Abstract
Targeted sequencing for somatic mutations across the hotspots of 50 cancer-related genes was performed using biopsy specimens to investigate whether clinicopathological factors and genomic alterations correlated with prognosis in locally advanced cervical cancer. Seventy patients diagnosed with International Federation of Obstetrics and Gynecology (FIGO) stage III to IVA cervical cancer underwent radiotherapy or concurrent chemoradiotherapy at the National Cancer Center Hospital between January 2008 and December 2017. Mutations were detected in 47 of 70 [67% of cases; frequency of genetic alterations was as follows: PIK3CA (51%), FBXW7 (10%), PTEN (7.1%), and TP53 (5.7%)]. The Cancer Genome Atlas (TCGA) datasets showed a similar distribution of somatic mutations, but PIK3CA mutation frequency was significantly higher in our cohort than in TCGA datasets (P = 0.028). Patients with TP53 mutation were significantly related to poor progression-free survival (PFS) (hazard ratio [HR] = 3.53, P = 0.042). Patients with tumor diameters > 70 mm were associated with poor prognosis (HR = 2.96, P = 0.0048). Patients with non-HPV16/18 genotypes had worse prognosis than those with HPV16/18 genotypes (HR = 2.15, P = 0.030). Hence, patients with locally advanced cervical cancer, TP53 mutation, large tumor diameter, and non-HPV16/18 genotype were independently correlated with poor PFS, despite concurrent chemoradiotherapy.
Subject terms: Cervical cancer, Cancer genomics
Introduction
Cervical cancer is the fourth most commonly diagnosed cancer, and the fourth leading cause of cancer-related deaths in women worldwide1. Cervical cancer is often caused by sexually transmitted infections with most human papillomavirus (HPV) types, especially high-risk HPV 16 and 182. Although screening for cervical cancer has improved over the past decade, more than 20% of cervical cancer patients were identified as International Federation of Obstetrics and Gynecology (FIGO) stage III–IV at initial diagnosis3. Most patients with locally advanced cervical cancer are treated with concurrent chemoradiotherapy (CCRT)4. Recently, the response rate for CCRT has increased due to the progress of irradiation technology, and complete response has been achieved in approximately 75% of patients5. However, some patients have a poor response to CCRT6; therefore, they relapse early after treatment and have a poor prognosis7. Clinical trials for providing additional treatment after CCRT to verify the increment of treatment effect, aiming to improve the prognosis in these patients, are currently underway8. Additionally, therapeutic drugs have been approved by the US Food and Drug Administration (FDA) for advanced cancer that use an immune checkpoint inhibitor, such as pembrolizumab9,10, and an angiogenesis inhibitor, such as bevacizumab11,12. However, the potential of benefiting from such molecular-targeting drugs is not well evaluated for patients with locally advanced cervical cancer.
In addition to tumor volume, performance status, treatment received, and prognostic factors for locally advanced cervical cancers, such as age, race, stage, histological type, grade, lymph node enlargement, and location, are associated with poor outcomes4. Several studies have focused on the prognostic factors for patients receiving CCRT in locally advanced cervical cancer, and it has been reported that larger tumor size and high-risk HPV were correlated with prognosis13,14. However, most studies have targeted operable cases. It is necessary to conduct a study on inoperable cases of locally advanced cervical cancer of FIGO stages III to IVA.
Comprehensive profiles of genomic alterations in cervical cancer have been published by The Cancer Genome Atlas (TCGA)15. PIK3CA mutations are one of the most frequently detected mutations in cervical cancer regardless of ethnicity15–17. Several studies have reported that cervical cancer patients with PIK3CA mutations are associated with worse prognosis than those without the mutation17–19. However, the results of some reports differ from these findings16,20, necessitating further discussion. In addition, most studies have focused on the association of single or multiple genetic mutations with prognosis using surgical specimens9,17,19–21; there are only a few reports on targeted sequencing using biopsy specimens from inoperable advanced cervical cancer22. Further studies are needed to elucidate the distribution of genomic alterations in locally advanced cervical cancer in order to identify novel therapeutic targets and biomarkers associated with prognosis. If poor prognostic factors or actionable mutation frequency are well understood, patients may opt for additional treatment after CCRT. Many previous studies have investigated the association of somatic mutations or clinicopathological factors, including high-risk HPV genotypes, with prognosis or response to adjuvant CCRT in early-stage cervical cancer patients receiving surgical operation. Only a few studies have suggested that these clinicopathological factors are associated with response to chemoradiotherapy in patients with locally advanced cervical cancer receiving CCRT.
In this study, we evaluated the pattern of genomic or actionable mutations in patients with FIGO stage III to IVA cervical cancer who received CCRT. We examined whether clinicopathological factors, including HPV genotypes and genomic alterations, were associated with prognosis in patients receiving CCRT.
