IL) equipment, with 10–20 electrode placement. EEGs performed at outside institutions were assumed to be collected in accordance with standard clinical practices; there was no evidence they were not. EEG systems from outside institutions included Nicolet, Easy Windows, XLTEK, Cadwell, and Stellate. The equipment manufacturer was not identifiable on 6 outside EEGs. This variability in equipment, personnel and recording environment is desirable in a biomarker validation study (Ewen et al., 2019), as it ecologically recapitulates the heterogeneous circumstances under which the test will be used in clinical practice.
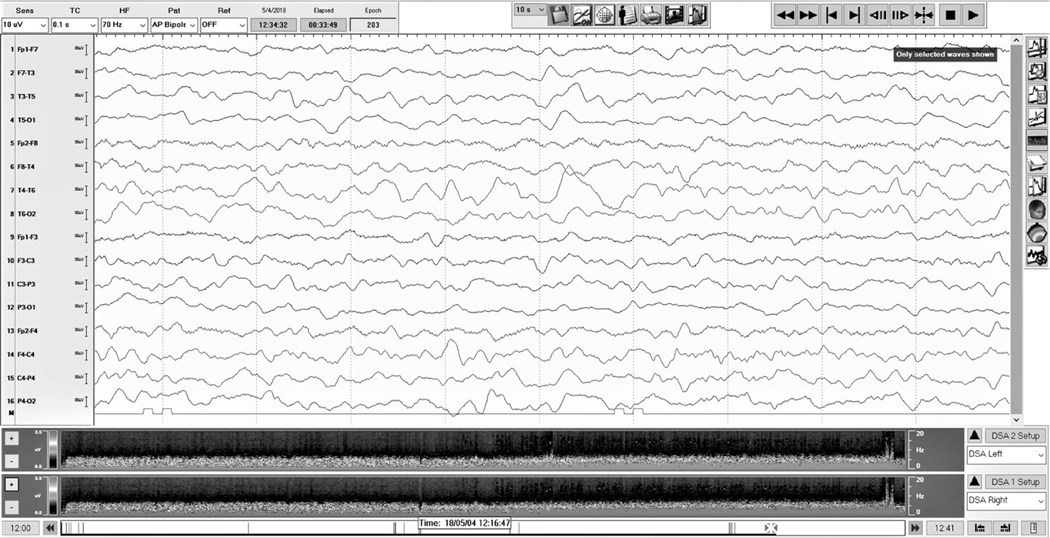
Figure 2.
Representative EEG of an infant with a later positive contrast-enhanced MRI. The quantitative EEG result was also positive. Decreased amplitude can be seen in the left-sided derivations of the tracing. Spectrograms for left and right scalp hemispheres are located below the tracing; although subtle, decreased amplitude can be seen in the spectrogram representing the left compared with the right.
qEEG Test Methods
qEEG analysis served as the index test. There were no adverse effects from the performance of this minimally invasive test. The methodology and diagnostic threshold did not change from Ewen et al. (Ewen et al., 2009). A single clinical neurophysiologist (JBE) selected the first 30 artifact-free, non-overlapping two-second epochs, prioritizing any sleep stage when available, using Persyst InsightII (Persyst Development Corporation, Prescott, AZ). If asychronous were seen, as is typical in this age range, an effort was made to balance epochs with spindles on the right and on the left. Transients (e.g., vertex waves, epileptiform discharges), when present, were avoided. Selection of epochs and exclusion of channels were the only operator-dependent steps. Channels were excluded when persistent artifact in a single channel or subset of channels prohibited identifying 30 artifact-free epochs, in the judgment of the rater. The neurophysiologist was blinded to subject identity, side and extent of PWB, clinical symptoms, and diagnostic group.
