Abstract
Mounting evidence reveals that dysregulation of circular RNAs (circRNAs) is involved in the development of glioblastoma. Leucine-rich repeat-containing 4 (LRRC4) has been shown to suppress tumors in glioblastoma. However, whether LRRC4 can regulate the formation of circRNA is not yet understood. In this study, LRRC4 was found to interact with SAM68. LRRC4 promoted the generation of circCD44 by inhibiting the binding between SAM68 and CD44 pre-mRNA. Moreover, downregulated expression of circCD44 was found in glioblastoma multiforme (GBM) tissues and GBM primary cells. Re-expression of circCD44 significantly suppressed the proliferation, colony formation, and invasion of GBM cells and inhibited tumor growth in vivo. Mechanistically, circCD44 could regulate the expression of SMAD6 via sponging miR-326 and miR-330-5p involved in the progression of GBM. Thus, the LRRC4/SAM68/circCD44/miR-326/miR-330-5p/SMAD6 signaling axis could be a potential target for GBM treatment.
Keywords: LRRC4, SAM68, circCD44, glioblastoma multiforme, miR-326/miR-330-5p, SMAD6
Graphical abstract
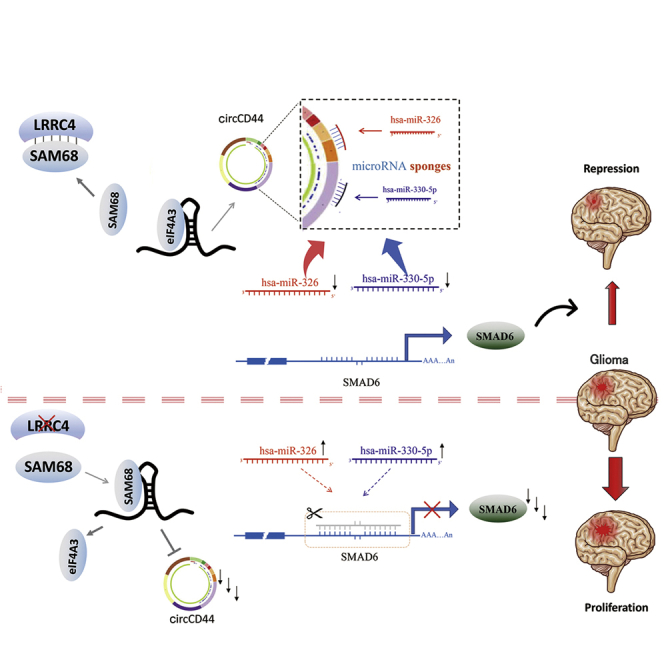
Studies demonstrate that leucine-rich repeat-containing 4 promotes the generation of a novel suppressive circCD44 by restraining the binding between SAM68 and CD44 pre-mRNA, inhibits glioblastoma multiforme cell proliferation through the SAM68/circCD44/miR-326/miR-330-5p/SMAD6 signaling axis, and therefore provides a potential therapeutic target for glioblastoma.
Introduction
Incidences and mortality rates of brain tumors rank in the top 10 among all malignant tumors in China, according to a 2019 national cancer epidemic report.1 The refractory malignant tumor seriously threatens individuals’ lives and health. Glioblastoma multiforme (GBM) is the most common malignant primary brain tumor, constituting 45% of all malignant central nervous system (CNS) tumors and 80% of all primary malignant CNS tumors.2 Current research shows that the main factors affecting the prognosis of patients with glioblastoma include the degree of surgical resection of tumor tissue and the molecular classification of tumors.3,4 With the improvement of surgical accuracy and the advancement of tumor imaging, it is easier to distinguish between glioblastoma tissue and normal brain tissue and to maximize the removal of tumor tissue.5 However, intensive invasive growth of tumor cells into surrounding normal brain tissue is a universal phenomenon of malignant glioma and is also the main reason for the recurrence of glioblastoma even when maximal surgical resection is performed.6 Thus, valid preventive and curative measures for this lethal disease are urgently needed.
Circular RNAs (circRNAs) are a class of isoform RNA molecules with a single-stranded, covalently closed loop structure. They largely feature high stability, abundance, and conservation and display tissue-type and developmental-stage-specific expression patterns,7,8 indicating that circRNAs have an important role in regulating gene expression and determining cell fate.9,10 CircRNAs are frequently implicated in different biological functions in carcinogenesis, including regulation of cell proliferation, invasion, metastasis, and autophagy.11, 12, 13, 14 In addition, mounting evidence reveals that dysregulation of circRNAs is involved in the development of glioma. For example, circCPA4 promotes the progression of glioblastoma via the let7/CPA4 axis,15 while CircMTO1 inhibits proliferation of glioblastoma cells via the miR-92/WWOX signaling pathway.16 Furthermore, on account of their stable accumulation in body fluids or exosomes, circRNAs also present favorable biomarkers and targets for the diagnosis and prognosis of cancers.7
Leucine-rich repeat-containing 4 (LRRC4), a member of the leucine-rich repeat (LRR) superfamily,17 is a relatively brain-enriched gene that was first cloned by our laboratory in 2000 (GenBank AF196976).18 Our team has investigated the role of LRRC4 in gliomagenesis over 15 years, since we first revealed the function of LRRC4 as a tumor suppressor for glioma in 2006.19 Over the past 5 years, we have reported that LRRC4 directly interacts with ERK1/2 through the D domain of LRRC4 and the CD domain of ERK1/2. Overexpression of LRRC4 anchors ERK1/2 in the cytoplasm and prevents the activation induced by MEK.20 Moreover, LRRC4 also takes part in regulating the GBM immune microenvironment. LRRC4 promotes nuclear factor κB (NF-κB) nuclear translocation and cytokine production in GBM cells by forming a triple-protein complex including PDPK1 and HSP90, then by modulating the infiltration of regulatory T (Treg) cells in the GBM microenvironment.21 In addition, we identified LRRC4 as a novel autophagy inhibitor that restores the sensitivity of GBMs to temozolomide (TMZ). Mechanistically, LRRC4 binds to DEPTOR, and this interaction results in autophagy inhibition, thus increasing the TMZ treatment response of GBM.22 Another report also found that LRRC4 binds to PIK3R1 and inhibits PIK3R1 activity, thereby suppressing epithelial ovarian cancer metastasis by targeting E-cadherin-dependent collective cell invasion and does so by inhibiting the PIK3R1-mediated AKT/GSK3β/β-catenin signaling pathway.23 However, whether LRRC4 can regulate the formation of circRNA is still not yet understood.
Here, we first identified that LRRC4 upregulated the expression of circRNA circCD44 by interacting with Sam68. Mechanistically, LRRC4 interacted with Sam68 and decreased the protein level of Sam68, then promoted the enrichment of eIF4A3 in conservative binding sites on pre-mRNA of CD44, thereby increasing the formation of circCD44. Furthermore, we also confirmed that circCD44 inhibits progression of glioblastoma via the miR-330-5p/miR-326/SMAD6 signaling pathway in vitro and in vivo. Our study provides novel insight into a mechanism by which LRRC4 mediates the formation and upregulation of tumor-suppressive circRNA circCD44, thereby inhibiting proliferation of glioblastoma cells.
