Abstract
Background
Global concerns have been raised due to upward trend of Multi-drug Resistant (MDR) Pseudomonas aeruginosa reports in ocular infections. Our aim was to characterize the virulence determinants of MDR P. aeruginosa causing ocular infections.
Methods
P. aeruginosa strains were isolated from 46 patients with conjunctivitis (2), endophthalmitis (11) and active keratitis (25) seen at our Institute, between 2016 and 2020. The isolates were identified by Vitek-2 and characterized based on growth kinetics, biofilm formation, motility, pyoverdine and pyocyanin production, phospholipase and catalase activity, urease production along with expression of exotoxins (exo-A, exo-U and exo-S) and correlated to its antibiotic profiles.
Results
Of the 46 P. aeruginosa isolates, 23 were MDR and were significantly (p = 0.03) associated with older (> 65) patients, along with higher production of pyoverdine (58.3%), pyocyanin (30.4%), phospholipase (91.6%) and protease (62.5%) activity, formed strong biofilms and exo-A (30.4%). No significant relation between motility, urease and catalase production with antibiotic susceptibility was observed. Heatmap and PCoA analysis confirmed this unique virulence profile associated with MDR-PA strains.
Conclusion
Phenotypic characteristics of P.aeruginosa might be responsible for increased colonization and antibiotic resistance observed in vivo and understanding these differences may lead to development of clinical guidelines for the management of MDR infections.
Keywords: Endophthalmitis, Multi-drug resistance, Pseudomonas aeruginosa, Virulence factors
Introduction
Pseudomonas aeruginosa is the most common Gram-negative isolate causing vision-threatening ocular infections including conjunctivitis, keratitis, orbital cellulitis and endophthalmitis [1–4]. P. aeruginosa has low nutritional requirements and can high tolerance to a range of physical conditions, thus making it more pathogenic. P. aeruginosa infections are very difficult to eradicate due to their intrinsic resistance to antibiotics [5], in addition, to various virulence factors like flagellin and lipopolysaccharide, as well as secreted products such as cytotoxins [6, 7], elastase [8, 9], alkaline protease [10, 11], protease IV [12], as well as its invasiveness and increased colonization has been reported to contribute to its pathogenicity [13–16]. The presence of these secretion toxin-encoding genes in clinical isolates from different infections is associated with differences in bacterial virulence [17, 18] and clinical outcomes [19]. Proteases is said to contribute to pathogenesis through destruction of connective tissue and degradation of host immunological factors [20] in patients with keratitis. Previous reports have also suggested that pyocyanin and pyoverdine not only contribute to the increased colonization in the lungs of patients with cystic fibrosis, it also interferes with cell respiration, calcium homeostasis and prostacyclin release from lung endothelial cells as well [21]. Another important factor is the ability of P. aeruginosa in biofilm formation beginning with the involvement of redundant planktonic cells in a complex and highly regulated physicochemical and biological signalling and thereby confers resistance and protects the bacteria against host immune responses [22–27]. Increasingly, clinical isolates of Pseudomonas are exhibiting multiple resistance to antibiotics and becoming pan drug resistant (XDR) [28]. The Emergence of resistant (MDR and XDR) strains in ocular settings, particularly in tropical countries like ours [29], has become a major problem, leaving few alternatives for treatment of these patients [30]. This increase in incidence of MDR and XDR infections is also associated with increased morbidity, mortality, and costs [31, 32]. The aim of our present study was to screen P. aeruginosa strains isolated from ocular infections, for their potential to produce various phenotypic virulence factors and correlating them with biofilm formation and their antibiotic susceptibility profile. The hypothesis tested was that MDR and XDR P. aeruginosa strains possess distinguishing virulence characteristics in comparison with P. aeruginosa that were susceptible to most antibiotics.
Materials and methods
The study was approved by the Institutional Review Board of the L V Prasad Eye Institute, and was performed in accordance with the ethical standards as laid down in the Declaration of Helsinki. Clinical and microbiology records were retrospectively reviewed for patients who were evaluated at our institute and diagnosed with culture-confirmed infections [keratitis, endophthalmitis, cellulitis, conjunctivitis] due to P. aeruginosa.
Bacterial isolates
Forty-six, Pseudomonas aeruginosa isolates from various clinical samples from patients with eye infections during the period of April 2016 to March 2020 were included in the study. The preserved isolates were revived and their identification was confirmed by standard microbiological and biochemical methods based on gram staining technique and colonial morphology in addition to Vitek 2 compact system testing using GN strips (bioMérieux).
Antibacterial susceptibility testing
For antibiotic susceptibility testing, minimum inhibitory concentration (MIC) was determined using E-test strips (Himedia) or VITEK® 2 AST cards according to the manufacturer’s protocol [33] and this included ciprofloxacin, moxifloxacin, gatifloxacin, ofloaxacin, ceftazidime, gentamicin, tetracycline, amikacin, tobramycin, piperacillin, norfloxacin, colistin and imipenem. All results were compared to the Clinical and Laboratory Standards Institute (CLSI) interpretative guidelines and the isolates were classified as susceptible (S), susceptible dose dependent (SDD), and resistant (R) [34]. However, for the purpose of analysis, SDD isolates were clubbed with susceptible isolates (S-PA). Multiple drug resistant (MDR) phenotype was assigned for strains that was resistant to ≥3 classes of antibiotics. The P. aeruginosa ATCC 27853 strain (American Type Culture Collection), was used as the quality control. The P. aeruginosa phenotype was defined as MDR and XDR according to the international expert proposal for interim standards guidelines [18].
