Abstract
Fimbriae (FimA) of Porphyromonas gingivalis are filamentous components on the cell surface and are thought to play an important role in the colonization and invasion of periodontal tissues. We previously demonstrated that fimA can be classified into four variants (types I to IV) on the basis of the nucleotide sequences of the fimA gene. In the present study, we attempted to detect the four different fimA genes in saliva and plaque samples isolated from patients with periodontitis using the PCR method. Four sets of fimA type-specific primers were designed for the PCR assay. These primers selectively amplified 392-bp (type I), 257-bp (type II), 247-bp (type III), and 251-bp (type IV) DNA fragments of the fimA gene. Positive PCR results were observed with reference strains of P. gingivalis in a type-specific manner. All other laboratory strains of oral and nonoral bacteria gave negative results. The sensitivity of the PCR assay for fimA type-specific detection was between 5 and 50 cells of P. gingivalis. Clinical samples were obtained from saliva and subgingival plaque from deep pockets (≥4 mm) of 93 patients with periodontitis. Bacterial genomic DNA was isolated from the samples, and the targeted fragments were amplified by PCR. The presence of P. gingivalis was demonstrated in 73 patients (78.5%), and a single fimA gene was detected in most patients. The distribution of the four fimA types among the P. gingivalis-positive patients was as follows: type I, 5.4%; type II, 58.9%; type III, 6.8%; type IV, 12.3%; types I and II, 6.8%; types II and IV, 2.7%; and untypeable, 6.8%. P. gingivalis with type II fimA was detected more frequently in the deeper pockets, and a significant difference of the occurrence was observed between shallow (4 mm) and deep (≥8 mm) pockets. These results suggest that P. gingivalis strains that possess type II fimA are significantly more predominant in periodontitis patients, and we speculate that these organisms are involved in the destructive progression of periodontal diseases.
Porphyromonas gingivalis is a gram-negative, black-pigmented anaerobe associated with several periodontal diseases including adult periodontitis, generalized juvenile periodontitis, periodontal abscesses, and refractory periodontitis (5, 31). This bacterium has most frequently been detected in deep periodontal pockets and has exhibited a low prevalence in healthy periodontal tissues without destructive inflammation (3, 10, 26).
It has become clear that heterogeneity exists in terms of virulence among various P. gingivalis strains, as assayed in experimental model systems. The encapsulated cells, which induced phlegmonous abscess and/or the necrosis frequently associated with death in experimental animals, are called virulent or invasive strains, and the nonencapsulated cells that induce pus formation and/or localized abscesses are classified into nonvirulent or noninvasive strains in animal models of subcutaneous infection (9, 24, 27, 29). It should be noted, however, that contradictory findings were obtained in another model with orally infected rats (7, 13). The encapsulated strains were less pathogenic than the noncapsulated strains. The animal model of subcutaneous infection measures the tissue invasiveness of the organisms, while the other model evaluates the capability of the organisms to attach to and colonize tissues in the mouth. Little information regarding the bacterial factors that determine the prevalence and distribution of P. gingivalis in patients with marginal periodontitis is available.
Studies have been performed to determine the distribution of the specific serotypes of P. gingivalis strains. In 63 periodontitis patients, all isolates from periodontal pockets were serotypeable with antisera raised against four strains representative of each type and 79.3% of the isolates reacted with the sera against their “type I” nonencapsulated strains (21). Other investigators used capsular K antigens for the serological typing of P. gingivalis. They showed that 45.4% of 185 strains from 185 patients were six K typeable (K1 to K6) and that K5 and K6 were predominant (14).
P. gingivalis fimbriae (FimA) are filamentous components on the cell surface and are thought to play an important role in the colonization and invasion of periodontal tissues (11, 19, 25). We previously demonstrated that the fimA gene can be classified into four variants (types I to IV) on the basis of the nucleotide sequences of the fimA gene (8, 12). It has been reported that strains 381, ATCC 33277, and HG565 expressing type I fimbriae strongly adhere to host proteins (2, 22, 23). The characterization of P. gingivalis fimbriae has been performed biochemically, genetically, and immunologically; however, nearly all of these studies were done with type I FimA (fimA) only (11, 25). The prevalence of strains that possess type I FimA (fimA) in humans is unknown. In this study, we developed a PCR assay to detect the four types of the fimA gene of P. gingivalis in saliva and plaque samples from patients, and we successfully identified the most predominant fimA type.