Results
Patient characteristics
The selection flowchart for the 70 patients used in this study is shown in Figure S1, and the patient characteristics are summarized in Table 1. Of the 70 patients, 65 (93%) were diagnosed with squamous cell carcinoma (SCC), and 68 cervical tissue samples (97%) were HPV-positive. Thirty-three patients (47%) had lymph node enlargement, and were suspected to have pelvic and/or para-aortic lymph node metastases. Fifty-five (79%) patients underwent CCRT, and the standard concurrent chemotherapy regimen was cisplatin-based. Patients over seventy-five years of age or those who had difficulty receiving chemotherapy according to the physician’s choice only underwent RT. The median tumor size was 52.5 mm (range, 30–100 mm). The tumor diameter was over 70 mm in 16 (23%) patients. The median follow-up period was 54 months (range, 6–135 months). Two-year overall survival (OS), progression-free survival (PFS), and locoregional relapse-free survival (LRFS) were 77.1% (95% confidence interval [CI], 65.4–85.3), 60.0% (95% CI, 47.6–70.4), and 71.2% (95% CI, 58.9–80.3), respectively.
Table 1.
Characteristics of locally advanced cervical cancer patients.
| Variable | n | (%) |
|---|---|---|
| Total patients | 70 | |
| Age, median years [range] | 63.5 [32–89] | |
| FIGO stage (2018) | ||
| IIIA | 6 | (8.6) |
| IIIB | 26 | (37.1) |
| IIIC1r | 18 | (25.7) |
| IIIC2r | 9 | (12.9) |
| IVA | 11 | (15.7) |
| Histology | ||
| Squamous cell carcinoma | 65 | (92.9) |
| Adenocarcinoma | 3 | (4.3) |
| Adenosquamous carcinoma | 1 | (1.4) |
| Neuroendocrine carcinoma | 1 | (1.4) |
| HPV genotype | ||
| Positive | 68 | (97.1) |
| HPV16 | 22 | (31.4) |
| HPV18 | 17 | (24.3) |
| HPV31 | 10 | (14.3) |
| HPV33 | 2 | (2.9) |
| HPV45 | 1 | (1.4) |
| HPV52 | 8 | (11.4) |
| HPV58 | 3 | (4.3) |
| HPV59 | 1 | (1.4) |
| HPV82 | 1 | (1.4) |
| HPV genotype not identified | 3 | (4.3) |
| Negative | 2 | (2.9) |
| Lymph node enlargement | ||
| No | 37 | (52.9) |
| Yes | 33 | (47.1) |
| Treatment | ||
| Radiation therapy | 15 | (21.4) |
| Concurrent chemoradiotherapy | 55 | (78.6) |
| Tumor size (mm) (median [range]) | 52.5 [30–100] | |
| < 70 | 54 | (77.1) |
| ≥ 70 | 16 | (22.9) |
| Median follow-up period, month [range] | 53.5 [6–135] | |
Correlation between clinicopathological factors and prognosis
We examined whether previously reported clinicopathological factors were correlated with prognosis in locally advanced cervical cancer. Univariate and multivariate analyses revealed that a tumor diameter larger than 70 mm was an unfavorable prognostic factor, especially for PFS (HR = 2.96, P = 0.0048) (Table 2, Table S1).
Table 2.
Correlation between clinico-pathological factors and progression free survival in locally advanced cervical cancer patients.
| Variable | Univariate | Multivariate* | ||
|---|---|---|---|---|
| Hazard ratio | P value | Hazard ratio | P value | |
| Age (≥ 60/< 60) | 1.08 (0.54–2.15) | 0.84 | 0.92 (0.43–1.98) | 0.84 |
| FIGO Stage (IV/III) | 0.74 (0.29–1.90) | 0.53 | 0.59 (0.22–1.60) | 0.30 |
| Histology (non-SCC/SCC) | 3.38 (1.29–8.88) | 0.013 | 2.70 (0.97–7.50) | 0.057 |
| Lymph node enlargement (positive/negative) | 1.56 (0.82–2.99) | 0.18 | 1.36 (0.68–2.74) | 0.38 |
| Treatment (RT**/CCRT***) | 1.48 (0.73–2.99) | 0.28 | 1.83 (0.84–3.97) | 0.13 |
| Tumor size (≥ 70 mm/< 70 mm) | 2.49 (1.24–5.00) | 0.01 | 2.96 (1.39–6.29) | 0.0048 |
*Cox proportional hazards regression analysis, **Radiation therapy, ***Concurrent chemoradiotherapy.