Growth curve analysis
The in vitro growth rate was assessed by diluting 1 × 106 CFU/ml of each isolate in BHI broth, on a rotary shaker at 160 rpm for 48 h. At periodic intervals, serial dilution of each culture was plated on an antibiotic-free Mueller Hinton agar and the number of CFUs [colony-forming units] was counted after 24 h incubation at 37 °C.
Phenotypic characterization of P. aeruginosa isolates
All phenotypic assays (described below) were performed as three independent biological replicates unless otherwise specified. The P. aeruginosa ATCC 27853 strain was used as the positive control to assay motility, biofilm formation and secreted virulence factors.
Measurement of Pyoverdine production and qualitative assessment of pyocyanin
Pyoverdine production by P. aeruginosa was assayed spectrophotometrically as previously described [35]. Briefly, the isolates were cultured in MHB to late stationary phase (OD of ‘3 was recorded at 600 nm). The cultures were then centrifuged at 10,000×g for 2 min and the supernatant were normalized for differences in cell density, and the absorbances measured at 405 nm. The concentration of pyoverdine was then calculated using the extinction coefficient as follows: Molar concentration = Absorbance/Extinction coefficient (1.9 X 10− 4 M− 1 cm− 1).
Pyocyanin production was assessed using a qualitative method as described earlier by Alonso et al. [36]. Inoculated plates were incubated at 37 °C for 24 h. Colonies appearing Dark green or blue in colour were considered to be pyocyanin producers.
In vitro microplate biofilm assay
The ability of P. aeruginosa strains to develop biofilm was determined on 96-well microtiter plates with crystal violet (CV) staining method as previously described [37]. Briefly, each P. aeruginosa strain was cultured on Mueller Hinton agar overnight, and a colony of each isolate was suspended in brain heart infusion (BHI) broth and incubated at 37 °C for 4 h. Bacterial suspensions were then adjusted to an optical density of 0.1 at 600 nm and added to flat-bottomed 96-well sterile culture plates. Following a 48 h incubation period at 37 °C, non-adherent bacteria were removed by washing and stained with 0.1% crystal violet. Following incubation at 30 min, the plates were again washed and ethyl alcohol was added each well, after which the absorbance was measured at 590 nm. The OD590 values were then normalized with initial OD600 values to account for differences in bacterial growth and biofilm was classified as weak, moderate or strong as described earlier [38].
Motility assay
Motility of the strains was determined in motility medium, which consists 1 (w/v) tryptone, 0.3% yeast extract, 0.5% NaCl and 0.3% agar. Plates were stab-inoculated from overnight cultures and incubated at 37 °C for 24 h. Each strain was assayed in triplicate. The diameter of the circular zone of growth was measured and expressed as a mean value in mm. An isolate showing a change of ≥ 10% was considered highly motile while the rest were categorized as weakly motile or non-motile.
Catalase activity
Determination of the enzyme’s presence in the bacterial strains was done using a sterile loop to place a small amount of growth onto the base of a Petri dish, followed by a drop of hydrogen peroxide and covering the Petri dish with a lid [39]. The development of effervescence indicates a positive result.
Extracellular protease activity
Qualitative protease activity was evaluated by streaking bacteria onto modified basal medium supplemented with 6.2 g/L skim milk protein and incubated for 48 h at 37 °C. Proteolytic activity was demonstrated by a clearing zone (> 12 mm) surrounding the bacterial growth.
Phospholipase [plc] activity assay
Haemolytic activity of the isolates was analysed by presence of clear halos around growth of the organism on blood agar plates after 24 h incubation at 37 °C [40].
Urease test
Urease activity was determined by inoculating the strains on to Christensen’s urea agar slope (Oxoid, Thebarton, South Australia) and incubated at 21 °C with protection from light for seven days [41]. Following incubation, the slopes were examined visually for a colour change to pink which was considered to be a positive result, and no change (or a yellow colour) was considered a negative result.
DNA extraction and genotypic detection of virulence genes
Strain DNA was extracted using the QIAamp DNA Mini Kit (50) (QIAGEN) following the manufacturer’s instructions. DNA was eluted in 30 μl of elution buffer. The genes, exotoxin S (exoS), exotoxin U (exoU), exotoxinA (exoA) were amplified using the specific primers as described earlier [41, 42]. The PCR protocol involved initial denaturation step at 95°C for 10 min, followed by 40 cycles of 94°C for 2 min, annealing (30s at 57 to 65 °C) and 72°C for 1 min and the final extension step at 72°C for 5 min.
Statistical analysis
The data were processed on spreadsheets and all statistical analyses were performed using GraphPad prism [5.0]. Phenotypes were treated as either categorical or continuous variables and analysed as appropriate. Wherever applicable all comparisons were evaluated using either χ2 test or unpaired student t test. The CFU were represented as mean CFU ± SE of 23 strains in each group at indicated time points and a P value of less than 0.05 was considered significant.
Results
Forty-six isolates of P. aeruginosa were obtained from various clinical specimens [corneal, conjunctival and scleral scraping, corneal buttons and vitreous fluids] during the study period and the clinical and demographic details are elaborated in Table 1. Clinical diagnosis included conjunctivitis [3], scleritis [1], endophthalmitis [12], cellulitis [2] and active keratitis [28] diagnosed at our institute, during the study period. Of these isolates, 23 [50%] were found to be MDR which also included 9 (19.5%) XDR strains. As expected, MDR-PA was significantly associated with poor visual outcome and prognosis (p = 0.03). Interestingly, these MDR strains were significantly (p = 0.03) associated with older (> 65) patients. Out the 23 patients infected with S-PA strains, 2 patients underwent evisceration who were diagnosed with post enucleation socket syndrome and microbial keratitis. Additionally, 2 patients with MDR-PA caused microbial keratitis and 2 patients with MDR-PA induced endophthalmitis cases lead to evisceration.
Table 1.
