Abstract
Background
Alzheimer's Disease (AD) is a complex and multifactorial disease and novel approaches are needed to illuminate the underlying pathology. Metabolites comprise the end-product of genes, transcripts, and protein regulations and might reflect disease pathogenesis. Blood is a common biofluid used in metabolomics; however, since extracellular vesicles (EVs) hold cell-specific biological material and can cross the blood-brain barrier, their utilization as biological material warrants further investigation. We aimed to investigate blood- and EV-derived metabolites to add insigts to the pathological mechanisms of AD.
Methods
Blood samples were collected from 10 AD and 10 Mild Cognitive Impairment (MCI) patients, and 10 healthy controls. EVs were enriched from plasma using 100,000×g, 1 h, 4 °C with a wash. Metabolites from serum and EVs were measured using liquid chromatography-mass spectrometry (LC-MS) and nuclear magnetic resonance (NMR) spectroscopy. Multivariate and univariate analyses were employed to identify altered metabolites in cognitively impaired individuals.
Results
While no significant EV-derived metabolites were found differentiating patients from healthy individuals, six serum metabolites were found important; valine (p = 0.001, fold change, FC = 0.8), histidine (p = 0.001, FC = 0.9), allopurinol riboside (p = 0.002, FC = 0.2), inosine (p = 0.002, FC = 0.3), 4-pyridoxic acid (p = 0.006, FC = 1.6), and guanosine (p = 0.004, FC = 0.3). Pathway analysis revealed branched-chain amino acids, purine and histidine metabolisms to be downregulated, and vitamin B6 metabolism upregulated in patients compared to controls.
Conclusion
Using a combination of LC-MS and NMR methodologies we identified several altered mechanisms possibly related to AD pathology. EVs require additional optimization prior to their possible utilization as a biological material for AD-related metabolomics studies.
Keywords: Alzheimer, Metabolites, Blood, Extracellular vesicles, Mass spectrometry, Nuclear magnetic resonance
Abbreviations: Aβ, Amyloid-β; ACE, Addenbrooke's cognitive examination; AD, Alzheimer's Disease; AUC, Area under the curve; BBB, Blood-brain barrier; BCAA, Branched-chain amino acid; CNS, Central nervous system; CSF, Cerebrospinal fluid; CV, Cross-validation; EVs, Extracellular vesicles; FAQ, Functional activities questionnaire; FDR, False discovery rate; MMSE, Mini-mental state examination; MCI, Mild cognitive impairment; PCA, Principal component analysis; p-tau, Phospho-tau; ROC, Receiver operating characteristics; sPLS-DA, Sparse partial least squared discriminant analysis; t-tau, Total-tau
1. Introduction
Alzheimer's Disease (AD) is a chronic neurodegenerative disease and comprises the largest part of dementia subtypes [1]. The amyloid hypothesis has for a long time been the main focus in AD research and the development of therapeutic interventions [2]. However, clinical trials are continuously failing [3], which has led researchers on new paths to uncover the complexity of this multifactorial disease [4]. Furthermore, with the current diagnostic methods, such as positron emission tomography and cerebrospinal fluid (CSF) proteins, some limitations hinder their use as first-line diagnostic or even screening tools. CSF provides for a biological fluid in close contact with the brain parenchyma, allowing identification of brain-related molecules in higher concentrations compared to that in peripheral blood samples. This biofluid may be useful for the analysis of brain metabolic alteration [5], but limitations associated with scanning methods and CSF sampling could be circumvented by blood-based biomarkers, as a minimally invasive diagnostic tool [6]. Blood is a versatile body fluid being in close connection with every organ, thus potentially reflecting their state [7]. The blood-brain barrier (BBB) becomes disrupted during AD pathogenesis, potentially allowing brain metabolites to be reflected in a blood sample [8].
With blood providing a complex matrix for biomarker investigations and AD being a multifactorial disease, integration of in-depth technologies and large data structures are needed. The term “systems biology” describes the understanding of the biological system as a whole, rather than paying attention to single factors in disease pathologies. This analytical power was realized by the omics-era [9]. Metabolomics is one of the newest fields in the omics family [9] being the study and exploration of all metabolites (<1,500 Da) in a cell, organ, or organism, and comprises lipids, amino acids, vitamins, peptides, and minerals, among others [10]. Since metabolites are the endpoints of genes, transcripts, and protein regulations, minor changes in the level of the upstream molecules can cause significant alterations in metabolites [11]. Not only disease progression can cause such changes, but also medication, nutrition, and environmental factors can affect metabolites [12]. Lastly, metabolic pathways are evolutionarily conserved across species, making them ideal targets for clinical studies [13].
The two most common methods for metabolomics studies are mass spectrometry (MS)-based metabolomics and nuclear magnetic resonance (NMR) spectroscopy [14]. Utilizing these two methods in combination can overcome several of their limitations, thus providing greater coverage of the metabolome, with MS-based metabolomics identifying low abundance metabolites and NMR identifying core metabolites in key metabolic pathways [12,13]. Interestingly, perturbations in metabolic pathways have been shown to be one of the first measurable changes to occur before manifestations of clinical symptoms [15]. Several studies have also examined metabolic alterations in AD, presenting different metabolic panels specific for AD [[16], [17], [18]].
Extracellular vesicles (EVs) are nano-sized particles, surrounded by a lipid-bilayer, and packed with active biomolecules such as proteins, lipids, and genetic material [19]. They are released by all cell types, including cells present within the central nervous system (CNS) [20], and hence, are important players in intercellular communication in physiological and pathological conditions. EVs can be identified in a wide variety of biofluids, including blood [19]. Several studies have investigated their role in AD in relation to the spreading [21] and clearing [22] of amyloid-β (Aβ). In addition, EVs have been shown to be able to bypass the BBB through various suggested mechanisms [23]. These features of EVs as potential sources of biomarkers have included them in the term “liquid biopsies” [24]. With both metabolomics and EVs being relatively new fields, the combination of the two in search for biomarkers is therefore also scarce. Few studies have examined the metabolome of EVs, mostly focused on biomarkers for various cancer types [25]. To the knowledge of the authors, no study has yet explored EV-derived metabolites in AD.
Therefore, this study aimed to explore metabolic perturbations related to pathological changes in AD, through the combined effort of MS- and NMR-based metabolomics approaches. Both serum and EV-derived metabolites were examined from patients with AD or Mild Cognitive Impairment (MCI) and compared to that of healthy controls. We identified several interesting metabolites distinguishing between diseased and healthy individual samples. Also, serum seemed to be a more suitable biological matrix for studying metabolic changes in AD; however, further optimization is needed for EVs in order to become a biological matrix of choice for future metabolomics studies.
