Abstract
In this review we bring together evidence that (i) RNA viruses are a cause of chromosomal instability and micronuclei (MN), (ii) those individuals with high levels of lymphocyte MN have a weakened immune response and are more susceptible to RNA virus infection and (iii) both RNA virus infection and MN formation can induce inflammatory cytokine production. Based on these observations we propose a hypothesis that those who harbor elevated frequencies of MN within their cells are more prone to RNA virus infection and are more likely, through combined effects of leakage of self-DNA from MN and RNA from viruses, to escalate pro-inflammatory cytokine production via the cyclic GMP–AMP synthase (cGAS), stimulator of interferon genes (STING) and the Senescence Associated Secretory Phenotype (SASP) mechanisms to an extent that is unresolvable and therefore confers high risk of causing tissue damage by an excessive and overtly toxic immune response. The corollaries from this hypothesis are (i) those with abnormally high MN frequency are more prone to infection by RNA viruses; (ii) the extent of cytokine production and pro-inflammatory response to infection by RNA viruses is enhanced and possibly exceeds threshold levels that may be unresolvable in those with elevated MN levels in affected organs; (iii) reduction of MN frequency by improving nutrition and life-style factors increases resistance to RNA virus infection and moderates inflammatory cytokine production to a level that is immunologically efficacious and survivable.
Abbreviations: ACE2, angiotensin-converting enzyme 2; ARDS, acute respiratory distress syndrome; cGAS, cyclic GMP–AMP synthase; DDR, DNA damage response; IRF3, interferon regulatory factor 3; MAVS, mitochondrial anti-viral signaling protein; MN, micronuclei or micronucleus; PRR, pattern recognition receptors; RIG-1, retinoic acid-inducible gene I; RONS, reactive oxygen and nitrogen species; SASP, senescence associated secretory phenotype; STING, cyclic GMP–AMP receptor stimulator of interferon genes; TMPRSS2, transmembrane protease serine 2
Keywords: Micronuclei, Aneuploidy, Viruses, COVID-19, cGAS-STING, Inflammation
1. Introduction
The SARS-CoV-2 virus that emerged recently belongs to the coronavirus family, a positive-sense single–stranded RNA virus [1]. COVID-19, caused by SARS-CoV-2, was first reported in China in December 2019. Angiotensin-converting enzyme 2 (ACE2) is the main receptor for SARS-CoV-2 and its entry into the cell is facilitated by the transmembrane protease serine 2 (TMPRSS2) [2,3]. Acute respiratory distress syndrome (ARDS) resulting from COVID-19 disease can rapidly lead to pro-inflammatory cytokine storms, multiple organ failure and death [4]. Age, male sex, inherited genetic background and co-morbidities (diabetes, cardio-vascular diseases, cancer, immune deficiencies) are important additional risk factors for COVID-19 and its severity [[5], [6], [7]].
Cytokine storms after viral infection result from the fight/competition between immune defense systems and viral strategies to escape the immune defense. Two complementary mechanisms are used by the immune system to fight against infections: (i) the innate immune system is the first line of defense, which on detection of invading pathogens activates the retinoic acid-inducible gene I (RIG-I) and mitochondrial antiviral signaling protein (MAVS), RIG-I-MAVS signaling axis and also the cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes (STING) cGAS-STING pathways which cause IRF3- and NFkB-mediated expression of interferons that, in turn, induce synthesis of pro-inflammatory cytokines and chemokines. The latter then attract inflammatory cells, such as neutrophils and macrophages that eliminate invading microbes, by phagocytosis and/or release of antimicrobial toxins. (ii) The second line of defense is the adaptive immune response system, which is mobilized to identify antigens specific to the pathogen, presenting them to specific lymphocyte subsets capable of making antibodies that can recognize these antigens and neutralize the pathogen. For more information on the details of the innate and adaptive immune response refer to recent expert reviews [8,9].
Viruses developed several strategies to escape or silence the immune system, leading to a chronic and rapidly growing infection, severe tissue damage and pro-inflammatory cytokine storms [4]. Moreover, it was suggested that free DNA in cytoplasm or blood from damaged cells may be a reason for a severe and escalating pro-inflammatory response to COVID-19 infection [10,11].
Genetic polymorphisms of the ACE2 genes crucial for SARS-CoV-2 entry into host cells [12,13], genes involved in immune related response [3,14] and ABO blood group genes [15] are considered potential modulators of the individual response to viral infection by COVID-19. Also, the greater COVID-19 risk for males relative to females, and the declining innate immunity and attenuated responsiveness of adaptive immunity cells with increasing age, may disrupt the delicate balance between innate and adaptive immunity and are considered important factors that play a role in severe COVID-19 infections [14,16,17].
The aim of our paper is to discuss the evidence and plausibility that RNA viruses, aging, gender, and environmental and/or lifestyle genotoxins may independently or interactively induce DNA damage and micronuclei (MN), and that entrapment of chromosomes in MN is an aggravating factor in a vicious pro-inflammatory cycle induced by RNA virus infection and self-DNA leaked from MN.
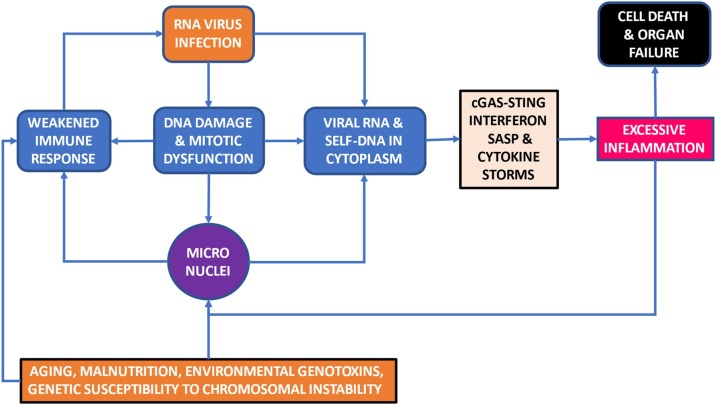
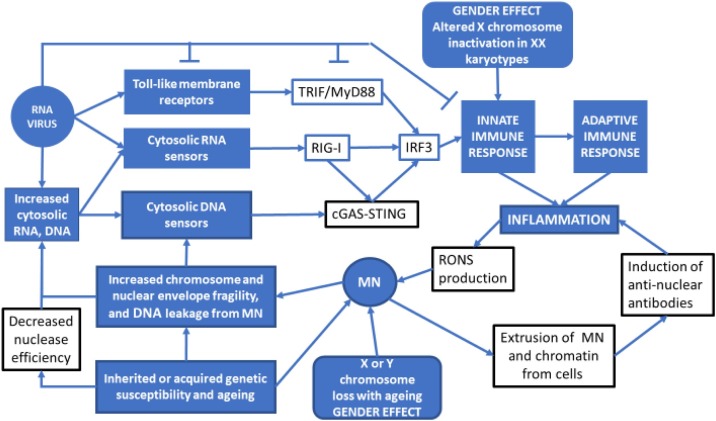
The basis of our central hypothesis is that DNA damage and mitotic errors, which lead to aneuploidy and MN formation, contribute significantly to the age and sex dependent aggravation of RNA virus disease (e.g. COVID-19) and induce cytokine storms. The overloading effect of MN may also be contributed by other factors including RNA virus-induced DNA damage and chromosome instability [18], malnutrition [19], environmental and lifestyle genotoxins [20] and genetic defects in DNA replication, DNA damage sensing and DNA repair [21]. Fig. 1 summarizes the central hypothesis and potential overloading effect of chromosome instability, in particular MN, on the interaction between RNA virus infection and the immune response. Fig. 2 is a schematic diagram explaining the origin of MN from chromosome fragments or whole chromosomes that are not segregated properly during mitosis due to various endogenous or exogenous genotoxic events or as a result of genetic defects affecting genome maintenance.
Fig. 1.
A visual summary of the central hypothesis that DNA damage and mitotic errors that lead to aneuploidy and MN formation contribute significantly to the age and sex dependent aggravation of RNA virus (e.g. COVID 19) infection and induced cytokine storms. The overloading effect of MN may also be contributed by other factors including RNA virus-induced DNA damage and chromosome instability, malnutrition, environmental and lifestyle genotoxins and genetic defects in DNA replication, DNA damage sensing and DNA repair.
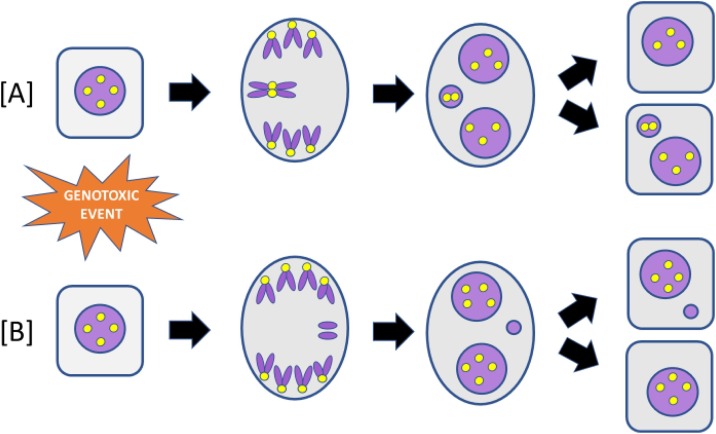
Fig. 2.
Diagram of MN formation caused by either a lagging [A] whole chromosome or [B] acentric chromosome fragment. The yellow dots represent centromeres. MN containing whole chromosomes can be identified by the presence of a centromere. MN containing acentric chromosome fragments do not contain a centromere.
In the following sections we will consider: (i) the cellular responses to viral infections, and to COVID-19 in particular, (ii) the vicious cycles of MN formation, RNA virus infection and inflammation, (iii) observations supporting our hypothesis/es and (iv) the implications for improvement of healthier immune responses to viral infections in aging populations.
2. The cellular responses to viral infections and COVID-19 in particular
2.1. Inflammation is governed by the balance between the host immune response and the viral strategies to escape them
The major steps from virus infection to inflammation are: attachment of the virus on the target cell, intracellularization of the viral nucleic acid (RNA or DNA), detection of the virus by membrane sensors and by cytosol RNA or DNA sensors, induction of the innate immune response through the RIG-I-MAVS signaling axis and/or the cGAS-STING DNA pathway, activation of IRF3- and NFkB-mediated expression of interferons, synthesis of inflammatory cytokines/chemokines and accumulation of immune cells at the infection site [4,11,22,23]. Cytopathologic viruses can induce mitotic disruption and a unique form of programmed cell death known as pyroptosis [22].
Two groups of pattern recognition receptors (PRR) detect and discriminate between molecules that are uniquely present in microbes. The first group consists of the membrane-bound Toll-like receptors (TLRs) that sense viral RNA or DNA in endosomes and phagosomes. [24]. The second group comprises sensors such as RIG-I and related helicases, melanoma differentiation association gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2), that sense RNA in cytosol [25,26], and the cytosolic DNA sensor, cGAS [24,27]. Inflammatory cytokines may also be induced directly by NFκB when it is triggered by immune receptors such as PRR [28]
After sensing and binding of viral DNA or RNA, the helicase RIG-1 releases caspase activation and recruitment domain (CARDS) which in association with MAVS undergoes cytosol-to-membrane relocalization at endoplasmatic derived membranes and aggregate into a “signalosome” [29]. The latter induces a signaling cascade leading to the expression of interferons, innate immune response genes, pro-inflammatory cytokines and chemokines that cooperate to limit virus infection [26].
cGAS plays a critical role in sensing/detecting self or foreign DNA in the cytoplasm. Upon binding DNA, cGAS catalyses the synthesis of cGAMP (cyclic GMP-AMP) which activates STING and then induces type 1 IFN production which leads to the secretion of inflammatory cytokines, such as tumor necrosis factor (TNF) and interleukin-6 (IL-6) [[27], [28], [29], [30]].
RNA viruses may also trigger STING signaling via RIG-I-dependent RNA sensing which induces STING expression, a process that is further facilitated by co-stimulation of TNF-α and type I interferons [31]. It is becoming increasingly evident that STING is required for host responses against both DNA and RNA viruses and for cross-talk between these mechanisms [32]. Furthermore, It is also plausible that RNA virus infection causes DNA damage in the host cells [33] resulting in leakage of self-DNA into the cytoplasm and activation of the cGAS-STING mechanism causing increased production of interferon (IFN) and inflammatory cytokines. In this regard it is relevant to note that bats, the only flying mammals, have an increased capacity to tolerate self DNA and viruses in the cytoplasm, including coronaviruses, by dampening STING activation via mutation of the functionally important S358 serine residue, which results in lower IFN-induced inflammation [34,35].
Viruses are able to evade cellular innate immune function by attenuating sensor responses [36]. The balance between the adequacy of immune defense mechanisms and the efficacy of the virus to escape them to ensure futher propagation ultimately affects the extent and duration of the inflammation. In the case of hyper-inflammation it might become locally destructive for the target tissue and, in worse cases, the damage may be systemic with lesions in several distal organs. In healthy young persons, the balance is in favor of the immune defense system and the inflammation resolving safely, leaving the host with “memory” cells that enable the host to react quickly in case of a new invasion.
An important consequence of RNA virus infection and COVID-19 disease is cell-free DNA (cfDNA) found in body fluids such as serum or plasma. cfDNA originates from nuclear or mitochondrial DNA released from dead/dying cells, DNA released from live cells, and foreign DNA from invading viruses or bacteria [37]. MN may also contribute to cfDNA when they are extruded from live cells as has been shown in live cell imaging studies in vitro but this has yet to be demonstrated in vivo [38,39]. Recent investigations showed that severity of COVID-19 disease and its progression is significantly associated with cfDNA in plasma, especially cfDNA identified as originating from lung, liver and erythroblasts identified using DNA methylation profiling [40,41]. Some of the cfDNA originates from neutrophil extracellular traps (NETs) that are released by neutrophils to destroy pathogens; it was reported that sera from patients with COVID-19 have elevated levels of myeloperoxidase-DNA complexes, and citrullinated histone H3 that are specific biomarkers of NETs [42]. Another important component of cfDNA is mitochondrial DNA (mtDNA). Recent studies suggest that SARS-CoV-2 infection may adversely alter mitochondrial function in host cells to favour viral replication, induce the release of pro-inflammatory mtDNA into the cytoplasm, which is ultimately ejected as cfDNA into body fluids following cell death [43,44]. Circulating mtDNA levels were shown to be highly elevated in patients who required intensive care or eventually died due to COVID-19 disease [45].
2.2. Genetics, sex and inflammation
Differences in immune response and inflammation among individuals may be due to genetic variation [46]. When considering the role of genetics of sex differences of immune responses, it is clear that genes encoded on the sex chromomes are important. The X-chromosome contains the largest number of immune-related genes, including genes that are involved in innate (e.g. Toll-like pattern recognition receptors TLR7 and TLR8 highly expressed in monocytes) and adaptive immune responses (e.g. chemokine receptor CXCR3 higly expressed on effector T-cells) [5]. Furthermore, the Toll-like receptors, encoded on the X-chromosome, may escape X inactivation resulting in higher expression levels than in males [5]. It is therefore plausible that the X chromosome plays an important role in the hyper-responsiveness of the female immune system, which may also explain the higher susceptibility for auto-immune diseases in women [47]. Why the specific balance in gene expression of X-chromosomes in females, including random inactivation and loss, increases risk for auto-immune diseases but is protective against several viral infections remains unclear. Sex also influences multiple aspects of adaptive response in humans, including lymphocyte subsets [48]. Furthermore, both innate and adaptive immunity decline with age but this trend is apparently more pronounced in males [17] who have a more rapid age-dependent increase in CD8 + T-cell senescence relative to females [49].
2.3. Genetics, sex and aging also influence the immune response and pathogenesis in COVID-19 patients
COVID-19 infection triggers release of inflammatory cytokines such as IL-6 but can also inhibit interferon production [50,51]. From early observations in different countries, it was clear that there is increased risk for death from COVID-19 for both sexes with advancing age, but at all stages above 30 years males have a significantly higher risk of death than females [5,[52], [53], [54]]. The sex dependent factors which may impact several steps of the immune response against COVID-19 infection and the pathogenesis of the disease are nicely analyzed in a recent review by Bunders and Altfeld [55]. The authors describe the sex differences at the different stages of the COVID-19 infection and host cellular responses: entry, immune responses, antibody responses, and T-cell responses. At all stages, women show stronger immune responses than men, which is evident in more robust vaccine response and increased susceptibility to auto-immune diseases [47]. Several immune response genes are encoded on the X-chromosome: such as ACE2, the principal receptor enabling SARS-COV-2 to enter human cells; TLR7, a RNA virus sensor; CD40 L and BTK, which regulate antibody responses; and IL-13RA1, the receptor for interleukin-13 [55,56]. Moreover, it is known that sex hormones can modulate the expression of some of the viral infection and immune response related genes. In particular for COVID-19, it was shown that estrogens downregulate the expression of ACE2 [57], and the TMPRSS2 promotion of virus entry into human host cells has been suggested to be enhanced by androgens [[58], [59], [60]].