MATERIALS AND METHODS
Bacterial strains.
P. gingivalis strains that possessed each of four fimA genotypes (12) were selected from our culture collections as follows: type I fimA, strains 381, BH18/10, and ATCC 33277; type II fimA, strains HW24D-1, OMZ314, and OMZ409; type III fimA, strains 6/26 and ATCC 49417; and type IV fimA, strains HG564 and W50. P. gingivalis strains representative of six different capsular serotypes (K types) (14, 30) were also used, i.e., strains W83 (K1), HG184 (K2), A7A1-28 (K3), ATCC 49417 (K4), HG1690 (K5), and HG1691 (K6). Organisms were grown in brain heart infusion broth as described previously (1). The specificities of the primers used were tested against the following organisms: Fusobacterium nucleatum ATCC 10953, Actinobacillus actinomycetemcomitans Y4, Prevotella intermedia ATCC 25261, Streptococcus mutans MT8148, Streptococcus sanguis ATCC 10556, and Escherichia coli NM522.
Clinical specimens.
The subjects were 93 systemically healthy Japanese adults who had marginal periodontitis associated with periodontal pocket formations deeper than 4 mm (33 males aged 19 to 74 years [mean age, 55.6 ± 11.5 years] and 60 females aged 21 to 79 years [mean age, 54.6 ± 14.8 years]) and who were referred to the Osaka University Dental Hospital for dental or periodontal treatment. The subjects had received neither professional cleaning nor an antibiotic medication within the 3 months before the study. The subjects were enrolled with informed consent. Subgingival plaque samples were taken from mesial and lingual subgingival sites of all molars with sterile Gracy curettes after supragingival plaque was gently removed. The samples were placed in sterile tubes containing 1 ml of phosphate-buffered saline (pH 7.4) on ice. The pocket probing depth was subsequently measured. The specimens from the two sites with the deepest probing depths were selected and were then vortex mixed and centrifuged at 12,000 × g for 1 min to pellet the bacterial cells. The bacterial genomic DNA was isolated from the samples with a DNA isolation kit (Puregene; Gentra Systems, Minneapolis, Minn.) according to the manufacturer’s instructions, and the isolated DNA was dissolved in 100 μl of distilled water. Expectorated whole saliva (ca. 1 ml) was also collected from each subject and placed into a sterile plastic tube on ice. The saliva samples were processed for the PCR assay by the method reported by Mättö et al. (20).
PCR primers and amplification.
Table 1 lists the PCR primers designed for this study. The fimA genotype-specific forward primers were selected from type-specific segments of nucleotide sequences of the four genotypes, and the reverse primer was common for all of the fimA types as a conserved and fimA-specific sequence. The specificities of the prospective primers were tested by the program Amplify (6), based on the DNA sequence information stored in GenBank-EMBL. A ubiquitous primer set that matches almost all bacterial 16S rRNA genes was used as a positive control (4), and P. gingivalis species-specific primers (16S rRNA specific) were used as described previously (28). The specificities and sensitivities of the two primer sets described above for the target organisms were investigated in the original studies. All of the primers were commercially synthesized (Amersham Pharmacia Biotech, Uppsala, Sweden).
TABLE 1.
fimA type-specific or 16S rRNA-specific primers used in this study
| Primer set | Sequence (5′ to 3′) | Size (bp)a |
|---|---|---|
| Universal primers for positive control | AGA GTT TGA TCC TGG CTC AG | 3,480 |
| GGC TAC CTT GTT ACG ACT T | ||
| P. gingivalis 16S rRNA | TGT AGA TGA CTG ATG GTG AAA ACC | 197 |
| ACG TCA TCC CCA CCT TCC TC | ||
| Type I fimA | CTG TGT GTT TAT GGC AAA CTT C | 392 |
| AAC CCC GCT CCC TGT ATT CCG A | ||
| Type II fimA | ACA ACT ATA CTT ATG ACA ATG G | 257 |
| AAC CCC GCT CCC TGT ATT CCG A | ||
| Type III fimA | ATT ACA CCT ACA CAG GTG AGG C | 247 |
| AAC CCC GCT CCC TGT ATT CCG A | ||
| Type IV fimA | CTA TTC AGG TGC TAT TAC CCA A | 251 |
| AAC CCC GCT CCC TGT ATT CCG A |
Expected size of PCR product.
