Summary
Little is known about the diversity and function of adipose tissue nerves, due in part to the inability to effectively visualize the tissue’s diverse nerve subtypes and the patterns of innervation across an intact depot. The tools to image and quantify adipose tissue innervation are currently limited. Here, we present a method of tissue processing that decreases tissue thickness in the z-axis while leaving cells intact for subsequent immunostaining. This was combined with autofluorescence quenching techniques to permit intact whole tissues to be mounted on slides and imaged by confocal microscopy, with a complementary means to perform whole tissue neurite density quantification after capture of tiled z-stack images. Additionally, we demonstrate how to visualize nerve terminals (the neuro-adipose nexus) in intact blocks of adipose tissue without z-depth reduction. We have included examples of data demonstrating nerve subtypes, neurovascular interactions, label-free imaging of collagen, and nerve bundle digital cross-sections.
Subject areas: Cell biology, Functional aspects of cell biology, Methodology in biological sciences
Graphical abstract
Highlights
-
•
Whole depot adipose tissue innervation was imaged and quantified by a novel method
-
•
Numerous aspects of adipose nerve heterogeneity were observed by microscopy
-
•
We have identified a nerve terminal in adipose, the neuro-adipose nexus
Cell biology; Functional aspects of cell biology; Methodology in biological sciences
Introduction
Historically overlooked as a location of diverse peripheral nerve innervation (Blaszkiewicz et al., 2019b), the adipose organ was most prominently inspected for innervation in the mid 1960s when sympathetic nerve fibers were visualized within brown adipose tissue (BAT) (Wirsen, 1964). BAT nerves were later comprehensively investigated by the Bartness research group (Cinti et al., 2016). Energy expending BAT was the first adipose tissue to be identified as being highly innervated due to its important role in thermogenesis (Fenzl and Kiefer, 2014), which requires significant sympathetic nervous system (SNS) input and release of norepinephrine (NE) (Bartness et al., 2010; Francois et al., 2019; Wirsen, 1964). Sensory innervation has also been documented in BAT, particularly around vasculature, and sensory neuropeptides have been proposed to play a role in lipolysis (Garretson et al., 2016). White adipose tissue (WAT) is typically associated with energy storage, but is a dynamic and plastic tissue capable of undergoing transformation during the “browning” process (stimulated by negative energy balance states like exercise or cold), which leads to increased presence of brown adipocytes and increased sympathetic neurite outgrowth. WAT is now appreciated to be highly innervated by both sympathetic (Foster et al., 1982; Foster and Bartness, 2006; Huesing et al., 2021) and sensory nerves (Fishman and Dark, 1987;Bartness et al., 2010), but not parasympathetic nerves (Giordano et al., 2006).
In order to visualize adipose innervation, it had been common practice to slice adipose tissue into 7–10μm thick sections on slides, and immunolabel for various neuronal markers (Murano et al., 2009; Giordano et al., 2006; Vargovic et al., 2011; Shi et al., 2005), which included the sympathetic nerve activation marker tyrosine hydroxylase (TH); the rate-limiting enzyme for the synthesis of catecholamine neurotransmitters such as NE. Several important findings emerged from this practice, but it was not without limitations. Thin sections of tissue reduced peripheral nerves to puncta, leaving investigators unable to accurately determine arborization or the ability to quantify innervation across an intact tissue. This is important, given our knowledge that adipose depot innervation patterns are heterogeneous, with denser innervation in certain regions of the tissue, such as around the central lymph node. Furthermore, due to the nature of thin sectioning, important neuro-vasculature and nerve-cell interactions within the diverse cell milieu of adipose tissue had been more difficult to discern.
Adipose tissue is notoriously difficult to image. Tissue resident lipids are autofluorescent and scatter light, and lipofuscin (especially prominent in unhealthy adipose) autofluoresces as well. Adipose tissue is also home to a dense vascular network including arteries, veins, capillaries, and lymphatics; all of which are highly autofluorescent. While autofluorescence is often exploited in clearing and lightsheet methods to visualize tissue architecture, it can also obscure signals from antibodies that fluoresce at a similar wavelength. Adipose depots can also be quite large in comparison to other organs, especially with the increased adiposity of obesity, making intact depot imaging more technically challenging. Metabolically healthy and lean mice may have inguinal subcutaneous white adipose tissue (ing-scWAT) depots that are several millimeters thick, and this increases drastically in obese mice. Because of these challenges, combating high light scatter and tissue autofluorescence has been a necessity for researchers conducting whole depot imaging.
Methods for optically clearing and delipidating whole organs for imaging have been developed, particularly for the brain, and now number dozens of options with slight variations on the clearing chemistry. Similar to the brain, the adipose tissue is full of light scattering lipids. Clearing and delipidating techniques are currently being applied to image adipose tissue and to image and quantify innervation within whole adipose depots (Zeng et al., 2015; Cao et al., 2018a, 2018b; Chi et al., 2018; Li et al., 2019; Jiang et al., 2017; Blaszkiewicz et al., 2019a). While useful for getting crisp images of the large nerve bundles (NBs) in adipose, this technique does have drawbacks. The current methods tend to image at low magnifications, with very large working distances (Cao et al., 2018b; Jiang et al., 2017; Chi et al., 2018; Zeng et al., 2015) to account for such large tissues. This can result in images that do not capture the small neurites that require high magnifications to be effectively visualized. Given the high likelihood that the small fiber nerves undergo the most remodeling in the tissue, and are the ones likely to die back in adipose neuropathy, it is important to capture the density of these small nerve fibers in adipose tissue investigations.
Our laboratory has revealed a new map of adipose anatomy in the ing-scWAT depot, and we and others have demonstrated that the pattern of innervation in scWAT is heterogeneous across the tissue (Blaszkiewicz et al., 2019a), similar to the heterogeneous browning potential also observed (Dichamp et al., 2019). In fact, regions of high innervation correlate with areas of browning. This plasticity and heterogeneity warrant a more comprehensive look at adipose innervation across an intact depot in order to assess anatomically specific regional changes that may occur with physiological or pathophysiological stimuli, especially in the anterior–posterior axis of the inguinal depot. The aforementioned clearing methods tend to rely on quantifying only a few small so-called ‘representative’ three-dimensional tissue sections, which can miss the heterogeneity and regional anatomy of the intact tissue. Until the regional variation in neurite density in scWAT (Blaszkiewicz et al., 2019a; Chi et al., 2018) is better understood, it cannot be accurately reduced to a few representative images or a small tissue block.
Furthermore, some of the most effective methods of tissue clearing use caustic reagents which, for optimal results, require immersion of specialized objective lenses (i.e., “BABB-safe”) into the clearing media for refractive index matching, and many of these shorten the fluorescence lifespan and can be detrimental to endogenous fluorophores. This can be costly and prevents tissues from being mounted onto slides for easy widefield epifluorescence imaging. Some clearing media can also solidify at room temperature which causes yet another hurdle for researchers. There is also the separate issue of obtaining homogeneous and consistent antibody or fluorophore staining through thick cleared tissue. Traditionally, poor antibody penetration has been combatted with drastically increased incubation times, or more recently with stochastic electrotransport (Kim et al., 2015) and barometric pressure (Fiorelli et al., 2020). While these solutions have resulted in better efficacy of immunolabeling, increased incubation times increased the risk of microbial growth during the staining process, and techniques such as stochastic electrotransport may not be easily implemented into current laboratory workflows. Finally, while many institutions do have light sheet microscopy capabilities, special large stages are required to image large tissues like adipose if they are visualized as an intact organ.
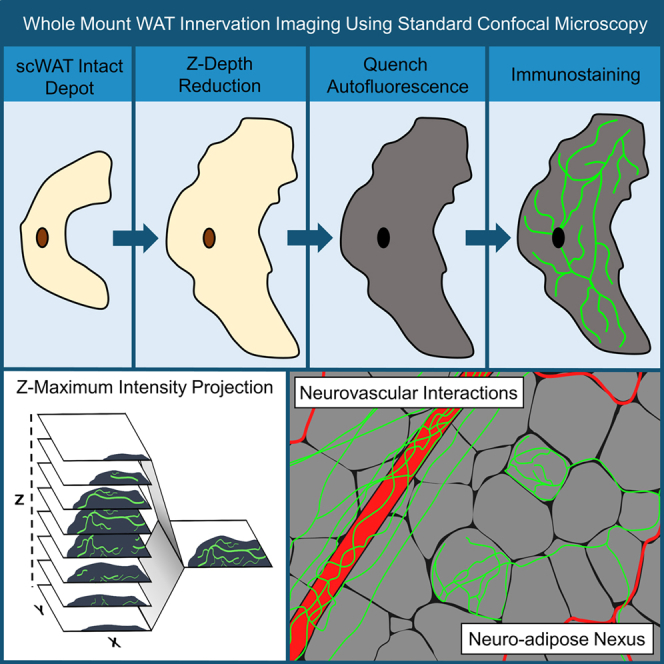
For these reasons, we have developed an alternative approach for adipose tissue processing that enables more researchers to utilize the whole depot imaging technique on any confocal microscope regardless of BABB-safe objectives, by utilizing mechanical pressure to homogeneously compress the intact depot to reduce tissue thickness. Z-depth reduction (ZDR) has been used for other tissues as a means of reducing tissue thickness to allow for faster image acquisition (Kim et al., 2018). Reducing tissue depth improves antibody penetration and consistent imaging in tiled z-stacks across the entire depot to ensure good resolution and comprehensive data collection, including of small nerve fibers.
This tissue processing and imaging workflow is accompanied by a simple option for quantification of adipose neurites using methods that we have made publicly available (see STAR Methods). Widefield microscopy is sufficient for many qualitative analyses of entire adipose depots, but for precise colocalization and quantitative analysis confocal imaging is required. In addition, the cost for light sheet microscopes can create a barrier to entry that we wish to ameliorate, allowing more research groups to investigate adipose innervation and the role of adipose nerves in physiology and pathophysiology.
We have tested this workflow using two scWAT depots (inguinal and axillary) which show slightly different responses to neurotrophic stimuli and browning potential (Blaszkiewicz et al., 2019a). This whole mount technique has been optimized for the pan-neuronal markers PGP9.5 (Thompson et al., 1983; Day and Thompson, 2010) and β3-tubulin (Draberova et al., 2008; Latremoliere et al., 2018); the sympathetic nerve marker, tyrosine hydroxylase (TH) (Shi et al., 2005); markers for sensory innervation that included advillin (AVIL) (Hasegawa et al., 2007; Hunter et al., 2018) and Nav 1.8 (Bird et al., 2013); the myelination marker myelin protein zero (MPZ) (D’Urso et al., 1990); as well as several others. This technique has also been optimized for non-antibody based fluorescent labeling approaches such as nuclear labeling with DAPI and vascular labeling with Isolectin IB4 (IB4). This has allowed us to further our understanding of scWAT in mice by characterizing the innervation that exists within this tissue with greater scrutiny, demonstrating neurovascular interactions, parenchymal innervation, neuroimmune interactions, and neuro-adipose connections. It has been demonstrated previously that the neurites innervating scWAT branch from the same NBs that lead to the skin (Huesing et al., 2021), but we and others have clearly demonstrated that axon endings do terminate inside the parenchyma of scWAT. For the first time, we reveal a terminal junction of axons in scWAT, which we call the neuro-adipose nexus.
Results and discussion
Development of the whole mount technique
Z-depth reduction and effects on tissue integrity
We have tested numerous clearing techniques with scWAT and BAT which included ScaleA2 (Hama et al., 2011), BABB (Dodt et al., 2007; Li et al., 2011), CUBIC (Susaki et al., 2015), CUBIC CB-perfusion (Susaki et al., 2015), iDISCO (Renier et al., 2014), uDISCO (Pan et al., 2016), UbasM (Chen et al., 2017), and a sucrose gradient method (Brantschen et al., 1989). Each clearing method had its own set of trade-offs, with some methods distorting tissue morphology, limiting fluorescence lifespan, and requiring costly objective lenses to image. Not all clearing techniques sufficiently clear adipose as well, and even a complete clearing of the tissue may result in loss of protein antigens needed for immunostaining. These pros and cons have been well documented in various tissues (Azaripour et al., 2016; Matryba et al., 2019; Yu et al., 2018; Seo et al., 2016). To circumvent these issues, we have developed a method of whole tissue processing, outlined in Figure 1A, that allows for whole adipose depots to be imaged on any widefield or confocal microscope without the need for immersion lenses or caustic clearing agents. The entire processing time for an intact adipose depot from a direct reporter mouse takes as little as 5 days. This is significantly shorter than most adipose clearing methods (10–14 days) and is comparable to the remainder.
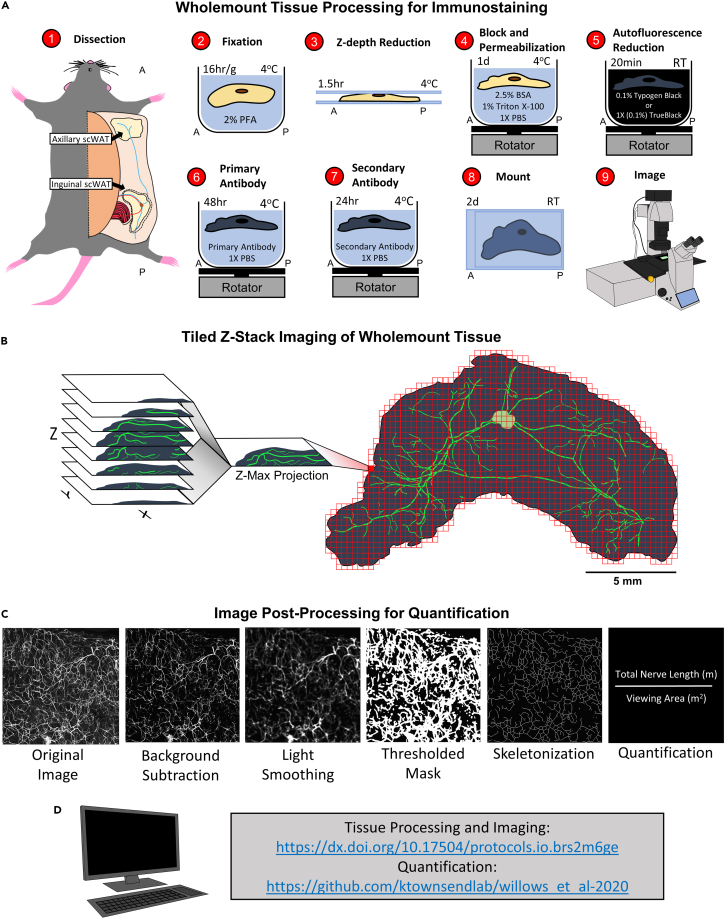
Figure 1.
