Abstract
Arteriosclerosis obliterans (ASO) of the lower extremities is identified as a kind of cardiovascular disease with aberrant proliferation and apoptosis of vascular smooth muscle cells (VSMCs). Accumulating studies have demonstrated the vital role of Yes1-associated transcriptional regulator (YAP1) in VSMCs, while its upstream regulatory mechanism in VSMCs in ASO of the lower extremities needs to be further elucidated. Herein, hsa_circ_0024093, a circular RNA (circRNA) from YAP1, was identified to positively regulate the protein level of YAP1 in VSMCs. Functionally, silencing of hsa_circ_0024093 obviously impeded cell proliferation and migration and promoted apoptosis in VSMCs in the in vitro model of ASO of the lower extremities. Mechanistically, it was found that hsa_circ_0024093 could regulate the expression of USP9X, which further induced YAP1 deubiquitination to stabilize YAP1 protein. In depth, it was revealed from mechanism experiments that hsa_circ_0024093 sequestered miR-889-3p or miR-4677-3p to enhance USP9X expression. Further, rescue assays validated that hsa_circ_0024093 regulated the miR-4677-3p/miR-889-3p/USP9X axis to accelerate the proliferation and migration of VSMCs in the in vitro model of ASO of the lower extremities. These findings may provide a novel perspective for better understanding of ASO of the lower extremities.
Keywords: arteriosclerosis obliterans, lower extremities, vascular smooth muscle cells, hsa_circ_0024093, YAP1, USP9X
Graphical abstract
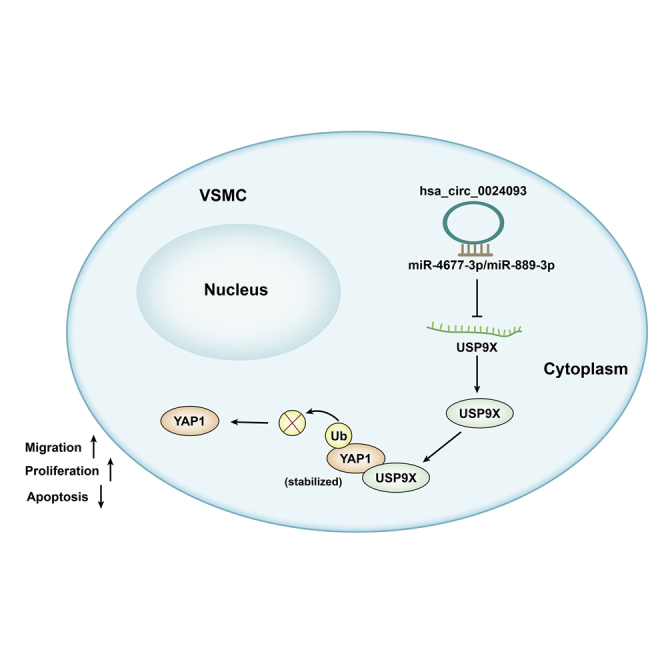
It was revealed that hsa_circ_0024093 regulated the miR-4677-3p/miR-889-3p/USP9X axis to promote YAP1 deubiquitination and enhance YAP1 protein stability, finally accelerating VSMC proliferation and migration in arteriosclerosis obliterans of the lower extremities. This may provide a novel understanding of arteriosclerosis obliterans of the lower extremities.
Introduction
Atherosclerosis is one of the most common vascular diseases leading to cerebral infarction, stroke, and cerebral ischemia-reperfusion injury.1, 2, 3 Arteriosclerosis obliterans (ASO) of the lower extremities is a disease causing limb dysfunction, and it may increase the risk of claudication, resting pain, and gangrene.4,5 The main treatments for ASO are open surgery, endovascular therapy, and physical therapy.6 In addition, the damage caused by ASO is connected with the abnormal proliferation and migration of vascular smooth muscle cells (VSMCs).7 Therefore, a better understanding of molecular mechanisms underlying the pathophysiological processes of VSMCs might supply a novel concept for ASO of the lower extremities.
Circular RNAs (circRNAs) are a class of non-coding RNAs with covalently closed loops generated from back-splicing of messenger RNAs (mRNAs).8,9 Emerging evidence has demonstrated that circRNAs serve as modulators in regulating the progression of numerous diseases. circUBXN7 inhibits cell growth and invasion by targeting miR-1247-3p to regulate B4GALT3 expression in bladder cancer.10 Hsa_circ_0020123 acts as an oncogene in non-small cell lung cancer through suppressing miR-144.11 Apart from the implication in cancers, circRNAs also play significant roles in cardiovascular diseases, such as myocardial ischemia/reperfusion injury12 and coronary atherosclerosis.13 However, the molecule mechanisms of circRNAs in VSMCs in ASO of the lower extremities remain rarely clear.
Yes-associated protein 1 (YAP1) is a key modulator of the Hippo signaling pathway and plays an important role in multiple biological processes, including cell proliferation, apoptosis, and differentiation.14 YAP1 facilitates cancer cell growth by inhibiting circRNA-000425.15 circFAT1 targets miR-375 to regulate the expression of YAP1 in osteosarcoma cells to facilitate proliferation, migration, and invasion.16 However, the regulatory mechanism upstream of YAP1 in VSMCs remains largely unknown in ASO of the lower extremities. Ubiquitination is a main post-translational modification that affects the degradation of proteins.17 It is well known that the deubiquitinase ubiquitin-specific peptidase 9 X-linked (USP9X) can stabilize YAP1 to promote tumor cell survival.18 Here, we also investigated the correlation between USP9X and YAP1 in VSMCs.
It is well known that circRNAs can function as a competing endogenous RNA (ceRNA) to sponge microRNAs (miRNAs) and then post-transcriptionally regulate messenger RNAs (mRNAs).19 circ-FOXM1 contributes to the progression of non-small cell lung cancer by targeting miR-1304-5p to regulate PPDPF and MACC1.20 Hsa_circ_0001368 inhibits the proliferation and invasion of gastric cancer cells by targeting the miR-6506-5p/FOXO3 axis.21 In our study, we also explored whether circRNA could act as a ceRNA in VSMCs in the in vitro model of ASO of the lower extremities.
We aimed to explore the role and regulatory mechanism of circRNA in VSMCs in the in vitro model of ASO of the lower extremities. Our study may contribute to the treatment of ASO of the lower extremities.