EEG data were down-sampled to a 200 Hz sampling rate, if needed. Prism:Spectrum (version 11; Persyst Development Corporation, Prescott, AZ) applied fast Fourier transformation to all of the channels of a bipolar longitudinal montage, in each epoch. There was no smoothing or filtering. We summed these power values into the classic EEG bands of delta (2.0–3.5 Hz), theta (4.0–7.5 Hz), alpha (8.0–12.5 Hz), beta (13.0–32.0 Hz), and total (2.0–32.0 Hz). For each band b, we generated a mean laterality score independently (LSb)(Hatfield et al., 2007) and calculated LSb for all bipolar longitudinal derivations (“Whole Hemisphere”) and, separately, only for those derivations that included electrodes O1 and O2 (“Occipital”; i.e., T5-O1/T6-O2 and P3-O1/P4-O2).
N = number of channel-pairs analyzed (i.e., 16 minus excluded channels for Whole Hemisphere analysis, and 2 minus excluded channels for Occipital), e = epoch identity, c = channel identity, b = EEG frequency band
A qEEG was deemed abnormal if any two bands in the “Whole Hemisphere” analysis or any two bands in the “Occipital” analysis had LSb ≥0.2 on the side ipsilateral to the PWB; this threshold was defined in Ewen et al. (Ewen et al., 2009). Internal controls confirm that the side of greatest brain involvement is almost always the side of greatest facial extent of PWB (see Supplementary Material-I). If a child had a bilateral PWB with a clear prominence, that side was used. If the child had a midline PWB or bilateral PWB without any clinically distinguishable laterality, an abnormal qEEG result on either side of the brain () was deemed to be clinically significant. The calculation of the final results was performed using a script in MATLAB (Mathworks, Natick, MA) by a research assistant (JHA).
History and Physical Exam
Each participant underwent a clinical evaluation by a pediatric neurologist with 20 years’ experience in the care of individuals with SWS (AMC). She prospectively assigned a Sturge-Weber Syndrome-Neurological Rating Score (SWS-NRS), which includes visual field cuts, seizure presence/frequency, hemiparesis, and cognitive atypicalities (Kelley et al., 2005); this score has been previously validated against SWS brain atrophy, perfusion MRI deficits, this current qEEG analysis and neuropsychological testing (Kelley et al., 2005, Lin et al., 2006, Hatfield et al., 2007, Kavanaugh et al., 2016). The total score ranges from 0 to 15, with higher scores representing greater degrees of neurological impairment (Kelley et al., 2005). This score was used to characterize the test sample and also to study probabilistic medical decision making within the specialty center (see Supplementary Material-II).
Statistical Analysis
Statistical analyses were performed only after the final results were locked. Our primary statistical inferences, to confirm the utility of qEEG in a community setting, were the comparison of the performance of qEEG to predict risk of developing SWS compared with a null distribution (H0:sensitivity/specificity of qEEG both=0.5), using a likelihood-ratio test (lrtest from the lmtest package v0.9–37 within R v3.6.1) to compare intercept only vs. qEEG (binarized). As a higher standard, we also determined whether qEEG provides incremental information beyond what would already be known by the stratified risk estimates provided by PWB extent, by testing PWB extent (ordinal variable) as the sole predictor vs. PWB extent plus binarized qEEG results. To quantify the amount of incremental information contained in the qEEG data, we examined the likelihood ratios (LR) of qEEG, collapsed across all levels of PWB extent. A LR is the ratio of the post-test probability over the pre-test probability. The positive LR (LR+) is the probability of developing SWS if the qEEG result is positive divided by the pre-test probability of developing SWS; negative LR (LR-) is the ratio of the post-test probability (given negative qEEG result) over the pre-test probability. The amount of information added by the test can be estimated by the magnitude of the LR (McGee, 2002).
Because this validation occurred in a test sample independent from the training sample in which a threshold was initially set (Ewen et al., 2009), the sensitivity and specificity estimates should be accurate for the application of the test in the population with similar inclusion/exclusion criteria and sampling methods as in our sample. However, the predictive model that includes was not previously thresholded in a different sample; we estimated model predictive ability (accuracy, sensitivity, specificity) using leave-one-out cross-validation (LOOCV). The qEEG term was binary, and the PWB term was binarized into lower (classes 1 and 2) and higher risk (classes 3 and 4) groups. The sensitivity/specificity of the PWB+qEEG risk-calculator derived from LOOCV are therefore rough estimates, as they are based on a diagnostic threshold that is recalculated for each run of the algorithm.