Adipose tissue processing, imaging, and quantification method
(A) Diagram depicting steps of whole mount tissue processing technique. See STAR Methods for detailed description of each step.
(B) Tiled Z-stacks were imaged for the entire tissue with either a 5X or 10X objective and a 2D maximum projection image was rendered.
(C) Post processing of single tile Zmax projections for neurite density quantification. Each Zmax projection tile was further processed by subtracting the background away and adding light smoothing. Next, a thresholded mask was applied and the image was skeletonized. Total neurite length was calculated for each tile and averaged for each tile and averaged for the entire viewing area.
(D) Link to whole mount protocols and script for quantification.
This approach has allowed us to image entire tissue depots and construct 2D visualizations from 3D data acquisition by tiling Zmax projection images taken at low magnifications (with 5X and 10X objectives) using a laser scanning confocal microscope (Figure 1B) and used for quantifying total nerve density (as described in STAR Methods section) (Figure 1C). Our quantification method is based on pixel intensity. By capturing data as a Zmax projection we retain the highest pixel intensities from each Z-plane to greatly decrease computation time for the quantitative analyses while keeping potentially lost data to a minimum. Z-axis spatial resolution is somewhat lost with this method, but the entirety of adipose nerve fiber density would be maintained if the goal is comprehensive quantification of small and large fibers and not spatial organization. We have developed our quantification method using open access software and Matlab and all code has been made publicly available in GitHub, where we will continue to deposit updates to any quantification methods we develop (Figure 1D).
To acquire a micrograph of an entire intact depot on a point scanning confocal microscope, imaging times are subject to the desired resolution, number of fluorescent markers, and size of the tissue. Acquisition time for an entire depot has averaged 13–16h when a single fluorophore is imaged at 10X objective magnification per tile at resolutions of 720 × 720 and a Z-step sampling size of 12μm. We also experimented with confocal resonant scanning and subsequent denoising with Denoise.ai (NIS-Elements AR software) to reduce image acquisition time at the cost of obtaining high resolution (Figure S1). A whole depot was imaged in only 2h 35min (Figure S1A). Unfortunately, we found that by reducing resolution to image faster, we lost most of the small neurites when compared to the slower point scanning approach (Figure S1B). Because of this we have determined that higher resolutions and longer imaging times are required to capture an accurate representation of parenchymal innervation. However, depending on an investigator’s needs resonant scanning can provide very fast information of gross morphological changes within the tissue.
Although the use of light sheet microscopy can decrease image acquisition time from hours to minutes, in our experience (using multiple platforms) imaging an intact whole adipose depot from various physiological states was extremely challenging with commercially available models. This led us to develop an adipose tissue processing technique that made possible imaging entire intact adipose depots (no need to bifurcate the depot) on a confocal microscope.
Whole mount processing of inguinal and axillary scWAT was achieved by first excising intact depots from the mouse and fixing in 2% PFA overnight. Tissue thickness was reduced to shorten incubation times, enhance antibody penetration, and reduce light scatter by flattening the fixed tissue between two large glass slides with binder clips to apply gentle pressure (Figure 1A) in a process we have termed ‘Z-depth reduction’ (ZDR.)
Tissues were then quenched for autofluorescence, immunolabeled and mounted on glass slides with a glycerol-based mountant. The glycerol-based mountant acts, in part, as a clearing agent, although its use was dictated by the increased coverslip adherence the high viscosity provided. Mounting in a non-clearing aqueous based mountant offers the same quality images, but we found that the coverslip adherence was poor, resulting in tissue oxidation and subsequent loss of signal, as well as increased autofluorescence.
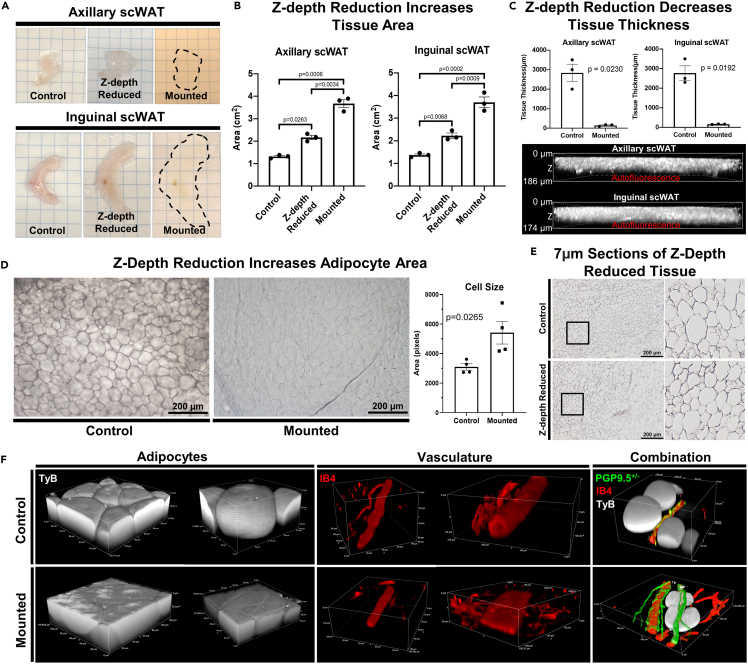
To investigate how ZDR altered tissue morphology, we first measured the change in tissue area that occurred by ZDR and subsequent mounting for axi-scWAT and ing-scWAT depots (Figure 2A). ZDR and mounting resulted in a significant increase to tissue area for both axi-scWAT (p = 0.0006) and ing-scWAT (p = 0.0002) depots (Figure 2B). In both cases, XY area was increased approximately by a factor of three with tissue area increasing after ZDR and again when mounted (Figure 2B). Tissue thickness in the Z-dimension was measured at the thickest point of each tissue before and after ZDR. Tissue thickness was reduced significantly (axi-scWAT p = 0.0230, ing-scWAT p = 0.0192) from approximately 2–3 mm before ZDR to less than 200 μm after mounting (Figure 2C). It is important to specify that the tissues used here are from metabolically healthy young adult mice. Tissues from obese or lipodystrophic mice will likely follow the same trend, but exact changes may be different and obese adipocytes on the surface of the tissue may be more prone to lysis (as we have observed in other experiments).
Figure 2.
Z-depth reduction (ZDR) and tissue morphology
(A) Axillary subcutaneous white adipose tissue (axi-scWAT) and inguinal (ing-scWAT) depots were Z-depth reduced and mounted on slides.
(B) Tissue area was measured before ZDR (control), after ZDR, and after mounting. N = 3, paired two-tailed Student’s t-test, alpha level 0.05, error bars are SEMs.
(C) Tissue thickness in the z-axis was measured at the thickest point before and after ZDR and mounting. Mounted tissues were measured by 3D projecting tissue autofluorescence captured on a Nikon A1R with a 20X objective. N = 3, paired two-tailed Student’s t-test, alpha level 0.05, error bars are SEMs.
(D) Ing-scWAT was either whole mount processed (Z-depth reduced) or left unaltered (control) and placed on a slide and imaged with transmitted light. Four representative 10X micrographs were captured for each tissue and the area for 30–120 cells was measured per micrograph and analyzed using a two-tailed Student’s t-test, Alpha level 0.05, error bars are SEMs.
(F) Hematoxylin staining of 7 μm ing-scWAT sections that either received ZDR before paraffin embedding or did not (control). Both ing-scWAT depots were excised from a PGP9.5+/− (green) direct reporter mouse (C57BL/6-Tg(Uchl1-EGFP)G1Phoz/J) and co-stained with IB4 (vasculature, red) and DAPI (nuclei, blue). One depot was Z-depth reduced and mounted whereas the other was left uncompressed to demonstrate morphological changes introduced by compressing the tissue; demonstrated through 3D reconstructions of similar structures.
Other details: Transmitted light micrographs were captured on a Nikon E400 upright microscope (D and E). Confocal micrographs captured on Nikon A1R (C and F). Scale bars are 186 μm and 174 μm (C), 200 μm (D and F) and 50 μm (E).
Next, we were interested in how adipocyte size and spacing was affected by the increased tissue area, and whether cells remained intact. Because tissue area and thickness were altered similarly between both inguinal and axillary depots, we narrowed our analysis to ing-scWAT moving forward. Unstained whole ing-scWAT depots were either Z-depth reduced and mounted or remained in their physiological state post-dissection and left unmounted. Transmitted light was used to visualize unstained cell structure prior to ZDR (Figure 2D, left panel). Tissue thickness was reduced by ZDR which increased light transmission and softened the cell outlines visible in the control tissues (Figure 2D, right panel). Cell size was averaged across four representative images of each tissue to quantify possible changes in cell size (30–120 cells counted per image). Only cells that had clear and in-focus boundaries (given this was not 3D imaging) were included in the quantification. The average cell area was increased in every instance (p = 0.0265) (Figure 2D). Hematoxylin staining of 7μm thick ing-scWAT sections showed that cells remained intact and structurally similar following ZDR (Figure 2E). A caveat of this analysis is that we were only able to investigate cell integrity following ZDR and not tissue mounting, which was demonstrated to cause additional strain to scWAT depots.
To further this investigation, we excised bilateral ing-scWAT depots from a PGP9.5-eGFP direct reporter mouse (here on out referred to as PGP9.5+/−), co-stained with the vascular marker isolectin IB4 (IB4) and used Typogen Black (TyB) which stains lipids and fluoresces in far red wavelengths to visualize adipocytes. One depot was Z-depth reduced while the other was not in an attempt to maintain in vivo tissue morphology (Figure 2F). Under closer inspection of individual adipocytes, we found that intracellular gaps were reduced to make room for the expanding cell area and that cells, due to being pressed against one another, adopted a polygonal circumference when viewed from above, and became wider in the x- and y-axis, and shorter in the z-axis (Figure 2E). Despite being pressed more closely together, the cells appeared intact in the ZDR samples. Blood vessels appeared fully intact as well, but as vessel diameter increased so too did the likelihood that the vessel would flatten out from the lumen being depressed (Figure 2E).
Using these same comparative methods, we looked for compression induced artifacts in innervation in whole mount tissue chunks. The tissues (>0.9mm thick) that required imaging in glass bottom dishes were difficult to image due to poor light penetrance (Figure S2A) and increased TyB far-red fluorescence which limited our observations to those tissues (<0.9mm thick) that could be mounted in depression slides without compression (Figure S2B). No structural variation in innervation was observed but without ZDR many of the small neurites were obscured and unable to be visualized effectively, further emphasizing the benefits of the ZDR technique.
In summary, our method of tissue processing increases tissue area in the X and Y directions to reduce tissue thickness in the Z direction, and understandably some morphological changes are introduced. This structural manipulation introduces slight changes to individual cells and tissue structures but leaves them fully intact. We believe the benefits of light penetrance and imaging ease outweigh these drawbacks, however, slight morphological changes to adipose using this approach cannot be ruled out and some dimensionality will be lost by design.