Demographic and microbiological data of the patients included in the study
| Categories | Susceptible(S-PA) N = 23(%) |
Multi-drug resistant (MDR-PA) N = 23(%) |
p-value |
|---|---|---|---|
| Gender | |||
| Male | 12 (52.17%) | 13 (56.52%) | a1 |
| Female | 11 (47.82%) | 10 (44.47%) | |
| Mean Age ± SD (years) | 39.29 ± 20.05 | 49.34 ± 18.67 | a0.11 |
| (Range) | (1–80) | (4–73) | |
| Disease | |||
| Microbial keratitis | 14 (60.86) | 11 (47.83) | |
| Endophthalmitis | 3 (13.04) | 8 (34.78) | |
| others | 6 (26.08) | 4 (17.39) | |
| VA | |||
| > (20/200) | 12 | 16 | 0.03 |
| > (20/20) - (20/200) < | 6 | 1 | |
| Evisceration | 2 | 4 | |
| Unknown | 3 | 2 | |
| Polymorphs | |||
| (0–1,0-3,0–5) | 8 | 5 | 0.2 |
| (0–10, plenty) | 13 | 17 | |
| Unknown | 2 | 1 | |
a: Pearson’s chi-squared test
Growth kinetics differences between MDR-PA and S-PA
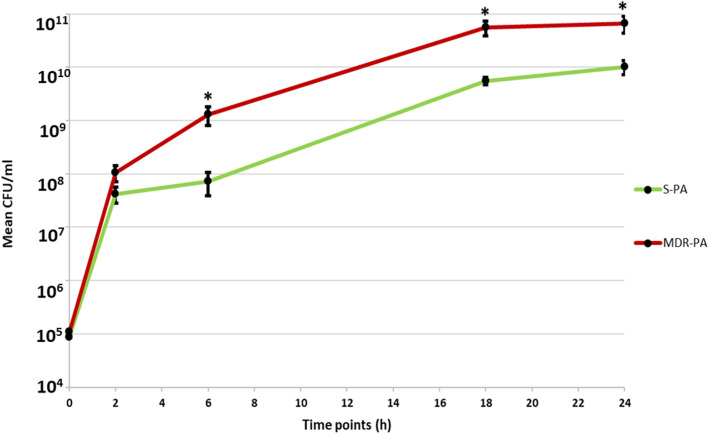
To determine potential differences in growth kinetics, the colony forming units on MHA plates were determined for each strain, by plotting the mean CFU of all S-PA and MDR-PA strains at different time points (0,2,6,18,24 h). At 6 h, 18 h and 24 h significant differences in colony counts were observed between the drug susceptible strains vs multi-drug resistant strains as shown in Fig. 1. The mean CFU at 6 h of S-PA Vs MDR-PA strains were (7.82 × 107 Vs 1.31 × 109, p = 0.01), at 18 h (5.22 × 109 Vs 5.6 × 1010, p = 0.01) and 24 h (1.01 × 1010 Vs 6.65 × 1010, p = 0.03). The plotted growth curve suggests that MDR-PA strains demonstrate significantly higher growth kinetics at later time points compared to S-PA strains.
Fig. 1.
Clinical isolates of P. aeruginosa were plated on MHA at indicated time points. Bacterial load and viability were enumerated by plate count method. Student’s test was used for the statistical analysis and data are represented as the mean colony forming units (CFU ± SD) from three sets of independent experiments. *p < 0.05
Phenotypic characterization of S-PA and MDR-PA strains
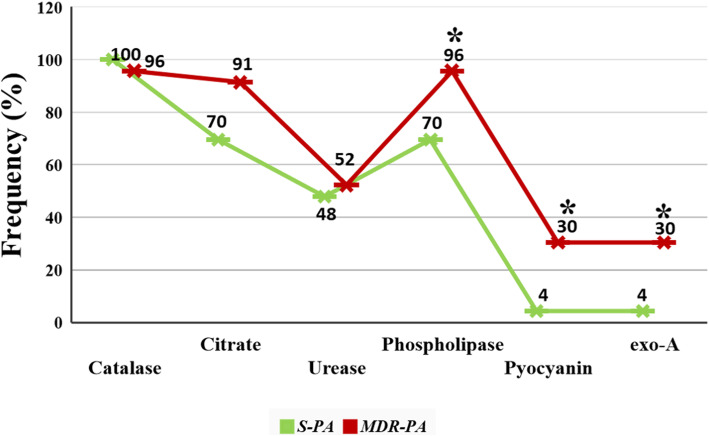
Almost all the strains in both S-PA and MDR-PA group were catalase positive. While 11/23 (48%) S-PA strains were urease positive and 12/23 (52%) of the MDR-PA group were positive for urease. Similarly, for citrate production, 91% of MDR-PA and 70% of the S-PA isolates were positive. Additionally, phospholipase C activity was significantly (chi square test, p = 0.02) higher in MDR-PA (96%) strains when compared with the S-PA strains (70%) as shown in Fig. 2, suggesting a potential role in strain virulence. We found that 7 (30.4%) out of the 23 MDR-PA isolates produced pyocyanin while only one S-PA (4.3%) isolate produced pyocyanin pigment and there is significant association of pyocyanin production with multidrug resistant (p = 0.02).
Fig. 2.