Results
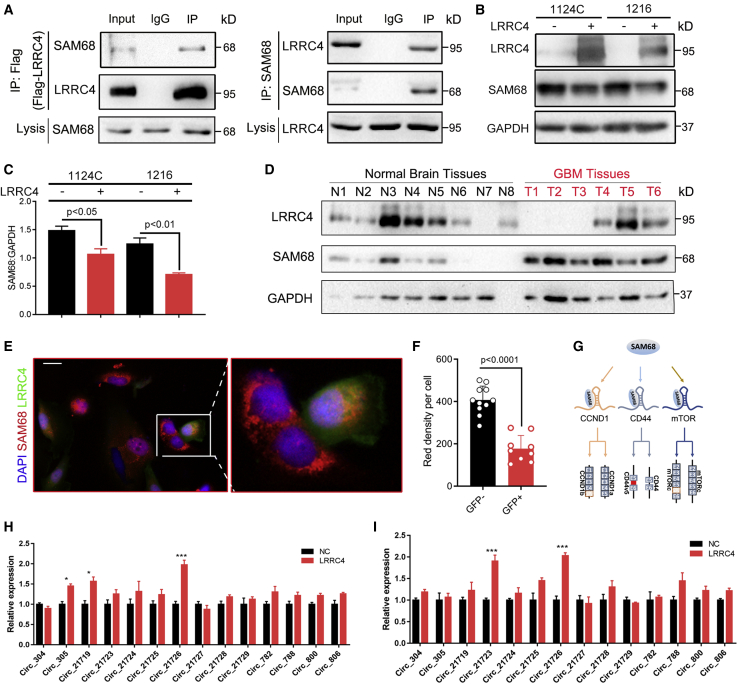
LRRC4 interacts with SAM68 and promotes expression of Circ_0021726
In our previous research, we generated a LRRC4 interactome based on mass spectrometry-based immunoprecipitation (IP) proteomics in order to identify LRRC4-binding proteins. The mass spectrometry results showed that multiple RNA binding proteins, including FUS, EWS, NONO, SAM68, and EWRS, were co-immunoprecipitated by LRRC4 antibody. The co-immunoprecipitation (coIP) assay was then used to explore the candidate proteins that interacted with LRRC4, and we confirmed that endogenous SAM68 was co-immunoprecipitated with LRRC4 from the cell extract in 1124C cells (Figure 1A). Consistently, LRRC4 was also co-immunoprecipitated with SAM68 antibody in 1124C cell extract (Figure 1A). We subsequently performed a glutathione S-transferase (GST) pull-down assay with LRRC4 and GST-fused SAM68. The results revealed that LRRC4 was pulled down by GST-fused SAM68 (Figure S1A). Next, we wanted to determine whether the interaction between LRRC4 and SAM68 could alter the expression of SAM68. We found that SAM68 expression was decreased in 1124C and 1216 primary glioma cells that were transfected with an LRRC4 plasmid (Figures 1B and 1C). We next examined whether LRRC4 affected the ubiquitin modification of SAM68, and we found that the ubiquitin modifications of SAM68 were also increased after LRRC4 transfection (Figure S1B). We further found that the inhibition effect of LRRC4 on SAM68 was blocked when LRRC4 overexpression cells were treated with MG132 (Figure S1C), indicating that LRRC4 induced the degradation of SAM68 by directly interacting with SAM68. Additionally, we examined LRRC4 and SAM68 protein expression in the total protein extraction of GBM specimens. As expected, LRRC4 expression was downregulated and SAM68 expression was upregulated in GBM tissues compared with normal brain tissue (Figure 1D). The expression of SAM68 was also investigated in The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) dataset. The results showed that the mRNA level of SAM68 was upregulated in both GBM and the low-grade glioma (LGG) group (Figures 1, S1D, and S1E). Moreover, the Kaplan-Meier assay showed that patients with a high SAM68 expression level had a worse prognosis than patients with a low SAM68 expression level (Figures 1, S1F, and S1G). This suggests that SAM68 acts as a pro-oncogene in GBM progression. We also used confocal fluorescence to examine the potential effects of LRRC4 on SAM68 localization or expression and revealed that SAM68 remained predominantly in the cytoplasm, but the SAM68 staining signal remarkably decreased in LRRC4 expression cells (Figures 1E and 1F). SAM68 belongs to the signal-transduction-activated RNA protein family. Previous research revealed that SAM68 regulates the variable splicing by directly binding to the pre-mRNA of CD44, mTOR, and cyclin D1 (Figure 1G),24, 25, 26 indicating that SAM68 plays an important role in the maturation of RNA. Recent studies also have shown that RNA binding proteins may directly bind to the flanking sequence of the precursor mRNA and then promote the formation of circRNA.27 These findings prompted us to speculate on whether LRRC4 can regulate the formation of circRNA by interacting with SAM68. First, we searched the candidate circRNA containing exon V5 of CD44 pre-mRNA, intron 5 of mTOR pre-mRNA, and intron 4 of cyclin D1 pre-mRNA in the circBase database. Then, we detected their expression in 1216, 1104, and 1124C primary glioma cells that were transfected with an LRRC4 plasmid or control plasmid by designing specific primers. The results revealed that hsa_circ_0021726 was upregulated in 1216, 1104, and 1124C cells that were transfected with an LRRC4 plasmid compared to those transfected with control plasmid (NC) (Figures 1H, 1I, and S1H).
Figure 1.
LRRC4 interacts with SAM68 and promotes expression of Circ_0021726
(A) 1124C cells were transfected with FLAG-LRRC4 plasmid. Co-immunoprecipitation with Flag antibody showed the interaction between LRRC4 and endogenous SAM68 in 1124C cells (left). 1124C cells were transfected with pCDNA3.1-LRRC4 plasmid, then co-immunoprecipitation with SAM68 antibody showed the interaction between SAM68 and LRRC4 in 1124C cells (right). (B) Representative western blot analysis of LRRC4, SAM68, and GAPDH in 1124C and 1216 cells, with or without transfection with LRRC4 plasmid. (C) Quantification of SAM68 protein expression in 1124C and 1216 cells, with or without transfection with LRRC4 plasmid. (D) Representative western blot analysis of LRRC4, SAM68, and GAPDH proteins in primary GBM tissues. (E) 1124C cells were transfected with green fluorescent protein (GFP)-LRRC4 plasmid. Cells were stained with anti-SAM68 antibody (red) as indicated. The nuclei were stained with DAPI (blue). Scale bar, 20 μm. (F) Quantitation of red fluorescence intensity between GFP-positive and GFP-negative cells. (G) The diagrams show how SAM68 regulated variable splicing by directly binding to the pre-mRNA of CD44, mTOR, and cyclin D1. (H) RT-qPCR analyses showed that LRRC4 increases the expression of hsa_circ_0021726 in 1124C cells. (I) RT-qPCR analyses showed that LRRC4 increases the expression of hsa_circ_0021726 in 1216 cells. ∗p<0.05; ∗∗p<0.01:∗∗∗p<0.001.
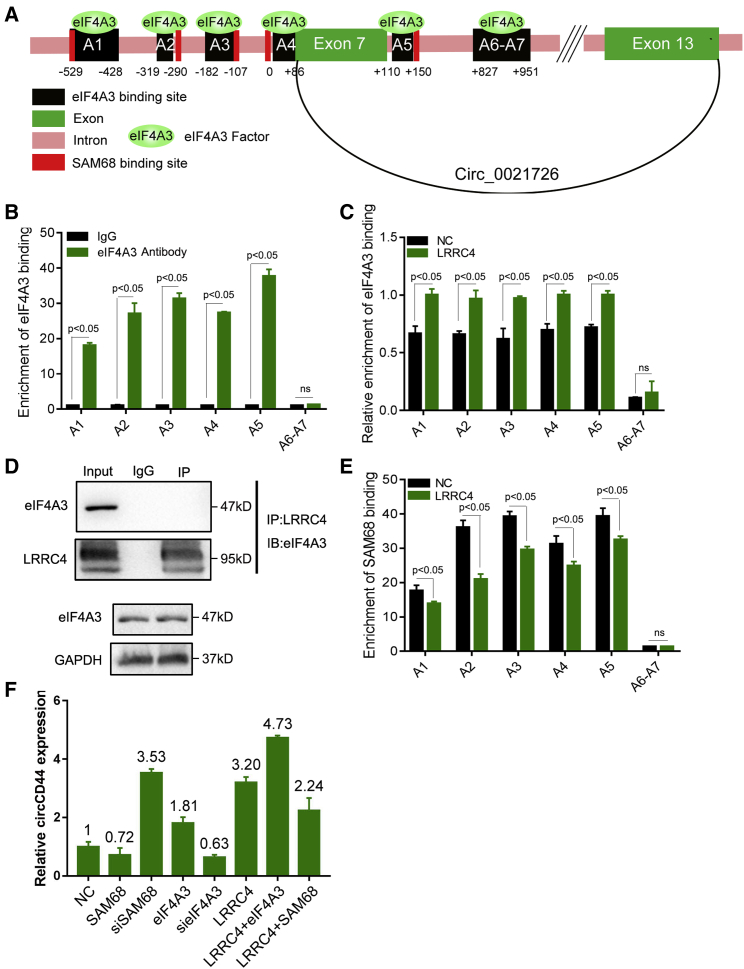
LRRC4 promotes the generation of circCD44 by inhibiting binding between SAM68 and CD44 pre-mRNA
To further reveal the mechanism by which LRRC4 regulates the expression of circ_0021726 in GBM, we explored hsa_circ_0021726 formation by using a bioinformatics method (circBase). We found that hsa_circ_0021726, with a molecular weight of 810 bp, was formed from exons 7 to 13 of CD44 (Figure 2A). Then, we named hsa_circ_0021726 as circCD44. By using another database (Circular RNA Interactome), we found that nine binding sites for eIF4A3 were present near the exon 7 region of the CD44 mRNA (Figure 2A). A previous study reported that eIF4A3 promotes circMMP9 expression by binding with circMMP9 mRNA.28 Our results also implied that eIF4A3 maybe regulate circCD44 formation by directly binding with binding sites in CD44 mRNA. We then conducted a RIP (RNA binding protein immunoprecipitation) assay to investigate the binding between eIF4A3 and binding sites in CD44 mRNA in 1124C cells. The results showed that eIF4A3 can bind with CD44 mRNA through the five putative binding sites, which we named A1, A2, A3, A4, and A5, but not A6 to A7 in the corresponding RNA-protein complex (Figure 2B). Furthermore, when compared to the control, overexpression of LRRC4 increased the binding between eIF4A3 and five putative binding sites (A1, A2, A3, A4, and A5) in GBM cells (Figure 2C). We next investigated whether LRRC4 regulates the expression of circCD44 by affecting eIF4A3. However, IP and western blotting assays showed that LRRC4 did not interact with eIF4A3 and expression of LRRC4 had no effect on protein-level alteration of eIF4A3 (Figure 2D), indicating that LRRC4 does not directly affect eIF4A3. In order to understand the molecular mechanism of LRRC4 promoting the generation of circCD44, we further analyzed the sequence of CD44 pre-mRNA through the UCSC database. The results showed that there were potential SAM68 binding sites near the eIF4A3 binding sites on CD44 pre-mRNA (Figure 2A, red box), which suggested that SAM68 and eIF4A3 bind competitively to the five putative binding sites. Data from an RIP assay using anti-SAM68 antibody indicated that SAM68 can bind with CD44 pre-mRNA through potential Sam68 binding sites. Overexpression of LRRC4 inhibited the binding between SAM68 and CD44 pre-mRNA (Figure 2E). These results revealed SAM68 binding with CD44 pre-mRNA, while expression of LRRC4 restrained the binding between SAM68 and CD44 pre-mRNA. We then knocked down the expression of eIF4A3 or overexpression of SAM68 and found a reduction in circCD44 expression, while overexpression of eIF4A3 and knocked-down expression of SAM68 increased circCD44 expression (Figure 2F). Furthermore, overexpression of LRRC4 and eIF4A3 significantly increased circCD44 expression, while overexpression of LRRC4 and SAM68 partially decreased circCD44 expression (Figure 2F).