Comparison of somatic mutation patterns in locally advanced cervical cancer using the present and TCGA datasets
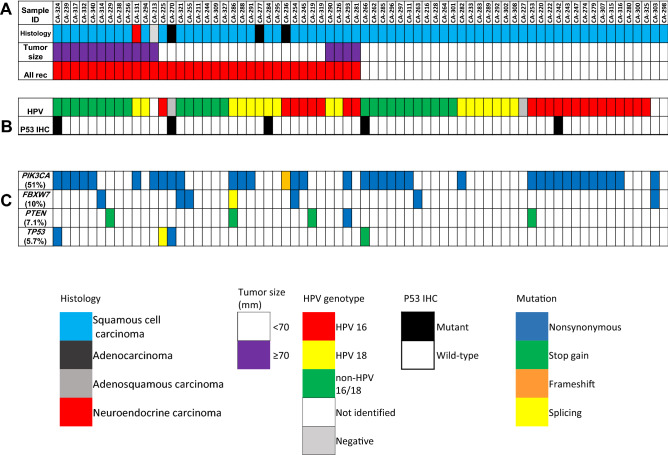
Targeted sequencing for the 70 specimens of locally advanced cervical cancer revealed pathogenic/oncogenic mutations in 47 cases (67%). PIK3CA was the most frequent genomic alteration detected in this study, with a frequency of 51%, followed by FBXW7 (10%), PTEN (7.1%), RB1 (13%) and TP53 (5.7%) (Fig. 1). There was no association between genomic mutation frequency and tumor size or lymph node enlargement. Twelve patients had local recurrence, ten of whom (83%) had pathogenic/oncogenic mutations. Further, the frequency of genomic alterations in patients with stage III to IVA cervical cancer in TCGA dataset was as follows: PIK3CA (30%), FBXW7 (15%), PTEN (15%), and TP53 (11%) (Figure S2). The mutation rates were 65% (42 of 65) in SCC and 100% (5 of 5) in non-SCC cases. The somatic mutations of locally advanced cervical cancer in TCGA datasets are summarized in Figure S2. There was no statistical difference in the distribution of somatic mutations between our cohort and TCGA datasets, except for the frequency of PIK3CA mutation in our cohort, which was higher than that in TCGA datasets (P = 0.028, Table S2).
Figure 1.
Clinicopathological factors and mutation profile (more than 5% frequency) in our cohort. (A) Clinical factors, histological types, and recurrence status; (B) HPV genotype and IHC staining pattern; and (C) mutation profile of the seventy patients with cervical cancer. Mutated genes are color-coded according to their mutation type. Data analysis was carried out using the Torrent Suite Software v5.0.4 (Thermo Fisher Scientific).
Actionable mutation in locally advanced cervical cancer
Actionable mutations registered with evidence levels of 1–3B in OncoKB were detected in 35 of 70 (50%) patients (Figure S3). Most somatic mutations were dominated by PIK3CA mutation, which is similar to TCGA datasets (Figure S3). These results indicated that 30–51% of patients with locally advanced cervical cancer may have benefited from mTOR/AKT/PI3K inhibitors.
Correlation between genomic alteration and prognosis
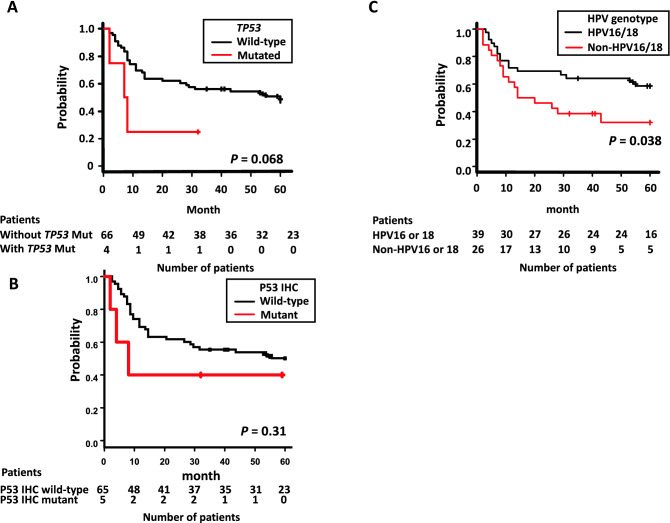
Genomic alterations in PIK3CA, FBXW7, and PTEN were not significantly correlated with PFS (Table S3). Although all patients with TP53 mutation (n = 4) received CCRT, and the tumor diameter was less than 70 mm in three out of four cases, TP53 mutants were independently correlated with poor survival (Fig. 2A). Most patients experienced recurrence within one year after the start of RT. Multivariate Cox proportional regression analysis indicated that patients with TP53 mutations were associated with poor PFS (HR = 3.53, P = 0.042, Table 3A, Table S4).
Figure 2.
Kaplan–Meier survival curves according to TP53 status and HPV genotypes. (A) Progression-free survival between TP53 wild-type (black line) and TP53 mutants (red line), (B) Progression-free survival between TP53 wild-type of IHC (black line) and TP53 mutant of IHC (red line), and (C) Progression-free survival between HPV16/18-positive (black line) and non-HPV16/18 (red line) patients.
Table 3.