Quenching tissue autoflourescence
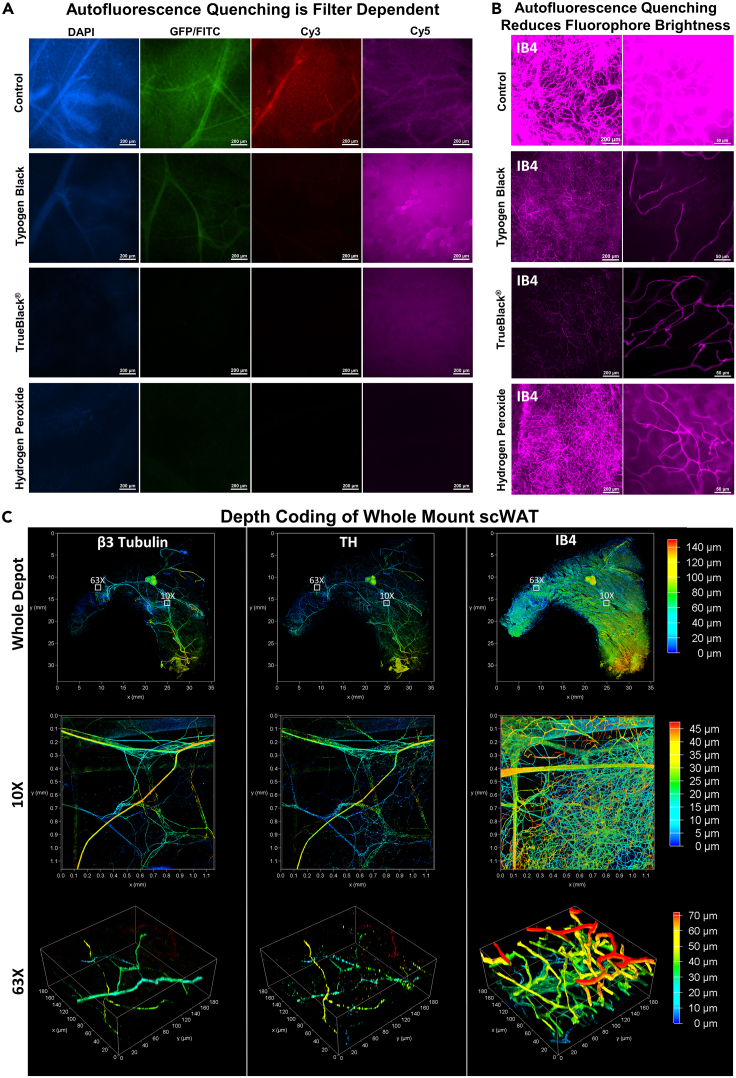
Tissue autofluorescence has always been a significant problem when imaging adipose due to the inherent autofluorescecent nature of lipids and lipofuscin that reside within it (Croce and Bottiroli, 2014). Sudan Black B (henceforth referred to by its historic and more appropriate name Typogen Black, abbreviated as TyB) staining has been used as a treatment to quench tissue autofluorescence for decades in 7–10μm thick sections (Schnell et al., 1999). We applied TyB and two other autofluorescence quenching techniques, TrueBlack® and hydrogen peroxide (used in many optical clearing protocols (Azaripour et al., 2016)), and compared quenching abilities against fluorescence filter cubes spanning the visible spectra: DAPI (blue), GFP/FITC (green), Cy3/TRITC (orange), and Cy5 (far-red; purple) (Figure 3A). All methods were able to reduce autofluorescence in the blue, green, and orange wavelengths similarly, but TyB was the least effective. The far-red Cy5 channel posed interesting barriers. Far-red excitation results in the least autofluorescence of unstained tissue. Unfortunately, this autofluorescence is intensified strongly by TyB staining, which itself is fluorescent in far-red wavelengths, and was left unquenched by TrueBlack®. In many instances the TyB far-red fluorescence can be taken advantage of, as it can act as a reliable stain for adipocytes (Figure 2F) and can even stain NBs, presumably by staining myelin lipids, though the extent and reliability of this has not been tested (Figure S3A).
Figure 3.
Whole mount adipose autofluorescence quenching and fluorescence staining
scWAT depots excised from C57BL/6J mice were whole mount processed and autofluorescecne quenched with either Typogen Black, TrueBlack®, or 5% hydrogen peroxide.
(A) Autofluorescence was evaluated for each blocking method for 4 different fluorescent filter cubes spanning the visible spectra: DAPI, GFP/FITC, Cy3/TRITC, and Cy5. Control tissue brightness was normalized via post processing across all four filters and these settings were then applied to each image for a given filter: DAPI (used as control brightness), GFP/FITC (+10% brightness), Cy3 (+20% brightness), Cy5 (+40% brightness).
(B) Quenching methods were further compared for the Cy5 filter by staining the tissues with IB4 conjugated to a 647nm fluorophore.
(C) Ing-scWAT depot was whole mount processed and stained with β3-Tubulin, Tyrosine Hydroxylase (TH), and IB4 and depth coded. Whole adipose depot was imaged as a tiled Z-maximum projection with a 10X objective, (0–140μm) (c, top row), a single tile captured at 10X (0–45μm) (C, middle row), and a high magnified section captured at 63X (0–70μm) (C, bottom row). Regions chosen for higher magnification images identified by white boxes with corresponding magnification. Captured on Leica TCS SP8 DLS microscope. Scale bars are 200 μm (A), 200 μm and 50 μm (B). X and Y axes are 35 mm (C, whole mount), 1.1 mm (C, 10X) and 180 μm (C, 63X).
Hydrogen peroxide was the only effective means of quenching autofluorescence in all 4 channels (Figure 3A), but added significant time to the overall protocol and could be utilized only if all channels are needed for a given experiment. We next wanted to see how 647nm fluorophores were affected by the various autofluorescence quenching techniques (Figure 3B). All methods appeared to reduce fluorophore brightness when compared to the control tissue. Hydrogen peroxide was the least masking, followed by TyB, and then TrueBlack®, which resulted in a very diminished signal (Figure 3B).
Although TyB had the least autofluorescence quenching potential it was found to be sufficient for our uses and was preferred over TrueBlack®, which tended to mask true signal, and preferred to hydrogen peroxide, which increased processing times significantly. For these reasons TyB was used when not staining with a 647nm fluorophore and TrueBlack® was used only when a 647nm fluorophore was used concurrently with other fluorophores.
Quenching dilutions were optimized in conjunction with pan-neuronal PGP9.5 staining to find the ideal balance between reduced tissue autofluorescence and undiminished true fluorescent signal (Figure S3B). Optimal concentrations, as determined by autofluorescence reduction and signal intensity, were found to be 0.1% TyB (in 70% EtOH) and 1X or 0.1% TrueBlack® (2% TrueBlack® dye in 98% dimethyl formamide, diluted from 20X to 1X with 70% EtOH.)
An additional means of reducing tissue autofluorescence was performed by washing with 1X PBS with 10U/mL Heparin, which removed highly autofluorescent red blood cells (Figure S3C). Alternatively, perfusion can be performed to remove blood cells prior to tissue collection.
Although the use of a glycerol-based mountant was decided upon due to its increased viscosity, it is also a known clearing agent (Song et al., 2015), and we found that adding only a couple of drops to the tissue was enough to provide some level of optical clarity (Figure 2A). This clearing was made more evident when tissues remained at room temperature for incubation and wash steps, prior to mounting, which resulted in reduced background autofluorescence (Figure S3D). It should be noted that autofluorescence when imaging with a widefield epifluorescent microscope is still quite present even after application of the aforementioned quenching techniques. Because of this, it is strongly recommended that confocal microscopy be used whenever possible to further reduce this background and non-specific signal.
To summarize: TyB staining should be avoided when using far red excitation but can be used as a reliable marker for adipocytes. TrueBlack can be an acceptable replacement but it can mask fluorescent signals, so avoid using it with dim fluorophores. Hydrogen peroxide yields the best results all around but adds significant time to the procedure. If only using one fluorophore, it may best to use far-red excitation and do all washes and incubations at room temperature to avoid the use of autofluorescence quenching reagents entirely.
Off-target antibody binding and antibody penetration
Choosing antibodies for whole tissue imaging is very important to determine target specificity. Secondary antibody non-specificity is likely to increase when immunostaining an entire tissue, and when primary-conjugated antibodies are not available or specific enough to the antigen of interest. We tested numerous secondary antibodies to confirm that we were only using those antibodies least likely to bind non-specifically (Figure S3E). Goat anti-rabbit IgG secondary antibodies, regardless of being highly cross-adsorbed or not, did not result in significant levels of non-specific binding. Because goat anti-mouse secondary antibodies had such high levels of non-specific binding, we decided to avoid the use of all mouse primary antibodies for whole mount immunostaining moving forward.
Uniform staining of large whole mount tissues has always been problematic due to the increased incubation times required for antibodies to completely diffuse throughout the tissue (Chung et al., 2013; Kim et al., 2015). Incubation times were reduced by 1–2 days by reducing tissue thickness with ZDR. Depth coding of immunostained scWAT following ZDR indicated homogeneous penetration of antibodies throughout the whole tissue (140μm total depth) (Figure 3C). This was confirmed for various fluorescent staining approaches, including directly labeled β3-tubulin, indirectly labeled TH, and IB4 staining. Depth information can even be somewhat retained following Z-maximum projecting by creating a height color coded projection, albeit with no accompanying color-coded depth scale provided at this time due to LASX software limitations (Figure S3F). We have not tried this ourselves, but the addition of a sucrose incubation step for tissues fixed with 4% paraformaldehyde can also improve axonal staining by decreasing the appearance of “beading” along the axon (Stradleigh et al., 2015).
Characterization of peripheral nerves in scWAT
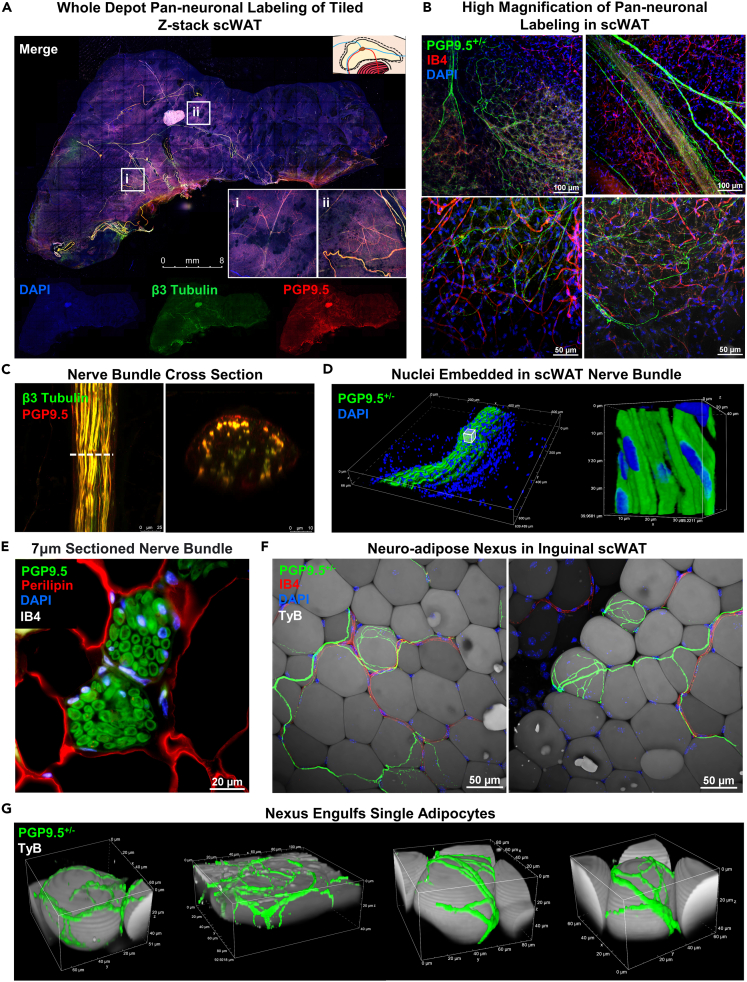
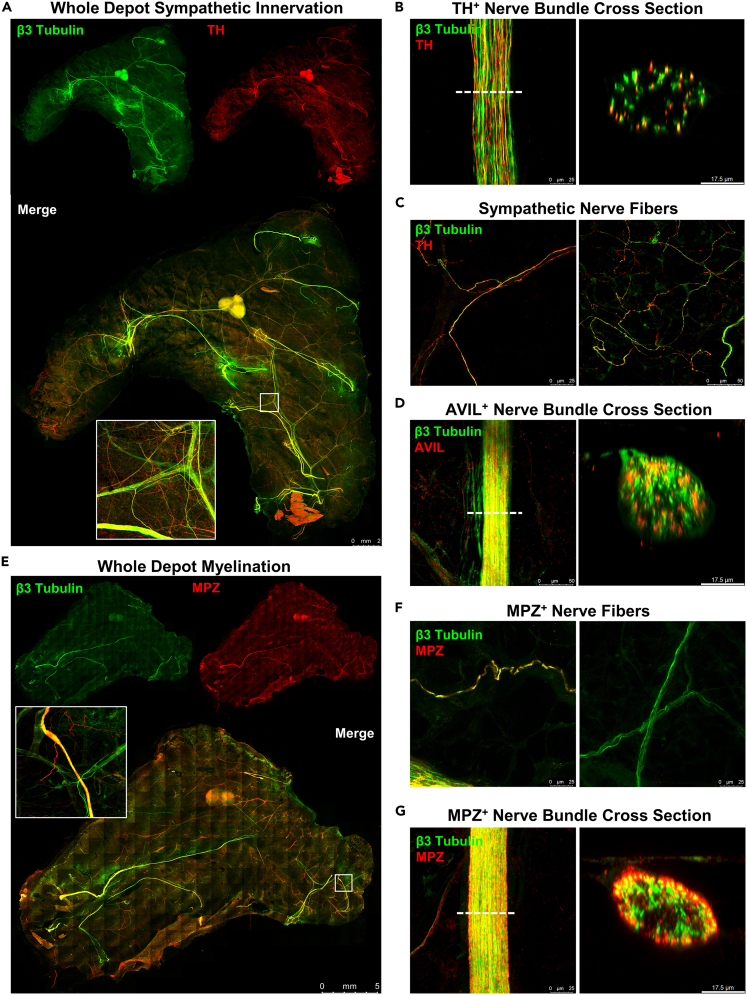
To demonstrate the versatility and capability of this technique we have imaged tissues in several ways. First, whole ing-scWAT was stained with pan-neuronal markers PGP9.5 and β3-tubulin and imaged at low magnification (5X) with limited resolution to observe overall morphology and fluorescence of the larger NBs (Figure 4A). Second, to visualize detailed innervation, areas of interest of a PGP9.5+/− ing-scWAT depot were imaged at 63X with much higher resolutions (Figure 4B). We also performed digital cross-sectioning of an ing-scWAT NB (Figure 4C).
Figure 4.
Peripheral innervation of scWAT
(A) Ing-scWAT depot from C57BL/6J mouse whole mount was processed and stained with DAPI (blue) and the pan-neuronal markers β3-Tubulin (green) and PGP9.5 (red) to show tissue morphology. Image captured as Zmax projection with 5X objective. White boxes show single tile expanded for visualization with 3.57X digital zoom-ins (i–ii).
(B) Representative images of PGP9.5+/− (green) small fiber innervation stained with IB4 (red) and DAPI (blue) imaged as Zmax projections with 20X and 40X objectives.