Graphic representation of frequency of enzymatic virulence factors (catalase, citrate, urease,phospholipase, pyocyanin and exo-A) expression. Pearson’s Chi-square test was used for the statistical analysis and data are represented as the mean from three sets of independent experiments. **p ≤ 0.01, *p < 0.05
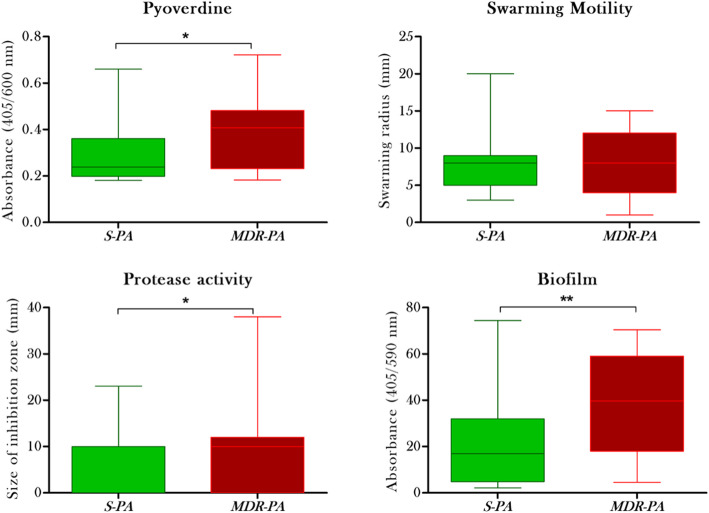
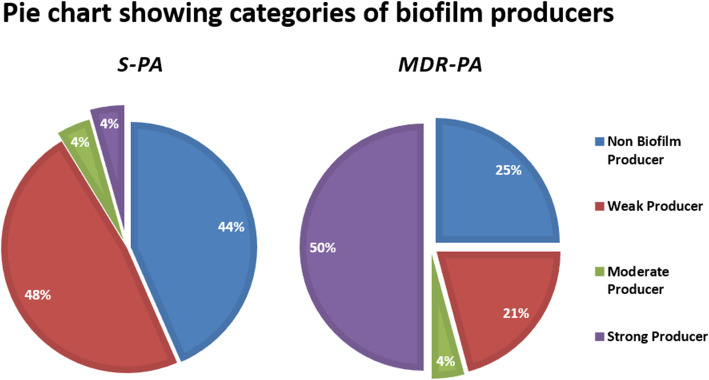
Looking at the pyoverdine production, the mean absorbance (Fig. 3A) was again significantly higher in MDR-PA strains, compared to the S-PA (0.39 ± 0.03 nm Vs 0.28 ± 0.02 nm, p = 0.01) indicating its role in virulence. Though the MDR-PA strains showed increased motility compared to the S-PA group, this difference (7.82 ± 0.90 mm vs 8.04 ± 0.04 mm, p = 0.8) was not statistically significant (Fig. 3B), however, 50% strains belonged to the XDR group. Similarly, protease activity was significantly higher in MDR-PA group when compared to S-PA group (9.48 ± 0.96 mm vs 5.17 ± 1.88 mm, p = 0.02) (Fig. 3C) of which, 80% strains belonged to the XDR phenotype. Analysis of biofilm ability revealed an increased production of biofilm by MDR-PA group (39.04 ± 4.88 nm vs 21.10 ± 4.19 nm, p = 0.007) as shown in Fig. 3D. Additionally, among the S-PA strains, 48% formed weak biofilms, 4% were moderate, 4% were strong biofilm formers (Fig. 4) and 44% were non biofilm producers. In comparison, in the MDR-PA group, 50% of the strains were strong biofilm producer, while 25% were weak biofilm producers. Next, we compared XDR-PA and MDR-PA group to find out if there is any association of XDR with the virulence traits. In our present study we didn’t find any significant association of XDR with virulence factors (biofilm; p = 0.51, urease; p = 0.13, protease; p = 0.42, motility; p = 0.40, pyoverdine; p = 0.71, citrate; p = 0.84) compared to the MDR-PA isolates.
Fig. 3.
(A) Quantitative analyses of the pyoverdine production by spectrophotometer analysis are shown in the box-plot. (B) Swarming activity were quantitively analysed by measuring the distance moved by the isolates (C) Quantitative analysis of protease production by S-PA and MDR-PA strains (D) Quantitative analyses of the results from crystal violet retention assays are shown in the box-plot. Unpaired Student’s test was used for the statistical analysis and data are represented as the mean ± SD from three sets of independent experiments. **p ≤ 0.01, *p < 0.05
Fig. 4.
Graphic representation of a biofilm production by S-PA and MDR-PA group. 50% of the MDR-PA isolates were strong producer whereas in case of S-PA only 4% isolates were strong biofilm producer
This trend suggests a significant association of biofilm production with multi-drug resistance. Summarizing the virulence factors, we observe that production of catalase, urease and swarming ability does not have any correlation with the level of antibiotic resistance (Table 2) whereas pyoverdine production, biofilm ability, protease and phospholipase activity were significantly associated with antibiotic resistance and virulence.
Table 2.
Frequency and Statistical correlation of different virulence factors among Resistant and susceptible P. aeruginosa strains
| Phenotypic character | S-PA | MDR-PA | P-value | OR (95% CI) |
|---|---|---|---|---|
| Pyoverdine | 4 | 14 | a0.01* | 6.65 (1.7–25.6) |
| Pyocyanin | 1 | 7 | a0.02* | 0.1 (0.01–0.93) |
| Biofilm | 13 | 18 | a0.007* | 2.23 (0.6–7.9) |
| Swarming | 4 | 8 | a0.12 | – |
| Catalase | 23 | 22 | – | |
| Protease | 5 | 15 | a0.02* | 3.8 (1.1–12.8) |
| Phospholipase | 16 | 22 | a0.01* | 5.8 (1.0–31.5) |
| Urease | 11 | 12 | a0.88 | – |
a:Pearson’s chi-squared test, P probability, OR Odds Ratio, CI confidence interval
Further, we compared association of anatomical site with the virulence factor. Out of the 25 samples 14(56%) were antibiotic susceptible and 11(44%) were multi-drug resistant. We did not observe any difference in the virulence factor (biofilm; p = 0.20, urease; p = 0.46, phospholipase; p = 0.12, protease; p = 0.32, motility; p = 0.57, pyoverdine; p = 0.30, citrate; p = 0.4) between the S-PA isolates and MDR-PA isolates.