Figure 2.
LRRC4 promotes the generation of circCD44 by inhibiting the binding between SAM68 and CD44 pre-mRNA
(A) Schematic sequence diagram of hsa_circ_0021726 in CD44 pre-mRNA. Black boxes represent eIF4A3 binding sites, and red boxes represent SAM68 binding sites. (B) The RIP assay in 1124C cells showed that eIF4A3 can bind with CD44 mRNA through five putative binding sites, which we named A1, A2, A3, A4, and A5, but not A6 to A7. (C) The RIP assay in 1124C cells showed that overexpression of LRRC4 enhanced eIF4A3 binding to the five putative binding sites in CD44 pre-mRNA. (D) Co-immunoprecipitation with LRRC4 antibody showed that LRRC4 does not interact with eIF4A3 (top). Immunoblotting showed that overexpression of LRRC4 does not alter the expression of eIF4A3 (bottom). (E) The RIP assay in 1124C cells showed that overexpression of LRRC4 decreased SAM68 binding to the five putative binding sites in CD44 pre-mRNA. (F) RT-qPCR analyses showed that the expression of hsa_circ_0021726 was regulated under the respective treatment condition in 1124C cells.
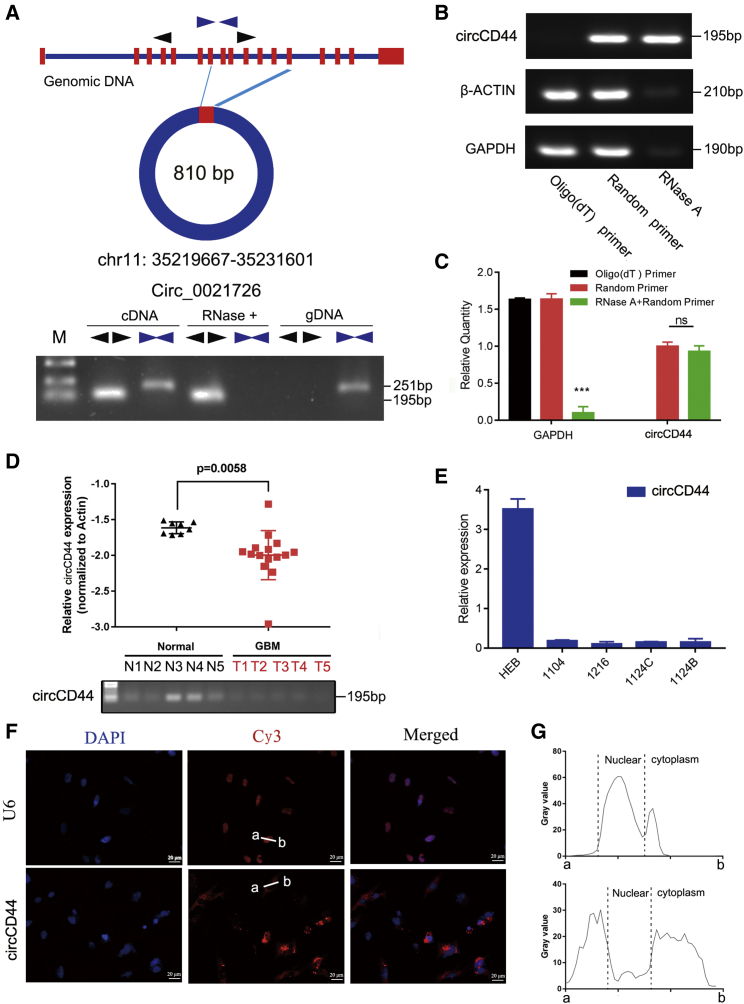
circCD44 is downregulated in GBM tissue and primary GBM cells
First, we aimed to verify that circCD44 is a circRNA. PCR analysis indicated that divergent primers could produce the circular isoform of CD44 with cDNA but not with genomic DNA (gDNA), while convergent primers could amplify the linear isoform of CD44 from both cDNA and gDNA in the primary GBM cells (Figure 3A). Furthermore, RT-PCR and qPCR showed that circCD44 cannot be reverse-transcripted using Olig(dt) primer, while it can be reverse-transcribed using random primers (Figures 3B and 3C). Also, circCD44 resists degradation of RNase R, while β-actin and gapdh mRNA can be degraded by RNase R (Figures 3B and 3C). These results indicated that circCD44 is a circRNA.
Figure 3.
circCD44 is a circular RNA and is downregulated in GBM
(A) Schematic representation of circCD44 formation (top). The RNAs were detected by PCR. Divergent primers could produce circRNAs in cDNA but not in genomic DNA (gDNA); convergent primers could produce cDNA and gDNA (bottom). (B) PCR and RT-qPCR showed that circCD44 cannot be reverse-transcribed using Olig(dt) primer, while it can be reverse-transcribed using random primers. (C) RT-qPCR showed that circCD44 resists degradation of RNase R, while gapdh mRNA can be degraded by RNase R. (D) The expression level of circCD44 was detected by RT-qPCR in GBM tissues (n = 14) and normal brain tissues (n = 8). (E) The expression level of circCD44 was detected by RT-qPCR in a human normal glial cell line (HEB) and primary glioma cells (1104, 1216, 1124C, 1124B). (F) Confocal FISH was performed to determine the location of circCD44 in 1124C cells. Scale bars, 20 μm. (G) Fluorescence signals were measured with ImageJ software. ∗p<0.05; ∗∗p<0.01:∗∗∗p<0.001.
We next explored the circCD44 expression level in GBM by using a qRT-PCR assay. A total of 14 patients with pathologically confirmed GBM were recruited and eight normal brain tissues were obtained for this assay. The results revealed that the circCD44 expression level was significantly decreased in GBM tissues compared with that in normal brain tissues (Figure 3D). We also tested circCD44 expression in a human normal glial cell line (HEB) and primary glioma cells (1104, 1216, 1124C, 1124B) using the qRT-PCR assay. The results showed that circCD44 was highly expressed in HEB cells but was lacking in 1104, 1216, 1124C, and 1124B primary glioma cells (Figure 3E). In addition, we investigated the location of circCD44 in 1124C cells with the fluorescence in situ hybridization (FISH) assay. Confocal results demonstrated that circCD44 was primarily expressed in the cytoplasm compared with the positive control (intranuclear RNA U6) (Figures 3F and 3G). Together, these data provided direct evidence showing that circCD44 is downregulated in GBM tissue and primary cells and suggested that circCD44 may act as a tumor suppressor in GBM.
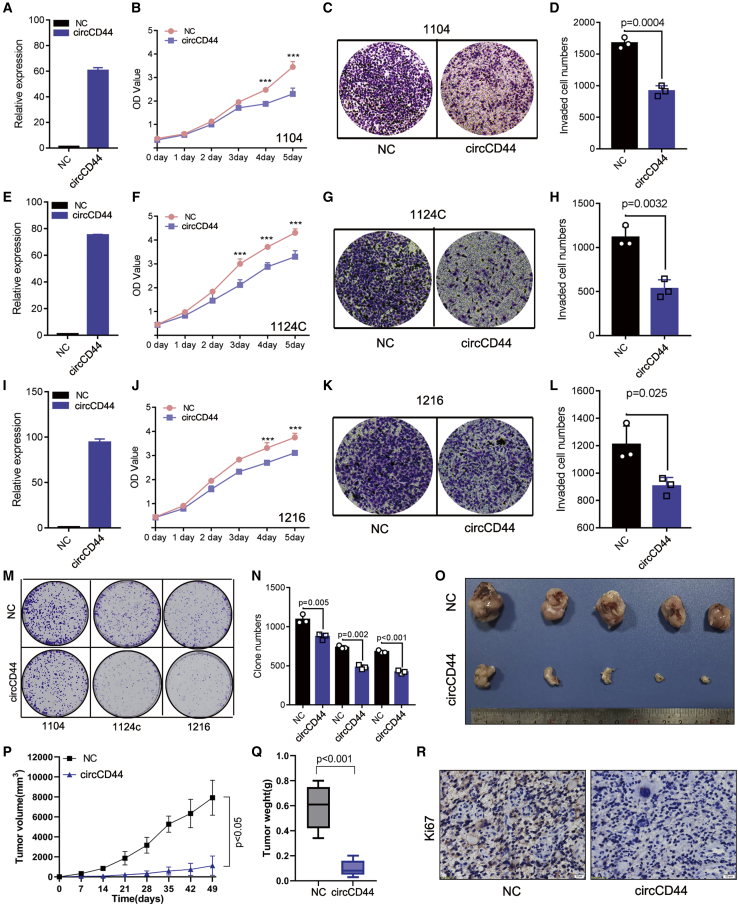
circCD44 is a tumor-suppressive circRNA in GBM
To explore the molecular mechanism by which circCD44 is associated with GBM progression, we conducted cell biology experiments to investigate the role of circCD44 in GBM cell proliferation, invasion, and clone-formation abilities. First, circCD44 and mock expression vectors were constructed and the successfully overexpressed circCD44 in 1104, 1124C, and 1216 cells was verified with RT-qPCR (Figures 4A, 4E, and 4I). Ectopic expression of circCD44 in 1104, 1124C, and 1216 cells suppressed GBM cell proliferation and invasion (Figures 4B–4L). To further test the tumor-suppressive function of circCD44, we analyzed the growth characteristics in GBM cells. The colony-formation assay showed that overexpression of circCD44 decreased the colony numbers compared to the control (Figures 4M and 4N). We next investigated the role of circCD44 in GBM development by means of a xenograft nude mouse model. In 1216 cells with stable expression of circCD44 or vector controls that were constructed and injected subcutaneously into mice, expression of circCD44 also significantly inhibited the growth of xenograft tumors (Figures 4O–4R). Overall, these data revealed that circCD44 is a tumor-suppressive circRNA in GBM and inhibited the tumorigenesis of GBM cells both in vitro and in vivo.