(C) Nerve bundle (NB) co-stained with β3-Tubulin (green) and PGP9.5 (red) imaged with 63X objective and digital cross-sectioning performed with a 2.54X zoom factor.
(D) PGP9.5+/− NB from an uncompressed ing-scWAT depot stained with DAPI and 3D projected demonstrating nuclei residing within NB captured with 40X objective.
(E) Paraffin embedded ing-scWAT NB co-stained with PGP9.5 (green), perilipin (red), DAPI (blue), and IB4 (white) also shows DAPI+ cells within an NB captured with 60X objective.
(F and G) PGP9.5+/− nerves branch from blood vessels (IB4, red) (F) and engulf single adipocytes (TyB, white) forming a neuro-adipose nexus (G). Captured with 40X objective (F) and reconstructed in 3D (G). Micrographs captured either on Leica TCS SP8 DLS (A and C), Nikon A1R Confocal (B, D, F, and G), or on Nikon E400 (E). Scale bars are 8 mm (A), 100 μm and 50 μm (B), 25 μm and 10 μm (C),20 μm (E), and 50 μm (F).
Whole tissue imaging exhibited near overlap of staining of the largest NBs by both pan-neuronal markers PGP9.5 and β3-tubulin (Figure 4A). Nerves are unevenly distributed throughout the tissue in the x- and y-axis, and in the anterior-posterior anatomical directions. Because the tissues are compressed and subsequently Zmax projected we were unable to accurately report nerve distribution within the z-axis. The highest concentration of large NBs were present in the centermost third of the tissue (in the x-y direction), surrounding the subiliac lymph node (SiLN), as reported previously (Blaszkiewicz et al., 2019a). At greater magnification, Zmax projection images showed an extensive network of neurites varying in diameter, running throughout the tissue and branching around adipocytes as well as vasculature (Figure 4B), also as previously noted(Blaszkiewicz et al., 2019a). Again, there appears to be almost complete fluorescence overlap of both pan-neuronal markers, with PGP9.5 being more readily visible on the smaller nerve fibers, which fits with the use of this antibody to assess small fiber neuropathy in the skin. Variation in immunofluorescence staining between the two pan-neuronal markers can also be attributed in part to β3-tubulin being directly labeled and PGP9.5 being indirectly labeled by a secondary antibody (and thus, some off-target binding of the secondary antibody, and variations in signal intensity can result). The use of a PGP9.5+/− mouse in lieu of antibodies was a direct result of this observation. However, using pan-neuronal antibodies is still a viable option, as we have demonstrated. The innate autofluorescent quality of adipose tissue, despite being greatly reduced by TyB staining, is still visible in most images and can help provide anatomical outlines. In fact, in most adipose clearing techniques in the published literature, autofluorescence of the tissue is exploited to reveal cellular architecture (Chi et al., 2018).
Digital cross-section imaging of large NBs demonstrated complete staining of the bundle, but with a gradation that appeared to result in a weaker signal at the bundle’s center (Figure 4C), which we have attributed to incomplete antibody penetration into the bundle perhaps due to the myelin barrier. We observed DAPI stained nuclei both surrounding the NB and residing between the numerous axons with our method. Because this was possibly an artifact introduced by compression, we also imaged NBs in ing-scWAT that did not receive ZDR (Figure 4D) (following the same methods used in Figures 2F and S2) and traditionally processed paraffin embedded tissues sectioned at 7μm. These DAPI stained nuclei were still present within and around the NBs regardless of how they were processed (Figures 4D and 4E). Since the nuclei of the neurons are located in the ganglia of the spine, the presence of nuclei within the NB suggests the presence of supporting cells (e.g., Schwann cells), neuroimmune cells (published previously, for example (Blaszkiewicz et al., 2020)), or potentially, perineural adipocytes residing within the bundles.
Most excitingly, for the first time, we have provided evidence of a neuro-adipose nexus (NAN) in ing-scWAT. These images were taken from blocks of ing-scWAT without z-depth reduction. While working to determine if adipose tissue nerve terminals formed a true synapse or tissue junction (like the neuromuscular junction), we observed this structure which begins with nerves traversing along blood vessels that more distally branch away from the vessel and engulf single adipocytes in a web of neurites (Figures 4F and 4G). We believe these may represent the elusive “synapse” or “junction” of nerve endings on terminal cells in adipose tissue since the wrapping neurites appear to be at nerve endings. This is distinct from the junctions formed by nerve endings on adipose tissue vasculature, where there is likely a different type of terminal structure. It is currently unknown if other cell types in WAT also possess NAN structures, such as cells in the stromovascular fraction. A previous demonstration of visualized neuro-adipose connections displayed a different anatomical organization (Zeng et al., 2015), and these images may have actually been an innervated blood vessel (similar to what we have shown in Figure S2B). Due to the dense innervation of adipose blood vessels, it can sometimes be challenging to distinguish nerves from blood vessels if the vessels do not have their own fluorescent marker.
In general, the understanding of adipose tissue innervation has not extended to characterizing the nerve ending cellular interactions, and whether or not the nerve terminal forms a true “synapse” or “junction” versus the diffusion of neuropeptides and neurotransmitters from free nerve endings. The discovery of a neuro-adipose nexus is a start in answering these questions, but further studies using nerve terminal or synaptic/junction markers will also be necessary.
To further characterize the nerves within scWAT, numerous immunostaining experiments were conducted by co-staining β3-tubulin with markers for either sympathetic nerves, sensory nerves, or myelination using the markers tyrosine hydroxylase (TH), advillin (AVIL), and myelin protein zero (MPZ) respectively. Whole mount imaging demonstrated fluorescence overlap of β3-tubulin and TH in the largest NBs with extensive TH+ axons that spanned throughout the tissue (Figure 5A). NB digital cross-sectioning showed that only some of the axons in each NB were TH+ (Figure 5B). The small individual axons branching through the tissue, however, were nearly all TH+ (Figure 5C), consistent with previous reports (Giordano et al., 2006; Jiang et al., 2017).
Figure 5.
Characterization of whole mount scWAT nerves.
(A–G) scWAT depots from C57BL/6J mice were whole mount processed and stained with β3-Tubulin (green) and either TH (red) (A–C), AVIL (red) (D), or MPZ (red) (E–G). Whole mount tiled Zmax projections of ing-scWAT (A and D) and axi-scWAT (G) imaged with 10X objective. White boxes show a single tile expanded for visualization with 5.75X digital zoom (A and E). NB cross sections imaged with 63X objective with a 2.54X zoom factor applied to the digital cross-section rendering (B and E). Representative images of small fiber innervation imaged as Zmax projections with 63X objective (C and F). All images captured on Leica TCS SP8 DLS confocal microscope (A–G). Scale bars are 2 mm (A), 25 μm and 10 μm (B), 25 μm and 50 μm (C), 50 μm and 10 μm (D), 5 mm (E), 25 μm (F), a 25 μm and 10 μm (G).
Digital cross-sectioning of AVIL+ sensory NBs revealed similar findings to cross-sectioning TH+ bundles, with only some of the axons within bundles presenting as AVIL+ (Figure 5D). This further suggested the presence of mixed nerves in adipose. It is worth noting that the AVIL antibody used here was observed to mark structural proteins in some large blood vessels as well, which accounted for the presence of AVIL+ regions that were β3-tubulin- when viewed as a tiled image. At greater magnification, these large blood vessels did not interfere with imaging, and the lack of AVIL+ nerve fibers became apparent. Due to this, AVIL would appear to be a less-than-ideal marker for quantifying sensory innervation in conditions that may also alter vascularity. It is thought at this time that an AVIL reporter mouse or a Nav 1.8 reporter mouse would be a far superior method of labeling sensory axons until superior antibodies or other techniques are developed. It is unclear at this time if myelin alone is a sufficient marker for sensory nerves, as there is a high likelihood thinly myelinated sympathetic nerves are also present in adipose tissues.
Therefore, Nav 1.8-Cre x tdTomato reporter mice were also used to investigate the presence of sensory innervation in scWAT. Nav 1.8 marks sodium channels specific to sensory nerves (Bird et al., 2013) and fluorescence imaging showed a number of Nav 1.8+ sensory nerves throughout the tissue (FigureS4A). These were both large bundles and smaller parenchymal fibers, which complements our own results with AVIL imaging and current literature suggesting sensory innervation plays a significant role in WAT metabolic function (Bartness and Bamshad, 1998; Shi et al., 2005). As observed with AVIL immunostaining, imaging of ing-scWAT from Nav 1.8-Cre x tdTomato reporter mice also suggest the presence of mixed NBs in this adipose depot (Figure S4A).
Whole mount staining with β3-tubulin and MPZ revealed that NBs 25μm and greater in diameter in scWAT tended to be myelinated (Figure 5E). In concordance with immunostaining, Luxol fast blue (myelin stain) staining of whole ing-scWAT depots was performed which showed that the most highly myelinated nerves traverse through or nearby the SiLN (Figure S4B). It is not yet clear if these are mainly sensory axons that are myelinated, or a mix of sensory and sympathetic. Zmax projection imaging of parenchymal nerve fibers revealed that the majority of the smaller nerve endings are unmyelinated (Figure 5F). Digital cross-sectioning of MPZ+ NBs showed that most axons within the bundle appeared myelinated, with the greatest fluorescence intensity being on the exterior of the bundle (Figure 5G). There appears to be incomplete colocalization at the bundle center which, at this time, is thought to be gradation caused by incomplete antibody penetrance, but it cannot be ruled out that the exterior axons of these bundles are more myelinated than the interior axons. This potent MPZ+ staining on the bundle periphery also acts to mask some of the β3-tubulin+ nerves that reside within.
Our whole mount imaging technique has validated the presence of mixed bundle nerves residing in WAT as shown previously (Zeng et al., 2015) and allowed for further investigation by creating digital cross sections of these mixed bundles. High magnification Zmax projections revealed that most neurites within the scWAT parenchyma are TH+, however, this was shown using TH-antibody which is known to be somewhat non-specific. Studies should be redone using a TH-reporter mouse to be certain. A relative minority of axons in the tissue are myelinated (the majority appear to be unmyelinated), and those that are myelinated reside mostly within or nearby large NBs, or in parallel to large blood vessels. The contribution of myelinated versus unmyelinated axons to tissue function is currently unknown, but these observations may provide clues. Quantified colocalization of the various neuronal markers outlined above (PGP9.5, β3-tubulin, TH, MPZ, AVIL) at the whole tissue level and on NB cross sections can serve as another route for continued investigation.
Neurovascular interaction in scWAT
The autonomic nervous system, comprised of sympathetic, parasympathetic, and sensory nerves, is required for regulating vascular tone (Sheng and Zhu, 2018) throughout the body, and WAT is no different. Generally, parasympathetic fibers are responsible for vasodilation and sympathetic nerve fibers are responsible for vasoconstriction, though this response is receptor specific and sympathetic nerves can mediate vasodilation as well (Sheng and Zhu, 2018). It has been suggested that WAT lacks parasympathetic innervation (Giordano et al., 2006), indicating that precise control of vasodilation is either not required in WAT or regulated entirely by sympathetic and sensory innervation.
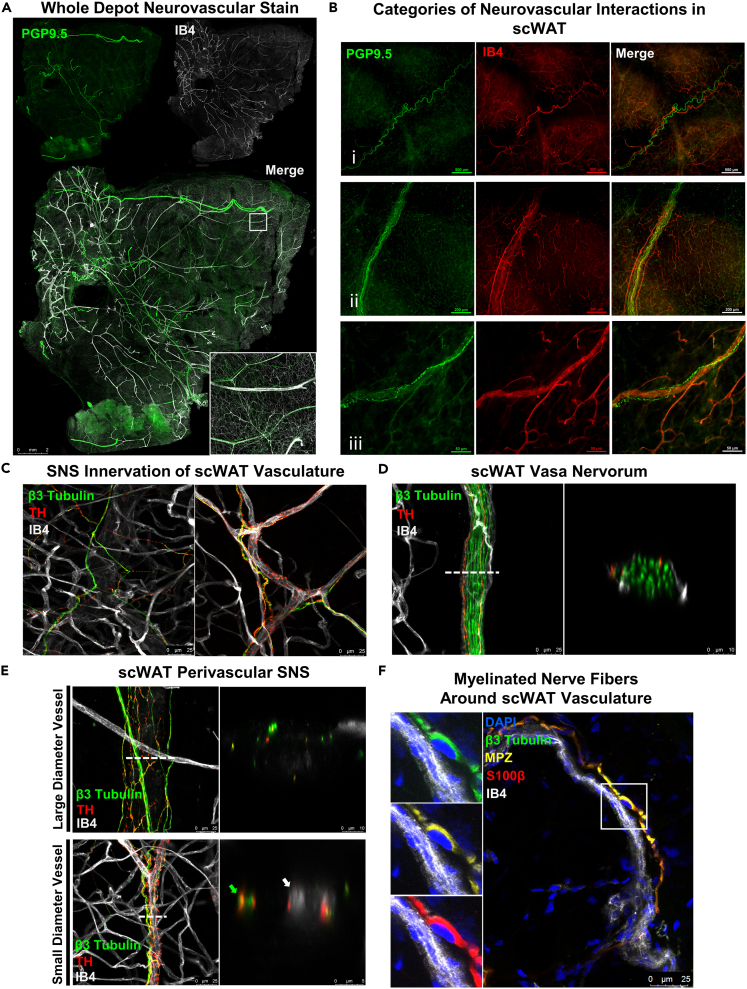
Neurovascular staining of whole mount scWAT was performed by co-staining with pan-neuronal markers and IB4, a marker for vasculature, as it binds to erythrocytes and endothelial cells (Martinez-Santibanez et al., 2014; Ernst and Christie, 2006; Peters and Goldstein, 1979; Ismail et al., 2003; Gorakshakar and Ghosh, 2016) and effectively marks vessels smaller than 50 μm in diameter. Whole mount axi-scWAT tiled Zmax projections exposed a dense vascular and lymphatic network residing in the scWAT depot (Figure 6A). In general, the highest concentration of NBs and blood vessels that are greater than 25μm in diameter are located within close proximity of one another. These observations are consistent with what we have observed in ing-scWAT (Blaszkiewicz et al., 2019a). Close inspection of the neurovascular interactions within scWAT revealed three specific and recurring types of interactions (Figure 6B): (1) Large NBs and blood vessels that run next to each other within the tissue but do not seem to interact or intersect; (2) large NBs that have a vascular supply, likely providing nutrients; or a vasa nervorum; and (3) blood vessels that are highly innervated by smaller neurites which may regulate vasoconstriction, like a perivascular sympathetic plexus.