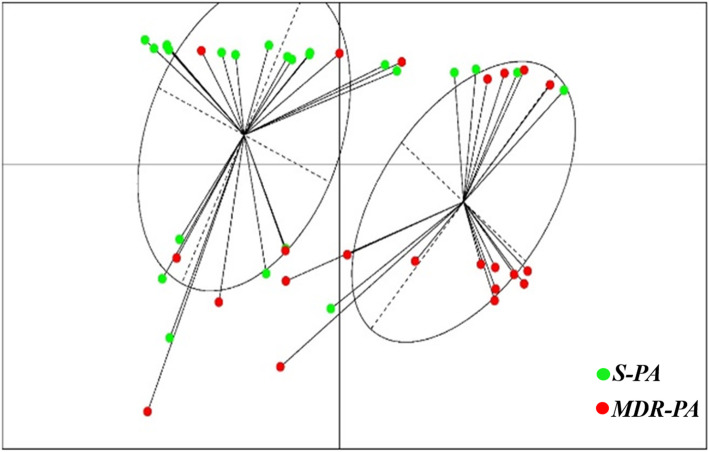
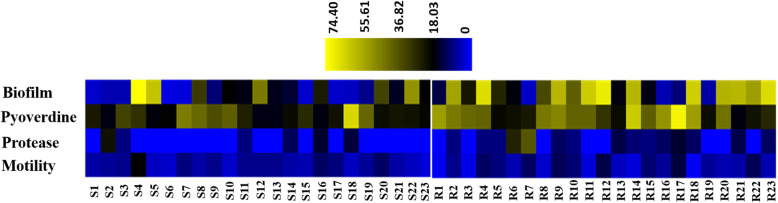
To further confirm our results, we carried out a principal Coordinate Analysis to explore and to visualize dissimilarities of these virulence factors amongst the S-PA and MDR-PA group. Application of this method to our data showed that the samples could be divided into two principal groups: one consisting primarily of S-PA strains (Fig. 5, cluster 1) and second independent clusters of mainly MDR-PA strains (Fig. 5, clusters 2). Red dot indicates MDR-PA and green indicates S-PA. The plot shows the Euclidian distance between the two groups based on how the two groups can be distinguished. We further constructed a heat map to depict the relative virulence factors expressed by each strain as shown in Fig. 6. The relative colour intensity of heat map gave a good overview of the profile differences between the two groups and demonstrates that the virulence factors are more strongly associated with the MDR-PA clinical isolates.
Fig. 5.
Principal coordinate analysis (PCoA) plot of S-PA (left) and MDR-PA (right) and the high-resolution display of red square indicates MDR-PA isolates while the green dot represents S-PA isolates. PCoA plot was generated from the values obtained from Pyoverdine, biofilm, protease and motility test (n = 23 in each group)
Fig. 6.
Heat map representing virulence factors of ocular clinical isolates of P. aeruginosa. The Virulence factors for 46 clinical isolates were tested by different methods specified earlier. Heat map was constructed to compare the S-PA and MDR-PA isolates. S1-S23 denotes drug susceptible strains, R1-R23 denotes MDR-PA strains. Blue represents a lower level of expression and yellow represents a higher level of expression
Detection of exotoxins in S-PA and MDR-PA strains
Our study found that the exoA was present in 1/23(4.3%) and exoS 2/23(8.69%) of the S-PA isolates while we couldn’t detect exoU gene in any of the S-PA isolates. In case of MDR-PA isolates, exoA was present in 7/23 (30.4%) of the isolates, being significantly associated with multidrug resistance (p = 0.02). However, exo-S was not detected in any of the MDR-PA strains and only 2 strains showed the presence of exo-U (8.69%).
Discussion
P. aeruginosa is a predominant Gram-negative bacterium causing ocular infections [43]. In the present study, P. aeruginosa strains were obtained from corneal ulcers, vitreous fluids and purulent discharge from patients diagnosed at our institute with various infectious conditions. Characterization of the armor of P. aeruginosa virulence factors is essential to understand the pathogenesis of this opportunistic pathogen as well as helps in exploring new antimicrobial strategies in MDR strains. P. aeruginosa is also known to secrete a number of extracellular products that aid in survival and increased virulence [44]. We hypothesized that distinct virulence traits of ocular P. aeruginosa strains would be associated with increased antibiotic resistance in patients with positive P. aeruginosa cultures, and as expected we found that 35 out of the 46 P. aeruginosa strains had more than two virulence factors. Factor analysis of the measured bacterial variables revealed interrelationships between several of these phenotypes. In our study, the growth curve analysis showed that the MDR-PA strains had shorter doubling time, especially at later time points, along with increased swarming motility, and though this association was not statistically significant, it explains its longer persistence in the host. Similarly, pyoverdine which is said to play a critical role in the pathogenesis of host infection by P. aeruginosa by removing ferric iron from the host causing mitochondrial damage and compromising ATP production [45] was significantly associated with MDR-PA group. This is in agreement with findings by Rodulfo et al. [6] and Finlayson [7] wherein pigment production was reported to be significantly associated with MDR expression along with elastase, protease, siderophore and DNase activity. To date, considerable number of studies have shown the potential importance of pyocyanin in the virulence and pathogenicity of pseudomonal infections [46, 47]. In the present study, ocular MDR-PA strains showed a significant association with pyocyanin production.