Results
Hsa_circ_0024093 promotes the protein level of YAP1 in PDGF-BB-treated VSMCs
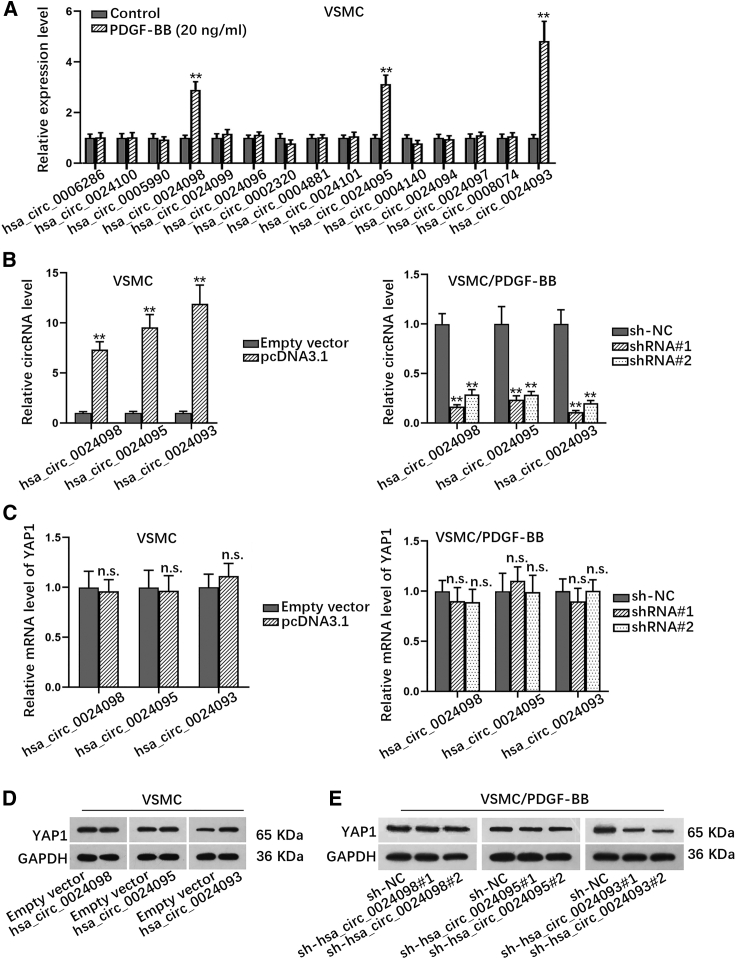
It is well known that circRNAs play crucial roles in regulating the biological behaviors of diverse cell types, including VSMCs.22 In addition, it has been reported that PDGF-BB is a pluripotent member of the PDGF family and can stimulate the proliferation and migration of perivascular cells.23 Based on this, we employed PDGF-BB to induce the proliferation and migration of VSMCs. It was manifested that the proliferation of VSMCs was facilitated by PDGF-BB in a dose-dependent manner, with a marked promotion on VSMC proliferation with treatment of at least 20 ng/mL PDGF-BB (Figure S1A). On this basis, we further tested the biological behaviors of VSMCs with treatment of 20 ng/mL PDGF-BB. As shown in Figure S1B, 5-ethynyl-29-deoxyuridine (EdU)-positive stained cell rate was significantly increased in VSMCs with PDGF-BB treatment. Meanwhile, we found through Cell Counting Kit-8 (CCK-8) assay that the proliferative ability was obviously promoted in PDGF-BB-treated VSMCs (Figure S1C). Simultaneously, we investigated whether PDGF-BB stimulated migration of VSMCs via Transwell assay. As expected, we found that the number of migrated cells was elevated in PDGF-BB-stimulated VSMCs (Figure S1D). Consistently, we observed that relative wound width was apparently narrowed under PDGF-BB induction (Figure S1E). In total, PDGF-BB induced the proliferation and migration of VSMCs. It has been reported that YAP1 is a transcriptional regulator of Hippo signaling pathway and can also promote cell proliferation in cancers.24 Through browsing the CircBase database: http://www.circbase.org/, 15 circRNAs were discovered to be originated from YAP1. To screen out the circRNA that could be upregulated by PDGF-BB in VSMCs, we applied total RNA extraction and real-time quantitative reverse-transcriptase polymerase chain reaction (real-time qRT-PCR) to detect the changes in the expression of these circRNAs. Results disclosed that the expression levels of three circRNAs (hsa_circ_0024098, hsa_circ_0024095, and hsa_circ_0024093) were apparently upregulated in VSMCs with PDGF-BB treatment (Figure 1A). Then we overexpressed these three circRNAs in VSMCs while silencing them in PDGF-BB-treated VSMCs (Figure 1B). Intriguingly, real-time qRT-PCR analyzed that all these circRNAs had no impact on the mRNA level of YAP1 in VSMCs (Figure 1C), while the results from western blot demonstrated that the protein level of YAP1 was increased in VSMCs with the overexpression of hsa_circ_0024093 and was decreased in PDGF-BB-treated VSMCs with hsa_circ_0024093 deficiency (Figures 1D and 1E; Figures S1F and S1G). These data could indicate that hsa_circ_0024093 positively regulates YAP1 protein in VSMCs.
Figure 1.
Hsa_circ_0024093 positively regulates YAP1 protein in VSMCs
(A) The expression of several circRNAs derived from YAP1 in VSMCs with or without 20 ng/mL PDGF-BB was detected. (B) The expression of three circRNAs (hsa_circ_0024098, hsa_circ_0024095, and hsa_circ_0024093) in VSMCs and PDGF-BB-treated VSMCs under different transfections was analyzed, respectively. (C) The mRNA level of YAP1 in VSMCs under different treatment or transfections was examined. (D and E) The protein level of YAP1 in indicated VSMCs was detected. ∗∗p < 0.01; n.s., no significance.
Hsa_circ_0024093 promotes cell proliferation and migration while inhibiting apoptosis of PDGF-BB-treated VSMCs
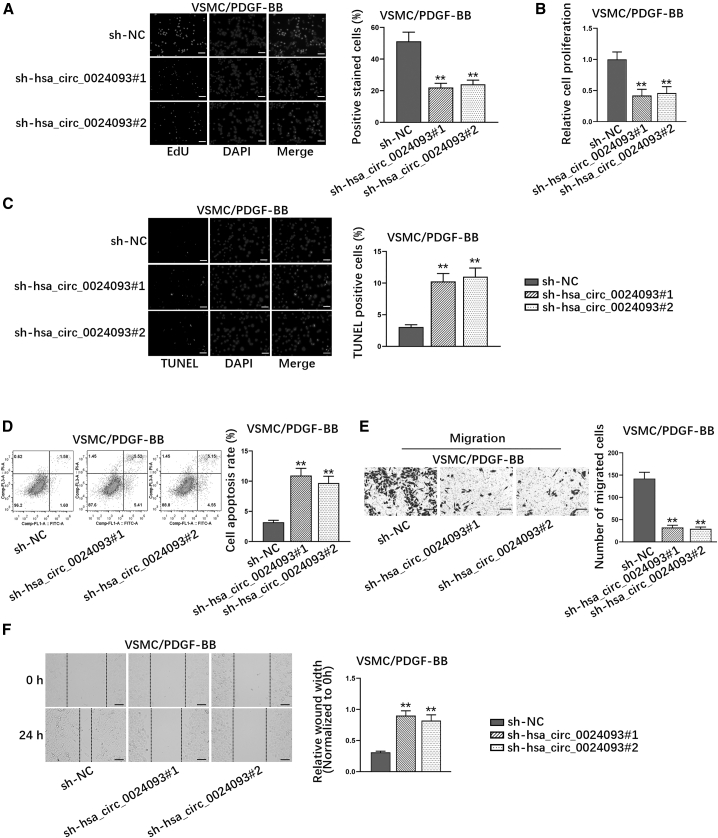
To further investigate the role of hsa_circ_0024093 in PDGF-BB-treated VSMCs, functional experiments were conducted. As indicated in Figure 2A, EdU-positive stained cell rate was cut down by hsa_circ_0024093 knockdown. Consistently, the results of CCK-8 experiments revealed that hsa_circ_0024093 knockdown obviously inhibited cell proliferation (Figure 2B). Furthermore, in the terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay, the percent of positive stained cells was increased by hsa_circ_0024093 silencing (Figure 2C), suggesting that hsa_circ_0024093 could suppress apoptosis of PDGF-BB-treated VSMCs. Similarly, the results from flow cytometry analysis also indicated that the apoptosis rate was enhanced by hsa_circ_0024093 knockdown in PDGF-BB-treated VSMCs (Figure 2D). In addition, we also assessed the impact of hsa_circ_0024093 knockdown on VSMC migration. It was suggested in Transwell assays that the number of migrated cells was markedly lessened by downregulated hsa_circ_0024093 (Figure 2E). Likewise, the results from wound-healing assays manifested that downregulating hsa_circ_0024093 led to an evident inhibition of the migration of PDGF-BB-treated VSMCs (Figure 2F). All these results could suggest that hsa_circ_0024093 promotes proliferation and migration while inhibiting apoptosis of PDGF-BB-treated VSMCs.
Figure 2.