Figure 6.
Neurovascular interaction within whole mount scWAT
(A) scWAT from C57BL/6J mice was whole mount processed using ZDR. Whole axi-scWAT depot stained with PGP9.5 (green) and IB4 (white) was captured as tiled Zmax projections imaged with 10X objective on Leica TCS SP8 DLS microscope. White box shows a single tile expanded for visualization 7.33X digital zoom.
(B) PGP9.5 (green) and IB4 (red) used to display the 3 categories of neurovascular interaction within scWAT, imaged on Nikon E400 with 4X, 10X, and 40X objectives respectively.
(C–D) Representative images of the innervation of blood vessels imaged as Zmax projections with 63X objective. β3-Tubulin (green), TH (red), IB4 (white). Sympathetic innervation of small blood vessels (C). Vasa nervorum of ing-scWAT NB imaged as Zmax projection digitally cross sectioned with a 2.54X zoom factor (D).
(E) Large and small diameter blood vessel digital cross sectioning. 2.54X zoom factor applied to large diameter vessel cross section, 6.96X zoom factor applied to small diameter vessel cross section.
(F) Representative image of myelinated nerve fiber around vasculature imaged as Zmax projection with 63X objective. Nucleated region expanded for visualization. DAPI (blue), β3-Tubulin (green), MPZ (yellow), S100β (red), IB4 (white). Scale bars are 2 mm (A), 500 μm (B-i), 200 μm (B-ii), 50 μm (B-iii), 25 μm (C), 25 μm and 10 μm (D), 25 μm, 10 μm, and 5 μm (E), and 25 μm (F).
Sympathetic innervation of scWAT blood vessels was also analyzed by co-staining β3-tubulin with TH and IB4. Small TH+ nerve fibers were found lining many of the vessels (Figure 6C). While small capillaries had significantly less innervation compared to larger arterioles, some TH+ nerves were found running along them. This supports the current literature that capillaries are relatively lacking in sympathetic innervation, whereas larger vessels have more sympathetic innervation (Rechthand et al., 1986; Klabunde, 2012; Thomas, 2011). Larger blood vessels likely also have sensory innervation as well (Westcott and Segal, 2013).
Not all nerves within the tissue are lining blood vessels; many can be found branching the gaps from one blood vessel to another or disassociated from the blood vessels entirely. The vasa nervorum, or the blood vessels supplying nutrients to the NB, were digitally cross-sectioned to show a blood vessel branching around an NB which contained 2 sympathetic axons (Figure 6D). Extensive innervation of arterioles could be found surrounding both large and small diameter vessels (Figure 6E). Of the nerves running throughout scWAT, the majority of MPZ+ nerves were found running along blood vessels and had nuclei directly contacting them, as indicated by co-staining with DAPI, β3-tubulin, MPZ, S100β (a Schwann cell marker), and IB4 (Figure 6F). Only one or two of the largest nerves lining each vessel tended to be myelinated. The smallest sympathetic projections that often engulfed many of the arterioles, when present, were found to be unmyelinated.
It is important to note several caveats with IB4 staining of vasculature. Mouse blood vessel diameter ranges from 250μm to 4μm, depending on type of vessel. Veins encompass the upper limit, while microvasculature such as capillaries encompass the lower limit (Müller et al., 2008). Large diameter blood vessels (>50um) often lose their erythrocytes during tissue preparation. However, erythrocytes tend to remain in the microvasculature. Because of this, the microvasculature is doubly stained (erythrocytes and endothelium) whereas arteries and veins present only with stained endothelium; notwithstanding, these vessels are still readily visible (Figure 6). Also, large sensory NBs, immunostained for AVIL, an actin binding protein expressed specifically in somatosensory neurons (Hunter et al., 2018; Hasegawa et al., 2007), tended to be co-stained with IB4. IB4 binding non-specifically to sensory neurons has been reported in the literature (Fang et al., 2006; Vulchanova et al., 2001). However, only large sensory NBs were observed to be labeled by IB4, eliminating the risk of misidentifying capillaries and, due to tissue morphology, positively stained NBs can be easily distinguished from the vasculature.
Whole mount imaging allows for quantification of total depot innervation and vascularity
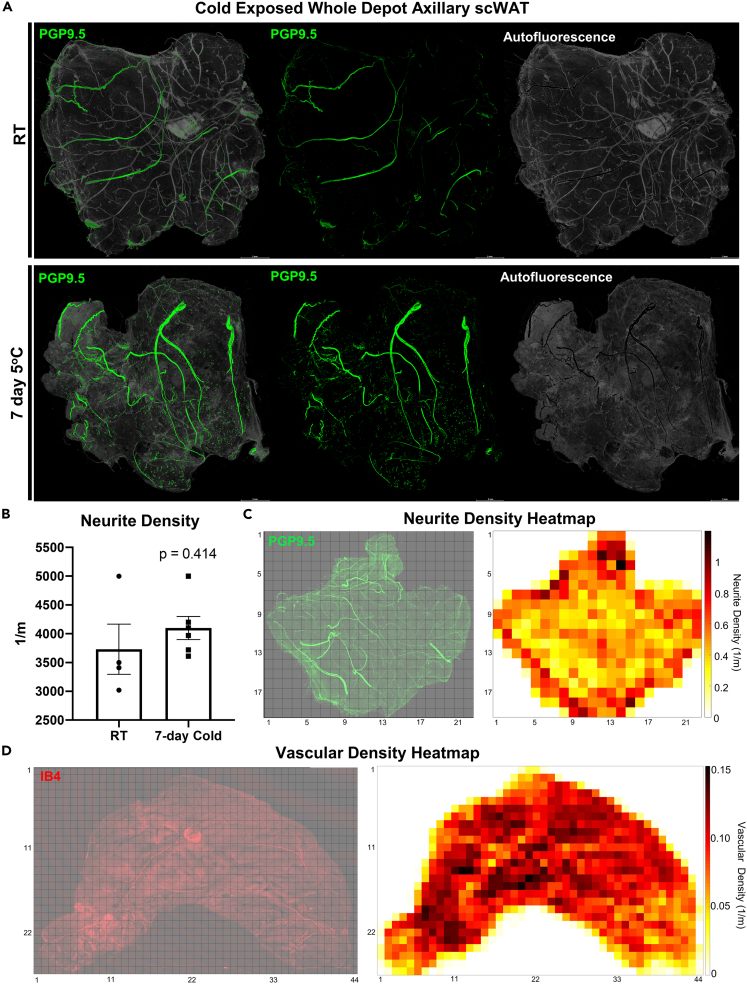
Axi-scWAT depots taken from male C57BL/6J mice, housed at room temperature (RT) or cold exposed at 5°C for 7-days, were processed for whole mount imaging using the pan-neuronal marker PGP9.5 (Figure 7A). The neurite densities per tile were then averaged for the entire area of the tissue for comparison among treatments. Quantification of images allowed for statistical analysis between experimental groups which showed that three of the four RT mice had less neurite density than all of the cold exposed mice but that overall, this difference was not statistically significant (p = 0.414) (Figure 7B). This is similar to our previous studies using ing-scWAT depots which used an alternative, but complementary, approach to whole tissue neurite density quantification (Blaszkiewicz et al., 2019a). Whole depot quantification might blunt regional differences in innervation.
Figure 7.
Whole depot neurite density quantification and heatmap generation
(A) Female C57BL/6J mice, aged 14–16 weeks, were either cold exposed (5°C) (n = 6) or maintained at room temperature (n = 4) for 7 days. Axi-scWAT depots were whole mount processed and stained with PGP9.5 and imaged with 10X objective. To demonstrate the difference between true signal and tissue autofluorescence pixel intensity less than 55 was considered to be autofluorescence (gray) and pixel intensity 55 or greater was considered true staining (green).
(B) Images were Zmax projected, tiled, and thresholded and neurite density quantification was performed, compared across the cohort, and analyzed using a two-tailed Student’s ttest. Alpha level 0.05, error bars are SEMs.
(C) Neurite density per Zmax tile was calculated and used to generate a heatmap.
(D) Ing-scWAT depot was stained with IB4 to mark vasculature, imaged at 10X, and vascular density per Zmax tile was calculated and used to generate heatmap. Each tile is comprised of a 0.12 cm × 0.12 cm area (C and D). All imaging was performed with a Leica TCS SP8 DLS laser scanning confocal microscope. Scale bars are 2 mm (A).
Skeletonizing and quantifying neurite density from Z-maximum projections, by definition, results in under sampling data. To verify the extent of this potential loss of spatial information, we calculated the neurite density on the individual Z-slices that make up the 3D confocal data cube and then synthesized these quantities in two different ways: by taking the average neurite density score across the confocal stack (Figure S5A), or by taking the maximum neurite density score across the confocal stack (Figure S5B). In both cases, there is a very high correlation between the synthesized neurite density score (either obtained by averaging or by taking the maximum) and the neurite density score obtained from the Zmax projection image (p<0.0001.) Of course, this does not allow us to conclude that no information is being lost by restricting the analysis to 2D Zmax projections. However, it does allow us to conclude that the potential gain that one might make by considering all the individual 2D slices instead of only the Zmax projection would have to be explored beyond simple statistical measures such as taking the average or the maximum. Additionally, this potential gain in information would have to be weighed against the additional computational analysis time and effort. Neurite density per Zmax tile was portrayed as a heatmap to better visualize region specific differences in neurite densities (Figure 7C). This quantification and analysis approach can also be applied to vasculature (Figure 7D).
Second harmonic generation imaging of obese ing-scWAT
We also visualized the changes to scWAT innervation of morbidly obese mice which we have demonstrated to have adipose neuropathy (BTBRob/ob) (Blaszkiewicz et al., 2019a). This proved technically difficult to accomplish using epifluorescence or standard confocal microscopy. Obese adipose tissue presents far more challenges; including fragility of adipocytes that can burst under pressure, autofluorescence due to fibrosis that is not ameliorated through TyB staining, and the sheer size of the sample which can often be too large to effectively compress with ZDR. Because of this, our technique is not yet tailored for obese tissues. However, with second harmonic generation label-free imaging using 2-photon microscopy, it was possible to visualize nerves and collagen within scWAT of BTBRob/ob animals (Figure S6). While collagen is abundant in both BTBR+/+ and BTBRob/ob animals, a greater degree of colocalization and apparent alignment between collagen fibers and nerves is visible in BTBRob/ob animals (Figure S6), likely contributing to our inability to image the adipose nerves successfully in this mouse model.
Discussion
The whole mount processing and imaging techniques presented here are intended to broaden the imaging capabilities currently available to researchers investigating the anatomy, structure, and function of adipose nerves. We do not present a method meant to fully replace the need for optically clearing adipose depots or the use of light sheet microscopy, but wish to provide an alternative and complementary method. We have presented data to support that more complex clearing techniques are not required to get high quality images of adipose innervation. We have demonstrated how our method can be applied to imaging on affordable widefield epifluorescence microscopes as well as laser scanning confocal microscopes, to illustrate that this method of tissue processing can be applicable to researchers regardless of the tools they have available. The ease and accessibility of being able to take an entire adipose tissue and mount it on a slide to be imaged on any microscope will ensure that novel discoveries improve our understanding of brain-adipose neural communication and tissue-level regulation of adipose innervation.
Although the time required to image entire tissues/depots for demonstrating gross neuroanatomy or to perform neurite density quantifications can be substantial, it takes only minutes to capture high-resolution single tile Z-stacks to demonstrate the finer structures and thin neurites abutting cells. Due to the broad adaptability of this technique, it is even possible to image whole tissues for aberrant gross anatomical changes in only a couple of hours, if desired, by lowering magnifications, decreasing resolutions, or by using a resonant scanning microscope.
The data collected herein reveal a rich and diverse neural innervation in mouse ing-scWAT, likely indicating physiological roles that are yet to be uncovered. For example, differences in myelinated versus unmyelinated axons, heterogeneity of TH+ nerves, and the contribution of sensory nerves in adipose are yet to be clarified. In addition, it is important to better understand the localization of true synapses or tissue junctions, versus release of nerve products from free nerve endings, and the role that the newly discovered neuro-adipose nexus plays in nerve function in ing-scWAT. The availability of new techniques allowing nerve visualization under various physiological and pathophysiological conditions will help the field advance an understanding of adipose innervation and brain-adipose communication, including neurovascular and neuroimmune contributions, and plasticity of neurites in adipose.