RESULTS
Participants
Of the 48 participants, six EEG showed interictal epileptiform discharges (3 per group) (Kossoff et al., 2014), of which one also showed a seizure. No record was excluded because of epileptiform findings (i.e., 30 epochs could be found which did not include epileptiform discharges). One EEG was excluded due to the exclusion of both occipital channel pairs due to artifact; this patient had a second EEG prior to the first birthday which was used instead. Twelve EEGs had channel pairs excluded. Three EEGs had 1 channel-pair excluded, 8 had 2 channel-pairs, and 1 had 4 channel-pairs excluded.
In 6 of these patients, there was no clear laterality to a bilateral PWB, and the PWB classification was recorded as “bilateral—symmetrical.” There was a significant difference in the distribution of PWB extent (Fisher’s exact p=0.0001). (mean±SD age at EEG for SWS-=2.9±2.4mos.; SWS+=4.3±2.9mos.; t-test p= 0.085) (Tables 1,2). (Tallman et al., 1991, Piram et al., 2012, Dutkiewicz et al., 2015)
Table 1.
Clinical characteristics of patients in study. PWB, port-wine birthmark; SWS, Sturge-Weber syndrome; SWS-NRS, SWS Neurological Rating Scale; SWS+, outcome group with Sturge-Weber syndrome diagnosed; SWS-, outcome group without SWS neurological involvement
| SWS+ (% of SWS+ group) | SWS- (% of SWS-group) | p-value | |
|---|---|---|---|
| Total | 32 | 16 | |
| Sex | 1 | ||
| Male | 14 (44%) | 7 (44%) | |
| Female | 18 (56%) | 9 (56%) | |
| PWB | <0.001 | ||
| Unilateral | 19 (59%) | 13 (81%) | |
| Bilateral | 12 (38%) | 2 (13%) | |
| Central | 1 (3%) | 1 (6%) | |
| Age at EEG | 0.085 | ||
| 0–5.9 mos | 24 (75%) | 14 (88%) | |
| 6–8.9 mos | 4 (12.5%) | 1 (6%) | |
| 9–11.9 mos | 4 (12.5%) | 1 (6%) | |
| SWS Brain Involvement | |||
| Unilateral | 24 (75%) | 0 | |
| Bilateral | 16(25%) | 0 | |
| None | 0 | 16 (100%) | |
| Mean SWS-NRS * | 2.61±3.48 (range: 0–12) | 0±0 | <0.001 |
Missing data from 6 participants: 2 in the SWS-group and 4 in the SWS+ group.
Table 2.
Extent of port-wine birthmark (PWB) in our patients with attributed Sturge-Weber syndrome (SWS) risk and in-sample SWS prevalence.
| PWB Extent | PWB Pattern | Number of participants | In-Sample SWS Prevalence | SWS Risk % from Literature | Reference |
|---|---|---|---|---|---|
| 1 | Unilateral (less than complete hemifacial) | 26 | 15 (58%) | 19–10 | (Tallman et al., 1991) |
| 2 | Central | 2 | 1 (50%) | 27 | (Dutkiewicz et al., 2015) |
| 3 | Bilateral (excluding Hemifacial) | 7 | 6 (86%) | 32–40 | (Tallman et al., 1991, Piram et al., 2012) |
| 4 | Hemifacial | 13 | 10 (78%) | 47 | (Dutkiewicz et al., 2015) |
qEEG Test Performance
This qEEG metric predicts the development of SWS brain involvement in infants with increased-risk PWB (LR p=0.031, intercept-only model vs. qEEG model). Sensitivity was 50%, and specificity was 81%. PPV/NPV are reported in Table 3, under multiple sets of assumptions. Subdividing the sample into younger infants (0–5.9 mos) and older infants (6–11.9 mos), younger infants showed a numerically smaller sensitivity and specificity (39% and 77%, respectively). Older infants showed a greater sensitivity and specificity (75% and 100%).