As far as genetic polymorphisms involved in genetic susceptibility to COVID-19 disease are concerned, the first studies concentrated on the ACE2 human receptor for cell invasion, and TMPRSS2 for S protein priming [13]. A study on ACE2 receptor polymorphism in an Italian cohort (183 females and 395 males) found no significant evidence that ACE2 is associated with disease severity/sex bias [12]. However, in the same study, the expression of TMPRSS2 levels and its genetic variants proved to be possible candidate disease modulators. Subsequent GWAS studies also identified a 3p21.31 gene cluster as a genetic susceptibility locus in patients with COVID-19 with respiratory failure and potential involvement of the ABO blood-group system [15]. These pioneering studies are now complemented by many others which were recently evaluated in an instructive review by Anastassopoulou et al. [61]. They reviewed the associations between specific human genetic variants and clinical disease severity or susceptibility to infection by COVID-19 that have been reported in the literature until mid-September 2020. They identified several human genes as associated with COVID-19 severity: ABO, ACE2, ApoE, HLA, IFITM3, SLC6A20, LZTFL1, CCR9, FYCO1, CXRR6, XRC1, TLR7, TMEM189-UBE2V1, TMPRSS2. The chromosome locations of these genes are well known and two of them, ACE2 and TLR7, are X-chromosome linked.
3. Vicious cycles of RNA virus infection, micronucleus formation and inflammation
3.1. Micronuclei (MN) can diminish immune function and induce inflammation
The main mechanisms for MN formation (Fig. 2) are lack of a functional centromere in the chromosome fragments or whole chromosomes, and/or defects in one or more of the proteins of the mitotic apparatus resulting in chromosome segregation failure. The mal-segregated and isolated whole chromosomes or chromosome fragments are subsequently surrounded by a micronuclear membrane which excludes them from the main nucleus.
MN frequency in peripheral blood lymphocytes increases due to aging, micronutrient deficiency or genotoxic stress and is associated with a reduction in the proliferation rate of lymphocytes and a lower circulating lymphocyte count [[62], [63], [64], [65], [66], [67]]. Furthermore, chromosome fragility syndromes, such as Immunodeficiency, Centromeric instability, Facial anomalies (ICF) syndrome, ataxia-telangiectasia, and Fanconi's anemia exhibit increased chromosome aberrations and MN in lymphocytes, lymphopenia and reduced antibody production [[68], [69], [70], [71], [72], [73], [74]]. Moreover, lymphopenia is an important risk factor for increased infection and mortality [75,76]. Lymphocytes are the cells of the adaptive immune response that provides a targeted response to invading pathogens by antibody production from B lymphocytes and/or selective killing of virally infected cells by cytotoxic T cells; T cells also play an important role by producing cytokines that stimulate the proliferation of B cells and T cells [77]. Therefore, a potential consequence of increased MN in lymphocytes is a diminished number and function of B and T cells resulting in reduced capacity to sustain and increase the adaptive immune response to the level required to suppress and eliminate the viral infection.
Therefore, on the one hand increased MN frequency in immune system cells can result in an inefficacious immune response allowing RNA viruses to establish themselves and proliferate causing organ failure. On the other hand, the presence of disrupted MN and their DNA leakage into the cytoplasm of cells infected by RNA viruses may amplify the cytokine storm to a level that they induce an exaggerated response by the innate immune cells (e.g. macrophages, neutrophils and eosinophils) causing excessive collateral damage to the infected organs resulting in their malfunction. Therefore, the combined effect of elevated MN in cells in the immune system and MN in the organs affected by RNA viral infection (e.g. lung in the case of COVID-19) is predicted to both weaken immune response to RNA virus infections and at the same time aggravate the proinflammatory effects of their infection in the affected organs.
Moreover, there is growing evidence that MN frequency in lymphocytes correlates with MN frequency in other tissues in the body [78] suggesting that increased MN may be induced systemically throughout the body. Furthermore, the induction of MN in cells of the immune system may cause the affected cells to become senescent and excrete pro-inflammatory chemokines [79,80] as they circulate throughout the body causing inflammation and DNA damage in cells of other tissues and organs. This possibility is supported by a recent study in mice showing that DNA damage induced selectively in the immune system caused immunosenescence, which in turn induced accelerated aging and senescence of solid organs throughout the body [81].
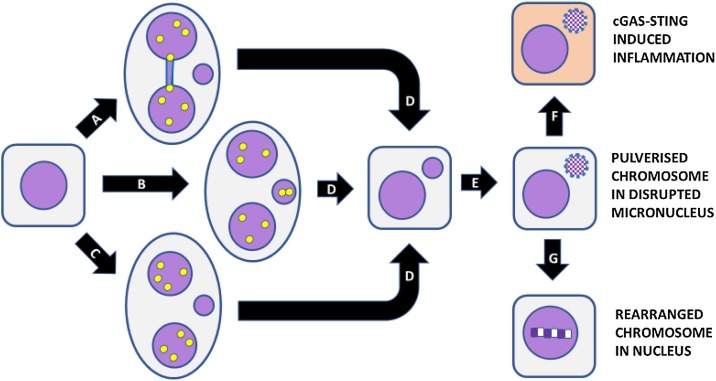
MN often have anomalies in their membrane that adversely affect metabolite transport and their metabolic capacity regarding DNA replication, transcription and repair is consequently impaired [[82], [83], [84]]. These defects may lead to massive amounts of DNA damage. Terradas et al. [85] reported that MN had defective DNA damage response (DDR) signaling, triggering genomic rearrangements. Furthermore, entrapment of chromosomes in MN can also lead to their fragmentation (Fig. 3 ). Fragmentation (or pulverization) of chromosomes trapped in MN may be due to inefficient DNA replication or repair caused by defective importation of the required enzymes into the MN. Impaired transport of enzymes and cofactors into MN could be caused by defective nuclear envelope which may also allow access of cytoplasmic DNAases [86]. The pulverization of chromosomes trapped in MN could also be due to their premature condensation before DNA synthesis is completed and the newly synthesized patches of DNA are ligated [87]. This catastrophic event, known as chromothripsis [88], has the added consequence that the fragments from the pulverized chromosome may be integrated within a nucleus and randomly ligated to form a hypermutated chromosome, a process known as chromoanasynthesis [89,90].
Fig. 3.
Consequences of MN formation. Micronuclei are formed at the binucleated cell stage as a result of (A) mis-repair of DNA breaks leading to nucleoplasmic bridge and acentric chromosome fragment, (B) mal-segregation of a whole chromosome, (C) lagging acentric chromosome fragment as a result of unrepaired DNA breaks. (D) Mononuclear cell with a micronucleus after completion of cytokinesis. (E) Shattering of chromosome trapped in micronucleus and disruption of nuclear envelope of micronucleus. (F) leakage of DNA from micronucleus leading to activation of cGAS-STING and induction of inflammatory cytokines. (G) Integration of pulverised chromosome within main nucleus and error-prone repair by non-homologous end joining resulting in a greatly rearranged mutant chromosome. Figure was reproduced from Genes (Basel). 2020;11(10):1203. doi: 10.3390/genes11101203, which was published by one of the authors (M. Fenech) who retains copyright under an open access Creative Commons CC BY 4.0 license with permission from MDPI.
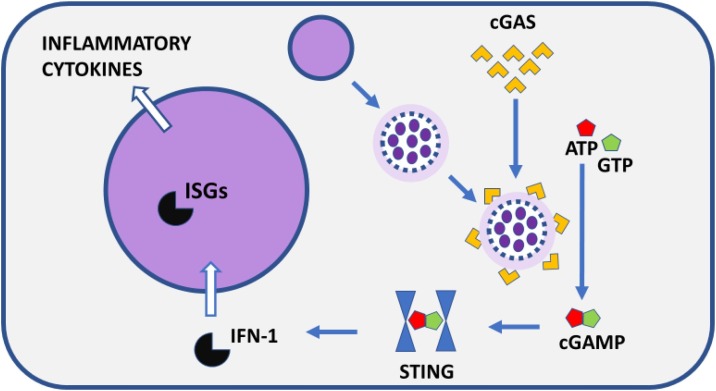
Another important consequence of pulverization of chromosomes in MN, especially in MN with defective membranes due to lack of proteins Rb and Lamin B1 [82], is leakage of DNA from the MN into the cytoplasm. The self-DNA is sensed by cGAS in the cytoplasm, cGAS is then activated to synthesize cGAMP which triggers STING and causes IRF3- and NFκB-mediated expression of type-1 interferons and proinflammatory cytokines [[91], [92], [93], [94], [95], [96]] (Fig. 4 ).
Fig. 4.
A diagrammatic outline of the process by which chromosome fragmentation in MN and disruption of the MN membrane leads to unravelling and leakage of DNA that is sensed by cGAS and triggers the cGAS-STING pathway of interferon-mediated innate immunity and inflammation. cGAS, Cyclic GMP-AMP Synthase; cGAMP, Cyclic guanosine monophosphate–adenosine monophosphate; STING, Stimulator of interferon genes; IFN-1, Type 1 interferon; ISGs, interferon stimulated genes. Figure was reproduced from Mutat Res. 2020;786:108342. doi: 10.1016/j.mrrev.2020.108342, with permission from Elsevier.
3.2. MN induction by RNA viruses
RNA viruses with single-stranded RNA as their genetic material use RNA polymerases of infected cells to replicate their genome; retroviruses, which have double-stranded RNA as their genetic code, use reverse transcriptase to make DNA copies of their genome and to integrate it into the host’s genome [97].
Furthermore, some RNA viruses such as HPV-E7 and hepatitis C virus (HCV) code for proteins that disable the function of proteins that control or are integral to the mitotic machinery (e.g. CDK2, tubulin, SAC, AURKB) of the host cell resulting in chromosome mal-segregation, aneuploidy and MN formation [98]. Furthermore, in 2006, it was shown that mammalian cells infected with avian RNA coronavirus infectious bronchitis virus (IBV) are inhibited from performing cytokinesis, which consequently facilitates IBV reproduction [99]. Cytokinesis inhibition leading to multinucleate polyploid cell formation is commonly observed in cancers and could be another mechanism by which RNA viruses could cause aneuploidy and MN formation. It was later shown that IBV also induces DNA strand breaks (measured using γH2AX assay) by causing DNA replication stress [100]. It is becoming increasingly apparent that RNA viruses may cause DNA damage or mitotic stress to activate DDR which may enhance viral reproduction [101].
The Zika virus, a single stranded RNA virus, has been shown to induce teratogenic effects in humans and it was hypothesized that this is possibly due to induction of mitotic errors leading to numerical chromosome aberrations [102]. Depending on the strain of the virus it was shown that Zika could also increase nuclear γH2AX and apoptosis or promote the p53-p21 signaling axis, driving cells toward cell cycle arrest [103,104]. Further in vitro molecular cytogenetic studies revealed that the Zika virus caused an increase in mitotic abnormalities, including multipolar spindle, lagging chromosomes, MN, aneuploidy and polyploidy [105]. Furthermore, similarly to the corona virus IBV, Zika induces cell cycle arrest via DDR to facilitate its replication [106].
Moreover, increased generation of reactive oxygen and nitrogen species (RONS) is a common feature of RNA virus infection [101]. This is because neutrophils and macrophages, recruited to the site of infection, release RONS to destroy the viruses and infected host cells and damage bystander normal cells. RONS generated by activated neutrophils and macrophages induce high levels of DNA damage, including MN, in healthy tissues [[107], [108], [109], [110]]. The RNA virus-induced MN in healthy tissues, in turn, may leak DNA in the cytoplasm and trigger the pro-inflammatory cGAS-STING that triggers INF-induced cytokines creating a vicious cycle of persistent RNA virus infection, MN formation and escalating cytokine storms that may exceed a threshold beyond which resolution becomes unachievable, resulting in tissue/organ damage and possibly organ failure as is the case with ARDS in severe COVID-19 cases.
3.3. Air pollution and malnutrition as risk factors for MN and COVID
It is well-recognized that ARDS is the major symptom of COVID-19 infection that may cause mortality [111]. Respiratory distress can also be caused by smoking and air pollution, both of which are associated with increased DNA damage in lung cells and/or peripheral blood leukocytes [[112], [113], [114]]. Evidence is emerging that exposure to air pollution increases the risk of COVID-19 infection, suggesting a 2-hit hypothesis that the combination of exposure to airborne genotoxins and COVID-19 virus further aggravates susceptibility to virus replication and lung inflammation [111,[115], [116], [117]]. The amplified inflammation is most likely triggered by (i) the persistent DNA damage response caused by chronic exposure to airborne genotoxicants leading to the activation of the Senescence Associated Secretory Phenotype (SASP) cytokines [[118], [119], [120], [121]] and/or (ii) the cGAS-STING inflammatory mechanism which senses self or foreign RNA and/or DNA in the cytoplasm originating either from viruses or from whole or shattered chromosomes that are trapped in disrupted MN following chromosome malsegregation due to mitotic catastrophe [91,122].
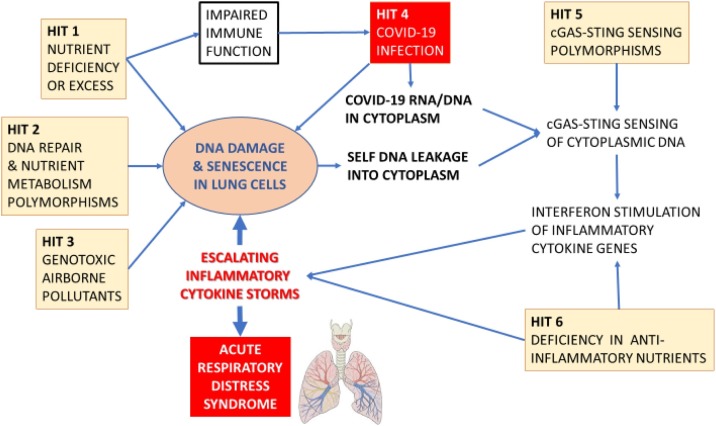
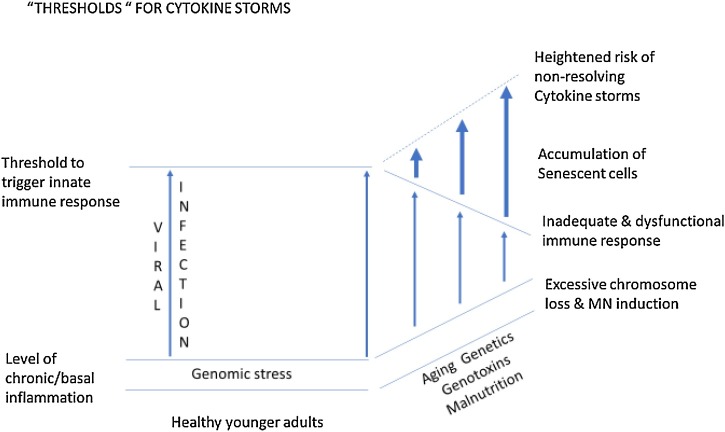
Apart from the double hit to genome integrity caused by RNA viruses such as COVID-19 and endogenous or exogenous genotoxins, there is a likely third deleterious hit to the genome caused by dietary deficiency or excesses. Evidence is mounting that obesity is a risk factor for COVID-19 disease risk and mortality and that micronutrient deficiencies that affect immune function such as deficiencies in vitamins A, B-6, B-9 i.e. folate, B-12, C, D, E, and inadequate intake of minerals such as zinc, selenium, iron and copper may also diminish resistance to viral infection [[123], [124], [125], [126], [127]]. Importantly, deficiencies in one or more of these micronutrients also increase DNA damage, MN and telomere attrition, with the latter being recently identified as a risk factor for COVID-19 [[128], [129], [130], [131], [132]]. Furthermore, deficiencies in micronutrients required for DNA replication and repair also increase sensitivity to endogenous or exogenous genotoxins [133,134]. Moreover, overweight and obesity has often been reported to be associated with increased DNA damage and elevated MN frequency [[135], [136], [137], [138], [139]]. Furthermore, a recent meta-analysis showed that patients with diabetes have significantly increased MN in lymphocytes and buccal cells, suggesting again the possibility that MN may contribute to or be a consequence of the susceptibility of diabetic cases to COVID-19 disease [140]. Moreover, those susceptible to RNA virus infections may already have underlying co-morbidities in several organs, such as the lung, heart, liver and kidney disease, all of which are associated with increased MN that may contribute to their pro-inflammatory status [[141], [142], [143], [144]]. Altogether it is plausible that the triple hit of COVID-19 infection, exposure to airborne genotoxicants and malnutrition may combine and interact to produce a perfect storm of inflammatory cytokines induced by the innate immune system’s response to foreign RNA from COVID-19 virus and self-DNA from damaged chromosomes in the cytoplasm of the target organ. Furthermore, it is plausible that susceptibility is further aggravated by inherited genetic defects in DNA repair and in nutrient absorption and metabolism. Fig. 5 summarises the common pathways by which RNA viruses, genomic instability, genetic factors, environmental stressors and the immune system may interact together to fuel escalating inflammatory cytokine storms. Fig. 6, Fig. 7 illustrate the multi-hit scenarios that our hypothesis proposes as the driving forces that accelerate the inflammatory processes beyond thresholds that become unresolvable and therefore detrimental to cellular and organ function.
Fig. 5.
A detailed map of the mechanistic interactions between RNA virus infection and self-DNA from disrupted micronuclei (MN), caused by ageing and genetic/gender effects, and their impacts on immune response and inflammation via the cGAS STING pathway.
Fig. 6.
The six acquired or inherited pathological hits that contribute to the vicious cycle of DNA damage and escalating inflammatory cytokine storms that ultimately lead to the ARDS in COVID-19.
Fig. 7.
A schematic diagram illustrating how increasing genotoxic stress caused by ageing, environmental, lifestyle and genetic factors interact with viral infection to go beyond the cellular sensing threshold that triggers a chronic non-resolving hyper-inflammatory response that may lead to severe tissue and organ damage.