The PCR amplification was performed in a total volume of 25 μl consisting of PCR beads (Ready-To-Go; Amersham Pharmacia Biotech), 0.8 μM each primer, and 2 to 5 μl of the template DNA solution (20 to 60 μg/ml) in sterile distilled water. The amplification reaction was performed in a thermal cycler (model 2400; Applied Biosystems, Branchburg, N.J.) with the following cycling parameters: an initial denaturation at 95°C for 5 min, 30 cycles consisting of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 7 min. Positive and negative controls were included in each PCR set and in the processing of all samples.
The PCR products were subjected to electrophoresis in a 2% agarose gel–Tris-acetate-EDTA buffer. The gel was stained with 0.5 μg of ethidium bromide per ml and photographed under UV illumination. A 100-bp DNA ladder (New England Biolabs, Beverly, MA) was used as a molecular size standard. The sensitivity of the PCR assay was studied with titrated cultures of P. gingivalis of the four fimA types (types I to IV; strains 381, HW24D-1, 6/26, and HG564, respectively; 109 cells/ml). The detection limit was determined for the simultaneous PCR by the use of known numbers of bacterial cells diluted with distilled water.
Periodontal examination.
The clinical parameters were measured by a single skilled examiner (A.A.) and included the pocket probing depth and bleeding on probing. The pocket probing depth was measured to the nearest millimeter at six points on the circumference of each tooth (mesio-, mid-, and distobuccal and disto-, mid-, and mesiolingual) from the gingival margin to the deepest probeable point with a round-ended probe tip (diameter, 0.4 mm).
Statistical analysis.
The chi-square test was used for the statistical analysis of the comparative frequencies of occurrence of the bacteria.
RESULTS
Specificity and sensitivity of the PCR assay.
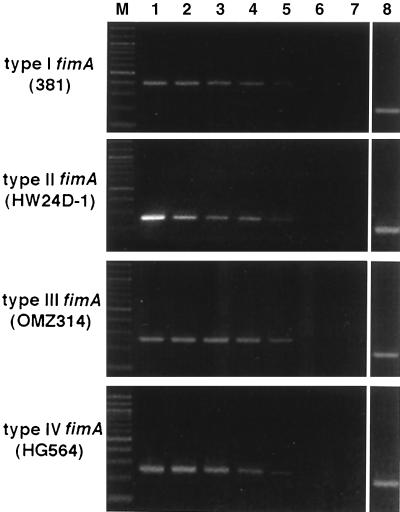
The fimA type-specific PCR products were selectively detected among strains of P. gingivalis and other species, as shown in Table 2. The positive PCR gave a single band with the expected sizes, as assessed by electrophoresis. No amplification was detected for any of the strains other than the prospective positive strains. The detection limit of the PCR assay was determined in the presence of titrated bacterial cells. As shown in Fig. 1, the PCR products were obtained as clear bands with the use of 50 cells in all fimA typings. No band was found in the lanes of PCR products with five or fewer cells including the negative control. Thus, the sensitivity of the PCR assay was shown to be between 5 and 50 cells for all fimA types. The sensitivities of the other two primer sets used for positive controls were in the same range as that of the fimA-specific primer (data not shown). No reaction was inhibited, even in the presence of 105 bacterial cells in a reaction mixture.
TABLE 2.
Specificities of primers against various target strains
| Strain | Amplification with the following primera:
|
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | Pg | U | |
| P. gingivalis | ||||||
| 381 | + | − | − | − | + | + |
| ATCC 33277 | + | − | − | − | + | + |
| BH18/10 | + | − | − | − | + | + |
| HW24D-1 | − | + | − | − | + | + |
| OMZ314 | − | + | − | − | + | + |
| OMZ409 | − | + | − | − | + | + |
| ATCC 49417 | − | − | + | − | + | + |
| 6/26 | − | − | + | − | + | + |
| HG564 | − | − | − | + | + | + |
| W50 | − | − | − | + | + | + |
| A. actinomycetemcomitans Y4 | − | − | − | − | − | + |
| F. nucleatum ATCC 10953 | − | − | − | − | − | + |
| P. intermedia ATCC 25611 | − | − | − | − | − | + |
| S. mutans MT8148 | − | − | − | − | − | + |
| S. sanguis ATCC 10556 | − | − | − | − | − | + |
| E. coli NM522 | − | − | − | − | − | + |
+, positive amplification; −, no PCR product; I to IV, fimA types; Pg, P. gingivalis species-specific 16S rRNA primers; U, universal primers for positive control.