Figure 4.
circCD44 inhibits GBM cell proliferation in vitro and in vivo
(A, E, and I) Successful overexpression of circCD44 in 1104 (A), 1124C (E), and 1216 (I) cells was detected by RT-qPCR. (B, F, and J) The effect of ectopic circCD44 expression on 1104 (B), 1124C (F), and 1216 (J) cell proliferation was assessed with the CCK-8 cell growth assay. (C, G, and K) The effect of ectopic circCD44 expression on 1104 (C), 1124C (G), and 1216 (K) cell invasion was assessed with the transwell assay. (D, H, and L) Quantification of the number of invading cells from (C), (G), and (K). (D, H, and L) Quantification of the invading cells number from invasion assay of 1104 (D), 1124C (H), and 1216 cells (L). (M) Cell proliferation ability was evaluated by colony formation. (N) Quantification of the clone numbers of (M). (O) The representative tumor images in each group are displayed (n = 5). (P) Tumor volumes were measured and the growth curves were drawn. (Q) Tumor weight was analyzed. (R) Ki67 expression in each group was detected via IHC. Scale bar, 20 μm. ∗p<0.05; ∗∗p<0.01:∗∗∗p<0.001.
The identification of SMAD6 as a circCD44-regulated gene in GBM cells
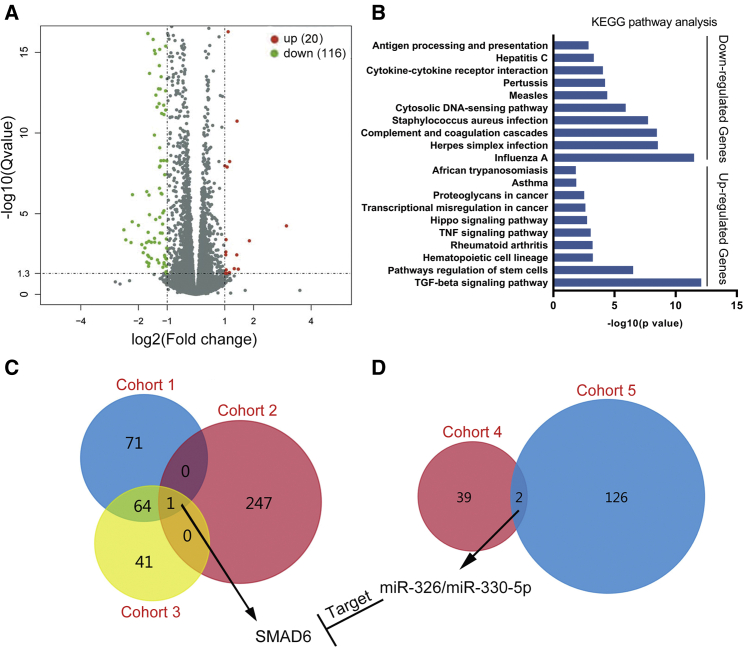
To determine the mechanisms that circCD44 inhibited the proliferation, invasion, and clone formation of GBM cells, we performed RNA sequencing analysis in paired 1124c and circCD44-overexpressed 1124c cell lines. DESeq2 was then used to analyze the differentially expressed genes. Compared to control cells, 20 genes were significantly upregulated and 116 genes were downregulated in circCD44-expressed 1124c cells (Figures 5A and 5C, cohort 1). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that upregulated genes were mainly involved in the transforming growth factor (TGF)-β signaling pathway and downregulated genes were mainly enriched in the cytokine-cytokine receptor interaction pathway (Figure 5B). How does circCD44 regulate these genes? An enormous body of evidence suggests that circRNAs play an important role in carcinogenesis by competitively binding to microRNAs (miRNAs), thereby regulating the expression level of tumor-related genes. Based on our finding that circCD44 is predominantly located in the cytoplasm in GBM cells (Figure 3F), we assumed that circCD44 functions as a competing endogenous RNA (ceRNA) by competitively binding to some miRNAs. First, by using the Circular RNA Interactome online database, a total of 41 miRNAs were found to potentially bind circCD44 (Table S1, cohort 4). Next, the target genes of these miRNAs were predicted and obtained by using FunRich software (named cohort 2). Subsequently, we analyzed differentially expressed genes between LRRC4-expressing cells and control cells (named cohort 3). Interestingly, only SMAD6 existed in all of the cohorts (Figure 5C). Thus, SMAD6 was chosen for further study. Furthermore, we also predicted and obtained miRNAs that potentially regulated differentially expressed genes between 1124c cells and circCD44-overexpressed 1124c cells (named cohort 5). Compared with 41 miRNAs that were found to potentially bind circCD44 in the Circular RNA Interactome online database, we found that both miR-330-5p and miR-326 exist in two cohorts (Figure 5D). To our surprise, the TargetScan database shows that SMAD6 is a common target gene of miR-330-5p and miR-326. Taken together, these results suggest that circCD44 may act as a glioma suppressor through the miR-330-5p/miR-326/SMAD6 signaling axis.
Figure 5.
Identification of SMAD6 as a circCD44-regulated gene in GBM cells
(A) Volcano plot and clustered heatmap showing that 20 genes were significantly upregulated and 116 genes were downregulated in circCD44-expressed 1124c cells. (B) KEGG pathways enriched among differentially expressed genes between circCD44-expressed 1124c cells and control cells. (C) Comparison of differentially expressed genes in three cohorts. Cohort 1 comprised differentially expressed genes between circCD44-expressed 1124c cells and control cells. Cohort 2 comprised the predicted target genes of miRNAs that were found to potentially bind circCD44. Cohort 3 comprised differentially expressed genes between LRRC4-expressing cells and control cells. (D) Comparison of differentially expressed miRNAs in two cohorts. Cohort 4 comprised miRNAs found to potentially bind circCD44 in the Circular RNA Interactome online database. Cohort 5 comprised miRNAs that potentially regulate differentially expressed genes between 1124c cells and circCD44-overexpressed 1124c cells.
circCD44 acts as a sponge and directly binds to miR-330-5p/miR-326
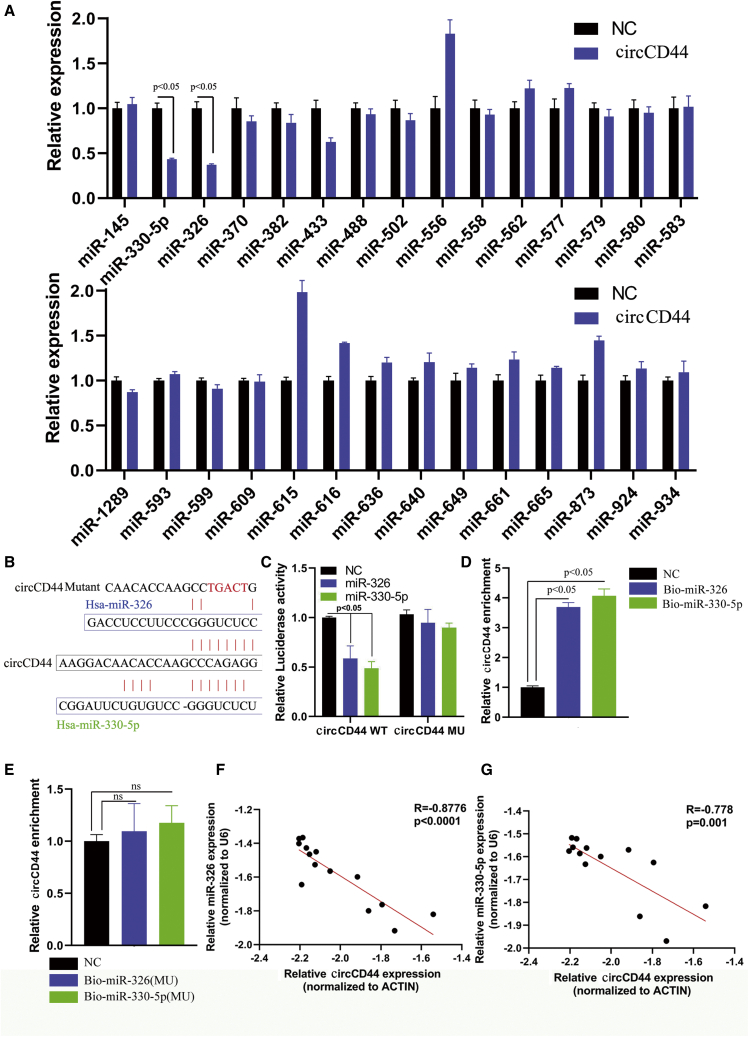
Next, RT-qPCR assays were conducted to investigate the impact of circCD44 on the expression of miRNAs that potentially bind to circCD44. The results showed that overexpression of ccircCD44 resulted in downregulation of miR-330-5p and miR-326 in GBM cells (Figure 6A). Does circCD44 directly bind to miR-330-5p/miR-326? Using bioinformatics analysis, we found that circCD44 had a putative binding site with miR-330-5p and miR-326 (Figure 6B). Subsequently, luciferase reporter assays were executed to verify the binding between circCD44 and miR-330-5p/miR-326. The results indicated that both miR-330-5p or miR-326 mimics obviously attenuated the luciferase activity of the circCD44-wild-type (WT) luciferase reporter compared with the scrambled control (Figure 6C), which suggests that circCD44 might directly combine with miR-330-5p or miR-326. In order to further confirm the binding of circCD44 with miR-330-5p/miR-326, RNA pull-down was conducted. Pull-down assay results also indicated that circCD44 was enriched in the biotin-labeled miR-330-5p-WT and miR-326-WT (Figure 6D) but not that of mutants (Figure 6E). In addition, Pearson correlation analysis revealed that the expression of circCD44 was reversely correlated with the level of miR-330-5p and miR-326 in collected clinical GBM samples (Figures 6F and 6G). All together, these results suggested that circCD44 could directly combine with miR-330-5p and miR-326 and act as a sponge of miR-330-5p and miR-326.