Table 3.
We calculated positive predictive value (PPV) and negative predictive value (NPV) under multiple assumptions in order to estimate performance in the community. We transformed prevalence of Sturge-Weber syndrome (SWS) to 28% based on prior work (Dutkiewicz et al., 2015) (second row). We then penalized the sensitivity under the assumption that individuals with SWS in the community would, on average, have a lower severity of brain involvement than in our sample. We conducted a sub-analysis in our sample of sensitivity in infants with SWS Neurological Rating Scale (SWS-NRS)≤4 (Kavanaugh et al., 2016) and estimated PPV/NPV based on that sensitivity and on a population prevalence of 28% (third row). SWS+, outcome group with Sturge-Weber syndrome diagnosed; SWS-, outcome group without SWS neurological involvement
| Assumpti ons | Accuracy | Sensitivity | Specificity | Derived PPV | Derived NPV | |
|---|---|---|---|---|---|---|
| Measured Values (in-sample prevalence = 67%) | N/A | 60% | 50% | 81% | 84% | 45% |
| Population Prevalence | Population prevalence = 28% (extrapolated from Zallmann et al., 2018b); in-sample sensitivity and specificity | 60% | 50% (in-sample measurement) | 81% (in-sample measurement) | 52% | 80% |
| Population Prevalence and Decreased Average Severity | Population prevalence = 28%; decreased clinical severity of SWS+ group in population | 50% | 37% (in-sample measurement for SWS-NRS ≤4 subsample) | 81% (in-sample measurement) | 42% | 76% |
The LR test contrasting the PWB vs. PWB+qEEG models (p=0.042) confirms that qEEG adds risk information beyond that information based on PWB extent alone. Using LOOCV, the estimated sensitivity and specificity are 72% and 56%, respectively. Assuming a population with similar prevalence and severity to our sample, the PPV and NPV are 77% and 50%, respectively. Correlation coefficients with SWS status for PWB were r=0.24; for qEEG, r=0.30.
The overall LR for the qEEG+PWB model over PWB alone was 7.97, or incremental information content of about 40% (McGee, 2002). LR+ for qEEG (collapsing across PWB strata) was 2.67; LR- was 0.62. This means that a positive qEEG imparts a 2.67× greater probability of being diagnosed with SWS than a pre-test probability based on PWB extent alone, and the post-test probability of a negative test is 0.64×the pre-test probability, equivalent to a 56% smaller probability of having SWS compared to the pre-test probability. Under a practice rubric early (pre-first-birthday) MRI referral was defined solely by qEEG results and excluded PWB extent, 2.5 infants in our sample would need to undergo qEEG testing to result in 1 referral for early MRI (or 2.7 infants, after transforming to literature-derived SWS prevalence figures). Similarly, 3 infants (or 3.3 in the broader population) would have to undergo qEEG testing to result in one who had a positive MRI and would be a candidate for prophylactic interventions (further assuming early MRI results are identical to post-first-birthday MRI results).
DISCUSSION
The primary finding was that qEEG helps predict the manifestation of SWS brain involvement in infants with facial PWB with a sensitivity of 50% and a specificity of 81%, within our consented referral sample. In the context of population estimates of the prevalence of SWS in infants with high-risk facial PWB (28%; extrapolated from Zallmann et al., 2018b), this would translate to a PPV of 52% and NPV of 81%. If we penalize the sensitivity based on an overly conservative assumption about the reduced average degree of brain involvement in the community vs. in our sample, the PPV and NPV would be as low as 42% and 76%, respectively. The actual PPV/NPV values in the community are likely to be between these values.