FIG. 1.
Sensitivity of PCR assay for detection of our fimA types of P. gingivalis. The sensitivity of the PCR assay was studied with titrated cultures of P. gingivalis of four fimA types (types I to IV; 109 cells of strains 381, HW24D-1, OMZ314, and HG564, respectively, per ml). The detection limit was determined for the simultaneous PCR by the use of known numbers of bacterial cells diluted in sterile distilled water. The following numbers of cells were added: 5 × 105 (lane 1), 5 × 104 (lane 2), 5 × 103 (lane 3), 5 × 102 (lane 4), 5 × 10 (lane 5), and 5 (lane 6). Lane 7, a negative control from the PCR without any bacterial cells; lane 8, a PCR product obtained with P. gingivalis species-specific 16S rRNA primers (5 × 103 cells); lane M, molecular size marker (a 100-bp DNA ladder).
Detection of four fimA types of P. gingivalis in clinical samples.
All six of the primer sets were simultaneously subjected to the PCR assay for each clinical specimen. The plaque and/or saliva samples of all 93 patients were positive for universal primers, and positive PCR results for P. gingivalis 16S rRNA primers were observed for 71 plaque samples and 66 saliva samples from a total of 73 patients (78.5% of the total subjects).
The distributions of the four fimA types among the 71 P. gingivalis-positive patients were successfully detected from plaque and saliva samples by the primers. A single fimA gene was detected in most of the samples, and the identical type was obtained with both plaque and saliva samples from a single subject. As shown in Table 3, type II fimA was detected at the highest frequency (58.9%) in the patients, and types I, III, and IV were found at very low frequencies of 5.4, 6.8, and 12.3%, respectively. In samples from seven patients, two different fimA types were simultaneously detected, i.e., five (6.8%) patients were each infected with types I and II and 2 (2.7%) patients were each infected with types II and IV. Five samples (6.8%) gave a negative PCR result with all of the fimA-specific primers (untypeable). The total incidence of type II fimA was 68.4%, indicating that more than two-thirds of the P. gingivalis-positive patients harbored type II fimA organisms. The four sets of type-specific fimA primers provided no positive PCR result for the samples which were negative for P. gingivalis with 16S rRNA primers. This result supports the specificity of the fimA primers.
TABLE 3.
Distribution of four fimA types among P. gingivalis-positive patients: relationship to pocket depth and age-gender
| Parameter (no. of patientsa) | Frequency of occurrence (%) of the following fimA typeb:
|
||||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | I and II | II and IV | UTc | |
| P. gingivalis-positive patients (73) | 5.4 | 58.9 | 6.8 | 12.3 | 6.8 | 2.7 | 6.8 |
| Pocket probing depth (mm) | |||||||
| 4 (9) | 22.2 | 44.4 | 0 | 11.1 | 0 | 0 | 22.2 |
| 5 (19) | 5.2 | 52.6 | 10.5 | 15.8 | 5.3 | 5.3 | 5.3 |
| 6 (22) | 0 | 54.5 | 13.6 | 9.1 | 9.1 | 4.6 | 9.1 |
| 7 (12) | 0 | 66.7 | 0 | 16.6 | 16.6 | 0 | 0 |
| 8–15 (11) | 9.1 | 90.9 | 0 | 0 | 0 | 0 | 0 |
| Age (yrs) | |||||||
| Male | |||||||
| 19–35 (3) | 0 | 33.3 | 33.3 | 0 | 0 | 0 | 33.3 |
| 36–57 (14) | 0 | 57.1 | 14.2 | 21.4 | 7.1 | 0 | 0 |
| 58–74 (13) | 0 | 53.8 | 7.7 | 15.4 | 7.7 | 0 | 15.4 |
| Female | |||||||
| 24–35 (6) | 0 | 33.3 | 16.7 | 16.7 | 16.7 | 16.7 | 0 |
| 36–57 (18) | 11.1 | 83.3 | 0 | 5.6 | 0 | 0 | 0 |
| 58–79 (19) | 10.5 | 52.6 | 0 | 10.5 | 10.5 | 5.3 | 10.5 |
Number of P. gingivalis-positive patients.