Our experiments also show that protease production had a role in increased virulence of resistant strains and the XDR group showed higher activity compared to MDR strains. While catalase activity has been demonstrated to be essential for the intracellular survival of bacteria such as Mycobacterium tuberculosis [48], our study did not find any association of catalase and urease activity with antibiotic profile. We did however, find a correlation between resistance patterns and phospholipases activity which is reported to play an important role in host cell penetration, cell lysis and are active component of bacterial toxins. On the contrary, other studies have shown association of catalase with virulence. This enzyme has been demonstrated to be an essential factor for the intracellular survival of few bacteria such as Mycobacterium tuberculosis [49], Campylobacter jejuni [50] and Helicobacter pylori [51]. Similarly, Urease activity has been shown to be an important pathogenic factor for the bacteria Helicobacter pylori and Proteus mirabilis [52–54]. It is also known to be involved in a series of processes that allow bacteria to colonize and induce a strong inflammatory response in the gastric epithelium [55]. Citrate is known important activator of master regulators expression of virulence factors, central metabolism, iron acquisition, and bacterial virulence of S. aureus [56].
Biofilm is another reported cause of multi-drug resistance in P. aeruginosa [57] and our results were in agreement with previous studies wherein production of biofilm was significantly associated with MDR and XDR strains. An earlier study on E.coli strains had suggested that although the virulence of an organism cannot be predicted accurately on the basis of its measurable phenotypes, the presence of multiple virulence factors increases the virulence of the organisms [58], and along with host conditions decrease the need for multiple virulence factors in the strains leading to serious infections [59]. Our study is in agreement with Subedi et al. [45] who reported that virulence factors, extracellular products including proteases, and the ability to produce biofilm may explain the poor visual prognosis in P. aeruginosa endophthalmitis despite rapid antibiotic therapy [60]. The most important extracellular factors of P. aeruginosa include exo-S, exo-U, exo-A. Each of the aforementioned factors are known to have toxic effect on mammalian cells [61, 62]. In the present study, we found that exo-A is associated with multidrug resistant not exo-S and exo-U. An earlier study by wolf et al. [63] has shown that the exoA-deficient mutants exhibit virulence 20 times less than the wild type strain in the mouse models.
Principal component analysis and heat map data reiterates the unique virulence profile of MDR-PA strains compared to S-PA group and these five factors (pyoverdine production, biofilm ability, protease and phospholipase activity) could thus be used as independent predictors of resistant profiles and virulence. Several limitations however, exist in our study. Firstly, these experiments were measured in vitro of P. aeruginosa isolated from primary clinical specimens, although these phenotypes were reproducible upon repeat assay, we only determined potential of expression of virulence factors under defined in vitro conditions, and not actually expressed in the host. Secondly, the visual outcome in resistant strains was not correlated with antibiotic resistance in our clinical practice. Nonetheless, our findings would be more representative of patients with positive cultures for P. aeruginosa seen in tertiary eye care hospitals and may aid in clinician decision-making in such a setting.
Conclusion
While antibiotic resistance is multifactorial, recognition of virulent strains by phenotypic characterization, is easier and allows immediate institution of appropriate therapy. To the best of our knowledge, this study was the first investigation regarding the phenotypic virulent characteristics amongst susceptible and resistant strains of P. aeruginosa in India. Further studies would focus to understand the genotypic characteristics involved in MDR-P. aeruginosa strains, which would aid in developing a rapid signature biomarker for resistant strains.
Acknowledgements
Hyderabad Eye Research Foundation (HERF), Hyderabad.
Financial disclosure
None of the authors have any other financial disclosures.
Authors’ contributions
conceptualization, J.J.; methodology, P. N, S. P, S. G, S.B.; software, P. N, S.P; validation, J.J., P.N.; Data analysis, P.N.; data curation, J.J.; writing—original draft preparation, P.N.; writing—review and editing, J.J; supervision, J.J.; project administration, J.J. All authors read and approved the final manuscript.
Funding
Hyderabad Eye Research Foundation (HERF).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the L V Prasad Eye Institute (LEC 09-18-125), and was performed in accordance with the ethical standards as laid down in the Declaration of Helsinki. Clinical and microbiology records were retrospectively reviewed for patients who were evaluated at our institute and diagnosed with culture-confirmed infections [keratitis, endophthalmitis, cellulitis, conjunctivitis] due to P. aeruginosa.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Galvis V, Tello A, Guerra A, Acuña MF, Villarreal D. Antibiotic susceptibility patterns of bacteria isolated from keratitis and intraocular infections at Fundación Oftalmológica de Santander (FOSCAL), Floridablanca. Colombia Biomedica. 2014;34(0):123–133. doi: 10.7705/biomedica.v34i0.1636. [DOI] [PubMed] [Google Scholar]
- 2.Iwalokun BA, Oluwadun A, Akinsinde KA, Niemogha MT, Nwaokorie FO. Bacteriologic and plasmid analysis of etiologic agents of conjunctivitis in Lagos, Nigeria. J Ophthalmic Inflamm Infect. 2011;1(3):395–103. doi: 10.1007/s12348-011-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhury R, Panda S, Sharma S, Sidh V. Staphylococcal infection, antibiotic resistance and therapeutics, antibiotic resistant bacteria – a continuous challenge in the new millennium. Dr. Marina Pana, editor. London: IntechOpen Limited; 2012. Staphylococcal Infection, Antibiotic Resistance and Therapeutics. [Google Scholar]