Correlation between TP53 mutation or HPV genotypes and progression-free survival in locally advanced cervical cancer patients.
| Variable | Univariate | Multivariate* | ||
|---|---|---|---|---|
| Hazard ratio | P value | Hazard ratio | P value | |
| (A) TP53 mutation (n = 70) | ||||
| Lymph node enlargement (Positive/negative) | 1.56 (0.82–2.99) | 0.18 | – | |
| Treatment (RT**/CCRT***) | 1.48 (0.73–2.99) | 0.28 | – | |
| Tumor size (≥ 70 mm/< 70 mm) | 2.49 (1.24–5.00) | 0.01 | 2.69 (1.33–5.44) | 0.0060 |
| TP53 mutation (n = 4) | 2.85 (0.86–9.44) | 0.061 | 3.53 (1.05–11.92) | 0.042 |
| (B) Non-HPV16/18 (n = 65) | ||||
| Lymph node enlargement (positive/negative) | 1.56 (0.82–2.99) | 0.18 | – | |
| Treatment (RT**/CCRT***) | 1.48 (0.73–2.99) | 0.28 | – | |
| Tumor size (≥ 70 mm/< 70 mm) | 2.40 (1.17–4.96) | 0.018 | 2.37 (1.14–4.93) | 0.021 |
| HPV genotype (non HPV16 or 18/HPV16 or 18) | 2.17 (1.10–4.29) | 0.026 | 2.15 (1.08–4.27) | 0.030 |
*Stepwise multiple regression analysis, **Radiation therapy, ***Concurrent chemoradiotherapy.
Association of p53 status with poor outcomes
We investigated the protein expression of p53 by IHC staining of the specimens of 70 patients. Representative results of IHC staining are shown in Figure S4. Five specimens presented with mutant p53 staining patterns, including three with TP53 mutations. p53 mutant staining patterns tended to be associated with poor survival of patients when compared with wild-type staining patterns (Fig. 2B).
Non-HPV16/18 patients had poorer survival than HPV16/18 patients
Sanger sequencing identified 65 cases of HPV genotypes, and we divided the HPV-positive patients into two groups—HPV16 or 18 (HPV16/18) group and non-HPV16/18 group. We investigated the correlation of the HPV16/18 and non-HPV16/18 groups with prognosis. Univariate and multivariate analyses showed that the non-HPV16/18 group had lower PFS compared with the HPV16/18 group (HR = 2.15, P = 0.030; Fig. 2C, Table 3B, Table S5). The non-HPV16/18 group was older than the HPV16/18 group, and the percentages of people over the age of 60 in both were 81% and 51% (P = 0.020), respectively. Owing to older age, more patients without HPV16/18 only received RT (15% vs. 31%, P = 0.22). Tumor diameter in the non-HPV16/18 group was higher than that in the HPV16/18 group; however, the difference was not significant. The percentage of patients whose tumor diameter was greater than 70 mm was 15% in the HPV16/18 group and 35% in the non-HPV16/18 group (P = 0.13).
Discussion
In this study, we identified the mutation profile and prognostic factors in patients with inoperable locally advanced cervical cancer who received CCRT or RT. Thirty to fifty percent of patients with locally advanced cervical cancer might have benefited from molecular-targeting drugs. Further, TP53 mutation, large tumor size, and non-HPV16/18 genotypes were indicative of poor prognosis.
In line with previous reports5,23, patients with tumor diameters over 70 mm had poor PFS in our cohort. In a phase II study of CCRT with brachytherapy in Japanese patients with locally advanced cervical cancer (JGOG1066)5, tumor diameter was correlated with PFS. In a prospective study, it was reported that a tumor diameter of ≥ 60 mm was an independent poor prognostic factor23. In our cohort, tumors ≥ 60 mm and ≥ 70 mm had an HR of 2.46 (95% CI, 1.28–4.71) and 2.49 (95% CI, 1.24–5.00) in univariate analysis, respectively. This suggests that the therapeutic effects of CCRT in patients with locally advanced cervical cancer are limited when the tumor diameter exceeds 60–70 mm. Therefore, these patients need additional treatments, such as molecular-targeting drugs based on actionable mutations.
In the past decade, next-generation sequencing has been performed for operable cervical cancer in patients receiving adjuvant CCRT or RT, and many genomic alterations have been identified. Although some genomic alterations, such as PIK3CA17,19,21, KRAS24, ERBB225, and STK1116, have been reported to correlate with prognosis, these mutations were not statistically associated with prognosis in our cohort of patients with locally advanced cervical cancer. In our cohort, among the somatic mutations identified in more than 5% cases, patients with TP53 mutation showed poor prognosis. Three out of four patients with TP53 mutation showed recurrence after CCRT within 10 months, and the tumors with this mutation were aggressive. Many studies suggest that TP53 regulates malignant phenotypes by gain-of-function mutations, including mutations detected in this study. A previous report showed that TP53 mutants had the worst OS when compared with wild-type or deletion-type TP53 variants26. Further, E6 and E7 oncoproteins secreted by high-risk HPV were expressed consistently. E6 and E7 proteins form complexes with p53 and retinoblastoma (Rb), respectively, and inhibit the activation of proteins in cell cycle regulation27. Interpretation of p53 IHC staining in cervical carcinoma has not been formally established26,28. In this study, we evaluated p53 IHC staining patterns as wild-type or mutant patterns using the p53 IHC staining patterns previously reported for vulvar SCC29. Three out of four patients with TP53 mutations had mutant type IHC staining pattern; patients with p53 IHC mutant type tended to be associated with poorer survival than the wild-type. These results indicate that p53 IHC staining can be used to evaluate cervical cancer. Although the correlation between TP53 mutation and radiosensitivity in patients with cervical cancer has not been fully studied, some studies have shown that TP53 mutation is correlated with poor prognosis after RT in SCC of the head and neck30–32. TP53 is the most frequently mutated gene across all cancer types33. TP53 mutation may also be associated with radioresistance or poor prognosis in cervical cancer.