We have recently demonstrated diminished innervation of adipose tissue under circumstances of obesity, diabetes, and aging, a condition we have called ‘adipose neuropathy’ (Blaszkiewicz et al., 2019a). Neuropathy of adipose tissue (i.e.. a reduction in neurite density that may represent pathological nerve die-back) could have effects beyond the removal of sympathetic nerves releasing NE. Given that other nerve products, including from adipose tissue sensory nerves (e.g., Substance P, VIP, CGRP, etc.) are found in adipose depots, the loss of proper innervation could also impact the physiological contributions of these neuromodulatory substances in adipose. Whether or not adipose sensory nerves directly communicate fuel status to the brain, perhaps in complement to endocrine factors such as leptin, is still an open question. Incoming sympathetic nerves may release NE to affect vasoconstriction, and impact local adipocytes and immune cells. We hope that by providing our method as a resource for investigators that these questions can begin to be answered.
Limitations of study
When implementing the techniques outlined in this manuscript there are certain limitations that should be considered. Potential changes to tissue architecture in the z-plane introduced by the ZDR method could occur. In this manuscript, we demonstrated tissue structural changes, or lack thereof, after ZDR, but our investigation was limited to cell types most relevant to this manuscript: subcutaneous WAT mature adipocytes, as well as adipose tissue blood vessels and nerves. We recommend that those who plan to adapt this technique for tissues not validated in this manuscript should conduct their own observational analysis to ensure that this technique does not create structural artifacts that may negatively impact their investigational goals. Additionally, ZDR by definition reduces z-axis spatial information. However, these tissues are not completely flattened, their depth has only been reduced by about 3-fold. This still retains some valuable z-axis information as demonstrated by the three dimensional and depth coded datasets presented here. The entirety of adipose nerve fiber density is also maintained, which is important if the goal is comprehensive quantification of tissue innervation. If exact spatial organization is required as an imaging endpoint, optical clearing may be a preferred method; however, clearing methods also have their limitations and are well known to cause tissue structural changes such as tissue expansion, shrinkage, and stiffening which could also alter spatial organization. A final option is to obtain a block of tissue in order to assess innervation without ZDR (as we did to observe the neuro-adipose nexus), but this would not capture the heterogeneity of innervation across an intact tissue.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Monoclonal Recombinant Anti-PGP9.5 [EPR4118] | Abcam | Cat#ab108986 |
| Rabbit Polyclonal Anti-PGP9.5 | Abcam | Cat#ab10404 |
| Rabbit Polyclonal Anti-Myelin Protein Zero | Abcam | Cat#ab31851 |
| Rabbit Polyclonal Anti-p92 (Advillin) | Abcam | Cat#ab72210 |
| Goat Polyclonal Anti-Perilipin-1 | Abcam | Cat#ab61682 |
| Rabbit Polyclonal Anti-Tyrosine Hydroxylase | Millipore | Cat#AB152 |
| Mouse Monolconal Anti-beta III Tubulin [2G10] Alexa Fluor 488 | Abcam | Cat#ab195879 |
| Rabbit Monolconal Recombinant Anti-S100 beta [EP1576Y] Alexa Fluor 647 | Abcam | Cat#ab196175 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-11001 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-11029 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-21121 |
| Goat anti-Mouse IgG, IgM (H+L) Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-10680 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-11055 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-11008 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermofisher Scientific | Cat#A-11034 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 532 | Thermofisher Scientific | Cat#A-11009 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermofisher Scientific | Cat#A-21428 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermofisher Scientific | Cat#A-11005 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermofisher Scientific | Cat#A-11012 |
| Chemicals, peptides, and recombinant proteins | ||
| Paraformaldehyde | Sigma-Aldrich | Cat#P6148 |
| 10X PBS Solution | Teknova | Cat#P0496 |
| Bovine Serum Albumin | Sigma-Aldrich | Cat#A4503 |
| Triton X-100 | Bio-Rad Laboratories | Cat#1610407 |
| TypogenBlack/Sudan Black B | Sigma-Aldrich | Cat#199664 |
| TrueBlackLipofuscin Autofluorescence Quencher | Biotium | Cat#23007 |
| Hydrogen Peroxide solution | Sigma-Aldrich | Cat#516813 |
| Dimethyl Sulfoxide | Sigma-Aldrich | Cat#D8418 |
| Methanol | Fischer Chemical | Cat#A452 |
| HEPES | Sigma-Aldrich | Cat#H7523 |
| Sodium Chloride | Sigma-Aldrich | Cat#S3014 |
| Calcium Chloride Dihyrdate | Sigma-Aldrich | Cat#C3306 |
| Isolectin GS-IB4 From Griffonia simplicifolia, Alexa Fluor 594 | Thermofisher Scientific | Cat#I21413 |
| Isolectin GS-IB4 From Griffonia simplicifolia, Alexa Fluor™ 647 Conjugate | Thermofisher Scientific | Cat#I32450 |
| DAPI, dilactate | Sigma-Aldrich | Cat#D9564 |
| Luxol Fast Blue Stain Kit | Abcam | Cat#ab150675 |
| Heparin Sodium Salt from Porcine Mucosa | Sigma-Aldrich | Cat#H3393 |
| EMS Glycerol Mounting Medium With DABCO | Electron Microscopy Sciences | Cat#17989-5 |
| Formalin | Protocol | Cat#245-684 |
| HistoChoiceClearing Agent | Sigma-Aldrich | Cat#H2779 |
| Ethanol | Sigma-Aldrich | Cat#E7023 |
| Mayers Hematoxylin Solution | Sigma-Aldrich | Cat#MHS16 |
| Blocking Reagent | EMD Millipore Corp | Cat#20773 |
| 20XRinse Buffer | EMD Millipore Corp | Cat#20845 |
| Antibody Diluent | Dako | Cat#S0809 |
| Light Diagnostics Mounting Fluid (non-permanent) | Sigma-Aldrich | Cat#5013 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664 |
| Mouse: PGP9.5+/-: C57BL/6-Tg(Uchl1-EGFP)G1Phoz/J | The Jackson Laboratory | Cat#022476 |
| Mouse: BTBR.Cg-Lepob/WiscJ | The Jackson Laboratory | Cat#004824 |
| Mouse: Nav 1.8cre x tdTomato | Dr. Ian Meng, University of New England | N/A |
| Software and algorithms | ||
| FIJI | Schindelin et al., 2012 | https://imagej.net/software/fiji/downloads |
| MATLAB x64 (version 2018b) | MathWorks | https://www.mathworks.com/products/matlab.html |
| Python (version 3.7) | Python Software Foundation | https://www.python.org/downloads/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
| LAS X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Nikon Elements Advanced Research Package | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| Microsoft PowerPoint 2019 | Microsoft | https://www.microsoft.com/en-us/download/office.aspx |
| Code generated for this manuscript has been made publicly available on GitHub | This paper, GitHub.com | https://github.com/ktownsendlab/willows_et_al-2020 |
| Other | ||
| 75 x 51 mm Glass Slide, 1.2 mm Thick | Electron Microscopy Sciences | Cat#71862-01 |
| Cover glass, 48x60mm, 1.5 thick | Brain Research Laboratories | Cat#4860-1.5D |
| Protocols used in this manuscript have been made publicly available on Protocols.io | This paper, Protocols.io | https://doi.org/10.17504/protocols.io.brs2m6ge |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kristy Townsend (kristy.townsend@osumc.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The following mouse strains were obtained from The Jackson Laboratory: C57BL/6J (Stock # 000664); BTBR+/+ (WT) and BTBRob/ob (MUT) (BTBR.Cg-Lepob/WiscJ, Stock # 004824), PGP9.5+/- (C57BL/6-Tg(Uchl1-EGFP)G1Phoz/J, Stock # 022476). Nav1.8 reporter mice (Nav 1.8cre x tdTomato) were a kind gift from Dr. Ian Meng, (University of New England, Biddeford, ME, USA). Animals were housed 3-5 to a cage (except for BTBR WT and MUT animals which were housed in pairs), in a monitored temperature and humidity-controlled environment with 12/12hr light/dark cycle. Cages were cleaned weekly, ad libitum access to standard chow diet and water was maintained. For all studies animals were sacrificed using CO2 followed by cervical dislocation.
Adult male and female C57BL/6J and PGP9.5+/- were used for majority of immunostaining studies. Adult male BTBR WT and MUT were used in second harmonic generation imaging. Adult female Nav1.8 reporter mice were used for whole mount immunostaining with PGP9.5 (GFP).
All procedures were performed in compliance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and were approved by an Institutional Animal Care and Use Committee.
Method details
Cold exposure experiments
All cold exposure was carried out in a diurnal incubator (Caron, Marietta, OH, USA) at 5°C. Adult female C57BL/6J mice were housed two to a cage and either maintained at room temperature or continuously cold exposed for 7 days. Axi-scWAT was collected for whole mount tissue processing.
Mouse adipose tissue collection and whole mount processing for immunofluorescence
Mice were euthanized using CO2 followed by cervical dislocation. Whole scWAT depots were carefully removed to remain fully intact and immediately fixed in 2% PFA at 4°C for 4–16hr depending on thickness of tissue. The tissues were then rinsed for 30 min with 1X PBS w/10U/mL heparin, twice at 4°C. Tissues next underwent ZDR in which tissues were flattened by being placed between two large glass slides bound tightly together with 2 large binder clips, for 1.5hr at 4°C. Tissues were incubated in blocking buffer (1XPBS/2.5% BSA/1% Triton X-100) at 4°C overnight but can be incubated up to 7 days if desired with blocking buffer replaced every 24hr. After blocking period tissues underwent one of three methods of autofluorescence quenching: 1) Incubation in 0.1% Typogen Black for 20 min at room temperature on a rotator, 2) incubation in 1X (0.1%) TrueBlack® Lipofuscin Autofluorescence Quencher for 10 min at room temperature on a rotator, or 3) stepwise dehydration and subsequent bleaching with 5% hydrogen peroxide in 20% DMSO/MeOH as described by Renier et al. (2014). Following quenching the tissues were washed with 1X PBS w/10U/mL heparin on rotating platform at 4°C replacing PBS every 1hr for a total of 4–6hr, or until all unbound stain was removed. Tissues were incubated with primary antibody for 48hr at 4°C. The following day tissues were washed with 1XPBS on a rotating platform at 4°C, replacing PBS every 1hr for a total of 4–6hr followed by incubation with secondary fluorescent antibodies overnight. Tissues were then again washed with 1XPBS on a rotating platform at 4°C, replacing PBS every 1hr for a total of 4–6hr. Following the immunostaining steps, when applicable, tissues were incubated with 1-5 μg/mL isolectin IB4 (IB4) (ThermoFischer, cat#:I32450 or cat#:I21413) diluted in HEPES buffer (1 mM CaCl2 dihydrate, 154 mOsm Na+, 25 mM HEPES, in H2O, pH 7.4), overnight at room temperature on a rotator. Tissues were then washed with 1XPBS on a rotating platform at room temperature for 1 hr twice. At this time, if DAPI co-staining was desired, the tissues were incubated in 100ng/mL DAPI for 1 hr at room temperature on a rotator. Tissues then received four 1-hr washes in 1XPBS at room temperature. At the conclusion of washing steps, tissues were placed on large glass slides medial side facing up. A few drops of glycerol based mounting media were added to the tissue and coverslip was placed on top. Glycerol-based mounting media was used because aqueous based mounting fluid lacks the required viscosity to fully adhere the coverslip when dealing with whole mount adipose. Slides were weighted down for 2 days and then sealed. Protocols for tissue dissection, processing for immunofluorescence, and confocal imaging can be found at https://dx.doi.org/10.17504/protocols.io.brs2m6ge.
Uncompressed whole mount tissue processing for immunofluorescence
When tissues were left uncompressed to visualize structural changes resulting from ZDR and mounting, small variations were made to the above whole mount procedure to ensure that tissues were handled equally but still able to be somewhat visualized without ZDR. Bilateral whole scWAT depots were excised fully intact and fixed as previously stated. One depot received ZDR whereas the other depot did not and remained in 1X PBS. Afterwards, each depot was diced into small 2 mm2 chunks and progressed through the above protocol until mounting. To avoid distortion introduced from traditional mounting approaches, the uncompressed control tissues were either imaged in a glass bottom dish which introduced no compression regardless of tissue thickness, and alternatively, depression slides were implemented for anterior and posterior tissue chunks, <0.9mm thick, distal to the SiLN located at the tissues thickest point. If any compression was observed when placing the coverslip, the tissue was excluded from further analysis.
Thin section adipose immunofluorescence
Whole ing-scWAT depots were excised from male C57BL/6J mice, fixed in 10% buffered formalin overnight at room temperature and embedded in paraffin. Paraffin embedded tissues were sectioned at 7μm, deparaffinized with HistoChoice Clearing Agent, and hydrated in decreasing concentrations of EtOH (100%, 95% 70%, 30%, 0%). Antigen unmasking was performed with Dako Target Retrieval Solution at boiling temperature for 20 min. Slides were incubated in 0.3% Typogen Black (in 70% EtOH) for 20 min at room temperature to reduce autofluorescence. Tissues were washed in Millipore Rinse Buffer twice for 5 min each at room temperature. Tissues were blocked for non-specific binding using Millipore Blocking Reagent for 20 min at 37°C. Primary antibody was added overnight at 4°C. The following day tissues were washed in rinse buffer and secondary antibody was added for 10 min at room temperature. Tissues were then rinsed, and the next primary antibody was added with steps repeated. Isolectin IB4 was added to the tissue for 1 hr at room temperature and then washed. Slides were incubated in DAPI (100 ng/mL) for 5 min at room temperature followed by rinses with dH2O. A drop of mounting fluid was applied to each tissue. Tissues were cover slipped, sealed, and imaged.