At NPV of 81% or even 76%, a negative test would lend reassurance and may provide enough information to allow families and clinicians to defer a diagnostic MRI until after 1 year of age. A PPV even of 42%, contrasted with a baseline risk of 28%, would be a signal that would push for early MRI to enable diagnosis and consideration of aspirin and anti-seizure presymptomatic treatment (Day et al., 2019). Even at these levels of PPV and NPV, we can already see that the qEEG results interact with risk estimates based on PWB extent alone: from the literature, we know that a hemifacial PWB already confers a 47% risk of SWS brain involvement (Dutkiewicz et al., 2015); this risk is greater than the 42% PPV of the qEEG (averaged over PWB strata). Nevertheless, we also know from our likelihood-ratio tests that the qEEG test adds information beyond prediction based on PWB extent alone. Taken together, these findings suggest the need for a risk-calculator predictive model that synthesizes both qEEG and PWB extent. The current data can serve as a training set on which a risk-score threshold is derived, and a future dataset would serve as a test set to derive solid PPV/NPV estimates. Such a dataset should be collected from the broad international community we hope to serve, using a sampling method that is similar to how the test would actually be applied—acquired from general pediatricians, ophthalmologists, dermatologists, and others who may come into contact with infants with PWB before child neurologists.
The presence of sharp waves in approximately 10% of infants without brain involvement was unanticipated. In reviewing all clinical information on these cases, including clinical follow-up as of the time of manuscript publication, there is no evidence of neurological involvement. In one case, two interpretations were provided by fellowship-trained, board-certified pediatric electroencephalographers: one for patient care purposes and one (as recorded here) for research purposes. The two electroencephalographers (unaware of each other’s interpretation) came to different conclusions about the presences of interictal epileptiform discharges, illustrating uncertainty in standard clinical interpretation and the benefit of relatively unsupervised techniques. In the SWS+ group, interictal discharges were only present in records that had a positive qEEG result.
The numerical decrease in both sensitivity and specificity in younger infants suggests both the potential and a need to improve signal-to-noise particularly in this sub-group; several signal-processing methodologies may help. Automated artifact correction/removal algorithms (Gabard-Durnam et al., 2018) and machine learning techniques, such as those more commonly used in MRI than EEG (Xiao et al., 2019) could reduce uncertainty due to human involvement. Frequency-based analysis could use an empiric definition of bands, as the traditional rhythmic structure developed in adults likely does not fully apply to infants, and particularly to younger infants. Other methods, such as ischemia-sensitive entropy (Bezerianos et al., 2003), occipital connectivity measures, or visual-evoked potentials (MacLean et al., 1975) could be examined. Because our clinical interpretations often relied on asymmetries in the posterior basic rhythm or spindles, and spindle detectors could be useful (Lacourse et al., 2019). A combination of measures could be developed (and subsequently validated) with sensitivity-specificity trade-offs made to optimize either PPV or NPV. Eventual clinical implementation of this technique could leverage (with appropriate validation) time-frequency-voltage analysis tools and asymmetry calculators that are increasingly available within commercial EEG products, or secure electronic transmission of EEG files to a central site for quality assurance and data analysis.
The clear next step is to validate the PWB+qEEG risk calculator using a sample from the broad community. As this step is important “implementation science” questions will arise and should be addressed empirically: does the accuracy of the test depend on patient factors (e.g., age, time of day) or on technical factors (e.g., equipment manufacturer, amplified frequency-response curves, and hardware filtering)? To the extent possible without compromising accuracy, more liberal requirements will allow more widespread implementation in the field.
Prioritizing PWB extent and qEEG assumes that there is low inter-rater reliability in the community in the history/physical exam (SWS-NRS) and in clinical EEG interpretation. Our anecdotal experience bears this out, but given the strong predictive contribution of SWS-NRS and clinical EEG interpretation (Supplementary Material-II), explicitly studying the reliability of these factors in the community may be warranted.