2. Methods
2.1. Study participants
A total of 30 participants were enrolled for this study, distributed in three groups with 10 AD patients, 10 MCI patients, and 10 healthy controls. The patient groups were consecutively included from the Department of Neurology at Aalborg University Hospital. Study inclusion was at their time of diagnosis and blood samples were drawn prior to initiation of treatment. The diagnosis of mild to moderate AD patients was based on the International Classification of Diseases and Related Health Problems 10th Edition (ICD10) criteria [26] and the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) [27]. For MCI patients, the diagnosis was made based on the Petersen criteria [28]. Paraclinical measurements of patients were performed when necessary for diagnostic certainty and included the Mini-Mental State Examination (MMSE), Addenbrooke's Cognitive Examination (ACE), Function Activities Questionnaire (FAQ), CSF Aβ, CSF phospho-tau (p-tau), and CSF total-tau (t-tau).
For comparison with patient groups, age- and sex-related donors were recruited from the blood bank at Aalborg University Hospital. Donors were required to be of age 65 or older and complete a questionnaire regarding physical and mental health, stating information about i.e. fatigue, chest pain, and memory impairment. All included participants signed a consent form prior to study inclusion. The study was conducted in accordance with the Declaration of Helsinki and approved by the local North Denmark Region Committee on Health Research Ethics (N-20150010).
2.2. Sample collection and routine analyses
Collection and handling of blood samples were performed as previously described [29]. Briefly, using the median cubital vein as access point and a 21-gauge needle, blood samples (plasma and serum) were collected in 9 mL 0.105 M (3.2%) trisodium citrate (Vacuette, Greiner Bio-One, Austria) and 10 mL clot activator tubes (BD Vacutainer, UK). After blood collection, samples were subjected to double centrifugation at 2,500×g for 15 min at room temperature to obtain either platelet-free plasma or serum. Plasma and serum were aspirated until 1 cm above the buffy coat or pellet. Aliquots of plasma and serum samples were snap-frozen in liquid nitrogen and stored at – 80 °C until further processing.
In addition, routine analyses were applied as previously described to ensure that study participants presented with no co-morbidities [29]. Briefly, measurements of normal system functioning and markers of organ function and damage were investigated by; alanine transaminase, albumin, carbamide, cholesterol, creatinine, C-reactive protein, glucose, high and low-density lipoprotein, haemoglobin, lactate dehydrogenase, and triglycerides.
2.3. Enrichment of extracellular vesicles
For MS, EVs were enriched from 1 mL plasma. A two-step centrifugation process at 100,000×g for 1 h at 4 °C was performed using an Avanti J-30i centrifuge together with a J A-30.50 fixed angle rotor, k-factor 280 (Beckman Coulter, Brea, CA, USA). In-between centrifugations pellets were washed with 1 mL 0.22 μm filtered buffer (10 mM ammonium acetate in HPLC grade water). The resulting final EV pellets were resuspended in 100 μL of the same buffer. For NMR analysis, EVs were enriched from 1 mL plasma in triplicates for one sample per group. A two-step centrifugation process at 100,000×g for 1 h at 4 °C was performed using a LKB 2331 Ultrospin 70 (LKB, Bromma, Sweden). In-between centrifugations pellets were washed with 1 mL 0.22 μm filtered phosphate-buffered saline. The resulting final EV pellets were resuspended in 150 μL of the same buffer, and due to the dilution of samples and relatively high metabolite concentration requested for NMR analysis (>1 μM), samples were pooled resulting in three samples in total, one EV isolate from each group. Furthermore, we have previously characterised pellets of EVs from the same enrichment, thus comfirming their presence in our samples [30].
2.4. Mass spectrometry analysis
For MS-based metabolomics analysis, both serum and EV samples were investigated. Samples were thawed on ice and four times volume extraction solvent was added, followed by vigorous vortexing. For serum, the extraction solvent comprised of methanol/acetonitrile/H2O (5:3:2), and for EVs methanol/acetonitrile (5:3) was used. Samples were then centrifuged at 16,000×g for 15 min at 4 °C. The supernatant was aspirated, lyophilized, and stored at – 20 °C. Prior to liquid chromatography-mass spectrometry (LC-MS) analysis, samples were dissolved in 0.1% formic acid (30 μL) and centrifuged at 16,000×g for 5 mintutes at room temperature.
The samples were analysed as in Dall et al. [31]. In brief, 5 μL was injected using a 400 μl/min and the following composition of eluent A (0.1% formic acid) and eluent B (0.1% formic acid, acetonitrile) solvents: 3% B from 0 to 1.5 min, 3%–40% B from 1.5 to 4.5 min, 40%–95% B from 4.5 to 7.5 min, 95% B from 7.5 to 10.1 min and 95%–3% B from 10.1 to 10.5 min before equilibration for 3.5 min with the initial conditions. The flow from the UPLC was coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) for mass spectrometric analysis in both positive and negative ion modes. The raw data were also processed as in Dall et al. [31] using MZmine (v 2.53) [32].
2.5. Nuclear magnetic resonance spectroscopy analysis
NMR spectroscopy was used to investigate serum and EV-derived metabolites. Samples were thawed for 30 min at 4 °C, vortexed, and centrifuged at 12,100×g for 5 min at 4 °C using a multifuge 3 S-R centrifuge (Heraeus, Hanau, Germany). A total of 400 μL serum supernatant or EV isolate was mixed with 200 μL buffer (0.2 M NaPO4, 99% D2O, pH 7.4), as previously described [33,34]. Samples were aliquoted in 5 mm NMR tubes and kept on ice until analysed. A PULCON sample consisting of glucose and buffer was used as an internal standard.
A Bruker AVANCE 800 MHz NMR spectrometer (Bruker BioSpin, Rheinstetten, Germany) equipped with a cryogenically cooled, triple-resonance (1H, 13C, 15N) CPP-TCI probe, and operated at 298.1 K (25 °C) was used to record 1H NMR spectra. T2 filtered Carr-Purcell-Meiboom-Gill (CPMG) experiments with water presaturation were obtained using the following parameters; 65536 data points covering a spectral width of 20 ppm using 256 scans for serum and 128 scans for plasma (EV) samples, with 32 dummy scans, a fixed receiver gain of 203, and a relaxation delay (D1) of 4 s. Presaturation of the water resonance was achieved during D1 by continuous irradiation at γB1/2π = 25 Hz. T2 filtering was then performed with a τ-180°-τ (τ = 300 μs) pulse sequence, which was repeated 256 times for 80 ms. The TopSpin 3.1 software (Bruker BioSpin, Rheinstetten, Germany) was used for spectral acquisition and processing, including enhancement of spectral resolution using artificial zero-filling by adding digital data points to the free induction decays, line broadening (0.3 Hz), Fourier transformation, phase and baseline correction, and calibration to the l-alanine methyl peak (1.48 ppm), as previously described [35]. Metabolite annotation was performed using 2D 1H–1H total correlation spectroscopy and 1H–13C heteronuclear single-quantum correlation spectra, the Human Metabolome Database (HMDB) [36], and literature [33,37,38], while quantification was based on integrating the sum of all points within a signal of interest, as previously described [38].