4. Our hypothesis/es
Considering that the severity of COVID-19 disease is influenced by aging and sex, it is important to search for factors that may interact with the SARS-CoV-2 virus to aggravate or reduce the pathogenesis. It is well known that hormonal status is a major contributer to sex differences. However, more recently, based on the pro-inflammatory consequences of MN formation, it was shown that MN are co-players in inflammation and immune–related abnormalities, such as auto-immune diseases [47,[92], [93], [94], [95]]. The following paragraphs summarize the associations between MN frequencies, aging, sex, lifestyle and exposure to environmental gentoxins, as support for our hypothesis/es that cytokine storm severity is fueled by interaction of MN and RNA viruses.
4.1. Constitutive, spontaneous and induced aneuploidy, inflammation and aging
In humans, genomic imbalance can be constitutive, such as trisomy 21, where all cells of the individual have the same abnormal genomic constitution, or the genomic aberration may be restricted to a small or large proportion of germ or somatic cells resulting from spontaneous or induced chromosome instability. The main mechanisms leading to aneuploidy are chromosome non-disjunction or chromosome gain or loss during meiotic or mitotic cell division [145,146].
The link between constitutive aneuploidy and inflammation is clear in Down syndrome caused by trisomy 21. People with Down syndrome have significantly elevated MN frequencies in both lymphocytes and buccal epithelial cells [[147], [148], [149], [150]], and show signs of chronic immune dysregulation, including a higher prevalence of auto-immune disorders, increased rates of hospitalization during respiratory viral infections and higher mortality rates from pneumonia and sepsis [151]. Several genes involved in immune control are encoded on chromosome 21, including 4 of the 6 interferons receptor units, and trisomy 21 was shown to dysregulate T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity [152]. Espinosa suggested that the immune system dysregulation caused by trisomy 21 may make Down syndrome cases more prone to an excessive pro-inflammatory response to RNA virus infections, therefore, causing them to be more susceptible to severe COVID-19 disease [153]. In fact a recent study, in a cohort of 8 million adults, reported a 4-fold and 10-fold higher risk for COVID-19 hospitalisation and death, respectively, in Down syndrome cases [154].
Spontaneous, age-dependent or chemically–induced aneuploidy can trigger inflammation depending on the mechanism reponsible of aneuploidisation. If the causal aneugenic event is chromosome non-disjunction giving rise to one monosomic and one trisomic daughter cell, the potential to induce inflammaton will depend on the genes present on the lost or gained chromosome(s) and on the cell type, in particular if immune cells are concerned; and if the aneugenic event leads to chromosome(s) lagging during mitosis and formation of a MN in the next interphase, it will also affect the fate of the chromosomes trapped in the MN [155]. Since DNA and chromatin may leak from disrupted MN, whole chromosomes trapped in MN can also directly trigger the innate immune cGAS-STING pathway and become pro-inflammatory [[91], [92], [93], [94], [95], [96]].
Increase of mitotic misregulation with aging was described by Ly et al. [156]. They found that, in contrast to fibroblasts from normal young or normal middle aged individuals, the normal old and Hutchinson-Gilford progeria groups had a greater proportion of cells with abnormally shaped nuclei, tetraploidy, and multiple nuclei which are events consistent with age-related increase in MN formation and down-regulation of genes involved in cell cycle G2/M transition, spindle assembly and chromosome segregation. More recently, confirmation of these findings was described by Macedo et al. [157] and Barroso-Vilares et al. [158]. Moreover, they observed that senescent fibroblasts isolated from elderly donor’s cultures are often aneuploid and exhibit increased secretion of pro-inflammatory cytokines [157]. In their second paper [158], they reported the increased presence of cGAS positive MN in cultures from the elderly and that these MN were often Rb-negative, suggesting the MN membrane was dysfunctional, which may facilitate cGAS sensing of self-DNA within MN.
4.2. MN/chromosome loss induced inflammation is age- and sex-dependent
Several studies have consistently reported that frequency of MN in lymphocytes in humans increases with age, and also with sex, being 1.2−1.6-fold higher in adult females relative to adult males [[159], [160], [161], [162], [163]]. A multitude of factors contribute to the increase in MN frequency in humans including malnutrition and exposure to occupational and environmental genotoxins [128,137,164]. Many studies reported a significant increase in MN frequencies in people diagnosed with degenerative diseases relative to healthy controls matched for age and sex [[165], [166], [167]]. The most robust evidence that MN are causal for a wide range of diseases comes from prospective cohort studies showing that above average MN frequency in lymphocytes predicts higher risk of (i) pre-eclampsia and intrauterine growth restriction in pregnancy [168], (ii) cardiovascular disease [169,170] and (iii) cancer risk [171,172] all of which are also linked with increased DNA damage and inflammation.
4.2.1. X-chromosome loss and/or X-chromosome entrapment in MN increases with aging in women and induces inflammation
Mal-segregation of the X chromosome during mitosis is a relatively common event in females that leads to its entrapment in MN and consequently to its loss from one of the daughter nuclei. Current evidence suggests that it is the inactivated X chromosome that tends to be malsegregated because of its high level of heterochromatization induced by the XIST RNA transcript [173]. Using FISH techniques, Bukvic et al. [174] and Russel et al. [175] observed that X chromosome loss in lymphocytes increases rapidly with age in women. Two other studies reported on the entrapment of mal-segregated X chromosomes into MN in women. Hando et al. [176] reported that up to 72 % of MN in peripheral blood lymphocytes of females contain an X chromosome. In a subsequent investigation the same group showed that 83 % of X chromosomes trapped in an MN were inactivated [177].
It is important to consider whether X-chromosome loss from the main nucleus and/or its entrapment in a MN is an initiating cause of inflammation in women because auto-immune diseases and other inflammation related disorders are elevated in females and increase with age [178,179]. Supporting this hypothesis is the increased susceptibility of Turner syndrome (X monosomy) cases to develop autoimmune disorders and the possibility that expression of immune system related genes on the solitary X chromosome may be abnormal due to lack of its counterpart [[180], [181], [182], [183]]. Furthermore, entrapment of the X chromosome in an MN may lead to (i) its shattering and massive rearrangement transforming it into a hypermutated abnormal X chromosome if re-integrated into the main nucleus or (ii) leakage of X-chromosome DNA into the cytoplasm and its sensing by cGAS if the micronuclear membrane is disrupted due to lack of Rb and lamin proteins.
4.2.2. Y-chromosome loss and/or its entrapment in MN increases with aging in men and induces inflammation
Loss of Y chromosome (LOY) in men has been reported in hematopoietic tissues and peripheral blood lymphocytes [184]. Similar to X chromosome loss in females, LOY in men was shown to be caused by its mal-segregation during mitosis leading to its entrapment in a MN and as a consequence its absence from one of the main nuclei [185,186]. In both of these molecular cytogenetic studies the frequency of Y-chromosome positive MN increased with age. A high-throughput technique to measure aneuploidy or loss of specific chromosomes is the SNP array method [186]. Using this method, it was possible to measure LOY in blood samples from thousands of men from three cohorts and show that LOY increases with age and smoking [[186], [187], [188]]. Furthermore, LOY is associated with increased risk of all-cause mortality, cancer, Alzheimer’s disease, and autoimmune diseases [[189], [190], [191]].
The mechanism linking LOY and immune dysfunction is not yet clear. Similar to the situation with loss of X chromosome, LOY and entrapment of the Y chromosome in a MN may lead to its hypermutation or activation of the cGAS-STING pro-inflammatory pathway. Using selective inactivation of the Y chromosome centromere, Ly et al. [192] showed that micronucleation of the Y chromosome does indeed lead to its shattering and rearrangement. LOY effectively generates X monosomy (Turner syndrome) genotype in males and, therefore, it seems plausible that similar immune dysfunction consequences might ensue that are akin to those in Turner syndrome females, if sufficient cells in a relevant organ are affected. In this regard it is intriguing that LOY in males and loss of X in females are both associated with increased risk of auto-immune thyroiditis [193].
4.3. Explaining apparent paradoxes
4.3.1. Higher frequencies of MN in females than males but lower risk of severe COVID-19 outcome in females than males
The fact that females have a higher MN frequency than males [194] does not appear to fit the proposed hypothesis that MN and COVID-19 interact to determine severity of inflammatory cytokine response because epidemiological data suggest women have less severe COVID-19 infection outcomes than men [195]. However, some possible explanations for this paradox are explained below:
-
1
Relative to females, men are more likely to have poor health behaviours such as smoking and alcohol consumption and higher number of co-morbidities associated with poor COVID-19 prognosis, including hypertension, cardiovascular disease and chronic obstructive pulmonary disease [195].
-
2
The excess MN in females relative to males can be explained by increased mitotic mal-segregation of the X chromosome. In fact, 72 % of MN in cytokinesis-blocked lymphocytes in females were reported to contain an X chromosome and 83 % of them contained the inactive X chromosome [176,177]. The inactive X chromosome is maintained in a condensed state in interphase in a nuclear bud to form a Barr body [196]. It is plausible that DNA of an inactive X chromosome in a MN may not be sensed by cGAS because of its unique condensed heterochromatic state, analogous to condensed chromosomes in metaphase that are not sensed by cGAS because of its phosphorylation and tethering to chromatin, which inhibits cGAS activation by chromatin-bound DNA [196,197]. If these mechanisms are also applicable to the inactive X chromosome, this means that the great majority of MN in females, i.e. those which contain the inactive X (iX) chromosome, may not be pro-inflammatory. Based on the figures above, we estimate that only 40 % of all MN in females do not contain the iX chromosome. Therefore, it is conceivable that females may have less MN that are potentially pro-inflammatory than males (see Table 1 . below).
Table 1.
Example of frequency of MN in males and females showing the estimated frequency of MN not containing the inactive X chromosome in two hypothetical cases.
| MALES | FEMALES |
||||
|---|---|---|---|---|---|
| ALL MN FR | ALL MN FR | X MN FR* | iX MN FR** | ALL MN FR minus iX MN FR | % MN not containing iX |
| 10 | 12 | 8.6 | 7.2 | 4.8 | 40 % |
| 10 | 16 | 11.5 | 9.6 | 6.4 | 40 % |
FR = frequency per 1000 BN cells. Note the “ALL MN FR” values shown reflect male:female ratios of either 1.0:1.2 or 1.0:1.6 as has been reported in several studies previously. *Assumes 72 % of baseline female MN are X chromosome; **assumes 83 % of female MN with an X chromosome contain an inactive X (iX) chromosome.
4.3.2. How can MN, which are rare events, contribute substantially to the inflammation process?
The mean frequency of lymphocytes containing MN in healthy adults ranges between a minimum of 2 per 1000 (binucleated) cells to 79 per 1000 (binucleated) cells [198] which may, arguably, be considered not large enough to contribute substantially to the inflammation process. However, cells with MN may just be the tip of the iceberg, in terms of representing several other genomic instability events that are mechanistically linked with MN formation, such as unrepaired telomere DNA breaks, telomere oxidation, telomere shortening, nucleoplasmic (chromatin bridges) and aneuploidy, that also induce senescence and the SASP [[199], [200], [201], [202], [203], [204]]. Furthermore, these additional pro-inflammatory DNA damage events may be present both in cells with or without a MN and their occurrence multiplied in subsequent mitotic divisions of their chromosomally unstable progeny.
Moreover, there is growing evidence that MN frequency in lymphocytes correlates with MN frequency in other tissues in the body [205], suggesting that increased MN may be induced systemically throughout the body. Furthermore, the induction of MN in cells of the immune system may cause the affected cells to become senescent and excrete pro-inflammatory chemokines as they circulate throughout the body causing a cascade of inflammation, DNA damage and senescence in cells of other tissues and organs via paracrine mechanisms [[206], [207], [208], [209], [210]]. This possibility is supported by a recent study in mice showing that DNA damage induced selectively in the immune system caused immune-senescence, which in turn induced accelerated aging and senescence of solid organs throughout the body [211].
5. Implications from our hypothesis/es
5.1. MN as biomarkers for increased risk of cytokine storms
The primary organ affected by the COVID-19 virus is the respiratory tract. Several studies have shown that MN are expressed in multiple sites in the respiratory tract including buccal mucosa, nasal epithelium and bronchial epithelial cells [[212], [213], [214]]. In humans, it is practical to collect buccal and nasal cells, which are also the cells most likely to come in contact with and be infected by the COVID-19 virus [215]. MN in nasal and buccal cells have been shown to be increased by exposure to airborne genotoxins, malnutrition, infectious diseases and, also, as a result of aging and genetic defects that predispose to chromosomal instability [[216], [217], [218], [219], [220], [221], [222]]. Furthermore, cells harboring MN have previously been shown to have increased expression of cGAS and STING in the cytoplasm making it plausible that MN, inflammatory cytokine production and COVID-19 virus progeny may co-exist within nasal and buccal cells [91,92]. If our hypothesis is correct, we would expect that the presence of COVID-19 virus and cytokine storms should be increased in those subjects with increased MN. The MN may either be pre-existing or elevated because of increased endogenous genotoxic RONS induced by the escalating cytokine storm inflammation caused by self-DNA and viral RNA in the cytoplasm. Measurement of MN and Nuclear Division Index (a biomarker of mitogen responsiveness) in lymphocytes using the cytokinesis-block micronucleus cytome assay [223] may also be informative because these biomarkers are associated with telomere loss (i.e. telomeres lost in MN originating from deleted terminal chromosome fragments or whole chromosome loss), lymphopenia and depressed immune response [[224], [225], [226]], all of which are risk factors for COVID-19.
5.2. Prevention of MN and cytokine storms
The causes of MN formation are well known and some of which may be preventable, such as malnutrition and exposure to environmental genotoxins. However, others, such as genetic predisposition to impaired uptake and metabolism of micronutrients required for genome integrity maintenance, particularly in those cases with rarer mutations that cause complete loss of function, may be insurmountable. Moderate genetic susceptibilities, however, may be rendered less hazardous if genotoxin exposure is minimized and nutrient deficiencies or excesses are curtailed by appropriate personalized dietary and life-style strategies [227,228]. Lowering the rate of DNA damage by these means could reduce the degree of inflammation triggered by the SASP and cGAS-STING mechanisms that sense the DNA damage insults incurred by cells in the body. In fact, several human nutritional intervention studies have demonstrated the feasibility of reducing the frequency of MN [229] which is one of the best validated DNA damage biomarkers known to induce the cGAS-STING pro-inflammatory pathway. Furthermore, improved genome integrity maintenance and metabolic function, facilitated by optimal nutrition, should also improve immune function to either (i) enable the efficient elimination of cells that have been rendered senescent due to their excessive DNA damage and/or viral RNA burden and/or (ii) dampen down the inflammatory response by pro-resolving mechanisms. Recent studies suggest that (a) certain phytonutrients, such as apigenin and methyl caffeate, can dampen down the extent of pro-inflammatory SASP cytokines induced by cGAS-STING response to self or foreign RNA/DNA [230,231], (b) neutrophil mediated cytotoxicity is moderated by increased synthesis of resolvins from ω-3 fatty acid precursors [232] and (c) senolytic phytonutrients, such as quercetin, can selectively induce death of senescent cells [233]. Therefore, it is conceivable that appropriate mixtures of anti-inflammatory and senolytic phytonutrients may also help prevent the deadly effects of acute of respiratory distress syndrome caused by cytokine storms.
6. Conclusions
Our central hypothesis is that chromosomal instability, which leads to aneuploidy and entrapment of chromosomes in MN, contributes significantly to the age and sex dependent aggravation of COVID-19 induced infection and cytokine storms. The interconnectedness of the key elements relating to this hypothesis indicates the complexity of the mechanisms involved that may tip the balance towards a vicious cycle of inflammation or towards resolution (Fig. 5, Fig. 6).
In the previous sections we discussed the key mechanisms and events underlying the proposed hypothesis: i) MN can induce inflammation, ii) constitutive, spontaneous and induced aneuploidy triggers inflammation and is age-dependent, iii) MN/chromosome loss induced inflammation is age- and sex-dependent, iv) MN caused by X-chromosome loss increases with aging in women and induces inflammation, v) MN caused by Y-chromosome loss increases with aging in men and induces inflammation. All these events have a common link: the triggering of pro-inflammatory pathways (cGAS-STING, SASP) by cytosolic DNA from disrupted MN in the target cells.
In the case of viral infection, having a large “spectrum “of DNA and/or RNA sensors (e.g. TLR3,TLR7,TLR9,hnRNP-A2B1,cGAS,AIM2,RIG-1) provides an important advantage to counteract the many strategies developed by viruses to escape immune response [234,235]. Furthermore, Emming and Schroder [234] hypothesized that having multiple DNA or RNA sensors also provides an organism with a more sensitive system to assess the level of pathogenic hazard and properly calibrate the intensity of the immune response in a manner that is efficacious in killing the pathogens and at the same time minimises collateral damage to normal tissue. If we accept this hypothesis, it is probable that increased levels of aneuploidy and MN frequencies add an extra pathogenic load in cells that are already fighting against viral infection that is further aggravated by the DNA damage and mitotic disruption caused by RNA viruses. Whether these effects are additive or synergistic (non-linear cummulative) requires further experimental evidence.