Widefield microscopy
All widefield micrographs were captured on a Nikon E400 with either a DS-fi2 color camera (Nikon, Japan) or Hamamatsu ORCA-Flash4.0 V2 Digital CMOS monochrome camera (Hamamatsu Photonics K.K., C11440-22CU) in conjunction with filter cubes for DAPI, GFP/FITC, Cy3/TRITC, and Cy5. Images captured with Hamamatsu ORCA-Flash4.0 V2 were pseudo-colored with NIS-Elements software (Nikon, Japan.) The following objectives were used: Nikon CFI Plan Apochromat Lambda 4X (NA 0.20, WD 20.0mm, dry), Nikon CFI Plan Apochromat Lambda 10X (NA 0.45, WD 4.00mm, dry), Nikon CFI Plan Fluor 20X (NA 0.50, WD 2.10mm, dry) and Nikon CFI Plan Fluor 40X (NA 0.75, WD 0.66mm, dry.)
Confocal microscopy
Confocal micrographs were captured on one of two laser scanning confocal microscopes: Leica TCS SP8 DLS (Leica Microsystems, Wetzler, Germany) and Nikon A1R (Nikon, Japan).
Leica TCS SP8 DLS - Fluorescent labels were excited with either a diode 405 nm laser (DAPI) or a white light laser (Alexa488, GFP, Alexa532, Alexa555, tdTomato, Alexa568, Alexa647) with excitation and emission spectra tuned specifically for each fluorophore or groups of fluorophores to reduce and eliminate crosstalk. All imaging was performed sequentially by frame. Photons were detected with standard PMTs and HyDs. The following objectives were used: HC PL FLUOTAR 5X (NA 0.15, WD 13.70mm, dry), HC PL APO CS2 10X (NA 0.40, WD 2.74mm, dry), HC PL APO CS2 20X (NA 0.75, WD 0.62mm, dry) and HC PL APO CS2 63X (NA 1.40, WD 0.14mm, oil.)
Nikon A1R - Fluorescent labels were excited with a 402 nm laser (DAPI), 487 nm (GFP), 561 nm (Alexa594), and 638 nm (TyB far-red autofluorescence). Imaged sequentially. Photons were detected with standard PMTs as well as GaAsPs. The following objectives were used: CFI Plan Apo Lambda 10X (NA 0.45, WD 4mm, dry), CFI Plan Apo Lambda 20X (NA 0.75, WD 1mm, dry), CFI Super fluor 40X (NA 1.3, WD 0.19mm, oil.) Used also for resonant scanning.
Intact scWAT depots were imaged by tiling Z-stacks across the full depth and area of the tissue. Whole depot micrographs in this manuscript were scanned bidirectionally at either 400Hz or 600Hz at 720x720 resolutions. Line averages ranged between 3-8 and Z-step size ranged between 5-16 μm. Tissue thickness, despite its variability, was easily captured within the Z-step range of the software (±250 μm of the focal point.) Identical image acquisition settings were applied for all tissues within cohorts that received neurite density quantifications. Anywhere between 4,000 and 65,000 individual images were captured per tissue which ranged from 100 to 900 tiles. These tiles were then merged and processed into a 2D maximum intensity projection either in LASX (Leica imaging software) or in Fiji (Schindelin et al., 2012) if being used for quantification. Whole depot image insets consisted of randomly chosen 5X or 10XZ-max tiles of the merged image expanded for improved visualization.
Representative images were captured at 1024x1024 resolution and were Z-max projected or 3D reconstructed with alpha blending in LASX and Nikon Elements Advanced Research software. Digital NB cross-sectioning was performed by using the XZY scanning function in the LASX software during image acquisition which applied a 2.54X zoom factor automatically to each image. All images were pseudo colored.
Neurite density quantification and heatmap generation
Maximum intensity projections through the Z-dimension (Z-max projection) were generated for each tile individually using Fiji (Schindelin et al., 2012). All single tile Z-max projections were further processed using MATLAB x64 software (version 2018b, MathWorks). To remove low-frequency background noise, a 2D Gaussian smoothing kernel was used to convolve each Z-max projection with a very large Gaussian blur using the imgaussfilt MATLAB command, with large standard deviation (∼150), which was then subtracted from its corresponding original Z-max projection. After background subtraction, a small gaussian blur (imgaussfilt with small standard deviation, ∼2-3) was used to broaden out the neurite signal slightly before thresholding the image. Next, a binary thresholded mask was generated from the processed image. In the mask, only regions having an area larger than 40 pixels were kept for further analysis (using the bwareaopen MATLAB command). Then the bwskel MATLAB command was used to perform the skeletonizing procedure, with the added feature of removing any branches less than 4 pixels long. Except for an experimental analysis of 2D Z-max projections vs. the individual 2D slices making up the 3D confocal data (Figure S5), total nerve length was calculated using values measured for each 2D Z-max projection. Total nerve arborization density was calculated as the ratio of total nerve length divided by the total viewing area, resulting in nerve length per square meter of tissue.
To generate heat maps a csv file was generated with the topological positions and the associated arborization quantity of each tile using the HeatMapChart function. Heatmap generation has multiple variables controlled by the user that could be explored further, such as the size of the subregion size and whether or not (and to what extent) the subregions may overlap. Code and instruction can be found at https://github.com/ktownsendlab/willows_et_al-2020.
It is also worth noting that the Python and MATLAB scripts developed for this study, provided above, allow the user to adjust the described parameters to account for batch differences due to variation in fluorescence intensity and autofluorescence. These adjustments can be made a priori or while looking at the skeletonized product of a single tile to quickly visualize how each adjustment changes the skeletonization.
Antibodies and fluorescing stains
Primary antibodies included: PGP9.5 (1:200, Abcam, ab108986 and ab10404); MPZ (1:250, Abcam, ab31851); Advillin (1:500, Abcam, ab72210); Perilipin-1 (1:100, Abcam, ab61682); TH (1:250, Millipore, AB152); β3-Tubulin conj. Alexa Fluor 488 (1:250, Abcam, ab195879); S100β conj. Alexa Fluor 647 (1:250, Abcam, ab196175).
Secondary antibodies (Thermofisher Scientific, Waltham, MA, USA): Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-11001), Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-11029), Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-21121), Goat anti-Mouse IgG, IgM (H+L) Secondary Antibody, Alexa Fluor 488 (1:500, A-10680), Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-11055) Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-11008), Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:500, A-11034), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 532 (1:500, A-11009) Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 (1:1000, A-21428), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (1:1000, A-11005), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (1:500, A-11012).
Fluorescent stains included: For vascular staining, tissues were incubated in isolectin IB4 stain conjugated to either Alexa Fluor 594 (1ug/ml, ThermoFisher Scientific, Waltham, MA, USA; Cat. #I21413) or 647 (1ug/ml, ThermoFisher Scientific, Waltham, MA, USA; Cat.#I32450). For nuclear staining, tissues were incubated in DAPI (100ng/ml, Sigma-Aldrich, St. Louis, MO, USA; Cat. #D9564).
Luxol fast blue myelin staining of whole adipose
Mice were euthanized, whole subcutaneous white adipose tissue (scWAT) depots were carefully removed to remain intact and fixed in 2% PFA at 4°C for 4hr-12hrs depending on thickness of tissue. The tissues were then rinsed for 10 min with 1X PBS, twice at 4°C. Tissues were incubated in Luxol Fast Blue (Abcam, Cambridge, U.K. Cat. #ab150675) overnight at room temperature, briefly rinsed with distilled water and differentiated in Lithium Carbonate Solution followed by 70% Alcohol Reagent until the solution ran clear and adipocytes were colorless while nerves remained blue. Tissue was then rinsed in distilled water, mounted, and imaged on a dissecting microscope.
Hematoxylin staining
Whole ing-scWAT depots were excised from male C57BL/6J, fixed in 10% buffered formalin overnight at room temperature and embedded in paraffin. Paraffin embedded tissues were sectioned at 7μm, deparaffinized in HistoChoice Clearing Agent, hydrated in EtOH at decreasing concentrations (100%, 95% 70%, 30%, 0%) and stained with hematoxylin to increase cell membrane contrast. A drop of mounting fluid was applied to each tissue. Tissues were then cover slipped, sealed, and imaged.
Cell size quantification
Whole adipose depots were excised from male C57BL/6J mice and fixed in 2% PFA overnight. Whole tissues were either Z-depth reduced and mounted on a glass slide (Z-depth reduced) or remained unmanipulated and placed on a glass slide (control). Representative images were taken of each tissue and cells with clear in-focus boundaries were outlined. Cell area was measured in FIJI (Schindelin et al., 2012).
Second harmonic generation imaging
All 2-photon microscopy studies used a modified Olympus FV300 system with an upright BX50WI microscopy stand (Olympus, Center Valley, Pennsylvania) and a mode-locked Ti:Sapphire laser (Chameleon, Coherent, Santa Clara, California). Laser power was modulated via an electro-optic modulator (ConOptics, Danbury, Connecticut). The fluorescence and SHG signals were collected in a non-descanned geometry using a single PMT (H7422 GaAsP, Hamamatsu, Hamamatsu City, Japan). Emission wavelengths were separated from excitation wavelengths using a 665 nm dichoric beamsplitter followed by 582/64 nm and 448/20 nm bandpass filters for Alexa 488 and SHG signals respectively (Semrock, Rochester, New York). Images were acquired using circular polarization with excitation power ranging from 1- 50 mW and a 40X (0.8 NA) water immersion objective with 3X optical zoom with scanning speeds of 2.71s/frame. All images were 515 x 512 pixels with a field of view of 85 μm.
Image processing and figure formatting
Images were compiled and formatted for figures in PowerPoint (Microsoft). In some cases, brightness was increased equally across the image to enhance small structure visibility. Figure brightness was adjusted identically when direct comparisons were made. Figures were each exported as a Tiff (1650x2200) and then reduced to 300x600 DPI and 8-bit depth for publication.
Quantification and statistical analysis
Cell size (N = 3–4) (Figure 2D) and whole depot neurite density (N = 4–6) (Figure 7B) was quantified using unpaired two-tailed t-tests with assumed Gaussian distribution and equal standard deviations in GraphPad Prism 9. Comparisons between Z-maximum projection, Z-slice averages, and Z-slice maximums (Figure S5) were made using an F-test on linear regression fits and calculated using the R language, version 3.4.4.
Additional resources
Protocols used for this publication can be found on Protocols.io at https://dx.doi.org/10.17504/protocols.io.brs2m6ge.
Acknowledgments
Dr. Benjamin Harrison of UNE for developing earlier iterations of whole depot innervation quantification method (published in Blaszkiewicz et al., 2019a). Morganne Robinson, Thomas Szewczyk and James Miller for technical assistance. Dr. Ian Meng of The University of New England for gifting Nav1.8 reporter mice adipose tissues. Dr. DustinUpdike of MDI Biological Laboratory for assistance with pilot imaging studies. This work was supported by NIH R01 (1R01DK114320-01A1) and American Heart Association (AHA) Collaborative Sciences Award (18CSA34090028).
Author contributions
JWW wrote the manuscript, created and optimized protocols, designed experiments, analyzed data, and conducted the majority of the microscopy studies and technical troubleshooting/optimization. MB wrote the manuscript, designed experiments, created and optimized protocols. EG conducted tissue processing. AL and ALD conducted clearing experiments. SB and AK developed neurite density quantification methods. WPB and KBT conducted 2-photon imaging. KLT wrote the manuscript, designed experiments, analyzed data, oversaw the project and serves as lead contact for this manuscript.
Declarations of interests
The authors declare no competing interests.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103127.