Hsa_circ_0024093 promotes proliferation and migration while inhibiting apoptosis of PDGF-BB-treated VSMCs
(A and B) EdU assay (scale bar, 100 μm) and CCK-8 assay illustrated the proliferation of PDGF-BB-treated VSMCs under hsa_circ_0024093 depletion. (C) TUNEL assay (scale bar, 80 μm) assessed the TUNEL-positive stained cell rate in PDGF-BB-treated VSMCs under hsa_circ_0024093 knockdown. (D) The apoptosis rate of PDGF-BB-treated VSMCs after hsa_circ_0024093 knockdown was examined by flow cytometry analysis. (E) Transwell assay (scale bar, 50 μm) detected the number of migrated cells in PDGF-BB-treated VSMCs with or without hsa_circ_0024093 downregulation. (F) Wound-healing assays (scale bar, 100 μm) tested the migration of PDGF-BB-treated VSMCs under indicated contexts. ∗∗p < 0.01.
Hsa_circ_0024093 enhances the stability of YAP1 protein
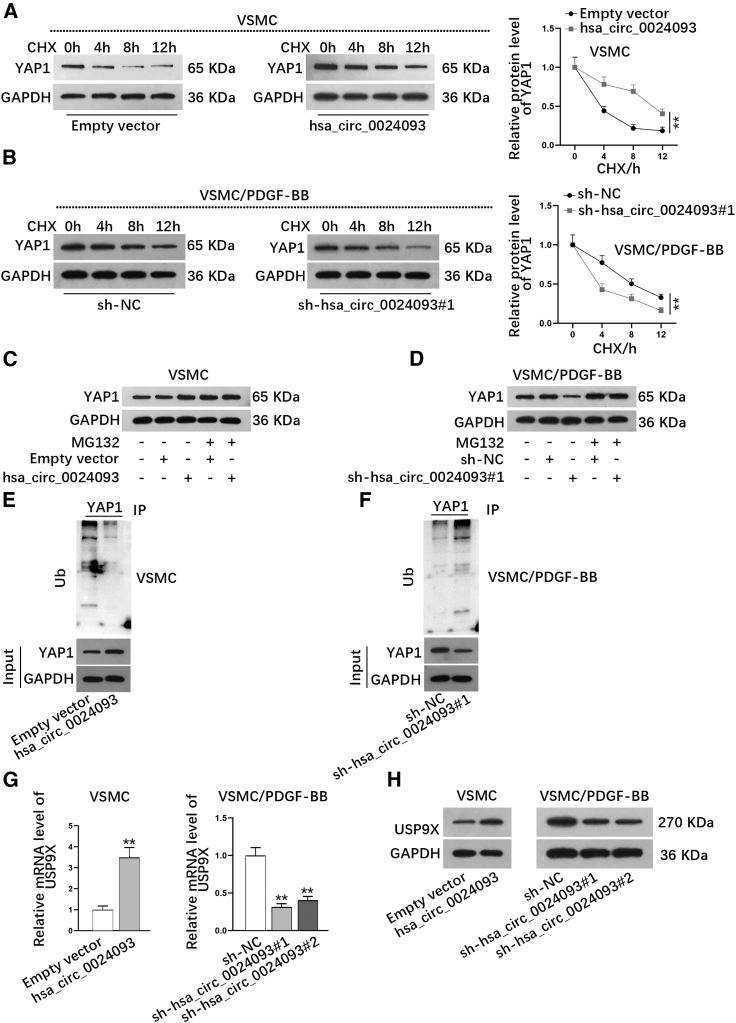
Previously, we discovered that hsa_circ_0024093 could regulate the protein level of YAP1. Here, we wanted to know the underlying mechanism in VSMCs. To test the influence of hsa_circ_0024093 on the stability of YAP1 protein, we treated VSMCs and PDGF-BB-treated VSMCs with the protein synthesis inhibitor cycloheximide (CHX). The data revealed that after inhibiting protein synthesis with CHX, the degradation rate of existing YAP1 protein was slowed down when hsa_circ_0024093 was overexpressed in VSMCs but accelerated when hsa_circ_0024093 was silenced in PDGF-BB-treated VSMCs (Figures 3A and 3B), indicating that YAP1 stability was positively affected by hsa_circ_0024093. Moreover, we also tested the regulation of hsa_circ_0024093 on YAP1 protein by the proteasome inhibitor MG132. The results from western blot indicated that MG132 treatment had no apparent effect on the protein level of YAP1 in hsa_circ_0024093-overexpressed VSMCs, but it markedly enhanced the YAP1 protein level in the control group (Figure 3C). Meanwhile, the reduced level of YAP1 protein induced by hsa_circ_0024093 deficiency was reversed under MG132 treatment (Figure 3D). These data implied that hsa_circ_0024093 hampered the degradation of the YAP1 protein level caused by the ubiquitin proteasome system. To further confirm this, we analyzed the impact of hsa_circ_0024093 on YAP1 ubiquitination. As anticipated, upregulated hsa_circ_0024093 decreased the ubiquitination level of YAP1 protein in VSMCs, while downregulated hsa_circ_0024093 accelerated YAP1 ubiquitination in PDGF-BB-treated VSMCs (Figures 3E and 3F; Figure S2A), with the accordingly enhanced free ubiquitin under hsa_circ_0024093 overexpression and lowered free ubiquitin upon hsa_circ_0024093 silence (Figure S2B). All these results could indicate that hsa_circ_0024093 hinders YAP1 ubiquitination to enhance the stability of YAP1 protein.
Figure 3.
Hsa_circ_0024093 stabilizes YAP1 protein
(A) YAP1 protein level was detected under CHX treatment in VSMCs with or without hsa_circ_0024093 overexpression. (B) The protein level of YAP1 was examined under CHX treatment in PDGF-BB-treated VSMCs under hsa_circ_0024093 inhibition. (C and D) YAP1 protein level was analyzed in VSMCs under diverse conditions. (E and F) IP assay assessed the ubiquitination level of YAP1 in indicated VSMCs. (G and H) The mRNA and protein levels of USP9X in VSMCs under different transfections were investigated. ∗∗p < 0.01.
Hsa_circ_0024093 regulates USP9X to stabilize YAP1 protein
It is well known that F-box and WD repeat domain containing 7 (FBXW7) and β-TRCP regulatory factors, which belong to F-box protein family, could affect the degradation of proteins by ubiquitination.25,26 Hence, we wondered whether hsa_circ_0024093 could regulate FBXW7 and β-TRCP to affect the ubiquitination of YAP1 protein. Data from real-time qRT-PCR and western blot showed that overexpressing or silencing hsa_circ_0024093 had no significant impacts on the mRNA and protein levels of FBXW7 and β-TRCP in VSMCs or PDGF-BB-treated VSMCs (Figures S2C and S2D), indicating that hsa_circ_0024093 did not depend on FBXW7 and β-TRCP to regulate YAP1 protein. In addition, it has been shown that USP9X induces YAP1 deubiquitination to enhance cell proliferation and metastasis in breast carcinoma.18 Thus, we further speculated whether hsa_circ_0024093 could affect USP9X to promote YAP1 deubiquitination and stabilize YAP1 protein. It was validated that the mRNA and protein levels of USP9X were obviously elevated due to hsa_circ_0024093 upregulation but notably lessened owing to hsa_circ_0024093 downregulation (Figures 3G and 3H). In addition, the results from RNA immunoprecipitation (RIP) assays attested that the enrichment of hsa_circ_0024093 had no difference between USP9X groups and IgG groups (Figure S2E), suggesting that hsa_circ_0024093 could not combine with USP9X in VSMCs or PDGF-BB-treated VSMCs. These data could mean that hsa_circ_0024093 positively modulates USP9X to hinder YAP1 ubiquitination in VSMCs.