Figure 6.
circCD44 acts as a sponge and directly binds to miR-330-5p/miR-326
(A) RT-qPCR assays were conducted to investigate the impact of circCD44 on the expression of miRNAs that potentially bind to circCD44. (B) The putative binding sites of miR-326 and miR-330-5p on the circCD44 wild-type (WT) or mutated sequence are shown. (C) The luciferase reporter assay was performed to detect the activity of circCD44 in 1124C cells cotransfected with miR-326, miR-330-5p, or scramble and circCD44 WT or circCD44 mutated (MU) vector. (D) RNA pull-down was performed to detect the binding of circCD44 and miR-326 or miR-330-5p. (E) RNA pull-down was performed to detect the binding of circCD44 and biotinylated miR-326 (MU) or biotinylated miR-330-5p (MU). (F) Correlation between the expression of miR-326 and circCD44 in collected GBM samples was evaluated with Pearson’s correlation test. (G) Correlation between the expression of miR-330-5p and circCD44 in collected GBM samples was evaluated with Pearson’s correlation test.
miR-330-5p/miR-326 reverses the tumor-suppressive effects of circCD44 in GBM cells
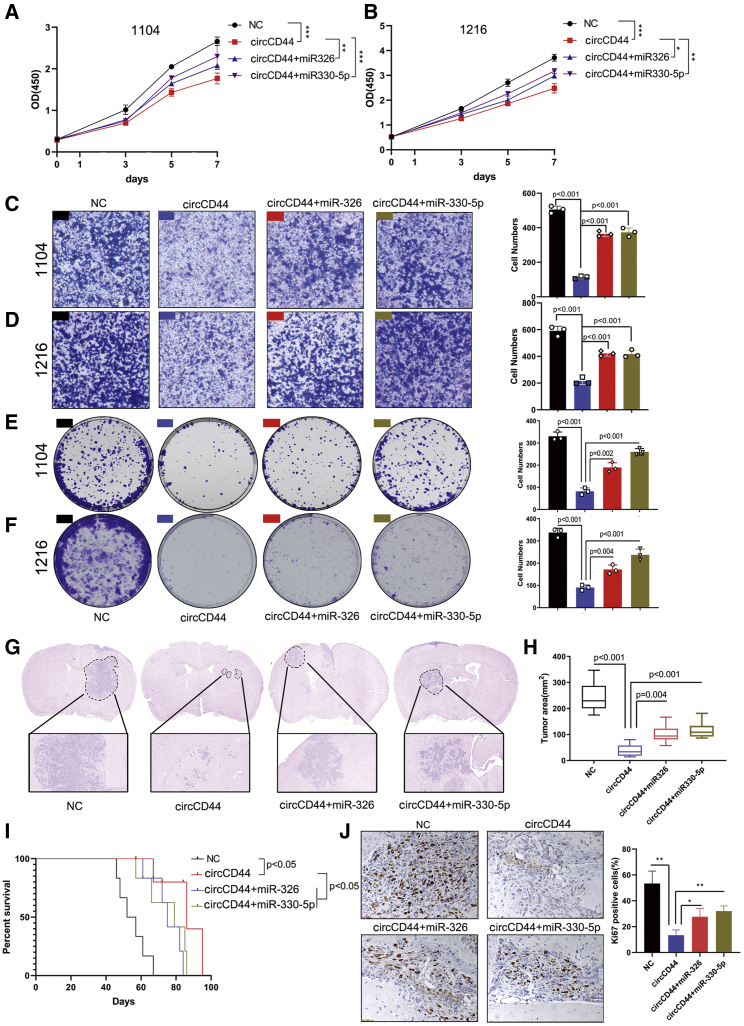
To further investigate the roles of miR-330-5p/miR-326 and circCD44 in GBM progression, we performed rescue assays to evaluate the effects of the circCD44/miR-330-5p/miR-326 axis on the proliferation, invasion, and clone-formation abilities of GBM cells. The results revealed that ectopic expression of miR-330-5p or miR-326 significantly attenuated the proliferation, invasion, and clone-formation-suppressing effects induced by upregulation of circCD44 in GBM cells (Figures 7A–7F). Furthermore, we also evaluated the effect of circCD44 and miR-330-5p/miR-326 in tumor growth in vivo by establishing orthotopic transplantation tumor models of glioma. The results showed that the section areas of tumors in the circCD44-expressing group were strikingly lower than those in the control group, while ectopic expression of miR-330-5p or miR-326 partly reversed the inhibitory roles of circCD44 in gliomagenesis (Figures 7G and 7H). Kaplan-Meier survival curves showed that nude mice injected with circCD44-overexpressing GBM cells had higher overall survival (OS) compared to the control group, while re-expression of miR-330-5p or miR-326 partly shortened OS relative to the circCD44-overexpressing group (Figure 7I). Immunohistochemical staining of excised tumors with Ki67 antibodies was also consistent with that mentioned earlier (Figure 7J). In summary, these results demonstrated that circCD44 could function as a sponge of miR-330-5p and miR-326 to inhibit glioma progression.
Figure 7.
miR-330-5p/miR-326 reverses the tumor-suppressive effects of circCD44 in GBM cells
(A and B) 1104 (A) and 1216 (B) cells were transfected with the indicated vectors and miRNAs, and CCK-8 assays were performed to assess the proliferation ability of the transfected cells. (C and D) 1104 (C) and 1216 (D) cells were transfected with the indicated vectors and miRNAs, and transwell assays were performed to assess the invasion ability of the transfected cells. (E and F) 1104 (E) and 1216 (F) cells were transfected with indicated vectors and miRNAs, and colony-formation assays were performed to assess the proliferation ability of the transfected cells. (G) H&E staining of the brain and tumors. Tumor areas are indicated by a dotted line. (H) Tumor areas (in mm3) were assessed with ImageJ software. (I) Kaplan-Meier survival curves of mice injected with transfected with indicated vectors and miRNAs. (J) Ki67 expression in each group was detected via IHC. Scale bar, 20 μm. ∗p<0.05; ∗∗p<0.01:∗∗∗p<0.001.
SMAD6 is a direct target of miR-330-5p and miR-326 and is regulated by the LRRC4/circCD44/miR-330-5p/miR-326 axis in GBM cells
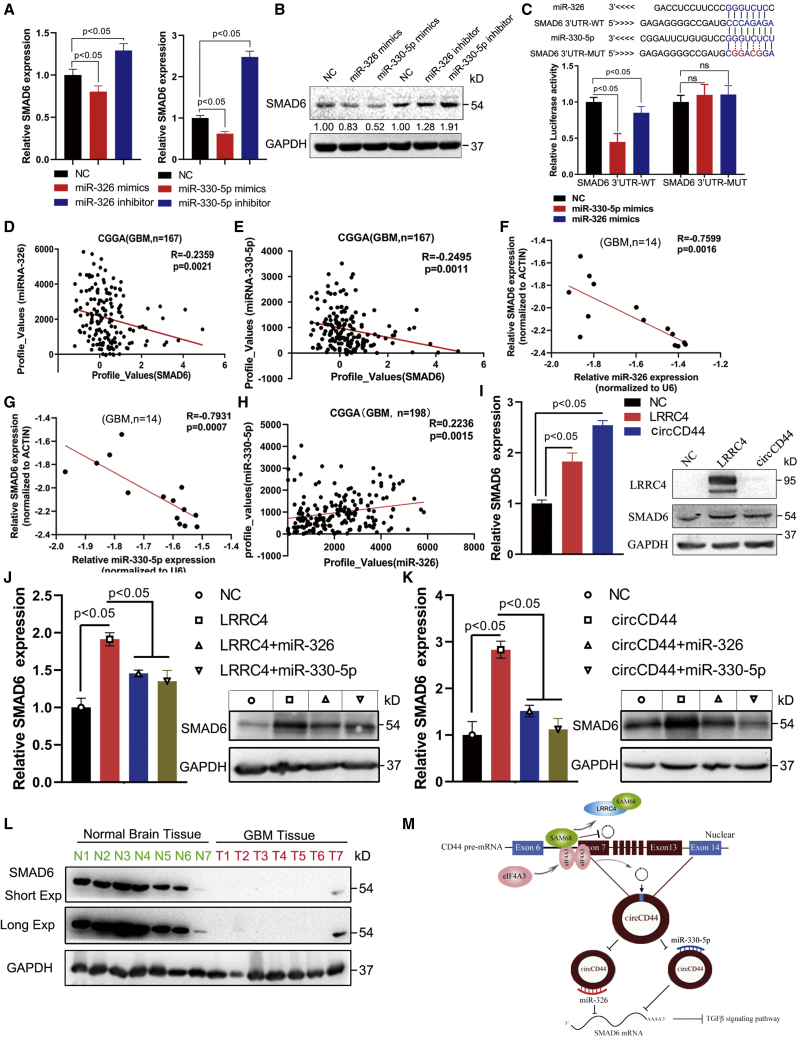
Based on TargetScan results showing that SMAD6 is common target gene of miR-330-5p and miR-326, we then determined whether SMAD6 is a direct target of miR-330-5p and miR-326. The qRT-PCR and western blotting experiments revealed that the expression of SMAD6 was decreased at both mRNA and protein levels in GBM cells transfected with miR-330-5p and miR-326 mimics, while SMAD6 expression was increased in GBM cells transfected with miR-330-5p and miR-326 inhibitors (Figures 8A and 8B). Furthermore, wild and mutant luciferase reporter plasmids containing SMAD6 3′ UTRs were constructed (Figure 8C). By measuring luciferase activity, we found that cells transfected with miR-330-5p and miR-326 mimics could significantly decrease the activity of the luciferase reporter carrying the WT 3′ UTR of SMAD6 but not that of the mutant 3′ UTR of SMAD6 (Figure 8C). Next, Pearson correlation analysis was used to show the relationship between SMAD6 and miR-330-5p/miR-326, and we found that SMAD6 was negatively correlated with the level of miR-330-5p and miR-326 in the CGGA database (Figures 8D and 8E). In addition, clinical GBM samples were collected and analyzed with correlation analysis, and the results also revealed that SMAD6 was negatively correlated with the expression level of miR-330-5p and miR-326 (Figures 8F and 8G). Interestingly, we also found that miR-326 was positively correlated with the expression of miR-330-5p (Figure 8H). Subsequently, we then measured whether SMAD6 is regulated by LRRC4 or circCD44. Overexpression of LRRC4 or circCD44 increased both mRNA and protein levels of SMAD6 (Figure 8I), and transfection with miR-330-5p or miR-326 mimics decreased both mRNA and protein levels of SMAD6 in LRRC4- or circCD44-expressing GBM cells (Figures 8J and 8K). Clinically, miR-330-5p and miR-326 were upregulated in high-grade gliomas compared with LGGs (Figures S2A and S2C), and patients with a high expression of miR-330-5p had a poorer prognosis relative to patients with gliomas that had a low expression level of miR-330-5p (Figure S2B). However, miR-326 was not a prognostic marker in glioma (Figure S2D). Furthermore, LRRC4 expression was positively correlated with SMAD6 in the CGGA GBM dataset (Figure S2E). Next, clinical GBM samples were collected and analyzed with qRT-PCR, and the results also revealed that circCD44 was negatively correlated with the expression level of SMAD6 (Figure S2F). We also examined the SMAD6 protein expression in total protein extraction of GBM specimens. As shown in Figure 8L, the expression of SMAD6 protein was significantly decreased in GBM samples compared with normal brain tissues, which also indicated that SMAD6 may be a tumor-suppressive gene in GBM. In addition, overexpression of SMAD6 impaired GBM cell growth and invasion in vitro (Figures S3A–S3C). Knockdown of SMAD6 partially reversed the tumor-suppressive effects of LRRC4 in GBM cells (Figures S3D–S3F). Taken together, these results indicate that SMAD6 is a direct target of miR-330-5p and miR-326 and is regulated via the LRRC4/Sam68/circCD44/miR-330-5p/miR-326 axis in GBM cells.