In our study, patients without HPV16/18 had poorer PFS than those with HPV16/18. Many previous studies have shown that early-stage cervical cancer patients with HPV16 and HPV18 have worse prognosis than patients without HPV16/1834. On the other hand, there are several conflicting results regarding the association between HPV type and survival due to differences in radiotherapy. Recent studies have reported better survival of patients with HPV16-positive or HPV16/18-positive genotypes who were administered CCRT for FIGO stage III/IV tumors35–37. Therefore, non-HPV16/18 status might be a poor prognostic factor, as it changes the response to chemotherapy. Although few reports have focused on the correlation between HPV genotypes and p53 status, non-HPV16/18 status might affect prognosis by exhibiting marked alterations in p53. It is also important to prevent cervical cancer caused by high-risk HPVs other than HPV16/18. A 9-valent HPV vaccine has been developed, which will provide protection against non-HPV16/18 infection and advanced tumors.
Using targeted sequencing, genomic alterations, such as PIK3CA, linked to molecular-targeting drugs were detected in locally advanced cervical cancer in both our cohort and TCGA dataset. PIK3CA is the most frequently mutated gene, playing a key role in the growth and differentiation of HPV-immortalized cells38. In addition, activation of the PI3K/AKT/mTOR pathway through PIK3CA regulates various transformed phenotypes38. The PI3K inhibitor alpelisib has been approved for treatment of hormone receptor-positive and human epidermal growth factor receptor 2-negative breast cancer by the US FDA39. Notably, therapeutic benefit from this drug has been observed in three of five cervical cancer patients harboring PIK3CA mutation in a phase I trial40. Therefore, this drug is a promising therapeutic option for locally advanced cervical cancer. There are several limitations in our study. First, this study was a single institution retrospective study and the number of cases participating this study was limited. Second, targeted sequencing for mutation analysis was performed in this study; therefore, we could analyze only hotspot mutations in 50 cancer-related genes. We will further perform genomic analysis for predicting prognosis of cervical cancer patients and outcomes of targeted therapies.
In conclusion, we presented the profile of genomic alterations of locally advanced cervical cancer in both our cohort and TCGA dataset. We identified that TP53 mutants were correlated with poor PFS in locally advanced cervical cancer. In addition, tumors with diameter greater than 70 mm and non-HPV16/18 genotype were associated with poor survival. Actionable mutations for molecular-targeting drugs were detected in more than half of our cohort. These prognostic factors may lead to the development of novel treatment approaches for patients with locally advanced cervical cancer.
Materials and methods
Patients and tumor samples
One hundred and thirteen patients underwent RT or CCRT at the National Cancer Center Hospital, Tokyo, between January 2008 and December 2017 (Figure S1). Seventy of the 113 Japanese patients had locally advanced cervical cancer with FIGO stage IIIA to IVA, and these patients were recruited for this study. Patients received external beam RT and brachytherapy41, and most of the chemotherapy regimen was cisplatin-based. Clinicopathological data, including age at histological diagnosis, FIGO stage, histological subtypes, status of pelvic/para-aortic lymph nodes, tumor size, treatment, and follow-up, were obtained from the electronic medical records. Cervical tumor specimens were collected by punch biopsy of the tumor before CCRT. The specimens were fixed in 10% neutral buffered formalin and embedded in paraffin (FFPE).
Treatment regimens for CCRT and RT
All patients, except one, received both external beam RT and brachytherapy (intracavitary brachytherapy or intracavitary/interstitial brachytherapy). The initial 20–40 Gray (Gy) was delivered to the whole pelvis using the 4-field box technique, followed by a 40 mm-wide midline block until pelvic side wall dose of 50 Gy. If enlarged lymph nodes were present, an additional 6–10 Gy was delivered with smaller fields. After the initiation of the midline block, a total of 3–4 sessions of brachytherapy were performed in 1–2 sessions per week, and the dose per fraction was 6 Gy. All brachytherapy was performed by an 192Iridium remote afterloading system (RALS, MicroSelectron, HDR™, Elekta, Veennendaal, The Netherlands). The concurrent chemotherapy regimen was usually 40 mg/m2/week of cisplatin, whereas some patients received other regimens, such as carboplatin, cisplatin plus tegafur, gimeracil, oteracil, and cisplatin plus fluorouracil.