Significant differences (P < 0.05) were found for the following comparisons: type II versus type I, III, IV, I-II, and II-IV, and untypeable infections; 4-mm versus 8- to 15-mm pocket probing depth male versus female sex; and age 24 to 35 years versus age 36 to 57 years for females.
UT, untypeable.
Relationship of fimA types to clinical parameters.
The relationship of some clinical parameters to the prevalence of fimA types was analyzed (Table 3). A linear relation was found between the pocket probing depth and the prevalence of type II fimA organisms, and a significant difference (P < 0.05) was obtained between the occurrence of type II organisms in shallow (4 mm) and deep (≥8 mm) pockets. In addition, the other fimA types seemed to occur more often in the pockets with shallow and moderate depths. The relationships between age/gender and the distribution of fimA types were also analyzed (Table 3). fimA type I was exclusively detected in female subjects (P < 0.05). The incidence of fimA type II was slightly greater in the middle age group (age, 36 to 57 years) than the other age groups for both the males and females, yet, a significant difference was observed only between the younger and middle age groups for females (P < 0.05).
Bleeding on probing was found in 77 patients (86%). There was no clear relationship between the patients with bleeding on probing and the prevalence of fimA.
fimA types of K-serotypeable strains.
The representative K-serotype strains were used in a PCR assay to determine their fimA types. Most of the K-typeable strains (the exceptions were the K1 and K6 strains) were shown to possess type II fimA, as follows: W83 (K1), type IV fimA; HG184 (K2), type II; A7A1-28 (K3), type II; ATCC 49417 (K4), type II; HG1690 (K5), type II; and HG1691 (K6), untypeable.
DISCUSSION
This is the first report of an investigation on the prevalence of P. gingivalis fimA genotypes in saliva and plaque samples from periodontitis patients. The present assay with the designed fimA type-specific primer sets gave clear PCR products, and the assay had sufficient sensitivities and specificities. The results obtained in the present study demonstrate that P. gingivalis strains that possess the type II fimA gene are most predominantly present in the oral flora of the periodontitis patients and that the type II fimA organisms might be involved in the etiology of advanced periodontitis. To date, only limited numbers of P. gingivalis strains have been shown to possess type II fimA, e.g., strains HW24D1, OMZ314, OMZ409, A7A2-10, AJW4, THUR28BM2, AJW3, JKG7, and A7A1-28 (ATCC 53977) (12, 16). A wide variety of studies regarding the adhesive functions and immunobiological activities of fimbriae have been performed (11). These studies suggested that P. gingivalis fimbriae are major virulence factors and are possible candidates for use in a vaccine. However, most investigators have dealt with type I FimA (fimA) in their studies; few studies with other types of fimbriae have been performed. Although the four types of fimbriae were previously purified from the organisms (15), no characterization of the functional heterogeneity of fimbriae has been reported yet. It should be noted that type II fimA strain A7A1-28 reportedly induced necrosis with death in an animal model (24), and strains of the virulent K types (types K2 to K5) also possess type II fimA, as revealed in this study. The virulent and invasive strains are all encapsulated, and these encapsulated strains were reported to be less adhesive to host proteins such as collagen types I and IV, fibronectin, laminin, and salivary proteins (2, 22, 23). To clarify the ecological factors that determine the colonization ability of P. gingivalis in humans, the characterization of the most prevalent type of fimbriae is required. It is also necessary to determine whether various type II fimA strains in the patients are virulent in animal models.
The prevalence and distribution of six K-typeable strains were previously examined by Laine et al. (14) in periodontitis patients, and the following prevalence ratios were reported: K1, 3.8%; K2, 2.2%; K3, 1.1%; K4, 3.2%; K5, 12.0%; and K6, 23.2%. The combined findings of their report and our own results suggest that type II fimA P. gingivalis may carry other antigens. Another serological study was performed with four different antisera against the reference strains to investigate the prevalence of serotypes of P. gingivalis in periodontitis patients (21). All of the isolated P. gingivalis strains were typeable as one of these four serotypes; 44.4% serotype I strains were reactive with anti-strain 381 serum, 11.1% serotype II strains were reactive with anti-JH4 serum, 6.3% serotype III strains were reactive with anti-W50 or anti-W83 serum, and 34.9% serotype IV strains were reactive with anti-ATCC 33277 serum. The serotype I and IV strains in that study possessed type I fimA, while the serotype III strains had type IV fimA. The fimA type of the serotype II strains was unknown. These results indicate that type I fimA strains are more frequently detected in periodontitis patients, which is not in agreement with our present findings. These serological studies were performed by the culture technique, and the method is not sensitive enough. The PCR assay can be expected to be more sensitive and specific than the culture method, as reported previously (4, 17).