In conclusion, RNA viruses generally, and specifically SARS-Cov-2 (COVID-19) disease, can generate cytokine storms that may be further accelerated by higher levels of disrupted MN in aged persons. If this hypothesis is confirmed, it may be beneficial to (i) conduct a MN assay to stratify patients for their risk of death by excessive cytokine storms and (ii) adjust their treatment to reduce their MN and inflammation load nutritionally and/or pharmacologically. Highly automated methods, such as image analysis, or imaging flow cytometry, are available to assess quickly and accurately MN frequencies in human lymphocytes [[236], [237], [238]]. If performed on T-lymphocytes, those frequencies will reflect chromosome loss accumulated during the last 6 months [239]. Genome sequencing might also be applied to identify relevant genetic polymorphisms (e.g. polymorphisms of cGAS-STING pathway) and specific chromosome imbalances, in particular loss of X and Y chromosomes. The combination of these tools in a systematic approach with other intersecting factors, such as inflammation biomarkers, sex hormone status, co-morbidities and previous exposure to environmental and/or genome stressors, might be critical to evaluate the biological factors contributing to heterogeneous COVID-19 outcomes, to assess the risk at the individual level, and to understand the genomic and cellular predictors of responsiveness to potential therapy.
Our central hypothesis can be verified by testing its predictions:
-
1
Those with high levels of lymphocyte MN have a weakened immune response and are more likely to be susceptible to RNA virus infection
-
2
The extent of inflammatory cytokine production in response to RNA virus infection is enhanced in those with higher MN frequency in the target tissue (e.g. respiratory tract and lungs in the case of COVID-19)
-
3
Reduction of MN frequency to its possible minimum (by improving nutrition, lifestyle factors and avoidance of environmental genotoxins) increases resistance to RNA virus infection and moderates inflammatory cytokine production to a level that is efficacious but not fatal.
Finally, it is important to note that the proposed hypotheses are also applicable to (i) DNA virus infections and (ii) the possibility of adverse inflammatory reactions resulting from the use of RNA vaccines, infectious DNA vaccines and attenuated adenovirus vaccines [[240], [241], [242]] but in the interest of brevity we have maintained the focus on RNA virus infections as this is more pertinent to the COVID-19 pandemic.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
None declared.
Acknowledgements
Not applicable.
References
- 1.Khan S., Siddique R., Shereen M.A., Ali A., Liu J., Bai Q., Bashir N., Xue M. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J. Clin. Microbiol. 2020;58(5 April):e00187–20. doi: 10.1128/JCM.00187-20. 23Erratum in: J Clin Microbiol. 2020 Jul 23;58(8): PMID: 32161092; PMCID: PMC7180238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunders M.J., Altfeld M. Implications of sex differences in immunity for SARS-CoV-2 pathogenesis and design of therapeutic interventions. Immunity. 2020;53(3 September):487–495. doi: 10.1016/j.immuni.2020.08.003. 15Epub 2020 Aug 17. PMID: 32853545; PMCID: PMC7430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choudhary S., Sreenivasulu K., Mitra P., Misra S., Sharma P. Role of genetic variants and gene expression in the susceptibility and severity of COVID-19. Ann. Lab. Med. 2021;41(2 March):129–138. doi: 10.3343/alm.2021.41.2.129. 1PMID: 33063674; PMCID: PMC7591285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the `Cytokine Storm’ in COVID-19. J. Infect. 2020;80(6 June):607–613. doi: 10.1016/j.jinf.2020.03.037. Epub 2020 Apr 10. PMID: 32283152; PMCID: PMC7194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20(7 July):442–447. doi: 10.1038/s41577-020-0348-8. Epub 2020 Jun 11. PMID: 32528136; PMCID: PMC7288618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautret P., Million M., Jarrot P.A., Camoin-Jau L., Colson P., Fenollar F., Leone M., La Scola B., Devaux C., Gaubert J.Y., Mege J.L., Vitte J., Melenotte C., Rolain J.M., Parola P., Lagier J.C., Brouqui P., Raoult D. Natural history of COVID-19 and therapeutic options. Expert Rev. Clin. Immunol. 2020;24(December):1–24. doi: 10.1080/1744666X.2021.1847640. Epub ahead of print. PMID: 33356661. [DOI] [PubMed] [Google Scholar]
- 7.Xie Y., Wang Z., Liao H., Marley G., Wu D., Tang W. Epidemiologic, clinical, and laboratory findings of the COVID-19 in the current pandemic: systematic review and meta-analysis. BMC Infect. Dis. 2020;20(1 August):640. doi: 10.1186/s12879-020-05371-2. 31PMID: 32867706; PMCID: PMC7457225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Netea M.G., Schlitzer A., Placek K., Joosten L.A.B., Schultze J.L. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell Host Microbe. 2019;25(1 January):13–26. doi: 10.1016/j.chom.2018.12.006. 9PMID: 30629914. [DOI] [PubMed] [Google Scholar]
- 9.Nikolich-Žugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat. Immunol. 2018;19(1 January):10–19. doi: 10.1038/s41590-017-0006-x. Epub 2017 Dec 14. Erratum in: Nat Immunol. 2018 Oct;19(10):1146. PMID: 29242543. [DOI] [PubMed] [Google Scholar]
- 10.Liu B. Free DNA, a reason for severe COVID-19 infection? Med. Hypotheses. 2020;142(September):109812. doi: 10.1016/j.mehy.2020.109812. Epub 2020 May 5. PMID: 32416412; PMCID: PMC7199685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthelot J.M., Lioté F. COVID-19 as a STING disorder with delayed over-secretion of interferon-beta. EBioMedicine. 2020;56(June):102801. doi: 10.1016/j.ebiom.2020.102801. Epub 2020 May 23. PMID: 32454408; PMCID: PMC7244443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging (Albany NY). 2020;12(11 June):10087–10098. doi: 10.18632/aging.103415. 5Epub 2020 Jun 5. PMID: 32501810; PMCID: PMC7346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53(3 June):425–435. doi: 10.1016/j.jmii.2020.04.015. Epub 2020 May 6. PMID: 32414646; PMCID: PMC7201239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16(10 October):626–638. doi: 10.1038/nri.2016.90. Epub 2016 Aug 22. PMID: 27546235. [DOI] [PubMed] [Google Scholar]
- 15.Severe Covid-19 GWAS Group, Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernández J., Prati D., Baselli G., Asselta R., Grimsrud M.M., Milani C., Aziz F., Kässens J., May S., Wendorff M., Wienbrandt L., Uellendahl-Werth F., Zheng T., Yi X., de Pablo R., Chercoles A.G., Palom A., Garcia-Fernandez A.E., Rodriguez-Frias F., Zanella A., Bandera A., Protti A., Aghemo A., Lleo A., Biondi A., Caballero-Garralda A., Gori A., Tanck A., Carreras Nolla A., Latiano A., Fracanzani A.L., Peschuck A., Julià A., Pesenti A., Voza A., Jiménez D., Mateos B., Nafria Jimenez B., Quereda C., Paccapelo C., Gassner C., Angelini C., Cea C., Solier A., Pestaña D., Muñiz-Diaz E., Sandoval E., Paraboschi E.M., Navas E., García Sánchez F., Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Blasi F., Téllez L., Blanco-Grau A., Hemmrich-Stanisak G., Grasselli G., Costantino G., Cardamone G., Foti G., Aneli S., Kurihara H., ElAbd H., My I., Galván-Femenia I., Martín J., Erdmann J., Ferrusquía-Acosta J., Garcia-Etxebarria K., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Terranova L., Moreira L., Santoro L., Scudeller L., Mesonero F., Roade L., Rühlemann M.C., Schaefer M., Carrabba M., Riveiro-Barciela M., Figuera Basso M.E., Valsecchi M.G., Wittig M., Ciccarelli M., Rodríguez-Gandía M., Bocciolone M., Miozzo M., Montano N., Braun N., Sacchi N., Martínez N., Özer O., Palmieri O., Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., Blandino Ortiz A., de Cid R., Ferrer R., Gualtierotti R., Nieto R., Goerg S., Badalamenti S., Marsal S., Matullo G., Pelusi S., Juzenas S., Aliberti S., Monzani V., Moreno V., Wesse T., Lenz T.L., Pumarola T., Rimoldi V., Bosari S., Albrecht W., Peter W., Romero-Gómez M., D’Amato M., Duga S., Banales J.M., Hov J.R., Folseraas T., Valenti L., Franke A., Karlsen T.H. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020;383(16 October):1522–1534. doi: 10.1056/NEJMoa2020283. 15Epub 2020 Jun 17. PMID: 32558485; PMCID: PMC7315890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karakas Celik S., Cakmak Genc G., Dursun A. A bioinformatic approach to investigating cytokine genes and their receptor variants in relation to COVID-19 progression. Int. J. Immunogenet. 2020;27(November) doi: 10.1111/iji.12522. doi: 10.1111/iji.12522. Epub ahead of print. PMID: 33246355; PMCID: PMC7753408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Márquez E.J., Trowbridge J., Kuchel G.A., Banchereau J., Ucar D. The lethal sex gap: COVID-19. Immun. Ageing. 2020;17(May):13. doi: 10.1186/s12979-020-00183-z. 21PMID: 32457811; PMCID: PMC7240166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryan E.L., Hollingworth R., Grand R.J. Activation of the DNA damage response by RNA viruses. Biomolecules. 2016;6(1 January):2. doi: 10.3390/biom6010002. 6PMID: 26751489; PMCID: PMC4808796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenech M.F. Nutriomes and personalised nutrition for DNA damage prevention, telomere integrity maintenance and cancer growth control. Cancer Treat. Res. 2014;159:427–441. doi: 10.1007/978-3-642-38007-5_24. PMID: 24114494. [DOI] [PubMed] [Google Scholar]
- 20.Wild C.P., Scalbert A., Herceg Z. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 2013;54(7 August):480–499. doi: 10.1002/em.21777. Epub 2013 May 16. PMID: 23681765. [DOI] [PubMed] [Google Scholar]
- 21.Luzhna L., Kathiria P., Kovalchuk O. Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front. Genet. 2013;4(July):131. doi: 10.3389/fgene.2013.00131. 11PMID: 23874352; PMCID: PMC3708156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6 June):363–374. doi: 10.1038/s41577-020-0311-8. Epub 2020 Apr 28. PMID: 32346093; PMCID: PMC7187672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berthelot J.M., Lioté F., Maugars Y., Sibilia J. Lymphocyte Changes in Severe COVID-19: Delayed Over-Activation of STING? Front. Immunol. 2020;11(December):607069. doi: 10.3389/fimmu.2020.607069. 1PMID: 33335532; PMCID: PMC7736628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallucci S., Maffei M.E. DNA sensing across the tree of life. Trends Immunol. 2017;38(10 October):719–732. doi: 10.1016/j.it.2017.07.012. Epub 2017 Sep 5. PMID: 28886908. [DOI] [PubMed] [Google Scholar]
- 25.Barral P.M., Sarkar D., Su Z.Z., Barber G.N., DeSalle R., Racaniello V.R., Fisher P.B. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 2009;124(2 November):219–234. doi: 10.1016/j.pharmthera.2009.06.012. Epub 2009 Jul 15. PMID: 19615405; PMCID: PMC3165056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow K.T., Gale M., Jr, Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36(April):667–694. doi: 10.1146/annurev-immunol-042617-053309. 26PMID: 29677479. [DOI] [PubMed] [Google Scholar]
- 27.Dhanwani R., Takahashi M., Sharma S. Cytosolic sensing of immuno-stimulatory DNA, the enemy within. Curr. Opin. Immunol. 2018;50(Febuary):82–87. doi: 10.1016/j.coi.2017.11.004. Epub 2017 Dec 13. PMID: 29247853; PMCID: PMC5916810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17(9 September):545–558. doi: 10.1038/nri.2017.52. Epub 2017 Jun 5. PMID: 28580957; PMCID: PMC5753586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esser-Nobis K., Hatfield L.D., Jr Gale M. Spatiotemporal dynamics of innate immune signaling via RIG-I-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2020;117(27 July):15778–15788. doi: 10.1073/pnas.1921861117. 7Epub 2020 Jun 22. PMID: 32571931; PMCID: PMC7354926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn J., Barber G.N. STING signaling and host defense against microbial infection. Exp. Mol. Med. 2019;51(12 December):1–10. doi: 10.1038/s12276-019-0333-0. 11PMID: 31827069; PMCID: PMC6906460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Lin R., Olagnier D. RIGulation of STING expression: at the crossroads of viral RNA and DNA sensing pathways. Inflamm. Cell Signal. 2017;4(1 January):e1491. doi: 10.14800/ics.1491. 1PMID: 28191486; PMCID: PMC5298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni G., Ma Z., Damania B. cGAS and STING: At the intersection of DNA and RNA virus-sensing networks. PLoS Pathog. 2018;14(8 August):e1007148. doi: 10.1371/journal.ppat.1007148. 16PMID: 30114241; PMCID: PMC6095619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan E.L., Hollingworth R., Grand R.J. Activation of the DNA damage response by RNA viruses. Biomolecules. 2016;6(1 January):2. doi: 10.3390/biom6010002. 6PMID: 26751489; PMCID: PMC4808796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie J., Li Y., Shen X., Goh G., Zhu Y., Cui J., Wang L.F., Shi Z.L., Zhou P. Dampened STING-Dependent interferon activation in bats. Cell Host Microbe. 2018;23(3 March):297–301. doi: 10.1016/j.chom.2018.01.006. e414Epub 2018 Feb 22. PMID: 29478775; PMCID: PMC7104992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang G., Cowled C., Shi Z., Huang Z., Bishop-Lilly K.A., Fang X., Wynne J.W., Xiong Z., Baker M.L., Zhao W., Tachedjian M., Zhu Y., Zhou P., Jiang X., Ng J., Yang L., Wu L., Xiao J., Feng Y., Chen Y., Sun X., Zhang Y., Marsh G.A., Crameri G., Broder C.C., Frey K.G., Wang L.F., Wang J. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118 January):456–460. doi: 10.1126/science.1230835. 25Epub 2012 Dec 20. PMID: 23258410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan Y.K., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14(6 June):360–373. doi: 10.1038/nrmicro.2016.45. Epub 2016 May 13. PMID: 27174148; PMCID: PMC5072394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabuschnig S., Bronkhorst A.J., Holdenrieder S., Rosales Rodriguez I., Schliep K.P., Schwendenwein D., Ungerer V., Sensen C.W. Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 2020;21(21 October):8062. doi: 10.3390/ijms21218062. 29PMID: 33137955; PMCID: PMC7662960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao X., Zhang Y., Yi Q., Hou H., Xu B., Chu L., Huang Y., Zhang W., Fenech M., Shi Q. Multiple origins of spontaneously arising micronuclei in HeLa cells: direct evidence from long-term live cell imaging. Mutat. Res. 2008;646(1-2 November):41–49. doi: 10.1016/j.mrfmmm.2008.09.004. 10Epub 2008 Sep 19. PMID: 18845170. [DOI] [PubMed] [Google Scholar]