2.6. Data analysis
For LC-MS, in serum and EV samples a total of 130 and 65 features were annotated on MS2 level, respectively and were subsequently corrected for signal drift using the statTarget R package [39]. For NMR spectroscopy a total of 38 metabolites were identified. Prior to multivariate analysis, data were generalized log-transformed and auto-scaled using MetaboAnalyst 5.0 (Xia Lab, Quebec, Canada) [40]. Supervised sparse-partial least squared discriminant analysis (sPLS-DA) was used to detect metabolites related to cognitive impairments. A five-fold cross-validation (CV) repeated 100 times was employed. Scores plots of sample grouping and loadings plots of selected metabolites are presented. Receiver operating characteristics (ROC) curves for group discrimination were created based on the CV adjusted models. The area under the curve (AUC) and 95% confidence intervals (CI) were used to report the sensitivity and specificity of the models. Multivariate analysis was performed using the mixOmics R package [41]. R script and files for sPLS-DA can be accessed in Supplementary Materials File S1 and Table S1 – S3.
Correlations were performed for metabolites identified by both MS and NMR to assess methods' compatibility using Pearson's ρ (Supplementary Material File Fig. S1). Data are presented as means ± standard deviations (SD). Group comparisons were performed using analysis of variance (ANOVA) with Tukey's honestly significant difference (HSD) post hoc test in IBM SPSS Statistics 27 (SPSS, Chicago, IL, USA). Benjamini-Hochberg false discovery rate (FDR) was applied for multiple correction. Fold changes (FC) were calculated for between-group comparisons. A p-value < 0.05 was considered statistically significant. GraphPad Prism 9.1.1 (GraphPad Software, La Jolla, CA, USA) was used for data visualization.
Network analysis was conducted using MetScape version 3.1.3 [42] for serum metabolites differentiating AD patients and healthy individuals. Networks were build using KEGG IDs. Log2 FC was used to indicate whether the affected metabolic pathways were altered in diseased compared to healthy individuals. Tables for MetScape analysis can be accessed in Supplementary Material File Table S4.
3. Results
3.1. Subject characteristics
The measured biochemical parameters, test results of cognitive performances, and paraclinical measurements have been presented in earlier studies by Nielsen et al. [29,30]. Briefly, most biochemical measurements were within standard reference intervals, however, with few individuals presenting elevated levels of LDL cholesterol and triglycerides. A small but significant difference in age was found between healthy and diseased individuals (p = 0.005). AD patients presented with significantly lower scores of cognitive testing based on MMSE (p = 0.04) and ACE (p = 0.007) tests, and higher scores on the FAQ test, compared to MCI patients. For paraclinical measurements of CSF markers, AD patients were observed to have slightly lower levels of Aβ and p-tau and higher levels t-tau (Table 1).
Table 1.
Demographics and clinical data of study groups. The presented values are shown as mean ± standard deviation. Abbreviations; Aβ: amyloid-β, ACE: Addenbrooke's Cognitive Examination, AD: Alzheimer's Disease, Con: healthy controls, CSF: cerebrospinal fluid, FAQ: Functional Activities Questionnaire, MCI: Mild Cognitive Impairment, MMSE: Mini-Mental State Examination, p-tau: phospho-tau, t-tau: total-tau. *Ages 51–70. Interval <500 for ages 71–90.
| Con (n = 10) | MCI (n = 10) | AD (n = 10) | p-value | Reference interval | |
|---|---|---|---|---|---|
| Demographics | |||||
| Female/male (n) | 6/4 | 8/2 | 6/4 | – | – |
| Age (years) | 65 ± 0.5 | 72 ± 5 | 70 ± 5 | 0.005 | – |
| Cognitive performance | |||||
| ACE | – | 85.0 ± 5.6 (n = 6) | 58.7 ± 16.5 (n = 3) | 0.007 | – |
| FAQ | – | 4.0 ± 2.0 (n = 3) | 10.4 ± 4.6 (n = 5) | 0.066 | – |
| MMSE | – | 27.4 ± 2.3 | 23.6 ± 4.6 | 0.041 | – |
| Paraclinical measurements | |||||
| CSF Aβ | – | 998. 5 ± 428.6 (n = 4) | 626.3 ± 260.9 (n = 6) | 0.148 | > 500 |
| CSF t-tau | – | 563.0 ± 363.9 (n = 4) | 628.2 ± 288.9 (n = 6) | 0.760 | < 61 |
| CSF p-tau | – | 98.0 ± 61.3 (n = 4) | 80.5 ± 29.5 (n = 6) | 0.556 | < 450* |
3.2. Metabolic signatures of cognitive impairments
For this study, serum samples were measured by both LC-MS and NMR, while EVs only by LC-MS due to samples being too diluted for NMR, resulting in few signals obtained on the NMR spectrum (Supplementary Material File Fig. S2).
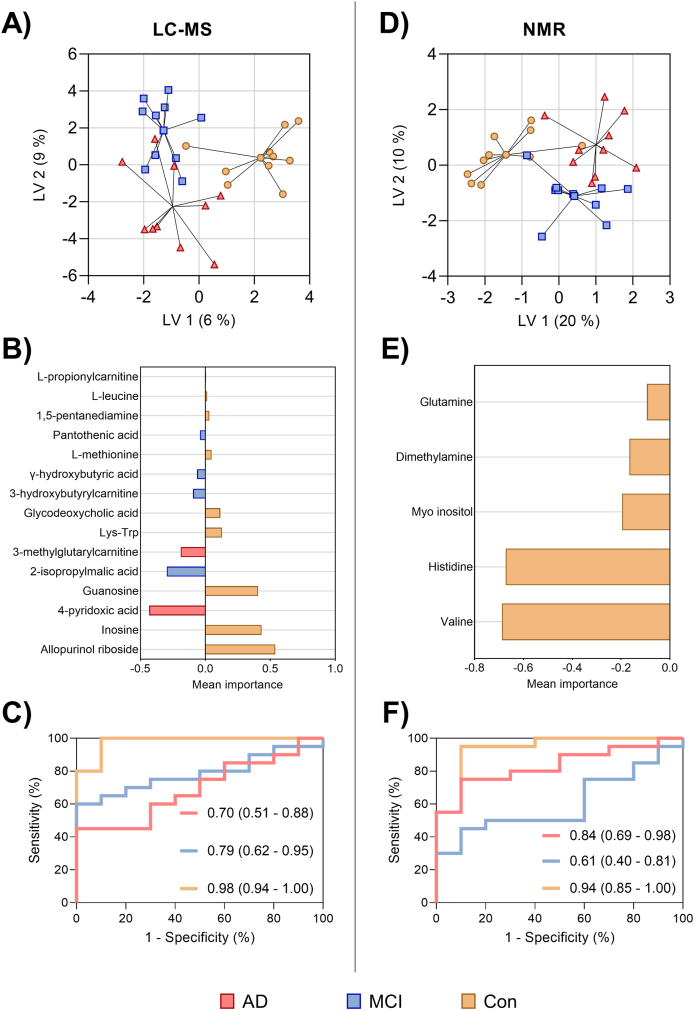
To facilitate the identification of possible metabolites associated with cognitive impairment sPLSDA was performed. A small overlap across the groupings could be observed for the serum samples measured by LC-MS and NMR (Fig. 1A – F). Among the 130 measured metabolites by LC-MS, 15 were found discriminating between groups by using 2 latent variables, with a classification error of 0.56 (Fig. 1A and B) and an AUC = 0.70 (95% CI = 0.51–0.88) for AD patients compared to MCI patients and controls, an AUC = 0.79 (95% CI = 0.62–0.95) for the MCI group compared to AD patients and controls, and an AUC = 0.98 (95% CI = 0.94–1.00) for healthy controls compared to patient groups (Fig. 1C). For NMR, five metabolites were found significantly contributing to sample grouping (Fig. 1D and E), with a classification error rate of 0.50, and an AUC = 0.84 (95% CI = 0.69–0.98) for AD compared to the MCI and control groups, an AUC = 0.61 (95% CI = 0.40–0.81) for MCI compared to the AD and healthy individuals, and an AUC = 0.94 (95% CI = 0.85–1.00) for healthy controls compared to patient groups (Fig. 1F). A less distinct separation was observed for the EV samples with a greater classification error rate of 0.62 (Supplementary Material File Fig. S3), indicating lower accuracy of EV-based metabolites in distinguishing patients from healthy individuals.