USP9X induces YAP1 deubiquitination to stabilize YAP1
A former report demonstrated that USP9X can modulate angiomotin (AMOT) family, including angiomotin like 2 (AMOTL2), to affect the phosphorylation and cytoplasmic sequestration of YAP1.27,28 To check whether USP9X could regulate AMOT or AMOTL2 to modulate YAP1, we then increased the expression of USP9X in VSMCs and decreased USP9X expression in PDGF-BB-treated VSMCs (Figures S3A and S3B). According to the data from real-time qRT-PCR and western blot, we found that the mRNA and protein levels of AMOT and AMOTL2 were not apparently changed when USP9X was overexpressed in VSMCs or silenced in PDGF-BB-treated VSMCs (Figures S3C and S3D), which verified that USP9X could not modulate AMOT and AMOTL2 to activate YAP1 phosphorylation. In other words, hsa_circ_0024093 promoted the stability of YAP1 not via the USP9X/AMOT/AMOT2 axis.
A previous report has demonstrated that deubiquitinase USP9X can regulate the stability of FBXW7 to affect the progression of colorectal cancer.29 Inconsistently, here we detected that changes in USP9X expression had no impact on the mRNA and protein levels of FBXW7 in VSMCs with or without PDGF-BB treatment (Figures S3E and S3F). Therefore, we then speculated that USP9X could directly affect YAP1 ubiquitination in VSMCs. As shown in Figures S4A and S4B, we discovered that under the treatment of CHX, YAP1 protein was degraded much more slowly in the face of USP9X upregulation in VSMCs but much faster in response to USP9X downregulation in PDGF-BB-treated VSMCs. Moreover, we found that the protein level of YAP1 in control VSMCs was obviously promoted under MG132 treatment, while no apparent changes in YAP1 level were discovered in USP9X-upregulated VSMCs after MG132 treatment (Figure S4C). However, the reduced YAP1 protein level due to USP9X depletion was apparently recovered owing to MG132 treatment (Figure S4D). Furthermore, the ubiquitination of YAP1 was reduced by USP9X upregulation in VSMCs and increased by USP9X knockdown in PDGF-BB-treated VSMCs (Figures S4E and S4F). Collectively, USP9X hampers YAP1 ubiquitination to stabilize YAP1 protein in VSMCs.
Hsa_circ_0024093 sequesters miR-889-3p and miR-4677-3p to upregulate USP9X
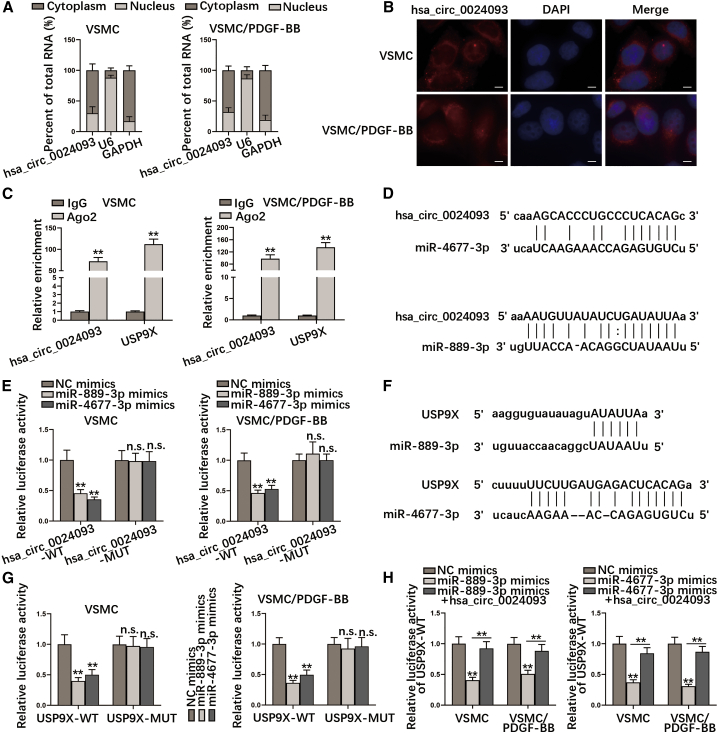
Next, we aimed to uncover the in-depth mechanism whereby hsa_circ_0024093 affected USP9X expression. As we know, the subcellular distribution of circRNAs is correlated with their regulatory mechanism.30 Through subcellular fractionation and fluorescence in situ hybridization (FISH) assays, we observed that hsa_circ_0024093 was mainly located in the cytoplasm of VSMCs with or without PDGF-BB treatment (Figures 4A and 4B). On this basis, we speculated that hsa_circ_0024093 might function in VSMCs or PDGF-BB-treated VSMCs through post-transcriptional regulation. The results from RIP assays indicated that both hsa_circ_0024093 and USP9X were highly precipitated in Ago2 groups (Figure 4C), suggesting that they co-existed in the Ago2-related RNA-induced silencing complexes (RISCs). Hence, we screened out miRNAs that were simultaneously associated with hsa_circ_0024093 and USP9X using the ENCORI database: http://starbase.sysu.edu.cn and found 22 miRNAs shared by both (Figure S5A). Furthermore, we examined the expression of these 22 miRNAs in VSMCs with or without PDGF-BB treatment. The results disclosed that the expression of 3 miRNAs (miR-663a, miR-889-3p, and miR-4677-3p) was obviously lessened under PDGF-BB treatment (Figure S5B). To verify the correlation between hsa_circ_0024093 and these three miRNAs, RNA pull-down assays were conducted. Results indicated that miR-889-3p and miR-4677-3p were abundantly precipitated by Bio-hsa_circ_0024093 (Figure S5C). In addition, we revealed the binding site between hsa_circ_0024093 and miR-4677-3p/miR-889-3p using ENCORI (Figure 4D). To determine the effectiveness of these binding sites, we constructed wild-type (WT) and mutant-type (MUT) hsa_circ_0024093 vectors for luciferase reporter assay. The data illustrated that the luciferase activity of hsa_circ_0024093-WT was definitely inhibited by upregulation of miR-4677-3p or miR-889-3p in VSMCs or PDGF-BB-treated VSMCs, while that of has_circ_0024093-MUT was hardly affected under the same condition (Figure 4E). Similarly, the binding sites of miR-889-3p or miR-4677-3p in the USP9X 3′ UTR were also predicted on ENCORI (Figure 4F). Also, overexpressed miR-889-3p or miR-4677-3p reduced the luciferase activity of USP9X-WT while exerting no effect on that of USP9X-MUT in VSMCs or PDGF-BB-treated VSMCs (Figure 4G). Further, the suppressive effect of miR-889-3p or miR-4677-3p upregulation on the luciferase activity of USP9X-WT was offset by hsa_circ_0024093 overexpression in VSMCs or PDGF-BB-treated VSMCs (Figure 4H). Together, hsa_circ_0024093 sponges miR-889-3p and miR-4677-3p to positively regulate USP9X.
Figure 4.
Hsa_circ_0024093 sponges miR-889-3p and miR-4677-3p to upregulate USP9X
(A and B) Subcellular fractionation and FISH assays (scale bar, 20 μm) monitored the cellular location of hsa_circ_0024093. (C) Enrichment of hsa_circ_0024093 and USP9X in anti-Ago2 in VSMCs and PDGF-BB-treated VSMCs was calculated. (D) The binding sites between hsa_circ_0024093 and miR-4677-3p/miR-889-3p were predicted on ENCORI. (E) Luciferase activity of hsa_circ_0024093-WT/MUT in indicated VSMCs with the transfection of miR-4677-3p mimics or miR-889-3p mimics was detected. (F) The binding sites of miR-889-3p/miR-4677-3p in the USP9X 3′ UTR were also predicted on ENCORI. (G) Luciferase activity of USP9X-WT/MUT in indicated VSMCs with overexpression of miR-889-3p or miR-4677-3p was detected. (H) Luciferase activity of USP9X-WT under different treatment and transfections was investigated. ∗∗p < 0.01; n.s., no significance.