- 39.Grabuschnig S., Bronkhorst A.J., Holdenrieder S., Rosales Rodriguez I., Schliep K.P., Schwendenwein D., Ungerer V., Sensen C.W. Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int. J. Mol. Sci. 2020;21(21 October):8062. doi: 10.3390/ijms21218062. 29PMID: 33137955; PMCID: PMC7662960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng A.P., Cheng M.P., Gu W., Sesing Lenz J., Hsu E., Schurr E., Bourque G., Bourgey M., Ritz J., Marty F.M., Chiu C.Y., Vinh D.C., De Vlaminck I. Cell-free DNA tissues of origin by methylation profiling reveals significant cell, tissue, and organ-specific injury related to COVID-19 severity. Med (N Y). 2021;2(4 April):411–422. doi: 10.1016/j.medj.2021.01.001. 9 e5 Epub 2021 Jan 16. PMID: 33521749; PMCID: PMC7836424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammad R., MAER Eldosoky, Fouad S.H., Elgendy A., Tawfeik A.M., Alboraie M., Abdelmaksoud M.F. Circulating cell-free DNA, peripheral lymphocyte subsets alterations and neutrophil lymphocyte ratio in assessment of COVID-19 severity. Innate Immun. 2021;27(3 April):240–250. doi: 10.1177/1753425921995577. Epub 2021 Mar 1. PMID: 33646058; PMCID: PMC8054149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., Woods R.J., Kanthi Y., Knight J.S. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5(11 June):e138999. doi: 10.1172/jci.insight.138999. 4PMID: 32329756; PMCID: PMC7308057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol., Cell Physiol. 2020;319(2 August):C258–C267. doi: 10.1152/ajpcell.00224.2020. 1Epub 2020 Jun 8. PMID: 32510973; PMCID: PMC7381712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kloc M., Ghobrial R.M., Kubiak J.Z. The role of genetic sex and mitochondria in response to COVID-19 infection. Int. Arch. Allergy Immunol. 2020;181(8):629–634. doi: 10.1159/000508560. Epub 2020 Jun 19. PMID: 32564017; PMCID: PMC7360490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scozzi D., Cano M., Ma L., Zhou D., Zhu J.H., O’Halloran J.A., Goss C., Rauseo A.M., Liu Z., Sahu S.K., Peritore V., Rocco M., Ricci A., Amodeo R., Aimati L., Ibrahim M., Hachem R., Kreisel D., Mudd P.A., Kulkarni H.S., Gelman A.E. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. 2021;6(4 Febuary):e143299. doi: 10.1172/jci.insight.143299. 22PMID: 33444289; PMCID: PMC7934921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol. Rev. 2020;296(1 July):205–219. doi: 10.1111/imr.12897. Epub 2020 Jul 13. PMID: 32658335; PMCID: PMC7404857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirsch-Volders M., Bolognesi C., Ceppi M., Bruzzone M., Fenech M. Micronuclei, inflammation and auto-immune disease. Mutat. Res. 2020;786(October-December):108335. doi: 10.1016/j.mrrev.2020.108335. Epub 2020 Sep 15. PMID: 33339583. [DOI] [PubMed] [Google Scholar]
- 48.Roved J., Westerdahl H., Hasselquist D. Sex differences in immune responses: hormonal effects, antagonistic selection, and evolutionary consequences. Horm. Behav. 2017;88(Febuary):95–105. doi: 10.1016/j.yhbeh.2016.11.017. Epub 2016 Dec 9. PMID: 27956226. [DOI] [PubMed] [Google Scholar]
- 49.Kverneland A.H., Streitz M., Geissler E., Hutchinson J., Vogt K., Boës D., Niemann N., Pedersen A.E., Schlickeiser S., Sawitzki B. Age and gender leucocytes variances and references values generated using the standardized ONE-Study protocol. Cytometry A. 2016;89(6 June):543–564. doi: 10.1002/cyto.a.22855. Epub 2016 May 3. PMID: 27144459. [DOI] [PubMed] [Google Scholar]
- 50.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2 March-April):327–331. doi: 10.23812/CONTI-E. PMID: 32171193. [DOI] [PubMed] [Google Scholar]
- 51.Birra D., Benucci M., Landolfi L., Merchionda A., Loi G., Amato P., Licata G., Quartuccio L., Triggiani M., Moscato P. COVID 19: a clue from innate immunity. Immunol. Res. 2020;68(3 June):161–168. doi: 10.1007/s12026-020-09137-5. PMID: 32524333; PMCID: PMC7286633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11(1 December):6317. doi: 10.1038/s41467-020-19741-6. 9PMID: 33298944; PMCID: PMC7726563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., Liu S., Yang J.K. Gender differences in patients with COVID-19: focus on severity and mortality. Front. Public Health. 2020;8(April):152. doi: 10.3389/fpubh.2020.00152. 29PMID: 32411652; PMCID: PMC7201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gebhard C., Regitz-Zagrosek V., Neuhauser H.K., Morgan R., Klein S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex Differ. 2020;11(1 May):29. doi: 10.1186/s13293-020-00304-9. 25PMID: 32450906; PMCID: PMC7247289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bunders M.J., Altfeld M. Implications of sex differences in immunity for SARS-CoV-2 pathogenesis and design of therapeutic interventions. Immunity. 2020;53(3 September):487–495. doi: 10.1016/j.immuni.2020.08.003. 15Epub 2020 Aug 17. PMID: 32853545; PMCID: PMC7430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsiambas E., Chrysovergis A., Papanikolaou V., Mastronikolis N., Ragos V., Kavantzas N., Lazaris A.C., Patsouris E., Riziotis C., Paschopoulos M., Kyrodimos E. Chromosome X riddle in SARS-CoV-2 (COVID-19) - related lung pathology. Pathol. Oncol. Res. 2020;26(4 October):2839–2841. doi: 10.1007/s12253-020-00878-0. Epub 2020 Jul 20. PMID: 32691391; PMCID: PMC7370877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J., Ji H., Zheng W., Wu X., Zhu J.J., Arnold A.P., Sandberg K. Sex differences in renal angiotensin converting enzyme 2 (ACE2) activity are 17β-oestradiol-dependent and sex chromosome-independent. Biol. Sex Differ. 2010;1(1 November):6. doi: 10.1186/2042-6410-1-6. 5PMID: 21208466; PMCID: PMC3010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020;10(6 June):779–782. doi: 10.1158/2159-8290.CD-20-0451. Epub 2020 Apr 10. PMID: 32276929; PMCID: PMC7437472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wambier C.G., Goren A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is likely to be androgen mediated. J. Am. Acad. Dermatol. 2020;83(1 July):308–309. doi: 10.1016/j.jaad.2020.04.032. Epub 2020 Apr 10. PMID: 32283245; PMCID: PMC7151476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wambier C.G., Goren A., Vaño-Galván S., Ramos P.M., Ossimetha A., Nau G., Herrera S., McCoy J. Androgen sensitivity gateway to COVID-19 disease severity. Drug Dev. Res. 2020;81(7 November):771–776. doi: 10.1002/ddr.21688. Epub 2020 May 15. PMID: 32412125; PMCID: PMC7273095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anastassopoulou C., Gkizarioti Z., Patrinos G.P., Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genomics. 2020;14(1 October):40. doi: 10.1186/s40246-020-00290-4. 22PMID: 33092637; PMCID: PMC7578581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Groarke E.M., Young N.S. Aging and hematopoiesis. Clin. Geriatr. Med. 2019;35(3 August):285–293. doi: 10.1016/j.cger.2019.03.001. Epub 2019 May 9. PMID: 31230730; PMCID: PMC8131033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fenech M. Important variables that influence base-line micronucleus frequency in cytokinesis-blocked lymphocytes-a biomarker for DNA damage in human populations. Mutat. Res. 1998;404(1-2 August):155–165. doi: 10.1016/s0027-5107(98)00109-2. 3PMID: 9729354. [DOI] [PubMed] [Google Scholar]
- 64.Fenech M., Baghurst P., Luderer W., Turner J., Record S., Ceppi M., Bonassi S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability--results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis. 2005;26(5 May):991–999. doi: 10.1093/carcin/bgi042. Epub 2005 Feb 10. PMID: 15705599. [DOI] [PubMed] [Google Scholar]
- 65.Catena C., Parasacchi P., Conti D., Sgura A., Trenta G., Righi E., Trinci M.M., Trinci M. Peripheral blood lymphocyte decrease and micronucleus yields during radiotherapy. Int. J. Radiat. Biol. 1997;72(5 November):575–585. doi: 10.1080/095530097143077. PMID: 9374437. [DOI] [PubMed] [Google Scholar]
- 66.Kimura M., Umegaki K., Higuchi M., Thomas P., Fenech M. Methylenetetrahydrofolate reductase C677T polymorphism, folic acid and riboflavin are important determinants of genome stability in cultured human lymphocytes. J. Nutr. 2004;134(1 January):48–56. doi: 10.1093/jn/134.1.48. PMID: 14704292. [DOI] [PubMed] [Google Scholar]
- 67.Courtemanche C., Elson-Schwab I., Mashiyama S.T., Kerry N., Ames B.N. Folate deficiency inhibits the proliferation of primary human CD8+ T lymphocytes in vitro. J. Immunol. 2004;173(5 September):3186–3192. doi: 10.4049/jimmunol.173.5.3186. 1PMID: 15322179. [DOI] [PubMed] [Google Scholar]
- 68.Gisselsson D., Shao C., Tuck-Muller C.M., Sogorovic S., Pålsson E., Smeets D., Ehrlich M. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma. 2005;114(2 July):118–126. doi: 10.1007/s00412-005-0343-7. Epub 2005 Apr 27. PMID: 15856360. [DOI] [PubMed] [Google Scholar]
- 69.Stacey M., Bennett M.S., Hulten M. FISH analysis on spontaneously arising micronuclei in the ICF syndrome. J. Med. Genet. 1995;32(7 July):502–508. doi: 10.1136/jmg.32.7.502. PMID: 7562960; PMCID: PMC1050540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weemaes C.M., The T.H., van Munster P.J., Bakkeren J.A. Antibody responses in vivo in chromosome instability syndromes with immunodeficiency. Clin. Exp. Immunol. 1984;57(3 Septenber):529–534. PMID: 6467678; PMCID: PMC1536265. [PMC free article] [PubMed] [Google Scholar]
- 71.Gennery A.R., Slatter M.A., Bredius R.G., Hagleitner M.M., Weemaes C., Cant A.J., Lankester A.C. Hematopoietic stem cell transplantation corrects the immunologic abnormalities associated with immunodeficiency-centromeric instability-facial dysmorphism syndrome. Pediatrics. 2007;120(5 November):e1341–4. doi: 10.1542/peds.2007-0640. Epub 2007 Oct 1. PMID: 17908720. [DOI] [PubMed] [Google Scholar]
- 72.Conley M.E., Spinner N.B., Emanuel B.S., Nowell P.C., Nichols W.W. A chromosomal breakage syndrome with profound immunodeficiency. Blood. 1986;67(5 May):1251–1256. PMID: 2421804. [PubMed] [Google Scholar]
- 73.Palazzo R.P., Jardim L.B., Bacellar A., de Oliveira F.R., Maraslis F.T., Pereira C.H.J., da Silva J., Maluf S.W. DNA damage and repair in individuals with ataxia-telangiectasia and their parents. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836(Pt B December):122–126. doi: 10.1016/j.mrgentox.2018.06.007. Epub 2018 Jun 1. PMID: 30442337. [DOI] [PubMed] [Google Scholar]
- 74.Maluf S.W., Erdtmann B. Genomic instability in Down syndrome and Fanconi anemia assessed by micronucleus analysis and single-cell gel electrophoresis. Cancer Genet. Cytogenet. 2001;124(1 January):71–75. doi: 10.1016/s0165-4608(00)00322-8. 1PMID: 11165325. [DOI] [PubMed] [Google Scholar]
- 75.Zidar D.A., Al-Kindi S.G., Liu Y., Krieger N.I., Perzynski A.T., Osnard M., Nmai C., Anthony D.D., Lederman M.M., Freeman M.L., Bonomo R.A., Simon D.I., Dalton J.E. Association of Lymphopenia With Risk of Mortality Among Adults in the US General Population. JAMA Netw Open. 2019;2(12 December):e1916526. doi: 10.1001/jamanetworkopen.2019.16526. 2PMID: 31790569; PMCID: PMC6902755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warny M., Helby J., Nordestgaard B.G., Birgens H., Bojesen S.E. Lymphopenia and risk of infection and infection-related death in 98,344 individuals from a prospective Danish population-based study. PLoS Med. 2018;15(11 November):e1002685. doi: 10.1371/journal.pmed.1002685. 1PMID: 30383787; PMCID: PMC6211632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 2004;4(1 January):11–22. doi: 10.1038/nrc1252. PMID: 14708024. [DOI] [PubMed] [Google Scholar]
- 78.Ceppi M., Biasotti B., Fenech M., Bonassi S. Human population studies with the exfoliated buccal micronucleus assay: statistical and epidemiological issues. Mutat. Res. 2010;705(1 July-September):11–19. doi: 10.1016/j.mrrev.2009.11.001. Epub 2009 Nov 20. PMID: 19932192. [DOI] [PubMed] [Google Scholar]
- 79.Callender L.A., Carroll E.C., Beal R.W.J., Chambers E.S., Nourshargh S., Akbar A.N., Henson S.M. Human CD8+ EMRA T cells display a senescence-associated secretory phenotype regulated by p38 MAPK. Aging Cell. 2018;17(1 Febuary):e12675. doi: 10.1111/acel.12675. Epub 2017 Oct 12. PMID: 29024417; PMCID: PMC5770853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frasca D., Diaz A., Romero M., Blomberg B.B. Human peripheral late/exhausted memory B cells express a senescent-associated secretory phenotype and preferentially utilize metabolic signaling pathways. Exp. Gerontol. 2017;87(Pt A January):113–120. doi: 10.1016/j.exger.2016.12.001. Epub 2016 Dec 6. PMID: 27931848. [DOI] [PubMed] [Google Scholar]
- 81.Yousefzadeh M.J., Flores R.R., Zhu Y., Schmiechen Z.C., Brooks R.W., Trussoni C.E., Cui Y., Angelini L., Lee K.A., McGowan S.J., Burrack A.L., Wang D., Dong Q., Lu A., Sano T., O’Kelly R.D., McGuckian C.A., Kato J.I., Bank M.P., Wade E.A., Pillai S.P.S., Klug J., Ladiges W.C., Burd C.E., Lewis S.E., LaRusso N.F., Vo N.V., Wang Y., Kelley E.E., Huard J., Stromnes I.M., Robbins P.D., Niedernhofer L.J. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594(7861 June):100–105. doi: 10.1038/s41586-021-03547-7. Epub 2021 May 12. PMID: 33981041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hatch E.M., Fischer A.H., Deerinck T.J., Hetzer M.W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154(1 July):47–60. doi: 10.1016/j.cell.2013.06.007. 3PMID: 23827674; PMCID: PMC3749778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terradas M., Martín M., Genescà A. Detection of impaired DNA replication and repair in Micronuclei as indicators of genomic instability and chromothripsis. Methods Mol. Biol. 2018;1769:197–208. doi: 10.1007/978-1-4939-7780-2_13. PMID: 29564826. [DOI] [PubMed] [Google Scholar]
- 84.Maciejowski J., Hatch E.M. Nuclear membrane rupture and its consequences. Annu. Rev. Cell Dev. Biol. 2020;36(October):85–114. doi: 10.1146/annurev-cellbio-020520-120627. 6Epub 2020 Jul 21. PMID: 32692592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terradas M., Martín M., Genescà A. Impaired nuclear functions in micronuclei results in genome instability and chromothripsis. Arch. Toxicol. 2016;90(11 November):2657–2667. doi: 10.1007/s00204-016-1818-4. Epub 2016 Aug 19. PMID: 27542123. [DOI] [PubMed] [Google Scholar]
- 86.Liu S., Kwon M., Mannino M., Yang N., Renda F., Khodjakov A., Pellman D. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 2018;561(7724):551–555. doi: 10.1038/s41586-018-0534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bell J.C., Straight A.F. Condensing chromosome condensation. Nat. Cell Biol. 2015;17(8):964–965. doi: 10.1038/ncb3212. [DOI] [PubMed] [Google Scholar]
- 88.Terzoudi G.I., Karakosta M., Pantelias A., Hatzi V.I., Karachristou I., Pantelias G. Stress induced by premature chromatin condensation triggers chromosome shattering and chromothripsis at DNA sites still replicating in micronuclei or multinucleate cells when primary nuclei enter mitosis. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;793:185–198. doi: 10.1016/j.mrgentox.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 89.Ly P., Teitz L.S., Kim D.H., Shoshani O., Skaletsky H., Fachinetti D., Page D.C., Cleveland D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017;19:68–75. doi: 10.1038/ncb3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ly P., Brunner S.F., Shoshani O., Kim D.H., Lan W., Pyntikova T., Flanagan A.M., Behjati S., Page D.C., Campbell P.J., Cleveland D.W. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet. 2019;51:705–715. doi: 10.1038/s41588-019-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Oliveira Mann C.C., Kranzusch P.J. cGAS conducts micronuclei DNA surveillance. Trends Cell Biol. 2017;27(10 October):697–698. doi: 10.1016/j.tcb.2017.08.007. Epub 2017 Sep 4. PMID: 28882413. [DOI] [PubMed] [Google Scholar]
- 92.Mackenzie K.J., Carroll P., Martin C.A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., Osborn R.T., Wheeler A.P., Nowotny M., Gilbert N., Chandra T., Reijns M.A.M., Jackson A.P. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548(7668 August):461–465. doi: 10.1038/nature23449. 24Epub 2017 Jul 24. PMID: 28738408; PMCID: PMC5870830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harding S.M., Benci J.L., Irianto J., Discher D.E., Minn A.J., Greenberg R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548(7668 August):466–470. doi: 10.1038/nature23470. 24Epub 2017 Jul 31. PMID: 28759889; PMCID: PMC5857357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Motwani M., Fitzgerald K.A. cGAS micro-manages genotoxic stress. Immunity. 2017;47(4 October):616–617. doi: 10.1016/j.immuni.2017.09.020. 17PMID: 29045895. [DOI] [PubMed] [Google Scholar]
- 95.Spektor A., Umbreit N.T., Pellman D. Cell biology: when your own chromosomes act like foreign DNA. Curr. Biol. 2017;27:R1228–R1231. doi: 10.1016/j.cub.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 96.Chunduri N.K., Storchová Z. The diverse consequences of aneuploidy. Nat. Cell Biol. 2019;21(1 January):54–62. doi: 10.1038/s41556-018-0243-8. Epub 2019 Jan 2. PMID: 30602769. [DOI] [PubMed] [Google Scholar]