We should also note that qEEG could be used to serve different roles, within SWS care, apart from risk estimation. We have already employed this same qEEG approach in a different context, using LSb as a continuous rather than binarized variable. In a recent, small clinical trial, we found pre-treatment qEEG differences in a group of children with SWS who suffered from stroke-like episodes (SLE) vs. those who did not. With sirolimus treatment, which is thought to change vascular function, the qEEG from the SLE group became more like the non-SLE group (Sebold et al., 2020). Similarly, qEEG analyses might be studied as monitoring biomarkers that could predict critical events, such as stroke-like episodes or an increase in seizures.
There were several limitations to investigation. SWS is a rare disease and our cohort was still comprised of a relatively small group of patients. The sampling was from a quaternary referral center rather than a sample based on the pattern of intended use. Further, the one human-supervised step—selection of epochs and exclusion of channels with persistent artifact—was performed by a pediatric electroencephalographer experienced in infant SWS clinical interpretation. Although he attempted to restrict selection of epochs based solely on the rules presented in the Methods section, it is possible that he was influenced by the gestalt of the clinical EEG. It will be beneficial in the future to study inter-rater reliability for this step.
Supplementary Material
Highlights
Quantitative EEG helps predict which infants with facial port-wine birthmark are most likely to develop Sturge-Weber syndrome.
Risk prediction is critical for emerging presymptomatic treatment of infants with port-wine birthmark.
Long-term collection of qEEG, clinical EEG, history and neurological exam data allow initial development of a predictive model.
Significance
qEEG can be used to help determine whether to obtain an MRI in the first year of life. The data collected can assist in developing a predictive model risk calculator that incorporates both PWB extent and qEEG results, which can be validated and then employed in the community.
ACKNOWLEDGEMENTS
We greatly appreciate the participation of our patients and their families.
FUNDING SOURCE
Research reported in this publication was supported by the Rare Diseases Clinical Research Network (RDCRN) of the National Institutes of Health under grant number U54NS065705-08 (Lawton, Comi, and Marchuk). The Brain Vascular Malformation Consortium [grant number: U54NS065705], which is a part of the NIH Rare Diseases Clinical Research Network (RDCRN), supported through the collaboration between the NIH Office of Rare Diseases Research (ORDR) at the National Center for Advancing Translational Science (NCATS) and the National Institute of Neurological Disorders and Stroke (NINDS). Support for Dr. Ewen’s effort came from came from NIH grant numbers K12 NS001696 and K23 NS073626 and the Kennedy Krieger Institute Intellectual Developmental Disabilities Research Center (IDDRC) [grant number: P50HD103538]. Additionally, support from the Faneca 66 Foundation (Comi) and Celebrate Hope Foundation (Comi) is acknowledged.
ROLE OF FUNDER/SPONSOR
No funding source had a role in the design of the study, collection or analysis of data, or in the writing of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Abbreviations: AUC, area under the curve; EEG, electroencephalogram; LR, likelihood ratio; KKI-SWSC, Kennedy Krieger Institute Sturge-Weber Syndrome Center; LOOCV, leave-one-out cross-validation; NPV, negative predictive value; PPV, positive predictive value; PWB, port-wine birthmark; qEEG, quantitative electroencephalogram; STARD, Standards for the Reporting of Diagnostic Accuracy Studies; SWS, Sturge-Weber syndrome; SWS-NRS, SWS-neurological rating scale.
CLINICAL TRIAL REGISTRY
None.