Fig. 1.
Metabolic signatures related to cognitive impairment through MS- and NMR-based approaches. Sparse-partial least squared discriminant analysis (sPLS-DA) models for serum metabolites, together with receiver operating characteristics (ROC) curves for each of the models. For LC-MS serum samples (A) scores plot, (B) loadings plot, and (C) ROC curves were shown. For NMR serum samples (D) scores plot, (E) loadings plot, and (F) ROC curves were shown. Each score represents a sample and the loadings represent the variation in a specific metabolite. The size of the bars indicates their importance for the sample grouping, and the color-coding indicate their importance for one of the groups. Based on the selected metabolites, the ROC curves compare their ability to distinguish the three study groups from each other for the datasets. The area under the curve (AUC) and the respective 95% CI are presented for each of the ROC curves. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
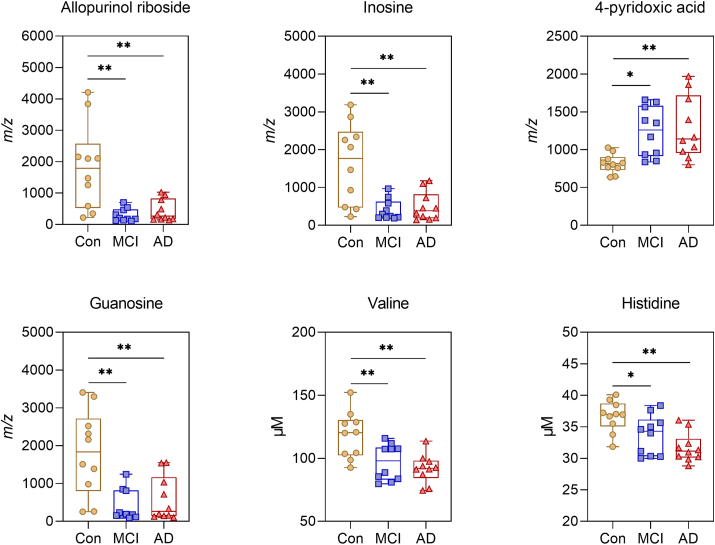
Several serum metabolites were found to be significantly altered, with 6 of these being significant after FDR correction (Table 2) for LC-MS analysis. For NMR, seven metabolites were significantly altered, with three of them being significant after FDR correction (Table 3). In contrast, EV samples only showed 5 significantly altered metabolites, and none of them being significant after FDR correction (Supplementary Material File Table S5). Fig. 2 depicts the most important serum metabolites found to differentiate between groups, including allopurinol ribosine, inosine, 4-pyridoxic acid, guanosine, valine, and histidine.
Table 2.
21 differentially altered metabolites in serum samples measured by mass spectrometry comparing all groups. Abbreviations; AD: Alzheimer's Disease, ANOVA: analysis of variance, Con: healthy controls, FC: fold change, FDR: false discovery rate, MCI: Mild Cognitive Impairment.
| Metabolites |
Mean ± SD [Intensities] |
ANOVA | FDR |
AD |Con |
MCI |Con |
AD |MCI |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Con | MCI | AD | FC | p | FC | p | FC | p | |||
| 1-Pentadecanoyl-sn-glycero-3-phosphocholine | 970.2 ± 282.5 | 1249.8 ± 478.9 | 507.9 ± 210.0 | 0.0004 | 0.020 | 0.5 | 0.021 | 1.3 | 0.213 | 0.4 | 0.0003 |
| Allopurinol riboside | 1829.6 ± 1293.6 | 296.5 ± 193.8 | 444.4 ± 330.8 | 0.0004 | 0.020 | 0.2 | 0.002 | 0.2 | 0.001 | 1.5 | 0.915 |
| Inosine | 1626.3 ± 1014.0 | 404.7 ± 258.1 | 496.4 ± 361.9 | 0.0005 | 0.020 | 0.3 | 0.002 | 0.2 | 0.001 | 1.2 | 0.950 |
| Guanosine | 1809.1 ± 1069.5 | 401.2 ± 387.7 | 590.5 ± 556.7 | 0.0006 | 0.020 | 0.3 | 0.004 | 0.2 | 0.001 | 1.5 | 0.848 |
| 1-Palmitoyl-sn-glycero-3-phosphocholine | 997.1 ± 246.8 | 1109.8 ± 283.7 | 625.4 ± 222.1 | 0.001 | 0.025 | 0.6 | 0.011 | 1.1 | 0.615 | 0.6 | 0.001 |
| 13-cis-Retinol | 876.4 ± 207.2 | 1244.9 ± 467.0 | 590.3 ± 293.7 | 0.001 | 0.032 | 0.7 | 0.194 | 1.4 | 0.073 | 0.5 | 0.001 |
| 4-Pyridoxic acid | 815.4 ± 119.9 | 1234.5 ± 309.8 | 1288.6 ± 393.5 | 0.004 | 0.074 | 1.6 | 0.006 | 1.5 | 0.016 | 1.0 | 0.921 |
| 1-Myristoyl-sn-glycero-3-phosphocholine | 1170.2 ± 527.6 | 1198.8 ± 534.9 | 548.9 ± 271.9 | 0.009 | 0.139 | 0.5 | 0.021 | 1.0 | 0.991 | 0.5 | 0.016 |
| 2-Isopropylmalic acid | 769.4 ± 209.3 | 1164.1 ± 319.1 | 1135.1 ± 311.0 | 0.011 | 0.147 | 1.5 | 0.029 | 1.5 | 0.017 | 1.0 | 0.974 |
| Leu-Leu | 842.5 ± 223.6 | 946.4 ± 319.7 | 1366.4 ± 488.3 | 0.011 | 0.147 | 1.6 | 0.013 | 1.1 | 0.816 | 1.4 | 0.051 |
| 7-Methylguanine | 1021.0 ± 141.6 | 1040.4 ± 165.1 | 1308.5 ± 298.9 | 0.013 | 0.156 | 1.3 | 0.021 | 1.0 | 0.980 | 1.3 | 0.033 |