- 97.Poltronieri P., Sun B., Mallardo M. RNA viruses: RNA roles in pathogenesis, coreplication and viral load. Curr. Genomics. 2015;16(5 October):327–335. doi: 10.2174/1389202916666150707160613. PMID: 27047253; PMCID: PMC4763971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taheri F., Goudarzi H., Faghihloo E. Aneuploidy and oncoviruses. Rev. Med. Virol. 2019;29(6 November):e2076. doi: 10.1002/rmv.2076. Epub 2019 Aug 12. PMID: 31407416. [DOI] [PubMed] [Google Scholar]
- 99.Dove B., Brooks G., Bicknell K., Wurm T., Hiscox J.A. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 2006;80(8 April):4147–4156. doi: 10.1128/JVI.80.8.4147-4156.2006. PMID: 16571830; PMCID: PMC1440480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu L.H., Huang M., Fang S.G., Liu D.X. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase δ. J. Biol. Chem. 2011;286(45 November):39546–39559. doi: 10.1074/jbc.M111.242206. 11Epub 2011 Sep 14. PMID: 21918226; PMCID: PMC3234778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ryan E.L., Hollingworth R., Grand R.J. Activation of the DNA damage response by RNA viruses. Biomolecules. 2016;6(1 January):2. doi: 10.3390/biom6010002. 6PMID: 26751489; PMCID: PMC4808796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bullerdiek J., Dotzauer A., Bauer I. The mitotic spindle: linking teratogenic effects of Zika virus with human genetics? Mol. Cytogenet. 2016;9(April):32. doi: 10.1186/s13039-016-0240-1. 19PMID: 27099632; PMCID: PMC4837584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghouzzi V.E., Bianchi F.T., Molineris I., Mounce B.C., Berto G.E., Rak M., Lebon S., Aubry L., Tocco C., Gai M., Chiotto A.M., Sgrò F., Pallavicini G., Simon-Loriere E., Passemard S., Vignuzzi M., Gressens P., Di Cunto F. ZIKA virus elicits P53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly. Cell Death Dis. 2016;7(10 October):e2440. doi: 10.1038/cddis.2016.266. 27Erratum in: Cell Death Dis. 2017 Jan 19;8(1):e2567. Erratum in: Cell Death Dis. 2018 Nov 20;9(12):1155. PMID: 27787521; PMCID: PMC5133962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Devhare P., Meyer K., Steele R., Ray R.B., Ray R. Zika virus infection dysregulates human neural stem cell growth and inhibits differentiation into neuroprogenitor cells. Cell Death Dis. 2017;8(10 October):e3106. doi: 10.1038/cddis.2017.517. 12PMID: 29022904; PMCID: PMC5682681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Souza B.S., Sampaio G.L., Pereira C.S., Campos G.S., Sardi S.I., Freitas L.A., Figueira C.P., Paredes B.D., Nonaka C.K., Azevedo C.M., Rocha V.P., Bandeira A.C., Mendez-Otero R., Dos Santos R.R., Soares M.B. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016;6(December):39775. doi: 10.1038/srep39775. 23PMID: 28008958; PMCID: PMC5180086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammack C., Ogden S.C., Madden J.C., Jr, Medina A., Xu C., Phillips E., Son Y., Cone A., Giovinazzi S., Didier R.A., Gilbert D.M., Song H., Ming G., Wen Z., Brinton M.A., Gunjan A., Tang H. Zika virus infection induces DNA damage response in human neural progenitors that enhances viral replication. J. Virol. 2019;93(20 September):e00638–19. doi: 10.1128/JVI.00638-19. 30PMID: 31375586; PMCID: PMC6798117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Umegaki K., Fenech M. Cytokinesis-block micronucleus assay in WIL2-NS cells: a sensitive system to detect chromosomal damage induced by reactive oxygen species and activated human neutrophils. Mutagenesis. 2000;15(3 May):261–269. doi: 10.1093/mutage/15.3.261. PMID: 10792021. [DOI] [PubMed] [Google Scholar]
- 108.Koehler C., Thielen S., Ginzkey C., Hackenberg S., Scherzed A., Burghartz M., Paulus M., Hagen R., Kleinsasser N.H. Nitrogen dioxide is genotoxic in urban concentrations. Inhal. Toxicol. 2013;25(6 May):341–347. doi: 10.3109/08958378.2013.788104. Epub 2013 May 23. PMID: 23701639. [DOI] [PubMed] [Google Scholar]
- 109.Han M., Guo Z., Li G., Sang N. Nitrogen dioxide inhalation induces genotoxicity in rats. Chemosphere. 2013;90(11 March):2737–2742. doi: 10.1016/j.chemosphere.2012.11.057. Epub 2013 Jan 16. PMID: 23332788. [DOI] [PubMed] [Google Scholar]
- 110.Dedon P.C., Tannenbaum S.R. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004;423(1 March):12–22. doi: 10.1016/j.abb.2003.12.017. 1PMID: 14989259. [DOI] [PubMed] [Google Scholar]
- 111.Frontera A., Cianfanelli L., Vlachos K., Landoni G., Cremona G. Severe air pollution links to higher mortality in COVID-19 patients: the "double-hit" hypothesis. J. Infect. 2020;81(2 August):255–259. doi: 10.1016/j.jinf.2020.05.031. Epub 2020 May 21. PMID: 32447007; PMCID: PMC7240268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Birch J., Anderson R.K., Correia-Melo C., Jurk D., Hewitt G., Marques F.M., Green N.J., Moisey E., Birrell M.A., Belvisi M.G., Black F., Taylor J.J., Fisher A.J., De Soyza A., Passos J.F. DNA damage response at telomeres contributes to lung aging and chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309(10 November):L1124–37. doi: 10.1152/ajplung.00293.2015. 15Epub 2015 Sep 18. PMID: 26386121; PMCID: PMC4652155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thaiparambil J., Dong L., Jasso D., Huang J.A., El-Zein R.A. Mitotic spindle apparatus abnormalities in chronic obstructive pulmonary disease cells: a potential pathway to lung Cancer. Cancer Prev. Res. (Phila) 2020;13(11 November):923–934. doi: 10.1158/1940-6207.CAPR-19-0557. Epub 2020 Jul 12. PMID: 32655004; PMCID: PMC7641916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karpman M.D., Eldridge R., Follis J.L., Etzel C.J., Shete S., El-Zein R.A. Chronic obstructive pulmonary disease among lung cancer-free smokers: the importance of healthy controls. Respir. Investig. 2018;56(1 January):28–33. doi: 10.1016/j.resinv.2017.11.002. Epub 2017 Dec 6. PMID: 29325677; PMCID: PMC5769708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhu Y., Xie J., Huang F., Cao L. Association between short-term exposure to air pollution and COVID-19 infection: evidence from China. Sci. Total Environ. 2020;727(July):138704. doi: 10.1016/j.scitotenv.2020.138704. 20Epub 2020 Apr 15. PMID: 32315904; PMCID: PMC7159846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fattorini D., Regoli F. Role of the chronic air pollution levels in the Covid-19 outbreak risk in Italy. Environ Pollut. 2020;264(September):114732. doi: 10.1016/j.envpol.2020.114732. Epub 2020 May 4. PMID: 32387671; PMCID: PMC7198142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Comunian S., Dongo D., Milani C., Palestini P. Air Pollution and Covid-19: The Role of Particulate Matter in the Spread and Increase of Covid-19’s Morbidity and Mortality. Int. J. Environ. Res. Public Health. 2020;17(12 June):4487. doi: 10.3390/ijerph17124487. 22PMID: 32580440; PMCID: PMC7345938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Woldhuis R.R., Heijink I.H., van den Berge M., Timens W., Oliver B.G.G., de Vries M., Brandsma C.A. COPD-derived fibroblasts secrete higher levels of senescence-associated secretory phenotype proteins. Thorax. 2020;(December):3. doi: 10.1136/thoraxjnl-2020-215114. thoraxjnl-2020-215114. Epub ahead of print. PMID: 33273021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. Epub 2012 Nov 8. PMID: 23140366; PMCID: PMC4166529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tchkonia T., Zhu Y., van Deursen J., Campisi J., Kirkland J.L. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest. 2013;123(3 March):966–972. doi: 10.1172/JCI64098. Epub 2013 Mar 1. PMID: 23454759; PMCID: PMC3582125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aoshiba K., Tsuji T., Yamaguchi K., Itoh M., Nakamura H. The danger signal plus DNA damage two-hit hypothesis for chronic inflammation in COPD. Eur. Respir. J. 2013;42(6 December):1689–1695. doi: 10.1183/09031936.00102912. Epub 2013 Feb 8. PMID: 23397294. [DOI] [PubMed] [Google Scholar]
- 122.Zierhut C., Funabiki H. Regulation and consequences of cGAS activation by Self-DNA. Trends Cell Biol. 2020;30(8 August):594–605. doi: 10.1016/j.tcb.2020.05.006. Epub 2020 Jun 13. PMID: 32546434; PMCID: PMC7368801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O’Rourke R.W., Lumeng C.N. Silver Spring; 2020. Pathways to Severe COVID-19 for People With Obesity. Obesity. Dec 3Epub ahead of print. PMID: 33270351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cuschieri S., Grech S. Obesity population at risk of COVID-19 complications. Glob Health Epidemiol Genom. 2020;5(November):e6. doi: 10.1017/gheg.2020.6. 6PMID: 33282327; PMCID: PMC7681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gombart A.F., Pierre A., Maggini S. A review of micronutrients and the immune system-working in harmony to reduce the risk of infection. Nutrients. 2020;12(1 January):236. doi: 10.3390/nu12010236. 16PMID: 31963293; PMCID: PMC7019735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Calder P.C. Nutrition, immunity and COVID-19. BMJ Nutr Prev Health. 2020;3(1 May):74–92. doi: 10.1136/bmjnph-2020-000085. 20PMID: 33230497; PMCID: PMC7295866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Briguglio M., Pregliasco F.E., Lombardi G., Perazzo P., Banfi G. The malnutritional status of the host as a virulence factor for new coronavirus SARS-CoV-2. Front Med (Lausanne). 2020;7(April):146. doi: 10.3389/fmed.2020.00146. 23PMID: 32391367; PMCID: PMC7191079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fenech M.F. Dietary reference values of individual micronutrients and nutriomes for genome damage prevention: current status and a road map to the future. Am. J. Clin. Nutr. 2010;91(5 May):1438S–1454S. doi: 10.3945/ajcn.2010.28674D. Epub 2010 Mar 10. PMID: 20219957. [DOI] [PubMed] [Google Scholar]
- 129.Dhillon V., Bull C., Fenech M. Academic Press; 2016. Chapter 10 - Telomeres, Aging, and Nutrition, Editor(s): Marco Malavolta, Eugenio Mocchegiani, Molecular Basis of Nutrition and Aging; pp. 129–140. ISBN 9780128018163. [DOI] [Google Scholar]
- 130.Froidure A., Mahieu M., Hoton D., Laterre P.F., Yombi J.C., Koenig S., Ghaye B., Defour J.P., Decottignies A. Short telomeres increase the risk of severe COVID-19. Aging (Albany NY). 2020;12(20 October):19911–19922. doi: 10.18632/aging.104097. 26Epub 2020 Oct 26. PMID: 33104521; PMCID: PMC7655194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aviv A. Telomeres and COVID-19. FASEB J. 2020;34(6 June):7247–7252. doi: 10.1096/fj.202001025. Epub 2020 May 19. PMID: 32427393; PMCID: PMC7276714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Benetos A., Lai T.P., Toupance S., Labat C., Verhulst S., Perret-Guillaume C., Gautier S., Ungeheuer M.N., Levy D., Susser E., Aviv A. A Mechanism for Severity of Disease in Older Patients with COVID-19: The Nexus between Telomere Length and Lymphopenia. medRxiv [Preprint]. 2020;4(October) doi: 10.1101/2020.10.01.20205393. 2020.10.01.20205393PMID: 33024983; PMCID: PMC7536885. [DOI] [Google Scholar]
- 133.Alimba C.G., Dhillon V., Bakare A.A., Fenech M. Genotoxicity and cytotoxicity of chromium, copper, manganese and lead, and their mixture in WIL2-NS human B lymphoblastoid cells is enhanced by folate depletion. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016;(March):35–47. doi: 10.1016/j.mrgentox.2016.02.002. 798-799Epub 2016 Feb 20. PMID: 26994492. [DOI] [PubMed] [Google Scholar]
- 134.Sharif R., Thomas P., Zalewski P., Graham R.D., Fenech M. The effect of zinc sulphate and zinc carnosine on genome stability and cytotoxicity in the WIL2-NS human lymphoblastoid cell line. Mutat. Res. 2011;720(1-2 Febuary):22–33. doi: 10.1016/j.mrgentox.2010.12.004. 28Epub 2010 Dec 15. PMID: 21167308. [DOI] [PubMed] [Google Scholar]
- 135.Kompella P., Vasquez K.M. Obesity and cancer: a mechanistic overview of metabolic changes in obesity that impact genetic instability. Mol. Carcinog. 2019;58(9 September):1531–1550. doi: 10.1002/mc.23048. Epub 2019 Jun 5. PMID: 31168912; PMCID: PMC6692207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Idolo A., Grassi T., Bagordo F., Panico A., De Giorgi M., Serio F., Guido M., Piscitelli P., De Filippis G., Raho A., De Donno A. Micronuclei in exfoliated buccal cells of children living in a cluster area of Salento (Southern Italy) with a high incidence of lung Cancer: the IMP.AIR study. Int. J. Environ. Res. Public Health. 2018;15(8 August):1659. doi: 10.3390/ijerph15081659. 5PMID: 30081588; PMCID: PMC6121539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scarpato R., Verola C., Fabiani B., Bianchi V., Saggese G., Federico G. Nuclear damage in peripheral lymphocytes of obese and overweight Italian children as evaluated by the gamma-H2AX focus assay and micronucleus test. FASEB J. 2011;25(2 Febuary):685–693. doi: 10.1096/fj.10-168427. Epub 2010 Nov 10. PMID: 21068397. [DOI] [PubMed] [Google Scholar]
- 138.Donmez-Altuntas H., Sahin F., Bayram F., Bitgen N., Mert M., Guclu K., Hamurcu Z., Arıbas S., Gundogan K., Diri H. Evaluation of chromosomal damage, cytostasis, cytotoxicity, oxidative DNA damage and their association with body-mass index in obese subjects. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014;771(September):30–36. doi: 10.1016/j.mrgentox.2014.06.006. 1Epub 2014 Jun 28. PMID: 25308439. [DOI] [PubMed] [Google Scholar]
- 139.Tamara A., Tahapary D.L. Obesity as a predictor for a poor prognosis of COVID-19: a systematic review. Diabetes Metab. Syndr. 2020;14(4 July-August):655–659. doi: 10.1016/j.dsx.2020.05.020. Epub 2020 May 12. PMID: 32438328; PMCID: PMC7217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Franzke B., Schwingshackl L., Wagner K.H. Chromosomal damage measured by the cytokinesis block micronucleus cytome assay in diabetes and obesity - A systematic review and meta-analysis. Mutat. Res. 2020;786(October-December):108343. doi: 10.1016/j.mrrev.2020.108343. Epub 2020 Nov 2. PMID: 33339574. [DOI] [PubMed] [Google Scholar]
- 141.Asanov M., Bonassi S., Proietti S., Minina V.I., Tomino C., El-Zein R. Genomic instability in chronic obstructive pulmonary disease and lung cancer: a systematic review and meta-analysis of studies using the micronucleus assay. Mutat. Res. 2021;787(January-June):108344. doi: 10.1016/j.mrrev.2020.108344. Epub 2020 Nov 10. PMID: 34083053. [DOI] [PubMed] [Google Scholar]
- 142.Andreassi M.G., Borghini A., Vecoli C. Micronucleus assay for predicting coronary artery disease: a systematic review and meta-analysis. Mutat. Res. 2021;787(January-June):108348. doi: 10.1016/j.mrrev.2020.108348. Epub 2020 Nov 16. PMID: 34083055. [DOI] [PubMed] [Google Scholar]
- 143.Wen C.H., Lin C.H., Tsao S.C., Su Y.C., Tsai M.H., Chai C.Y. Micronucleus scoring in liver fine needle aspiration cytology. Cytopathology. 2013;24(6 December):391–395. doi: 10.1111/cyt.12009. Epub 2012 Sep 14. PMID: 22974178. [DOI] [PubMed] [Google Scholar]
- 144.Stopper H., Bankoglu E.E., Marcos R., Pastor S. Micronucleus frequency in chronic kidney disease patients: a review. Mutat. Res. 2020;786(October-December):108340. doi: 10.1016/j.mrrev.2020.108340. Epub 2020 Oct 17. PMID: 33339580. [DOI] [PubMed] [Google Scholar]
- 145.Kirsch-Volders M., Pacchierotti F., Parry E.M., Russo A., Eichenlaub-Ritter U., Adler I.D. Risks of aneuploidy induction from chemical exposure: twenty years of collaborative research in Europe from basic science to regulatory implications. Mutat. Res. 2019;779(January-March):126–147. doi: 10.1016/j.mrrev.2018.11.002. Epub 2018 Dec 27. PMID: 31097149. [DOI] [PubMed] [Google Scholar]