DNA preparation and next-generation sequencing
Genomic DNA was extracted from FFPE tumor tissues using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Purified genomic DNA (50 ng) obtained from tumor tissues was used for library construction using the Ion AmpliSeq™ Cancer Hotspot Panel v2 (Thermo Fisher Scientific, Waltham, MA, USA), which targets approximately 2800 COSMIC mutational hotspot regions of 50 cancer-related genes. An Ion AmpliSeq™ Custom Panel, designed for the TP53 gene (coverage: all coding regions) using Ion AmpliSeq™ Designer (https://www.ampliseq.com), was also used. Sequencing was performed on the Ion Proton platform (Thermo Fisher Scientific). For quality control, samples with a mean read depth of coverage over 1000 and a base quality score of 20 (with ≤ 1% probability of being incorrect), which accounted for 90% of the total reads, were selected.
Locally advanced cervical cancer in TCGA database
We selected 54 cases with locally advanced cervical cancer registered in TCGA database. Somatic mutations called from whole genome sequencing and whole exome sequencing data available in TCGA database were downloaded as a mutation annotation format (MAF) file via the cBioPortal for Cancer Genomics (http://www.cbioportal.org).
Classification of oncogenic/pathogenic mutations
Data analysis was carried out using the Torrent Suite Software v5.0.4 (Thermo Fisher Scientific). We selected mutations that met the following criteria: the frequency of variant alleles was more than 4% in tumor tissues; single nucleotide polymorphisms were excluded if they showed a threshold allele frequency ≥ 0.01 in either the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project (ESP6500; http://evs.gs.washington.edu/EVS/) or the integrative Japanese Genome Variation Database (iJGVD, 20181105; https://ijgvd.megabank.tohoku.ac.jp/). The variants have been registered as “pathogenic/likely pathogenic variants” in ClinVar42 or “oncogenic/likely oncogenic variants” in OncoKB (http://oncokb.org) databases using the OncoKB annotator commit 8910b65 (accessed on June 29, 2019). All selected variants were validated using the Integrative Genomics Viewer (IGV; http://www.broadinstitute.org/igv/).
Definition of actionable mutations
OncoKB is a precision oncology knowledge database that contains information on the effects and treatment implications of specific genomic alterations in cancer patients. Somatic mutations and copy number alterations have been categorized into four evidence levels. In the present study, genetic aberrations with evidence levels 1–3B according to OncoKB level of evidence V2 were designated as actionable mutations for molecular-targeting drugs43.
Immunohistochemical (IHC) staining of p53
IHC staining was performed on FFPE specimens. Representative whole 4 μm-thick sections were analyzed. After deparaffinization, the protein expression of p53 was evaluated using a monoclonal antibody against human p53 protein (clone DO-7, Dako, Glostrup, Denmark). IHC staining was performed using a Dako autostainer (Dako, CA, USA) and visualized using EnVision Detection System (Dako), according to the manufacturer’s instructions. The slides were counterstained with hematoxylin. Staining for p53 expression was evaluated as wild-type or mutant29. Scattered, mosaic, mid-epithelial p53 expression was considered to represent the wild-type staining pattern. Mutant staining pattern was characterized by diffuse strong nuclear positivity in the basal and upper layers of the tumor cells, or complete absence of p53 staining with appropriate positive internal control.
Identification of HPV genotyping by Sanger sequencing
HPV genotyping was performed for the 70 cases. Genomic DNA (10 ng) was amplified via polymerase chain reaction (PCR) using TaKaRa Taq DNA polymerase (Takara Bio Inc., Shiga, Japan) for two distinct HPV genomic regions. The HPV E6/E7 homologous region was amplified using the pU-1M/pU2R (HPVpU-1M: 5′-TGTCAAAAACCGTTGTGTCC-3′, and HPVpU-2R: 5′-GAGCTGTCGCTTAATTGCTC-3′) primer set, and the region containing the HPV L1 gene was amplified using the GP5+/GP6+ (GP5+: 5′-TTTGTTACTGTGGTAGATACTAC-3′, and GP6+: 5′-GAAAAATAAACTGTAAATCATATTC-3′) primer set. PCR reactions were performed using the TaKaRa PCR Human Papillomavirus Typing Set (TakaRa Bio Inc.). PCR products were purified using the NucleoSpin Gel (Takara Bio Inc.) or PCR Clean-up kit (Takara Bio Inc.). Sanger sequencing was performed using an ABI 3130xl DNA Sequencer (Applied Biosystems, Foster City, California, USA), according to the manufacturer’s instructions. Similarity between the obtained sequences and various HPV genotypes in the GenBank database was determined using Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Detection of high-risk HPV types in cervical cancer tissues
To clarify the frequency of HPV-positive results in these samples, we performed in situ hybridization assay for HPV detection (HPV-ISH) using HPV-III High Risk probes (Roche Diagnostics, Mannheim, Germany), according to the manufacturer’s instructions. This assay can detect high-risk HPV genotypes, including HPV-16, 18, 31, 33, 35, 45, 52, 56, 58, and 66, in cervical cancer specimens16.