| 2-Phenylethanol,sulfate | 724.1 ± 390.3 | 845.6 ± 503.7 | 1570.6 ± 882.6 | 0.017 | 0.164 | 2.2 | 0.021 | 1.2 | 0.912 | 1.9 | 0.054 |
| 1-Oleoyl-sn-glycero-3-phosphocholine | 971.3 ± 289.9 | 1070.1 ± 270.7 | 674.4 ± 289.6 | 0.017 | 0.164 | 0.7 | 0.086 | 1.1 | 0.742 | 0.6 | 0.017 |
| l-Lysine | 1138.1 ± 312.3 | 795.2 ± 150.8 | 1085.9 ± 280.5 | 0.019 | 0.164 | 1.0 | 0.904 | 0.7 | 0.023 | 1.4 | 0.060 |
| 5-Androsten-3β,17β-diol-3-sulfate | 777.0 ± 490.1 | 696.2 ± 513.6 | 1478.9 ± 775.1 | 0.020 | 0.164 | 1.9 | 0.053 | 0.9 | 0.957 | 2.1 | 0.028 |
| 1-Palmitoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine | 892.4 ± 397.8 | 1094.5 ± 336.2 | 633.7 ± 217.4 | 0.020 | 0.164 | 0.7 | 0.230 | 1.2 | 0.399 | 0.6 | 0.015 |
| 9(10)-Epoxy-12Z-octadecenoic acid | 694.8 ± 412.9 | 1342.5 ± 685.4 | 768.1 ± 379.0 | 0.023 | 0.179 | 1.1 | 0.950 | 1.9 | 0.032 | 0.6 | 0.061 |
| 3-Methylglutarylcarnitine | 600.8 ± 190.9 | 995.2 ± 420.7 | 1214.7 ± 695.4 | 0.037 | 0.251 | 2.0 | 0.031 | 1.7 | 0.211 | 1.2 | 0.604 |
| D-erythro-Sphingosine-1-phosphate | 905.0 ± 247.9 | 1156.4 ± 454.3 | 732.4 ± 255.8 | 0.039 | 0.251 | 0.8 | 0.523 | 1.3 | 0.263 | 0.6 | 0.031 |
| Glycodeoxycholic acid | 1370.1 ± 869.5 | 837.6 ± 691.7 | 527.0 ± 314.6 | 0.039 | 0.251 | 0.4 | 0.032 | 0.6 | 0.226 | 0.6 | 0.590 |
| Lys-Trp | 827.9 ± 106.5 | 736.1 ± 47.4 | 774.3 ± 47.9 | 0.041 | 0.251 | 0.9 | 0.278 | 0.9 | 0.032 | 1.1 | 0.515 |
Table 3.
Seven altered metabolites in serum samples measured by nuclear magnetic resonance spectroscopy comparing all groups. Abbreviations; AD: Alzheimer's Disease, ANOVA: analysis of variance, Con: healthy controls, FC: fold change, FDR: false discovery rate, MCI: Mild Cognitive Impairment.
| Metabolites |
Mean ± SD [μM] |
ANOVA | FDR |
AD |Con |
MCI |Con |
AD |MCI |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Con | MCI | AD | FC | p | FC | p | FC | p | |||
| Valine | 118.4 ± 17.6 | 96.6 ± 13.5 | 91.8 ± 10.7 | 0.001 | 0.016 | 0.8 | 0.001 | 0.8 | 0.009 | 0.9 | 0.730 |
| Histidine | 36.7 ± 2.4 | 33.7 ± 3.0 | 31.7 ± 2.2 | 0.001 | 0.016 | 0.9 | 0.001 | 0.9 | 0.048 | 0.9 | 0.232 |
| Formate | 4.6 ± 0.9 | 3.0 ± 0.8 | 4.1 ± 0.8 | 0.001 | 0.016 | 0.9 | 0.415 | 0.7 | 0.001 | 1.4 | 0.026 |
| Myo-inositol | 40.2 ± 5.9 | 33.6 ± 3.8 | 34.6 ± 4.2 | 0.013 | 0.121 | 0.9 | 0.044 | 0.8 | 0.016 | 1.0 | 0.902 |
| Glutamine | 308.4 ± 33.3 | 277.0 ± 18.0 | 283.0 ± 14.9 | 0.020 | 0.154 | 0.9 | 0.075 | 0.9 | 0.023 | 1.0 | 0.852 |
| Dimetylamine | 5.6 ± 2.2 | 4.0 ± 1.3 | 3.6 ± 0.8 | 0.025 | 0.159 | 0.6 | 0.027 | 0.7 | 0.092 | 0.9 | 0.834 |
| Isoleucine | 44.8 ± 8.9 | 37.3 ± 7.0 | 36.5 ± 6.0 | 0.046 | 0.248 | 0.8 | 0.062 | 0.8 | 0.096 | 1.0 | 0.974 |
Fig. 2.
Important serum metabolites identified for discrimination of cognitively affected and healthy individuals. Boxplots representation with medians, interquartile ranges, and whiskers for minimum and maximum measurements. Significance is indicated by ** <0.01 and * <0.05. Allopurinol riboside, inosine, 4-pyridoxic acid, and guanosine were measured by LC-MS, while valine and histidine were measured by NMR.
3.3. Dysregulated pathway analysis related to cognitive impairment
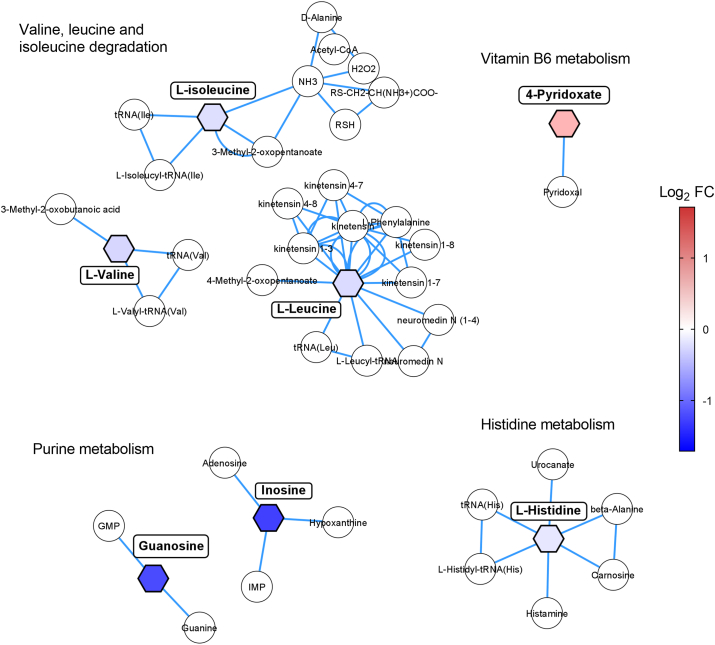
To extrapolate metabolic changes possibly related to disease pathology, pathway analysis was performed (Fig. 3). 4-pyridoxic acid involved in vitamin B6 metabolism (log2 FC = 0.7, p = 0.003) was found to be the most elevated metabolite, while inosine (log2 FC = −1.7, p = 0.006) and guanosine (log2 FC = −1.6, p = 0.007), both from the purine metabolism were the most decreased metabolites. Other impaired pathways included histidine and branch-chained amino acids (BCAAs, valine, leucine, and isoleucine) metabolisms.