Figure 8.
SMAD6 is a direct target of miR-330-5p and miR-326 and is regulated by the LRRC4/circCD44/miR-330-5p/miR-326 axis in GBM cells
(A) RT-qPCR detected the relative expression of SMAD6 after transfection with miR-326, miR-326 inhibitor, miR-330-5p, and miR-330-5p inhibitor. (B) Western blotting detected the relative expression of SMAD6 after transfection with miR-326, miR-326 inhibitor, miR-330-5p, and miR-330-5p inhibitor. (C) The luciferase reporter assay was performed to detect the activity of the SMAD6 3′ UTR in 1124C cells cotransfected with miR-326, miR-330-5p, or scramble and SMAD6 3′ UTR wild-type (WT) or SMAD6 3′ UTR mutated (MU) vector. (D) Correlation between the expression of miR-326 and SMAD6 in the CGGA dataset was evaluated with Pearson’s correlation test. (E) Correlation between the expression of miR-330-5p and SMAD6 in the CGGA dataset was evaluated with Pearson’s correlation test. (F and G) Correlation between the expression of miR-326 (F) or miR-330-5p (G) and SMAD6 in collected GBM samples was evaluated with Pearson’s correlation test. (H) Correlation between the expression of miR-330-5p and miR-326 in the CGGA dataset was evaluated with Pearson’s correlation test. (I) The relative expression of SMAD6 was detected after transfection with LRRC4 and circCD44 by qRT-PCR and western blotting. (J) The relative expression of SMAD6 was detected after transfection with the indicated vectors and miRNAs by qRT-PCR and western blotting. (K) The relative expression of SMAD6 was detected after transfection with the indicated vectors and miRNAs by qRT-PCR and western blotting. (L) Representative western blot analysis of SMAD6 protein expression in primary GBM tissues. (M) A model demonstrating that LRRC4 mediates the formation of tumor-suppressive circCD44 by directly interacting with SAM68 in GBM.
Discussion
LRRC4 belongs to the LRR superfamily of synaptic adhesion proteins. LRRC4 is composed of a C-terminal PDZ-binding domain, through which it can bind to PDZ domain-containing proteins, the N-terminal LRR domain, and the immunoglobulin (Ig)-like domain. Functionally, LRRC4 is a synaptic adhesion protein that plays an important role in regulating synaptic transmission as well as glioma tumorigenesis.29,30 Xu et al.31 revealed that LRRC4 is a polarity regulator that localizes asymmetrically in rat hippocampal neurons and is required for differentiation of the future axon. Meanwhile, an extensive study reported by our group has been devoted to the capacity of the Lrrc4 gene to inhibit glioma cell proliferation and invasion and increase the TMZ treatment response of glioblastoma through different signaling pathways.18, 19, 20, 21, 22, 23 Here, we explore novel tumor-suppressive roles and molecular mechanisms of LRRC4 in gliomagenesis. This study not only increases our knowledge of the tumor-suppressive function of LRRC4, but it also offers a number of significant findings.
First, we identified SAM68 as a new interactive partner for LRRC4. Our group reported previously that LRRC4 could bind with some proteins, including Erk1/2 (altering the cellular location of Erk1/2),20 PDPK1 (enhancing the interaction between PDPK1 and IKKβ),21 and DEPTOR (promoting the degradation of DEPTOR), in glioma cells.22 In this study, by conducting coIP combined with mass spectrometry experiments, we identified SAM68 as a new interactive partner for LRRC4. SAM68 belongs to the STAR (signal transduction and activation of RNA metabolism) family of RNA binding proteins. It is widely expressed in all kinds of cells but performs different functions under different environments.32 In general, the main functions of SAM68 are as follows. First, in gene transcription regulation, SAM68 could interact with transcription factors such as CBP and appears to regulate CBP-dependent gene expression.33 In addition, studies have shown that SAM68 interacts with androgen receptors to enhance its transcriptional activity.34 Second, in the regulation of alternative splicing, SAM68 interacts with alternative splicing factor U2AF65 and the expression of SAM68 results in enhanced binding of the U2AF65 subunit to an alternatively spliced pre-mRNA sequence, including the pre-mRNA of the cell-surface molecule CD44.35,36 Paronetto et al.37 found that SAM68 can bind with the AU sequence on intron 4 of the cyclin D1 pre-mRNA, promote the inclusion of intron 4 in the D1b mRNA, and thereby enhance splicing of cyclin D1b. In addition, Paronetto et al.38 found that SAM68 binds the mRNA for Bcl-x and affects its alternative splicing. Beyond its known functions, we first discovered a novel function of SAM68 in gliomagenesis; that is, SAM68 also plays an important role in the generation and formation of RNA circCD44. Mechanistically, SAM68 shares the same binding sites with eIF4A3 on CD44 pre-mRNA. A previous study reported that eIF4A3 promotes circMMP9 expression by binding with circMMP9 mRNA, indicating that eIF4A3 is a key regulator in the generation of circRNA. In this study, we found that the enrichment of eIF4A3 on pre-mRNA of CD44 was repressed when SAM68 was present in the mRNA for CD44, and the depletion of SAM68 promoted eIF4A3 binding of the mRNA for CD44 and increased the formation of circCD44. SAM68 is upregulated in GBM tissues and high expression of SAM68 is strongly associated with poor survival (Figures 1 and S1), indicating that SAM68 plays a protumorigenic role in glioma progression. Our findings revealed that LRRC4 decreased the protein level of Sam68. In a previous report, we confirmed that LRRC4 induces the degradation of DEPTOR by directly interacting with DEPTOR and promoting the binding between DEPTOR and CUL3.22 The explanation of why LRRC4 could reduce the protein level of SAM68 is that LRRC4 may also act as a bridge of SAM68 and E3 ubiquitin ligase CUL3. Additionally, we also confirmed that LRRC4 promoted the formation of circCD44 in glioma cells by interacting with SAM68 and reducing its protein expression. This suggests that LRRC4 hinders glioma cell growth partly through both decreasing the protein level of SAM68 and increasing the formation of circCD44.