Statistical analysis
The Kaplan–Meier method was applied to estimate survival, PFS, and LRFS. Differences in outcomes were compared using the log-rank test. PFS was defined as the interval from the start of the first RT to either disease progression or death. OS was defined as the interval from the start of the first RT to death. LRFS was defined as the interval from the start of first RT to either locoregional disease progression or death. PFS, OS, and LRFS were determined at the last contact date for each patient. Cox regression analysis was used to assess the univariate prognostic significance of survival. Using multivariate Cox proportional-hazards models, we considered each mutation status, histological subtype, para-aortic lymph node metastasis, and tumor size. The data cut-off date was January 29, 2020. Statistical analyses were performed with EZR version 1.3744, which is based on R and R commander.
Ethics declarations
This retrospective study was approved by the Institutional Review Board of National Cancer Center Hospital (approval number 2017-136) and follows the ethical standards laid down in the Declaration of Helsinki. Informed consent was obtained from all patients.
Supplementary Information
Acknowledgements
This work was supported by grants-in-aid from the Mitsui Life Social Welfare Foundation, Public Foundation of the Vaccination Research Center, Grant-in-Aid for Young Scientists (B) Number 18K15654, and National Cancer Center Research and Development Fund (29-A-2, NCC Biobank, and NCC Core Facility). The authors are grateful to Hitoshi Ichikawa, Sachiyo Mitani, Aya Kuchiba, and to other physicians and staff at the National Cancer Center and other hospitals for their help and support. We would like to thank Editage (https://www.editage.jp) for providing English language editing.
Author contributions
I.K., K.S., H.Y., and T.K. designed the study. I.K. wrote the initial draft of the manuscript. K.S., M.M., S.H., Y.A., D.T., and H.Y. contributed to the analysis and interpretation of data, and assisted in manuscript preparation. All other authors contributed to data collection, interpretation, and critical manuscript review. All authors approved the final version of the manuscript and agree to be accountable for all aspects of the study. We shall ensure that questions related to the accuracy or integrity of any part of the study will be appropriately investigated and resolved.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kouya Shiraishi, Email: kshirais@ncc.go.jp.
Tomoyasu Kato, Email: tokato@ncc.go.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98527-2.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382:889–899. doi: 10.1016/S0140-6736(13)60022-7. [DOI] [PubMed] [Google Scholar]
- 3.Wright JD, et al. Population-level trends in relative survival for cervical cancer. Am. J. Obstet. Gynecol. 2015;213(670):e671–e677. doi: 10.1016/j.ajog.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 5.Toita T, et al. Phase II study of concurrent chemoradiotherapy with high-dose-rate intracavitary brachytherapy in patients with locally advanced uterine cervical cancer: Efficacy and toxicity of a low cumulative radiation dose schedule. Gynecol. Oncol. 2012;126:211–216. doi: 10.1016/j.ygyno.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 6.Lanciano R, et al. Randomized comparison of weekly cisplatin or protracted venous infusion of fluorouracil in combination with pelvic radiation in advanced cervix cancer: A gynecologic oncology group study. J. Clin. Oncol. 2005;23:8289–8295. doi: 10.1200/JCO.2004.00.0497. [DOI] [PubMed] [Google Scholar]
- 7.Waggoner SE. Cervical cancer. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- 8.Vora C, Gupta S. Targeted therapy in cervical cancer. ESMO Open. 2018;3:e000462. doi: 10.1136/esmoopen-2018-000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung HC, et al. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: Results from the phase II KEYNOTE-158 study. J. Clin. Oncol. 2019;37:1470–1478. doi: 10.1200/JCO.18.01265. [DOI] [PubMed] [Google Scholar]
- 10.Duska LR, et al. Results of an early safety analysis of a study of the combination of pembrolizumab and pelvic chemoradiation in locally advanced cervical cancer. Cancer. 2020;126:4948–4956. doi: 10.1002/cncr.33136. [DOI] [PubMed] [Google Scholar]
- 11.Tewari KS, et al. Improved survival with bevacizumab in advanced cervical cancer. N. Engl. J. Med. 2014;370:734–743. doi: 10.1056/NEJMoa1309748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K, et al. Phase II trial of paclitaxel, carboplatin, and bevacizumab for advanced or recurrent cervical cancer. Gynecol. Oncol. 2019;154:554–557. doi: 10.1016/j.ygyno.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 13.Queiroz ACM, et al. Risk factors for pelvic and distant recurrence in locally advanced cervical cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019;235:6–12. doi: 10.1016/j.ejogrb.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, et al. A new marker based on risk stratification of human papillomavirus DNA and tumor size to predict survival of locally advanced cervical cancer. Int. J. Gynecol. Cancer. 2019;29:459–465. doi: 10.1136/ijgc-2018-000095. [DOI] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Research Network Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirose S, et al. Genomic alterations in STK11 can predict clinical outcomes in cervical cancer patients. Gynecol. Oncol. 2020;156:203–210. doi: 10.1016/j.ygyno.