Fig. 3.
Dysregulated metabolic pathways related to AD pathology. Hexagonal nodes indicate altered metabolites in the study and circular nodes represent metabolites involved in the pathway not identified in the study. Color codes represent the log2 FC values, with red representing upregulated metabolites and blue representing downregulated metabolites. Mapped metabolites are based on KEGG IDs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we investigate serum and EV-derived metabolites possibly related to cognitive impairments, including MCI and AD. Using multivariate and univariate statistics, we identified altered metabolic signatures in serum able to differentiate cognitive affected from healthy individuals.
For serum samples, we investigated the metabolome using both LC-MS and NMR to obtain a broader coverage of the metabolome. Although LC-MS has a higher sensitivity and coverage (130 identified metabolites), the reproducibility and robustness of NMR present great clinical applicablitity [43] since the 38 identified metabolites could be quantified (μM concentration).
Important serum metabolites revolved around BCAAs (valine, leucine, and isoleucine), purine (inosine and guanosine), and histidine metabolisms, which were found decreased, while 4-pyridoxic acid was increased in AD patients. BCAAs have previously been associated with increased risk for AD and dementia [44], hence, our study confirms previous findings. Valine is the most extensively studied of the BCAAs in relation to AD, and its levels were previously been found decreased in AD patients [45]. Also, valine levels showed correlation with the cognitive decline in patients [17]. A function of BCAAs is part of the glutamate metabolism [46]. Glutamine is converted to glutamate, acting as principal excitatory neurotransmitters in the CNS [47]. Decreased levels of BCAAs, as we observed, could affect this conversion of glutamine and glutamate, thereby decreasing neurotransmission. In line with this observation, we found lower levels of glutamine in the cognitively affected individuals, and a previous study has also reported decreased levels of glutamate in AD patients [48]. In addition, a study found a correlation between peripheral and CSF glutamine levels [49].
Guanosine possesses protective effects for neurons by i.e. modulating neurochemical processes, reducing oxidative stress, and regulate glutamate excitotoxicity and inflammation [50]. Inosine also provides benefical effects on the CNS, by improving memory and learning, as well as providing anti-inflammatory effects [51]. We also found decreased levels of allopurinol riboside, a metabolite which inhibits the effects of purine nucleoside phosphorylase on inosine [52]. This could be a response to the already observed lower levels of the purines inosine and guanosine, thereby partially preventing the conversion of inosine to hypoxanthine and guanosine to guanine.
Histidine, a precursor to an important component in the immune response histamine, was found decreased in AD in our study. Histidine has been shown to possess multiple neuroprotective functions in relation to cerebral hypoperfusion, promoting neurogenesis and BBB integrity post disruption [53]. Furthermore, treatment with this amino acid reduced glial scarring and promoted migration of astrocytes towards the core, thus providing long-term neuroprotection [54]. Hence, our findings may indicate possible derangements in the overall immune response mechanisms.
Hyperhomocysteinaemia is a modifiable risk factor of AD and dementia. Studies have shown that dietary supplementation with B vitamins, such as folate, B12, and B6 have the ability to lower homocysteine levels, improve upon cognition, and decrease the progression of MCI and AD [55]. 4-pyridoxic acid is the catabolic product of vitamin B6 [56], and thus, our observations of an increased level in AD and MCI patients could be due to preventive measurements by the study participants taking these dietary supplements.
As part of their biogenesis, EVs are packed with molecular components from their parental cell, including metabolites at possibly sub-nanomole concentrations. Thus, even with the use of the more sensitive MS methodology, adequate detection levels might still be difficult to achieve [57]. This was evident in our study, both through a more limited number of metabolites measured by MS when compared to serum, but also when performing NMR analysis, since we were unable to recover adequate metabolite information not even after pooling of several samples per measurement. According to Gézsi and co-workers [58], the detection limit, as well as an adequate volume for analysis complicates the usage of EVs in omics studies. Despite our limited findings, few studies in cancer research have investigated the EV metabolome, confirming that EVs are able to modify the metabolome of the recipient cell [59]. Hence, several challenges remain to be resolved, including preanalytical and analytical steps related to enrichment procedure and global consensus and standard operating procedures, before their usage in metabolomics can be harnessed [25,60]. Thus, in terms of usage for metabolic investigations, our study indicates serum as preferable compared to EVs.
Although our study showed promising indications of metabolic signatures in blood for AD, some limitations are to be mentioned. Firstly, our study population consisted of small cohorts of patients and healthy controls. However, even with small sample subset, clear differences between diseased and healthy individuals were observed. Secondly, although patient groups were verified clinally, not all patients had paraclinical measurements or neuropsychological tests, as it was deemed unnecessary for the diagnosis made by the physician. Thirdly, the healthy individuals were on average younger compared to AD and MCI patients, since recruitment of older blood donors was not feasible. The difference in age was found to be significant, however, this was possibly also due to the narrow age span in the control group, compared to that of patients, although their age spans overlapped. In addition, including CSF samples for disease characterisation using metabolomics alongside blood samples could possibly strengthen the identification of CNS-related affected pathways reflected in the peripheral system, but CSF sampling was not available in most of the patients and controls. Lastly, characterising metabolic changes might provide targets for the implicated molecular processes, which in turn could aid in alleviating AD-related pathological processes such as neuro-inflammation and –transmission.
Our findings aided in the search for molecular signatures related to AD using untargeted metabolomics strategies, although, for such discovery-based approaches, further validations are needed to confirm these candidates in larger independent cohorts.
5. Conclusions
In this study, we identified several serum metabolite alterations in AD and MCI patients related to BCAAs, purine, histidine, and 4-pyridoxate metabolisms. Also, serum provides a more suitable matrix for investigating metabolic alterations in relation to AD pathology compared to that of EVs, however, additional optimization is needed for EVs to confirm this finding.
Data availability
Data is available upon request.
CRediT authorship contribution statement
Jonas Ellegaard Nielsen: Conceptualization, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration. Raluca Georgiana Maltesen: Formal analysis, Investigation, Resources, Data curation, Writing – review & editing. Jesper F. Havelund: Formal analysis, Investigation, Resources, Data curation, Writing – review & editing. Nils J. Færgeman: Resources, Writing – review & editing. Charlotte Held Gotfredsen: Formal analysis, Investigation, Data curation, Writing – review & editing. Karsten Vestergård: Resources, Writing – review & editing, All authors have read and agreed to the published version of the manuscript. Søren Risom Kristensen: Conceptualization, Resources, Writing – review & editing, Supervision, Project administration. Shona Pedersen: Conceptualization, Resources, Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
None.