Our present findings indicated the involvement of type II fimA strains in advanced periodontitis. The type II strains are also predominant in the middle age group, especially in females. Thus, it might be possible that the periodontitis patients infected or colonized with type II fimA strains could lose their dentition earlier due to the destructive nature of the harbored strains compared to the time to the loss of dentition for patients infected or colonized with other fimA type strains.
Five of the patients in the present study were shown to harbor fimA untypeable strains, indicating that more than four genotypes may exist in P. gingivalis. A restriction fragment length polymorphism (RFLP) analysis showed that the majority of fimA loci of 38 P. gingivalis strains could be classified into four groups, and the other seven strains were divided into three minor groups with Southern blots probed with fimA of strain 381 (18). Although those results may reflect the diversities of fimA flanking regions, they might also support the existence of other fimA genotypes. The P. gingivalis strains divided in the major four RFLP groups probably correspond to those classified by present fimA genotypes I to IV. The minor three RFLP groups might indicate the unknown fimA types. The cloning and sequencing of novel fimA genes isolated from the untypeable samples are under investigation in our laboratories.
ACKNOWLEDGMENTS
We thank A. J. van Winkelhoff (ACTA, Vrije University, Amsterdam, The Netherlands) for his generous gift of K-type representative strains. We also thank S. Shizukuishi, S. Akiyama, H. Daikoku, T. Kato, Y. Mori, T. Murayama, T. Kishima, and M. Kuboniwa (Osaka University Dental Hospital) for help in the collection of clinical samples.
This work was supported by grants-in-aid C-10671933 and C-10671934 from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Shizukuishi S. Plaque formation: involvement of salivary protein molecules. Dent Outlook. 1996;87:217–227. . (In Japanese.) [Google Scholar]
- 3.Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Conrads G, Mutters R, Fischer J, Brauner A, Lutticken R, Lampert F. PCR reaction and dot-blot hybridization to monitor the distribution of oral pathogens within plaque samples of periodontally healthy individuals. J Periodontol. 1996;67:994–1003. doi: 10.1902/jop.1996.67.10.994. [DOI] [PubMed] [Google Scholar]
- 5.Darveau R P, Tanner A, Page R C. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Engels W R. Contributing software to the internet: the Amplify program. Trends Biochem Sci. 1993;18:448–450. doi: 10.1016/0968-0004(93)90148-g. [DOI] [PubMed] [Google Scholar]
- 7.Evans R T, Klausen B, Ramamurthy N S, Golub L M, Sfintescu C, Genco R J. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch Oral Biol. 1992;37:813–819. doi: 10.1016/0003-9969(92)90115-o. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara T, Nakagawa I, Morishima S, Takahashi I, Hamada S. Inconsistency between the fimbrillin gene and the antigenicity of lipopolysaccharides in selected strains of Porphyromonas gingivalis. FEMS Microbiol Lett. 1994;124:333–342. doi: 10.1111/j.1574-6968.1994.tb07305.x. [DOI] [PubMed] [Google Scholar]
- 9.Genco C A, Cutler C W, Kapczynski D, Maloney K, Arnold R P A. Novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect Immun. 1991;59:1255–1263. doi: 10.1128/iai.59.4.1255-1263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffen A L, Becker M R, Lyons S R, Moeschberger M L, Leys E J. Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol. 1998;36:3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and etiology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–138. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 12.Hamada S, Fujiwara T, Morishima T, Takahashi I, Nakagawa I, Kimura S, Ogawa T. Molecular and immunological characterization of the fimbriae of Porphyromonas gingivalis. Microbiol Immunol. 1994;38:921–930. doi: 10.1111/j.1348-0421.1994.tb02148.x. [DOI] [PubMed] [Google Scholar]