Hsa_circ_0024093 facilitates proliferation and impedes apoptosis of PDGF-BB-treated VSMCs through the miR-889-3p/miR-4677-3p/USP9X axis
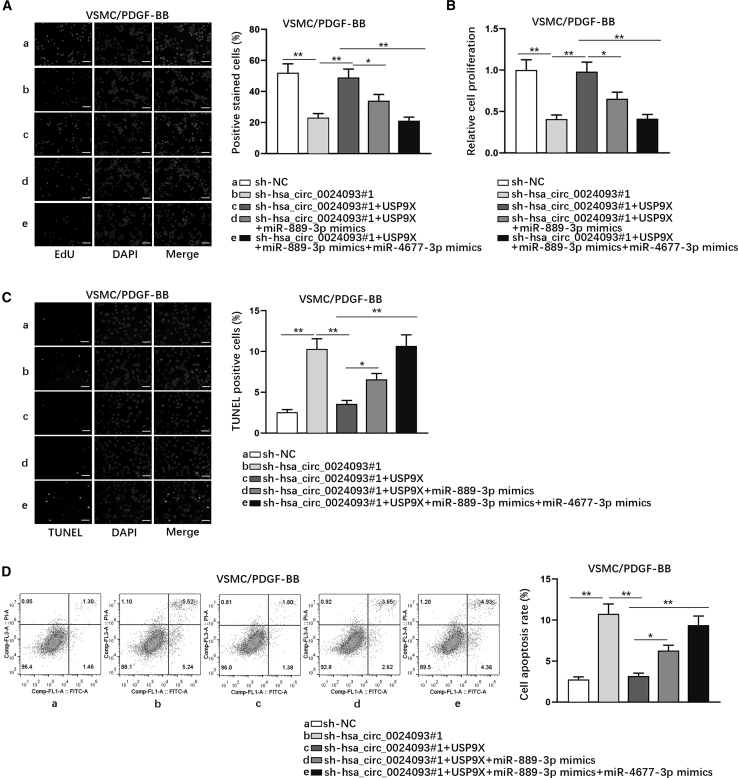
To further determine whether hsa_circ_0024093 functioned through miR-889-3p/miR-4677-3p/USP9X signaling, rescue experiments were conducted in PDGF-BB-treated VSMCs. As observed from the data of EdU assays, the reduced number of EdU-positive stained cells due to hsa_circ_0024093 knockdown was reversed by co-transfection of USP9X, while such recovery was partially mitigated after miR-889-3p overexpression and fully offset upon co-upregulation of miR-889-3p and miR-4677-3p (Figure 5A). Likewise, such trends were also discovered based on the results from CCK-8 assay (Figure 5B). In the meantime, the results from TUNEL assay and flow cytometry analysis manifested that cell apoptosis stimulated by the silencing of hsa_circ_0024093 was fully abolished after USP9X upregulation, whose effect was then partly counteracted by elevated miR-889-3p and completely countervailed under co-elevation of miR-889-3p and miR-4677-3p (Figures 5C and 5D). Overall, hsa_circ_0024093 promotes proliferation and impedes apoptosis of PDGF-BB-treated VSMCs through the miR-889-3p/miR-4677-3p/USP9X axis.
Figure 5.
Hsa_circ_0024093 affects proliferation and apoptosis of PDGF-BB-treated VSMCs through the miR-889-3p/miR-4677-3p/USP9X axis
(A and B) EdU assays (scale bar, 100 μm) and CCK-8 assays detected the proliferation of PDGF-BB-treated VSMCs responding to different transfections. (C and D) TUNEL assay (scale bar, 80 μm) and flow cytometry analysis revealed cell apoptosis under diverse conditions. ∗p < 0.05, ∗∗p < 0.01.
Hsa_circ_0024093 induces migration of PDGF-BB-treated VSMCs by the miR-889-3p/miR-4677-3p/USP9X axis
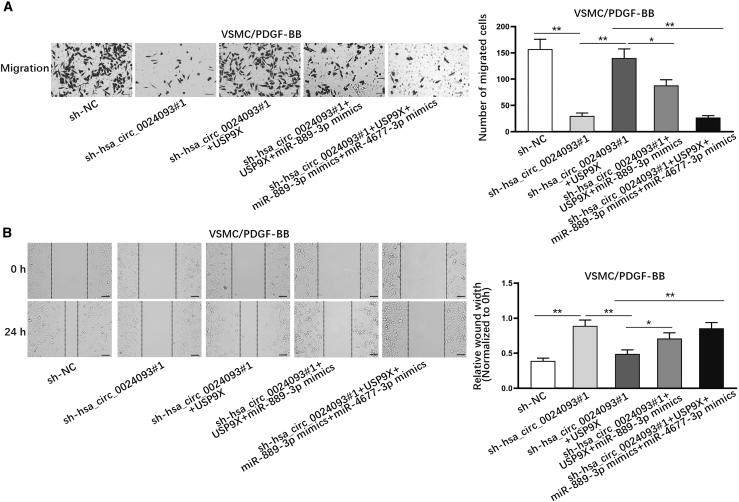
In addition, we also would like to know whether hsa_circ_0024093 affected migration of PDGF-BB-treated VSMCs by regulating the miR-889-3p/miR-4677-3p/USP9X pathway. The outcomes of Transwell assays showed that hsa_circ_0024093 knockdown weakened the cell migration, which was restored in response to an augment in USP9X expression level; moreover, the restoration of cell migration induced by USP9X overexpression was partly offset by augmented expression of miR-889-3p but fully counteracted under co-upregulation of miR-889-3p and miR-4677-3p (Figure 6A). Consistently, the results from wound-healing assay indicated that the reduced wound-healing rate induced by hsa_circ_0024093 knockdown was reversed by USP9X overexpression, whereas the reversion was partly mitigated in response to miR-889-3p upregulation but fully offset under the elevation of both miR-889-3p and miR-4677-3p (Figure 6B). All these results suggested that hsa_circ_0024093 mediated the miR-889-3p/miR-4677-3p/USP9X pathway to accelerate migration of PDGF-BB-treated VSMCs. In summary, the present work discovered that hsa_circ_0024093 mediated USP9X to enhance YAP1 protein stability by promoting YAP1 deubiquitination and regulated the miR-4677-3p/miR-889-3p/USP9X axis to accelerate VSMC proliferation and migration in ASO of the lower extremities.
Figure 6.
Hsa_circ_0024093 affects migration of PDGF-BB-treated VSMCs by miR-889-3p/miR-4677-3p/USP9X signaling
(A and B) Transwell assay (scale bar, 50 μm) and wound-healing assay (scale bar, 100 μm) examined the migration of PDGF-BB-treated VSMCs with different transfections. ∗p < 0.05, ∗∗p < 0.01.
Discussion
ASO of the lower extremities is a type of cardiovascular disease. The proliferation and migration of VSMCs are important symbols for arteriosclerosis development.31 Increasing evidence has proved that circRNAs play significant roles in regulating the function of VSMCs. circ-SATB2 promotes STIM1 expression to regulate VSMC proliferation and differentiation through targeting miR-939.32 circRNA WDR77 promotes VSMC proliferation and migration by targeting miR-124 to regulate FGF2 expression.33 circ_Lrp6 affects the proliferation, migration, and differentiation of VSMCs through targeting miRNA-145.34 Interestingly, despite numerous supports for the implication of circRNAs in the function of VSMCs, few circRNAs have been recognized in ASO of the lower extremities. YAP1 is the core effector of the Hippo signaling pathway, and reports have indicated that YAP1 can facilitate cell proliferation and migration while suppressing apoptosis in VSMCs.35,36 In this study, we focused on YAP1-derived circRNAs, which could be the upstream of YAP1 in VSMCs of ASO of the lower extremities. As a result, we discovered that hsa_circ_0024093, a kind of circRNA originated from YAP1, was the positive regulator of YAP1 protein in VSMCs. By means of loss-of-function assays, we further confirmed that hsa_circ_0024093 facilitated cell proliferation and migration and impeded apoptosis in VSMCs, which was consistent with the role of YAP1 in VSMCs. All these findings supported the promoting role of hsa_circ_0024093 in VSMCs in the in vitro model of ASO of the lower extremities.