All authors reviewed and approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
CONFLICT OF INTEREST DISCLOSURES
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bezerianos A, Tong S, Thakor N. Time-dependent entropy estimation of EEG rhythm changes following brain ischemia. Ann Biomed Eng. 2003;31:221–32. [DOI] [PubMed] [Google Scholar]
- Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. BMJ Open. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi AM, Sahin M, Hammill A, Kaplan EH, Juhasz C, North P, et al. Leveraging a Sturge-Weber Gene Discovery: An Agenda for Future Research. Pediatr Neurol. 2016;58:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AM, Hammill AM, Juhasz C, Pinto AL, Roach ES, McCulloch CE, et al. Hypothesis: Presymptomatic treatment of Sturge-Weber Syndrome With Aspirin and Antiepileptic Drugs May Delay Seizure Onset. Pediatr Neurol. 2019;90:8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz A-S, Ezzedine K, Mazereeuw-Hautier J, Lacour J-P, Barbarot S, Vabres P, et al. A prospective study of risk for Sturge-Weber syndrome in children with upper facial port-wine stain. J Am Acad Dermatol. 2015;72:473–80. [DOI] [PubMed] [Google Scholar]
- Dymerska M, Kirkorian AY, Offermann EA, Lin DD, Comi AM, Cohen BA. Size of Facial Port-Wine Birthmark May Predict Neurologic Outcome in Sturge-Weber Syndrome. J Pediatr. 2017;188:205–9 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen JB. The eternal promise of EEG-based biomarkers: Getting closer? Neurology. 2016;87:2288–9. [DOI] [PubMed] [Google Scholar]
- Ewen JB, Beniczky S. Validating biomarkers and diagnostic tests in clinical neurophysiology: Developing strong experimental designs and recognizing confounds. In: Schomer DL, Lopes da Silva FH, editors. Niedermeyer’s Electroencephalography. 7th ed. New York: Oxford University Press; 2018. [Google Scholar]
- Ewen JB, Kossoff EH, Crone NE, Lin DD, Lakshmanan BM, Ferenc LM, et al. Use of quantitative EEG in infants with port-wine birthmark to assess for Sturge-Weber brain involvement. Clin Neurophysiol. 2009;120:1433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen JB, Sweeney JA, Potter WZ. Conceptual, Regulatory and Strategic Imperatives in the Early Days of EEG-Based Biomarker Validation for Neurodevelopmental Disabilities. Front Integr Neurosci. 2019;13:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration. Drug Safety and Availability - FDA Drug Safety Communication: FDA review results in new warnings about using general anesthetics and sedation drugs in young children and pregnant women. 2016. [Google Scholar]
- Gabard-Durnam LJ, Mendez Leal AS, Wilkinson CL, Levin AR. The Harvard Automated Processing Pipeline for Electroencephalography (HAPPE): Standardized Processing Software for Developmental and High-Artifact Data. Front Neurosci. 2018;12:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools). September25. 2017. [Google Scholar]
- Hatfield LA, Crone NE, Kossoff EH, Ewen JB, Pyzik PL, Lin DD, et al. Quantitative EEG asymmetry correlates with clinical severity in unilateral Sturge-Weber syndrome. Epilepsia. 2007;48:191–5. [DOI] [PubMed] [Google Scholar]
- Jacobs AH, Walton RG. The incidence of birthmarks in the neonate. Pediatrics. 1976;58:218–22. [PubMed] [Google Scholar]
- Kanda T, Ishii K, Kawaguchi H, Kitajima K, Takenaka D. High signal intensity in the dentate nucleus and globus pallidus on unenhanced T1-weighted MR images: relationship with increasing cumulative dose of a gadolinium-based contrast material. Radiology. 2013;270:834–41. [DOI] [PubMed] [Google Scholar]
- Kavanaugh B, Sreenivasan A, Bachur C, Papazoglou A, Comi A, Zabel TA. Intellectual and adaptive functioning in Sturge-Weber Syndrome. Child Neuropsychol. 2016;22:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley TM, Hatfield LA, Lin DD, Comi AM. Quantitative analysis of cerebral cortical atrophy and correlation with clinical severity in unilateral Sturge-Weber syndrome. J Child Neurol. 2005;20:867–70. [DOI] [PubMed] [Google Scholar]