Supplemental information
Data and code availability
Neurite density quantification and heatmap generation code developed for this publication can be found on GitHub at https://github.com/ktownsendlab/willows_et_al-2020. All other data is included in this manuscript and any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Azaripour A., Lagerweij T., Scharfbillig C., Jadczak A.E., Willershausen B., van Noorden C.J. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog.Histochem.Cytochem. 2016;51:9–23. doi: 10.1016/j.proghi.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Bartness T.J., Bamshad M. Innervation of mammalian white adipose tissue: implications for the regulation of total body fat. Am. J. Physiol. 1998;275:R1399–R1411. doi: 10.1152/ajpregu.1998.275.5.R1399. [DOI] [PubMed] [Google Scholar]
- Bartness T.J., Vaughan C.H., Song C.K. Sympathetic and sensory innervation of brown adipose tissue. Int. J. Obes. 2010;34:S36–S42. doi: 10.1038/ijo.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird E.V., Christmas C.R., Loescher A.R., Smith K.G., Robinson P.P., Black J.A., Waxman S.G., Boissonade F.M. Correlation of Nav1.8 and Nav1.9 sodium channel expression with neuropathic pain in human subjects with lingual nerve neuromas. Mol. Pain. 2013;9:52. doi: 10.1186/1744-8069-9-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszkiewicz M., Willows J.W., Dubois A.L., Waible S., Dibello K., Lyons L.L., Johnson C.P., Paradie E., Banks N., Motyl K. Neuropathy and neural plasticity in the subcutaneous white adipose depot. PLoS One. 2019;14:e0221766. doi: 10.1371/journal.pone.0221766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszkiewicz M., Willows J.W., Johnson C.P., Townsend K.L. The importance of peripheral nerves in adipose tissue for the regulation of energy balance. Biology. 2019;8:10. doi: 10.3390/biology8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszkiewicz M., Wood E., Koizar S., Willows J., Anderson R., Tseng Y.H., Godwin J., Townsend K.L. The involvement of neuroimmune cells in adipose innervation. Mol. Med. 2020;26:126. doi: 10.1186/s10020-020-00254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantschen S., Gauchat J.F., de Weck A.L., Stadler B.M. Regulatory effect of recombinant interleukin (IL)3 and IL4 on cytokine gene expression of bone marrow and peripheral blood mononuclear cells. Eur. J. Immunol. 1989;19:2017–2023. doi: 10.1002/eji.1830191108. [DOI] [PubMed] [Google Scholar]
- Cao Y., Wang H., Wang Q., Han X., Zeng W. Three-dimensional volume fluorescence-imaging of vascular plasticity in adipose tissues. Mol. Metab. 2018;14:71–81. doi: 10.1016/j.molmet.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang H., Zeng W. Whole-tissue 3D imaging reveals intra-adipose sympathetic plasticity regulated by NGF-TrkA signal in cold-induced beiging. Protein Cell. 2018;9:527–539. doi: 10.1007/s13238-018-0528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li G., Li Y., Li Y., Zhu H., Tang L., French P., Mcginty J., Ruan S. UbasM: an effective balanced optical clearing method for intact biomedical imaging. Sci. Rep. 2017;7:12218. doi: 10.1038/s41598-017-12484-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J., Wu Z., Choi C.H.J., Nguyen L., Tegegne S., Ackerman S.E., Crane A., Marchildon F., Tessier-Lavigne M., Cohen P. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 2018;27:226–236.e3. doi: 10.1016/j.cmet.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S.Y., Kalyanasundaram S., Andalman A.S., Davidson T.J., Mirzabekov J.J., Zalocusky K.A., Mattis J., Denisin A.K. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti S., Garretson J.T., Grill H.J., Levine A.S., Trayhurn P. Tim Bartness, Ph.D. (1953-2015) Temperature (Austin) 2016;3:31–38. doi: 10.1080/23328940.2016.1150699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A.C., Bottiroli G. Autofluorescence spectroscopy and imaging: a tool for biomedical research and diagnosis. Eur. J. Histochem. 2014;58:2461. doi: 10.4081/ejh.2014.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso D., Brophy P.J., Staugaitis S.M., Gillespie C.S., Frey A.B., Stempak J.G., Colman D.R. Protein zero of peripheral nerve myelin: biosynthesis, membrane insertion, and evidence for homotypic interaction. Neuron. 1990;4:449–460. doi: 10.1016/0896-6273(90)90057-m. [DOI] [PubMed] [Google Scholar]
- Day I.N., Thompson R.J. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog.Neurobiol. 2010;90:327–362. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Dichamp J., Barreau C., Guissard C., CARRIERE A., Martinez Y., Descombes X., Penicaud L., Rouquette J., Casteilla L., Plouraboue F., Lorsignol A. 3D analysis of the whole subcutaneous adipose tissue reveals a complex spatial network of interconnected lobules with heterogeneous browning ability. Sci. Rep. 2019;9:6684. doi: 10.1038/s41598-019-43130-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt H.U., Leischner U., Schierloh A., Jahrling N., Mauch C.P., Deininger K., Deussing J.M., Eder M., Zieglgansberger W., Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Draberova E., del Valle L., Gordon J., Markova V., Smejkalova B., Bertrand L., de Chadarevian J.P., Agamanolis D.P., Legido A., Khalili K. Class III beta-tubulin is constitutively coexpressed with glial fibrillary acidic protein and nestin in midgestational human fetal astrocytes: implications for phenotypic identity. J. Neuropathol. Exp. Neurol. 2008;67:341–354. doi: 10.1097/NEN.0b013e31816a686d. [DOI] [PubMed] [Google Scholar]
- Ernst C., Christie B.R. Isolectin-IB 4 as a vascular stain for the study of adult neurogenesis. J. Neurosci. Methods. 2006;150:138–142. doi: 10.1016/j.jneumeth.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Fang X., Djouhri L., Mcmullan S., Berry C., Waxman S.G., Okuse K., Lawson S.N. Intense isolectin-B4 binding in rat dorsal root ganglion neurons distinguishes C-fiber nociceptors with broad action potentials and high Nav1.9 expression. J. Neurosci. 2006;26:7281–7292. doi: 10.1523/JNEUROSCI.1072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenzl A., Kiefer F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin.Investig. 2014;19:25–37. doi: 10.1515/hmbci-2014-0022. [DOI] [PubMed] [Google Scholar]
- Fiorelli R., Sidhu G.S., Cebrián-Silla A., Melendez E.L., Mehta S., Garcia-Verdugo J.M., Sanai N. Enhanced tissue penetration of antibodies through pressurized immunohistochemistry. bioRxiv. 2020 doi: 10.1101/2020.09.25.311936. [DOI] [Google Scholar]
- Fishman R.B., Dark J. Sensory innervation of white adipose tissue. Am. J. Physiol. 1987;253:R942–R944. doi: 10.1152/ajpregu.1987.253.6.R942. [DOI] [PubMed] [Google Scholar]
- Foster D.O., Depocas F., Zuker M. Heterogeneity of the sympathetic innervation of rat interscapular brown adipose tissue via intercostal nerves. Can. J. Physiol. Pharmacol. 1982;60:747–754. doi: 10.1139/y82-104. [DOI] [PubMed] [Google Scholar]
- Foster M.T., Bartness T.J. Sympathetic but not sensory denervation stimulates white adipocyte proliferation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R1630–R1637. doi: 10.1152/ajpregu.00197.2006. [DOI] [PubMed] [Google Scholar]
- Francois M., Torres H., Huesing C., Zhang R., Saurage C., Lee N., Qualls-Creekmore E., Yu S., Morrison C.D., Burk D. Sympathetic innervation of the interscapular brown adipose tissue in mouse. Ann. N. Y Acad. Sci. 2019;1454:3. doi: 10.1111/nyas.14119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garretson J.T., Szymanski L.A., Schwartz G.J., Xue B., Ryu V., Bartness T.J. Lipolysis sensation by white fat afferent nerves triggers brown fat thermogenesis. Mol.Metab. 2016;5:626–634. doi: 10.1016/j.molmet.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano A., Song C.K., Bowers R.R., Ehlen J.C., Frontini A., Cinti S., Bartness T.J. White adipose tissue lacks significant vagal innervation and immunohistochemical evidence of parasympathetic innervation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R1243–R1255. doi: 10.1152/ajpregu.00679.2005. [DOI] [PubMed] [Google Scholar]
- Gorakshakar A.C., Ghosh K. Use of lectins in immunohematology. Asian J. Transfus. Sci. 2016;10:12–21. doi: 10.4103/0973-6247.172180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Abbott S., Han B.X., Qi Y., Wang F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J. Neurosci. 2007;27:14404–14414. doi: 10.1523/JNEUROSCI.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesing C., Qualls-Creekmore E., Lee N., Francois M., Torres H., Zhang R., Burk D.H., Yu S., Morrison C.D., Berthoud H.R. Sympathetic innervation of inguinal white adipose tissue in the mouse. J. Comp. Neurol. 2021;529:1465–1485. doi: 10.1002/cne.25031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D.V., Smaila B.D., Lopes D.M., Takatoh J., Denk F., Ramer M.S. Advillinis expressed in all adult neural crest-derived neurons. eNeuro. 2018;5 doi: 10.1523/ENEURO.0077-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail J.A., Poppa V., Kemper L.E., Scatena M., Giachelli C.M., Coffin J.D., Murry C.E. Immunohistologic labeling of murine endothelium. Cardiovasc. Pathol. 2003;12:82–90. doi: 10.1016/s1054-8807(02)00166-7. [DOI] [PubMed] [Google Scholar]
- Jiang H., Ding X., Cao Y., Wang H., Zeng W. Dense intra-adipose sympathetic arborizations are essential for cold-induced beiging of mouse white adipose tissue. Cell Metab. 2017;26:686–692 e3. doi: 10.1016/j.cmet.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim H.J., Jang M.J., Kim J.H., Lee J.H., Lee E., Park K., Kim H., Lee J., Kwag J. BrainFilm, a novel technique for physical compression of 3D brain slices for efficient image acquisition and post-processing. Sci. Rep. 2018;8:8531. doi: 10.1038/s41598-018-26776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., Cho J.H., Murray E., Bakh N., Choi H., Ohn K., Ruelas L., Hubbert A., Mccue M., Vassallo S.L. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde R.E. Lippincott Williams & Wilkins/Wolters Kluwer; 2012. Cardiovascular Physiology Concepts. [Google Scholar]
- Latremoliere A., Cheng L., Delisle M., Wu C., Chew S., Hutchinson E.B., Sheridan A., Alexandre C., Latremoliere F., Sheu S.H. Neuronal-specific TUBB3 is not required for normal neuronal function but is essential for timely axon regeneration. Cell Rep. 2018;24:1865–1879.e9. doi: 10.1016/j.celrep.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Rutlin M., Abraira V.E., Cassidy C., Kus L., Gong S., Jankowski M.P., Luo W., Heintz N., Koerber H.R. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Mao Z., Yang L., Sun K. Co-staining blood vessels and nerve fibers in adipose tissue. J. Vis.Exp. 2019 doi: 10.3791/59266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Santibanez G., Cho K.W., Lumeng C.N. Imaging white adipose tissue with confocal microscopy. Methods Enzymol. 2014;537:17–30. doi: 10.1016/B978-0-12-411619-1.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matryba P., Kaczmarek L., Gołąb J. Advances in ex situ tissue optical clearing. Laser Photon. Rev. 2019;13:1800292. [Google Scholar]
- Müller B., Lang S., Dominietto M., Rudin M., Schulz G., Deyhle H., Germann M., Pfeiffer F., David C., Weitkamp T. High-resolution tomographic imaging of microvessels. Proc. SPIE. 2008;7078:70780B. [Google Scholar]
- Murano I., Barbatelli G., Giordano A., Cinti S. Noradrenergic parenchymal nerve fiber branching after cold acclimatisation correlates with brown adipocyte density in mouse adipose organ. J. Anat. 2009;214:171–178. doi: 10.1111/j.1469-7580.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Cai R., Quacquarelli F.P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Erturk A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Peters B.P., Goldstein I.J. The use of fluorescein-conjugated Bandeiraea simplicifolia B4-isolectin as a histochemical reagent for the detection of alpha-d-galactopyranosyl groups. Their occurrence in basement membranes. Exp.Cell Res. 1979;120:321–334. doi: 10.1016/0014-4827(79)90392-6. [DOI] [PubMed] [Google Scholar]
- Rechthand E., Hervonen A., Sato S., Rapoport S.I. Distribution of adrenergic innervation of blood vessels in peripheral nerve. Brain Res. 1986;374:185–189. doi: 10.1016/0006-8993(86)90409-9. [DOI] [PubMed] [Google Scholar]
- Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell S.A., Staines W.A., Wessendorf M.W. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J. Histochem. Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- Seo J., Choe M., Kim S.Y. Clearing and labeling techniques for large-scale biological tissues. Mol.Cells. 2016;39:439–446. doi: 10.14348/molcells.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Zhu L. The crosstalk between autonomic nervous system and blood vessels. Int. J. Physiol. Pathophysiol. Pharmacol. 2018;10:17–28. [PMC free article] [PubMed] [Google Scholar]
- Shi H., Song C.K., Giordano A., Cinti S., Bartness T.J. Sensory or sympathetic white adipose tissue denervation differentially affects depot growth and cellularity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R1028–R1037. doi: 10.1152/ajpregu.00648.2004. [DOI] [PubMed] [Google Scholar]
- Song E., Seo H., Choe K., Hwang Y., Ahn J., Ahn S., Kim P. Optical clearing based cellular-level 3D visualization of intact lymph node cortex. Biomed. Opt. Express. 2015;6:4154–4164. doi: 10.1364/BOE.6.004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradleigh T.W., Greenberg K.P., Partida G.J., Pham A., Ishida A.T. Moniliform deformation of retinal ganglion cells by formaldehyde-based fixatives. J. Comp. Neurol. 2015;523:545–564. doi: 10.1002/cne.23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- Thomas G.D. Neural control of the circulation. Adv. Physiol. Educ. 2011;35:28–32. doi: 10.1152/advan.00114.2010. [DOI] [PubMed] [Google Scholar]
- Thompson R.J., Doran J.F., Jackson P., Dhillon A.P., Rode J. PGP 9.5--a new marker for vertebrate neurons and neuroendocrine cells. Brain Res. 1983;278:224–228. doi: 10.1016/0006-8993(83)90241-x. [DOI] [PubMed] [Google Scholar]
- Vargovic P., Ukropec J., Laukova M., Cleary S., Manz B., Pacak K., Kvetnansky R. Adipocytes as a new source of catecholamine production. FEBSLett. 2011;585:2279–2284. doi: 10.1016/j.febslet.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Vulchanova L., Olson T.H., Stone L.S., Riedl M.S., Elde R., Honda C.N. Cytotoxic targeting of isolectin IB4-binding sensory neurons. Neuroscience. 2001;108:143–155. doi: 10.1016/s0306-4522(01)00377-3. [DOI] [PubMed] [Google Scholar]
- Westcott E.B., Segal S.S. Perivascular innervation: a multiplicity of roles in vasomotor control and myoendothelial signaling. Microcirculation. 2013;20:217–238. doi: 10.1111/micc.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsen C. Adrenergic innervation of adipose tissue examined by fluorescence microscopy. Nature. 1964;202:913. doi: 10.1038/202913a0. [DOI] [PubMed] [Google Scholar]
- Yu T., Qi Y., Gong H., Luo Q., Zhu D. Optical clearing for multiscale biological tissues. J. Biophotonics. 2018;11 doi: 10.1002/jbio.201700187. [DOI] [PubMed] [Google Scholar]
- Zeng W., Pirzgalska R.M., Pereira M.M., Kubasova N., Barateiro A., Seixas E., Lu Y.H., Kozlova A., Voss H., Martins G.G. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell. 2015;163:84–94. doi: 10.1016/j.cell.2015.08.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Neurite density quantification and heatmap generation code developed for this publication can be found on GitHub at https://github.com/ktownsendlab/willows_et_al-2020. All other data is included in this manuscript and any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.