In addition, our study found that hsa_circ_0024093 could stabilize YAP1 protein through promoting YAP1 deubiquitination. As we know, β-TRCP and FBXW7 are members of F-box protein family, which can facilitate the ubiquitination and degradation of proteins.37 Of note, a former study has reported that β-TRCP could increase YAP1 ubiquitination and degradation.38 Nevertheless, our study disclosed that hsa_circ_0024093 had no impact on either β-TRCP or FBXW7. Instead, we found that hsa_circ_0024093 stabilized YAP1 through targeting USP9X, a deubiquitinating enzyme that hampers protein ubiquitination mediated by the ubiquitin proteasome system.39 Previously, USP9X has been reported to affect YAP1 in two ways: it can directly promote YAP1 deubiquitination,18 and it can stimulate YAP1 phosphorylation through modulating AMOT or AMOTL2.40 Intriguingly, our study found that USP9X could not regulate AMOT or AMOTL2 to affect YAP1 phosphorylation but stabilized YAP1 by hindering YAP1 ubiquitination. Previously, Khan et al.29 pointed out that USP9X can also modulate FBXW7 stability in colorectal cancer. Currently, our study proved that USP9X could not regulate FBXW7 expression in VSMCs. Conclusively, all these results verified that hsa_circ_0024093 regulated USP9X to directly hinder YAP1 ubiquitination, therefore stabilizing YAP1 protein in VSMCs.
We also investigated the mechanism whereby hsa_circ_0024093 regulated USP9X in VSMCs. Our study found that hsa_circ_0024093 was mainly located in the cytoplasm, implying the potential regulation of hsa_circ_0024093 on protein-coding genes at the post-transcriptional level.41 Further, miR-889-3p and miR-4677-3p were screened out as the shared miRNAs for hsa_circ_0024093 and USP9X in VSMCs. As we know, miR-889-3p has been reported to facilitate the proliferation of osteosarcoma cells through targeting MNDA.42 In addition, Zhong et al.43 found that miR-4677-3p targets ZEB1 to inhibit the progression of lung adenocarcinoma. Interestingly, there are both similarities and differences between our study and these previous reports. The present study revealed that miR-889-3p and miR-4677-3p were sponged by hsa_circ_0024093, and, in this manner, hsa_circ_0024093 served as a ceRNA of USP9X in VSMCs. Through rescue experiments, we further found that hsa_circ_0024093 affected the proliferation and migration of VSMCs via targeting the miR-889-3p/miR-4677-3p/USP9X axis.
In conclusion, our study specified the functional role of hsa_circ_0024093 in VSMCs in the in vitro model of ASO of the lower extremities. The findings may provide a promising target for better understanding of ASO of the lower extremities.
Materials and methods
Cell culture and treatment
VSMCs purchased from ATCC (Manassas, VA, USA) were cultured with 10% fetal bovine serum (FBS) and 1% antibiotics in F-12K medium (Thermo Fisher Scientific, Rockford, IL, USA) at 37°C and 5% CO2. VSMCs were divided into two groups, including the control group (without any treatment) and the PDGF-BB (20 ng/mL; Thermo Fisher Scientific) group, and cells in both groups were subsequently incubated at 37°C.
Real-time qRT-PCR
Total RNA was extracted from VSMCs with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), for producing cDNA with PrimeScript RT Reagent Kit (Promega, Madison, WI, USA). Following the user guide, qPCR was conducted by using the SYBR Green PCR Kit (Takara) with StepOnePlus Real-time PCR Systems (Applied Biosystems, Foster City, CA, USA). Gene expression was calculated with the 2−ΔΔCt method, standardized to GAPDH or U6. Each experiment was repeated at least three times.
Cell transfection
The pcDNA3.1 circRNA vectors and short hairpin RNAs (shRNAs) designed for circRNAs were acquired from GenePharma (Shanghai, China), as well the control vectors and control shRNAs. In addition, miR-889-3p mimics, miR-4677-3p mimics, NC mimics, and shRNAs specific to USP9X and pcDNA3.1/USP9X were also synthesized by GenePharma. Cells were transfected for 48 h using Lipofectamine 3000 reagent (Invitrogen). Each experiment was repeated at least three times.
EdU assay
EdU assay was undertaken in VSMCs after PDGF-BB treatment or transfection using the BeyoClick EdU Cell Proliferation Kit (Beyotime, Shanghai, China) with Alexa Fluor 594. Cells were counterstained with EdU and DAPI and then assayed with inverted microscope (Olympus, Tokyo, Japan). Each experiment was repeated at least three times.
CCK-8 assay
After PDGF-BB treatment or indicated transfections, VSMCs in 96-well plates were seeded at the density of 1 × 105 cells/well all night. 10 μL of CCK-8 reagent (MedChem Express, Monmouth Junction, NJ, USA) was then added to each well for 3 h. The absorbance at 450 nm was detected. Each experiment was repeated at least three times.
TUNEL assay
The treated or transfected VSMCs were washed in PBS and fixed using 4% paraformaldehyde (PFA). Then, TUNEL reagent (Merck, Darmstadt, Germany) was used for staining apoptotic cells, and DAPI was employed to stain cell nuclei. After that, cells in five random non-overlapping microscopic fields were counted under optical microscopy (Olympus). Each experiment was repeated at least three times.
Flow cytometry analysis
Cell apoptosis was also estimated with double Annexin V/propidium iodide (PI) staining kit (Invitrogen). After staining with fluorescein isothiocyanate (FITC)-Annexin V and PI for 15 min, cells were assayed by use of a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Each experiment was repeated at least three times.
Transwell assay
After PDGF-BB treatment or transfections, cells in serum-free medium were added to upper Transwell chambers, and complete culture medium was added to lower chambers. The Transwell chamber was fixed after 24 h, and then cell migration was assessed after staining cells with crystal violet solution. Cells in five random fields were chosen for counting. Each experiment was repeated at least three times.
Wound-healing assay
Cell samples were cultured until 90% confluence in 6-well plates, and then a scratch was created using a 200 μL pipette tip. After culturing in serum-free medium for 0 and 24 h, the wound closure was imaged under the microscope (Olympus). Each experiment was repeated at least three times.
Western blot
Cells were lysed with RIPA lysis buffer after treatment or transfections, and then proteins in cell lysates were separated via 12% SDS-PAGE and shifted to polyvinylidene fluoride (PVDF) membranes. After being sealed in 5% nonfat milk, membranes were probed with primary antibodies against loading control GAPDH (ab8245, 1/10,000; Abcam, Cambridge, MA, USA) and YAP1 (ab52771, 1/5,000; Abcam), β-TRCP (#4394, 1/1,000; Cell Signaling Technology, Boston, MA, USA), FBXW7 (ab109617, 1 μg/mL; Abcam), USP9X (#14898, 1/1,000; Cell Signaling Technology), AMOT (16870-1-AP, 1/1,000; Proteintech, Chicago, IL, USA), and AMOTL2 (23351-1-AP, 1/1,000; Proteintech) all night. After that, membranes were subjected to incubation with horseradish peroxidase (HRP)-tagged secondary antibodies for 2 h. The protein signals were examined after washing in TBST, by use of enhanced chemiluminescence (ECL) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Each experiment was repeated at least three times.