- Kossoff EH, Bachur CD, Quain AM, Ewen JB, Comi AM. EEG evolution in Sturge-Weber syndrome. Epilepsy Res. 2014;108:816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani J, Pearl PL, Sullivan L, Riel-Romero RM, Cheek J, Stecker M, et al. American Clinical Neurophysiology Society Guideline 5: Minimum Technical Standards for Pediatric Electroencephalography. J Clin Neurophysiol. 2016;33:320–3. [DOI] [PubMed] [Google Scholar]
- Lacourse K, Delfrate J, Beaudry J, Peppard P, Warby SC. A sleep spindle detection algorithm that emulates human expert spindle scoring. J Neurosci Methods. 2019;316:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DD, Barker PB, Hatfield LA, Comi AM. Dynamic MR perfusion and proton MR spectroscopic imaging in Sturge-Weber syndrome: correlation with neurological symptoms. J Magn Reson Imaging. 2006;24:274–81. [DOI] [PubMed] [Google Scholar]
- Lopez J, Yeom KW, Comi A, Van Haren K. Case report of subdural hematoma in a patient with Sturge-Weber syndrome and literature review: questions and implications for therapy. J Child Neurol. 2013;28:672–5. [DOI] [PubMed] [Google Scholar]
- MacLean C, Appenzeller O, Cordaro JT, Rhodes J. Flash evoked potentials in migraine. Headache. 1975;14:193–8. [DOI] [PubMed] [Google Scholar]
- McGee S. Simplifying likelihood J Gen Intern Med. 2002;17:646–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piram M, Lorette G, Sirinelli D,D, Giraudeau B, Maruani A. Sturge-Weber syndrome in patients with facial port-wine stain. Pediatr Dermatol. 2012;29:32–7. [DOI] [PubMed] [Google Scholar]
- Radermecker J. L’électroencéphalographie dans l’angiomatose encéphalo-trigéminée de Sturge-Weber-Krabbe. Acta Neurol Psychiatr Belg. 1951;51:427–51. [PubMed] [Google Scholar]
- Sahin M, Jones SR, Sweeney JA, Berry-Kravis E, Connors BW, Ewen JB, et al. Discovering translational biomarkers in neurodevelopmental disorders. Nat Rev Drug Discov. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebold AJ, Day AM, Ewen J, Adamek J, Byars A, Cohen B, et al. Sirolimus Treatment in Sturge-Weber Syndrome. Pediatr Neurol. 2020;115:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley MD, Tang H, Gallione CJ, Baugher J, Frelin LP, Cohen B, et al. Sturge-Weber Syndrome and Port-Wine Stains Caused by Somatic Mutation in GNAQ. N Engl J Med. 2013;368:1971–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujansky E, Conradi S. Sturge-Weber syndrome: age of onset of seizures and glaucoma and the prognosis for affected children. J Child Neurol. 1995;10:49–58. [DOI] [PubMed] [Google Scholar]
- Tallman B, Tan OT, Morelli JG, Piepenbrink J, Stafford TJ, Trainor S, et al. Location of port-wine stains and the likelihood of ophthalmic and/or central nervous system complications. Pediatrics. 1991;87:323–7. [PubMed] [Google Scholar]
- Ville D, Enjolras O, Chiron C, Dulac O. Prophylactic antiepileptic treatment in Sturge-Weber disease. Seizure. 2002;11:145–50. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Yan Z, Zhao Y, Tao B, Sun H, Li F, et al. Support vector machine-based classification of first episode drug-naive schizophrenia patients and healthy controls using structural MRI. Schizophr Res. 2019;214:11–7. [DOI] [PubMed] [Google Scholar]
- Zallmann M, Leventer RJ, Mackay MT, Ditchfield M, Bekhor PS, Su JC. Screening for Sturge-Weber syndrome: A state-of-the-art review. Pediatr Dermatol. 2018a;35:30–42. [DOI] [PubMed] [Google Scholar]
- Zallmann M, Mackay MT, Leventer RJ, Ditchfield M, Bekhor PS, Su JC. Retrospective review of screening for Sturge-Weber syndrome with brain magnetic resonance imaging and electroencephalography in infants with high-risk port-wine stains. Pediatr Dermatol. 2018b;35:575–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.