- 146.Lynch A.M., Eastmond D., Elhajouji A., Froetschl R., Kirsch-Volders M., Marchetti F., Masumura K., Pacchierotti F., Schuler M., Tweats D. Targets and mechanisms of chemically induced aneuploidy. Part 1 of the report of the 2017 IWGT workgroup on assessing the risk of aneugens for carcinogenesis and hereditary diseases. Mutat. Res. 2019;847(November):403025. doi: 10.1016/j.mrgentox.2019.02.006. Epub 2019 Mar 2. PMID: 31699346. [DOI] [PubMed] [Google Scholar]
- 147.Ferreira F.L., Prá D., Martino-Roth M.G., Garcias G.L. Buccal micronucleus frequency is associated with age in Down syndrome. Genet. Mol. Res. 2009;8(4 October):1231–1237. doi: 10.4238/vol8-4gmr636. 13PMID: 19876863. [DOI] [PubMed] [Google Scholar]
- 148.Thomas P., Harvey S., Gruner T., Fenech M. The buccal cytome and micronucleus frequency is substantially altered in Down’s syndrome and normal ageing compared to young healthy controls. Mutat. Res. 2008;638(1-2 Febuary):37–47. doi: 10.1016/j.mrfmmm.2007.08.012. 1Epub 2007 Sep 1. PMID: 17920640. [DOI] [PubMed] [Google Scholar]
- 149.George A., Venkatesan S., Ashok N., Saraswathy R., Hande M.P. Assessment of genomic instability and proliferation index in cultured lymphocytes of patients with Down syndrome, congenital anomalies and aplastic anaemia. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836(Pt A December):98–103. doi: 10.1016/j.mrgentox.2018.06.015. Epub 2018 Jun 8. PMID: 30389169. [DOI] [PubMed] [Google Scholar]
- 150.Maluf S.W., Erdtmann B. Genomic instability in Down syndrome and Fanconi anemia assessed by micronucleus analysis and single-cell gel electrophoresis. Cancer Genet. Cytogenet. 2001;124(1 January):71–75. doi: 10.1016/s0165-4608(00)00322-8. 1PMID: 11165325. [DOI] [PubMed] [Google Scholar]
- 151.Malle L., Gao C., Hur C., Truong H.Q., Bouvier N.M., Percha B., Kong X.F., Bogunovic D. Individuals with Down syndrome hospitalized with COVID-19 have more severe disease. Genet. Med. 2021;23(3 March):576–580. doi: 10.1038/s41436-020-01004-w. Epub 2020 Oct 16. PMID: 33060835; PMCID: PMC7936948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Araya P., Waugh K.A., Sullivan K.D., Núñez N.G., Roselli E., Smith K.P., Granrath R.E., Rachubinski A.L., Enriquez Estrada B., Butcher E.T., Minter R., Tuttle K.D., Bruno T.C., Maccioni M., Espinosa J.M. Trisomy 21 dysregulates T cell lineages toward an autoimmunity-prone state associated with interferon hyperactivity. Proc. Natl. Acad. Sci. U. S. A. 2019;116(48 November):24231–24241. doi: 10.1073/pnas.1908129116. 26Epub 2019 Nov 7. PMID: 31699819; PMCID: PMC6883781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Espinosa J.M. Down syndrome and COVID-19: a perfect storm? Cell Rep Med. 2020;1(2 May):100019. doi: 10.1016/j.xcrm.2020.100019. 19Epub 2020 May 1. PMID: 32501455; PMCID: PMC7252041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Clift A.K., Coupland C.A.C., Keogh R.H., Hemingway H., Hippisley-Cox J. COVID-19 mortality risk in down syndrome: results from a cohort study of 8 million adults. Ann. Intern. Med. 2020;21(October):M20–4986. doi: 10.7326/M20-4986. Epub ahead of print. PMID: 33085509; PMCID: PMC7592804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kirsch-Volders M., Plas G., Elhajouji A., Lukamowicz M., Gonzalez L., Vande Loock K., Decordier I. The in vitro MN assay in 2011: origin and fate, biological significance, protocols, high throughput methodologies and toxicological relevance. Arch. Toxicol. 2011;85(8 August):873–899. doi: 10.1007/s00204-011-0691-4. Epub 2011 May 3. PMID: 21537955. [DOI] [PubMed] [Google Scholar]
- 156.Ly D.H., Lockhart D.J., Lerner R.A., Schultz P.G. Mitotic misregulation and human aging. Science. 2000;287(5462 March):2486–2492. doi: 10.1126/science.287.5462.2486. 31 PMID: 10741968. [DOI] [PubMed] [Google Scholar]
- 157.Macedo J.C., Vaz S., Bakker B., Ribeiro R., Bakker P.L., Escandell J.M., Ferreira M.G., Medema R., Foijer F., Logarinho E. FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nat. Commun. 2018;9(1 July):2834. doi: 10.1038/s41467-018-05258-6. 19PMID: 30026603; PMCID: PMC6053425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Barroso-Vilares M., Macedo J.C., Reis M., Warren J.D., Compton D., Logarinho E. Small-molecule inhibition of aging-associated chromosomal instability delays cellular senescence. EMBO Rep. 2020;21(5 May):e49248. doi: 10.15252/embr.201949248. 6Epub 2020 Mar 5. PMID: 32134180; PMCID: PMC7202060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fenech M., Neville S., Rinaldi J. Sex is an important variable affecting spontaneous micronucleus frequency in cytokinesis-blocked lymphocytes. Mutat. Res. 1994;313(2-3 October-December):203–207. doi: 10.1016/0165-1161(94)90050-7. PMID: 7523905. [DOI] [PubMed] [Google Scholar]
- 160.Fenech M. Important variables that influence base-line micronucleus frequency in cytokinesis-blocked lymphocytes-a biomarker for DNA damage in human populations. Mutat. Res. 1998;404(1-2 August):155–165. doi: 10.1016/s0027-5107(98)00109-2. 3PMID: 9729354. [DOI] [PubMed] [Google Scholar]
- 161.Bonassi S., Fenech M., Lando C., Lin Y.P., Ceppi M., Chang W.P., Holland N., Kirsch-Volders M., Zeiger E., Ban S., Barale R., Bigatti M.P., Bolognesi C., Jia C., Di Giorgio M., Ferguson L.R., Fucic A., Lima O.G., Hrelia P., Krishnaja A.P., Lee T.K., Migliore L., Mikhalevich L., Mirkova E., Mosesso P., Müller W.U., Odagiri Y., Scarffi M.R., Szabova E., Vorobtsova I., Vral A., Zijno A. HUman MicroNucleus project: international database comparison for results with the cytokinesis-block micronucleus assay in human lymphocytes: I. Effect of laboratory protocol, scoring criteria, and host factors on the frequency of micronuclei. Environ. Mol. Mutagen. 2001;37(1):31–45. PMID: 11170240. [PubMed] [Google Scholar]
- 162.Bolognesi C., Lando C., Forni A., Landini E., Scarpato R., Migliore L., Bonassi S. Chromosomal damage and ageing: effect on micronuclei frequency in peripheral blood lymphocytes. Age Ageing. 1999;28(4 July):393–397. doi: 10.1093/ageing/28.4.393. PMID: 10459794. [DOI] [PubMed] [Google Scholar]
- 163.Wojda A., Zietkiewicz E., Witt M. Effects of age and gender on micronucleus and chromosome nondisjunction frequencies in centenarians and younger subjects. Mutagenesis. 2007;22(3 May):195–200. doi: 10.1093/mutage/gem002. Epub 2007 Feb 6. PMID: 17284771. [DOI] [PubMed] [Google Scholar]
- 164.Kirsch-Volders M., Fenech M., Bolognesi C. Validity of the Lymphocyte Cytokinesis-Block Micronucleus Assay (L-CBMN) as biomarker for human exposure to chemicals with different modes of action: a synthesis of systematic reviews. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;836(Pt A December):47–52. doi: 10.1016/j.mrgentox.2018.05.010. Epub 2018 May 7. PMID: 30389162. [DOI] [PubMed] [Google Scholar]
- 165.Bonassi S., El-Zein R., Bolognesi C., Fenech M. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: evidence from human studies. Mutagenesis. 2011;26(1 January):93–100. doi: 10.1093/mutage/geq075. PMID: 21164188. [DOI] [PubMed] [Google Scholar]
- 166.Andreassi M.G., Barale R., Iozzo P., Picano E. The association of micronucleus frequency with obesity, diabetes and cardiovascular disease. Mutagenesis. 2011;26(1 January):77–83. doi: 10.1093/mutage/geq077. PMID: 21164186. [DOI] [PubMed] [Google Scholar]
- 167.Migliore L., Coppedè F., Fenech M., Thomas P. Association of micronucleus frequency with neurodegenerative diseases. Mutagenesis. 2011;26(1 January):85–92. doi: 10.1093/mutage/geq067. PMID: 21164187. [DOI] [PubMed] [Google Scholar]
- 168.Furness D.L., Dekker G.A., Hague W.M., Khong T.Y., Fenech M.F. Increased lymphocyte micronucleus frequency in early pregnancy is associated prospectively with pre-eclampsia and/or intrauterine growth restriction. Mutagenesis. 2010;25(5 September):489–498. doi: 10.1093/mutage/geq032. Epub 2010 Jun 25. PMID: 20581221; PMCID: PMC2925157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Federici C., Botto N., Manfredi S., Rizza A., Del Fiandra M., Andreassi M.G. Relation of increased chromosomal damage to future adverse cardiac events in patients with known coronary artery disease. Am. J. Cardiol. 2008;102(10 November):1296–1300. doi: 10.1016/j.amjcard.2008.07.024. 15Epub 2008 Sep 5. PMID: 18993144. [DOI] [PubMed] [Google Scholar]
- 170.Murgia E., Maggini V., Barale R., Rossi A.M. Micronuclei, genetic polymorphisms and cardiovascular disease mortality in a nested case-control study in Italy. Mutat. Res. 2007;621(1-2 August):113–118. doi: 10.1016/j.mrfmmm.2007.02.015. 1Epub 2007 Mar 2. PMID: 17448506. [DOI] [PubMed] [Google Scholar]
- 171.Murgia E., Ballardin M., Bonassi S., Rossi A.M., Barale R. Validation of micronuclei frequency in peripheral blood lymphocytes as early cancer risk biomarker in a nested case-control study. Mutat. Res. 2008;639(1-2 March):27–34. doi: 10.1016/j.mrfmmm.2007.10.010. 1Epub 2007 Nov 13. PMID: 18155071. [DOI] [PubMed] [Google Scholar]
- 172.Bonassi S., Znaor A., Ceppi M., Lando C., Chang W.P., Holland N., Kirsch-Volders M., Zeiger E., Ban S., Barale R., Bigatti M.P., Bolognesi C., Cebulska-Wasilewska A., Fabianova E., Fucic A., Hagmar L., Joksic G., Martelli A., Migliore L., Mirkova E., Scarfi M.R., Zijno A., Norppa H., Fenech M. An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis. 2007;28(3 March):625–631. doi: 10.1093/carcin/bgl177. Epub 2006 Sep 14. PMID: 16973674. [DOI] [PubMed] [Google Scholar]
- 173.Disteche C.M., Berletch J.B. X-chromosome inactivation and escape. J. Genet. 2015;94(4 December):591–599. doi: 10.1007/s12041-015-0574-1. PMID: 26690513; PMCID: PMC4826282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bukvic N., Gentile M., Susca F., Fanelli M., Serio G., Buonadonna L., Capurso A., Guanti G. Sex chromosome loss, micronuclei, sister chromatid exchange and aging: a study including 16 centenarians. Mutat. Res. 2001;498(1-2 November):159–167. doi: 10.1016/s1383-5718(01)00279-0. 15PMID: 11673081. [DOI] [PubMed] [Google Scholar]
- 175.Russell L.M., Strike P., Browne C.E., Jacobs P.A. X chromosome loss and ageing. Cytogenet. Genome Res. 2007;116(3):181–185. doi: 10.1159/000098184. PMID: 17317957. [DOI] [PubMed] [Google Scholar]
- 176.Hando J.C., Nath J., Tucker J.D. Sex chromosomes, micronuclei and aging in women. Chromosoma. 1994;103(3 June):186–192. doi: 10.1007/BF00368011. PMID: 7924621. [DOI] [PubMed] [Google Scholar]
- 177.Tucker J.D., Nath J., Hando J.C. Activation status of the X chromosome in human micronucleated lymphocytes. Hum. Genet. 1996;97(4 April):471–475. doi: 10.1007/BF02267069. PMID: 8834245. [DOI] [PubMed] [Google Scholar]
- 178.Howcroft T.K., Campisi J., Louis G.B., Smith M.T., Wise B., Wyss-Coray T., Augustine A.D., McElhaney J.E., Kohanski R., Sierra F. The role of inflammation in age-related disease. Aging (Albany NY). 2013;5(1 January):84–93. doi: 10.18632/aging.100531. PMID: 23474627; PMCID: PMC3616233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gubbels Bupp M.R., Potluri T., Fink A.L., Klein S.L. The confluence of sex hormones and aging on immunity. Front. Immunol. 2018;9:1269. doi: 10.3389/fimmu.2018.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ranke M.B., Saenger P. Turner’s syndrome. Lancet. 2001;358:309–314. doi: 10.1016/S0140-6736(01)05487-3. [DOI] [PubMed] [Google Scholar]
- 181.Elsheikh M., Wass J.A.H., Conway G.S. Autoimmune thyroid syndrome in women with Turner’s syndrome—the association with karyotype. Clin. Endocrinol. 2001;55(2):223–226. doi: 10.1046/j.1365-2265.2001.01296.x. [DOI] [PubMed] [Google Scholar]
- 182.Invernizzi P., Miozzo M., Selmi C., Persani L., Battezzati P.M., Zuin M., Lucchi S., Meroni P.L., Marasini B., Zeni S., Watnik M., Grati F.R., Simoni Gershwin M.E., Podda M. X Chromosome Monosomy: A Common Mechanism for Autoimmune Diseases. J. Immunol. 2005;175(1 July):575–578. doi: 10.4049/jimmunol.175.1.575. 1. [DOI] [PubMed] [Google Scholar]
- 183.Kamikawa Y., Donohoe M.E. The dynamics of X-chromosome inactivation in mouse development Mol. Reprod. Dev. 2014;81:141–147. doi: 10.1002/mrd.22282. [DOI] [PubMed] [Google Scholar]
- 184.Stone F.F., Sandberg A.A. Sex chromosome aneuploidy and aging. Mutat. Res. 1995;338:107–113. doi: 10.1016/0921-8734(95)00016-y. [DOI] [PubMed] [Google Scholar]
- 185.Nath J., Tucker J.D., Hando J.C. Y chromosome aneuploidy, micronuclei, kinetochores and aging in men. Chromosoma. 1995;103:725–731. doi: 10.1007/BF00344234. [DOI] [PubMed] [Google Scholar]
- 186.Catalan J., Autio K., Kuosma E., Norppa H. Age-dependent inclusion of sex chromosomes in lymphocyte micronuclei of man. Am. J. Hum. Genet. 1998;63:1464–1472. doi: 10.1086/302092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dumanski J.P., Lambert J.C., Rasi C., Giedraitis V., Davies H., Grenier-Boley B., Lindgren C.M., Campion D., Dufouil C., Pasquier F., Amouyel P.P., Lannfelt L., Ingelsson M., Kilander L., Lind L., Forsberg L.A. European Alzheimer’s disease initiative i mosaic loss of chromosome y in blood is associated with Alzheimer disease. Am. J. Hum. Genet. 2016;98:1208–1219. doi: 10.1016/j.ajhg.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dumanski J.P., Rasi C., Lönn M., Davies H., Ingelsson M., Giedraitis V., Lannfelt M., Magnusson P.K., Lindgren C.M., Morris A.P., Cesarini D., Johannesson M., Tiensuu Janson E., Lind L., Pedersen N.L., Ingelsson E., Forsberg L.A. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhou W., Machiela M.J., Freedman N.D., Rothman N., Malats N., Dagnall C., Caporaso N., Teras L.T., Gaudet M.M., Gapstur S.M., Stevens V.L., Jacobs K.B., Sampson J., Albanes D., Weinstein S., Virtamo J., Berndt S., Hoover R.N., Black A., Silverman D., Figueroa J., Garcia-Closas M., Real F.X., Earl J., Marenne G., Rodriguez-Santiago B., Karagas M., Johnson A., Schwenn M., Wu X., Gu J., Ye Y., Hutchinson A., Tucker M., Perez-Jurado L.A., Dean M., Yeager M., Chanock S.J. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat. Genet. 2016;48:563–568. doi: 10.1038/ng.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Forsberg L.A. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum. Genet. 2017;136(5):657–663. doi: 10.1007/s00439-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Guo X., Dai X., Zhou T., Wang H., Ni J., Xue J., Wang X. Mosaic loss of human Y chromosome: what, how and why. Hum. Genet. 2020;139:421–446. doi: 10.1007/s00439-020-02114-w. [DOI] [PubMed] [Google Scholar]
- 192.Ly P., Teitz L.S., Kim D.H., Shoshani O., Skaletsky H., Fachinetti D., Page D.C., Cleveland D.W. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 2017;19(1 January):68–75. doi: 10.1038/ncb3450. Epub 2016 Dec 5. PMID: 27918550; PMCID: PMC5539760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Persani L., Bonomi M., Lleo A., Pasini S., Civardi F., Bianchi I., Campi I., Finelli P., Miozzo M., Castronovo C., Sirchia S., Gershwin M.E., Invernizzi P. Increased loss of the Y chromosome in peripheral blood cells in male patients with autoimmune thyroiditis. J. Autoimmun. 2012;38(2-3 May):J193–6. doi: 10.1016/j.jaut.2011.11.011. Epub 2011 Dec 22. PMID: 22196921. [DOI] [PubMed] [Google Scholar]
- 194.Fenech M., Bonassi S. The effect of age, gender, diet and lifestyle on DNA damage measured using micronucleus frequency in human peripheral blood lymphocytes. Mutagenesis. 2011;26(1 January):43–49. doi: 10.1093/mutage/geq050. PMID: 21164181. [DOI] [PubMed] [Google Scholar]