Acknowledgements
The authors are greatly indebted to Mette Ullits Thomsen, Helle Dalsgaard Holst, Helle Hylander, and Mette Jespersgaard for their assistance in enrolment and blood sample collection from patients and controls. The NMR Center DTU and the Villum Foundation are acknowledged for access to the 800 MHz spectrometer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metop.2021.100125.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Abeysinghe A.a.DT., Deshapriya R.D.U.S., Udawatte C. Alzheimer's disease; a review of the pathophysiological basis and therapeutic interventions. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117996. [DOI] [PubMed] [Google Scholar]
- 2.Mullane K., Williams M. Alzheimer's disease beyond amyloid: can the repetitive failures of amyloid-targeted therapeutics inform future approaches to dementia drug discovery? Biochem Pharmacol. 2020;177 doi: 10.1016/j.bcp.2020.113945. [DOI] [PubMed] [Google Scholar]
- 3.Panza F., Lozupone M., Logroscino G., Imbimbo B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat Rev Neurol. 2019;15:73–88. doi: 10.1038/s41582-018-0116-6. [DOI] [PubMed] [Google Scholar]
- 4.Räisänen U., Bekkers M.-J., Boddington P., Sarangi S., Clarke A. The causation of disease - the practical and ethical consequences of competing explanations. Med Health Care Philos. 2006;9:293–306. doi: 10.1007/s11019-006-9007-5. [DOI] [PubMed] [Google Scholar]
- 5.Korecka M., Shaw L.M. Mass spectrometry‐based methods for robust measurement of Alzheimer's disease biomarkers in biological fluids. J Neurochem. 2021 doi: 10.1111/jnc.15465. jnc.15465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lista S., O'Bryant S.E., Blennow K., Dubois B., Hugon J., Zetterberg H. Biomarkers in sporadic and familial Alzheimer's disease. J Alzheimers Dis. 2015;47:291–317. doi: 10.3233/JAD-143006. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs J.M., Adkins J.N., Qian W.-J., Liu T., Shen Y., Camp D.G. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 8.Baird A.L., Westwood S., Lovestone S. Blood-based proteomic biomarkers of Alzheimer's disease pathology. Front Neurol. 2015;6:236. doi: 10.3389/fneur.2015.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampel H., O'Bryant S.E., Castrillo J.I., Ritchie C., Rojkova K., Broich K. Precision medicine - the golden gate for detection, treatment and prevention of Alzheimer's disease. J Prev Alzheimers Dis. 2016;3:243–259. doi: 10.14283/jpad.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamichhane S., Sen P., Dickens A.M., Hyötyläinen T., Orešič M. In: Jaumot J., Bedia C., Tauler R., editors. vol. 82. Elsevier; 2018. Chapter fourteen - an overview of metabolomics data analysis: current tools and future perspectives; pp. 387–413. (Data analysis for omic sciences: methods and applications). [DOI] [Google Scholar]
- 11.Nielsen J., Oliver S. The next wave in metabolome analysis. Trends Biotechnol. 2005;23:544–546. doi: 10.1016/j.tibtech.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Stringer K.A., McKay R.T., Karnovsky A., Quémerais B., Lacy P. Metabolomics and its application to acute lung diseases. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins J.M., Trushina E. Application of metabolomics in Alzheimer's disease. Front Neurol. 2018;8:1–20. doi: 10.3389/fneur.2017.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart D.S. Applications of metabolomics in drug discovery and development. Drugs R. 2008;9:307–322. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- 15.Pagani M., Nobili F., Morbelli S., Arnaldi D., Giuliani A., Öberg J. Early identification of MCI converting to AD: a FDG PET study. Eur J Nucl Med Mol Imag. 2017;44:2042–2052. doi: 10.1007/s00259-017-3761-x. [DOI] [PubMed] [Google Scholar]
- 16.Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., MacArthur L.H. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toledo J.B., Arnold M., Kastenmüller G., Chang R., Baillie R.A., Han X. Metabolic network failures in Alzheimer's disease: a biochemical road map. Alzheimers Dement. 2017;13:965–984. doi: 10.1016/j.jalz.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiandaca M.S., Zhong X., Cheema A.K., Orquiza M.H., Chidambaram S., Tan M.T. Plasma 24-metabolite panel predicts preclinical transition to clinical stages of Alzheimer's disease. Front Neurol. 2015;6:237. doi: 10.3389/fneur.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sardar Sinha M., Ansell-Schultz A., Civitelli L., Hildesjö C., Larsson M., Lannfelt L. Alzheimer's disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018;136:41–56. doi: 10.1007/s00401-018-1868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuyama K., Sun H., Sakai S., Mitsutake S., Okada M., Tahara H. Decreased amyloid-β pathologies by intracerebral loading of glycosphingolipid-enriched exosomes in Alzheimer model mice. J Biol Chem. 2014;289:24488–24498. doi: 10.1074/jbc.M114.577213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Record M., Subra C., Silvente-Poirot S., Poirot M. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol. 2011;81:1171–1182. doi: 10.1016/j.bcp.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Zheng H., Wu X., Yin J., Wang S., Li Z., You C. Clinical applications of liquid biopsies for early lung cancer detection. Am J Cancer Res. 2019;9:2567–2579. [PMC free article] [PubMed] [Google Scholar]
- 25.Williams C., Palviainen M., Reichardt N.-C., Siljander P.R.-M., Falcón-Pérez J.M. Metabolomics applied to the study of extracellular vesicles. Metabolites. 2019;9 doi: 10.3390/metabo9110276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. International classification of diseases (ICD). n.d.