Second, our studies identify hsa_circ_0021726 as circCD44, which is a novel GBM-suppressive circRNA. circCD44, with a molecular weight of 810 bp, was formed from exons 7 to 13 of CD44 pre-mRNA. To our knowledge, before this study, there were no published articles about the function of circCD44. We found that circCD44 expression was significantly decreased in GBM. The overexpression of circCD44 inhibits glioma cell growth in vitro and in vivo. To date, circRNAs have mainly been attributed to active roles in the cytosol, where they can be translated into peptides39, 40 or act as ceRNA to regulate the expression of oncogenes or tumor-suppressor genes via sponging miRNAs.41 In this study, we found that circCD44 has predicted binding sites for miR-326 and miR-330-5p, and we confirmed the direct binding between circCD44 and miR-326 and miR-330-5p. miR-326 and miR-330-5p may act as oncogenes in tumorigenesis. Kim et al.42 confirmed that miR-326 enhanced the invasion and migration potential of cancer cells. Xiao et al.43 reported that miR-330-5p expression is upregulated in HCC tissues and cell lines, and miR-330-5p knockdown inhibits proliferation and growth of HCC cells in vitro and in vivo. Feng et al.44 reported that miR-330-5p suppresses glioblastoma cell proliferation and invasiveness, and this finding was based on only the U251 cell line. However, we did confirm that miR-330-5p acts as an oncogene in primary glioma cell lines 1104, 1124c, and 1216. We thought that tumor heterogeneity may play an important role in different glioma cells. Furthermore, based on the ceRNA hypothesis, we also found that circCD44 regulated the expression of SMAD6, which is the target gene of both miR-326 and miR-330-5p. SMAD6, one of the inhibitory Smads, plays an important role in TGF-β1-mediated negative regulation of proinflammatory signaling.45 Jiao et al.46 found that ectopic SMAD6 is predominately expressed in the cytoplasm and can act as a tumor suppressor in inhibiting migration, invasion, and metastasis. Our study also confirmed that overexpression of SMAD6 inhibited glioma cell growth and invasion, indicating that SMAD6 can act as a glioma suppressor in vitro. Subsequently, the tumor-suppressive effects might be reversed by SMAD6 knockdown. These results further confirm our hypothesis that circCD44 serves as a ceRNA for miR-326 and miR-330-5p to enhance SMAD6 expression and regulate the TGF-β signaling pathway in GBM progression. However, detailed mechanisms in the regulatory process need further elucidation.
In conclusion, our study revealed that LRRC4, which has been proven to be a tumor-suppressor gene, directly interacts with SAM68 and decreases the protein level of SAM68, thus promoting eIF4A3 binding of the mRNA for CD44 and increasing the formation of circCD44. Moreover, circCD44 serves as a ceRNA for miR-326 and miR-330-5p to enhance SMAD6 expression and regulate the TGF-β signaling pathway in GBM progression. Thus, the LRRC4/Sam68/circCD44/miR-326/miR-330-5p/SMAD6 signaling axis could be a potential target for GBM treatment.
Materials and methods
Tissue samples and primary cell culture
GBM tissues were collected from the Department of Neurosurgery at Xiangya Hospital Central South University (Hunan, China). All of the experimental schemes were approved by the Ethics Committee of Xiangya Medical College of Central South University and were performed following national guidelines. Primary cell (1104, 1124c, and 1216) culture protocols were described in our previous report.47 Briefly, primary glioma tissues were minced with a GentleMACS Dissociator (Miltenyi Biotec, Gladbach, Germany) and digested with 0.25% trypsin at 37°C for 30 min. Digestion was stopped by adding trypsin inhibitor, and cells were passed through a 40-μm nylon cell strainer (352340; Corning) to obtain single-cell suspensions.
Cell lines and culture conditions
Primary cell lines 1104, 1124c, and 1216 as well as normal glial cell line HEB were conserved in our laboratory. Cells were cultured in DMEM (GIBCO, Carlsbad, CA) supplemented with 10% fetal bovine serum and antibiotics. Cells were maintained in a humidified incubator at 37°C with 5% CO2.
Antibodies and reagents
Antibodies against SAM68 (10222-1-AP, WB, 1:1,500; IP, 1:200; RIP, 1:200), GAPDH (60004-1-Ig, WB, 1:2,000), HA (66006-2-Ig, WB, 1:2,000), GST (66001-2-Ig, WB, 1:5,000; IP, 1:500), and eIF4A3 (17504-1-AP, WB, 1:1,500; IP, 1:150; RIP, 1:200) were purchased from Proteintech (Wuhan, Hubei, China). Antibodies against Flag (F3165, IP, 1:200) was purchased from Sigma-Aldrich (St. Louis, MO). Antibodies against LRRC4 (bs-1974R, WB, 1:1,000) and SMAD6 (bs-23329R, WB, 1:1,000) was purchased from Boaosen Biotechnology (Beijing, China). Anti-Ki67 antibody (PB9026, immunohistochemistry [IHC], 1:1,500) was purchased from Boster Biological Technology (Pleasanton, CA). Cy3-labeled circCD44 and U6 were used to observe the localization of circCD44. FISH analysis was performed using a FISH kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions.
Cell proliferation and invasion assays
CCK-8, EDU, colony formation, and transwell invasion assays were performed according to our previous report.47
RNA isolation and qRT-PCR
Total RNA was extracted from tissues or harvested cells with TRIzol reagent (Invitrogen, Carlsbad, CA) and then used for first-strand cDNA synthesis using RevertAid RT Reverse Transcription Kit (Thermo Fisher). Real-time PCR reactions were performed with TB Green Premix Ex Taq II (Takara, Dalian, China) in a CFX-96 real-time PCR system (Bio-Rad, Hercules, CA), and the relative expression of genes was normalized using GAPDH as a housekeeping gene.
RNA pull-down assay
To pull down the circRNA by miRNA, biotinylated miR-326 and miR-330-5p mimics or their mutants were synthesized by RiboBio. 1124c cells that transfected with biotinylated miR-326 and miR-330-5p mimics or their mutants were lysed in IP buffer. Next, the biotin-coupled RNA complex was pull downed by streptavidin-coated magnetic bead adsorption. The immunoprecipitated RNA complex was collected by using TRIzol and ethanol precipitation. The enrichment of circCD44 was measured with qRT-PCR.
RIP assay
After crosslinking with 0.4% formaldehyde for 15 min at room temperature, cells were lysed in IP buffer (25 mM of Tris, pH 7.5, 150 mM of NaCl, 1% Triton X-100, and 1 mM of EDTA) with RNasin inhibitor and cocktail. Cell lysates were incubated with the indicated antibodies at 4°C overnight. Protein A/G magnetic beads were washed twice with IP buffer and were then added to the reaction mixtures and incubated for 2 h at 4°C. Then the immunoprecipitated RNAs were collected by using TRIzol and ethanol precipitation and subjected to qRT-PCR analysis.
IP and immunoblotting
Our previous report provides a detailed description of this protocol.22 For the IP assay, GBM cells were washed with cold PBS and lysed in coIP buffer. Cell lysates containing 500 μg of total protein were incubated with Flag or antibodies at 4°C overnight. Protein A/G magnetic beads were moderately washed twice with coIP buffer and were then added to the reaction mixtures and subjected to incubation for 2 h at 4°C. After magnetic separation, the magnetic beads were washed four times with coIP buffer and boiled for 10 min after the addition of 2× SDS loading buffer. The immunoprecipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and 50 μg of total protein was used as the input control. For immunoblotting, cells were lysed in RIPA buffer. Protein lysates were fractionated by SDS-PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Germany), and then incubated with the indicated primary antibodies, washed, and probed with HRP (horseradish peroxidase)-conjugated secondary antibodies.
Xenograft tumor model
Animal experiments were approved by the Animal Care and Use Committee of Central South University. Briefly, for subcutaneous tumor formation, 2 × 106 cells with 200 μL of serum-free medium were injected subcutaneously in the left flank of nude mice. Tumor volume was calculated in accordance with the formula (length × width2/2) and measured weekly, and tumor weight was determined. For the intracranial implantation mouse model, 4 × 105 cells in 4 μL of serum-free medium were stereotactically injected into the striatum (1.0 mm anterior and 1.0 mm lateral from the Bregma suture and 3.0 mm below the pial surface) of nude mice.
Immunohistochemistry
Tumor xenograft paraffin sections were deparaffinized and then rehydrated through an alcohol series, followed by antigen retrieval with sodium citrate buffer. Sections were blocked with 3% hydrogen peroxide for 10 min and with normal goat serum for 30 min at room temperature. Then, the sections were incubated with Ki67 antibodies (1:1,500) overnight at 4°C and were incubated with biotinylated secondary antibody (Maxim Biotechnologies) for 30 min at room temperature, then detected by HRP-conjugated DAB.
Statistical analysis
Data are presented as the mean ± SD from at least three separate experiments, and the data were analyzed with GraphPad Prism 8 software. Differences between variables of two groups were examined with the Student’s t test, and one-way ANOVA was used to evaluate the differences among variables of multiple groups. OS curves for xenograft tumor model mice were calculated with the Kaplan-Meier method. p < 0.05 was considered statistically significant.
Acknowledgments
We thank the Department of Neurological Surgery, Xiangya Hospital, Central South University, Hunan, China, for providing the GBM tissue samples. This work was supported by grants from the National Natural Science Foundation of China (82073096, 81874850, 81502185, 82103171), the Key Research and Development Plan of Hunan Province (2020SK2053), and the Natural Science Foundation of Hunan Province (2021JJ30612).
Author contributions
M.W. and J.F. designed the study; J.F., X.R., H.F., and D.L. performed experiments; X.C., X.Z., and Q.L. analyzed and interpreted the data; and M.W., Q.L., and J.F. were the major contributors in writing and revising the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.08.026.
Contributor Information
Qing Liu, Email: liuqingdr@csu.edu.cn.
Minghua Wu, Email: wuminghua554@aliyun.com.