- 4.Summaiya M, Sangita R, Neeta K. Ocular infections: rational approach to antibiotic therapy. Natl J Med Res. 2012;2:122–124. [Google Scholar]
- 5.Lister PD, Wolter DJ, Hanson ND. Antibacterial–resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:4582–4610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleiszig SM, Wiener-Kronish JP, Miyazaki H, Vallas V, Mostov KE, Kanada D, Sawa T, Yen TS, Frank DW. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65(2):579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology (Reading) 2001;147(10):102659–102669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 8.Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13(8):8389–8397. doi: 10.1016/j.tim.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171(11):111209–111223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman AL, Lory S. Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr Opin Microbiol. 2004;7:139–144. doi: 10.1016/j.mib.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, Manos J, Elkins M, Bye P, Willcox M, Bell S, Wainwright C, Harbour C. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol. 2007;45(6):61697–61704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobden JA. Pseudomonas aeruginosa proteases and corneal virulence. DNA Cell Biol New York: Mary Ann Liebert, Inc. 2002;21:56391–56396. doi: 10.1089/10445490260099674. [DOI] [PubMed] [Google Scholar]
- 13.Rodulfo H, Arcia A, Hernández A, Michelli E, Martinez DDV, Guzman M, Sharma A, Donato M. Virulence factors and integrons are associated with MDR and XDR phenotypes in nosocomial strains of Pseudomonas aeruginosa in a Venezuelan university hospital. Rev Inst Med Trop Sao Paulo. 2019;61:e20. doi: 10.1590/S1678-9946201961020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlayson EA, Brown PD. Comparison of antibiotic resistance and virulence factors in pigmented and non–pigmented Pseudomonas aeruginosa. West Indian Med J. 2011;60:124–132. [PubMed] [Google Scholar]
- 15.Minandri F, Imperi F, Frangipani E, Bonchi C, Visaggio D, Facchini M, Pasquali P, Bragonzi A, Visca P. Role of Iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun. 2016;84(8):82324–82335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang D, Kirienko DR, Webster P, Fisher AL, Kirienko NV. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a activates a distinct host response. Virulence. 2018;9(1):804–817. doi: 10.1080/21505594.2018.1449508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleiszig SM, Lee EJ, Wu C, Andika RC, Vallas V, Portoles M, Frank DW. Cytotoxic strains of Pseudomonas aeruginosa can damage the intact corneal surface in vitro. CLAO J. 1998;24:141–147. [PubMed] [Google Scholar]
- 18.Hauser AR, Cobb E, Bodi M, Mariscal D, Vallés J, Engel JN, et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator–associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30(3):3521–3528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Engel LS, Hill JM, Moreau JM, Green LC, Hobden JA, O'Callaghan RJ. Pseudomonas aeruginosa protease IV produces corneal damage and contributes to bacterial virulence. Invest Ophthalmol Vis Sci. 1998;39:3662–3665. [PubMed] [Google Scholar]
- 20.Iglewski BH. Pseudomonas. In: Baron S, editor. Medical Microbiology. 4. Galveston (TX): University of Texas Medical Branch. Galveston; 1996. [PubMed] [Google Scholar]
- 21.Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10(6):6644–6648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Høiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PØ, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. The clinical impact of bacterial biofilms. Int J Oral Sci. 2011;3(2):255–265. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):54181318–54181322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 24.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183(18):185395–185401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186(14):144449–144456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:9276135–9276138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 27.Harris A, Torres-Viera C, Venkataraman L, DeGirolami P, Samore M, Carmeli Y. Epidemiology and clinical outcomes of patients with multiresistant Pseudomonas aeruginosa. Clin Infect Dis. 1999;28(5):1128–1133. doi: 10.1086/514760. [DOI] [PubMed] [Google Scholar]
- 28.National Nosocomial Infections Surveillance System National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32(8):8470–8485. doi: 10.1016/j.ajic.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Pathengay A, Moreker MR, Puthussery R, Ambatipudi S, Jalali S, Majji AB, Mathai A, Husssain N, Dave V, Sharma S, Das T. Clinical and microbiologic review of culture–proven endophthalmitis caused by multidrug–resistant bacteria in patients seen at a tertiary eye care center in southern India. Retina. 2011;31(9):91806–91811. doi: 10.1097/IAE.0b013e31820f4b9d. [DOI] [PubMed] [Google Scholar]
- 30.Chatterjee M, Anju CP, Biswas L, Anil Kumar V, Gopi Mohan C, Biswas R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol. 2016;306(1):148–158. doi: 10.1016/j.ijmm.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Carmeli Y, Troillet N, Karchmer AW, Samore MH. Health and economic outcomes of antibiotic resistance in Pseudomonas aeruginosa. Arch Intern Med. 1999;159(10):101127–101132. doi: 10.1001/archinte.159.10.1127. [DOI] [PubMed] [Google Scholar]
- 32.Fritsche TR, Sader HS, Toleman MA, Walsh TR, Jones RN. Emerging metallo–beta–lactamase–mediated resistances: a summary report from the worldwide SENTRY antimicrobial surveillance program. Clin Infect Dis. 2005;41(s4):4276–4278. doi: 10.1086/430790. [DOI] [PubMed] [Google Scholar]
- 33.Novick WJ. Development of in vitro susceptibility testing criteria and quality control parameters, 2nd ed. Clin Microbiol Newsl. 1989;Vol 11(Issue 8):60–62. doi: 10.1016/0196-4399(89)90068-8. [DOI] [Google Scholar]