CHX and MG132 treatments
To test the impact of hsa_circ_0024093 on the stability of YAP1 protein, VSMCs (treated with or without PDGF-BB) were subjected to 25 μg/mL CHX treatment, the protein synthesis inhibitor, for 0, 4, 8, or 12 h. For assessing the influence of hsa_circ_0024093 on YAP1 ubiquitination, indicated VSMCs were processed with 20 μM of MG132, the proteasome inhibitor that inhibits protein ubiquitination mediated by proteasomes. After treatments for indicated times, protein levels were analyzed via western blot. Each experiment was repeated at least three times.
Immunoprecipitation (IP)
After indicated treatment or transfection, the VSMC samples were lysed with RIPA buffer and washed in PBS. The lysates were collected for IP assay. Then, 1 mg protein in the lysates was subjected to YAP1 antibody (#14074; Cell Signaling Technology); ubiquitin antibody (10201-2-AP; Proteintech) was used in control group. After incubation for 4 h at 4°C, proteins in the precipitates were eluted and then detected using western blot. Each experiment was repeated at least three times.
RIP
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was applied for RIP assay with human Ago2 antibody, USP9X antibody, and control IgG antibody. Cells were cultured in RIP buffer containing antibody-bound magnetic beads. One night later, RNAs in the precipitates were analyzed by real-time qRT-PCR. Each experiment was repeated at least three times.
Subcellular fractionation
PARIS Kit (Invitrogen) was applied for subcellular fractionation as per the user guide. Cell samples were lysed with cell fractionation buffer and then centrifuged. The isolated RNAs were extracted and purified for real-time qRT-PCR, with GAPDH and U6 as respective cytoplasmic and nuclear indicators. Each experiment was repeated at least three times.
FISH
For RNA FISH assay, the hsa_circ_0024093-specific probe was designed and synthesized by Ribobio (Guangzhou, China). Cells in the slides were hybridized with FISH probe overnight and then counterstained in DAPI solution. After that, cells were visualized using an Olympus fluorescence microscope. Each experiment was repeated at least three times.
RNA pull-down assay
RNA pull-down assay was undertaken in VSMCs treated with or without PDGF-BB by use of Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific). Cells were lysed by using lysis buffer, and then the lysates were mixed with Bio-hsa_circ_0024093-WT/MUT probes and streptavidin-coated magnetic beads. Following overnight incubation, RNAs in the pulled-down complexes were studied by real-time qRT-PCR. Each experiment was repeated at least three times.
Luciferase reporter assay
The full-length sequences of hsa_circ_0024093 or USP9X 3′ UTR fragments covering wild-type or mutant miR-889-3p/miR-4677-3p binding sites were separately inserted into the pmirGLO dual-luciferase vectors for luciferase reporter assay. The acquired constructs were co-transfected with NC mimics, miR-889-3p mimics, or miR-4677-3p mimics, or together with vectors covering hsa_circ_0024093, in VSMCs or PDGF-BB-treated VSMCs for 48 h. Relative luciferase activities were studied using dual-luciferase reporter assay system (Promega, Madison, WI, USA). Each experiment was repeated at least three times.
Statistical analyses
Data were presented as the mean ± standard deviation (SD) of three individually conducted experiments. Statistical analyses of data collected from this study were accomplished by Student’s t test or one-way ANOVA using GraphPad Prism 7 (La Jolla, CA, USA), with p < 0.05 as the threshold of statistical significance.
Acknowledgment
Thanks for all supports from our laboratory. The study was supported by the Project of Shanghai Municipal Health Commission (no. 20194Y0358), the Health Commission of Minhang District of Shanghai (no. 2020MW08), the Science and Technology Commission of Shanghai Municipality (STCSM) (no. 17401934800), the Health and Family Planning Commission of Minhang District of Shanghai (no. 2017MW23), and the Cultivation Project of the National Natural Science Foundation from South Campus, Renji Hospital, School of Medicine, Shanghai Jiao Tong University (no. H0215).
Author contributions
X.Z. and P.W. provided administration. K.Y. and M.R.L. performed the experiments. Y.T.S. and H.F.Q. analyzed data. X.Z. and Y.F.W. wrote the draft. W.L. revised the draft. All authors have read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.026.
Contributor Information
Yunfei Wang, Email: yunfeiww@126.com.
Wei Liang, Email: weiliang3003@163.com.
Supplemental information
References
- 1.Kanuri S.H., Ipe J., Kassab K., Gao H., Liu Y., Skaar T.C., Kreutz R.P. Next generation MicroRNA sequencing to identify coronary artery disease patients at risk of recurrent myocardial infarction. Atherosclerosis. 2018;278:232–239. doi: 10.1016/j.atherosclerosis.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sukhanov S., Higashi Y., Shai S.Y., Snarski P., Danchuk S., D’Ambra V., Tabony M., Woods T.C., Hou X., Li Z. SM22α (Smooth Muscle Protein 22-α) Promoter-Driven IGF1R (Insulin-Like Growth Factor 1 Receptor) Deficiency Promotes Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018;38:2306–2317. doi: 10.1161/ATVBAHA.118.311134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y., Zhang Y., Han B., Yang L., Chen X., Huang R., Wu F., Chao J., Liu P., Hu G. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J. Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Criqui M.H., Vargas V., Denenberg J.O., Ho E., Allison M., Langer R.D., Gamst A., Bundens W.P., Fronek A. Ethnicity and peripheral arterial disease: the San Diego Population Study. Circulation. 2005;112:2703–2707. doi: 10.1161/CIRCULATIONAHA.105.546507. [DOI] [PubMed] [Google Scholar]
- 5.Diehm C., Allenberg J.R., Pittrow D., Mahn M., Tepohl G., Haberl R.L., Darius H., Burghaus I., Trampisch H.J., German Epidemiological Trial on Ankle Brachial Index Study Group Mortality and vascular morbidity in older adults with asymptomatic versus symptomatic peripheral artery disease. Circulation. 2009;120:2053–2061. doi: 10.1161/CIRCULATIONAHA.109.865600. [DOI] [PubMed] [Google Scholar]
- 6.Spronk S., Bosch J.L., den Hoed P.T., Veen H.F., Pattynama P.M., Hunink M.G. Intermittent claudication: clinical effectiveness of endovascular revascularization versus supervised hospital-based exercise training--randomized controlled trial. Radiology. 2009;250:586–595. doi: 10.1148/radiol.2501080607. [DOI] [PubMed] [Google Scholar]
- 7.Schachter M. Vascular smooth muscle cell migration, atherosclerosis, and calcium channel blockers. Int. J. Cardiol. 1997;62(Suppl 2):S85–S90. doi: 10.1016/s0167-5273(97)00245-3. [DOI] [PubMed] [Google Scholar]
- 8.Fan H., Li Y., Liu C., Liu Y., Bai J., Li W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem. Biophys. Res. Commun. 2018;507:178–184. doi: 10.1016/j.bbrc.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Fang J., Hong H., Xue X., Zhu X., Jiang L., Qin M., Liang H., Gao L. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 10.Liu H., Chen D., Bi J., Han J., Yang M., Dong W., Lin T., Huang J. Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging (Albany NY) 2018;10:2606–2623. doi: 10.18632/aging.101573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu D., Yan B., Xin R., Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 2018;8:1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C.L., Wang J.P., Xue X., Liu N., Zhang X.H., Zhao Z., Liu J.G., Zhang C.P., Piao Z.H., Liu Y. Effect of Circular ANRIL on the Inflammatory Response of Vascular Endothelial Cells in a Rat Model of Coronary Atherosclerosis. Cell. Physiol. Biochem. 2017;42:1202–1212. doi: 10.1159/000478918. [DOI] [PubMed] [Google Scholar]