- 13.Klausen B, Evans R T, Genco R J. Vaccination against P. gingivalis in experimental animals. In: Shah N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 341–349. [Google Scholar]
- 14.Laine M L, Appelmelk B J, van Winkelhoff A J. Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J Dent Res. 1997;76:1840–1844. doi: 10.1177/00220345970760120601. [DOI] [PubMed] [Google Scholar]
- 15.Lee J-Y, Sojar H T, Amano A, Genco R J. Purification of major fimbrial proteins of Porphyromonas gingivalis. Protein Expr Purif. 1995;6:496–500. doi: 10.1006/prep.1995.1066. [DOI] [PubMed] [Google Scholar]
- 16.Lee J-Y, Sojar H T, Bedi G S, Genco R J. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun. 1991;59:383–389. doi: 10.1128/iai.59.1.383-389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loesche W J. DNA probe and enzyme analysis in periodontal diagnostics. J Periodontol. 1992;63:1102–1106. doi: 10.1902/jop.1992.63.12s.1102. [DOI] [PubMed] [Google Scholar]
- 18.Loos B G, Dyer D W. Restriction fragment length polymorphism analysis of the fimbrillin locus, fimA, of Porphyromonas gingivalis. J Dent Res. 1992;71:1173–1181. doi: 10.1177/00220345920710050901. [DOI] [PubMed] [Google Scholar]
- 19.Malek R, Fisher J G, Caleca A, Stinson M, VanOs C J, Lee J-Y, Cho M-I, Genco R J, Evans R T, Dyer D W. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mättö J, Saarela M, Alaluusua S, Oja V, Jousimies-Somer H, Asikainen S. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J Clin Microbiol. 1998;36:157–160. doi: 10.1128/jcm.36.1.157-160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagata A, Man-yoshi T, Sato M, Nakamura R. Serological studies of Porphyromonas (Bacteroides) gingivalis and correlation with enzyme activity. J Periodontal Res. 1991;26:184–190. doi: 10.1111/j.1600-0765.1991.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 22.Naito Y, Tohda H, Okuda K, Takazoe I. Adherence and hydrophobicity of invasive and noninvasive strains of Porphyromonas gingivalis. Oral Microbiol Immunol. 1993;8:195–202. doi: 10.1111/j.1399-302x.1993.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 23.Naito Y, Gibbons R J. Attachment of Bacteroides gingivalis to collagenous substrata. J Dent Res. 1988;67:1075–1080. doi: 10.1177/00220345880670080301. [DOI] [PubMed] [Google Scholar]
- 24.Neiders M E, Chen P B, Suido H, Reynolds H S, Zambon J J, Shlossman M, Genco R J. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–198. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 25.Okuda K. Attachment mechanisms and colonization. In: Shah H N, Mayrand D, Genco R J, editors. Biology of the species Porphyromonas gingivalis. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 139–158. [Google Scholar]
- 26.Riviere G R, Smith K S, Tzagaroulaki E, Kay S L, Zhu X, DeRouen T A, Adams D F. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J Periodontol. 1996;67:109–115. doi: 10.1902/jop.1996.67.2.109. [DOI] [PubMed] [Google Scholar]
- 27.Sundqvist G, Figdor D, Hanstrom L, Sorlin S, Sandstrom G. Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand J Dent Res. 1991;99:117–129. doi: 10.1111/j.1600-0722.1991.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 28.Tran S D, Rudney J D. Multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol. 1996;34:2674–2678. doi: 10.1128/jcm.34.11.2674-2678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Steenbergen T J, Delemarre F G, Namavar F, De Graaff J. Differences in virulence within the species Bacteroides gingivalis. Antonie Leeuwenhoek. 1987;53:233–244. doi: 10.1007/BF00393930. [DOI] [PubMed] [Google Scholar]
- 30.van Winkelhoff A J, Appelmelk B J, Kippuw N, de Graaff J. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol. 1993;8:259–265. doi: 10.1111/j.1399-302x.1993.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 31.Zambon J J. Microbiology of periodontal disease. In: Genco R J, Goldman H M, Cohen D W, editors. Contemporary periodontics. St. Louis, Mo: The C. V. Mosby Co.; 1990. pp. 147–160. [Google Scholar]