- 27.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group* under the auspices of department of health and human services task force on Alzheimer's disease. Neurology. 1984;34 doi: 10.1212/WNL.34.7.939. 939–939. [DOI] [PubMed] [Google Scholar]
- 28.Winblad B., Palmer K., Kivipelto M., Jelic V., Fratiglioni L., Wahlund L.-O. Mild cognitive impairment - beyond controversies, towards a consensus: report of the international working group on mild cognitive impairment. J Intern Med. 2004;256:240–246. doi: 10.1111/j.1365-2796.2004.01380.x. [DOI] [PubMed] [Google Scholar]
- 29.Ellegaard Nielsen J., Sofie Pedersen K., Vestergård K., Georgiana Maltesen R., Christiansen G., Lundbye-Christensen S. Novel blood-derived extracellular vesicle-based biomarkers in Alzheimer's disease identified by proximity extension assay. Biomedicines. 2020;8 doi: 10.3390/biomedicines8070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen J.E., Honoré B., Vestergård K. Shotgun-based proteomics of extracellular vesicles in Alzheimer’s disease reveals biomarkers involved in immunological and coagulation pathways. Sci Rep. 2021;11:18518. doi: 10.1038/s41598-021-97969-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dall K.B., Havelund J.F., Harvald E.B., Witting M., Faergeman N.J. HLH-30-dependent rewiring of metabolism during starvation in C. elegans. Aging Cell. 2021;20 doi: 10.1111/acel.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maltesen R.G., Hanifa M.A., Kucheryavskiy S., Pedersen S., Kristensen S.R., Rasmussen B.S. Predictive biomarkers and metabolic hallmark of postoperative hypoxaemia. Metabolomics. 2016;12:87. doi: 10.1007/s11306-016-1018-5. [DOI] [Google Scholar]
- 34.Simonsen C., Magnusdottir S.O., Andreasen J.J., Wimmer R., Rasmussen B.S., Kjærgaard B. Metabolic changes during carbon monoxide poisoning: an experimental study. J Cell Mol Med. 2021 doi: 10.1111/jcmm.16522. jcmm.16522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maltesen R.G., Wimmer R., Rasmussen B.S. A longitudinal serum NMR-based metabolomics dataset of ischemia-reperfusion injury in adult cardiac surgery. Sci Data. 2020;7:198. doi: 10.1038/s41597-020-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R. Hmdb 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanifa M.A., Skott M., Maltesen R.G., Rasmussen B.S., Nielsen S., Frøkiær J. Tissue, urine and blood metabolite signatures of chronic kidney disease in the 5/6 nephrectomy rat model. Metabolomics. 2019;15:112. doi: 10.1007/s11306-019-1569-3. [DOI] [PubMed] [Google Scholar]
- 38.Maltesen R.G., Rasmussen B.S., Pedersen S., Hanifa M.A., Kucheryavskiy S., Kristensen S.R. Metabotyping patients' journeys reveals early predisposition to lung injury after cardiac surgery. Sci Rep. 2017;7 doi: 10.1038/srep40275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luan H., Ji F., Chen Y., statTarget Cai Z. A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data. Anal Chim Acta. 2018;1036:66–72. doi: 10.1016/j.aca.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karnovsky A., Weymouth T., Hull T., Tarcea V.G., Scardoni G., Laudanna C. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saigusa D., Matsukawa N., Hishinuma E., Koshiba S. Identification of biomarkers to diagnose diseases and find adverse drug reactions by metabolomics. Drug Metabol Pharmacokinet. 2021;37:100373. doi: 10.1016/j.dmpk.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Tynkkynen J., Chouraki V., van der Lee S.J., Hernesniemi J., Yang Q., Li S. Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer's disease: a prospective study in eight cohorts. Alzheimers Dement. 2018;14:723–733. doi: 10.1016/j.jalz.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basun H., Forssell L.G., Almkvist O., Cowburn R.F., Eklöf R., Winblad B. Amino acid concentrations in cerebrospinal fluid and plasma in Alzheimer's disease and healthy control subjects. J Neural Transm Park Dis Dement Sect. 1990;2:295–304. doi: 10.1007/BF02252924. [DOI] [PubMed] [Google Scholar]
- 46.Fernstrom J.D. Branched-chain amino acids and brain function. J Nutr. 2005;135 doi: 10.1093/jn/135.6.1539S. 1539S-46S. [DOI] [PubMed] [Google Scholar]
- 47.Chaudhry F.A., Schmitz D., Reimer R.J., Larsson P., Gray A.T., Nicoll R. Glutamine uptake by neurons: interaction of protons with system a transporters. J Neurosci. 2002;22:62–72. doi: 10.1523/JNEUROSCI.22-01-00062.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fayed N., Modrego P.J., Rojas-Salinas G., Aguilar K. Brain glutamate levels are decreased in Alzheimer's disease: a magnetic resonance spectroscopy study. Am J Alzheimers Dis Other Demen. 2011;26:450–456. doi: 10.1177/1533317511421780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niedzwiecki M.M., Walker D.I., Howell J.C., Watts K.D., Jones D.P., Miller G.W. High‐resolution metabolomic profiling of Alzheimer's disease in plasma. Ann Clin Transl Neurol. 2020;7:36–45. doi: 10.1002/acn3.50956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tasca C.I., Lanznaster D., Oliveira K.A., Fernández-Dueñas V., Ciruela F. Neuromodulatory effects of guanine-based purines in health and disease. Front Cell Neurosci. 2018;12:376. doi: 10.3389/fncel.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang N., Yan X., Zhou W., Zhang Q., Chen H., Zhang Y. NMR-based metabonomic investigations into the metabolic profile of the senescence-accelerated mouse. J Proteome Res. 2008;7:3678–3686. doi: 10.1021/pr800439b. [DOI] [PubMed] [Google Scholar]
- 52.Ray A.S., Olson L., Fridland A. Role of purine nucleoside phosphorylase in interactions between 2’,3’-dideoxyinosine and allopurinol, ganciclovir, or tenofovir. Antimicrob Agents Chemother. 2004;48:1089–1095. doi: 10.1128/AAC.48.4.1089-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song J., Yang L., Nan D., He Q., Wan Y., Guo H. Histidine alleviates impairments induced by chronic cerebral hypoperfusion in mice. Front Physiol. 2018;9:662. doi: 10.3389/fphys.2018.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao R., Jiang L., Wang R., Zhao H., Chen Y., Li Y. Histidine provides long-term neuroprotection after cerebral ischemia through promoting astrocyte migration. Sci Rep. 2015;5:15356. doi: 10.1038/srep15356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhargava S., Bhandari A., Choudhury S. Role of homocysteine in cognitive impairement and Alzheimer's disease. Indian J Clin Biochem. 2018;33:16–20. doi: 10.1007/s12291-017-0646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherwood R.A. Laboratory Assessment of Vitamin Status; 2019. Methods for assessment of vitamin B6; pp. 181–191. Elsevier. [DOI] [Google Scholar]
- 57.Chitoiu L., Dobranici A., Gherghiceanu M., Dinescu S., Costache M. Multi-omics data integration in extracellular vesicle biology—utopia or future reality? Int J Math Stat. 2020;21:8550. doi: 10.3390/ijms21228550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gézsi A., Kovács Á., Visnovitz T., Buzás E.I. Systems biology approaches to investigating the roles of extracellular vesicles in human diseases. Exp Mol Med. 2019;51:1–11. doi: 10.1038/s12276-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopes-Rodrigues V., Di Luca A., Mleczko J., Meleady P., Henry M., Pesic M. Identification of the metabolic alterations associated with the multidrug resistant phenotype in cancer and their intercellular transfer mediated by extracellular vesicles. Sci Rep. 2017;7:44541. doi: 10.1038/srep44541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dudzik D., Macioszek S., Struck-Lewicka W., Kordalewska M., Buszewska-Forajta M., Waszczuk-Jankowska M. Perspectives and challenges in extracellular vesicles untargeted metabolomics analysis. Trac Trends Anal Chem. 2021;143 doi: 10.1016/j.trac.2021.116382. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available upon request.