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 17.McIntyre JB, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol. Oncol. 2013;128:409–414. doi: 10.1016/j.ygyno.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Lachkar B, et al. Prognostic significance of PIK3CA mutation in stage IIB to IVA cervical cancers treated by concurrent chemoradiotherapy with weekly cisplatin. Medicine (Baltimore) 2018;97:e11392. doi: 10.1097/MD.0000000000011392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright AA, et al. Oncogenic mutations in cervical cancer: Genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013;119:3776–3783. doi: 10.1002/cncr.28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigsby P, et al. Clinical outcomes and differential effects of PI3K pathway mutation in obese versus non-obese patients with cervical cancer. Oncotarget. 2018;9:4061–4073. doi: 10.18632/oncotarget.23664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang L, et al. PIK3CA mutation analysis in Chinese patients with surgically resected cervical cancer. Sci. Rep. 2015;5:14035. doi: 10.1038/srep14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimoto Y, et al. Mutation profiling of uterine cervical cancer patients treated with definitive radiotherapy. Gynecol. Oncol. 2020;159:546–553. doi: 10.1016/j.ygyno.2020.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Endo D, et al. Prognostic factors for patients with cervical cancer treated with concurrent chemoradiotherapy: A retrospective analysis in a Japanese cohort. J. Gynecol. Oncol. 2015;26:12–18. doi: 10.3802/jgo.2015.26.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, et al. Mutational analysis of KRAS and its clinical implications in cervical cancer patients. J. Gynecol. Oncol. 2018;29:e4. doi: 10.3802/jgo.2018.29.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang L, et al. ERBB2 mutation: A promising target in non-squamous cervical cancer. Gynecol. Oncol. 2018;148:311–316. doi: 10.1016/j.ygyno.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 26.Halle MK, et al. Clinicopathologic and molecular markers in cervical carcinoma: A prospective cohort study. Am. J. Obstet. Gynecol. 2017;217(432):e431–432 e417. doi: 10.1016/j.ajog.2017.05.068. [DOI] [PubMed] [Google Scholar]
- 27.Lowy DR, Schiller JT. Human papillomavirus biology. J. Natl. Cancer Inst. Monogr. 1996;21:141–143. [PubMed] [Google Scholar]
- 28.Zhou R, Wei C, Liu J, Luo Y, Tang W. The prognostic value of p53 expression for patients with cervical cancer: A meta analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015;195:210–213. doi: 10.1016/j.ejogrb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Kortekaas KE, et al. Performance of the pattern-based interpretation of p53 immunohistochemistry as a surrogate for TP53 mutations in vulvar squamous cell carcinoma. Histopathology. 2020;77:92–99. doi: 10.1111/his.14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinhofer I, et al. Targeted next-generation sequencing identifies molecular subgroups in squamous cell carcinoma of the head and neck with distinct outcome after concurrent chemoradiation. Ann. Oncol. 2016;27:2262–2268. doi: 10.1093/annonc/mdw426. [DOI] [PubMed] [Google Scholar]
- 31.Alsner J, Sorensen SB, Overgaard J. TP53 mutation is related to poor prognosis after radiotherapy, but not surgery, in squamous cell carcinoma of the head and neck. Radiother. Oncol. 2001;59:179–185. doi: 10.1016/S0167-8140(01)00301-2. [DOI] [PubMed] [Google Scholar]
- 32.Perri F, et al. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck. 2015;37:763–770. doi: 10.1002/hed.23837. [DOI] [PubMed] [Google Scholar]
- 33.Bailey MH, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;174:1034–1035. doi: 10.1016/j.cell.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai CH, et al. Role of human papillomavirus genotype in prognosis of early-stage cervical cancer undergoing primary surgery. J. Clin. Oncol. 2007;25:3628–3634. doi: 10.1200/JCO.2007.11.2995. [DOI] [PubMed] [Google Scholar]
- 35.Onuki M, et al. Human papillomavirus genotype and prognosis of cervical cancer: Favorable survival of patients with HPV16-positive tumors. Papillomavirus Res. 2018;6:41–45. doi: 10.1016/j.pvr.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hang D, et al. Independent prognostic role of human papillomavirus genotype in cervical cancer. BMC Infect. Dis. 2017;17:391. doi: 10.1186/s12879-017-2465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuschieri K, et al. Influence of HPV type on prognosis in patients diagnosed with invasive cervical cancer. Int. J. Cancer. 2014;135:2721–2726. doi: 10.1002/ijc.28902. [DOI] [PubMed] [Google Scholar]
- 38.Henken FE, et al. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol. Cancer. 2011;10:71. doi: 10.1186/1476-4598-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andre F, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 40.Juric D, et al. Phosphatidylinositol 3-kinase alpha-selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: Results from the first-in-human study. J. Clin. Oncol. 2018;36:1291–1299. doi: 10.1200/JCO.2017.72.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chargari C, et al. Brachytherapy: An overview for clinicians. CA Cancer J. Clin. 2019;69:386–401. doi: 10.3322/caac.21578. [DOI] [PubMed] [Google Scholar]
- 42.Landrum MJ, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunami K, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019;110:1480–1490. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.