Supplemental information
References
- 1.Feng R.M., Zong Y.N., Cao S.M., Xu R.H. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. (Lond.) 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom Q.T., Gittleman H., Stetson L., Virk S.M., Barnholtz-Sloan J.S. Epidemiology of gliomas. Cancer Treat. Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 3.Goldbrunner R., Ruge M., Kocher M., Lucas C.W., Galldiks N., Grau S. The treatment of gliomas in adulthood. Dtsch. Arztebl. Int. 2018;115:356–364. doi: 10.3238/arztebl.2018.0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borisov N., Sorokin M., Tkachev V., Garazha A., Buzdin A. Cancer gene expression profiles associated with clinical outcomes to chemotherapy treatments. BMC Med. Genomics. 2020;13(Suppl 8):111. doi: 10.1186/s12920-020-00759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Höhne J., Schebesch K.M., de Laurentis C., Akçakaya M.O., Pedersen C.B., Brawanski A., Poulsen F.R., Kiris T., Cavallo C., Broggi M. Fluorescein sodium in the surgical treatment of recurrent glioblastoma multiforme. World Neurosurg. 2019;125:e158–e164. doi: 10.1016/j.wneu.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Yu T., Wang X., Zhi T., Zhang J., Wang Y., Nie E., Zhou F., You Y., Liu N. Delivery of MGMT mRNA to glioma cells by reactive astrocyte-derived exosomes confers a temozolomide resistance phenotype. Cancer Lett. 2018;433:210–220. doi: 10.1016/j.canlet.2018.06.041. [DOI] [PubMed] [Google Scholar]
- 7.Sun J., Li B., Shu C., Ma Q., Wang J. Functions and clinical significance of circular RNAs in glioma. Mol. Cancer. 2020;19:34. doi: 10.1186/s12943-019-1121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei M., Zheng G., Ning Q., Zheng J., Dong D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 10.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji F., Du R., Chen T., Zhang M., Zhu Y., Luo X., Ding Y. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol. Ther. Nucleic Acids. 2020;19:413–420. doi: 10.1016/j.omtn.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 14.Gan X., Zhu H., Jiang X., Obiegbusi S.C., Yong M., Long X., Hu J. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol. Cancer. 2020;19:45. doi: 10.1186/s12943-020-01163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng H., Qin C., Zhang C., Su J., Xiao Q., Xiao Y., Xiao K., Liu Q. circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J. Cell. Mol. Med. 2019;23:6658–6665. doi: 10.1111/jcmm.14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Zhong B., Zhang W., Wu J., Wang Y. Circular RNA CircMTO1 inhibits proliferation of glioblastoma cells via miR-92/WWOX signaling pathway. Med. Sci. Monit. 2019;25:6454–6461. doi: 10.12659/MSM.918676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ru W.J., Qian J., Li D., Ling L.X., Chen T., Jiang L., Zhang B.C., Zhou J., Li G.Y. Identification of LRRC4, a novel member of leucine-rich repeat (LRR) superfamily, and its expression analysis in brain tumor. Prog. Biochem. Biophysics. 2002;29:233–239. [Google Scholar]
- 18.Zhang Q.H., Wang L.L., Cao L., Peng C., Li X.L., Tang K., Li W.F., Liao P., Wang J.R., Li G.Y. Study of a novel brain relatively specific gene LRRC4 involved in glioma tumorigenesis suppression using the Tet-on system. Acta Biochim. Biophys. Sin. (Shanghai) 2005;37:532–540. doi: 10.1111/j.1745-7270.2005.00079.x. [DOI] [PubMed] [Google Scholar]
- 19.Wu M., Huang C., Gan K., Huang H., Chen Q., Ouyang J., Tang Y., Li X., Yang Y., Zhou H. LRRC4, a putative tumor suppressor gene, requires a functional leucine-rich repeat cassette domain to inhibit proliferation of glioma cells in vitro by modulating the extracellular signal-regulated kinase/protein kinase B/nuclear factor-kappaB pathway. Mol. Biol. Cell. 2006;17:3534–3542. doi: 10.1091/mbc.E05-11-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Guo Q., Wang R., Xu G., Li P., Sun Y., She X., Liu Q., Chen Q., Yu Z. The D domain of LRRC4 anchors ERK1/2 in the cytoplasm and competitively inhibits MEK/ERK activation in glioma cells. J. Hematol. Oncol. 2016;9:130. doi: 10.1186/s13045-016-0355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., Feng J., Liu Y., Liu Q., Fan L., Liu Q., She X., Liu C., Liu T., Zhao C. Novel therapy for glioblastoma multiforme by restoring LRRC4 in tumor cells: LRRC4 inhibits tumor-infiltrating regulatory T cells by cytokine and programmed cell death 1-containing exosomes. Front. Immunol. 2017;8:1748. doi: 10.3389/fimmu.2017.01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J., Zhang Y., Ren X., Li D., Fu H., Liu C., Zhou W., Liu Q., Liu Q., Wu M. Leucine-rich repeat containing 4 act as an autophagy inhibitor that restores sensitivity of glioblastoma to temozolomide. Oncogene. 2020;39:4551–4566. doi: 10.1038/s41388-020-1312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao C., She X., Zhang Y., Liu C., Li P., Chen S., Sai B., Li Y., Feng J., Liu J. LRRC4 suppresses E-cadherin-dependent collective cell invasion and metastasis in epithelial ovarian cancer. Front. Oncol. 2020;10:144. doi: 10.3389/fonc.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matter N., Herrlich P., König H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 25.Huot M.É., Vogel G., Zabarauskas A., Ngo C.T., Coulombe-Huntington J., Majewski J., Richard S. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol. Cell. 2012;46:187–199. doi: 10.1016/j.molcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Bielli P., Busà R., Paronetto M.P., Sette C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer. 2011;18:R91–R102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 27.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Wang R., Zhang S., Chen X., Li N., Li J., Jia R., Pan Y., Liang H. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol. Cancer. 2018;17:166. doi: 10.1186/s12943-018-0911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soto F., Watkins K.L., Johnson R.E., Schottler F., Kerschensteiner D. NGL-2 regulates pathway-specific neurite growth and lamination, synapse formation, and signal transmission in the retina. J. Neurosci. 2013;33:11949–11959. doi: 10.1523/JNEUROSCI.1521-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P., Xu G., Li G., Wu M. Function and mechanism of tumor suppressor gene LRRC4/NGL-2. Mol. Cancer. 2014;13:266. doi: 10.1186/1476-4598-13-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G., Wang R., Wang Z., Lei Q., Yu Z., Liu C., Li P., Yang Z., Cheng X., Li G., Wu M. NGL-2 is a new partner of PAR complex in axon differentiation. J. Neurosci. 2015;35:7153–7164. doi: 10.1523/JNEUROSCI.4726-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bielli P., Busà R., Paronetto M.P., Sette C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer. 2011;18:R91–R102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 33.Hong W., Resnick R.J., Rakowski C., Shalloway D., Taylor S.J., Blobel G.A. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol. Cancer Res. 2002;1:48–55. [PubMed] [Google Scholar]
- 34.Rajan P., Gaughan L., Dalgliesh C., El-Sherif A., Robson C.N., Leung H.Y., Elliott D.J. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 2008;215:67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 35.Tisserant A., König H. Signal-regulated pre-mRNA occupancy by the general splicing factor U2AF. PLoS ONE. 2008;3:e1418. doi: 10.1371/journal.pone.0001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naor D., Nedvetzki S., Golan I., Melnik L., Faitelson Y. CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 2002;39:527–579. doi: 10.1080/10408360290795574. [DOI] [PubMed] [Google Scholar]
- 37.Paronetto M.P., Cappellari M., Busà R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K.E., Sette C. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70:229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paronetto M.P., Achsel T., Massiello A., Chalfant C.E., Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu P., Mo Y., Peng M., Tang T., Zhong Y., Deng X., Xiong F., Guo C., Wu X., Li Y. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X., Huang M., Xing L., Yang R., Wang X., Jiang R., Zhang L., Chen J. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol. Cancer. 2020;19:73. doi: 10.1186/s12943-020-01183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y., Kim H., Park H., Park D., Lee H., Lee Y.S., Choe J., Kim Y.M., Jeoung D. miR-326-histone deacetylase-3 feedback loop regulates the invasion and tumorigenic and angiogenic response to anti-cancer drugs. J. Biol. Chem. 2014;289:28019–28039. doi: 10.1074/jbc.M114.578229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao S., Yang M., Yang H., Chang R., Fang F., Yang L. miR-330-5p targets SPRY2 to promote hepatocellular carcinoma progression via MAPK/ERK signaling. Oncogenesis. 2018;7:90. doi: 10.1038/s41389-018-0097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng L., Ma J., Ji H., Liu Y., Hu W. miR-330-5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170019. BSR20170019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Kim E.Y., Kim B.C. Lipopolysaccharide inhibits transforming growth factor-beta1-stimulated Smad6 expression by inducing phosphorylation of the linker region of Smad3 through a TLR4-IRAK1-ERK1/2 pathway. FEBS Lett. 2011;585:779–785. doi: 10.1016/j.febslet.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 46.Jiao J., Zhang R., Li Z., Yin Y., Fang X., Ding X., Cai Y., Yang S., Mu H., Zong D. Nuclear Smad6 promotes gliomagenesis by negatively regulating PIAS3-mediated STAT3 inhibition. Nat. Commun. 2018;9:2504. doi: 10.1038/s41467-018-04936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng J., Zhang Y., She X., Sun Y., Fan L., Ren X., Fu H., Liu C., Li P., Zhao C. Hypermethylated gene ANKDD1A is a candidate tumor suppressor that interacts with FIH1 and decreases HIF1α stability to inhibit cell autophagy in the glioblastoma multiforme hypoxia microenvironment. Oncogene. 2019;38:103–119. doi: 10.1038/s41388-018-0423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.