- 14.Zhao L., Guan H., Song C., Wang Y., Liu C., Cai C., Zhu H., Liu H., Zhao L., Xiao J. YAP1 is essential for osteoclastogenesis through a TEADs-dependent mechanism. Bone. 2018;110:177–186. doi: 10.1016/j.bone.2018.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z., Huang S., Cao Y., Yao Y., Li J., Chen J., Jiang B., Yuan X., Xiang X., Xiong J., Deng J. YAP1 inhibits circRNA-000425 expression and thus promotes oncogenic activities of miR-17 and miR-106. Biochem. Biophys. Res. Commun. 2018;503:2370–2375. doi: 10.1016/j.bbrc.2018.06.163. [DOI] [PubMed] [Google Scholar]
- 16.Liu G., Huang K., Jie Z., Wu Y., Chen J., Chen Z., Fang X., Shen S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol. Cancer. 2018;17:170. doi: 10.1186/s12943-018-0917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T., Jing B., Sun B., Liao Y., Song H., Xu D., Guo W., Li K., Hu M., Liu S. Stabilization of PTGES by deubiquitinase USP9X promotes metastatic features of lung cancer via PGE2 signaling. Am. J. Cancer Res. 2019;9:1145–1160. [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Liu T., Li Y., Wu C., Luo K., Yin Y., Chen Y., Nowsheen S., Wu J., Lou Z., Yuan J. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 2018;37:2422–2431. doi: 10.1038/s41388-018-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R., Ni J., Lu S., Jiang S., You L., Liu H., Shou J., Zhai C., Zhang W., Shao S. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging (Albany NY) 2019;11:8484–8501. doi: 10.18632/aging.102334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu G., Shi H., Deng L., Zheng H., Kong W., Wen X., Bi H. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019;513:207–212. doi: 10.1016/j.bbrc.2019.03.213. [DOI] [PubMed] [Google Scholar]
- 21.Lu J., Zhang P.Y., Li P., Xie J.W., Wang J.B., Lin J.X., Chen Q.Y., Cao L.L., Huang C.M., Zheng C.H. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem. Biophys. Res. Commun. 2019;512:29–33. doi: 10.1016/j.bbrc.2019.02.111. [DOI] [PubMed] [Google Scholar]
- 22.Xu J.Y., Chang N.B., Rong Z.H., Li T., Xiao L., Yao Q.P., Jiang R., Jiang J. circDiaph3 regulates rat vascular smooth muscle cell differentiation, proliferation, and migration. FASEB J. 2019;33:2659–2668. doi: 10.1096/fj.201800243RRR. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Andersson P., Hosaka K., Zhang Y., Cao R., Iwamoto H., Yang X., Nakamura M., Wang J., Zhuang R. The PDGF-BB-SOX7 axis-modulated IL-33 in pericytes and stromal cells promotes metastasis through tumour-associated macrophages. Nat. Commun. 2016;7:11385. doi: 10.1038/ncomms11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García P., Rosa L., Vargas S., Weber H., Espinoza J.A., Suárez F., Romero-Calvo I., Elgueta N., Rivera V., Nervi B. Hippo-YAP1 Is a Prognosis Marker and Potentially Targetable Pathway in Advanced Gallbladder Cancer. Cancers (Basel) 2020;12:778. doi: 10.3390/cancers12040778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J., Zhou W., Yang F., Chen G., Li H., Zhao Y., Liu P., Li H., Tan M., Xiong X., Sun Y. The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat. Commun. 2017;8:14002. doi: 10.1038/ncomms14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King B., Trimarchi T., Reavie L., Xu L., Mullenders J., Ntziachristos P., Aranda-Orgilles B., Perez-Garcia A., Shi J., Vakoc C. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M., Kim M., Park S.J., Lee C., Lim D.S. Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 2016;17:64–78. doi: 10.15252/embr.201540809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mana-Capelli S., McCollum D. Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling. J. Biol. Chem. 2018;293:18230–18241. doi: 10.1074/jbc.RA118.004187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan O.M., Carvalho J., Spencer-Dene B., Mitter R., Frith D., Snijders A.P., Wood S.A., Behrens A. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J. Clin. Invest. 2018;128:1326–1337. doi: 10.1172/JCI97325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Zhang X., Li C., Yue L., Ding N., Riordan T., Yang L., Li Y., Jen C., Lin S. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 2019;16:220–232. doi: 10.1080/15476286.2019.1565284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao H., Gabbiani G., Bochaton-Piallat M.L. Arterial smooth muscle cell heterogeneity: implications for atherosclerosis and restenosis development. Arterioscler. Thromb. Vasc. Biol. 2003;23:1510–1520. doi: 10.1161/01.ATV.0000090130.85752.ED. [DOI] [PubMed] [Google Scholar]
- 32.Mao Y.Y., Wang J.Q., Guo X.X., Bi Y., Wang C.X. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem. Biophys. Res. Commun. 2018;505:119–125. doi: 10.1016/j.bbrc.2018.09.069. [DOI] [PubMed] [Google Scholar]
- 33.Chen J., Cui L., Yuan J., Zhang Y., Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017;494:126–132. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 34.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 35.Liu T., Xu J., Guo J.L., Lin C.Y., Luo W.M., Yuan Y., Liu H., Zhang J. YAP1 up-regulation inhibits apoptosis of aortic dissection vascular smooth muscle cells. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4632–4639. [PubMed] [Google Scholar]
- 36.Xie C., Guo Y., Zhu T., Zhang J., Ma P.X., Chen Y.E. Yap1 protein regulates vascular smooth muscle cell phenotypic switch by interaction with myocardin. J. Biol. Chem. 2012;287:14598–14605. doi: 10.1074/jbc.M111.329268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren H., Koo J., Guan B., Yue P., Deng X., Chen M., Khuri F.R., Sun S.Y. The E3 ubiquitin ligases β-TrCP and FBXW7 cooperatively mediates GSK3-dependent Mcl-1 degradation induced by the Akt inhibitor API-1, resulting in apoptosis. Mol. Cancer. 2013;12:146. doi: 10.1186/1476-4598-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Q., Quan M., Dong J., Bai J., Wang J., Han R., Wang W., Cai Y., Lv Y.Q., Chen Q. The WW domains dictate isoform-specific regulation of YAP1 stability and pancreatic cancer cell malignancy. Theranostics. 2020;10:4422–4436. doi: 10.7150/thno.42795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox J.L., Wilder P.J., Wuebben E.L., Ouellette M.M., Hollingsworth M.A., Rizzino A. Context-dependent function of the deubiquitinating enzyme USP9X in pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2014;15:1042–1052. doi: 10.4161/cbt.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanh Nguyen H., Andrejeva D., Gupta R., Choudhary C., Hong X., Eichhorn P.J., Loya A.C., Cohen S.M. Deubiquitylating enzyme USP9x regulates hippo pathway activity by controlling angiomotin protein turnover. Cell Discov. 2016;2:16001. doi: 10.1038/celldisc.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greene J., Baird A.M., Brady L., Lim M., Gray S.G., McDermott R., Finn S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge D., Chen H., Zheng S., Zhang B., Ge Y., Yang L., Cao X. Hsa-miR-889-3p promotes the proliferation of osteosarcoma through inhibiting myeloid cell nuclear differentiation antigen expression. Biomed. Pharmacother. 2019;114:108819. doi: 10.1016/j.biopha.2019.108819. [DOI] [PubMed] [Google Scholar]
- 43.Zhong Y., Wang J., Lv W., Xu J., Mei S., Shan A. LncRNA TTN-AS1 drives invasion and migration of lung adenocarcinoma cells via modulation of miR-4677-3p/ZEB1 axis. J. Cell. Biochem. 2019;120:17131–17141. doi: 10.1002/jcb.28973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.