- 34.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug–resistant, extensively drug–resistant and pandrug–resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):3268–3281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 35.Merritt JH, Kadouri DE, O'Toole GA (2005) Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. 10.1002/9780471729259.mc01b01s00 [DOI] [PMC free article] [PubMed]
- 36.Alonso B, Fernández-Barat L, Di Domenico EG, Marín M, Cercenado E, Merino I, de Pablos M, Muñoz P, Guembe M. Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect Dis. 2020;20(1):909. doi: 10.1186/s12879-020-05534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choy MH, Stapleton F, Willcox MDP, Zhu H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens– and non–contact lens–related keratitis. J Med Microbiol. 2008;57(12):121539–121546. doi: 10.1099/jmm.0.2008/003723-0. [DOI] [PubMed] [Google Scholar]
- 38.Oka N, Suzuki T, Ishikawa E, Yamaguchi S, Hayashi N, Gotoh N, Ohashi Y. Relationship of virulence factors and clinical features in keratitis caused by Pseudomonas aeruginosa. Invest Ophthalmol Vis Sci. 2015;56(11):116892–116898. doi: 10.1167/iovs.15-17556. [DOI] [PubMed] [Google Scholar]
- 39.Wargo MJ, Ho TC, Gross MJ, Whittaker LA, Hogan DA. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect Immun. 2009;77(3):31103–31111. doi: 10.1128/IAI.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradbury RS, Reid DW, Inglis TJ, Champion AC. Decreased virulence of cystic fibrosis Pseudomonas aeruginosa in Dictyostelium discoideum. Microbiol Immunol. 2011;55(4):4224–4230. doi: 10.1111/j.1348-0421.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- 41.Dave A, Samarth A, Karolia R, Sharma S, Karunakaran E, Partridge L, MacNeil S, Monk PN, Garg P, Roy S. Characterization of ocular clinical isolates of Pseudomonas aeruginosa from non–contact Lens related keratitis patients from South India. Microorganisms. 2020;8(2):2260. doi: 10.3390/microorganisms8020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanomand A, Farajnia S, Najar Peerayeh S, Majidi J. Cloning, expression and characterization of recombinant exotoxin A-flagellin fusion protein as a new vaccine candidate against Pseudomonas aeruginosa infections. Iran Biomed J. 2013;17(1):1–7. doi: 10.6091/ibj.22.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teweldemedhin M, Gebreyesus H, Atsbaha AH, Asgedom SW, Saravanan M. Bacterial profile of ocular infections: a systematic review. BMC Ophthalmol. 2017;17(1):212. doi: 10.1186/s12886-017-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subedi D, Vijay AK, Willcox M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: an ocular perspective. Clin Exp Optom. 2018;101(2):2162–2171. doi: 10.1111/cxo.12621. [DOI] [PubMed] [Google Scholar]
- 46.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72(7):4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16(3):463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manca C, Paul S, Barry CE, III, Freedman VH, Kaplan G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun. 1999;67:74–79. doi: 10.1128/IAI.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day WA, Jr, Sajecki JL, Pitts TM, Joens LA. Role of catalase in campylobacter jejuni intracellular survival. Infect Immun. 2000;68(11):6337–6345. doi: 10.1128/IAI.68.11.6337-6345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basu M, Czinn SJ, Blanchard TG. Absence of catalase reduces long-term survival of Helicobacter pylori in macrophage phagosomes. Helicobacter. 2004;9(3):211–216. doi: 10.1111/j.1083-4389.2004.00226.x. [DOI] [PubMed] [Google Scholar]
- 52.Eaton K, Brooks C, Morgan D, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59(7):2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones B, Lockatell CV, Johnson D, Warren J, Mobley HLT. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990;58(4):1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuda M, Karita M, Morsted MG, Okita K, Nakasaki T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62(8):3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olivera-Severo D, Uberti AF, Marques MS, Pinto MT, Gomez-Lazaro M, Figueiredo C, Leite M, Carlini CR. A New Role for Helicobacter pylori Urease: Contributions to Angiogenesis. Front Microbiol. 2017;8:1883. doi: 10.3389/fmicb.2017.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding Y, Liu X, Chen F, di H, Xu B, Zhou L, Deng X, Wu M, Yang CG, Lan L. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2014;111(46):E4981–E4990. doi: 10.1073/pnas.1411077111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karami P, Khaledi A, Mashoof RY, Yaghoobi MH, Karami M, Dastan D, et al. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Reports. 2020;Volume 18:100561. doi: 10.1016/j.genrep.2019.100561. [DOI] [Google Scholar]
- 58.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4(1):180–128. doi: 10.1128/CMR.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raksha R, Srinivasa H, Macaden RS. Occurrence and characterisation of uropathogenic Escherichia coli in urinary tract infections. Indian J Med Microbiol. 2003;21(2):2102–2107. doi: 10.1016/S0255-0857(21)03130-3. [DOI] [PubMed] [Google Scholar]
- 60.Abo EA, Negm S, Ismail A, Mostafa AE. An outbreak of post cataract Pseudomonas aeruginosa acute endophthalmitis in Egypt. Bull Natl Res Cent. 2019;43:13. doi: 10.1186/s42269-019-0055-4. [DOI] [Google Scholar]
- 61.Khalifa ABH, Moissenet D, Thien HV, Khedher MA. Virulence factors in Pseudomonas aeruginosa: mechanisms and modes of regulation. Ann Biol Clin. 2011;69(4):393–403. doi: 10.1684/abc.2011.0589. [DOI] [PubMed] [Google Scholar]
- 62.de Bentzmann S, Polette M, Zahm J-M, Hinnrasky J, Kileztky C, Bajolet O, Klossek J-M, Filloux A, Lazdunski A, Puchelle E. Pseudomonas aeruginosa virulence factors delay airway epithelial wound repair by altering the actin cytoskeleton and inducing overactivation of epithelial matrix metalloproteinase-2. Lab Investig. 2000;80(2):209–219. doi: 10.1038/labinvest.3780024. [DOI] [PubMed] [Google Scholar]
- 63.Wolf P, Beile U. Pseudomonas exotoxin a: Fromvirulence factor to anti-cancer agent. Int J Med Microbiol. 2009;299(3):161–176. doi: 10.1016/j.ijmm.2008.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.