- 195.Haitao T., Vermunt J.V., Abeykoon J., Ghamrawi R., Gunaratne M., Jayachandran M., Narang K., Parashuram S., Suvakov S., Garovic V.D. COVID-19 and sex differences: mechanisms and biomarkers. Mayo Clin. Proc. 2020;95(10 October):2189–2203. doi: 10.1016/j.mayocp.2020.07.024. Epub 2020 Aug 4. PMID: 33012349; PMCID: PMC7402208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Galupa R., Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 2018;52(November):535–566. doi: 10.1146/annurev-genet-120116-024611. 23Epub 2018 Sep 26. PMID: 30256677. [DOI] [PubMed] [Google Scholar]
- 197.Li T., Huang T., Du M., Chen X., Du F., Ren J., Chen Z.J. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 2021;371(6535 March) doi: 10.1126/science.abc5386. 19eabc5386Epub 2021 Feb 4. PMID: 33542149; PMCID: PMC8171060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fenech M. Important variables that influence base-line micronucleus frequency in cytokinesis-blocked lymphocytes-a biomarker for DNA damage in human populations. Mutat. Res. 1998;404(1-2 August):155–165. doi: 10.1016/s0027-5107(98)00109-2. 3PMID: 9729354. [DOI] [PubMed] [Google Scholar]
- 199.Fouquerel E., Barnes R.P., Uttam S., Watkins S.C., Bruchez M.P., Opresko P.L. Targeted and persistent 8-Oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell. 2019;75(1 July):117–130. doi: 10.1016/j.molcel.2019.04.024. 11e6Epub 2019 May 14. PMID: 31101499; PMCID: PMC6625854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fumagalli M., Rossiello F., Clerici M., Barozzi S., Cittaro D., Kaplunov J.M., Bucci G., Dobreva M., Matti V., Beausejour C.M., Herbig U., Longhese M.P. d’Adda di Fagagna F. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012;14(4 March):355–365. doi: 10.1038/ncb2466. 18Erratum in: Nat Cell Biol. 2012 May;14(5):555. PMID: 22426077; PMCID: PMC3717580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rossiello F., Herbig U., Longhese M.P., Fumagalli M. d’Adda di Fagagna F. Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 2014;26(June):89–95. doi: 10.1016/j.gde.2014.06.009. Epub 2014 Aug 11. PMID: 25104620; PMCID: PMC4217147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Barroso-Vilares M., Logarinho E. Chromosomal instability and pro-inflammatory response in aging. Mech. Ageing Dev. 2019;182(September):111118. doi: 10.1016/j.mad.2019.111118. Epub 2019 May 15. PMID: 31102604. [DOI] [PubMed] [Google Scholar]
- 203.Andriani G.A., Almeida V.P., Faggioli F., Mauro M., Tsai W.L., Santambrogio L., Maslov A., Gadina M., Campisi J., Vijg J., Montagna C. Whole chromosome instability induces senescence and promotes SASP. Sci. Rep. 2016;6(October):35218. doi: 10.1038/srep35218. 12PMID: 27731420; PMCID: PMC5059742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. Epub 2012 Nov 8. PMID: 23140366; PMCID: PMC4166529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ceppi M., Biasotti B., Fenech M., Bonassi S. Human population studies with the exfoliated buccal micronucleus assay: statistical and epidemiological issues. Mutat. Res. 2010;705(1 July-September):11–19. doi: 10.1016/j.mrrev.2009.11.001. Epub 2009 Nov 20. PMID: 19932192. [DOI] [PubMed] [Google Scholar]
- 206.Xu W., Larbi A. Markers of t cell senescence in humans. Int. J. Mol. Sci. 2017;18(8 August):1742. doi: 10.3390/ijms18081742. 10PMID: 28796199; PMCID: PMC5578132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Frasca D., Diaz A., Romero M., Blomberg B.B. Human peripheral late/exhausted memory B cells express a senescent‐associated secretory phenotype and preferentially utilize metabolic signaling pathways. Exp. Gerontol. 2017;87:113–120. doi: 10.1016/j.exger.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 208.Callender L.A., Carroll E.C., Beal R.W.J., Chambers E.S., Nourshargh S., Akbar A.N., Henson S.M. Human CD8+ EMRA T cells display a senescence-associated secretory phenotype regulated by p38 MAPK. Aging Cell. 2018;17(1 Febuary):e12675. doi: 10.1111/acel.12675. Epub 2017 Oct 12. PMID: 29024417; PMCID: PMC5770853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Acosta J.C., Banito A., Wuestefeld T., Georgilis A., Janich P., Morton J.P., Athineos D., Kang T.W., Lasitschka F., Andrulis M., Pascual G., Morris K.J., Khan S., Jin H., Dharmalingam G., Snijders A.P., Carroll T., Capper D., Pritchard C., Inman G.J., Longerich T., Sansom O.J., Benitah S.A., Zender L., Gil J. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15(8 August):978–990. doi: 10.1038/ncb2784. Epub 2013 Jun 16. PMID: 23770676; PMCID: PMC3732483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gonzalez-Meljem J.M., Apps J.R., Fraser H.C., Martinez-Barbera J.P. Paracrine roles of cellular senescence in promoting tumourigenesis. Br. J. Cancer. 2018;118(10 May):1283–1288. doi: 10.1038/s41416-018-0066-1. Epub 2018 Apr 19. PMID: 29670296; PMCID: PMC5959857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yousefzadeh M.J., Flores R.R., Zhu Y., Schmiechen Z.C., Brooks R.W., Trussoni C.E., Cui Y., Angelini L., Lee K.A., McGowan S.J., Burrack A.L., Wang D., Dong Q., Lu A., Sano T., O’Kelly R.D., McGuckian C.A., Kato J.I., Bank M.P., Wade E.A., Pillai S.P.S., Klug J., Ladiges W.C., Burd C.E., Lewis S.E., LaRusso N.F., Vo N.V., Wang Y., Kelley E.E., Huard J., Stromnes I.M., Robbins P.D., Niedernhofer L.J. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594(7861 June):100–105. doi: 10.1038/s41586-021-03547-7. Epub 2021 May 12. PMID: 33981041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Thomas P., Holland N., Bolognesi C., Kirsch-Volders M., Bonassi S., Zeiger E., Knasmueller S., Fenech M. Buccal micronucleus cytome assay. Nat. Protoc. 2009;4(6):825–837. doi: 10.1038/nprot.2009.53. Epub 2009 May 7. PMID: 19444240. [DOI] [PubMed] [Google Scholar]
- 213.Knasmueller S., Holland N., Wultsch G., Jandl B., Burgaz S., Misík M., Nersesyan A. Use of nasal cells in micronucleus assays and other genotoxicity studies. Mutagenesis. 2011;26(1 January):231–238. doi: 10.1093/mutage/geq079. PMID: 21164207. [DOI] [PubMed] [Google Scholar]
- 214.Lippman S.M., Peters E.J., Wargovich M.J., Stadnyk A.N., Dixon D.O., Dekmezian R.H., Loewy J.W., Morice R.C., Cunningham J.E., Hong W.K. Bronchial micronuclei as a marker of an early stage of carcinogenesis in the human tracheobronchial epithelium. Int. J. Cancer. 1990;45(5 May):811–815. doi: 10.1002/ijc.2910450503. 15PMID: 2335384. [DOI] [PubMed] [Google Scholar]
- 215.Zhong M., Lin B., Pathak J.L., Gao H., Young A.J., Wang X., Liu C., Wu K., Liu M., Chen J.M., Huang J., Lee L.H., Qi C.L., Ge L., Wang L. ACE2 and furin expressions in oral epithelial cells possibly facilitate COVID-19 infection via respiratory and fecal-oral routes. Front Med. (Lausanne). 2020;7(December):580796. doi: 10.3389/fmed.2020.580796. 10PMID: 33363183; PMCID: PMC7758442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bonassi S., Coskun E., Ceppi M., Lando C., Bolognesi C., Burgaz S., Holland N., Kirsh-Volders M., Knasmueller S., Zeiger E., Carnesoltas D., Cavallo D., da Silva J., de Andrade V.M., Demircigil G.C., Domínguez Odio A., Donmez-Altuntas H., Gattas G., Giri A., Giri S., Gómez-Meda B., Gómez-Arroyo S., Hadjidekova V., Haveric A., Kamboj M., Kurteshi K., Martino-Roth M.G., Montero Montoya R., Nersesyan A., Pastor-Benito S., Favero Salvadori D.M., Shaposhnikova A., Stopper H., Thomas P., Torres-Bugarín O., Yadav A.S., Zúñiga González G., Fenech M. The HUman MicroNucleus project on eXfoLiated buccal cells (HUMN(XL)): the role of life-style, host factors, occupational exposures, health status, and assay protocol. Mutat. Res. 2011;728(3 November-December):88–97. doi: 10.1016/j.mrrev.2011.06.005. Epub 2011 Jul 8. PMID: 21763453. [DOI] [PubMed] [Google Scholar]
- 217.Shah S., Singaraju S., Bertin E.T., Singaraju M., Sharma A. Quantification of micronuclei in exfoliated cells of human immunodeficiency virus/AIDS-infected female patients. J. Oral Maxillofac. Pathol. 2019;23(2 May-August):301. doi: 10.4103/jomfp.JOMFP_251_17. PMID: 31516242; PMCID: PMC6714255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Drummond R.L., Rhoden C.R., Lubianca Neto J.F., Fleck A.D.S., RCPK Padoin, Amantéa S.L. Micronucleus count in nasal epithelial cells from patients with chronic rhinosinusitis and polyps. Braz. J. Otorhinolaryngol. 2020;86(6 November-December):743–747. doi: 10.1016/j.bjorl.2019.05.004. Epub 2019 Jun 18. PMID: 31285184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ramírez M.J., Minguillón J., Loveless S., Lake K., Carrasco E., Stjepanovic N., Balmaña J., Català A., Mehta P.A., Surrallés J. Chromosome fragility in the buccal epithelium in patients with Fanconi anemia. Cancer Lett. 2020;472(March):1–7. doi: 10.1016/j.canlet.2019.12.008. 1Epub 2019 Dec 10. PMID: 31830560. [DOI] [PubMed] [Google Scholar]
- 220.Ferreira F.L., Prá D., Martino-Roth M.G., Garcias G.L. Buccal micronucleus frequency is associated with age in Down syndrome. Genet. Mol. Res. 2009;8(4 October):1231–1237. doi: 10.4238/vol8-4gmr636. 13PMID: 19876863. [DOI] [PubMed] [Google Scholar]
- 221.Thomas P., Harvey S., Gruner T., Fenech M. The buccal cytome and micronucleus frequency is substantially altered in Down’s syndrome and normal ageing compared to young healthy controls. Mutat. Res. 2008;638(1-2 Febuary):37–47. doi: 10.1016/j.mrfmmm.2007.08.012. 1Epub 2007 Sep 1. PMID: 17920640. [DOI] [PubMed] [Google Scholar]
- 222.Rosin M.P., Ochs H.D. In vivo chromosomal instability in ataxia-telangiectasia homozygotes and heterozygotes. Hum. Genet. 1986;74(4 December):335–340. doi: 10.1007/BF00280482. PMID: 3793095. [DOI] [PubMed] [Google Scholar]
- 223.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007;2(5):1084–1104. doi: 10.1038/nprot.2007.77. PMID: 17546000. [DOI] [PubMed] [Google Scholar]
- 224.Aviv A. Telomeres and COVID-19. FASEB J. 2020;34(6 June):7247–7252. doi: 10.1096/fj.202001025. Epub 2020 May 19. PMID: 32427393; PMCID: PMC7276714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Conrad A., Boccard M., Valour F., Alcazer V., Tovar Sanchez A.T., Chidiac C., Laurent F., Vanhems P., Salles G., Brengel-Pesce K., Meunier B., Trouillet-Assant S., Ader F. Lyon HEMINF Study Group. VaccHemInf project: protocol for a prospective cohort study of efficacy, safety and characterisation of immune functional response to vaccinations in haematopoietic stem cell transplant recipients. BMJ Open. 2019;9(2 Febuary):e026093. doi: 10.1136/bmjopen-2018-026093. 15PMID: 30772864; PMCID: PMC6398679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang G.H., Ji B.Q., Li Y., Zheng G.Q., Ye L.L., Hao Y.H., Ren J.C., Zhou L.F., Xu X.W., Zhu Y., Xia Z.L. Benchmark doses based on abnormality of WBC or micronucleus frequency in Benzene-Exposed chinese workers. J. Occup. Environ. Med. 2016;58(2 Febuary):e39–44. doi: 10.1097/JOM.0000000000000639. PMID: 26849270. [DOI] [PubMed] [Google Scholar]
- 227.Fenech M.F. Nutriomes and personalised nutrition for DNA damage prevention, telomere integrity maintenance and cancer growth control. Cancer Treat. Res. 2014;159:427–441. doi: 10.1007/978-3-642-38007-5_24. PMID: 24114494. [DOI] [PubMed] [Google Scholar]
- 228.van Ommen B., van den Broek T., de Hoogh I., van Erk M., van Someren E., Rouhani-Rankouhi T., Anthony J.C., Hogenelst K., Pasman W., Boorsma A., Wopereis S. Systems biology of personalized nutrition. Nutr. Rev. 2017;75(8 August):579–599. doi: 10.1093/nutrit/nux029. 1Erratum in: Nutr Rev. 2017 Aug 1;75(8):672. PMID: 28969366; PMCID: PMC5914356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Thomas P., Wu J., Dhillon V., Fenech M. Effect of dietary intervention on human micronucleus frequency in lymphocytes and buccal cells. Mutagenesis. 2011;26(1 January):69–76. doi: 10.1093/mutage/geq072. PMID: 21164185. [DOI] [PubMed] [Google Scholar]
- 230.Lim H., Park H., Kim H.P. Effects of flavonoids on senescence-associated secretory phenotype formation from bleomycin-induced senescence in BJ fibroblasts. Biochem. Pharmacol. 2015;96(4 August):337–348. doi: 10.1016/j.bcp.2015.06.013. 15Epub 2015 Jun 18. PMID: 26093063. [DOI] [PubMed] [Google Scholar]
- 231.Perrott K.M., Wiley C.D., Desprez P.Y., Campisi J. Apigenin suppresses the senescence-associated secretory phenotype and paracrine effects on breast cancer cells. Geroscience. 2017;39(2 April):161–173. doi: 10.1007/s11357-017-9970-1. Epub 2017 Apr 4. PMID: 28378188; PMCID: PMC5411372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Serhan C.N., Levy B.D. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J. Clin. Invest. 2018;128(7 July):2657–2669. doi: 10.1172/JCI97943. 2Epub 2018 May 14. PMID: 29757195; PMCID: PMC6025982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li W., Qin L., Feng R., Hu G., Sun H., He Y., Zhang R. Emerging senolytic agents derived from natural products. Mech. Ageing Dev. 2019;181(July):1–6. doi: 10.1016/j.mad.2019.05.001. Epub 2019 May 8. PMID: 31077707. [DOI] [PubMed] [Google Scholar]
- 234.Emming S., Schroder K. Tiered DNA sensors for escalating responses. Science. 2019;365(6460 September):1375–1376. doi: 10.1126/science.aay2701. 27. [DOI] [PubMed] [Google Scholar]
- 235.Chow K.T., Gale M., Jr, Loo Y.M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36(April):667–694. doi: 10.1146/annurev-immunol-042617-053309. 26PMID: 29677479. [DOI] [PubMed] [Google Scholar]
- 236.Decordier I., Papine A., Vande Loock K., Plas G., Soussaline F., Kirsch-Volders M. Automated image analysis of micronuclei by IMSTAR for biomonitoring. Mutagenesis. 2011;26(1 January):163–168. doi: 10.1093/mutage/geq063. [DOI] [PubMed] [Google Scholar]
- 237.Fenech M., Kirsch-Volders M., Rossnerova A., Sram R., Room H., Bolognesi C., Ramakumar A., Soussaline F., Schunck C., Elhajoujii A., Anwar W., Bonassi S. HUMN project initiative and review of validation, quality control and prospects for further development of automated micronucleus assays using image cytometry systems. Int. J. Hyg. Environ. Health. 2013;216:541–552. doi: 10.1016/j.ijheh.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 238.Wang Q., Rodrigues M.A., Repin M., Pampou S., Beaton-Green L.A., Perrier J., Garty G., Brenner D.J., Turner H.C., Wilkins R.C. Automated triage radiation biodosimetry: integrating imaging flow cytometry with high-throughput robotics to perform the cytokinesis-block micronucleus assay. Radiat. Res. 2019;191(4 April):342–351. doi: 10.1667/RR15243.1. Epub 2019 Feb 19. PMID: 30779694; PMCID: PMC6483078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Fenech M., Knasmueller S., Bolognesi C., Holland N., Bonassi S., Kirsch-Volders M. Micronuclei as biomarkers of DNA damage, aneuploidy, inducers of chromosomal hypermutation and as sources of pro-inflammatory DNA in humans. Mutat. Res. 2020;786(October-December):108342. doi: 10.1016/j.mrrev.2020.108342. Epub 2020 Oct 28. PMID: 33339572. [DOI] [PubMed] [Google Scholar]
- 240.Lam E., Stein S., Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J. Virol. 2014;88(2 January):974–981. doi: 10.1128/JVI.02702-13. Epub 2013 Nov 6. PMID: 24198409; PMCID: PMC3911663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Anghelina D., Lam E., Falck-Pedersen E. Diminished innate antiviral response to adenovirus vectors in cGAS/STING-Deficient mice minimally impacts adaptive immunity. J. Virol. 2016;90(13 June):5915–5927. doi: 10.1128/JVI.00500-16. 10PMID: 27076643; PMCID: PMC4907218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sharma S., Schmid M.A., Sanchez Felipe L., Grenelle J., Kaptein S.J.F., Coelmont L., Neyts J., Dallmeier K. Small-molecule inhibitors of TBK1 serve as an adjuvant for a plasmid-launched live-attenuated yellow fever vaccine. Hum. Vaccin. Immunother. 2020;16(9 September):2196–2203. doi: 10.1080/21645515.2020.1765621. 1Epub 2020 Jun 23. PMID: 32574095; PMCID: PMC7553677. [DOI] [PMC free article] [PubMed] [Google Scholar]